Abstract
Shortly after the discovery of endothelial progenitor cells (EPCs) in 1997, many clinical trials were conducted using EPCs as a cellular based therapy with the goal of restoring damaged organ function by inducing growth of new blood vessels (angiogenesis). Results were disappointing, largely because the cellular and molecular mechanisms of EPC-induced angiogenesis were not clearly understood. Following injection, EPCs must migrate to the target tissue and engraft prior to induction of angiogenesis. In this study EPC migration was investigated in response to tumor necrosis factor α (TNFα), a pro-inflammatory cytokine, to test the hypothesis that organ damage observed in ischemic diseases induces an inflammatory signal that is important for EPC homing. In this study, EPC migration and incorporation were modeled in vitro using a co-culture assay where TNFα treated EPCs were tracked while migrating towards vessel-like structures. It was found that TNFα treatment of EPCs increased migration and incorporation into vessel-like structures. Using a combination of genomic and proteomic approaches, NF-kB mediated upregulation of CADM1 was identified as a mechanism of TNFα induced migration. Inhibition of NF-kB or CADM1 significantly decreased migration of EPCs in vitro suggesting a role for TNFα signaling in EPC homing during tissue repair.
Keywords: Endothelial Progenitor Cells, CADM1, TNFα, Cellular Migration
Introduction
The discovery of endothelial progenitor cells (EPCs) and the first demonstration that a cellular based therapy could be used as a regenerative therapeutic was published by Asahara and colleagues in 1997.[1] Successful use of EPCs in animal models to regenerate lost organ function prompted the initiation of human trials.[2, 3] Conflicting results from human trials raised notable concern regarding the efficacy of using EPCs to restore organ function.[4, 5] The prevailing explanation of these results was based on the observation that the cell population that was used to induce regeneration was derived from autologous donors. It was hypothesized that because the cells were isolated from individuals with cardiovascular disease, the cells were therapeutically incompetent as a result of the underlying pathophysiology. Very little data exists supporting this hypothesis as the molecular mechanisms of EPC-induced angiogenesis are not well understood. Therefore, development of an effective cellular-based therapy requires a better understanding of the cellular mechanisms of regeneration.
EPCs are primarily found in the bone marrow, peripheral circulation, and vessel walls with the majority being in the bone marrow.[6] It has been demonstrated that EPCs are mobilized out of the bone marrow to sites of ischemia in order to repair damage.[7] Many groups have studied the mobilization of EPCs, associating acute inflammation with increased EPC mobilization.[8, 9] The majority of human disease states induce an inflammatory response.[10] It is well understood that immune cells are recruited to an active site of disease, illness, or infection and can induce apoptosis in infected cells, collect debris, and provide support for repair. EPCs and immune cells share a common precursor, so it is likely they play an innate regenerative role.[1] For example, acute events such as myocardial infarction, stroke, and dilated cardiomyopathy increase the number of peripheral EPCs.[11-13] In these studies, patients observed with a higher number of peripheral EPCs typically exhibited better outcomes than patients with a lower number of peripheral EPCs. In contrast, patients with chronic diseases typically have decreased numbers of peripheral EPCs and poorer outcomes.[14-16]
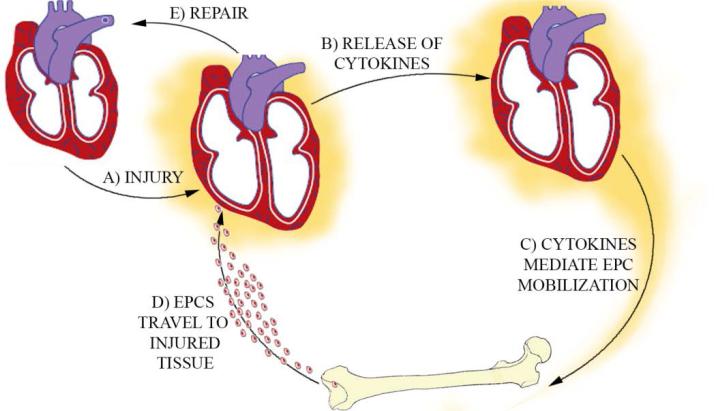
We hypothesized that repair of organ damage by endogenous EPCs functions as a feedback controlled system to maintain tissue integrity (Figure 1). In such a system, an acute injury would induce the production of pro-inflammatory cytokines that would act to both mobilize EPCs from the bone marrow and recruit EPCs from the circulation to the target tissue. Recruitment of cells and repair of damaged tissue would thereby decrease the injury signal completing the feedback loop. In the context of the published human data, acute stimulation of EPC mobilization would be classified as an adaptive regenerative response as regeneration would be followed by a decrease in the mobilization signal. Cells with impaired regenerative function would lead to a buildup of mobilizing cytokines resulting in chronic stimulation of EPC mobilization. This would be considered a maladaptive response.[17, 18]
Figure 1. Bone Marrow Stem Cell Repair Axis.
EPCs are transported from the bone marrow to the myocardium. Injured myocardium releases inflammatory signals that mobilize EPCs from the bone marrow. Cells are released into the circulation and blood flow through damaged organs induces recruitment of cells to target tissue. These cells can then repair the damaged myocardium and suppress the inflammatory signal, completing the feedback loop.
In this study, Tumor Necrosis Factor (TNFα) was used as the prototypic pro-inflammatory cytokine to induce EPC migration as others have demonstrated the role of TNFα in activation of EPC function.[19-21] Kelly et. al. demonstrated that TNFα receptors play a key role in stem cell-mediated regeneration of cardiac function.[22] TNFα is a pro-inflammatory cytokine that acts primarily to regulate immune cell function including rolling, adhesion, proliferation, and apoptosis.[23][24-30] To increase localized recruitment of leukocytes during an inflammatory response, TNFα simulation of the endothelium increases the number of adhesive molecules expressed on the cell surface.[31] In addition to production of TNFα in response to a foreign antigen, TNFα is produced in response to other stimuli, for example, during exercise or in traumatic injury.[32-35] Because TNFα has known roles in cellular recruitment, adhesion and remodeling, it was hypothesized that acute, low-dose treatment would increase the migratory properties of EPCs to vascular endothelial cells.
The EPC mobilization process has been very well studied and characterized. The process by which EPCs migrate to a damaged tissue following an insult or injection (Figure 1) has not been well studied. Elucidation of mechanisms that control migration of EPCs to damaged tissue could provide insight into approaches to augment the repair process induced by endogenous EPCs. Additionally, assessment of cells prior to treatment could help predict the likelihood of successful therapy.
While the importance of TNF signaling in tissue regeneration has been demonstrated, studies have typically quantified high-level phenotypes such as cardiac function in response to sequentially deleting TNF receptors. The goal of this manuscript was to investigate the cellular functions and associated molecular mechanisms that occur after injection of EPCs, specifically those that control EPC migration to target areas of the vasculature in response to TNFα. To accomplish this, a comprehensive approach was developed where TNFα-induced signaling was analyzed in 3 parts: (1) receptors, (2) effectors, and (3) intracellular signaling. Experiments were designed to elucidate the molecular mechanisms at each level. At the receptor level, TNFα signaling is a well characterized process; however, there are multiple TNF receptors and pathway effects. To determine the mechanism of TNF receptor signaling (Part 1) in EPCs, gene expression of key TNFα signaling components was following treatment. To determine potential effector proteins (Part 2), liquid chromatography tandem-mass spectrometry (LC/MS-MS) was used to identify candidate proteins that were differentially regulated in response to TNFα stimulation and that were consistent with observed receptor signaling. To determine the specific mechanisms of intracellular signaling, candidate proteins and signaling pathway members were inhibited and EPC migration was evaluated in response to TNFα (Part 3). In conducting these studies, a new molecular mechanism of TNFα-induced migration was proposed, tested, and validated.
