Abstract
A complex interplay of signaling events, including the Wnt pathway, regulates sprouting of blood vessels from preexisting vasculature during angiogenesis. Here we show that two distinct mutations in the (uro)chordate-specific Gumby/Fam105b gene cause an embryonic angiogenic phenotype in gumby mice. Gumby interacts with Disheveled 2 (Dvl2), is expressed in canonical Wnt-responsive endothelial cells and encodes an Ovarian Tumor Domain (OTU) class of deubiquitinase (DUB) that specifically cleaves linear ubiquitin linkages. A crystal structure of Gumby in complex with linear di-ubiquitin reveals how the identified mutations adversely impact substrate binding and catalytic function in line with the severity of their angiogenic phenotypes. Gumby interacts with HOIP/Rnf31, a key component of the linear ubiquitin assembly complex (LUBAC), decreases linear ubiquitination and activation of NFκB dependent transcription. This work provides support for the biological importance of linear (de)ubiquitination in angiogenesis, craniofacial and neural development and in modulating Wnt signaling.
Introduction
During angiogenesis, new blood vessels sprout from pre-existing vasculature to form a microcapillary network and become uniquely adapted to the physiology and function of organs they infiltrate (reviewed in 1). Several well-characterized molecular pathways direct vascular patterning, but contributions of Wnt pathways are just emerging. Wnt signaling pathways control a broad spectrum of events, including cell-fate specification, proliferation and migration (reviewed in 2), and are grouped into the canonical, β-catenin-dependent and non-canonical pathways. Both canonical and non-canonical Wnt pathways influence angiogenesis (reviewed in 3,4). Canonical Wnt signaling is required for angiogenesis in the central nervous system (CNS) 5,6, while noncanonical Wnt signaling has been implicated in global early embryonic angiogenesis 7. The events that occur in Wnt-responsive cells depend, in part, on Dishevelled and its associated proteins to transmit stimuli to the appropriate pathway(s) (reviewed in 8).
Protein modifications, such as ubiquitination, offer a rapid mechanism, by which cells integrate inputs from signaling pathways. Ubiquitin is an evolutionarily conserved 76-amino acid protein used to label and alter the fate of cellular proteins. During protein ubiquitination, a covalent bond is generated most commonly between the most C-terminal amino acid residue, a glycine, and a lysine in the modified protein. Ubiquitin itself contains seven lysine residues that can undergo ubiquitination to form polyubiquitin oligomers (“chains”). These chains serve to direct protein localization, stability and activity. The linear ubiquitin assembly complex (LUBAC), comprised of HOIL-1/1b, HOIP(Rnf31) and Sharpin proteins, mediates formation of an atypical ubiquitin chain topology 9 involving the linear linkage of the carboxy-terminus of glycine 76 in one ubiquitin protomer to the free amino-terminus of methionine 1 in a second protomer (reviewed in 10–12). To date, linear ubiquitination has been shown to regulate NF-kB dependent inflammation and adaptive immunity 13–16. Deubiquitination, the selective removal of ubiquitins, is equally important and is performed by members of five distinct deubiquitinase (DUB) families, one of which is the Ovarian Tumor (OTU) domain containing protease family 17. Currently, no DUB has been shown to exclusively cleave linear ubiquitin chains.
Here our studies of the recessive, embryonically lethal gumby mouse mutant and its angiogenic phenotype have lead to identifying the affected (uro)chordate-specific Gumby/Fam105b gene. We show that Gumby encodes a linear ubiquitin-specific DUB that structurally belongs to the OTU family. Gumby can associate with LUBAC and counteract known LUBAC functions. We identify a role for the Gumby-LUBAC axis in regulating canonical Wnt signaling. Our findings highlight the importance of linear (de)ubiquitination in angiogenesis, craniofacial and neuronal development and Wnt signaling.
The gumby mutation causes embryonic angiogenic deficits
gumby/gumby mice were identified based on abnormal sprouting of the facial nerve at embryonic day (E)10.5 18, appear normal before E11.5, but die between E12.5–E14. Because shared molecular mechanisms can guide axons and blood vessel branching, we examined vascular development in E10.0–11.0 +/+, gumby/+ and gumby/gumby embryos by whole-mount immunohistochemistry with platelet endothelial cell adhesion molecule-1 (PECAM-1) antibody (Supplementary Fig. 1). The major structures of the vascular system appeared similar in controls and mutants. However, branching vascular networks in the head and trunk were improperly organized and less complex in gumby homozygotes. In the medial region of the embryonic head, several large diameter cranial vessels branch to form a hierarchical vascular network (Supplementary Fig. 1a, b). In gumby/gumby embryos, large cranial vessels were dilated, branching reduced and endothelial cells (ECs) accumulated at branchpoints (Supplementary Fig. 1e, f). Normally, a ‘capillary’ network, the perineural vascular plexus (PNVP), forms in the trunk between intersomitic vessels and extends into the neurectoderm (Supplementary Fig. 1c, d) 19. In gumby/gumby embryos, fewer and less elaborate vessel extensions formed between the somites and the PNVP (Supplementary Fig. 1g, h; Fig. 1l).
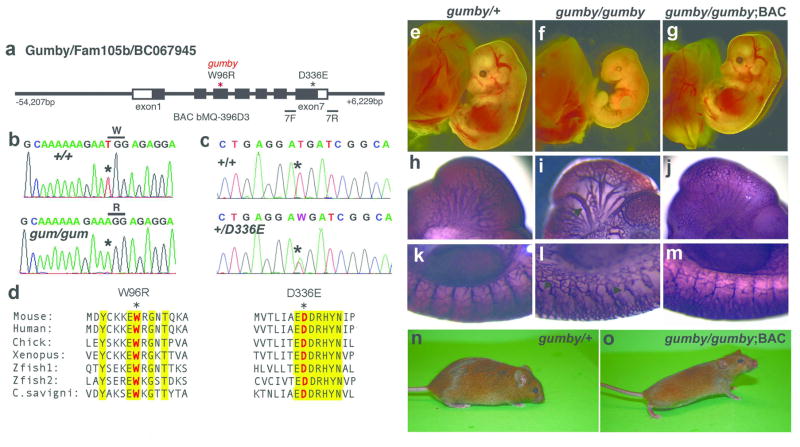
Figure 1. Identification of the gumby (GumW96R) causative mutation and the new GumD336E allele.
a, Schematic diagram of the Gumby/Fam105b/BC067945 gene and BAC bMQ-396D3. Primers 7F and 7R used to identify mice carrying GumD336E flank exon 7. Sequencing traces from b, +/+ and gumby/gumby (gum/gum) and c, +/+ and GumD336E/+ (+/D336E) mice. d, Amino acid sequence alignment spanning GumW96R and GumD336E mutations. Trp96 and Asp336 are shown in red, marked with asterisks. Yellow highlights interspecies amino acid sequence identity. (e–o) BAC rescue of lethality and vascular abnormalities of gumby/gumby mice. Morphological appearance of e, gumby/+, f, gumby/gumby, and g, gumby/gumby embryos carrying the BAC transgene (gumby/gumby;BAC) at E14.0. h–m, PECAM-1 immunohistochemistry of h, k, gumby/+, i, l, gumby/gumby and j, m, gumby/gumby;BAC embryos at E10.5. h–j, cranial vasculature and k–m, trunk vasculature shown at the forelimb bud level. Arrow indicates dilated cranial vessels; arrowheads highlight stunted branches in the trunk. n, adult gumby/+ and o, gumby/gumby;BAC littermates.
The GumbyW96R mutation in the Gumby/Fam105b gene causes gumby phenotypes
Meiotic mapping of 154 progeny from gumby/+ intercrosses refined the critical interval to 1.7Mb between D15Mit18 (26.7Mb) and SNP rs13482490 (28.4Mb) on mouse chromosome 15 (Ensembl assembly v35) (Supplementary Fig. 2). Sequencing genes within this interval identified a T to A transversion (T285A) in the third exon of the Gumby/Fam105b/BC067945 gene, which substitutes tryptophan at position 96 to arginine, and is referred to as GumbyW96R (Fig. 1b). Tryptophan 96 is conserved in all known orthologues (Fig. 1d). While Ciona intestinalis and savigyni genomes each carry a copy, related genes are absent in non-chordates. Gumby/Fam105b/BC067945 is henceforth referred to as the Gumby gene.
