Abstract
Protein engineering holds the potential to transform the metabolic drug landscape through the development of smart, stimulus-responsive drug systems. Protein therapeutics are a rapidly expanding segment of Food and Drug Administration approved drugs that will improve clinical outcomes over the long run. Engineering of protein therapeutics is still in its infancy, but recent general advances in protein engineering capabilities are being leveraged to yield improved control over both pharmacokinetics and pharmacodynamics. Stimulus-responsive protein therapeutics are drugs which have been designed to be metabolized under targeted conditions. Protein engineering is being utilized to develop tailored smart therapeutics with biochemical logic. This review focuses on applications of targeted drug neutralization, stimulus-responsive engineered protein prodrugs, and emerging multicomponent smart drug systems (e.g., antibody-drug conjugates, responsive engineered zymogens, prospective biochemical logic smart drug systems, drug buffers, and network medicine applications).
Keywords: biochemical logic, drug buffer, engineered zymogen, network medicine, protein engineering, stimulus-responsive, targeted drug activation, targeted drug neutralization
INTRODUCTION
The outcome of current drug therapies can vary widely between individuals, and balancing safety and efficacy is particularly challenging for narrow therapeutic window drugs [1, 2] and highly cytotoxic cancer therapies [3]. Smart, stimulus-responsive drug systems for enhanced control of drug metabolism and drug action are a possible solution to these challenges. Smart drugs that respond appropriately to varying physiological and pathological signals will be more effective and cause fewer side effects than existing treatments [4]. Non-protein components of smart drug systems, such as hydrogels, are in active development, but control of protein function (input and output) will be central to achieving advanced smart drug systems in vivo. Proteins are the ideal material for key biosensing and functional responses because they are capable of exquisitely sensitive stimulus-responsive behavior, and protein engineering capabilities are rapidly advancing [5–7]. Although protein engineering is often described as being important for future therapeutics, the role of protein engineering is frequently described in a narrow scope: focused primarily on alternative scaffolds which act as antibody substitutes, fusion proteins for increased half-life, PEGylation [8, 9], glycosylation [9, 10], or mutagenesis for reduced immunogenicity [9, 11–13]. In reality, the impact of protein engineering will be much broader. The tuning and design of protein stimulus-responsive behavior is now possible through protein engineering and this has the potential to transform the drug metabolism landscape. In particular, the use of engineered proteins for targeted drug activation or neutralization offers the potential for significantly enhanced control of pharmacokinetics and pharmacodynamics, and for reduced side effects when used in combination with powerful anticancer therapeutics. Many recent successes in related areas of biosensing, biocatalysis, and synthetic biology hold great potential for application in the emerging area of engineered drug metabolism [14–17].
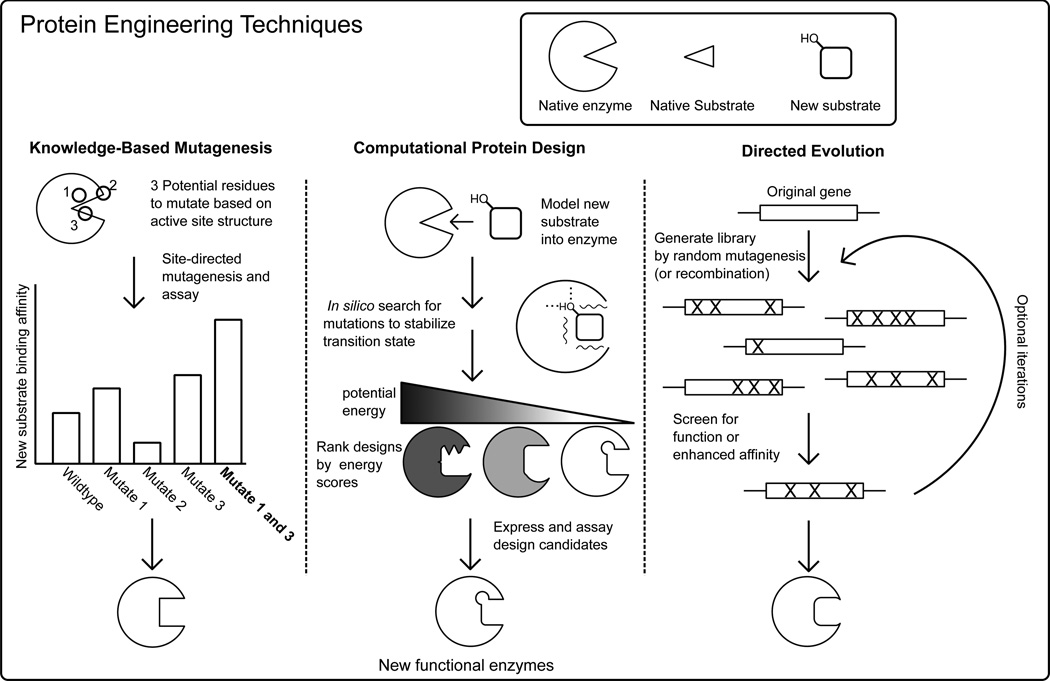
In addition to the modification of proteins via derivatization, for the purposes of this review, protein engineering consists of three major strategies: knowledge-based mutagenesis (KBM), computational protein design (CPD), and directed evolution (DE) (Fig. 1). The simplest form of rational protein engineering, knowledge-based mutagenesis (KBM), applies general biochemical principles and knowledge gained from prior studies to guide mutagenesis of native proteins with the goal of achieving improved or novel structural and/or functional properties. Computational protein design uses molecular modeling programs to predict amino acid sequences that will fold into a desired structure. This acts as a rigorous test of our understanding of the structure-function relationship [18]. CPD often entails generating protein design candidates by mutating residues on an existing high-resolution structure and then energetically evaluating the designs to find variants that are optimized for certain physicochemical properties such as protein stability or enzymatic activity [19]. Directed evolution introduces desired properties (e.g., enzymatic activity) into proteins via random mutation or gene recombination [20]. Functional variants with desired properties are then identified from these libraries through screening or selection. In its generic form, directed evolution lacks some of the de novo potential of computational design. However, DE can be applied to a protein without detailed knowledge of its structure or the detailed molecular mechanism required for its function [21]. Frequently, KBM, CPD, and DE methods are merged, allowing investigators to confer desired physicochemical properties efficiently and accelerate discovery [22]. Together, these approaches form a powerful toolset allowing us to manipulate an enzyme’s input and output sensitivity by either changing substrate specificity and binding affinity [23–25], conferring adaptive catalytic function [6], or creating novel activity [26]. A comprehensive review of KBM, CPD and DE are beyond the scope of this article; recent reviews of directed evolution and computational design achievements are covered elsewhere [5, 27, 28].
Fig. (1).
Simplified schematic of using protein engineering to redesign an enzyme’s substrate specificity highlighting knowledge-based mutagenesis, computational protein design, and directed evolution.
This review will focus on the application of recent advances in protein engineering to the development of stimulus-responsive protein therapeutics. Protein therapeutic drug delivery methods including gene therapy [29], and intracellular protein delivery [30] have been reviewed recently elsewhere, and are not covered in this review. Additionally, protein engineering efforts to increase in vivo stability such as PEGylation [31], and methods to reduce immunogenicity and antigenicity [32], while of great clinical significance, are beyond the scope of this review.
TARGETED DRUG NEUTRALIZATION
Specific antidotes that neutralize drugs or toxins in a selective manner are an important class of therapeutics that are increasingly available for a wide range of targets due to recent advances in the development of antibody therapeutics [33]. For narrow therapeutic window drugs such as the cardiac glycoside digoxin and the anti-coagulant warfarin, careful monitoring is required to maintain drug levels within a safe and effective range [34, 35]. Even with careful medical care, accidental overdoses of these medicines occur because an individual patient’s response to a drug may vary. The availability of an antidote can be lifesaving and contributes to the success of a therapeutic. The ability to counteract the effects of warfarin with vitamin K shots and digoxin with immune globulins has been important for the management of both drugs [35, 36]. Antibody-based antidotes have been developed for a growing number of drugs and toxins such as colchicine, desipramine, methamphetamine, cocaine, anthrax, and botulinum toxins [37, 38]. Alternative engineered protein scaffolds such as anticalins are also currently being developed as antidotes. For example, Eyer and colleagues described the testing of an anticalin as a digoxin antidote [39]. New oral anticoagulants have become available with more predictable pharmacokinetics than warfarin, but because no specific antidote was originally available some physicians were reluctant to prescribe them [38, 40]. However, this problem is in the process of being resolved, as Litzenburger and colleagues have recently reported an antibody fragment based antidote for the oral anticoagulant dabigatran [38].
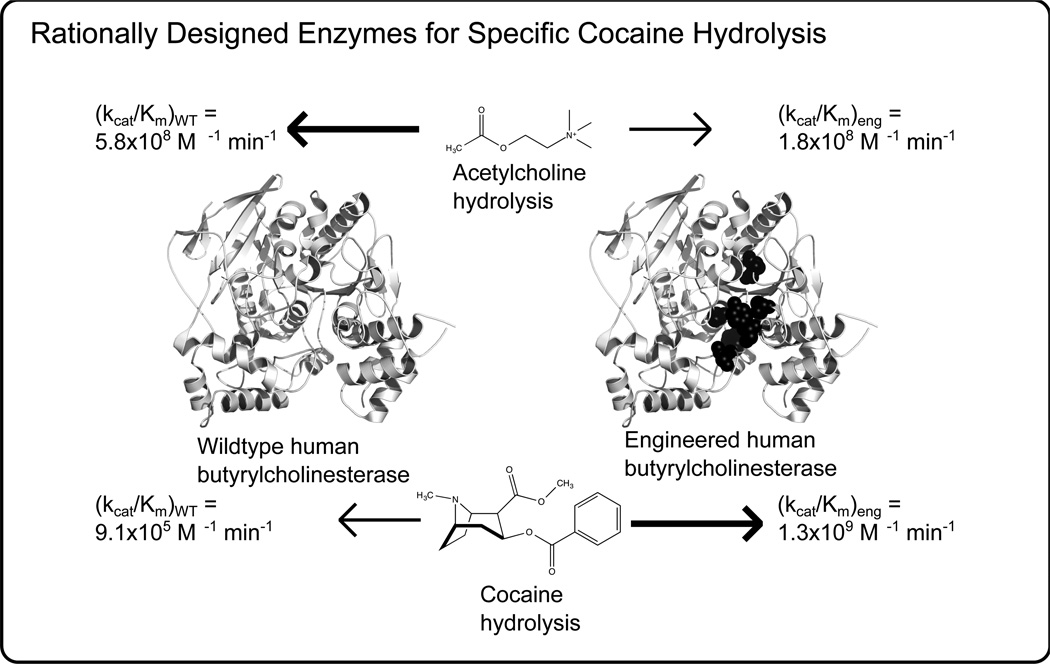
In addition to the more established class of antidotes which rely on molecular recognition for their activity, protein engineering is now being applied to develop enzymatic antidotes for therapeutic purposes. In particular, enzymatic antidotes are targeted for the treatment of overdoses of abused drugs and for addiction control [41]. Most noteworthy has been the development of hydrolases for the neutralization of cocaine (Fig. 2). For example, Xue et al. reported the CPD based engineering of human butyrylcholinesterase which yielded a variant with kcat/KM for (−)-Cocaine 1390 times larger than the wild type enzyme and also less reactive to its natural substrate acetylcholine [42]. Also, Lui designed nanocomplexes containing alcohol oxidase and catalase which reduced blood alcohol levels in mice [43]. The ability to significantly modify the function of a protein through protein engineering, such as the substrate specificity of human butyrylcholinesterase, highlights the growing potential of protein engineering for a wider impact on drug metabolism and medicine in general.