Methods
Electrically-Stimulated Model of Hindlimb Angiogenesis
The Medical College of Wisconsin (MCW) Institutional Animal Care and Use Committee approved all animal protocols. Sprague Dawley rats were placed on 4% salt diet one week before surgery to suppress angiogenesis.[36] Nine to ten week old rats were anesthetized with an intramuscular injection mixture of ketamine (70 mg/kg), Xylazine (4 mg/kg) and Acepromazine (1 mg/kg). Aseptically, a battery-powered stimulator was implanted on the medial right limb.[37] Electrodes induced contractions of the tibialis anterior (TA) and extensor digitorum longus (EDL) muscles for eight consecutive hours, daily. The contralateral leg was used as a control. All animals were euthanized after seven days of stimulation and expression of TNFα was measured via PCR and ELISA.
EPC Isolation and In Vivo Expansion
EPCs were isolated from Sprague Dawley (SD) rat (Harlan) tibia/femur bone marrow as previously described.[19, 38] SD rats were euthanized, the tibia/femur were isolated, and the bone marrow was flushed using a 20 gauge needle. The RBCs were separated from the bone marrow using a polysucrose solution and differential centrifugation (Histopaque 1083; Sigma-Aldrich 10831). The mononuclear fraction was collected, washed, and seeded at a density of 1×107 cells/dish onto fibronectin coated (10μg/mL) cell culture dishes. Cells were cultured in MCDB131 (E3000-01B; US Biological; Swampscott, MA) supplemented with 10% FBS and the EGM-2 supplement pack (Lonza). Cells were grown at 37°C, 20% O2 and 5% CO2.
Cell Culture
Rat cardiac microvascular endothelial cells (RCMVECs) were plated, expanded (P4-P6) in 100 mm dishes (R1111; Cell Biologics; Chicago, IL) and cultured to confluence in endothelial cell media (MCDB131, E3000-01B; US Biological; Swampscott, MA) with the EGM-MV pack (CC-4147; Lonza; Basel, Switzerland).
Migration Assay
Tube migration assay was conducted as described previously.[38] RCMVEC-tubes were grown in four-well slides (Nunc Lab-Tek) coated with 250μL of Growth Factor Reduced Matrigel (BD Biosciences). Mature EPCs were treated with TNFα (1ng/mL)/control (PBS) for 3 hrs. EPCs were stained with DAPI (30 min, 40μg/mL) and lifted using an enzyme free dissociation buffer. 10,000 EPCs were added to each well. Brightfield/fluorescent micrographs were taken using a Nikon TS-100 micrograph 2 hours and 14 hours following EPC seeding (initial studies revealed that assessment at 2 hours is the earliest time point repeatable results can be demonstrated, assessment at 14 hours is the latest time point before results become inconsistent). The number and locations of fluorescent EPCs was detected/recorded. These locations were registered against the bright field images of tube-like structures and the number of cells that had migrated to tubes was determined.[39] The ratio of incorporated versus total EPCs was calculated and compared across groups.
qPCR Results and Analysis
Expression of genes in the TNFα pathway was measured using the RT2 Proflier™ PCR Array (Qiagen; PARN-063Z). EPCs were treated with TNFα (1ng/mL) or vehicle (PBS) for 3 hours. RNA was isolated using the RNeasy Mini-Kit (QIAGEN, Cat. # 74104) and converted to cDNA using the RT2 First Strand Kit (QIAGEN 330401). Samples were run on a 7900-HT Real-Time PCR Thermocycler (Invitrogen) and analyzed using QIAGEN software.
CADM-1 qPCR was done by loading 50ng of RNA/well into a 1X solution of TaqMan Fast Virus 1-Step Master Mix (Invitrogen 4444432) with validated CADM-1 primers (Life Technologies Rn00457556_m1). 18S ribosomal subunit (Invitrogen 4333760F) was used as the control gene, data were analyze via the Livak and Schmittgen method and expressed as relative transcript abundance.
TNFα ELISA
TNFα levels in unstimulated/stimulated skeletal muscle homogenates were measured with an ELISA kit (Invitrogen KRC3011). Skeletal muscle was disrupted in 500 μL buffer with a protease inhibitor (Roche 11697498001) and 0.5% Triton X-100 in PBS with a TissueLyser II (Qiagen). Total protein concentration was determined using Bio-Rad DC protein assay and lysates were analyzed for TNFα.
Proteomics Results
Cell surface proteins were identified following chemical isolation using LC-MS/MS. EPC surface proteins were isolated using a combination of two separate chemical isolation methods, both as described previously, a glycoprotein isolation (Cell Surface Capture, CSC)[40] and a cell membrane associated biotinylation isolation (CMABI).[41]
To isolate using the CSC method, surface glycoproteins were oxidized by adding 4mL of 1mM sodium-meta-periodate (Fisher, PI20504) at 4°C for 15 minutes. Cells were lifted using an enzyme free dissociation buffer (Milipore, S-014-B). Oxidized glycans were labeled at 4°C for 60 minutes using 4mL of 10mM biocytin hydrazide (Biotium, 90060). EPCs were lysed and the membrane fraction was pelleted at 35,000×g. The membrane fraction was reduced for 30 minutes (100mM, NaHCO3; 0.1% RapiGest, 5mM TCEP) and alkylated for 30 minutes in iodoacetamide (10mM). Samples were digested for 18 hours and peptides labeled with the oxidized glycan modification were purified and analyzed using an LTQ Orbitrap Velos Mass Spectrometer
To isolate using the CMABI method, EPCs were lifted from culture dishes using an enzyme free dissociation buffer (Milipore, S-014-B) at 4°C for 45 minutes. Cells were washed, pelleted, resuspended and labeled using 4mL of 1mM NHS-SS-Biotin for 60 minutes. Cells were lysed and membrane fractions were separated at 55,000×g for 2 hours at 4°C. Samples were digested using trypsin for 18 hours. Peptides labeled with the NHS-SS-Biotin modification were isolated using streptavidin coated beads (Pierce, 53117). Samples were then washed, reduced, (100mM, NaHCO3; 0.1% RapiGest, 5mM TCEP), alkylated (10mM iodoacetamide) and eluted in 0.1% trifluroacetic acid. Peptides were purified using C18 columns and analyzed using an LTQ Orbitrap Velos Mass Spectrometer.
Immunoblot to confirm CADM-1 differential regulation
Cultured EPCs were treated with TNFα (1ng/mL)/control (PBS) for 3 hours and RCMVECs were treated with TNFα/control for 12 hours. Cells were isolated in a membrane prep buffer (255mM sucrose, 20mM HEPES, 1mM EDTA) supplemented with protease inhibitor (Roche) and lysed using a 24 gauge needle. 10μg of protein from each sample was loaded onto a 10% polyacrylamide gel (BioRad) and transferred to a PVDF membrane. The membranes were blocked with nonfat dry milk and probed overnight with the CADM1 antibody (1:1000, abcam ab3910). Secondary probing was done using HRP conjugated goat anti-rabbit (BioRad #170-6515, 1:5000) for 2 hours at 4°C. Visualization and development was done with chemiluminescence (Pierce). The 55kD bands were quantified based upon integrated optical density (Molecular Devices). Results were normalized by staining the blot protein using a coomassie stain (BioRad).
NF-kB Inhibition
EPC migration experiments were repeated using an NF-kB inhibitor. 1 hour prior to TNFα/vehicle treatment, a synthetic peptide NF-kB inhibitor (sc-3060; Santa Cruz Biotechnology) or a control, scrambled peptide inhibitor (sc-3061; Santa Cruz Biotechnology) was place into cell culture media at 1μg/mL. Following treatment, TNFα (1ng/mL)/control (PBS) was administered for 3 hours. EPCs were harvested and used for the migration assay or RNA analysis.
siRNA-mediated CADM-1 Knockdown Experiments
EPCs were isolated and cultured in 100mm dishes. After 10 days of expansion, cells were transfected with ON-TARGETplus CADM1 siRNA (J-101011-12) or the scrambled control (D-001810-02-05; Thermo Scientific) using DharmaFECT3 transfection reagent (T-2003-03; Thermo Scientific) according to the manufacturer's instructions. Briefly, for a single dish, 6.3μL of siRNA (20μM) was mixed with 193.7μL of opti-MEM reduced serum media (Invitrogen 31985062) in one tube while 40μL of DharmaFECT3 and 160μL of opti-MEM reduced serum media (Invitrogen 31985062) were mixed in a second. The two solutions were combined, incubated at room temperature for 20 minutes, and added to 4.6mL of MCDB131 + EGM-2 media (without antibiotics). The transfection-ready mixture was added to the cell culture, yielding a final concentration of 25nM. After 24 hours of transfection, the media on the transfecting EPCs was changed to MCDB131 + EGM-2 (with antibiotics). After a further 24 hours, the cells were treated with TNFα (1ng/mL) or vehicle (PBS) for 3 hours for use in the migration assay or RNA was isolation to assess knockdown efficiency.