To test whether this is the gumby causative gene, we performed rescue experiments with bacterial artificial chromosome (BAC) bMQ-396D that spans the Gumby gene and ~60kb of its flanking region (Fig. 1a, e–o, Supplementary Fig. 3). One founder BAC transgenic line rescued the lethality (Fig. 1 g, o) and vascular deficits (Fig. 1j, m) of gumby/gumby mice. Thus, this mutation causes the gumby phenotype, and we refer to this gumby allele as GumW96R.
Identification and analysis of an additional Gumby allele, GumD336E
We identified mice carrying another allele of Gumby, GumD336E, by screening a bank of cryoperserved sperm and genomic DNA from over 6000 G1 male progeny from N-ethyl-N-nitrosourea mutagenized G0 males (http://www.brc.riken.jp/lab/gsc/mouse/index.html) 20. In the GumD336E allele, a T to A transversion in exon 7 changes conserved aspartate 336 to glutamate (Fig. 1c). Both GumW96R and GumD336E homozygotes show reduced branchial arches and embryonic lethality after E12.5 (data not shown). Using anti-PECAM-1 whole-mount immunofluorescence, we quantified the relative deficits in the cranial vasculature of GumD336E and GumW96R homozygotes at E10.5. (Fig. 2a–f). We found decreased numbers of secondary and tertiary vessels branching off the internal carotid artery (ICA) in GumW96R (Fig. 2c, g) and GumD336E (Fig. 2e, g) homozygotes relative to +/+ littermates (Fig. 2a, g). We examined vessel dilation by measuring the diameter of the ICA prior to its migration to the posterior head (Fig. 2b, d, f, h) and secondary branch dilation by measuring the diameter of the first branch off the ICA (Fig. 2i). GumW96R homozygotes had larger dilated ICAs (Fig. 2h) and secondary branches (Fig. 2i) compared to +/+ and GumD336E homozygotes. Immunoblot and immnofluourescence experiments indicate that the GumW96R and GumD336E mutations do not detectably compromise Gumby protein level or cytoplasmic localization (Supplementary Fig. 4). These findings further support an angiogenic requirement for Gumby and predict that the GumW96R mutation impacts protein function more severely than GumD336E.
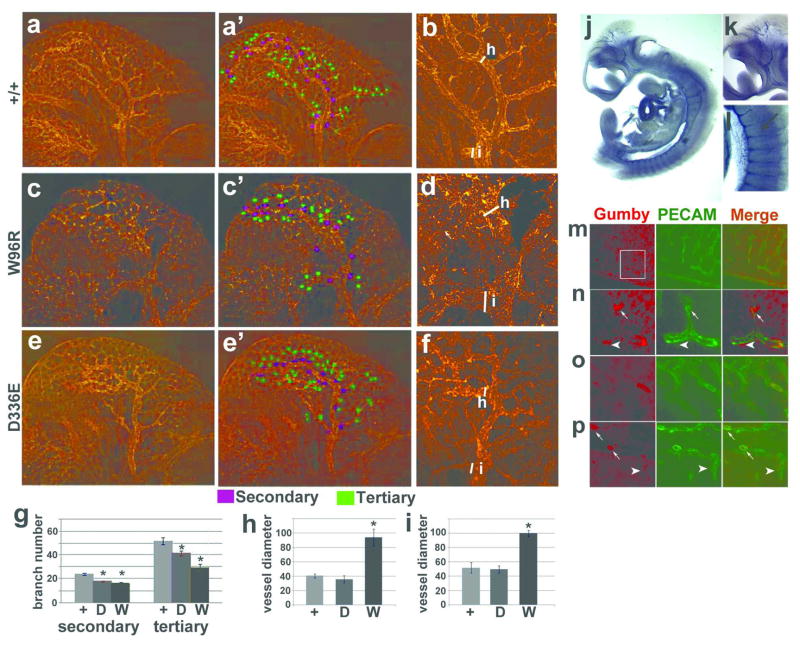
Figure 2. Analyses of vascular phenotypes and Gumby expression.
a–f, Whole mount anti-PECAM-1 immunofluorescence showing cranial vasculature of E10.5 a, a′, b, +/+, c, c′, d, GumW96R/GumW96R (W96R) and e, e′, f, GumD336E/GumD336E (D336E) embryos. a, c, e, Composite whole head images. a′, c′, e′, Secondary and tertiary vessels are marked in purple and green, respectively. b, d, f, Analyses of cranial vessel dilation of intercranial artery (ICA). White bars indicate the areas of the ICA (h) and first branch (i) measured. g, Quantification of secondary and tertiary vessels branching off the primary ICA per side of head. h, Quantification of h, ICA and i, first ICA branch diameters. * indicates statistical significance relative to +/+ embryos (n = 3 or 6, p < 0.05, Student’s t-test).
j–l, RNA in situ hybridization of E10.5 +/+ embryos detects Gumby RNA in the vasculature. Magnified views from k, head and l, trunk. An intersomitic vessel (ISV) is marked with an arrow.
m–p, Immunofluorescence with anti-PECAM-1 (green) and anti-Gumby (red) antibodies detects Gumby in endothelial cells in cross-sections of the trunk in +/+ E11.5 embryos. n High magnification of boxed area in m. Gumby protein is enriched near presumptive tips of vessels (arrows) and vascular buds (arrowheads).
Gumby mRNA is expressed in the developing vasculature and other regions affected in mutants, including branchial arches and neural crest cells (Fig. 2j–l, Supplementary Fig. 5). The walls of blood vessels are composed of ECs, which line the luminal vessel surface, and perivascular cells, which encircle the outside of the vascular endothelium 21. In immunofluorescence experiments on E10.5–11.5 embryonic sections with an anti-Gumby antibody (Supplementary Fig. 6), Gumby protein colocalized with the endothelial-specific marker PECAM-1, but not perivascular markers Smooth Muscle Actin and Desmin (Fig. 2m–p, data not shown). Gumby protein is enriched in a subset of ECs. Gumby-enriched ECs were present throughout ISVs and, in the PVNP and cranial vasculature, often located near possible leading regions of vessels (arrows, Fig. 2n, p) or at ‘vascular buds’ (arrowheads, Fig. 2n, p).
Gumby is an OTU-fold protease with selectivity for linear ubiquitin chains
Gumby/FAM105B was identified in the NCBI database as a putative member of the OTU-fold deubiquitinating enzymes (DUBs), a prediction we confirmed by x-ray crystallographic analysis to 1.7Å resolution (Supplementary Table 1). With 14.9% sequence identity (Supplementary Fig. 7) and a superimposable catalytic triad (Cys129, His339, and Asn341), Gumby most closely resembled the catalytic domain of OTUB1 (RMSD = 2.09 Å) 22–24, a DUB that specifically cleaves K48 linked ubiquitin chains 22,25, (Fig. 3a). As such, we examined DUB activity against all eight di-ubiquitin chain linkages in vitro (Fig. 3b right panel). In contrast to OTUB1, Gumby displayed selectivity for linear di-ubiquitin (Fig. 3b left panel). DUB activity was dependent on Cys129, was not measurable against the ubiquitin-AMC, and was insensititive to inhibition by ubiquitin-vinyl sulfone (Supplementary Fig. 8a,b,c). Unlike OTUB1 22,25, residues preceding the catalyic domain were not required for DUB activity (compare GumbyM55 vs GumbyR79 in Supplementary Fig. 8d). The Gumby mutations Trp96Arg and Asp336Glu mapped to a surface of the OTU-fold, 14.1Å and 11.4Å respectively from the catalytic cysteine (Fig. 3a), positions expected to impact on catalytic function. In confirmation, kinetic analysis of the W96R mutant revealed ~10,000 and 5 fold perturbations in kcat and KM respectively while the D336E mutant revealed ~50 fold perturbation in kcat (Supplementary Fig. 8d). These effects on enzymatic function paralleled the relative severity of mutant phenotypes in vivo (Fig. 2a–i).
