Abstract
Interferon-induced proteins with tetratricopeptide repeats (IFITs) are innate immune effector molecules that are thought to confer antiviral defence through disruption of protein–protein interactions in the host translation-initiation machinery. However, it was recently discovered that IFITs can directly recognize viral RNA bearing a 5′-triphosphate group (PPP-RNA), which is a molecular signature that distinguishes it from host RNA. Here we report crystal structures of human IFIT5, its complex with PPP-RNAs, and an amino-terminal fragment of IFIT1. The structures reveal a new helical domain that houses a positively charged cavity designed to specifically engage only single-stranded PPP-RNA, thus distinguishing it from the canonical cytosolic sensor of double-stranded viral PPP-RNA, retinoic acid-inducible gene I (RIG-I, also known as DDX58). Mutational analysis, proteolysis and gel-shift assays reveal that PPP-RNA is bound in a non-sequence-specific manner and requires a 5′-overhang of approximately three nucleotides. Abrogation of PPP-RNA binding in IFIT1 and IFIT5 was found to cause a defect in the antiviral response by human embryonic kidney cells. These results demonstrate the mechanism by which IFIT proteins selectively recognize viral RNA, and lend insight into their downstream effector function.
Supplementary information
The online version of this article (doi:10.1038/nature11783) contains supplementary material, which is available to authorized users.
Subject terms: X-ray crystallography, Biochemistry, Innate immunity, Virology
Crystal structures reveal insight into how interferon-induced proteins with tetratricopeptide repeats (IFITs) selectively recognize viral RNA.
Supplementary information
The online version of this article (doi:10.1038/nature11783) contains supplementary material, which is available to authorized users.
IFIT protein recognition of viral RNA
IFITs — interferon-induced proteins with tetratricopeptide repeats — are antiviral components of the innate immune system. Recent work suggested that they may act as RNA sensors, and in this study Bhushan Nagar and colleagues elucidate the structural basis for the specific recognition of triphosphorylated viral RNA (PPP-RNA). The crystal structures of human IFIT5, its complex with PPP-RNAs, and an amino-terminal fragment of IFIT1 reveal that the triphosphate group fits neatly into a deep pocket within the protein, which also accommodates the first three nucleotides of the RNA.
Supplementary information
The online version of this article (doi:10.1038/nature11783) contains supplementary material, which is available to authorized users.
Main
The innate immune system relies on several germ-line-encoded receptors to distinguish self from non-self molecules in order to mount an appropriate early defence response. During viral infection, non-self molecules are derived from viral genomes generally in the form of double-stranded RNA (dsRNA) or PPP-RNA that is not protected by a 5′-cap. The canonical host proteins responsible for sensing or interacting with these foreign nucleic acids include the Toll-like receptors, RIG-I-like receptors and nucleotide oligomerization domain (NOD)-like receptors1. Recently, an unbiased proteomics approach discovered that the IFITs could also directly engage PPP-RNA2.
IFITs are among the most potently expressed proteins of a group of interferon-stimulated genes (ISGs)3, which are the culmination of virally triggered signalling pathways that lead to the production of interferon (IFN)-α, IFN-β and other cytokines. They are evolutionarily conserved from mammals to fish, with four well-characterized paralogues in humans: IFIT1 (also known as p56 and ISG56), IFIT2 (also known as p54 and ISG54), IFIT3 (also known as p60 and ISG60) and IFIT5 (also known as p58 and ISG58), ranging in mass from 54 to 56 kDa. IFITs are composed of tetratricopeptide repeats (TPRs), degenerate helix–turn–helix motifs of 34 amino acids in length, which are usually present in multiple copies as tandem arrays that generate solenoid-type scaffolds well-suited for mediating protein–protein interactions4 (Supplementary Figs 1 and 2).
IFITs have been implicated in modulating several biological processes, including inhibition of translation initiation, cell proliferation, and migration, in addition to mediating antiviral effects3. Most of these functions are thought to occur through disruptive protein–protein interactions between IFITs and host cellular factors. Through their TPRs, human IFIT1 and IFIT2 were shown to inhibit key steps during translation initiation by interacting with the ‘e’ or ‘c’ subunits of eIF3 (refs 5, 6). However, the unexpected finding that IFITs can bind RNA suggested a more direct role: after infection or interferon stimulation, it was found that IFITs form large multiprotein complexes with other family members and several different RNA-binding proteins, leading to viral clearance2. Like RIG-I, productive binding of both IFIT1 and IFIT5 were shown to depend on the presence of cytosolic PPP-RNAs2,7,8. However, crystallographic and biochemical analyses of RIG-I bound to RNA revealed that it is a dsRNA-specific translocase9, which optimally interacts with blunt-ended PPP-RNA10,11,12,13,14,15. The mechanism by which IFITs recognize PPP-RNA is unknown.