Results
Expression of TNFα is increased in an in vivo model of electrically stimulated angiogenesis
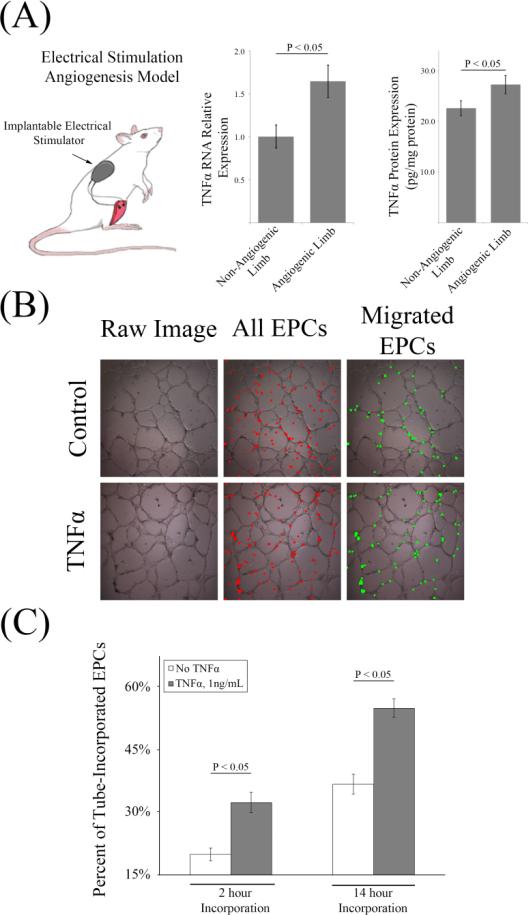
Previous work from both our group and others has demonstrated that electrically stimulating nerves innervating the hind-limb musculature induces angiogenesis in normal animals.[42, 43] In angiogenesis-incompetent animals, stimulating hind-muscle contractions after injecting EPCs restores impaired angiogenesis.[36, 44] The molecular mechanisms of angiogenesis in this model are not fully understood. It was hypothesized that TNFα was involved in this process. To test this hypothesis, Sprague Dawley rats were surgically implanted with hind limb stimulators that induced muscle twitching for 8 hours/day for 1 week. After 1 week, the tibialis anterior and extensor digitorum longus were harvested and expression of TNFα was measured using qPCR and ELISA. It was found that electrical stimulation significantly increased TNFα RNA by 65% and protein by 25% (Figure 2, A). The finding of increased TNFα expression in this model suggests that TNFα plays a key role in mediating EPC-induced angiogenesis. Based upon the role of TNFα in leukocyte recruitment, we hypothesized that TNFα increases the migratory activity of EPCs to the endothelium.
Figure 2. TNFα Produced in an In Vivo Model of Angiogenesis Increases the Migratory Activity of EPCs In Vitro.
(A) TNFα expression is increased in an in vivo model of angiogenesis. A hind limb muscle stimulator that has previously been shown to induce angiogenesis was implanted and run for 7 days. After 7 days, RNA expression of TNFα was significantly increased in the stimulated limb vs the non-stimulated control (n = 6 per group). TNFα protein expression in homogenized hindlimb muscle (pg/mg of homogenized protein) was also found to be significantly upregulated in the stimulated vs the non-stimulated control (n = 6 per group). (B) In vitro EPC migration assay. Because TNFα expression was found to be increased in the angiogenic limb, EPCs were treated with TNFα/control and their locations were tracked with respect to capillary-like tubes in vitro. (C) 3hr, 1ng/mL TNFα/vehicle pre-treatment of EPCs increased the fraction that migrated towards tubes (P < 0.05) at 2 hours and 14 hours (n = 16 per group).
TNFα increases the preferential migration of EPCs towards vessel-like structures in vitro
To measure the migratory activity of EPCs, vessel-like structures were grown on a Matrigel™ based substrate using rat microvascular endothelial cells (RCMVECs). EPCs were pre-treated with TNFα or vehicle for 3 hours prior to suspending 1×104 DAPI stained EPCs in each well containing fully mature tubes. EPCs were pretreated in order to keep conditions consistent with proteomic and gene expression experiments. Each well was imaged (brightfield and fluorescence in order to track both cell types) at 2 hours and 14 hours. The number and locations of each EPC was assessed to determine if it had migrated to a tube. It was found that TNFα pretreatment of EPCs significantly increased the fraction of EPCs that migrated to vessel-like structures at both 2 hours and 14 hours (Figure 2, B and C). The mechanism underlying this phenotype was further investigated with qPCR and proteomic analyses.
Low dose, acute TNFα treatment of EPCs signals through the TNFR2 (p75) pathway, not the TNFR1 (p55) pathway
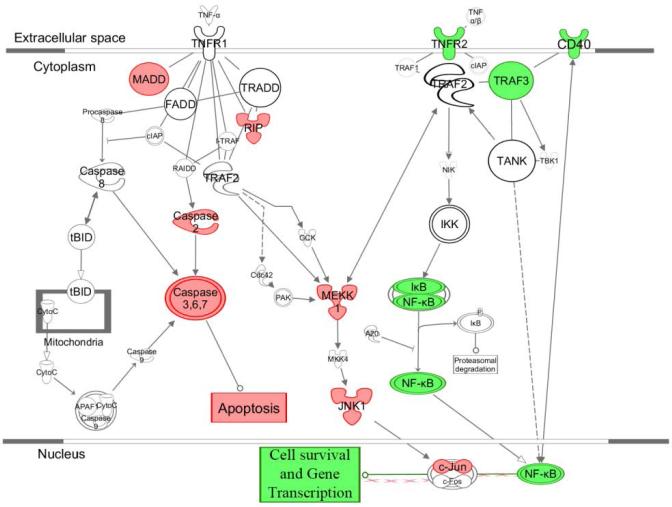
TNFα is known to signal through two pathways, TNFR1 (p55) and TNFR2 (p75).[45, 46] EPCs were treated with TNFα or vehicle and gene expression of key pathway components was measured using qPCR. In the p55 pathway, MADD, RIP, Caspase 2, and Caspase 3 were found to be significantly down-regulated indicating attenuated apoptosis signaling. In the p75 pathway, TNFR2, TRAF3, and NF-kB were all found to be significantly upregulated. The canonical TNFα receptor pathway members were imported from Ingenuity Pathway Analysis and genes with significantly increased expression in response to TNFα were colored green whereas genes detected with significantly decreased expression were colored red (Figure 3). As it has been demonstrated that NF-kB is activated through the TNFR2 pathway,[47, 48] these results suggest increased migration of EPCs through TNFα treatment occurs primarily through the TNFR2 (p75) pathway. This is further investigated through inhibition of NF-kB experiments.
Figure 3. TNFα Pathway Analysis Results by qPCR.
To determine which TNF receptor was driving the migratory phenotype, expression of key pathway genes was measured in response to TNFα. Significantly upregulated genes are displayed in green and significantly down regulated genes are displayed in red. Key genes down regulated in the TNFR1 family are members of the caspase family. Key genes upregulated in the TNFR2 family are NF-kB and TNFR2 suggesting that the migratory phenotype is being signaled through the TNFR2 family via NF-kB (n = 6 per group).