Fig. (2).
Human butyrylcholinesterase (represented by PDB code 1P0I) has been transformed into an effective cocaine hydrolysis enzyme by protein engineering. The best variant had a kcat/KM for (−)-Cocaine 1390 times larger than the wild type enzyme and also less reactive to its natural substrate acetylcholine [42]. The engineered enzyme contains six mutations in the active site, the locations of which are highlighted as black spheres, which accommodate the bulkier substrate and form more favorable hydrogen bonding interactions.
TARGETED DRUG ACTIVATION
One advantage of protein therapeutics over traditional small molecule drugs is their naturally stimulus-responsive behavior. Although, “smart” behavior was not originally engineered, its presence contributed to the success of several early protein therapeutics. For instance, tissue-type plasminogen activator (t-Pa) binds specifically to fibrin (a molecular recognition stimulus) and only then efficiently converts plasminogen to plasmin (a proteolytic response). Therefore, clot breakdown occurs locally in the vicinity of thrombi (where fibrin is deposited) in response to t-PA administration, instead of systemically, resulting in fewer side effects [44]. The most basic form of a stimulus-responsive protein therapeutic is one that is capable of targeted, biosensing-based activation. This activation can also be considered a specific pharmacokinetic metabolism of the protein therapeutic, for which it has been deliberately engineered. The engineering of protein therapeutics for targeted activation is still in an early stage, but is progressing rapidly. In this section, discussion begins with protein-based prodrugs, which includes both protein-small molecule conjugates in which the small molecule is activated upon reception of a signal by the protein component — as well as protein-protein systems in which protein components act as both biosensor and effector. Then, this sub-section of the review will move up in complexity to two component therapy systems in which engineered proteins activate prodrugs that are delivered separately, and then finally into light responsive systems in which enzymatic activity is controlled exogenously.
Antibody-Drug Conjugates
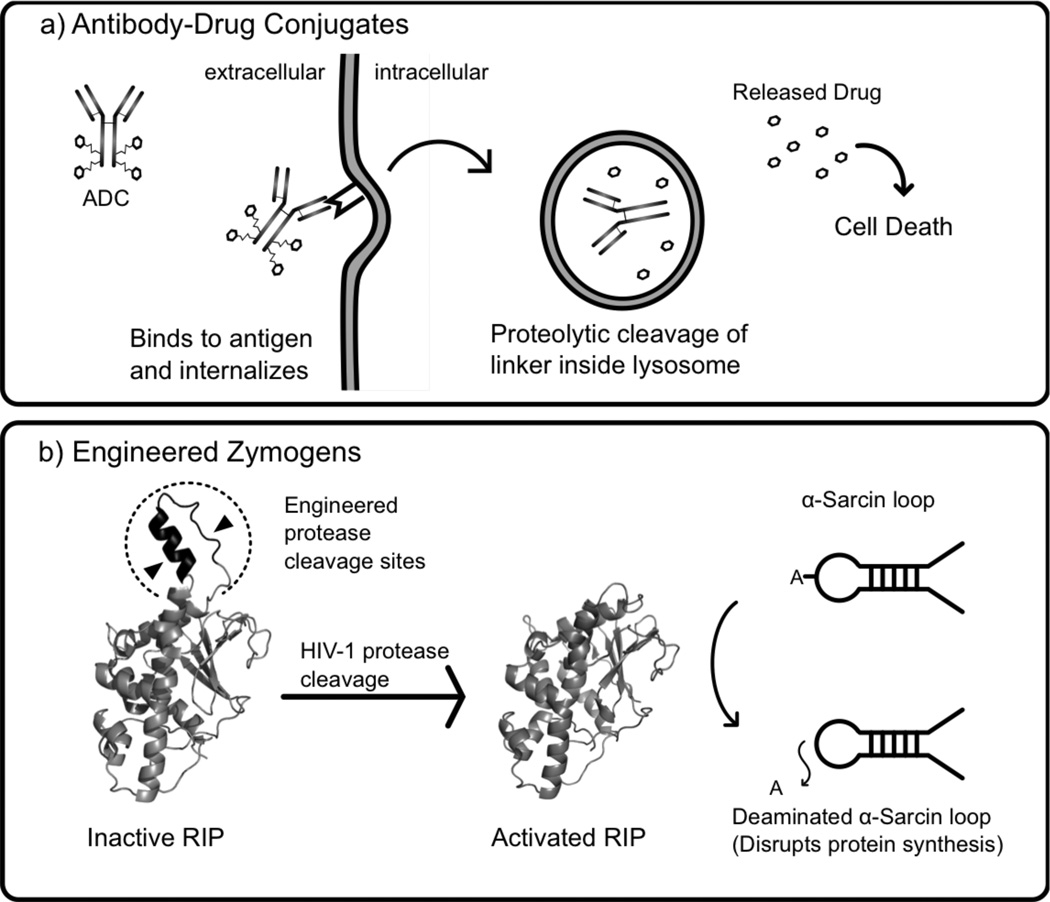
Antibody-drug conjugates (ADC) are the simplest form of stimulus-responsive protein therapeutics in which a monoclonal antibody is used to target the conjugated drug to specific locations, such as cancer cells [45, 46]. A sensitive linker connecting the antibody and drug allows selective release of the drug (Fig. 3a). High selectivity of binding to tumor cells and low cross-reactivity with healthy tissues are important parameters for selection of the monoclonal antibody. Due to the low quantity of ADC per administered dose that reaches the target location, high potency drugs are required and the linker needs to be sufficiently stable to avoid premature release of the drug. Most current ADCs are designed for cell internalization via receptor-mediated endocytosis prior to release of the drug, although non-internalizing and vascular targeting ADCs are also in development. Several strategies for responsive linkers are currently used including hydrazones, which are cleaved in the low pH environment of lysosomes and endosomes, disulfides, which allow drug release in the reducing environment of the cytosol, and cancer-specific protease liable peptides. Additionally, “non-cleavable” linkers, such as thioethers, are very stable and require enzymatic degradation of the internalized antibody in lysosomes or endosomes to release the drug [45–47]. Approximately 35 ADCs are currently being investigated in clinical studies, and two ADCs, brentuximab vedotin and adotrastuzumab emtansine, were recently approved for marketing by the US Food and Drug Administration (FDA) [48]. Brentuximab vedotin is composed of a monoclonal antibody which targets CD30, a protein expressed selectively in Hodgkin lymphoma, connected by cathepsin cleavable linkers to the anti-mitotic drug monomethyl auristatin E [45]. Adotrastuzumab emtansine is a conjugate of the anti-Her2-mAb Trastuzumab and the cytotoxic drug mertansine, linked by thioether linkages, and has shown promising results against advanced HER2 positive breast cancer [45]. Due to the conjugation methods currently employed, each monoclonal antibody may be conjugated to a variable number of drugs at different locations on the antibody in a heterogeneous manner, but a recently developed nonnative amino acid based selective labeling method reports the achievement of chemically homogenous ADCs with improved half-life, efficacy, and safety [49].
Fig. (3).
Illustrations of select classes of protein therapeutics engineered for targeted drug activation. A) Antibody-drug conjugates (ADCs) are composed of a monoclonal antibody securely linked to several molecules of a cytotoxic drug. The antibody selectively binds to a receptor on a cancer cell and is internalized via receptor mediated endocytosis. Once delivered to a lysosome, the linker is proteolytically cut and the drug is released leading to cell death [45, 137]. B) Engineered zymogens are proteins which have been engineered to be in an inactive state until activated by a specific signal, such as proteolytic cleavage. Law et al. engineered a zymogen variant of maize ribosome-inactivating protein (RIP) (represented by PDB code 2PQG) which is activated by HIV-1 protease for anti-HIV therapy [56]. Once activated, maize RIP (PDB code 2PQI) removes an adenine from the α-sarcin site on the large (28S) ribosomal subunit, disrupting protein synthesis [138].
Engineered Zymogens
Engineered zymogens are a class of stimulus-responsive protein therapeutics in which the protein acts as both the biosensor and effector. Like natural zymogens, engineered zymogens are in an inactive configuration prior to receiving an activating signal, such as proteolytic cleavage (Fig. 3b). A variety of engineered zymogens are currently in development including engineered coagulation cascade proteins, zymoxins, and dendronized proteases [50–52]. Zymoxins (engineered zymogen toxins via KBM) are currently an active area of research with significant emphasis on activation by viral proteases for selective killing of virally infected cells. Raines and collaborators pioneered this area of research by developing circularly permuted zymogens of bovine pancreatic ribonuclease (RNase A) with the natural termini connected with HIV or Hepatitus C protease cleavable linkers [53]. The potential of unmodified mammalian RNase A as a therapeutic is limited due to the presence of cytosolic ribonuclease inhibitor which binds mammalian RNase A homologs with high affinity, but variants of RNase A that are less sensitive to ribonuclease inhibitor have been engineered. An alternative solution has been presented recently by Callís et al. who used a similar circular permutation strategy to create an Onconase zymogen activated by HIV-1 protease [54]. Onconase is an amphibian homolog of Ribonuclease A, which is more highly cytotoxic, and less sensitive to ribonuclease inhibitor.
Using alternative toxins, the Benhar Lab recently developed several zymoxin fusion proteins formed by linking a diphtheria, ricin, or MazF toxin catalytic domain to an inhibitory peptide or domain via a Hepatitus C NS3 cleavable linker. The zymoxins were delivered to the cytosol as a transgene by an adenoviral vector or as a fusion protein containing the binding and translocation domain of Pseudomonas exotoxin A to facilitate entry of the zymogen into the cytosol of the target cells [51, 55]. The MazF system had the most promising results, being well tolerated by healthy cells, while eradicating Hepatitus C virus infected cells [55]. In related work, Shaw and collaborators have engineered HIV protease activation into several Ribosome-inactivating proteins (RIPs) for anti-HIV therapy. First, they redesigned the natural zymogen Maize RIP by inserting HIV-1 protease recognition sequences into the inactivation loop, creating several HIV-1 protease-activated Maize RIP zymogens (Fig. 3b) [56]. Additionally, they recently engineered zymogen-like variants of ricin by addition of HIV-1 protease recognition sequences to the C-terminus region [57]. In addition to anti-viral zymoxins, anti-cancer zymogens are also under development. Mühlebach et al. report the development of an oncolytic measles virus preferentially activated in liver tumor tissue through the engineering of matrix metalloproteinase cleavage sites into the measles virus F protein (a natural zymogen, normally activated by the protease furin) which mediates fusion of the viral and cellular membranes [58].