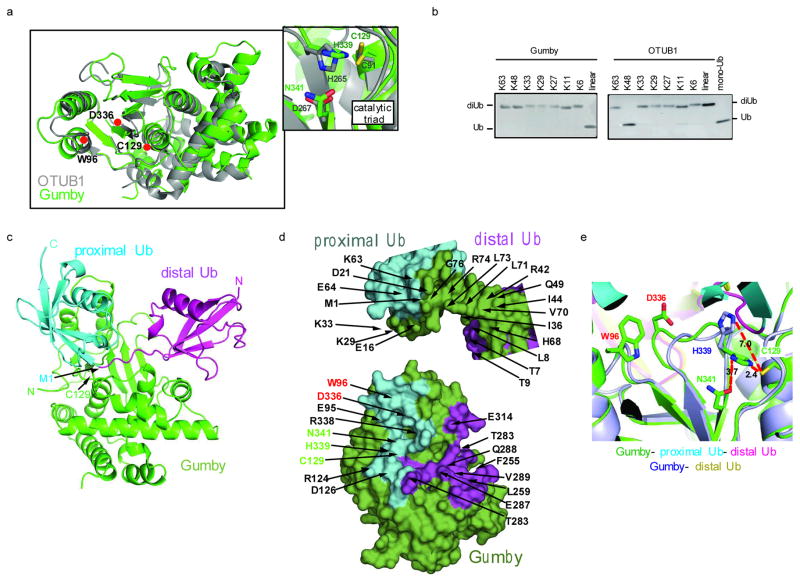
Figure 3. Structural and biochemical analysis of Gumby.
a, Ribbons view of apo-Gumby (green) superimposed on OTUB1 (PDB 4DDG) (grey). Inset shows catalytic triad. b, Cleavage specificity towards di-ubiquitin chains c, Ribbons view of the Gumby-linear di-ubiquitin complex. d, Peel-away surface views of linear di-ubiquitin (top) bound to Gumby (bottom). Domains colored as in c, with residues involved in inter-molecular contacts colored according to the domains contacted. Catalytic triad and mutant positions labeled green and red respectively. e, Active site comparison of Gumby-di-ubiquitin and Gumby-mono ubiquitin complexes. A productive orientation of His339 is observed only in the Gumby-di-ubiquitin complex.
We solved the 2.4Å and 2.8Å crystal structures of Gumby in the presence of free ubiquitin and linear di-ubiquitin substrate, respectively (Supplementary Table 1). Linear di-ubiquitin engaged an extensive surface (total buried surface area = 3267 Å2) on Gumby with the active site Cys129 centrally positioned at the scissile bond linking Gly76 and Met1 of distal and proximal ubiquitin moieties (Fig. 3a). Mono ubiquitin engaged the distal ubiquitin-binding site of Gumby similar to the mode observed in the di-ubiquitin (Supplementary Fig. 9a) co-structure, but different from modes observed in yeast OTU1, viral OTU, and human OTUB5 co-structures, possibly reflecting differences in substrate specificities (Supplementary Fig. 9b).
In the di-ubiquitin complex, distally bound ubiquitin engaged Gumby through a combination of hydrophobic and polar interactions burying a large total surface area of 1771 Å2 (Fig. 3c,d). The contact surface on distal ubiquitin was composed by strands β3, β4, loops connecting β1–β2, β3–β4, and the C-terminal tail, while the reciprocal surface on Gumby was composed by helix α8, loops between β2–α3, α9–α10, and β3–β4 (Fig. 3c, Supplementary Fig. 7). Proximally bound ubiquitin engaged Gumby primarily through hydrophilic interaction burying a total surface area of 1496 Å2. The interaction surface on proximally bound ubiquitin was composed by stands β1, β2, helix α1, and loops between β2–α1 and β3–β4, while the reciprocal contact surface on Gumby was composed of helices α1, α2, loop regions β2–α3, α9–α10, and β5–β6. A detailed list of contact residues and side chain interactions between linear ubiquitin and Gumby are shown in Fig. 3d and Supplementary Fig. 9c,d.
The prominent position of Trp96 and Asp336 in the general vicinity of the active site and on the direct contact surface with di-ubiquitin substrate allowed rationalization of the differential effects of the Trp96Arg and Asp336Glu Gumby mutations. As a generally conserved hydrophobic position in OTU sequences, Trp96 contributes to enzyme structure in addition to mediating a direct contact with Lys33 of the proximally bound ubiquitin (Fig. 3d and Supplementary Fig. 9d). Mutation of the partially buried Trp side chain to Arg would perturb both local structure and substrate binding affinity manifesting in the observed changes to both kcat, and KM. Consistent with this inference, mutation of the topologically equivalent hydrophobic position in OTUB1 (Tyr61Arg) abolished all trace of catalytic function (Supplementary Fig. 8e) and direct binding experiments showed a ~20 fold loss in substrate affinity for the Trp96Arg Gumby mutant (Supplementary Fig. 8f). From its highly solvent exposed position in the apo structure, Asp336 contributes less to enzyme structure while forming a direct salt interaction with Lys29 of the proximal site ubiquitin. Thus the conservative Asp336Glu mutation would impact less on active site structure while preserving a favourable salt interaction with Lys29 on ubiquitin manifesting in the observed smaller change to kcat and no change to KM.
Comparison of Gumby with OTUB1 (PDB 4DDG) 23 allows for a rationalization of its unique substrate specificity and regulation (Supplementary Fig. 9e). Despite employing equivalent topological surfaces on their OTU-folds for substrate engagement (center of mass positions for distal and proximal ubiquitins differ by only 1.6 and 3.9 Å, whereas angles of rotation differ more greatly by 60° and 135°, respectively), only 2 of 45 total contact residues on Gumby, namely Cys129 and His339, were conserved in OTUB1. Since both Gumby and OTUB1 engage continuous and extensive surfaces spanning both ubiquitin molecules in their respective di-ubiquitin substrates, centered on the site of cleavage, this would greatly constrain the ability of each enzyme to recognize substrates with alternate linkage topologies, which present vastly different interaction surfaces. Similar to OTUB1, a productive orientation of the catalytic triad is only apparent for Gumby bound to di-ubiquitin substrate. The coupling of catalytic activation with the binding of preferred substrate (Fig. 3e) would further accentuate specificity by prohibiting cleavage of partially and/or suboptimally engaged substrates.
Gumby interacts with and can counteract LUBAC
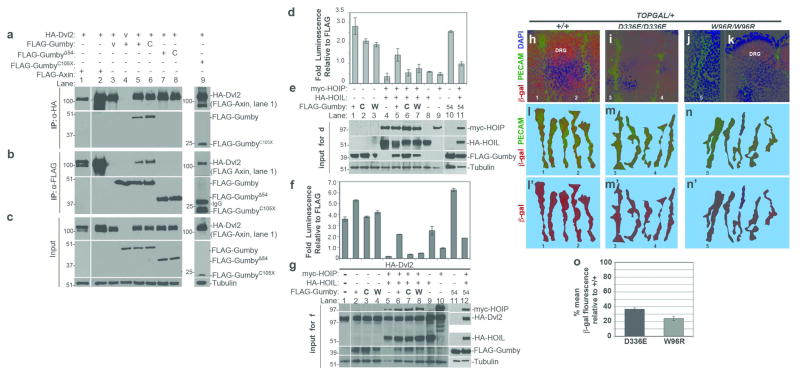
We identified the LUBAC component HOIP as a Gumby interactor by affinity purification of FLAG-tagged Gumby and its associated proteins followed by mass spectrometry (AP-MS). While full-length Gumby possesses a PDZ binding motif (PBM) and interacts with multiple PDZ-domain containing proteins when expressed in HEK293 cells (data not shown), a Gumby construct containing a mutated PBM consisting of four sequential alanine residues (GumbyΔPBM) (Fig. 4a) interacted with HOIP (Fig. 4b; Supplementary Table 2). No other LUBAC components were detected, suggesting that the interaction with HOIP may be independent of other delineated LUBAC components. By performing immunoprecipitation coupled to immunoblotting using FLAG-Gumby expression constructs, we confirmed this interaction and mapped it to the amino-terminus of Gumby. A construct truncated at position 105 (GumbyC105X) sufficed to mediate the interaction, while one missing the first amino-terminal 54 residues (GumbyΔ54) could not interact with HOIP. HOIP interacted with wild type Gumby and GumbyC129S proteins equivalently (Fig. 4c–e). Thus, Gumby interacts through its N-terminus with HOIP independently of its PBM or catalytic DUB domain.