NF-kB mediates TNFα-stimulated migration to in vitro tubes
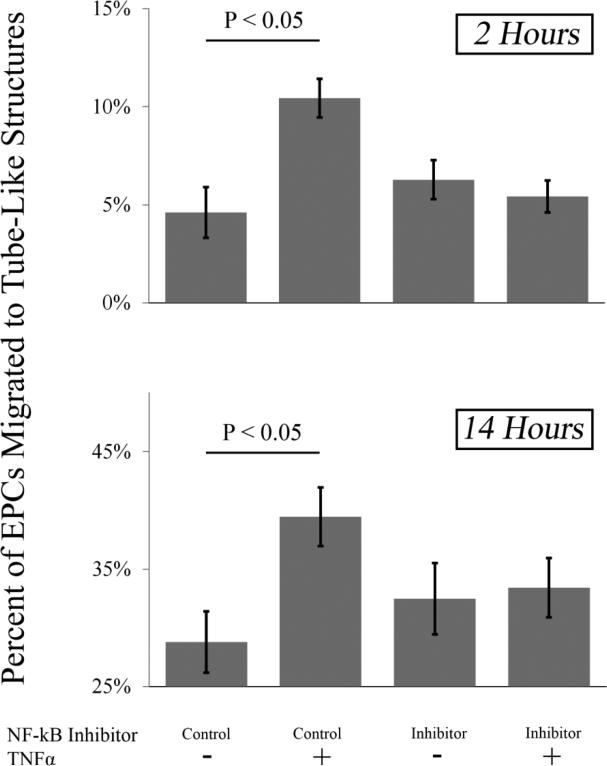
The results of Figure 3 suggested that TNFα-induced migration occurs through NF-kB. To test the hypothesis that NF-kB regulated genes transcription increases EPC migration, the migration assay was repeated using a peptide inhibitor of NF-kB or a scrambled-peptide control (Figure 4). When using a control, it was found after both 2 and 14 hours that TNFα increased EPC migration to tube-like structures in vitro (P < 0.05). In the presence of the targeting NF-kB inhibitor, there was no change in migration towards tube-like structures at 2 hours and 14 hours. However, the increased migration induced by TNFα treatment was inhibited in EPCs pre-treated with the NF-kB inhibitor compared to vehicle-treated cells, supporting the hypothesis that increased migration in response to TNFα treatment is mediated through NF-kB. As other studies have shown that NF-kB signaling in the TNF pathway is through the TNFR2 receptor, these data, combined with those in figure 3, support the hypothesis that TNF signaling in this system is occurring through the TNFR2 receptor.
Figure 4. Inhibition of NF-kB Reduces TNFα Induced Migration.
Prior to TNFα administration, EPCs were pretreated for 1 hour with a 1μg/mL dose of a peptide inhibitor of NF-kB or a scrambled control. The TNFα induced migratory phenotype was still observed in the presence of the scrambled inhibitor at 2 hour and 14 hours (P < 0.05) but the phenotype was abolished at 2 hours and 14 hours when the NF-kB inhibitor was administered (n = 16 per group).
LC-MS/MS identification of unique membrane proteins that mediate migration
To identify effectors mediating the migratory phenotype induced by TNFα, EPC and RCMVEC surface proteins were isolated and analyzed using LC-MS/MS. Four separate groups were analyzed (1. RCMVECs, control 2. RCMVECs, TNFα treated 3. EPCs, control 4. EPCs, TNFα treated). Relative protein abundance was quantified using spectral counting as previously described [36, 38].
Prior to candidate filtering using Visualize 1.58,[49] 6000 unique proteins were identified. Approximately 1000 of these proteins passed quality filters. The list was further filtered by eliminating proteins that did not participate in a relevant function (adhesion, incorporation, recruitment), were not differentially regulated in response to TNFα (P < 0.05), or where a binding partner was not identified in the complimentary cell type. In accordance with results from Figures 3 and 4, proteins had to be predicted in silico to be regulated by NF-kB using Genomatix Genome Analyzer.[50] These criteria narrowed the candidate list to 7 candidate protein pairs (Table 1). Cell Adhesion Molecule 1 (CADM1) was chosen as the final candidate protein as it was demonstrated to have the largest up-regulation in response to TNFα treatment and deficiencies in rodent models functionally matched the observed phenotype most closely.[51-53]
Table 1. Candidate Proteins from LC/MS-MS Experiments.
Candidate Migration Protein Pairs. After chemically isolating cell surface proteins and completing an LC-MS/MS analysis, over 6000 proteins in total were identified. Quality filters narrowed the identified candidate list to 1000 potential targets. Functional filters described in the text further narrowed down the candidate list to 7 potential targets. Cell Adhesion Molecule 1 (CADM1) was chosen as the primary candidate because it met all criteria, had the largest increase in response to TNFα, and had the most biologically relevant function.
| Protein | Differentially Regulated In | Observed Binding Partner | Binding Partner Differential Regulation? | Normalized Ratio | Normalized P-Value |
|---|---|---|---|---|---|
| Cell Adhesion Molecule 1 | EPCs | CADM1-4 | Upregulated, Not Significant | 8.54 | 9.409E-08 |
| FRAS1-Related Extracellular Matrix Protein 3 | RCMVECs | Basement Membrane (Laminins) | Yes, Various forms detected | 6.40 | 0.0029273 |
| PDZ Domain-Containing Protein 3 | EPCs | Many | No | 2.99 | 0.032395 |
| Junctional Adhesion Molecule A | RCMVECs | MPDZ | No | 2.24 | 2.144E-05 |
| Neural Cell Adhesion Molecule 1 | RCMVECs | NCAM1 | No | 2.09 | 2.482E-04 |
| Neuroplastin | EPCs | Neuroplastin | Yes | TNFα Only | 2.839E-11 |
| Afadin | EPCs | Various Actins | Yes | TNFα Only | 4.113E-13 |
Confirmation of LC-MS/MS results that CADM1 is differentially regulated in response to TNFα
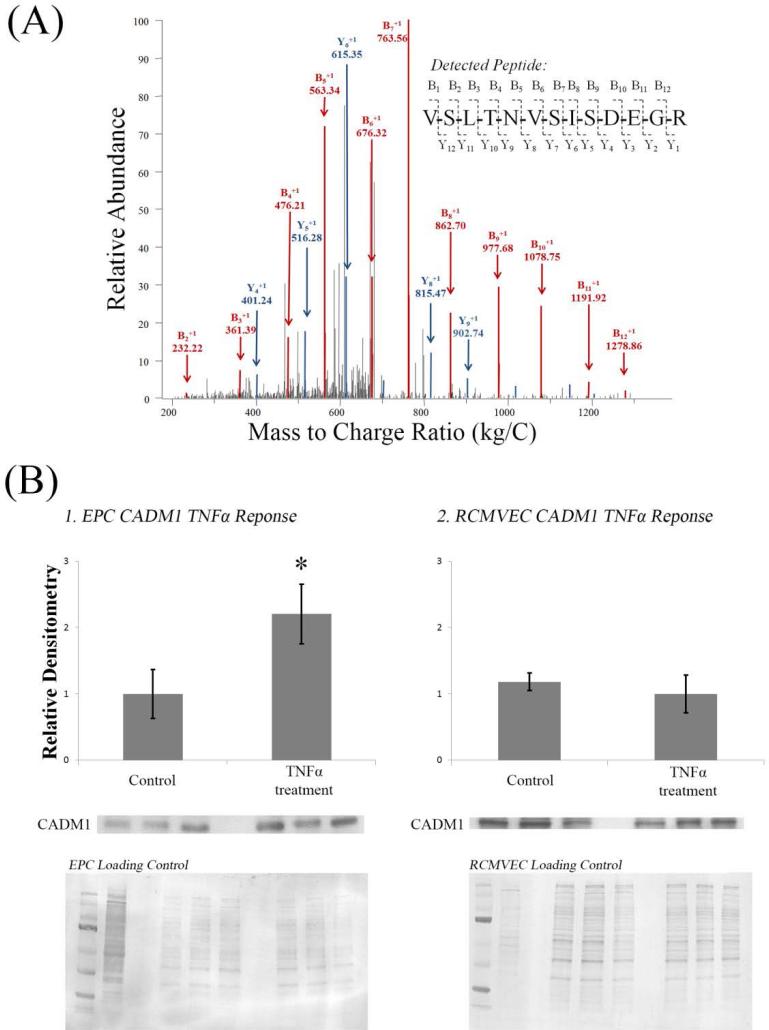
A sample LC-MS/MS spectra of a CADM1 specific peptide is shown in Figure 5, A. Immunoblots of CADM1 in EPCs and RCMVECs were completed to validate CADM1 as a candidate protein to mediate the migratory process of EPCs (Figure 5, B). In EPCs, CADM1 expression was found to be significantly increased in response to TNFα, whereas in RCMVECs CADM1 expression was detected, but no differential regulation was found in response to TNFα.