In addition to zymogens activated by proteolytic cleavage, proteins have been engineered via CPD, which respond to other stimuli including changes in temperature, pH, or exposure to reactive oxygen species (ROS). Many natural enzymes show significant activity only over a narrow temperature range. Through protein engineering, this active range can be tuned to create a temperature sensitive zymogen, which is activated within a designed range of temperature. As a proof of principle, the Wilson group recently reported the computational design of 100 adenylate kinase variants using a multi-state approach with a twenty degree range of melting temperatures and variable activity ranges [6]. Also, synthetic pH sensitive zymogens have been constructed by self-assembly of dendrimers onto the surface of trypsin, papain, and DNase I. At near neutral pH, the dendrimers sterically block the normal action of the enzymes, while at reduced pH the dendrimers dissociate, allowing activation of the zymogens [52]. There is growing realization of the importance of ROS signaling for a variety of cellular processes, and a large number of proteins have been identified that are responsive to ROS second messengers such as H2O2 through reversible cysteine oxidation mechanisms [59]. The possibility of engineering proteins to respond to ROS signals is appealing, and recently Callahan et al. engineered via CPD a reversibly redox regulated intein through the addition of a cysteine residue which neutralizes the protein’s intein activity under oxidizing conditions by the formation of a disulfide bond with a cysteine in the active site [60]. However, because other amino acids, such as lysine, arginine, and histidine can also be oxidized by ROS which can lead to loss of protein function [59], a better understanding of the interaction of ROS species with proteins is required for wider application of engineered ROS responsive proteins. With this goal in mind, the Wilson group recently used computational protein design to engineer a functional, lysine free adenylate kinase [61]. This lysine free construct was used to test the role of lysine in protein modification due to ROS exposure, and the Wilson group is in the process of extending this work to design of proteins that resistant to ROS degradation. Overall, protein engineering has proved useful for creating zymogens that activate in a specific cellular or environmental context. The next stage of therapeutic complexity involves further controlling a drug’s activity through two component systems such as exogenous light responsive control or directed enzyme prodrug therapy.
Directed Enzyme Prodrug Therapy
In directed enzyme prodrug therapy (DEPT), an enzyme is delivered in a targeted manner to the desired site of action. Then a non-bioactive prodrug is administered systemically, and activated locally by the previously delivered enzyme. In order to ensure that the enzymatic activation of the prodrug only occurs at the targeted location, both the prodrug and the activating enzyme typically need to be orthogonal to natural human enzymes. Several classes of DEPT are currently under development including Antibody Directed (ADEPT), Gene Directed (GDEPT), Bacterial Directed (BDEPT), and Substrate Mediated (SMEPT) with different requirements for engineering of the prodrug activating enzymes. ADEPT utilizes an antibody-enzyme conjugate where the antibody serves as the biomolecular recognition (sensing) component and the conjugated enzyme is the effector [62]. A clinical trial of a bacterial carboxypeptidase conjugated to a single chain variable domain directed against carcinoembryonic antigen (CEA) showed promise in stabilizing patients with CEA-expressing tumors, but treatment was limited due to immunogenicity. Next generation ADEPT efforts are focused on overcoming this hurdle through the use of humanized antibodies and bacterial prodrug activating enzymes with removed B- and T-cell epitopes [62, 63].
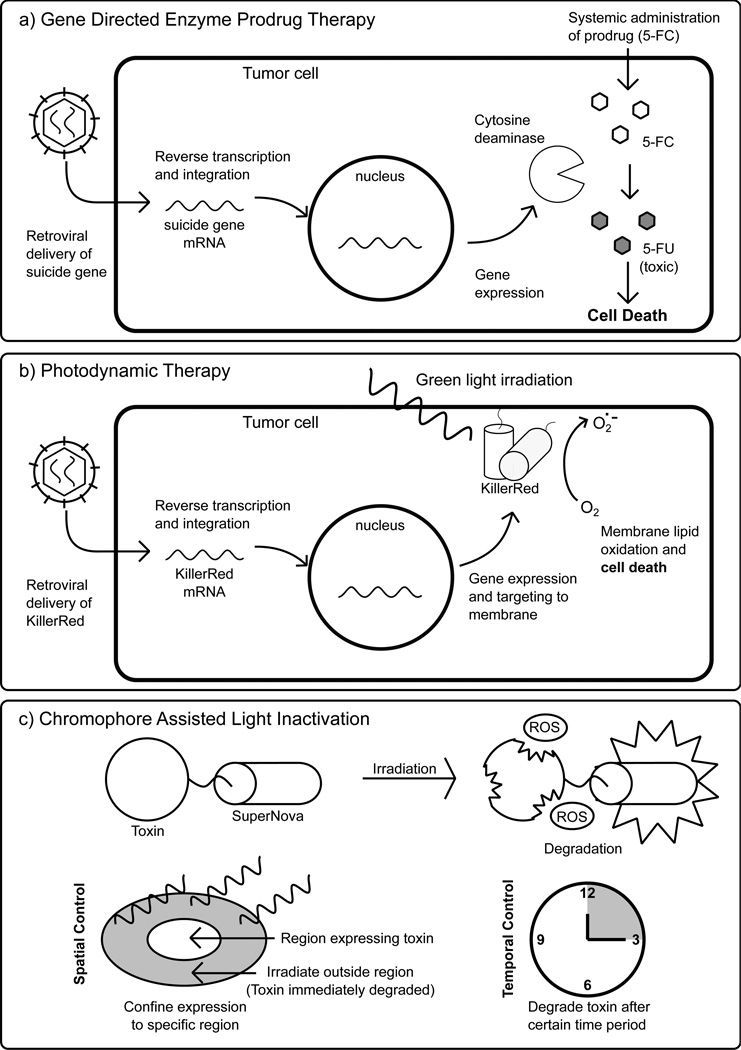
GDEPT, sometimes called suicide gene therapy, is a second class of DEPT that is rapidly developing as a cancer therapy and typically uses viral vectors (gene therapy) to deliver the therapeutic gene(s) (Fig. 4a) [64, 65]. An interesting variation on GDEPT, BDEPT, uses bacteria (typically armed with therapeutic genes), which preferentially colonize cancer cells to activate prodrugs locally [66]. Because current gene delivery methods are unlikely to reach all cancer cells, leakage of activated drug to neighboring cells (the so-called “bystander effect”) is often considered a positive aspect of GDEPT systems [65]. The most commonly used enzyme prodrug pairs are herpes simplex virus thymidine kinase activation of Ganciclovir, and cytosine deaminase activation of 5-Fluorocytosine (5-FC) (Fig. 4a), but there is currently active discovery of additional enzyme/prodrug systems [64]. For example, the use of type I nitroreductase from Leishmania major as an activator of leishmanicidal prodrugs has recently been proposed [67]. Protein engineering is required to move beyond naturally available enzyme systems, and progress has recently been achieved in this area. The active site of a thymidine-active deoxycytidine kinase was engineered via KBM for multiple prodrug activation so that multiple prodrugs could be administered and activated yielding synergistically enhanced bystander effect killing of cancer cells [68]. In many GDEPT systems, DNA-based control of expression of the prodrug activating enzyme serves as the biosensing component, but engineered stimulus-responsive prodrug activating enzymes are also in development. For example, the Ostermeier Lab engineered a fusion protein via KBM containing a CH1 domain from the human p300 protein as a HIF-1a recognition input domain (increased in certain cancer cells) and yeast cytosine deaminase as the prodrug activation output domain [69]. Finally, in addition to cancer therapy, Substrate Mediated Enzyme Prodrug Therapy is emerging as a component of tissue engineering and anti-inflammatory therapy in which a prodrug activating enzyme is embedded in a hydrogel or other biomaterial for increased control of surface mediated drug delivery [70].
Fig. (4).
A) In Gene Directed Enzyme Prodrug Therapy (GDEPT), a viral vector is used to deliver the gene for a prodrug activating enzyme to specific cells, such as cancer cells. Then, a non-bioactive prodrug is administered systemically, but only activated locally in the cancer cells where the gene is expressed [64]. In this example, the enzyme represented is cytosine deaminase which catalyzes the conversion of non-toxic 5-Fluorocytosine (5-FC) to cytotoxic 5-Fluorouracil (5-FU). B) In this application of photodynamic therapy, a protein photosensitizer, KillerRed, is delivered into tumor cells by a viral vector and targeted to the cell membrane. Irradiation of KillerRed leads to production of ROS, which can oxidize the cell membrane and result in cell death. C) A potential use of Chromophore Assisted Light Inactivation (CALI) in which the photosensitizer SuperNova is fused to a protein toxin. Due to the light responsive nature of CALI, the toxin’s activity can be limited to a confined location and duration.
Light Responsive On/Off Control of Drug Activity
Photodynamic therapy is a method for selectively killing cancer cells that involves systematic administration of a photosensitizer followed by local irradiation at tumors using fiber optic cables. When activated, the photosensitizers can generate reactive oxygen species (ROS), which can induce cell death. A number of small molecule photosensitizers have been FDA approved and are clinically used to treat certain tumors [71]. Protein-based photosensitizers have also been under investigation [72]. The most notable example is KillerRed, a dimeric Green Fluorescent Protein homolog that produces ROS by a Type I photosensitization mechanism [73, 74]. KillerRed can be localized to the mitochondria or cell membrane and induce cell death in response to green light [75](Fig. 4b). The advantages of KillerRed are that it is fully genetically encoded, it doesn’t require any additional co-factors, and since it is biodegradable it has a faster clearance time than small molecule photosensitizers [76].
Chromophores Assisted Light Inactivation (CALI) is another exciting venue of light controlled activity. CALI used photosensitizers fused to a target molecule of interest. When excited, the photosensitizers produce ROS that predominantly damage and inactivate the target molecule (and potentially any other closely interacting molecules). This effectively allows for more finely localized and controlled knockout experiments. KillerRed is a promising photosensitizer for CALI; however, its tendency to dimerize can interfere with the target molecule’s normal function. To rectify this, Takemoto et al. engineered a monomeric KillerRed variant known as SuperNova [77]. This was achieved by first using KBM to disrupt the dimerization interface and then applying DE to restore phototoxicity. While CALI shows great promise for studying cellular pathways and molecular mechanisms in vivo, it also can potentially be used therapeutically such as by fusing SuperNova to protein toxins. This would allow the expression of toxins to be confined in a spatially and temporally controlled manner (Fig. 4c).
In addition to the production of ROS, the ability to switch protein activity (on or off) with light opens up a wide range of clinically relevant effector functions. For example, the Wooley Lab used KBM and CPD to pioneer the use of engineered red light switchable proteins through the conjugation of photoisomerizable azobenzene groups to a variety of proteins including papain, RNase S, and a Fyn SH3 domain. This work has been extended by others to include the restriction enzyme scPvuII, [78], a cadherin [79], and most recently a naturally ATP-driven type II chaperonin was converted to a light-gated nanocage using the same method [80].