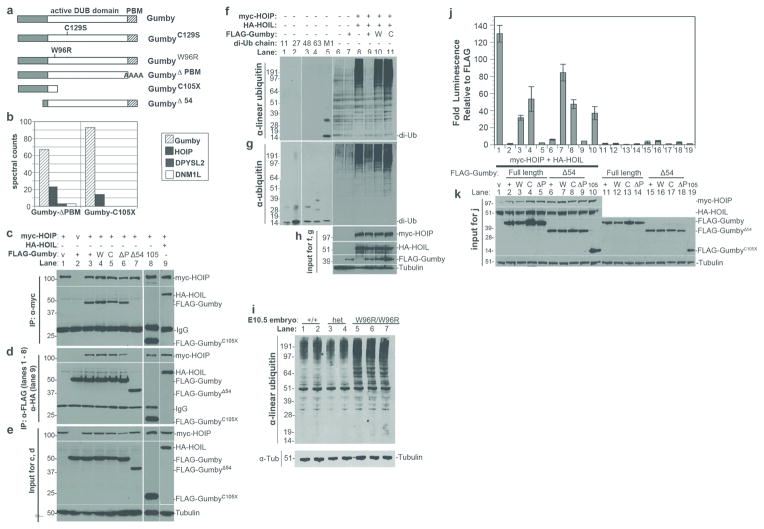
Figure 4. Gumby interacts with HOIP/RNF31 and counteracts LUBAC activity.
a, FLAG-tagged Gumby constructs used. b, Representative mass spectromety results for FLAG-tagged GumbyΔPBM and GumbyC105X immunoprecipitations. HOIP was identified as the most significant interacting protein in HEK293 cells expressing FLAG-tagged GumbyΔPBM or GumbyC105X. Significance threshold was set at a protein probability of greater than 90% (determined using SAINT analysis), less than 10% project frequency and a minimum of 3 peptides.
c–e, Immunoprecipitation coupled to immunoblotting using FLAG-Gumby expression constructs, HA-HOIL and myc-HOIP determined that HOIP interacts with wild type Gumby (+), GumbyC129S (C), GumbyΔPBM (P) and GumbyC105X (105) proteins, but not GumbyΔ54 (54). c, Immunoblot of immunoprecipitation using anti-myc antibody for myc-HOIP. d, Immunoblot of immunoprecipitation using anti-FLAG antibody for FLAG-tagged Gumby constructs. e, inputs for immunoprecipitation experiments.
f–h, Immunoblot using anti-linear ubiquitin antibody detected decreased levels of linear ubiquitinated proteins in HEK293T cells, when Gumby, but not GumbyC129S or GumbyW96R, was co-expressed with HOIL and HOIP. The anti-linear ubiquitin antibody recognizes linear diubiquitin (M1), but not control diubiquitin chains K11 (11), K48 (48), K63 (63). g, Equivalent levels of ubiquitination were detected in HOIL-HOIP co-expressing cells by anti-pan-ubiquitin antibody. h, Inputs for f and g.
i, Immunoblot with anti-linear ubiquitin antibody detects increased levels of linear ubiquitinated protein in individual E10.5 GumW96R/GumW96R embryos (W96R/W96R) relative to +/+ and /+ (het) littermates.
j, Catalytically active Gumby counteracts HOIP-HOIL dependent stimulation of luciferase expression from an NF-κB reporter in HEK293T cells. Wildtype, GumbyΔ54, GumbyΔPBM or GumbyΔ54ΔPBM fully suppress this stimulation. Data are presented as means +/− s.e.m. (n=3; P<0.05, Student’s t-test).
k, Immunoblot of inputs for luciferase assays.
Next we tested whether Gumby could affect linear ubiquitination of proteins in vivo. Co-expressing HOIL and HOIP in HEK293T cells increased the level of linearly ubiquitinated proteins in immunoblot and immunofluorescence experiments using an anti-linear ubiquitin antibody 26 (Fig. 4f–h, Supplementary Fig. 10). Co-transfection of FLAG-Gumby, but not FLAG-GumbyC129S and FLAG-GumbyW96R, with HOIL and HOIP decreased overall levels of proteins modified with linear ubiquitin (Fig. 4f, Supplementary Fig. 10). In mice, levels of linearly ubiquitinated proteins are elevated in E10.0 GumW96R/GumW96R embryos relative to their +/+ and heterozygous littermates, as detected by immunoblot analyses using an anti-linear ubiquitin antibody (Fig. 4i). Thus, the GumW96R mutation compromises linear deubiquitination in cell culture and animals.
Finally, we examined whether Gumby could oppose LUBAC-dependent activation of NFκB directed transcription (Fig. 4j, k). Co-transfecting an NFκB-dependent reporter construct with Gumby or GumbyΔPBM caused ~100-fold inhibition of LUBAC dependent activation in HEK293T cells. GumbyC129S, GumbyW96R and GumbyC105X could not repress this activation to an equivalent level. Neither LUBAC components nor Gumby in combination or alone affect AP2 dependent transcription (data not shown), suggesting that these are not effects on general transcription. While Gumby interacts with HOIP via its N-terminal region, in these assays, GumbyΔ54 can still antagonize LUBAC-dependent activation. Thus, in an overexpression system, the ability of Gumby to fully offset LUBAC function requires its catalytic activity.
Gumby and LUBAC modulate Wnt signaling
Recovery of Gumby as a Dishevelled 2 (Dvl2) interactor in a yeast two hybrid screen suggested a role for Gumby in Wnt signaling 27. We confirmed the Gumby-Dvl2 interaction in HEK293T cells by immunoprecipitation of HA-tagged Dvl2, which recovered FLAG-tagged Gumby and GumbyC129S equivalently and reciprocally (Fig. 5a–c). GumbyΔ54 could not interact with Dvl2, while GumbyC105X could. Thus, Gumby interacts with Dvl2 via its amino terminal region.
Figure 5. Gumby interacts with Dvl2 and can modulate Wnt signaling.
a, Immunoprecipitation (IP) with anti-HA antibody of HA-tagged Dvl2 (HA-Dvl2) recovered FLAG-tagged Gumby, GumbyC129S (C), GumbyC105X (105) and FLAG-Axin (positive control), but not GumbyΔ54 (54). FLAG-vector is designated as “v”. b, In immunoprecipitation with anti-FLAG antibody, all FLAG-Gumby constructs except for FLAG-GumbyΔ54 recovered HA-Dvl2. c, Input amounts and tubulin levels. d, f, In assays for Wnt3a activation of luciferase expression from the TOPFLASH reporter performed in d,e, the absence or f, g, presence of HA-Dvl2, FLAG-Gumby enhanced TOPFLASH expression more than FLAG-GumbyW96R (W) and FLAG-GumbyC129S. HA-HOIL and myc-HOIP co-expression inhibited TOPFLASH. Gumby, but not GumbyC129S or GumbyW96R, reversed HOIL-HOIP dependent inhibition. e, g, Immunoblots show inputs for luciferase assays in d and f, respectively. Data are presented as means +/− s.e.m. n=3 or more. (P<0.05, Student’s t-test).
h–o Analysis of β-galactosidase (β-gal) expression in intersomitic vessels (ISVs) of E10.5 embryos. h, Co-immunofluorescence with anti-β-gal (red) and anti-PECAM-1 (green) antibodies detects high βgal within ISVs and dorsal root ganglia (DRG) of +/+;TOPGAL/+ embryos. In i, GumD336E/GumD336E;TOPGAL/+ and j,k, GumW96R/GumW96R;TOPGAL+ embryos, β-gal signal is lower in ISVs and in DRG. k–m′, representative traces of ISVs scored for β-gal fluorescence intensity are shown for l,l′, +/+;TOPGAL/+, m,m′, GumD336E/GumD336E;TOPGAL/+ and n,n′, GumW96R/GumW96R;TOPGAL+ embryos. Numbers in k–m′ mark specific ISVs traced in h, i and j. n, Graph of percentage mean fluorescence in ISVs of GumD336E/GumD336E;TOPGAL/+ (D336E) and GumW96R/GumW96R;TOPGAL+ (W96R) embryos. (36.6 +/− 1.8% and 24.0 +/− 2.8%, respectively). Data are presented as means +/− s.e.m. Effect of Gumby genotype on anti-β-gal mean fluorescence intensity was highly significant (Kruskal-Wallis test (k=3); H=43.69; p<0.0001).