Figure 5. Validation of CADM1 as a Candidate Protein Pair.
(A) Example LC-MS/MS Spectra. 16 scans of the peptide sequenced ‘VSLTNVSISDEGR’ were observed in the CSC TNFα treated EPC data set. The peptide has 12 potential peptide fragmentation sites and therefore 12 potential ‘B’ and ‘Y’ ions. Following peptide fragmentation, the following m/z spectra was obtained. 11 ‘B’ ions were observed (11 shown) and 9 ‘Y’ ions were observed (5 shown).
(B) Western blot analysis was conducted to confirm quantitation via spectral counting of CADM1 in EPCs (1) and RCMVECs (2). Statistically significant regulation was observed in EPCs, but not RCMVECs confirming proteomics results. Quantitation was performed by normalizing CADM1 bands to total protein detected on the membrane via coomassie staining.
qPCR Analysis of CADM1 expression in response to TNFα
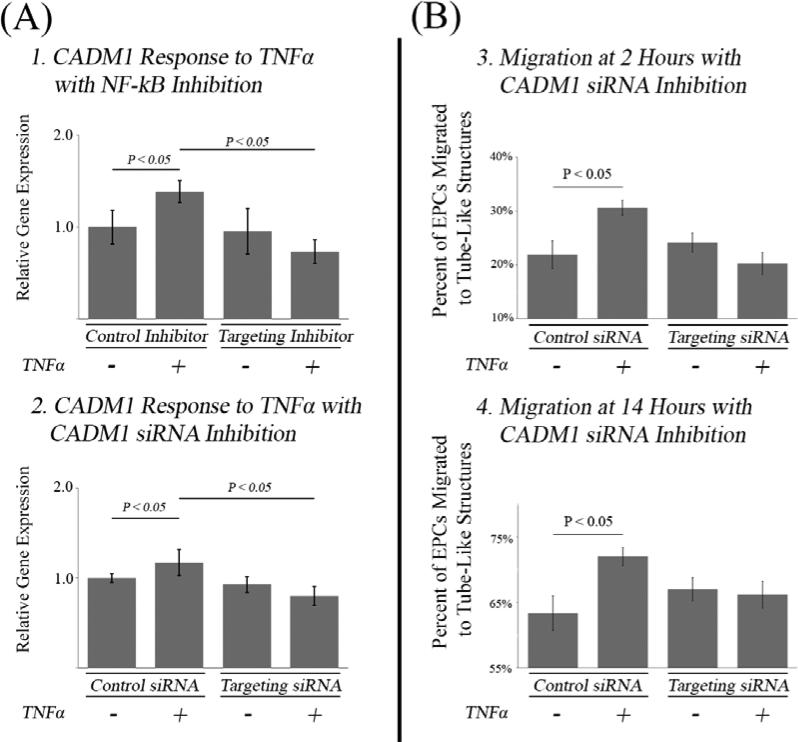
To test the hypothesis that TNFα differentially regulates CADM1 via NF-kB, expression of CADM1 was assessed via qPCR in the presence or absence of an NF-kB synthetic peptide inhibitor (Figure 6, Panel A). When EPCs were treated with an NF-kB or a matched control (scrambled peptide) inhibitor, CADM1 expression was found to be differentially upregulated in response to TNFα (P < 0.05). When EPCs were treated with TNFα and the NF-kB inhibitor, the increased CADM1 expression was eliminated confirming the hypothesis that CADM1 is differentially regulated by TNFα through NF-kB signaling.
Figure 6. Analysis of CADM1 for a Function Role in EPC Migration in Response to TNFα.
(A) PCR Analysis of CADM1 Expression in Response to Inhibitors. (1) CADM1 gene expression was measured in response to TNFα in the presence of a synthetic peptide inhibitor of NF-kB or the scrambled control. In presence of the scrambled control TNFα induced up regulation of CADM1 (P < 0.05) whereas in presence of the inhibitor this response was abolished. (2) In the presence of scrambled siRNA, TNFα was able to increase gene expression of CADM1 (P < 0.05) whereas in the presence of targeting siRNA this phenotype was abolished (n = 16 per group).
(B) EPC Migration with CADM1 Knockdown. Prior to TNFα administration, EPCs were transfected with a CADM1 siRNA or a scrambled control. The TNFα induced migratory phenotype was still observed in the presence of the scrambled inhibitor at 2 hour and 14 hours (P < 0.05) but the phenotype was abolished at 2 hours and 14 hours when the targeted siRNA was transfected (n = 16 per group).
EPC Migration with CADM1 Knockdown
To test the hypothesis that TNFα induced up-regulation of CADM1 increases the migration of EPCs towards tube-like structures in vitro, the migration experiment was repeated using EPCs transfected with CADM1 siRNA or a scrambled control (Figure 6, B). In EPCs transfected with the scrambled control it was found after both 2 and 14 hours that TNFα still preferentially increased EPC migration towards tube-like structures in vitro (P < 0.05). In EPCs transfected with targeting siRNA, migration towards tube-like structures was still observed at 2 hours and 14 hours, but a preferential increase from TNFα treatment was inhibited, supporting the hypothesis that increased migration in response to TNFα treatment is mediated through CADM1.
Discussion
In this study, the molecular mechanism driving increased EPC migration in response to TNFα was investigated. Signaling was grouped into three categories: 1) receptors, 2) effectors, and 3) intracellular processing. It was found that TNFα signaling in EPCs was occurring through NF-kB stimulation from TNFR2. To study effectors, a proteomic study identified CADM1 as the highest priority candidate and its role in the migratory process was directly tested and confirmed. To study intracellular processing downstream of TNFR2, NF-kB blockade demonstrated an attenuated increase in migration in response to TNFα treatment. Finally, TNFα regulation of CADM1 through an NF-kB mediated mechanism was tested and confirmed by inhibiting NF-kB and measuring CADM1 gene expression.
Migratory/Incorporation phenotype
It has been shown that very few cells are required to induce angiogenesis in animal models suggesting that a small sub-fraction of injected cells are responsible for the majority of observed therapeutic effects. Kaczorowski et. al. demonstrated an injection of 500 EPCs induced angiogenesis in a model of hind limb ischemia, far too few cells to track with current imaging modalities.[36] In vivo tracking studies have shown that the cellular signal is cleared within hours of injection. In spite of this, regenerative effects are still observed.[54] Detection of little or no signal supports the finding that it takes very few cells to observe significant regenerative effects. In absence of a signal it is difficult to ascertain where these cells are migrating and when they are engrafting. Therefore, studies that utilize in vivo imaging of labeled EPCs do not directly observe EPC migration. To directly study EPC migration an in vitro system was utilized that allowed for direct observation of migrating cells.
Several in vitro migration assays exist. Two examples are 1) the scratch assay, where a cellular monolayer is disrupted and an index of repair is measured and 2) the Boyden chamber assay, where cells are stimulated to migrate across a barrier with small pores.[55] The scratch assay is optimally suited for proliferating cells and the Boyden chamber assay is optimally suited for cells that migrate in response to a chemokine gradient. Because the EPCs in the present study were not proliferating and were pre-treated (as opposed to following a chemokine gradient), it was necessary to use a different assay. Asahara et. al. demonstrated in a model of hind limb ischemia that injected EPCs preferentially migrated to the ischemic limb.[7, 56] The assay used in the present study mimicked in vivo migration quantifying the fraction of cells that had preferentially settled on a vessel-like structure. Similar to the in vivo work by Asahara et. al., our results suggest that inflammatory cytokines secreted by the hind limb activate circulating EPCs increasing vessel incorporation (Figure 2).