We describe here the crystal structure of full-length human IFIT5 with and without PPP-RNAs, as well as an N-terminal, protease-resistant fragment of human IFIT1 (nIFIT1). The structures reveal a novel arrangement of TPR domains that directly bind PPP-RNA in a non-sequence-specific manner and, to our knowledge, represent the first example of a TPR protein bound to a nucleic acid ligand. Structure-guided biochemical analysis of IFIT5 indicated that only single-stranded RNA (ssRNA) can be accommodated within the protein, which undergoes a compaction upon binding. Finally, functional analysis in human embryonic kidney (HEK) cells reveals a reduction of viral replication only in the presence of proper PPP-RNA binding by IFIT1 or IFIT5.
Crystal structures of IFIT5 and nIFIT1
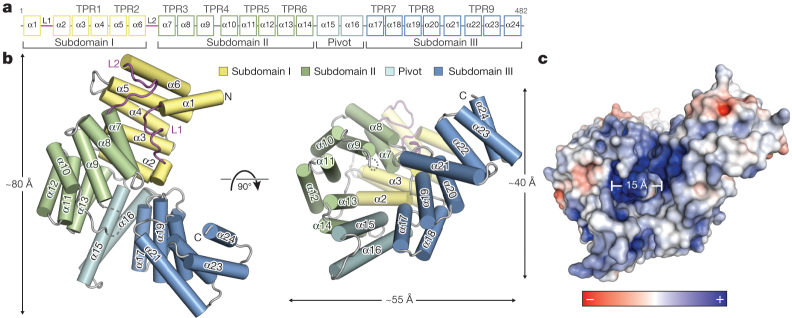
We crystallized and determined the structures of full-length human IFIT5 (residues 1–482) at 2.1 Å resolution and an N-terminal fragment of IFIT1 (residues 7–279) at 1.9 Å resolution using single-wavelength anomalous diffraction. The structure of IFIT5 reveals a helical domain with approximate dimensions of 80 Å × 55 Å × 40 Å (Fig. 1a, b). In most multi-TPR-containing proteins, such as O-linked N-acetylglucosamine transferase, the relationship between successive TPRs is regular and repeating, such that they form open-ended superhelical structures with distinct convex and concave surfaces4,16. In IFIT5, of its total 24 α-helices, 18 form canonical TPRs (TPRs 1–9; Fig. 1a), whereas the remaining 6 helices intervene between the TPRs such that the regular repeating relationship between them is disrupted. This results in the formation of three distinct bundles of TPRs (subdomains I, II and III) oriented with respect to one another to give the overall protein a relatively closed clamp-shaped structure (Fig. 1a and Supplementary Fig. 5a).
Figure 1. Structural overview of human IFIT5.
a, Secondary structure, TPR motif and subdomain organization of IFIT5. b, Orthogonal views of IFIT5 with helices represented as cylinders. c, Surface representation of IFIT5 coloured by electrostatic potential (using APBS) from negative ( −5kTe−1; red) to positive (+5kTe−1; blue).
The topology of subdomain I is unusual in that its two canonical TPRs (α3 to α6) are capped off on both ends by helices α1 and α2, preventing its further propagation into a superhelix. This is facilitated by a connecting 17-residue loop (L1) containing a highly conserved Cys-His-Phe-Thr-Trp pentapeptide motif that is invariant among nearly all of the IFIT proteins (Supplementary Fig. 2), and forms a single turn of a helix that packs against the concave inner face of subdomain I (Supplementary Fig. 4). This same arrangement of subdomain I is also found in the structure of nIFIT1 (root mean squared deviation, 1.4 Å; Supplementary Fig. 3) and is probably a defining characteristic of all IFIT proteins given the high TPR and sequence conservation in subdomains I and II (Supplementary Figs 1 and 2).