Additionally, in the last few years there has been an explosion in the development of engineered proteins for optogenetics, a technology in which engineered light sensitive proteins are genetically encoded and delivered by gene therapy [81]. Although simple light based targeted activation [82] and targeted neutralization methods [83] have been developed from engineered light sensitive protein domains, the greatest benefit comes from systems with light based on/off switchability. Light based on/off control can be achieved through short-lived reversibly activated states or through two wavelength control of activation and neutralization. The on/off capability of these systems allows true spatial and temporal control of protein activity, since diffusion or leakage of the protein outside the target area will cause it to be turned off [84]. Two of the most successful reversible light responsive protein components developed via KBM include the Light, Oxygen, Voltage or LOV domain, and the phytochrome B (PhyB) and phytochrome interacting factor 6 (Pif6) system. In particular, LOV domains are a subset of the larger PAS family of sensor proteins found in modular combination with a variety of effector domains. In response to blue light, LOV domains undergo a reversible allosteric shift due to the formation of a covalent bond between a conserved cysteine residue in the core of the protein and a carbon atom of the flavin cofactor’s isoalloxazine ring [85, 86]. LOV domains have been used to engineer many on/off light switchable fusion proteins including TrpR, Rac1, the histidine kinase FixL, and dihydrofolate reductase [85, 87]. Also, both LOV domains and the Pif6 system have been used to engineer light switchable protein-protein interactions [84, 87–89].
In addition to light-induced allostery and protein-protein interactions, protein engineering for photoinduced energy and electron transfer is an emerging research area with potential application to stimulus responsive protein therapeutics. Tran et al. recently reported a cytochrome P450 BM3 variant labeled with a Ruthenium polypyridine photosensitizer that demonstrated efficient light-driven hydroxylation of lauric acid [90]. Cryptochromes are an additional class of light sensing proteins which are being explored for biotechnological uses including light activated protein-protein interactions [91]. Cryptochromes are related to phytolases and frequently contain an antenna domain containing a light-harvesting chromophore which subsequently transfers excitation energy to a reduced flavin in the catalytic domain via energy transfer. In phytolases, electron transfer then proceeds from the excited FADH−* through three conserved tryptophan residues to a pyrimidine dimer, repairing the DNA by splitting it back into monomers [92]. Cryptochromes undergo a conformational change upon light absorption, which has been shown to be involved in their natural signaling response, but the role of their electron transfer functionality is not yet fully understood [93, 94]. Parallel to the Cheruzel group’s demonstration with cytochrome p450, photoinduced energy and electron transfer could allow localized activity of engineered therapeutic oxidoreductases (perhaps of non-human derivation) without their natural partner, as well as light induced activation or neutralization of engineered redox proteins to therapeutically control ROS signaling or other redox sensitive cellular pathways. However, better understanding of intraprotein energy and electron transfer will be required to engineer therapeutic proteins that utilize these mechanisms. Toward this end the Wilson group recently reported a new energy transfer mechanism which resolved confounding observations in ruthenium polypyridine labeled azurin, which has been one of the main model systems for studying intramolecular electron transfer in proteins [95] and Wilson et al. are currently using computational protein design to tune energy and electron transfer rates in the same model system.
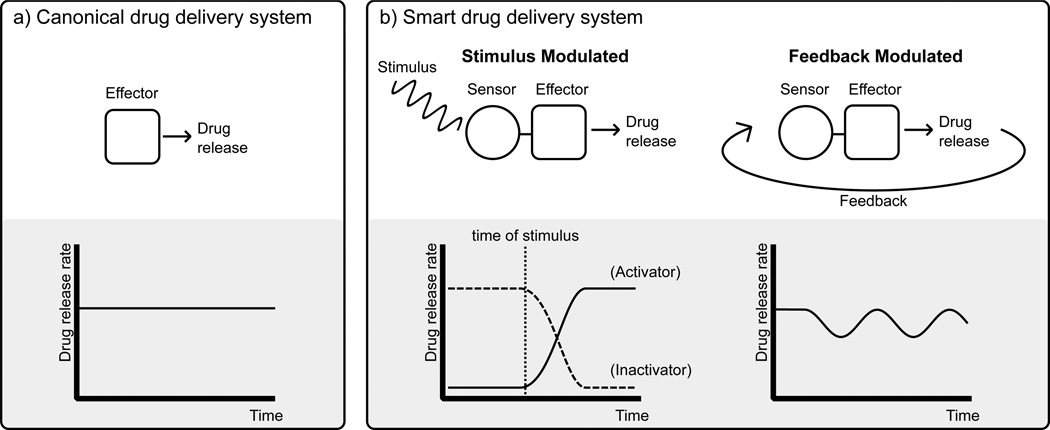
SMART RESPONSE DRUG SYSTEM DESIGN
Smart response drug systems are an exciting frontier of drug development, which can greatly benefit from increased protein engineering capabilities. Ideally, smart response drug systems utilize feedback mechanisms to intelligently modulate a therapeutic effect in response to biomarkers or other relevant stimuli. In many existing delayed release systems, a drug is released slowly at an approximately constant rate. In contrast, smart drug delivery systems require the presence of external stimuli to either turn on the release of a drug in activation-modulated release systems or to modulate the extent of a drug that is released in feedback-modulated release systems [96] (Fig. 5). Smart response systems are frequently multicomponent and may include multiple proteins in biosensing, logical, and effector roles. This section of the review begins by highlighting current work related to engineering protein allostery, which is a key technology for general development of the stimulus-responsive proteins needed for smart response systems. Then we review recent progress toward three emerging areas where engineered proteins can play critical roles: biochemical logic based stimulus-responsive delayed release systems, drug buffers for narrow therapeutic window drugs, and network medicine. Although most work so far on smart response drug systems is related to drug delivery, the recent protein engineering work related to network medicine highlights the possibility of using stimulus-responsive proteins in other complex therapeutic applications.
Fig. (5).
Drug delivery system strategies. A) Current drug delivery methods typically involve releasing a drug at a relatively constant rate. B) Smart drug delivery systems have both a sensing and effector component. This can allow drug release to either be activated (or inhibited) in response to a specific stimulus (e.g., pH, light, or specific biomarkers) or regulated in a feedback-dependent manner to provide a consistent free drug concentration.
Engineering of Protein Allostery
The ability to engineer allosteric responses in proteins for the linkage of disparate biosensing and effector functions is a key technology that will enable more advanced protein based smart response drug systems. Although much work remains to be done in this area, in addition to the LOV-based allostery work previously mentioned, there have been a variety of recent successes in this area, which highlight the increasing ability of protein engineers to modulate and design allosteric protein systems. Allostery can be engineered in several ways — e.g., through the introduction of mutations within an existing protein, or by the addition of a novel domain, which provides allosteric regulation.
In the first category, Deckert et al. used KBM to introduce allosteric control into β-glycosidase and β-glucuronidase enzymes by the introduction of tryptophan-to-glycine mutations which caused loss of function which could be rescued by the addition of indole [97]. Rana et al. used loop mutagenesis to switch thrombin catalytic activation specificity from Na+ to K+ [98]. Also, Wu et al. used an allostery inspired approach to modify the substrate specificity of a thermostable Baeyer–Villiger monooxygenase through directed evolution involving mutants away from the active site which conferred the desired altered substrate specificity as a result of mutationally induced large domain movements [99]. Additionally, the Wilson group recently used directed evolution to engineer alternate cooperative-communication in the lactose repressor (LacI) [21]. Starting with an allosterically ‘dead’ LacI mutant (D88A) that binds and represses DNA in both the absence and presence of its inducer (IPTG), error-prone PCR was used to introduce compensatory mutations. Screening yielded five new functional variants: three with wildtype-like repressor phenotype, and two with functionally inverted co-repressor phenotype (similar to the related purine repressor). One of the best recent analyses of allosteric communication came from the Ranganathan lab. Using statistical coupling analysis, they were able to predict the residues involved in allosteric communication in both the PDZ domain. These positions were then experimentally validated through a full saturation mutagenesis study [100].
Several groups have also recently conferred allostery through the addition of a regulatory domain to a non-allosteric protein. Cross et al. used KBM to introduce allosteric control into a 3-deoxy-Darabino-heptulosonate 7-phosphate synthase by the addition of an ACT domain [101]. Rizk et al. used a directed evolution-based phage display strategy to engineer synthetic antigen binders (sABs) that recognized the bound form of maltose-binding protein (MBP) to rescue a binding-deficient mutant of MBP [102]. Finally, Dagliyan et al. rationally designed via KBM a single chain regulatory element by “rewiring” the rapamycin binding complex of FK506-binding protein and FKBP12-rapamycin binding protein into a single polypeptide chain and inserting this new sequence called “uniRapR” into Src kinase, yielding a system which demonstrated functional allostery in HeLa cells and zebrafish tissue [103]. In general, the engineering of allostery remains challenging, but the plasticity of allosteric pathways demonstrated by the reports described in this section is encouraging because this plasticity should allow the tuning of protein allostery needed for engineering therapeutic systems.
Stimulus-Responsive Drug Release Using Biochemical Logic
Although there are increasing developments in microelectronic systems, which can interface with biosensors and regulate drug release systems, purely biochemical logic systems are advantageous for certain applications, such as smart pills or implantable drug release systems for which full biodegradability after their service lifetime is desired. Peppas and colleagues pioneered the use of proteins in a stimulus-responsive delayed release system for the feedback regulated control of insulin release from an engineered hydrogel. The incorporation of glucose oxidase and catalase into a pH sensitive P(DEAEM-g-EG) hydrogel created a system in which enzyme catalyzed conversion of glucose caused a drop in pH, causing the hydrogel to expand and release insulin. Additionally, feedback control was achieved due to reversible swelling in response to pulsatile variations in glucose [4, 104, 105].