Canonical Wnt signaling activates T cell-specific transcription factor/lymphoid enhancer factor (TCF/LEF) to induce downstream target gene expression 2. We tested whether Gumby might modulate canonical Wnt3a signaling by assaying TCF/LEF dependent luciferase expression of the TOPFLASH reporter in HEK293T cells (Fig. 5d–g). FLAG-Gumby could enhance luciferase expression in the presence of Wnt3a. FLAG-GumbyW96R or FLAG-GumbyC129S enhanced Wnt3a-induced TOPFLASH expression to a lesser degree. When we asked if Gumby and LUBAC might play opposing roles in Wnt signaling, we found that co-expressing HA-HOIL and myc-HOIP together inhibited TOPFLASH expression over tenfold. Addition of FLAG-Gumby or GumbyΔ54 reversed HOIL-HOIP dependent inhibition, but FLAG-GumbyC129S and GumbyW96R could not. The same trends were seen in the presence of Dvl2 (Fig. 5 f, g). These data suggest that linear (de)ubiquitination via the Gumby-LUBAC axis can modulate canonical Wnt signaling.
To determine whether Gumby compromised Wnt-response in ECs, we took advantage of canonical Wnt pathway reporter mice (TOPGAL) that carry a transgene with LEF/TCF binding sites directing β-galactosidase (β-gal) expression 28. In triple immunofluorescence experiments using antibodies against β-gal, Gumby and PECAM-1 at signal saturation for β-gal, Gumby protein is expressed exclusively in ECs expressing β-gal in E10.5 +/+;TOPGAL/+, GumW96R/GumW96R;TOPGAL/+ and GumD336E/GumD336E;TOPGAL/+ embryos (Supplementary Fig. 11). Gumby/β-gal co-localization was also seen in the dorsal root ganglia and telencephalon (Fig. 5h, k, Supplementary Fig. 11, data not shown). To test whether canonical Wnt signaling read-out might be affected in mutant ECs, we analyzed relative β-gal levels in ISVs, because, in these, all ECs co-express β-gal and Gumby (Supplementary Fig 11) and their stereotypic organization facilitates comparative analyses. Under conditions where we detect intermediate β-gal signal in PECAM-1-labeled ISVs of E10.5 +/+;TOPGAL/+ embryos, β-gal immunofluorescence was significantly reduced in GumW96R/GumW96R;TOPGAL/+ and GumD336E/GumD336E;TOPGAL/+ (Fig. 5 h–o). Levels were also reduced in other Gumby expressing regions, including the dorsal root ganglia and telencephalon (data not shown). We note that GumW96R and GumD336E homozygotes show a CNS-specific angiogenic phenotype similar to that reported upon disruption of canonical Wnt signaling 5,6. Thus, these findings suggest a role for Gumby in canonical Wnt signaling during angiogenesis.
Discussion
Here we have shown that loss of the linear DUB activity of Gumby compromises angiogenesis, neuronal and craniofacial development. So far, biological roles identified for LUBAC components in the mouse have been milder. Mutations in Sharpin lead to chronic proliferative dermatitis (cpdm) 29,30. HOIL-1/1l mouse mutants show no overt pathology unless challenged 31. HOIP mutant phenotypes remain to be reported. Explanations for these observations include existence of other LUBAC complexes or components or unappreciated functional redundancy between Sharpin and HOIL. Also, in the absence of Gumby, constitutive linear ubiquitination of proteins could be more damaging than absence of linear ubiquitination. For instance, in cases where linear ubiquitination is inhibiting Wnt signaling, Wnt signaling could not be (re)activated by deubiquitination.
Our work reveals that Gumby via its amino-terminus interacts with HOIP and Dvl2. In overexpression experiments, the ability of Gumby to fully counteract LUBAC function requires its catalytic activity. However, in vivo at endogenous protein levels the amino-terminal interaction may have important functions - perhaps to regulate activity of the overall complex, guide substrate selection, direct subcellular localization or direct Gumby to modulate specific pathways.
In humans, deletions of the Gumby-containing interval on chromosome 5p15.2 are associated with mental retardation and craniofacial anomalies observed in Cri du Chat Syndrome (CdCS) patients 32–34. While not the only gene deleted, Gumby might contribute to some CdCS symptoms via its effects on linear deubiquitination.
In summary, our work has identified Gumby as a linear ubiquitin-specific DUB with roles in angiogenesis, other developmental processes and Wnt signaling. This opens the gateway to future investigations towards identification of Gumby substrates, understanding of the roles and regulation of the linear deubiquitination-ubiquitination balance and possibly development of anti-angiogenic therapies.
Complete Online Methods
Method Summary
All mouse husbandry and handling was performed in conformity with the Canadian Council of Animal Care recommendations (AUP 0024a-00H). For all experiments, unless otherwise stated, a minimum of six gumby homozygotes and six +/+ littermates were examined. X-ray structure analyses, binding studies using ITC, deubiquitination assays, RNA and protein expression analyses, immunoprecipitations and Wnt reporter expression analyses in vivo and in cell culture were performed by standard methods and are described in detail in Supplementary Methods.
Mouse husbandry
All mouse husbandry and handling was performed in conformity with the Canadian Council of Animal Care recommendations (AUP 0024a-00H). For all experiments, unless stated otherwise, a minimum of six gumby (GumW96R) and GumD336E homozygotes and six +/+ littermates were examined. Both mutations arose and were ultimately maintained on the C57BL6/J background.
For fine-mapping the gumby (GumW96R) mutation, heterozygous carriers and homozygous mutants were identified with D15Mit130 and D15Mit111. Recombinant embryos were scored with D15Mit252, D15Mit280, D15Mit18, D15Mit201 from www-genome.wi.mit.edu/cgi-bin/mouse/sts-info and SNP markers rs3707949 and rs13482490 from www.ncbi.nlm.nih.gov/projects/SNP. Once identified, a tetra-primer ARMS-PCR assay was used to identify the GumW96R and GumD336E mutations essentially as described in 35.
All primer sequences are listed in Supplementary Table 3.
Mice transgenic for bMQ-396D3 BAC (Chr.15: 27,544,413-27,629,541bp) from AB2.2 embryonic stem cell DNA (129S7/SvEvBrd-Hprtb-m2) 36 were generated by the Michigan University Transgenic Core into (C57BL/6 X SJL)F2 females. The breeding strategy to produce GumW96R/GumW96R;BAC transgenic mice is shown in Supplementary Figure 3.
To assess canonical Wnt signaling, homozygous TOPGAL mice 37 were crossed with GumW96R/+ and GumD336E/+ mice. GumW96R/+;TOPGAL/+ mice and GumD336E/+;TOPGAL/+ were backcrossed to their respective heterozygous mutant backgrounds to obtain GumW96R/GumW96R;TOPGAL/+ and. GumD336E/GumD336E;TOPGAL/+ embryos.
Sequencing
Coding sequences of candidate genes were amplified by PCR of exons and flanking introns from genomic DNA or cDNA from gumby/gumby E11.5 embryos. For expression constructs, wildtype Gumby coding region was amplified from I.M.A.G.E. clone 4430188 and GumbyW96R from GumW96R/GumW96R E10.5 embryonic cDNA using the Superscript™ First-Strand Synthesis System (Invitrogen). All clones were verified by sequencing.
Expression analyses
Northern Blot analyses were performed using total RNA purified with Trizol™ (Invitrogen) at 20μg per lane and resolved as described in 38.
Whole-mount RNA in situ hybridization was performed, as previously described 39, using Gumby cDNA region 459-1059bp, corresponding to AA153-352, cloned into SacI/EcoRI sites of pBluescript SKII (Stratagene). Templates for antisense probes were generated by XhoI digestion and transcription with T7 polymerase, and sense probes by ClaI digestion and transcription with T3 polymerase.
Anti-Gumby polyclonal antibody
Glutathione S-transferase (GST)-tagged Gumby protein (Gumby-GST) was used to generate anti-Gumby antibody in rabbits by the Laboratory Division of Comparative Medicine, University of Toronto. Antibody was purified with Affi-Gel15 coupled to maltose binding protein-tagged Gumby fusion protein (MBP-Gumby). Vectors used were pGEX-HTa (GE Healthcare) and pMAL-C2 vector (New England Biolabs).