The remainder of the IFIT5 structure forms a superhelix encompassing subdomains II and III, as well as a pair of extended non-TPR helices (α15 and α16) that form a pivot point between the latter two subdomains (Fig. 1b). Subdomain II forms a canonical four-TPR-repeat domain in which, notably, its first helix (α7) interacts with subdomain I in a manner reminiscent of TPR protein–ligand interactions observed previously17 (Supplementary Fig. 4). This leads to the concave surface of subdomain II forming one wall of a large cavity in the centre of the protein closed off at its base by helix α2 (Fig. 1b). The same TPR–ligand relationship between subdomains I and II is also maintained in the nIFIT1 structure (Supplementary Fig. 3).
The rest of the cylindrical cavity is created by the intervening pivot helices and the N-terminal TPRs from subdomain III. Subdomain III begins with two typical TPRs followed by an interrupting helix (α21), which inverts the direction of the final TPR9 such that it forms an S-shaped appendage at the carboxy terminus with two potential ligand-interacting concave surfaces (Fig. 1b and Supplementary Fig. 5b). The deep pocket formed by this atypical arrangement of TPRs is approximately 28 Å deep by 15 Å wide, and is lined with an expansive collection of positively charged residues well-suited for the accommodation of nucleic acid (Fig. 1c).
IFIT5 specifically binds PPP-RNAs
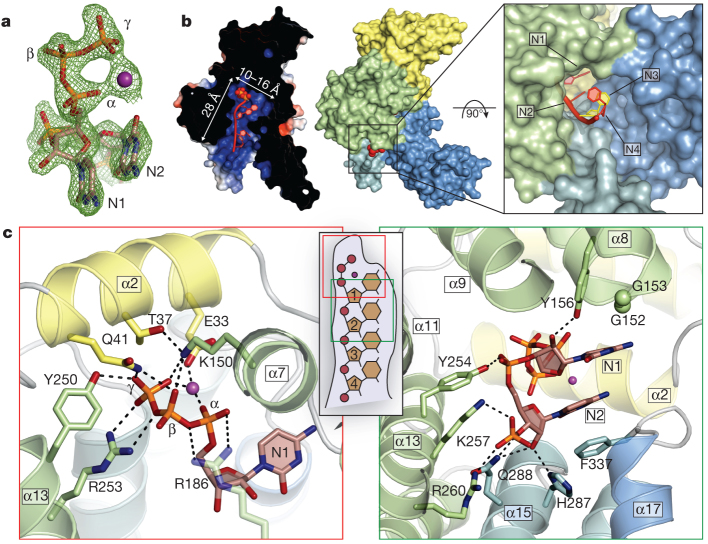
To understand the structural basis for RNA binding by IFIT5, we in vitro transcribed 5′-triphosphate-bearing, short oligonucleotides of cytidine, uridine and adenosine, purified each PPP-RNA in complex with IFIT5 (Supplementary Fig. 6) and determined their structures at resolutions of 1.86 Å (oligo-C), 2.0 Å (oligo-U) and 2.5 Å (oligo-A) using molecular replacement with the unliganded structure. All of the structures were similar, and therefore we initially describe the general features of the IFIT5-oligo-C complex as it was the highest resolution structure. Difference Fourier maps revealed strong positive electron density within the central positively charged pocket from which the 5′-triphosphate and the first four nucleotides of the RNA could be reliably modelled (Fig. 2a and Supplementary Fig. 10a). The 5′-triphosphate group is nestled deep within the pocket and makes a multitude of electrostatic interactions with protein side chains from helix α2 (Glu 33, Thr 37 and Gln 41) located at the very base of the pocket, and residues from the concave inner surface of subdomain II (Lys 150, Tyr 250 and Arg 253) (Fig. 2c). Arg 187 in IFIT1 was previously identified to be required for RNA interaction; the homologous residue in IFIT5 (Arg 186) makes a weak salt-bridge with the α- and β-phosphates, and van der Waals contacts with the first ribose moiety. These RNA-interacting residues are for the most part conserved in sequence and structure between IFIT5 and IFIT1 (Supplementary Figs 2 and 7), the only IFITs that have been shown to bind PPP-RNA with strong affinity2. One notable exception is Thr 37, which is replaced in IFIT1 by Arg 38, suggesting slight differences in RNA recognition between IFIT1 and IFIT5. Conversely, in IFIT3, which is known to not bind PPP-RNA2, Tyr 250 is substituted with a negatively charged residue, Asp 242, and Arg 186 with His 182 (Supplementary Fig. 2), both of which would interfere directly with RNA binding.