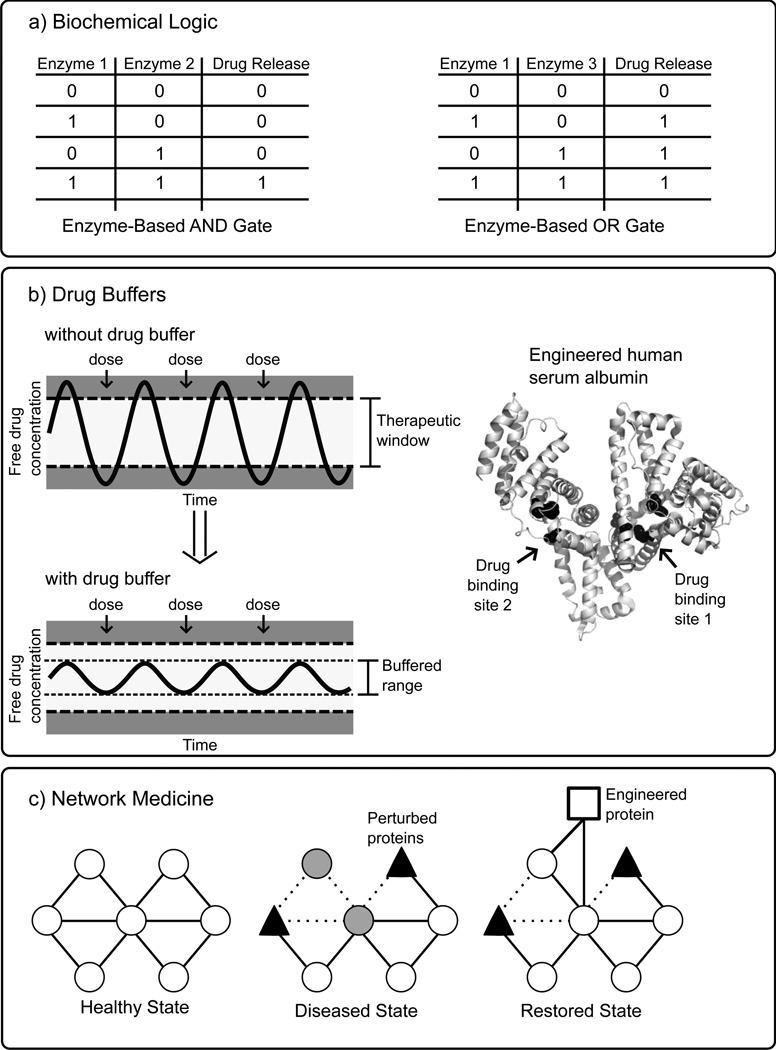
Recently, this work has been extended utilizing proteins as molecular recognition elements which regulate release of a drug from an engineered hydrogel [105], and the Katz, Privman, and Wang labs have developed a variety of biochemical logic systems (Fig. 6a) utilizing cascades of enzymes to process multiple biomarkers, and ultimately trigger the release of a drug in the presence of certain input combinations [106]. Tokarev et al. reported the combination of a pH sensitive stimulus-responsive hydrogel with biochemical logic systems. In these systems, a set of enzymes served as the inputs and only an output that corresponded to a drop in pH would activate the hydrogel. For example, an AND gate required the combined action of invertase and glucose oxidase to convert sucrose to gluconic acid to lower the pH. Alternatively, an OR gate was formed by swapping out the invertase with an esterase and adding ethyl butyrate to the system, so that the presence of either enzyme would cause a drop in pH. In both cases, the inclusion of urea in the system allowed a reset functionality by the input of urease which could restore the original pH value [107]. This work was extended by Privman et al. to construct a biochemical AND gate with required inputs of alanine transaminase and lactate dehydrogenase, biomarkers characteristic of liver injury, which activated a cascade through output of NAD+ allowing glucose dehydrogenase to produce gluconic acid yielding a drop in pH which switched on a polymer-modified electrode [108]. A similar system described by Zhou et al. was designed as a biochemical logic “Sense-Act-Treat” system which responded to biomarkers characteristic of abdominal trauma with the release of a drug [109]. Finally, more complex biochemical logic networks with multiple inputs, including both enzymes and their substrates, were reported which culminated in a simulated drug release [110]. So far, these biochemical logic systems have relied on combinations of natural enzymes, but future systems will likely be further tailored and expanded through the inclusion of engineered proteins.
Fig. (6).
Illustrations of protein engineering enabled smart response drug systems. A) Stimulus-responsive delayed release drug systems have been proposed which utilize combinations of enzymes which can integrate multiple inputs according to biochemical (typically Boolean) logic to release an appropriate amount of one or several drugs under specified conditions [106]. B) A drug buffer is an engineered multivalent ligand-binding protein therapeutic which has been designed to bind or release a narrow therapeutic window drug in such a manner that the free drug concentration is maintained within a safe and effective range. For illustrative purposes, human serum albumin (PDB code 1O9X) is shown with potential engineered drug binding sites at Sudlow’s site 1 and site 2 (black spheres) [116]. C) Network medicine is new therapeutic approach which aims to restore health by targeting and correcting aberrant signaling networks associated with cancer and other diseases. In this highly simplified schematic, a disease state is the result of perturbed proteins which disrupt the organization of the healthy signaling network. The introduction of an engineered signaling protein “re-wires” the network around the perturbed proteins, restoring health.
Drug Buffer
A drug buffer is a second therapeutic component, administered along with a narrow therapeutic window drug, which actively maintains the serum free drug concentration within a safe and effective range (Fig. 6b). Natural biological systems use many classes of ligand-binding proteins for transport and controlled release of small molecules including lipid binding proteins, periplasmic binding proteins, lectins, and serum albumins, and recent progress in protein engineering of ligand binding proteins holds great promise for responsive control of drug transport and delivery [111]. There has been significant development in recent years in the use of human serum albumin (HSA) as a drug carrier to increase the half-life of both small molecule and protein therapeutics either by direct conjugation or through the attachment of fatty acids or other moieties known to bind to HSA. In particular, fatty acid derivatives of insulin such as insulin detemir have been very successful, and Abraxane®, an albumin-bound form of paclitaxel, was recently approved by the FDA for use against certain metastatic lung and prostate cancers [112]. Additionally, engineering of albumin binding to the neonatal Fc receptor has been demonstrated as an additional mechanism for increased half-life [113, 114].
Although drug binding to HSA is well-studied [115–117] and new information continues to accumulate [118], use of this data for engineering of HSA for tuning of drug binding has so far been limited. Baker and colleagues recently reported a significant advance in the engineering of ligand-binding proteins with the successful development of a digoxin binding protein with picomolar affinity, on par with anti-digoxin antibody therapeutics [119]. This advance was realized through improved computational design methods incorporating hydrogen-bonding interactions, followed by directed evolution refinement utilizing high-throughput methods including yeast surface display, fluorescence-activated cell sorting, and next generation deep sequencing. Their selection of digoxin is particularly encouraging for the resolution of ongoing problems with dosing of narrow therapeutic index drugs like digoxin. In the near future, these improved protein engineering methods may allow the construction of engineered ligand-binding proteins based on scaffolds such as HSA which can act as drug buffers for narrow therapeutic index drugs. These engineered drug buffers would contain multiple drug binding sites with tailored affinities to maintain free drug plasma concentration within a safe and effective range. For a bivalent drug buffer, when the free drug concentration is within the designed range site 1 (Fig. 6b) would bind the drug, and if the free drug concentration fell below level 1, drug would be released from site 1, helping to maintain free drug concentration within an effective range. If free drug concentration instead rose above level 2, lower affinity site 2 would bind the drug, keeping the free drug concentration within a safe range.
Network Medicine and Cell Therapy
Network medicine is a new therapeutic approach, which aims to restore health by targeting and correcting aberrant signaling networks associated with cancer and other diseases (Fig. 6c) [120, 121]. Many existing drugs target a specific protein associated with a disease, but the clinical results from many of these drugs have been less promising and more variable than expected. Network medicine is a complex endeavor, which builds on recent evidence linking multiple genes to disease states. It uses new quantitative methods to measure signaling network status, computational methods to model the signaling network, and multistage, multicomponent drug treatment programs based on measured network status and model predictions [122]. In 2012, Lee et al. reported pioneering results of a network medicine approach to fight triple-negative breast cancer (TNBC). In this study, a systematic time and dose dependent approach to identifying drug combinations which were most effective at killing TNBC was evaluated using multiple types of quantitative data in combination with computational network models. The authors found that sequential treatment with the EGF receptor kinase inhibitor erlotinib, followed by the DNA damaging drug doxorubicin was more effective that simultaneous treatment because several hours of treatment with erlotinib was required to modulate the EGFR pathway sufficiently to induce a TNBC phenotype with increased susceptibility to doxorubicin [123, 124].
One challenge to implementing network medicine is the limited number of drug targets accessible with FDA approved drugs, and the promiscuity of many of these drugs [125]. The use of multiple small molecule drugs at low doses is one possible solution [125, 126], but in the long run engineered proteins introduced into the diseased cells to reprogram aberrant cell signaling networks holds the greatest promise for specific control [120]. The use of engineered proteins to reprogram cell signaling networks is in fact a biomimetic approach, which can leverage increasing scientific knowledge of similar methods used by pathogens to disrupt and usurp signaling pathways [127, 128]. The modularity of signaling proteins, which are typically composed of different combinations of conserved domain types, holds great potential for engineering, and recent efforts to engineer cell signaling proteins for synthetic biology may be applied towards network medicine [129–131]. For example, Lim and colleagues have previously developed libraries of synthetic signaling proteins with tunable input/output control through the combination of modular autoinhibitory domains [132], and recently developed an orthogonal (non-crossreacting) Intersectin/GTPase Cdc42 signaling system by computational redesign of the interface between these two proteins [133]. Delivery of multiple engineered signaling proteins into target cells will likely be challenging, but in addition to gene therapy approaches, Carleton et al. have recently developed an engineered type III secretion system for use in vaccine delivery [134], and this technology is also amenable for use in network medicine. Finally, cells are the ultimate “smart” therapeutic vehicle, and the use of cell-based therapeutics will depend on engineering of signaling pathway proteins for tailored stimulus-responsive properties [135]. Park et al. have made initial steps in this direction by demonstrating that a T lymphocyte expressing a G protein-coupled receptor engineered to respond to a bioinert drug-like small molecule, clozapine N-oxide, will migrate to the site of clozapine N-oxide-releasing beads implanted in a live mouse [136].
CONCLUSIONS
Medicine and in particular pharmacology are currently on the verge on an exciting transition from small molecule chemical therapy to biological therapy encompassing protein therapeutics, gene therapy, and cellular therapy. Because of the centrality of proteins to biological function, protein engineering is a common and critical enabling technology for these emerging biological therapies. For protein therapeutics, control of half-life and immunogenicity are essential for clinical use, and protein engineering in these areas will continue to be important. However, the full potential of protein engineering for future therapeutics is much broader, and is centered on the modulation of protein function, and especially stimulus-responsive function. This will likely become increasingly apparent as gene therapy and other intracellular protein delivery methods improve, and protein design constraints related to traditional delivery methods of purified protein are reduced or eliminated.
As the recent work highlighted in this review demonstrates, protein engineering capabilities are rapidly advancing, and of particular relevance for protein therapeutics are the varied efforts to introduce and tune switchable protein activity. For select applications, such as enzyme replacement therapy, natural proteins may be sufficient to achieve a positive clinical outcome, but in general novel stimulus-responsive protein behaviors are desired and so these must be engineered. Also, therapeutics frequently need to be potent molecules in order to achieve clinical results, but this potency increases the likelihood of deleterious side effects. In the body, naturally potent proteins have their expression carefully regulated and often are further post-translationally controlled, being produced as inactive zymogens, and quickly degraded or neutralized when no longer needed. Leveraging and expanding on these natural modules through engineering protein therapeutics to be stimulus-responsive for targeted activation and/or targeted neutralization is a new frontier with tremendous growth potential. Current research toward better understanding of protein molecular recognition, allostery, and catalysis, and further development of computational protein design methods will speed this transition.