Antibody specificity was tested by diluting in either 5% non-fat instant milk powder/TBST for Western blot (immunoblot) or 10% Normal Goat Serum/PBST (immunofluorescence) and incubating it with either GST-Gumby or GST alone, immobilized to glutathione-Sepharose™ 4B beads (GE Healthcare). After 1.5 h of rocking at RT beads were spun down, and the supernatant was used as a primary antibody (Supplementary Fig. 6).
Immunofluorescence
Cells, embryos and tissues were fixed in 4% paraformaldehyde (Sigma). Embryos and tissues were embedded in Tissue-Tek OCT (Miles laboratories) and sectioned at 16–20μm. Samples were washed in PBS, blocked in PBST (PBS-0.3% Triton-X 100) with 10% NGS for 2.5 h at RT, incubated with primary antibodies diluted in PBST-1% NGS overnight at 4°C, washed in PBS, incubated with fluorescently labeled secondary antibodies for 1 hour at RT, washed in PBS and mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories). Whole mount immunohistochemistry with anti-CD31 antibody (1:1000) was performed as previously described 40.
Antibodies used were: rabbit anti-mouse Gumby (1:500); mouse monoclonal anti-human Smooth Muscle Actin (Clone 1A4 DAKO, 1:1,000); monoclonal mouse anti-human Desmin (Clone D33 DAKO, 1:1,000); mouse anti-FLAG M2 (Clone F1804 Sigma, 1:1000); hamster anti-mouse CD31 (PECAM-1) (Millipore 1398Z, 1:500); chick anti-β-galactosidase (Abcam); anti-linear ubiquitin (Genentech, 1 μg/ml) antibodies. Secondary antibodies were goat anti-rat FITC, donkey anti-chick Cy3, donkey anti-hamster Cy2, donkey anti-rabbit Cy3, (all from Jackson Immunoresearch); donkey anti-mouse AlexaFluor 488, goat anti-mouse AlexaFluor 594, goat anti-rabbit AlexaFluor 488, goat anti-rabbit AlexaFluor 594 (all from Molecular Probes); goat anti-chick Alexa 488, goat anti-hamster Alexa 647 and goat anti-chick Alexa 488 (all from Invitrogen).
Immunofluorescence was visualized using a Nikon D-eclipse C1 confocal microscope system, Leica MZFIII microscope equipped with Qimaging 1300C digital camera or Leica spinning disc confocal microscope and analyzed with Adobe Photoshop and Volocity or ImageJ.
Anti-β-gal immunofluorescence
Immunofluorescence was performed on matched sections blocked for 1 hour with 5% NGS, 5% normal donkey serum (NDS), 1% blocking solution (prepared from a 10% solution in maleate buffer), 20mM MgCl2, 0.3% Tween-20 in PBS. Chick anti-β-galactosidase, Cy3-anti-Chicken and Cy2-anti-Hamster were each preincubated for 1 hour at room temperature with 3mg mouse embryo powder in 2% NGS, 2% NDS, 1% blocking solution, 20mM MgCl2, 0.3% Tween-20 in PBS, centrifuged at 10,000g for 10 minutes at 4 °C, then applied to sections at appropriate dilution. Three lots of chick anti-β-gal antibody were tested to identify one optimal preparation. Sections were incubated with anti–β-gal and hamster anti-PECAM-1 antibodies for 14–18 hours at 4° C, washed five times with PBS/0.2% Tween. Secondary antibodies were applied at 1:400 dilutions for 45 minutes. Sections were washed in PBS three times and mounted as usual. Using serial dilutions ranging from 1:100 to 1:4000, we determined that immunofluorescent anti-β-gal signal was saturated at 1:200 to 1:500, not detectable at 1:2000, and present at intermediate levels at 1:1000 dilution on +/+; TOPGAL/+ sections.
To establish Gumby and β-gal colocalization in ECs within ISVs, we used the 1:500 anti-β-gal dilution, analyzed more than three animals per genotype and found complete co-localization in ISVs. For quantification purposes, we counted >100 cells from >10 ISVs per genotype. Gumby and β-gal expression in other embryonic ECs is less uniform. Given the phenotypes we observed in blood vessels within the neural tissue, we focused analyses on these by sampling 394 ECs in 20 transverse sections through an E10.5 +/+; TOPGAL/+ embryo.
We used the 1:1000 dilution in subsequent quantification experiments. Experiments were performed in parallel on a minimum of six sections from three animals per genotype. Images were collected as 3.5 μm stacks under identically set conditions for all samples. For quantification, at least 15 ISVs located between the fore- and hindlimbs were sampled per genotype. Immunofluorescence was analyzed using ImageJ software 41. PECAM positive endothelial cells in ISVs were traced as a group and the mean fluorescent intensity of ISVs (i.e. of endothelial cells within ISVs) was measured. A Kruskal-Wallis test (k=3) based on ranks was performed on the data, since they did not meet the parametric assumptions regarding normality and equivalence of variance. Effect of Gumby genotype on anti-β-gal mean fluorescence intensity was highly significant (H=43.69; p<0.0001).
Immunoblot analysis
Transfected cells and mouse tissues were lysed in RIPA buffer (1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, in PBS pH 7.4 with protease inhibitor cocktail tablet (Roche). SDS-polyacrylamide gel electrophoresis was performed with 10–15μg of total lysate and immunoblot analysis was performed as described. To control for loading, membranes were stripped and probed for Tubulin.
Antibodies used were rabbit Gumby (1:5,000); mouse Tubulin (Clone DM1A Sigma, 1:2000); goat HRP-IgG (anti-rabbit, rat, human or mouse) (Jackson Immunoresearch, 1:10,000); mouse FLAG (Sigma M2, 1:1000); rat HA (Roche, 1:1000); mouse myc (Santa Cruz Biotechnology, 1:500); human linear ubiquitin (Genentech, 1:1000) and rabbit pan-ubiquitin (Dako, 1:1000)
Immunoprecipitation
Human HEK293T cells were transfected using Effectene transfection reagent (Qiagen). Cells were lysed 18–24 hours after transfection in 50mM Tris pH 7.4, 100mM EDTA, 150mM NaCl and 0.5% Triton X-100 and protease inhibitor cocktail (Roche). Aliquots containing 400μg total protein were incubated for 1 hour with 1μg Flag antibody (Mouse M2, Sigma), 2μg myc antibody (mouse, Santa Cruz) or 2μg HA antibody (Rat, High Affinity, Roche). 20μl of Protein A/G plus agarose beads (Santa Cruz) were added and co-immunoprecipitation was performed as in 42. For Dvl2 immunoprecipitation, 4μM MG132 was also added to media 18 hrs post-transfection and during lysis and IP. Samples were analyzed by immunoblot.
In vitro luciferase assays
Wnt signaling was assayed in HEK293T cells cotransfected with various expression constructs and with TOPFLASH or FOPFLASH reporters 43 and the normalizing transfection efficiency control PRL vector using Effectene (Qiagen), incubated for 18–20 hours and then stimulated for 4 hours with Wnt3a conditioned or control media. Similarly, NFκB dependent transcription was assayed with a reporter containing 6 NFκB binding sites and a control plasmid with 6 AP1 binding sites that drive firefly luciferase expression in Dual luciferase assays (Promega) 15. Values were normalized relative to FLAG-pcDNA3.1. Constructs never activated the FOPFLASH or AP1 reporters. At least three independent experiments were performed with samples in triplicate for each construct.
Analysis of Linear ubiquitinated proteins
Analyses of linear ubiquitinated proteins by immunoblot and immunofluorescence were performed as described in 26. For embryonic lysates, Embryos were harvested at embryonic day (E) 10.5 and washed twice with PBS. Individual embryos and 293T cells were lysed in 8M Urea, 50mM Tris pH 7.5, 25 mM NaCl, 2mM EDTA, 2mM N-ethylmaleimide and complete mini protease inhibitors (Roche) and were disrupted by passing through an 18 gauge needle.
Protein expression and purification
Human FAM105B residues 79-352 (GumbyR79) corresponding to the catalytic domain and residues 55-352 (GumbyM55) corresponding to the catalytic domain with an additional 24 residue N-terminal extension, were amplified by PCR from a full-length cDNA clone (IMAGE: 4430188) and cloned into pGEX-TEV downstream of GST and pProEx-TEV downstream of His6. W96R, C129A, C129S and D336E mutations were introduced using standard techniques and all clones were sequence verified. Linear diubiquitin was generated by PCR amplification of residues 1-152 from Ubc (IMAGE:4076286) and cloned into pProEx.