Figure 2. Structure of IFIT5 bound to PPP-RNA.
a, Fo– Fc electron-density map of the triphosphate and first two nucleotides contoured at 3.5σ before inclusion of RNA into the model. The metal ion is indicated with a purple sphere. b, Left, cross-section of the complex coloured by surface electrostatic potential. The triphosphate is shown as spheres and RNA nucleotides are shown in red. Middle, surface representation of IFIT5 bound to PPP-RNA coloured by subdomain. Protruding RNA is shown as red spheres. Right, close-up view looking down the axis of the RNA-binding pocket. c, Close-up view of the residues making specific contacts with the triphosphate group (left) and the first two nucleotides, N1 and N2 (right). Helices are coloured according to the subdomain to which they belong. Hydrogen bond and salt-bridge interactions are indicated with black dashed lines.
Interestingly, a metal ion that bridges the α- and γ-phosphates also seems to be an integral part of PPP-RNA recognition as it neutralizes the negative charge in this region from Glu 33 (Fig. 2c and Supplementary Fig. 8). On the basis of ligand distances and geometry, the ion is probably Mg2+ from the in vitro transcription reactions, but could potentially also be Na+ (a component of the crystallization buffer). It is unlikely that capped messenger RNA (mRNA) can be accommodated within this pocket owing to size constraints. In addition, given the critical interactions made with the γ-phosphate and the metal ion, the pocket is unlikely to accept 5′-monophosphorylated or 5′-hydroxylated RNA with considerable affinity. Thus, the structure of the IFIT5 TPR domains have evolved to specifically engage PPP-RNA, and in doing so, distinguish between self and non-self nucleic acids.
Following the 5′-triphosphate end of the RNA, the first two nucleotides (N1, N2) are stably bound along the pocket before the third and fourth nucleotides (N3, N4) begin to protrude from the mouth of the pocket (Fig. 2b). Well-defined density is observed for the phosphodiester backbone and ribose sugars (Supplementary Fig. 10a), which also form several specific interactions with the protein (Fig. 2c). In particular, the 5′-phosphate of N2 hydrogen bonds with Tyr 254, and the 5′-phosphate of N3 makes a salt-bridge with Arg 260 and Lys 257, and hydrogen bonds with Gln 288 (Fig. 2c). The 5′-phosphate of N4 interacts with Arg 294, and weak electron density was observed for the 5′-phosphate of a fifth nucleotide (Supplementary Figs 10a and 11a). The 2′-hydroxyl of the ribose sugars also make specific interactions with the protein, but in this case, interactions that are dependent on the sugar pucker. N1 adopts a C2′-endo conformation (commonly found in B-form double-stranded DNA; Supplementary Fig. 9) and hydrogen bonds with Tyr 156 (Fig. 2c), whereas N2 and N3 are C3′-endo (as found in A-form dsRNA) and interact through their 2′-hydroxyls with His 287 and Gln 288, and Arg 294 and Asp 343, respectively (Fig. 2c and Supplementary Fig. 11a).
PPP-RNA recognition is non-sequence specific
To investigate the potential for sequence-specific interactions at the 5′-end, we compared the crystal structures of IFIT5 in complex with the different RNAs. In both the oligo-C and oligo-U complexes, the pyrimidine base at position 1 is abutted from the top by van der Waals interactions with Tyr 156 and two glycine residues from the loop of TPR3 (between α7 and α8), and from the bottom by non-specific stacking interactions against the second base, which in turn stacks with Phe 337 (Fig. 2c). Notably, the first two bases do not make any specific hydrogen bonds with protein residues and there is ample space adjacent to the pyrimidine ring edges, suggesting that the larger purine bases can also be easily accommodated (Fig. 3). The structure of the oligo-A complex confirms this notion and reveals that the adenine rings reach further out into the periphery making additional non-specific van der Waals contacts with Thr 371, His 374 and Phe 339, which were absent with the pyrimidine bases (Fig. 3b–d).
Figure 3. The interaction between IFIT5 and PPP-RNA is non-sequence specific.