ABBREVIATIONS
- DE
Directed Evolution
- CPD
Computational Protein Design
- KBM
Knowledge-based Mutagenesis
- t-PA
Tissue-type plasminogen activator
- ADC
Antibody-drug conjugate
- FDA
US Food and Drug Administration
- RNaseA
Ribonuclease A
- RIP
Ribosome-inactivating protein
- ROS
Reactive oxygen species
- CALI
Chromophore Assisted Light Inactivation
- DEPT
Directed Enzyme Prodrug Therapy
- ADEPT
Antibody Directed Enzyme Prodrug Therapy
- GDEPT
Gene Directed Enzyme Prodrug Therapy
- BDEPT
Bacterial Directed Enzyme Prodrug Therapy
- SMEPT
Substrate Mediated Enzyme Prodrug Therapy
- CEA
Carcinoembryonic antigen
- 5-FC
5-Fluorocytosine
- 5-FU
5-Fluorouracil
- PDT
Photodynamic therapy
- LOV
Light, Oxygen, or Voltage Domain
- PhyB
Phytochrome B
- Pif6
Phytochrome interacting factor 6
- HSA
Human Serum Albumin
- TNBC
Triple-negative breast cancer
REFERENCES
- 1.Burns M. Management of narrow therapeutic index drugs. Journal of thrombosis and thrombolysis. 1999;7(2):137–143. doi: 10.1023/a:1008829403320. [DOI] [PubMed] [Google Scholar]
- 2.Blix HS, Viktil KK, Moger TA, Reikvam A. Drugs with narrow therapeutic index as indicators in the risk management of hospitalised patients. Pharmacy Practice (18863655) 2010;8(1) doi: 10.4321/s1886-36552010000100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Litvak-Greenfeld D, Benhar I. Risks and untoward toxicities of antibody-based immunoconjugates. Advanced drug delivery reviews. 2012;64(15):1782–1799. doi: 10.1016/j.addr.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Wanakule P, Roy K. Disease-Responsive Drug Delivery: The Next Generation of Smart Delivery Devices. Curr Drug Metab. 2012;13(1):42–49. doi: 10.2174/138920012798356880. [DOI] [PubMed] [Google Scholar]
- 5.Kiss G, Celebi-Olcum N, Moretti R, Baker D, Houk KN. Computational Enzyme Design. Angew Chem Int Edit. 2013;52(22):5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- 6.Howell SC, Inampudi KK, Bean DP, Wilson CJ. Understanding thermal adaptation of enzymes through the multistate rational design and stability prediction of 100 adenylate kinases. Structure. 2014;22(2):218–229. doi: 10.1016/j.str.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Lutz S. Beyond directed evolution-semi-rational protein engineering and design. Curr Opin Biotech. 2010;21(6):734–743. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelegri-O'Day EM, Lin EW, Maynard HD. Therapeutic Protein-Polymer Conjugates: Advancing Beyond PEGylation. J Am Chem Soc. 2014;136(41):14323–14332. doi: 10.1021/ja504390x. [DOI] [PubMed] [Google Scholar]
- 9.Carter PJ. Introduction to current and future protein therapeutics: A protein engineering perspective. Exp Cell Res. 2011;317(9):1261–1269. doi: 10.1016/j.yexcr.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Costa AR, Rodrigues ME, Henriques M, Oliveira R, Azeredo J. Glycosylation: impact, control and improvement during therapeutic protein production. Critical reviews in biotechnology. 2014;34(4):281–299. doi: 10.3109/07388551.2013.793649. [DOI] [PubMed] [Google Scholar]
- 11.Kimchi-Sarfaty C, Schiller T, Hamasaki-Katagiri N, Khan MA, Yanover C, Sauna ZE. Building better drugs: developing and regulating engineered therapeutic proteins. Trends in pharmacological sciences. 2013;34(10):534–548. doi: 10.1016/j.tips.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Caravella J, Lugovskoy A. Design of next-generation protein therapeutics. Current opinion in chemical biology. 2010;14(4):520–528. doi: 10.1016/j.cbpa.2010.06.175. [DOI] [PubMed] [Google Scholar]
- 13.Ryu JK, Kim HS, Nam DH. Current status and perspectives of biopharmaceutical drugs. Biotechnol Bioproc E. 2012;17(5):900–911. [Google Scholar]
- 14.Nestl BM, Nebel BA, Hauer B. Recent progress in industrial biocatalysis. Current opinion in chemical biology. 2011;15(2):187–193. doi: 10.1016/j.cbpa.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Bommarius AS, Blum JK, Abrahamson MJ. Status of protein engineering for biocatalysts: how to design an industrially useful biocatalyst. Current opinion in chemical biology. 2011;15(2):194–200. doi: 10.1016/j.cbpa.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Blackstock D, Park M, Sun Q, Tsai S-L, Chen W. Engineering protein modules for diagnostic applications. Current Opinion in Chemical Engineering. 2013;2(4):416–424. [Google Scholar]
- 17.Zhang FZ, Keasling J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011;19(7):323–329. doi: 10.1016/j.tim.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Alvizo O, Allen BD, Mayo SL. Computational protein design promises to revolutionize protein engineering. BioTechniques. 2007;42(1):31, 33, 35. doi: 10.2144/000112336. passim. [DOI] [PubMed] [Google Scholar]
- 19.Das R, Baker D. Macromolecular modeling with rosetta. Annual review of biochemistry. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 20.Kuchner O, Arnold FH. Directed evolution of enzyme catalysts. Trends in biotechnology. 1997;15(12):523–530. doi: 10.1016/S0167-7799(97)01138-4. [DOI] [PubMed] [Google Scholar]
- 21.Meyer S, Ramot R, Kishore Inampudi K, Luo B, Lin C, Amere S, Wilson CJ. Engineering alternate cooperative-communications in the lactose repressor protein scaffold. Protein engineering, design & selection : PEDS. 2013;26:433–443. doi: 10.1093/protein/gzt013. [DOI] [PubMed] [Google Scholar]
- 22.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453(7192):190-U4. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 23.Boas FE, Harbury PB. Design of protein-ligand binding based on the molecular-mechanics energy model. Journal of molecular biology. 2008;380(2):415–424. doi: 10.1016/j.jmb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang JH, Dawes G, Stemmer WP. Directed evolution of a fucosidase from a galactosidase by DNA shuffling and screening. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(9):4504–4509. doi: 10.1073/pnas.94.9.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CY, Georgiev I, Anderson AC, Donald BR. Computational structure-based redesign of enzyme activity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3764–3769. doi: 10.1073/pnas.0900266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St Clair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329(5989):309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutz S. Beyond directed evolution--semi-rational protein engineering and design. Curr Opin Biotechnol. 2010;21(6):734–743. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson CJ. Rational protein design: developing next-generation biological therapeutics and nanobiotechnological tools. Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology. 2014 doi: 10.1002/wnan.1310. [DOI] [PubMed] [Google Scholar]
- 29.Sheridan C. Gene therapy finds its niche. Nature biotechnology. 2011;29(2):121–128. doi: 10.1038/nbt.1769. [DOI] [PubMed] [Google Scholar]
- 30.Du J, Jin J, Yan M, Lu Y. Synthetic nanocarriers for intracellular protein delivery. Curr Drug Metab. 2012;13(1):82–92. doi: 10.2174/138920012798356862. [DOI] [PubMed] [Google Scholar]
- 31.Milla P, Dosio F, Cattel L. PEGylation of proteins and liposomes: a powerful and flexible strategy to improve the drug delivery. Curr Drug Metab. 2012;13(1):105–119. doi: 10.2174/138920012798356934. [DOI] [PubMed] [Google Scholar]
- 32.Liu W, Onda M, Lee B, Kreitman RJ, Hassan R, Xiang L, Pastan I. Recombinant immunotoxin engineered for low immunogenicity and antigenicity by identifying and silencing human B-cell epitopes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(29):11782–11787. doi: 10.1073/pnas.1209292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferner R. Raising more antibodies. Bmj-Brit Med J. 2013;347 doi: 10.1136/bmj.f5969. [DOI] [PubMed] [Google Scholar]
- 34.Lenzini P, Wadelius M, Kimmel S, Anderson JL, Jorgensen AL, Pirmohamed M, Caldwell MD, Limdi N, Burmester JK, Dowd MB, Angchaisuksiri P, Bass AR, Chen J, Eriksson N, Rane A, Lindh JD, Carlquist JF, Horne BD, Grice G, Milligan PE, Eby C, Shin J, Kim H, Kurnik D, Stein CM, McMillin G, Pendleton RC, Berg RL, Deloukas P, Gage BF. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clinical pharmacology and therapeutics. 2010;87(5):572–578. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehle M, Patel C, Giugliano RP. Digoxin: clinical highlights: a review of digoxin and its use in contemporary medicine. Critical pathways in cardiology. 2011;10(2):93–98. doi: 10.1097/HPC.0b013e318221e7dd. [DOI] [PubMed] [Google Scholar]
- 36.Levi M, Eerenberg E, Kamphuisen PW. Bleeding risk and reversal strategies for old and new anticoagulants and antiplatelet agents. Journal of thrombosis and haemostasis : JTH. 2011;9(9):1705–1712. doi: 10.1111/j.1538-7836.2011.04432.x. [DOI] [PubMed] [Google Scholar]
- 37.Rainey GJA, Young JAT. Antitoxins: Novel strategies to target agents of bioterrorism. Nat Rev Microbiol. 2004;2(9):721–726. doi: 10.1038/nrmicro977. [DOI] [PubMed] [Google Scholar]
- 38.Schiele F, van Ryn J, Canada K, Newsome C, Sepulveda E, Park J, Nar H, Litzenburger T. A specific antidote for dabigatran: functional and structural characterization. Blood. 2013;121(18):3554–3562. doi: 10.1182/blood-2012-11-468207. [DOI] [PubMed] [Google Scholar]
- 39.Eyer F, Steimer W, Nitzsche T, Jung N, Neuberger H, Muller C, Schlapschy M, Zilker T, Skerra A. Intravenous application of an anticalin dramatically lowers plasma digoxin levels and reduces its toxic effects in rats. Toxicology and applied pharmacology. 2012;263(3):352–359. doi: 10.1016/j.taap.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Vílchez JA, Gallego P, Lip GY. Safety of new oral anticoagulant drugs: a perspective. Therapeutic Advances in Drug Safety. 2014;5(1):8–20. doi: 10.1177/2042098613507945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorelick DA. Pharmacokinetic strategies for treatment of drug overdose and addiction. Future medicinal chemistry. 2012;4(2):227–243. doi: 10.4155/fmc.11.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue L, Ko MC, Tong M, Yang W, Hou S, Fang L, Liu J, Zheng F, Woods JH, Tai HH, Zhan CG. Design, preparation, and characterization of high-activity mutants of human butyrylcholinesterase specific for detoxification of cocaine. Molecular pharmacology. 2011;79(2):290–297. doi: 10.1124/mol.110.068494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Du JJ, Yan M, Lau MY, Hu J, Han H, Yang OO, Liang S, Wei W, Wang H, Li JM, Zhu XY, Shi LQ, Chen W, Ji C, Lu YF. Biomimetic enzyme nanocomplexes and their use as antidotes and preventive measures for alcohol intoxication. Nat Nanotechnol. 2013;8(3):187–192. doi: 10.1038/nnano.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craik CS, Page MJ, Madison EL. Proteases as therapeutics. Biochem J. 2011;435:1–16. doi: 10.1042/BJ20100965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casi G, Neri D. Antibody-drug conjugates: basic concepts, examples and future perspectives. Journal of controlled release : official journal of the Controlled Release Society. 2012;161(2):422–428. doi: 10.1016/j.jconrel.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 46.Alley SC, Okeley NM, Senter PD. Antibody-drug conjugates: targeted drug delivery for cancer. Current opinion in chemical biology. 2010;14(4):529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 47.Teicher BA, Doroshow JH. The promise of antibody-drug conjugates. The New England journal of medicine. 2012;367(19):1847–1848. doi: 10.1056/NEJMe1211736. [DOI] [PubMed] [Google Scholar]