Native and selenomethionine-labelled His6-tagged Gumby proteins, ubiquitin and linear di-ubiquitin were expressed in E.coli BL21(DE3) Codon+ (Agilent Technologies) and purified using a HiTrap nickel chelting HP column (GE Healthcare) using standard lab protocols. Tag free proteins in 20 mM HEPES pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol were concentrated to 10–25 mg/ml and stored frozen at −80°C.
5-Iodoacetamidofluorescein (5-IAF, Molecular Probes)-labeled linear di-ubiquitin was generated by incubating 100 μM purified diUbCys0 (modified by inclusion of a non-native cysteine residue at the ubiquitin amino-terminus) with 500 μM of 5-IAF in a buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl, and 1 mM DTT for 3 hours at 20°C followed by size exclusion chromatography.
Purified USP2, USP5, and USP21 were obtained as described in 44.
Structure determination by x-ray crystallography
Crystals of GumbyM55 were grown at 20°C by mixing equal volumes of 600 μM protein with solution containing 100 mM MES pH 6.0, 200mM MgCl2, 19% PEG3350 and cryo-protected with 20% glycerol. Diffraction to 1.60Å resolution (λ=0.97917) was collected at NE-CAT 24-ID-C beamline at 100K and processed using HKL2000. Phased electron density maps from 9 seleno-methionine positions were generated by SHELX. Protein structure was built using Arp/Warp and refined using Phenix and Coot. 96.4%, 3.6% and 0% of the residues resided in the most favoured, allowed and disallowed regions of the Ramachandran plot (PROCHECK). Crystals of an equimolar mixture of GumbyM55-C129S and ubiquitin (900 μM final concentration) were grown at 20°C by mixing equal volumes of protein with solution containing 100 mM Bis Tris pH 5.5, 200 mM ammonium sulfate, 20% PEG3350 and cryo-protected with 20% glycerol. Diffraction data was collected at 100K at a home source (λ=1.5418) with Rigaku Saturn944+ detector and processed using HKL3000. Crystals of an equimolar complex of GumbyR79-C129A and linear di-ubiquitin were grown at 20°C by mixing equal volumes of protein with a solution of 100 mM acetate pH 5.5, 100 mM CaCl2, 100 mM Glycine, 2.5 M sodium formate and 24% PEG 3350 and cryoprotected with Paratone-N. Diffraction data was collected at NE-CAT 24-ID-C beam line (λ=0.97917) at 100K and processed using HKL2000. The Gumby-ubiquitin and Gumby-linear di-ubiquitin co-structures were determined by molecular replacement using Phaser with GumbyM55 and ubiquitin as search models. 94.8%, 4.7% and 0.5% of residues in the Gumby-ubiquitin complex and 96.8%, 2.4% and 0.8% of residues in the Gumby-diubiquitin complex reside in most favoured, allowed and disallowed regions respectively. Statistics for data collection, phasing and structure refinement are presented in Supplementary Table 1.
Deubiquitination assay
Substrate specificity analysis of Gumby and OTUB1 shown in Figure 3b was determined by incubating 1 μg of the indicated di-ubiquitin substrates (Boston Biochem) with 1 μg of the indicated DUB in a 20 μl reaction with 20 mM HEPES pH 7.5, 300 mM NaCl, 1 mM DTT at 37°C for 18 hours. Reactions were stopped by the addition of 6x Laemmli buffer. Reaction products were subjected to 4–12% PAGE and stained with coomassie brilliant blue.
For enzyme titration experiments in Supplementary Figure 8a and e, 1 μg of linear di-ubiquitin or K48-linked di-ubiquitin was incubated with the indicated concentration of Gumby or OTUB1 in 20 mM HEPES pH 7.5, 300 mM NaCl, 1 mM DTT at 37°C for 30 minutes in a 10 μl volume. Reactions were stopped by the addition of 6x Laemmli buffer, subjected to 15% PAGE and gel stained with coomassie brilliant blue.
Kinetic analyses of GumbyM55, GumbyR79, GumbyD336E and GumbyW96R DUB activity were performed by determining the initial reaction rates from a series of di-ubiquitin cleavage reactions containing 0.08 to 150 μM of fluorescein-labelled linear di-ubiquitin substrate. Reactions were performed at 37°C in 20 mM HEPES pH 7.5, 300 mM NaCl, 1 mM DTT by mixing Gumby (0.5 nM GumbyM55, 0.4 nM GumbyR79, 30 nM GumbyD336E or 25 μM GumbyW96R) and fluorescein-labelled linear di-ubiquitin at 0.08, 0.4, 2, 10, 25, 50, 100 or 150 μM concentration. 35 μl aliquots were obtained at various time points from a 600 μl reaction volume, stopped by the addition of 6x Laemmli buffer, and followed by 15% PAGE analysis. The gel was imaged with a Typhoon FLA 9500 (GE Healthcare) using the Alexa-Fluor 488nM protocol. Bands on the gel corresponding to fluorescein-labelled mono-ubiquitin and standards of fluorescein-labelled di-ubiquitin were used to quantitate the amount of reaction product at each time point using ImageQuant (GE Healthcare). Initial rates for each reaction performed in duplicate were calculated from the linear portions of each reaction profile and fitted to a non-linear Michaelis-Menten equation using Prism v5 (GraphPad Software Inc.) to calculate KM, Vmax, and Kcat values. (See Supplementary Microsoft Excel spreadsheet for replicate data).
For ubiquitin-vinyl sulfone inhibition assays shown in Supplementary Figure 8b, 0.6 μM Gumby, USP2, USP5 and USP21 were incubated with indicated amounts of ubiquitin-vinyl sulfone (Boston Biochem) for 30 minutes at 37°C and subsequently used in a DUB assay as described above. Ub-AMC assays shown in Supplementary Figure 8c were carried out with 5, 50, and 500 nM of the indicated DUBs and 600 nM of Ub-AMC (Boston Biochem) in a buffer containing 50 mM HEPES pH 7.5, 100 mM NaCl, 0.03% Brij-35, and 0.1 mg/ml BSA for 2 hours at 30°C. Fluorescence was measured using an EnSpire 2300 plate reader (PerkinElmer) at 380 nm excitation and 480 nm emission wavelengths.
Isothermal Titration Calorimetry
Calorimetric titrations were carried out on a Microcal VP-ITC titration calorimeter. Protein samples were prepared in 20 mM HEPES, pH 7.5, 300 mM NaCl, 1 mM DTT. 0.2 to 0.5 mM linear di-ubiquitin within the syringe was titrated into 20 to 50 μM concentrations of GumbyC129S or GumbyC129S/W96R proteins residing in the sample cell. Experiments were carried out mimimally in triplicate at 30°C. Data analysis was performed using Origin software (Microcal) (See Supplementary Microsoft Excel spreadsheet for replicate data).
Mass spectrometric analysis
For mass spectrometry, generation of stable inducible cell lines, induction of protein expression and affinity purification on FLAG M2 magnetic beads was performed as described in (REF 22710030). Proteins digested with trypsin on beads were loaded onto reversed-phase capillary columns and analyzed by LC-MS/MS on a LTQ mass spectrometer, as described previously (REF 21630450). RAW files were saved in our local interaction proteomics LIMS, ProHits45, searched with Mascot version 2.3 (Matrix Science) against the human and adenovirus complements of the RefSeq protein database (version 45; 34604 entries), enabling one missed cleavage site, and Met oxidation and Asn/Gln deamidation as variable modifications. Search results were further analyzed within ProHits45, using the SAINT statistical tool 46,47. SAINT analysis was done using two biological replicates per bait. Bait protein samples were analyzed alongside 8 negative control runs (FLAG alone), using the following parameters: 8 negative controls compressed into 6 (nControl:6); nburn: 2000, niter: 5000, lowMode: 0; minFold: 1, normalize: 1 46. After SAINT analysis, results were further filtered to show only those proteins identified in with an average SAINT probability of 0.9, at least 2 unique peptides, and with a frequency of identification less than in 10% of all the samples annotated in a database consisting of > 2000 FLAG AP-MS experiments.