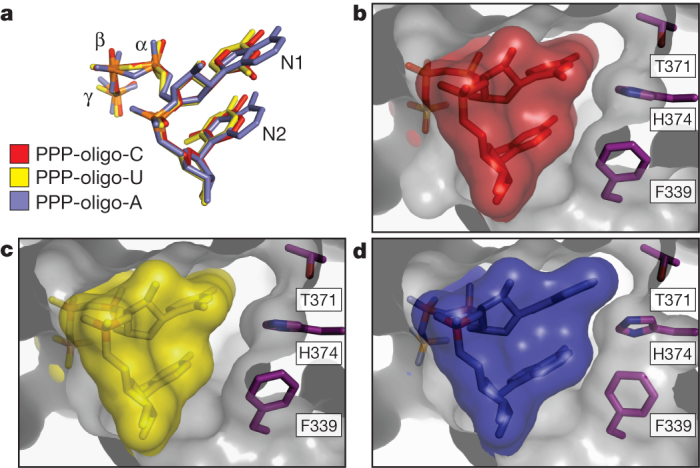
Close-up of the RNA-binding pocket in an orientation similar to that shown in Fig. 2b, right panel. a, Alignment of the first two nucleotides from the three IFIT5-RNA complexes. b, c, Surface and stick representation of the first two nucleotides within the IFIT5-oligo-C and IFIT5-oligo-U complexes. The protein surface is depicted as a transparent grey cutaway. d, IFIT5-oligo-A complex. Detailed binding between the protein and all nucleotides is depicted in Supplementary Fig. 12.
The remaining bases stack against Phe 339 in a manner analogous to that observed for the first two bases (Fig. 2b and Supplementary Fig. 11) and interact with a mobile loop from TPR4. Thus, IFIT5 seems to have evolved the capacity to accommodate any 5′-PPP-RNA sequence that may potentially be present in a viral genome.
PPP-RNA binding involves a conformational change
Because the RNA-binding site in IFIT5 is a deep and narrow pocket, the means by which RNA enters is unclear. Superposition of the RNA-bound and -free forms of IFIT5 reveal that the RNA-bound state is more compact, with the largest motions occurring at the pivot helices between subdomain III and the rest of the protein (Fig. 4a and Supplementary Fig. 13). These motions position several key residues from the different subdomains for optimal interaction with the RNA. Moreover, limited proteolysis of IFIT5 in the presence and absence of RNA supports the notion of compaction and stabilization of the protein in the RNA-bound form (Fig. 4b and Supplementary Fig. 15).
Figure 4. IFIT5 undergoes a conformational change upon binding PPP-RNA.
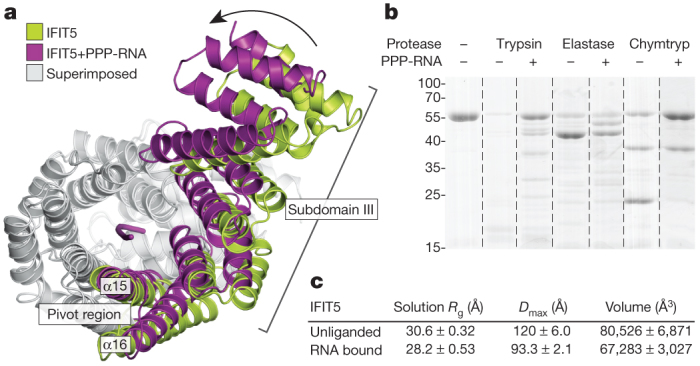
a, Comparison of IFIT5 bound to PPP-RNA (magenta) and the unbound form (green). Superimposed regions are coloured light grey. b, SDS–polyacrylamide gel electrophoresis of limited protease digestion of IFIT5 in the presence and absence of RNA taken from each experiment at the 15-min time point (see also Supplementary Fig. 15). Chymtryp, chymotrypsin. c, Summary of SAXS results. Measurements are the average from three concentrations, with the corresponding standard deviation.