TNFα signaling
TNFα signaling occurs through two receptors. Signaling through TNFR1 (p55) induces apoptosis, whereas signaling through TNFR2 (p75) induces NF-kB mediated gene transcription.[45, 57-59] Downstream signaling converges on several transcription factors, and the effect TNFα has on cellular activity has been shown to be a function of TNFα concentration, duration of exposure and cell type.[60-65] Kelly et al. studied the role of the TNF receptors in mesenchymal stem cell (MSC) mediated cardiac regeneration. Sequential knock-out of the TNF receptors showed that p55 signaling was detrimental and p75 was beneficial when using MSCs as a cellular-based therapy to improve cardiac function following myocardial infarction. Even more interesting was the finding that when the p75 receptor was mutated, cardiac recovery was reduced to levels observed with a vehicle injection.[22] These results demonstrate that TNFα signaling through the p75 receptor is necessary for MSC based regenerative therapy. Results of the present study suggest that TNFα activation through p75 signaling induces increased EPC migration in vitro.
The results of the current study demonstrate that a low, acute dose of TNFα to EPCs serves to increase the migratory activity of EPCs through p75 activation of NF-kB. Clinically, these results are important in patients with diseases of increased or decreased angiogenesis. Increased angiogenesis is a feature of several types of cancer as increased levels of EPCs have been found in tumors in vivo.[66, 67] Disruption of the EPC migratory process would be beneficial to cancer patients as EPCs have been shown to enhance tumors proliferation. Decreased angiogenesis is a feature of ischemic diseases including angina pectoris and heart failure.[68] To translate these findings clinically, in diseases of increased angiogenesis it would be beneficial to maximize p55 signaling and in diseases of decreased angiogenesis it would be beneficial to maximize p75 signaling. This could be done by tailoring anti-inflammatory treatments for the individual patient or through the development of drugs that selectively block TNFα signaling at either the p55 or p75 receptor.
Kishore et al. demonstrated the importance of TNFα signaling through the p75 vs the p55 receptor in recovery from myocardial infarction (MI).[45] To test the hypothesis that p75 signaling would be protective whereas p55 signaling would be harmful in MI recovery, the p55 and p75 receptors were knocked out (KO) in separate strains of mice. Left anterior descending arteries were occluded to induce an MI and metrics of recovery were measured. In p55/KO mice, myocardial recovery was improved and in p75/KO mice, myocardial recovery was impaired. In that study the distribution of the p55/p75 receptors was measured in tissues as a function of age and it was found that cardiac p75 receptor density decreased with age, suggesting that the ability to recover from an MI reduces with age due to reduced p75 signaling.
Kishore et al. also measured pro-angiogenic gene expression in EPCs isolated from elderly and young humans. It was found that EPCs isolated from younger humans expressed more pro-angiogenic genes. A follow up study by Sasi et. al. demonstrated that EPCs isolated from elderly vs younger humans had less p75 expression. This finding was offered as explanation for the expression of lower angiogeneic gene expression in cells isolated from elderly patients.[69] The results of these studies and the current study suggest that increased mortality from MI observed in the elderly [70] is due to attenuated EPC repair activity due to decreased angiogenic gene expression in EPCs. In Figure 1 it was proposed that cardiac damage is repaired through a negative-feedback system. These findings would suggest that aging and/or disease impairs the function of this regenerative system through differential expression of the p55/p75 receptors.
CADM1: An additional role for a versatile protein?
Cell Adhesion Molecule 1 is a protein that has been found to be strongly expressed in neurons,[71-75] spermatogenic cells,[52, 76-78] immune cells,[79-87] and endothelial cells.[88, 89] Functionally, CADM1 has been shown to participate in homophilic binding, and paradoxically, tumor suppression. It would be reasonable to hypothesize that CADM1-mediated adhesion of EPCs would increase tumor growth. However, members of the CADM family have an immune component, effectively suppressing tumor growth once in the bound form.[90]
There exists an extensive body of literature describing CADM1 as a tumor suppressor protein, but only two studies that describe “CADM1” and its role in the vascular system.[88, 89] Hasstedt et al. was the follow-up on a human linkage study that had the goal of identifying single nucleotide polymorphisms (SNPs) in genes of the Kindred Vermont II family.[89] The Kindred Vermont II is a family of a couple born in Vermont in the 1830s that have increased susceptibility to venous thrombosis (VT).[91] Prior to the study by Hasstedt et al. in 2009, a linkage analysis narrowed the VT causing region to an area of 109 genes.[91, 92] After analyzing the DNA sequences of the 109 genes identified in the disease-causing region, it was found that 8 CADM1 SNPs correlated with a patients’ risk for VT. These SNPs were shown to mutate the amino acids within the CADM1 protein, likely altering function. Either an increase or decrease in CADM1 binding kinetics could lead to improper recruitment of EPCs ultimately causing the development of vascular malformations and generation of clots.[93]
Kaczorowski et al. compared angiogenic-competent and angiogenic-incompetent EPCs derived from two genetically distinct rat strains in an in vivo model of hind limb stimulation.[36] The angiogenic-incompetent EPCs were isolated from a salt-sensitive (SS/MCWi) rat and the angiogenic-competent EPCs were isolated from an SS-13BN/MCWi rat.[94, 95] In this study, it was found that 5×102 EPCs from the angiogenic-competent strain had the same therapeutic efficacy as 5×105 from the angiogenic-incompetent EPCs, indicating a 1000-fold difference in therapeutic potency between the two strains. A proteomic analysis of surface proteins was conducted, investigating differential expression between the two strains. The conclusion was that the phenotype of impaired angiogenesis was due to increased immune reactivity towards EPCs derived from the SS/MCWi strain. In the proteomic analysis by Kaczorowski et. al., CADM1 was found to be increased 10 fold in the SS/MCWi (impaired angiogenesis) compared to the SS-13BN/MCWi strain. These results indicate an important point, that successful adhesion and migration of EPCs are not the sole processes responsible for induction of angiogenesis. Successful migration is necessary for induction of angiogenesis, but additional steps with distinct mechanisms are also required for angiogenesis. In the context of human studies, it is important to note that EPCs isolated from a genetically heterogeneous population are likely to have large differences in therapeutic potency, illustrating the need for further research to develop clinical assays that can measure EPC potency prior to therapy.
Conclusions
In summary, EPC therapies are a promising clinical treatment. Prior to becoming a mainstream therapy, further research identifying the regenerative mechanisms is necessary to obtain consistent clinical results. To achieve this, studies need to be conducted to better understand the mechanisms of EPC-induced migration, binding, and regeneration. Results from this study indicate that TNFα signaling through TNFR2 that induces NF-kB and CADM1 transcription likely plays a role in the process of EPC migration to a site of organ damage in vivo. Our results suggest that the expression of molecular biomarkers (TNFR2, NF-kB, and CADM1) in EPCs could serve to effectively screen EPCs pre-therapy and potentially increase the success rates in human EPC trials.
Significance Statement.
Endothelial progenitor cells (EPCs) have been shown to regenerate blood vessels in animal studies. Human trials have not yet demonstrated therapeutic efficacy as the molecular regenerative mechanisms are poorly understood. In this study, it was discovered that EPC migration towards vessels is increased in the presence of the inflammatory signal, TNFα. A combined genomic and proteomic analysis was used to propose and validate a mechanism of TNFα-mediated migration. These results suggest that the protein CADM1 mediates TNFα-mediated EPC migration towards vessels. In humans, CADM1 SNPs have been associated with vascular disease/dysfunction, confirming the importance of this mechanism clinically.
Acknowledgements
The authors would like to thank SZP for her help with manuscript preparation.
Grants
This work was supported by NIH grant HL082798 and a gift from the Kern Family Foundation. AP was supported in part by an MSTP Institutional Training Fellowship from NIGMS (T32 GM80202). BH was supported by a post-doctoral training fellowship from NHLBI (T32 HL94273). CCK was supported by K99/00-AG-039511.