Supplementary Material
Acknowledgments
We thank Thom Saunders and the Michigan University Transgenic Core facility for generating Gumby BAC transgenic mice, Kazuhiro Iwai for HOIL and HOIP constructs, Koichi Nakajima for NFκB and AP2 reporter constructs, Ben Alman and C.C. Hui for TOPGal reporter mice, Jim Woodgett for TOPFlash, FOPFlash and PRL vectors, Tony Pawson Wnt3a expressing L-cells, Miriam Barrios-Ramos for help with fluorometry, Joe Culotti and C.C. Hui for critical feedback and I. Kourinov and staff at NE-CAT for collecting diffraction dataset. B.R. holds the Canada research chair in proteomics and molecular medicine. AC.G. holds the Canada research chair in Functional Proteomics. F.S. holds the Canada research chair in Structural Principles of Signal Transduction. S.M.A. and T.S. were supported by CIHR predoctoral student fellowships. This work was supported by operating funds from the JSPS KAKENHI (Grant Numbers 15200032 to Y.G. and 21240043 to Y.G. and R.F.), Canadian Institutes for Health Research to B.R. (MOP119289), A-C.G. (MOP123433), F.S. (MOP57795), and S.P.C. (MOP 97966, IHO 94384 and MOP 111199).
Footnotes
Contributions:
E.R. designed genetic experiments with S.P.C., identified and confirmed the Gumby causative mutation, performed expression analyses and characterized the GumW96R angiogenic phenotype; S.M.A. performed protein interaction assays and analyzed the Gumby-LUBAC connection; D.C. and Y-C.J. performed X-ray crystallographic analyses and DUB assays; T.A.M. analyzed angiogenesis and Wnt signaling; T.S. performed DUB chain profiling assay; H.H. and Y-C.J. performed ITC analyses; Y.G. supervised R.F.; R.F. identified the GumD336E mutant, W.H.D. ran the Mass Spectrometry, A-C.G. supervised W.H.D.; G.X. helped with SNP mapping; B.R. supervised T.S. and F.S. supervised D.C., H.H., Y-C.J. and together they designed experiments and wrote manuscript; S.P.C. conceived and coordinated project, designed and performed experiments, supervised E.R., S.M.A. and T.A.M. and wrote manuscript.
The coordinates and structure factors have been deposited in the Protein Data bank under the accession numbers 4KSJ (apo-Gumby/Fam105b), 4KSK (Gumby-ubiquitin complex) and 4KSL (Gumby-linear diubiquitin complex).
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Risau W. Mechanisms of angiogenesis. Nature. 1997;386(6626):671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 2.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 3.Liebner S, Plate KH. Differentiation of the brain vasculature: the answer came blowing by the Wnt. Journal of angiogenesis research. 2010;2:1. doi: 10.1186/2040-2384-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zerlin M, Julius MA, Kitajewski J. Wnt/Frizzled signaling in angiogenesis. Angiogenesis. 2008;11(1):63–69. doi: 10.1007/s10456-008-9095-3. [DOI] [PubMed] [Google Scholar]
- 5.Daneman R, et al. Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc Natl Acad Sci U S A. 2009;106(2):641–646. doi: 10.1073/pnas.0805165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenman JM, et al. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322(5905):1247–1250. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- 7.Cirone P, et al. A role for planar cell polarity signaling in angiogenesis. Angiogenesis. 2008;11(4):347–360. doi: 10.1007/s10456-008-9116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wharton KA., Jr Runnin’ with the Dvl: proteins that associate with Dsh/Dvl and their significance to Wnt signal transduction. Dev Biol. 2003;253(1):1–17. doi: 10.1006/dbio.2002.0869. [DOI] [PubMed] [Google Scholar]
- 9.Kirisako T, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25(20):4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18(5):520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 11.Walczak H, Iwai K, Dikic I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi: 10.1186/1741-7007-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwai K. Linear polyubiquitin chains: a new modifier involved in NFkappaB activation and chronic inflammation, including dermatitis. Cell Cycle. 2011;10(18):3095–3104. doi: 10.4161/cc.10.18.17437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471(7340):591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471(7340):637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokunaga F, et al. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471(7340):633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 16.Niu J, Shi Y, Iwai K, Wu ZH. LUBAC regulates NF-kappaB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 2009;30(18):3741–3753. doi: 10.1038/emboj.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Mar L, Rivkin E, Kim DY, Yu JY, Cordes SP. A genetic screen for mutations that affect cranial nerve development in the mouse. J Neurosci. 2005;25(50):11787–11795. doi: 10.1523/JNEUROSCI.3813-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogan KA, Ambler CA, Chapman DL, Bautch VL. The neural tube patterns vessels developmentally using the VEGF signaling pathway. Development. 2004;131(7):1503–1513. doi: 10.1242/dev.01039. [DOI] [PubMed] [Google Scholar]
- 20.Gondo y. Trends in large-scale mouse mutagenesis: from genetics to functional genomics. Nature Reviews Genetics. 2008;9(10):803–810. doi: 10.1038/nrg2431. [DOI] [PubMed] [Google Scholar]
- 21.Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007;21(20):2511–2524. doi: 10.1101/gad.1589207. [DOI] [PubMed] [Google Scholar]
- 22.Edelmann MJ, et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. The Biochemical journal. 2009;418(2):379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 23.Juang YC, et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Molecular cell. 2012;45(3):384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483(7391):618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, et al. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. Journal of molecular biology. 2009;386(4):1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto ML, et al. Engineering and structural characterization of a linear polyubiquitin-specific antibody. J Mol Biol. 2012;418(3–4):134–144. doi: 10.1016/j.jmb.2011.12.053. [DOI] [PubMed] [Google Scholar]
- 27.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 28.Maretto S, et al. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(6):3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.HogenEsch H, et al. A spontaneous mutation characterized by chronic proliferative dermatitis in C57BL mice. Am J Pathol. 1993;143(3):972–982. [PMC free article] [PubMed] [Google Scholar]
- 30.Seymour RE, et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 2007;8(5):416–421. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- 31.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136(6):1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Mainardi PC, et al. The natural history of Cri du Chat Syndrome. A report from the Italian Register. Eur J Med Genet. 2006;49(5):363–383. doi: 10.1016/j.ejmg.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Mainardi PC, et al. Clinical and molecular characterisation of 80 patients with 5p deletion: genotype-phenotype correlation. J Med Genet. 2001;38(3):151–158. doi: 10.1136/jmg.38.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, et al. High-resolution mapping of genotype-phenotype relationships in cri du chat syndrome using array comparative genomic hybridization. Am J Hum Genet. 2005;76(2):312–326. doi: 10.1086/427762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29(17):E88–88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams DJ, et al. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics. 2005;86(6):753–758. doi: 10.1016/j.ygeno.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 37.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126(20):4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 38.Ambulos NP, Jr, Duvall EJ, Lovett PS. Method for blot-hybridization analysis of mRNA molecules from Bacillus subtilis. Gene. 1987;51(2–3):281–286. doi: 10.1016/0378-1119(87)90317-9. [DOI] [PubMed] [Google Scholar]
- 39.Kim FA, et al. The vHNF1 homeodomain protein establishes early rhombomere identity by direct regulation of Kreisler expression. Mech Dev. 2005;122(12):1300–1309. doi: 10.1016/j.mod.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Vecchi A, et al. Monoclonal antibodies specific for endothelial cells of mouse blood vessels. Their application in the identification of adult and embryonic endothelium. Eur J Cell Biol. 1994;63(2):247–254. [PubMed] [Google Scholar]
- 41.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 42.Torban E, Wang HJ, Groulx N, Gros P. Independent mutations in mouse Vangl2 that cause neural tube defects in looptail mice impair interaction with members of the Dishevelled family. J Biol Chem. 2004;279(50):52703–52713. doi: 10.1074/jbc.M408675200. [DOI] [PubMed] [Google Scholar]
- 43.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5(3):367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 44.Ernst A, et al. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339(6119):590–595. doi: 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu G, et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol. 28(10):1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi H, et al. Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr Protoc Bioinformatics. Chapter 8(Unit8):15. doi: 10.1002/0471250953.bi0815s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi H, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 8(1):70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30(2):144–154. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.