To better understand the nature of the conformational change upon RNA binding, we used small-angle X-ray scattering (SAXS) measurements, which provide information on macromolecular size, state and flexibility directly in solution18,19. SAXS analysis revealed reductions in the radius of gyration (Rg, ∼2.5 Å), the maximum dimension (Dmax, ∼25 Å) and the volume (∼14,000 Å3) of the protein upon addition of RNA (Fig. 4c and Supplementary Fig. 14). The scattering curves show good agreement between solution (Rg, 28.2 Å) and crystal structure (Rg, 27.5 Å) for the RNA-bound form (Supplementary Fig. 14h), in contrast to the unliganded form, which displays considerable differences (solution Rg, 30.6 Å; crystal structure Rg, 28.3 Å). This suggests that in solution, the unliganded protein is either more open or possibly flexible. To discern between these possibilities, we subjected the SAXS data to a Porod–Debye analysis, which provides information on the degree of flexibility present in the scattering sample19. For both unliganded and RNA-bound IFIT5, the Porod–Debye plot showed characteristic plateaus that indicate the presence of distinct conformations for both species (Supplementary Fig. 14k, l). Thus, unliganded IFIT5 probably exists in a more open conformation in solution than that observed in the crystal structure, facilitating RNA entry.
IFIT5 and IFIT1 bind only PPP-ssRNAs
The internal diameter of the RNA-binding pocket in IFIT5 is roughly 15 Å, leaving no room to accommodate dsRNA, which would require a diameter of greater than 21 Å. Moreover, at least three bases are necessary to span the length of the pocket, suggesting that IFIT5 is potentially a sensor for PPP-ssRNA, or base-paired PPP-RNA with a minimum three-nucleotide overhang. By contrast, foreign PPP-RNA species in the cytosol that optimally activate RIG-I seem to require blunt-ended RNAs, which are thought to be the most potent immune-stimulant of RIG-I (ref. 15).
To assess the recognition of distinct PPP-RNA species by IFITs, we used gel-shift assays. A 44-nucleotide ssRNA with no predicted secondary structure within the 5′ 22 nucleotides was in vitro transcribed, to which complementary RNA strands of 15–20 nucleotides were annealed to generate dsRNA with blunt ends and various 5′-overhangs. Consistent with the crystal structure, we found that IFIT5 could shift both PPP-ssRNA and PPP-dsRNA with at least three-nucleotide overhangs, but could not efficiently shift blunt-ended PPP-RNA or PPP-dsRNA with 1–2-nucleotide overhangs (Fig. 5a and Supplementary Fig. 16). Similarly, IFIT1 could only shift PPP-ssRNA or PPP-dsRNA with at least five-nucleotide overhangs (Supplementary Fig. 16). As a negative control we used IFIT3, which could not shift any species of RNA (Fig. 5a and Supplementary Fig. 16d). Thus, owing to the limitations imposed by their RNA-binding pockets, IFIT5 and IFIT1 can engage only PPP-RNAs that have single-stranded 5′-ends.
Figure 5. Functional analyses of IFIT binding to PPP-RNA.
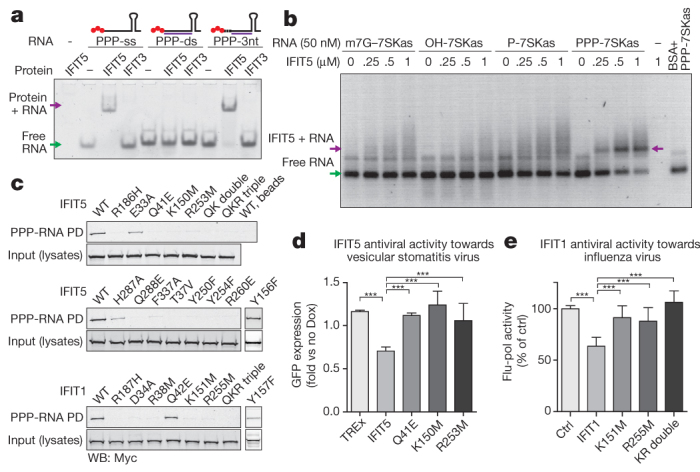
a, Mobility shift assay between IFIT5 or IFIT3 and ssRNA, dsRNA with blunt ends, or dsRNA with a three-nucleotide (3nt) overhang as indicated by the schematics above each set of lanes. PPP is indicated by red spheres, in vitro transcribed top strand is indicated by the black line, and synthetic complementary RNA is shown in purple. b, Agarose gel-shift assay between IFIT5 and the various RNAs indicated. 7SKas, 7SKantisense; BSA, bovine serum albumin; m7G; 7-methyl-guanosine cap. c, Biotinylated RNA pull-downs (PD) of wild-type (WT) and mutant IFIT1 and IFIT5 from HEK293 cell lysates. QK double indicates Q41E/K150M and QKR triple indicates Q41E/K150M/R253M. Y156F and Y157F were carried out separately, and the appropriate positive and negative controls are described in Supplementary Fig. 17. d, e, PPP-RNA binding is required for antiviral activities of IFIT5 and IFIT1. d, Replication of vesicular stomatitis virus expressing green fluorescent protein (GFP) in doxycycline (Dox)-inducible HEK Flp-In TREx cells expressing IFIT5 (and mutants). Average fold change (± s.d.) in doxycycline-treated versus untreated cells of ten measurements. e, Influenza virus in 293T cells transfected with IFIT1 (and mutants). Average percentage (± s.d.) of influenza polymerase (flu-pol) activity as compared to control (ctrl) of four independent experiments done in duplicate measurements. ***P < 0.001 (one-way analysis of variance, Tukey’s multiple comparison test).