Footnotes
Author Contributions:
AP – Conception/design, Collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
BH – Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
CC – Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
CR – Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
TS – Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
EE – Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript
AG – Conception/design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
Disclosures
None of the authors have any disclosures to report
References
- 1.Asahara T, Murohara T, Sullivan A, et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 2.Zuk PA, Zhu M, Mizuno H, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 3.Kawamoto A, Gwon HC, Iwaguro H, et al. Therapeutic potential of ex vivo expanded endothelial progenitor cells for myocardial ischemia. Circulation. 2001;103:634–637. doi: 10.1161/01.cir.103.5.634. [DOI] [PubMed] [Google Scholar]
- 4.Wollert KC, Meyer GP, Lotz J, et al. Intracoronary autologous bone-marrow cell transfer after myocardial infarction: the BOOST randomised controlled clinical trial. Lancet. 2004;364:141–148. doi: 10.1016/S0140-6736(04)16626-9. [DOI] [PubMed] [Google Scholar]
- 5.Kang HJ, Kim HS, Zhang SY, et al. Effects of intracoronary infusion of peripheral blood stem-cells mobilised with granulocyte-colony stimulating factor on left ventricular systolic function and restenosis after coronary stenting in myocardial infarction: the MAGIC cell randomised clinical trial. Lancet. 2004;363:751–756. doi: 10.1016/S0140-6736(04)15689-4. [DOI] [PubMed] [Google Scholar]
- 6.Foley L, Whitaker M. Concise Review: Cell Therapies: The Route to Widespread Adoption. Stem Cells Translational Medicine. 2012;1:438–447. doi: 10.5966/sctm.2011-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asahara T, Masuda H, Takahashi T, et al. Bone marrow origin of endothelial progenitor cells responsible for postnatal vasculogenesis in physiological and pathological neovascularization. Circ Res. 1999;85:221–228. doi: 10.1161/01.res.85.3.221. [DOI] [PubMed] [Google Scholar]
- 8.Volaklis KA, Tokmakidis SP, Halle M. Acute and chronic effects of exercise on circulating endothelial progenitor cells in healthy and diseased patients. Clin Res Cardiol. 2013;102:249–257. doi: 10.1007/s00392-012-0517-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Yan H. Simvastatin increases circulating endothelial progenitor cells and reduces the formation and progression of diabetic retinopathy in rats. Exp Eye Res. 2012;105:1–8. doi: 10.1016/j.exer.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Ozkok A, Aktas E, Yilmaz A, et al. Decrease in endothelial progenitor cells associated with inflammation, but not with endothelial dysfunction in chronic hemodialysis patients. Clin Nephrol. 2013;79:21–30. doi: 10.5414/CN107318. [DOI] [PubMed] [Google Scholar]
- 11.Grundmann M, Woywodt A, Kirsch T, et al. Circulating endothelial cells: a marker of vascular damage in patients with preeclampsia. Am J Obstet Gynecol. 2008;198:317, e311–315. doi: 10.1016/j.ajog.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, Zhu J, Zou L, et al. Changes in number and biological function of endothelial progenitor cells in hypertension disorder complicating pregnancy. J Huazhong Univ Sci Technolog Med Sci. 2008;28:670–673. doi: 10.1007/s11596-008-0612-9. [DOI] [PubMed] [Google Scholar]
- 13.Zhou WJ, Zhu DL, Yang GY, et al. Circulating endothelial progenitor cells in Chinese patients with acute stroke. Hypertens Res. 2009;32:306–310. doi: 10.1038/hr.2009.16. [DOI] [PubMed] [Google Scholar]
- 14.Wang HY, Gao PJ, Ji KD, et al. Circulating endothelial progenitor cells, C-reactive protein and severity of coronary stenosis in Chinese patients with coronary artery disease. Hypertens Res. 2007;30:133–141. doi: 10.1291/hypres.30.133. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Ayala E, Yao Q, Holmen C, et al. Imbalance between detached circulating endothelial cells and endothelial progenitor cells in chronic kidney disease. Blood Purif. 2006;24:196–202. doi: 10.1159/000090519. [DOI] [PubMed] [Google Scholar]
- 16.Nonaka-Sarukawa M, Yamamoto K, Aoki H, et al. Circulating endothelial progenitor cells in congestive heart failure. Int J Cardiol. 2007;119:344–348. doi: 10.1016/j.ijcard.2006.07.191. [DOI] [PubMed] [Google Scholar]
- 17.Bakogiannis C, Tousoulis D, Androulakis E, et al. Circulating endothelial progenitor cells as biomarkers for prediction of cardiovascular outcomes. Curr Med Chem. 2012;19:2597–2604. doi: 10.2174/092986712800492995. [DOI] [PubMed] [Google Scholar]
- 18.Dome P, Halmai Z, Dobos J, et al. Investigation of circulating endothelial progenitor cells and angiogenic and inflammatory cytokines during recovery from an episode of major depression. J Affect Disord. 2012;136:1159–1163. doi: 10.1016/j.jad.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Prisco AR, Prisco MR, Carlson BE. TNF-α increases endothelial progenitor cell adhesion to the endothelium by increasing bond expression and affinity. 2015:H1368–H1381. doi: 10.1152/ajpheart.00496.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrich D, Seebach C, Wilhelm K, et al. High dosage of simvastatin reduces TNF-alpha-induced apoptosis of endothelial progenitor cells but fails to prevent apoptosis induced by IL-1beta in vitro. J Surg Res. 2007;142:13–19. doi: 10.1016/j.jss.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Ablin JN, Boguslavski V, Aloush V, et al. Effect of anti-TNFalpha treatment on circulating endothelial progenitor cells (EPCs) in rheumatoid arthritis. Life Sci. 2006;79:2364–2369. doi: 10.1016/j.lfs.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 22.Kelly ML, Wang M, Crisostomo PR, et al. TNF receptor 2, not TNF receptor 1, enhances mesenchymal stem cell-mediated cardiac protection following acute ischemia. Shock. 2010;33:602–607. doi: 10.1097/SHK.0b013e3181cc0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carswell EA, Old LJ, Kassel RL, et al. An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci U S A. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerszten RE, Garcia-Zepeda EA, Lim Y-C, et al. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature. 1999;398:718–723. doi: 10.1038/19546. [DOI] [PubMed] [Google Scholar]
- 25.Luscinskas PD, Francis W, Gimbrone M, Jr, Michael A. Endothelial-dependent mechanisms in chronic inflammatory leukocyte recruitment. Annu Rev Med. 1996;47:413–421. doi: 10.1146/annurev.med.47.1.413. [DOI] [PubMed] [Google Scholar]
- 26.Luscinskas FW, Kansas GS, Ding H, et al. Monocyte rolling, arrest and spreading on IL-4-activated vascular endothelium under flow is mediated via sequential action of L-selectin, beta 1-integrins, and beta 2-integrins. J Cell Biol. 1994;125:1417–1427. doi: 10.1083/jcb.125.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ley K, Baker J, Cybulsky M, et al. Intravenous interleukin-8 inhibits granulocyte emigration from rabbit mesenteric venules without altering L-selectin expression or leukocyte rolling. The Journal of Immunology. 1993;151:6347–6357. [PubMed] [Google Scholar]
- 28.Wyble CW, Hynes KL, Kuchibhotla J, et al. TNF-α and IL-1 upregulate membrane-bound and soluble E-selectin through a common pathway. Journal of Surgical Research. 1997;73:107–112. doi: 10.1006/jsre.1997.5207. [DOI] [PubMed] [Google Scholar]