- 48.Beck A, Reichert JM. Antibody-drug conjugates: present and future. mAbs. 2014;6(1):15–17. doi: 10.4161/mabs.27436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian F, Lu Y, Manibusan A, Sellers A, Tran H, Sun Y, Phuong T, Barnett R, Hehli B, Song F, DeGuzman MJ, Ensari S, Pinkstaff JK, Sullivan LM, Biroc SL, Cho H, Schultz PG, DiJoseph J, Dougher M, Ma D, Dushin R, Leal M, Tchistiakova L, Feyfant E, Gerber HP, Sapra P. A general approach to site-specific antibody drug conjugates. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(5):1766–1771. doi: 10.1073/pnas.1321237111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivanciu L, Toso R, Margaritis P, Pavani G, Kim H, Schlachterman A, Liu JH, Clerin V, Pittman DD, Rose-Miranda R, Shields KM, Erbe DV, Tobin JF, Arruda VR, Camire RM. A zymogen-like factor Xa variant corrects the coagulation defect in hemophilia. Nature biotechnology. 2011;29(11):1028–1033. doi: 10.1038/nbt.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shapira A, Gal-Tanamy M, Nahary L, Litvak-Greenfeld D, Zemel R, Tur-Kaspa R, Benhar I. Engineered toxins "zymoxins" are activated by the HCV NS3 protease by removal of an inhibitory protein domain. PloS one. 2011;6(1):e15916. doi: 10.1371/journal.pone.0015916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng DYW, Arzt M, Wu YZ, Kuan SL, Lamla M, Weil T. Constructing Hybrid Protein Zymogens through Protective Dendritic Assembly. Angew Chem Int Edit. 2014;53(1):324–328. doi: 10.1002/anie.201308533. [DOI] [PubMed] [Google Scholar]
- 53.Raines RT. Chembiomolecular Science. Springer; 2013. Enzymes as Chemotherapeutic Agents; pp. 281–291. [Google Scholar]
- 54.Callis M, Serrano S, Benito A, Laurents DV, Vilanova M, Bruix M, Ribo M. Towards Tricking a Pathogen's Protease into Fighting Infection: The 3D Structure of a Stable Circularly Permuted Onconase Variant Cleavedby HIV-1 Protease. PloS one. 2013;8(1) doi: 10.1371/journal.pone.0054568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shapira A, Shapira S, Gal-Tanamy M, Zemel R, Tur-Kaspa R, Benhar I. Removal of Hepatitis C Virus-Infected Cells by a Zymogenized Bacterial Toxin. PloS one. 2012;7(2) doi: 10.1371/journal.pone.0032320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Law SK, Wang RR, Mak AN, Wong KB, Zheng YT, Shaw PC. A switch-on mechanism to activate maize ribosome-inactivating protein for targeting HIV-infected cells. Nucleic acids research. 2010;38(19):6803–6812. doi: 10.1093/nar/gkq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Au KY, Wang RR, Wong YT, Wong KB, Zheng YT, Shaw PC. Engineering a switch-on peptide to ricin A chain for increasing its specificity towards HIV-infected cells. Biochimica et biophysica acta. 2014;1840(3):958–963. doi: 10.1016/j.bbagen.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 58.Muhlebach MD, Schaser T, Zimmermann M, Armeanu S, Hanschmann KM, Cattaneo R, Bitzer M, Lauer UM, Cichutek K, Buchholz CJ. Liver cancer protease activity profiles support therapeutic options with matrix metalloproteinase-activatable oncolytic measles virus. Cancer research. 2010;70(19):7620–7629. doi: 10.1158/0008-5472.CAN-09-4650. [DOI] [PubMed] [Google Scholar]
- 59.Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nature chemical biology. 2011;7(8):504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Callahan BP, Topilina NI, Stanger MJ, Van Roey P, Belfort M. Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nature structural & molecular biology. 2011;18(5):630–633. doi: 10.1038/nsmb.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sivey JD, Howell SC, Bean DJ, McCurry DL, Mitch WA, Wilson CJ. Role of lysine during protein modification by HOCl and HOBr: halogen-transfer agent or sacrificial antioxidant? Biochemistry. 2013;52(7):1260–1271. doi: 10.1021/bi301523s. [DOI] [PubMed] [Google Scholar]
- 62.Sharma S, Bagshawe K. Antibody-Directed Enzyme Prodrug Therapy (ADEPT) for Cancer. In: Reddy LH, Couvreur P, editors. Macromolecular Anticancer Therapeutics. New York: Springer; 2010. pp. 393–406. [Google Scholar]
- 63.Weidle UH, Tiefenthaler G, Georges G. Proteases as activators for cytotoxic prodrugs in antitumor therapy. Cancer genomics & proteomics. 2014;11(2):67–79. [PubMed] [Google Scholar]
- 64.Duarte S, Carle G, Faneca H, de Lima MC, Pierrefite-Carle V. Suicide gene therapy in cancer: where do we stand now? Cancer letters. 2012;324(2):160–170. doi: 10.1016/j.canlet.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 65.Dachs GU, Tupper J, Tozer GM. From bench to bedside for gene-directed enzyme prodrug therapy of cancer. Anti-cancer drugs. 2005;16(4):349–359. doi: 10.1097/00001813-200504000-00001. [DOI] [PubMed] [Google Scholar]
- 66.Lehouritis P, Springer C, Tangney M. Bacterial-directed enzyme prodrug therapy. Journal of Controlled Release. 2013;170(1):120–131. doi: 10.1016/j.jconrel.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 67.Voak AA, Gobalakrishnapillai V, Seifert K, Balczo E, Hu LQ, Hall BS, Wilkinson SR. An Essential Type I Nitroreductase from Leishmania major Can Be Used to Activate Leishmanicidal Prodrugs. J Biol Chem. 2013;288(40):28466–28476. doi: 10.1074/jbc.M113.494781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neschadim A, Wang JCM, Lavie A, Medin JA. Bystander killing of malignant cells via the delivery of engineered thymidine-active deoxycytidine kinase for suicide gene therapy of cancer. Cancer Gene Ther. 2012;19(5):320–327. doi: 10.1038/cgt.2012.4. [DOI] [PubMed] [Google Scholar]
- 69.Wright CM, Wright RC, Eshleman JR, Ostermeier M. A protein therapeutic modality founded on molecular regulation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(39):16206–16211. doi: 10.1073/pnas.1102803108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fejerskov B, Zelikin AN. Substrate Mediated Enzyme Prodrug Therapy. PloS one. 2012;7(11) doi: 10.1371/journal.pone.0049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agostinis P, Berg K, Cengel KA, Foster TH, Girotti AW, Gollnick SO, Hahn SM, Hamblin MR, Juzeniene A, Kessel D, Korbelik M, Moan J, Mroz P, Nowis D, Piette J, Wilson BC, Golab J. Photodynamic therapy of cancer: an update. CA: a cancer journal for clinicians. 2011;61(4):250–281. doi: 10.3322/caac.20114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wojtovich AP, Foster TH. Optogenetic control of ROS production. Redox biology. 2014;2:368–376. doi: 10.1016/j.redox.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nature biotechnology. 2006;24(1):95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 74.Vegh RB, Solntsev KM, Kuimova MK, Cho S, Liang Y, Loo BLW, Tolbert LM, Bommarius AS. Reactive oxygen species in photochemistry of the red fluorescent protein "Killer Red". Chem Commun. 2011;47(17):4887–4889. doi: 10.1039/c0cc05713d. [DOI] [PubMed] [Google Scholar]
- 75.Bulina ME, Lukyanov KA, Britanova OV, Onichtchouk D, Lukyanov S, Chudakov DM. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat Protoc. 2006;1(2):947–953. doi: 10.1038/nprot.2006.89. [DOI] [PubMed] [Google Scholar]
- 76.Liao ZX, Li YC, Lu HM, Sung HW. A genetically-encoded KillerRed protein as an intrinsically generated photosensitizer for photodynamic therapy. Biomaterials. 2014;35(1):500–508. doi: 10.1016/j.biomaterials.2013.09.075. [DOI] [PubMed] [Google Scholar]
- 77.Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, Kotera I, Arai Y, Horiuchi M, Fukui K, Ayabe T, Inagaki F, Suzuki H, Nagai T. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci Rep-Uk. 2013;3 doi: 10.1038/srep02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beharry AA, Woolley GA. Azobenzene photoswitches for biomolecules. Chem Soc Rev. 2011;40(8):4422–4437. doi: 10.1039/c1cs15023e. [DOI] [PubMed] [Google Scholar]
- 79.Ritterson RS, Kuchenbecker KM, Michalik M, Kortemme T. Design of a Photoswitchable Cadherin. J Am Chem Soc. 2013;135(34):12516–12519. doi: 10.1021/ja404992r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoersch D, Roh SH, Chiu W, Kortemme T. Reprogramming an ATP-driven protein machine into a light-gated nanocage. Nat Nanotechnol. 2013;8(12):928–932. doi: 10.1038/nnano.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bacchus W, Fussenegger M. The use of light for engineered control and reprogramming of cellular functions. Curr Opin Biotech. 2012;23(5):695–702. doi: 10.1016/j.copbio.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 82.Mills E, Chen X, Pham E, Wong S, Truong K. Engineering a photoactivated caspase-7 for rapid induction of apoptosis. ACS synthetic biology. 2012;1(3):75–82. doi: 10.1021/sb200008j. [DOI] [PubMed] [Google Scholar]