Functional validation of PPP-RNA binding to IFITs
To examine the functional relevance of residues involved in binding PPP-RNA, we used PPP-RNA-coated beads to pull down c-Myc-tagged wild-type and mutant IFIT5 and IFIT1 expressed in HEK293 cells. We began by first corroborating that IFIT5, like IFIT1, could be pulled down by RNA only when it is triphosphorylated at the 5′-end (Supplementary Fig. 17a), and that replacing the triphosphate with 5′-cap, 5′-monophosphate or 5′-hydroxyl diminishes the binding (Fig. 5b). The affinity of PPP-RNA for IFIT5 is between 250–500 nM (Fig. 5b), similar to that found previously for IFIT1 (ref. 2).
Next, we mutated key RNA contacts within the pocket and found that in most cases, a single-residue substitution was sufficient to abolish RNA binding in vitro (Fig. 5c). All residues recognizing the PPP-RNA were critical for binding, with the exception of Glu 33, Tyr 156 and His 287 (see Supplementary Fig. 17). Homologous mutations in IFIT1 also lead to abrogation of RNA binding. Thus, the PPP-RNA-binding pocket identified here is probably involved in a similar mode of recognition in other IFIT family members.
Finally, to investigate whether the RNA-interacting residues are important for the antiviral activity of IFIT5 against virus infection, we used HEK293 Flp-In TREx cells that inducibly express IFIT5, and IFIT5 mutants that have lost their ability to bind PPP-RNA. Consistent with the mutational analysis, IFIT5 mutants were impaired in their ability to restrict growth of vesicular stomatitis virus compared to wild-type IFIT5 (Fig. 5d). Similarly, IFIT1 lacking the ability to bind PPP-RNA was not able to inhibit the activity of an influenza virus polymerase (Fig. 5e), consistent with the notion that binding to PPP-RNA is critical for the antiviral activity of IFIT1.
Discussion
The structural basis for IFIT recognition of foreign RNA described here validates the new paradigms put forth for how this family of interferon-stimulated genes carry out their effector functions, and brings to the forefront the versatility of the TPR motif in recognizing diverse ligands, paralleling established receptors of the innate immune system such as those containing leucine-rich repeats1.
In addition to using protein–protein interactions to confer downstream antiviral activity2,3,5,6, the principal molecular role of IFITs seems to be initiated by direct recognition of foreign PPP-ssRNAs. PPP-RNAs are found within the genome of negative-sense ssRNA viruses such as influenza and vesicular stomatitis virus. Other RNA viruses, such as positive-sense viruses which have 5′-capped genomes, can also generate cytosolic PPP-RNAs as replicative intermediates during their life cycle. Hence, the evolution of a binding site to specifically recognize PPP-RNA allows IFITs to distinguish self from non-self RNAs, as cytosolic host ssRNAs bear a 5′-monophosphate (on ribosomal RNA and transfer RNA)20,21 or are 5′-capped (in the case of mRNA). In doing so, one possible mechanism for IFIT function may be to latch onto the ends of viral RNA, preventing it from being properly replicated or packaged into progeny virions.
Recent studies have suggested a role for IFIT proteins in sensing the 5′-cap methylation status of some viral RNA (for example, West Nile virus, poxvirus and coronavirus)22,23,24. A 5′-cap is present on positive-sense virus genomes, but most viruses also have the ability to either hijack a cap from host mRNA or encode machinery to add a 5′-cap structure to their mRNA, thereby potentially circumventing IFIT recognition25. Although a 5′-cap cannot be accommodated within the RNA-binding pocket of IFIT5 identified here, we do not preclude the possibility that higher order complexes of IFITs may be able to recognize capped viral RNAs.
Finally, given that bacterial mRNAs also bear a free 5′-PPP and can access the cytosol during infection to potentiate induction of IFN-β26, it is plausible that IFITs could also have a role in anti-bacterial innate immunity. Taken together, it is clear that unravelling the structural details that underlie IFIT biology will improve our understanding of the complex interplay between pathogens and host innate immunity, and hopefully pave the way for the development of new immunotherapeutics.
Note added in proof: While this paper was under review, the crystal structure of unliganded IFIT2 was published27.
Methods Summary
For crystallization, all constructs were cloned into a modified pET28a vector containing an N-terminal, Ulp1-cleavable His6-SUMO tag28 and expressed in Escherichia coli. Proteins were purified using standard Nickel-affinity, ion-exchange, and size-exclusion chromatography. PPP-RNAs were generated by in vitro transcription. Crystals of nIFIT1, IFIT5 and IFIT5 in complex with PPP-RNA were grown from a sparse matrix screen (see Supplementary Information). The structures of nIFIT1 and IFIT5 were solved by single-wavelength anomalous diffraction phasing of selenomethionine-derivatized protein, and the complexes with PPP-RNA were solved by molecular replacement using unliganded IFIT5 as a search model. Pull-downs with PPP-RNA and functional assays with HEK Flp-in TREx cells are described elsewhere2. Full methods can be found in Supplementary Information.
Supplementary information
This file contains Supplementary Text, Supplementary Tables 1-6, Supplementary Figures 1-17 and Supplementary References. (PDF 21428 kb)
Acknowledgements
We thank S. Labiuk and P. Grochulski for X-ray data collection performed on beamline 08ID-1 at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research (CIHR), the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. Thanks also to all laboratory members for discussions and technical assistance. B.N. is supported by a Canada Research Chair, a Career Development Award from the Human Frontiers Science Program (CDA 0018/2006-C/1) and an operating grant from the CIHR (CIHR grant MOP-82929). Work in the G.S.-F. laboratory is funded by the Austrian Academy of Sciences and the i-FIVE ERC grant. A.P. is an EMBO Long-Term Fellowship recipient (ATLF 463-2008). Y.M.A. was supported by the CIHR Strategic Training Initiative in Chemical Biology.
PowerPoint slides
Author Contributions
B.N., G.S.-F., Y.M.A., M.W.G. and A.P. designed the project. B.N., Y.M.A., A.P., M.W.G. and G.S.--shift experiments. M.W.G. performed the mutagenesis, pull-down and gel-shift experiments and A.P. performed functional experiments and pull-down experiments.
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 4HOQ, 4HOR, 4HOS, 4HOT and 4HOU. Reprints and permissions information is available at www.nature.com/reprints.
Competing interests
The authors declare no competing financial interests.
Footnotes
Andreas Pichlmair and Maria W. Górna: These authors contributed equally to this work.
Contributor Information
Giulio Superti-Furga, Email: gsuperti@cemm.oeaw.ac.at.
Bhushan Nagar, Email: bhushan.nagar@mcgill.ca.
References
- 1.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 2.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nature Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 3.Fensterl V, Sen GC. The ISG56/IFIT1gene family. J. Interferon Cytokine Res. 2011;31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Main ERG, Xiong Y, Cocco MJ, D’Andrea L, Regan L. Design of stable α-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/S0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 5.Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terenzi F, Hui DJ, Merrick WC, Sen GC. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- 7.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 8.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 9.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nature Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C, et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jínek M, et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nature Struct. Mol. Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc. Natl Acad. Sci. USA. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 19.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao S, Scott F, Fierke CA, Engelke DR. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/S0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 22.Daffis S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nature Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szretter KJ, et al. 2′- O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8:e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nature Rev. Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sander LE, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, et al. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 2012;22:1328–1338. doi: 10.1038/cr.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/S1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Text, Supplementary Tables 1-6, Supplementary Figures 1-17 and Supplementary References. (PDF 21428 kb)
Data Availability Statement
Primary accessions
Protein Data Bank
Data deposits
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 4HOQ, 4HOR, 4HOS, 4HOT and 4HOU. Reprints and permissions information is available at www.nature.com/reprints.