- 83.Bonger KM, Rakhit R, Payumo AY, Chen JK, Wandless TJ. General method for regulating protein stability with light. ACS chemical biology. 2014;9(1):111–115. doi: 10.1021/cb400755b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toettcher JE, Voigt CA, Weiner OD, Lim WA. The promise of optogenetics in cell biology: interrogating molecular circuits in space and time. Nat Methods. 2011;8(1):35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krauss U, Lee J, Benkovic SJ, Jaeger KE. LOVely enzymes - towards engineering light-controllable biocatalysts. Microb Biotechnol. 2010;3(1):15–23. doi: 10.1111/j.1751-7915.2009.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taylor BL, Zhulin IB. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol R. 1999;63(2):479-+. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olson EJ, Tabor JJ. Post-translational tools expand the scope of synthetic biology. Current opinion in chemical biology. 2012;16(3–4):300–306. doi: 10.1016/j.cbpa.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 88.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9(4):379-U92. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lungu OI, Hallett RA, Choi EJ, Aiken MJ, Hahn KM, Kuhlman B. Designing Photoswitchable Peptides Using the AsLOV2 Domain. Chem Biol. 2012;19(4):507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tran NH, Nguyen D, Dwaraknath S, Mahadevan S, Chavez G, Nguyen A, Dao T, Mullen S, Nguyen TA, Cheruzel LE. An Efficient Light-Driven P450 BM3 Biocatalyst. J Am Chem Soc. 2013;135(39):14484–14487. doi: 10.1021/ja409337v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7(12):973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annual review of plant biology. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 93.Ozturk N, Selby CP, Zhong D, Sancar A. Mechanism of photosignaling by Drosophila cryptochrome: role of the redox status of the flavin chromophore. J Biol Chem. 2014;289(8):4634–4642. doi: 10.1074/jbc.M113.542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu HT, Liu B, Zhao CX, Pepper M, Lin CT. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16(12):684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tobin PH, Wilson CJ. Examining Photoinduced Energy Transfer in Pseudomonas aeruginosa Azurin. J Am Chem Soc. 2014;136(5):1793–1802. doi: 10.1021/ja412308r. [DOI] [PubMed] [Google Scholar]
- 96.Alvarez-Lorenzo C, Concheiro A. Smart drug delivery systems: from fundamentals to the clinic. Chem Commun (Camb) 2014 doi: 10.1039/c4cc01429d. [DOI] [PubMed] [Google Scholar]
- 97.Deckert K, Budiardjo SJ, Brunner LC, Lovell S, Karanicolas J. Designing allosteric control into enzymes by chemical rescue of structure. J Am Chem Soc. 2012;134(24):10055–10060. doi: 10.1021/ja301409g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rana S, Pozzi N, Pelc LA, Di Cera E. Redesigning allosteric activation in an enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(13):5221–5225. doi: 10.1073/pnas.1018860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu S, Acevedo JP, Reetz MT. Induced allostery in the directed evolution of an enantioselective Baeyer-Villiger monooxygenase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):2775–2780. doi: 10.1073/pnas.0911656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491(7422):138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cross PJ, Allison TM, Dobson RC, Jameson GB, Parker EJ. Engineering allosteric control to an unregulated enzyme by transfer of a regulatory domain. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(6):2111–2116. doi: 10.1073/pnas.1217923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rizk SS, Paduch M, Heithaus JH, Duguid EM, Sandstrom A, Kossiakoff AA. Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins. Nature structural & molecular biology. 2011;18(4):437–442. doi: 10.1038/nsmb.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dagliyan O, Shirvanyants D, Karginov AV, Ding F, Fee L, Chandrasekaran SN, Freisinger CM, Smolen GA, Huttenlocher A, Hahn KM, Dokholyan NV. Rational design of a ligand-controlled protein conformational switch. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(17):6800–6804. doi: 10.1073/pnas.1218319110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Podual K, Doyle FJ, Peppas NA. Glucose-sensitivity of glucose oxidase-containing cationic copolymer hydrogels having poly(ethylene glycol) grafts. Journal of Controlled Release. 2000;67(1):9–17. doi: 10.1016/s0168-3659(00)00195-4. [DOI] [PubMed] [Google Scholar]
- 105.Mohammed JS, Murphy WL. Bioinspired Design of Dynamic Materials. Adv Mater. 2009;21(23):2361–2374. [Google Scholar]
- 106.Katz E, Privman V. Enzyme-based logic systems for information processing. Chem Soc Rev. 2010;39(5):1835–1857. doi: 10.1039/b806038j. [DOI] [PubMed] [Google Scholar]
- 107.Tokarev I, Gopishetty V, Zhou J, Pita M, Motornov M, Katz E, Minko S. Stimuli-Responsive Hydrogel Membranes Coupled with Biocatalytic Processes. Acs Appl Mater Inter. 2009;1(3):532–536. doi: 10.1021/am800251a. [DOI] [PubMed] [Google Scholar]
- 108.Privman M, Tam TK, Bocharova V, Halamek J, Wang J, Katz E. Responsive Interface Switchable by Logically Processed Physiological Signals: Toward "Smart" Actuators for Signal Amplification and Drug Delivery. Acs Appl Mater Inter. 2011;3(5):1620–1623. doi: 10.1021/am200165m. [DOI] [PubMed] [Google Scholar]
- 109.Zhou M, Zhou ND, Kuralay F, Windmiller JR, Parkhomovsky S, Valdes-Ramirez G, Katz E, Wang J. A Self-Powered "Sense-Act-Treat" System that is Based on a Biofuel Cell and Controlled by Boolean Logic. Angew Chem Int Edit. 2012;51(11):2686–2689. doi: 10.1002/anie.201107068. [DOI] [PubMed] [Google Scholar]
- 110.Mailloux S, Halamek J, Katz E. A model system for targeted drug release triggered by biomolecular signals logically processed through enzyme logic networks. Analyst. 2014;139(5):982–986. doi: 10.1039/c3an02162a. [DOI] [PubMed] [Google Scholar]
- 111.De Wolf FA, Brett GM. Ligand-binding proteins: Their potential for application in systems for controlled delivery and uptake of ligands. Pharmacol Rev. 2000;52(2):207–236. [PubMed] [Google Scholar]
- 112.Kratz F. A clinical update of using albumin as a drug vehicle — A commentary. Journal of Controlled Release. (0) doi: 10.1016/j.jconrel.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 113.Andersen JT, Dalhus B, Viuff D, Thue Ravn B, Gunnarsen KS, Plumridge A, Bunting K, Antunes F, Williamson R, Athwal S, Allan E, Evans L, Bjoras M, Kjaerulff S, Sleep D, Sandlie I, Cameron J. Extending Serum Half-life of Albumin by Engineering FcRn Binding. J Biol Chem. 2014 doi: 10.1074/jbc.M114.549832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Andersen JT, Dalhus B, Cameron J, Daba MB, Plumridge A, Evans L, Brennan SO, Gunnarsen KS, Bjoras M, Sleep D, Sandlie I. Structure-based mutagenesis reveals the albumin-binding site of the neonatal Fc receptor. Nature communications. 2012;3:610. doi: 10.1038/ncomms1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fasano M, Curry S, Terreno E, Galliano M, Fanali G, Narciso P, Notari S, Ascenzi P. The extraordinary ligand binding properties of human serum albumin. IUBMB life. 2005;57(12):787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- 116.Fanali G, di Masi A, Trezza V, Marino M, Fasano M, Ascenzi P. Human serum albumin: from bench to bedside. Molecular aspects of medicine. 2012;33(3):209–290. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 117.Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, Curry S. Structural basis of the drug-binding specificity of human serum albumin. Journal of molecular biology. 2005;353(1):38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 118.Zhang F, Xue J, Shao J, Jia L. Compilation of 222 drugs' plasma protein binding data and guidance for study designs. Drug discovery today. 2012;17(9–10):475–485. doi: 10.1016/j.drudis.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 119.Tinberg CE, Khare SD, Dou JY, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, Baker D. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501(7466):212-+. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pawson T, Linding R. Network medicine. FEBS letters. 2008;582(8):1266–1270. doi: 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 121.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature reviews. Genetics. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kitano H. A robustness-based approach to systems-oriented drug design. Nature reviews. Drug discovery. 2007;6(3):202–210. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- 123.Erler JT, Linding R. Network medicine strikes a blow against breast cancer. Cell. 2012;149(4):731–733. doi: 10.1016/j.cell.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 124.Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149(4):780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Robin X, Creixell P, Radetskaya O, Santini CC, Longden J, Linding R. Personalized network-based treatments in oncology. Clinical pharmacology and therapeutics. 2013;94(6):646–650. doi: 10.1038/clpt.2013.171. [DOI] [PubMed] [Google Scholar]
- 126.Lotsch J, Geisslinger G. Low-dose drug combinations along molecular pathways could maximize therapeutic effectiveness while minimizing collateral adverse effects. Drug discovery today. 2011;16(23–24):1001–1006. doi: 10.1016/j.drudis.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 127.Munter S, Way M, Frischknecht F. Signaling during pathogen infection. Science's STKE : signal transduction knowledge environment. 2006;2006(335):re5. doi: 10.1126/stke.3352006re5. [DOI] [PubMed] [Google Scholar]
- 128.Wei P, Wong WW, Park JS, Corcoran EE, Peisajovich SG, Onuffer JJ, Weiss A, Lim WA. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012;488(7411):384-+. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lim WA. Designing customized cell signalling circuits. Nature reviews. Molecular cell biology. 2010;11(6):393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annual review of biochemistry. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 131.Kiel C, Yus E, Serrano L. Engineering signal transduction pathways. Cell. 2010;140(1):33–47. doi: 10.1016/j.cell.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 132.Dueber JE, Mirsky EA, Lim WA. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nature biotechnology. 2007;25(6):660–662. doi: 10.1038/nbt1308. [DOI] [PubMed] [Google Scholar]
- 133.Kapp GT, Liu S, Stein A, Wong DT, Remenyi A, Yeh BJ, Fraser JS, Taunton J, Lim WA, Kortemme T. Control of protein signaling using a computationally designed GTPase/GEF orthogonal pair. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(14):5277–5282. doi: 10.1073/pnas.1114487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carleton HA, Lara-Tejero M, Liu X, Galan JE. Engineering the type III secretion system in non-replicating bacterial minicells for antigen delivery. Nature communications. 2013;4:1590. doi: 10.1038/ncomms2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Science translational medicine. 2013;5(179):179ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Park JS, Rhau B, Hermann A, McNally KA, Zhou C, Gong D, Weiner OD, Conklin BR, Onuffer J, Lim WA. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(16):5896–5901. doi: 10.1073/pnas.1402087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Teicher BA, Chari RVJ. Antibody Conjugate Therapeutics: Challenges and Potential. Clin Cancer Res. 2011;17(20):6389–6397. doi: 10.1158/1078-0432.CCR-11-1417. [DOI] [PubMed] [Google Scholar]
- 138.Stirpe F. Ribosome-inactivating proteins. Toxicon : official journal of the International Society on Toxinology. 2004;44(4):371–383. doi: 10.1016/j.toxicon.2004.05.004. [DOI] [PubMed] [Google Scholar]