- 29.Walsh LJ, Trinchieri G, Waldorf HA, et al. Human dermal mast cells contain and release tumor necrosis factor alpha, which induces endothelial leukocyte adhesion molecule 1. Proc Natl Acad Sci U S A. 1991;88:4220–4224. doi: 10.1073/pnas.88.10.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugarman BJ, Aggarwal BB, Hass PE, et al. Recombinant human tumor necrosis factor-alpha: effects on proliferation of normal and transformed cells in vitro. Science. 1985;230:943–945. doi: 10.1126/science.3933111. [DOI] [PubMed] [Google Scholar]
- 31.McHale JF, Harari OA, Marshall D, et al. TNF-alpha and IL-1 sequentially induce endothelial ICAM-1 and VCAM-1 expression in MRL/lpr lupus-prone mice. J Immunol. 1999;163:3993–4000. [PubMed] [Google Scholar]
- 32.McClain CJ, Cohen DA. Increased tumor necrosis factor production by monocytes in alcoholic hepatitis. Hepatology. 1989;9:349–351. doi: 10.1002/hep.1840090302. [DOI] [PubMed] [Google Scholar]
- 33.Shohami E, Novikov M, Bass R, et al. Closed head injury triggers early production of TNFα and IL-6 by brain tissue. Journal of Cerebral Blood Flow & Metabolism. 1994;14:615–619. doi: 10.1038/jcbfm.1994.76. [DOI] [PubMed] [Google Scholar]
- 34.Gleeson M. Immune function in sport and exercise. Journal of Applied Physiology. 2007;103:693–699. doi: 10.1152/japplphysiol.00008.2007. [DOI] [PubMed] [Google Scholar]
- 35.Redlich CA, Gao X, Rockwell S, et al. IL-11 enhances survival and decreases TNF production after radiation-induced thoracic injury. The Journal of Immunology. 1996;157:1705–1710. [PubMed] [Google Scholar]
- 36.Kaczorowski CC, Stodola TJ, Hoffmann BR, et al. Targeting the endothelial progenitor cell surface proteome to identify novel mechanisms that mediate angiogenic efficacy in a rodent model of vascular disease. Physiol Genomics. 2013;45:999–1011. doi: 10.1152/physiolgenomics.00097.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linderman JR, Kloehn MR, Greene AS. Development of an implantable muscle stimulator: measurement of stimulated angiogenesis and poststimulus vessel regression. Microcirculation. 2000;7:119–128. [PubMed] [Google Scholar]
- 38.Hoffmann BR, Wagner JR, Prisco AR, et al. Vascular endothelial growth factor-A signaling in bone marrow-derived endothelial progenitor cells exposed to hypoxic stress. Physiol Genomics. 2013;45:1021–1034. doi: 10.1152/physiolgenomics.00070.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prisco AR, Bukowy JD, Hoffmann BR, et al. Automated Quantification Reveals Hyperglycemia Inhibits Endothelial Angiogenic Function. PLoS One. 2014;9:e94599. doi: 10.1371/journal.pone.0094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wollscheid B, Bausch-Fluck D, Henderson C, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prisco A, Hoffmann BR, Kaczorowski CC, et al. Targeted Proteomics: Endothelial Cell Membrane Response to TNF-{alpha}. The FASEB Journal. 2013;27:737, 736. [Google Scholar]
- 42.Amaral SL, Linderman JR, Morse MM, et al. Angiogenesis induced by electrical stimulation is mediated by angiotensin II and VEGF. Microcirculation. 2001;8:57–67. [PubMed] [Google Scholar]
- 43.Linderman JR, Kloehn MR, Greene AS. Development of an implantable muscle stimulator: measurement of stimulated angiogenesis and poststimulus vessel regression. Microcirculation. 2000;7:119–128. [PubMed] [Google Scholar]
- 44.Karcher JR, Greene AS. Bone marrow mononuclear cell angiogenic competency is suppressed by a high salt diet. Am J Physiol Cell Physiol. 2013 doi: 10.1152/ajpcell.00164.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kishore R, Tkebuchava T, Sasi SP, et al. Tumor necrosis factor-alpha signaling via TNFR1/p55 is deleterious whereas TNFR2/p75 signaling is protective in adult infarct myocardium. Adv Exp Med Biol. 2011;691:433–448. doi: 10.1007/978-1-4419-6612-4_45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neumann B, Machleidt T, Lifka A, et al. Crucial role of 55-kilodalton TNF receptor in TNF-induced adhesion molecule expression and leukocyte infiltration. J Immunol. 1996;156:1587–1593. [PubMed] [Google Scholar]
- 47.Munroe ME, Bishop GA. Role of tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) in distinct and overlapping CD40 and TNF receptor 2/CD120b-mediated B lymphocyte activation. Journal of Biological Chemistry. 2004;279:53222–53231. doi: 10.1074/jbc.M410539200. [DOI] [PubMed] [Google Scholar]
- 48.Rauert H, Wicovsky A, Müller N, et al. Membrane tumor necrosis factor (TNF) induces p100 processing via TNF receptor-2 (TNFR2). Journal of Biological Chemistry. 2010;285:7394–7404. doi: 10.1074/jbc.M109.037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halligan BD, Greene AS. Visualize: a free and open source multifunction tool for proteomics data analysis. Proteomics. 2011;11:1058–1063. doi: 10.1002/pmic.201000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werner T. Bioinformatics applications for pathway analysis of microarray data. Current opinion in biotechnology. 2008;19:50–54. doi: 10.1016/j.copbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Takayanagi Y, Fujita E, Yu Z, et al. Impairment of social and emotional behaviors in Cadm1-knockout mice. Biochem Biophys Res Commun. 2010;396:703–708. doi: 10.1016/j.bbrc.2010.04.165. [DOI] [PubMed] [Google Scholar]
- 52.Wakayama T, Iseki S. Role of the spermatogenic–Sertoli cell interaction through cell adhesion molecule-1 (CADM1) in spermatogenesis. Anatomical science international. 2009;84:112–121. doi: 10.1007/s12565-009-0034-1. [DOI] [PubMed] [Google Scholar]
- 53.Ito A, Ichiyanagi N, Ikeda Y, et al. Adhesion molecule CADM1 contributes to gap junctional communication among pancreatic islet α-cells and prevents their excessive secretion of glucagon. Islets. 2012;4:49–55. doi: 10.4161/isl.18675. [DOI] [PubMed] [Google Scholar]
- 54.Fadini GP, Losordo D, Dimmeler S. Critical reevaluation of endothelial progenitor cell phenotypes for therapeutic and diagnostic use. Circulation research. 2012;110:624–637. doi: 10.1161/CIRCRESAHA.111.243386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kramer N, Walzl A, Unger C, et al. In vitro cell migration and invasion assays. Mutat Res. 2013;752:10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Krasinski K, Spyridopoulos I, Asahara T, et al. Estradiol accelerates functional endothelial recovery after arterial injury. Circulation. 1997;95:1768–1772. doi: 10.1161/01.cir.95.7.1768. [DOI] [PubMed] [Google Scholar]
- 57.Sasi S, Yan X, Enderling H, et al. Breaking the ‘harmony’of TNF-α signaling for cancer treatment. Oncogene. 2011;31:4117–4127. doi: 10.1038/onc.2011.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goukassian DA, Qin G, Dolan C, et al. Tumor necrosis factor-α receptor p75 is required in ischemia-induced neovascularization. Circulation. 2007;115:752–762. doi: 10.1161/CIRCULATIONAHA.106.647255. [DOI] [PubMed] [Google Scholar]
- 59.Wang C, Deng L, Hong M, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y, Zhou BP. TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br J Cancer. 2010;102:639–644. doi: 10.1038/sj.bjc.6605530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeller CN, Wang Y, Markel TA, et al. Role of tumor necrosis factor receptor 1 in sex differences of stem cell mediated cardioprotection. Ann Thorac Surg. 2009;87:812–819. doi: 10.1016/j.athoracsur.2008.12.033. [DOI] [PubMed] [Google Scholar]
- 62.Kim YS, Park HJ, Hong MH, et al. TNF-alpha enhances engraftment of mesenchymal stem cells into infarcted myocardium. Front Biosci (Landmark Ed) 2009;14:2845–2856. doi: 10.2741/3417. [DOI] [PubMed] [Google Scholar]
- 63.Sethi G, Sung B, Aggarwal BB. TNF: a master switch for inflammation to cancer. Front Biosci. 2008;13:5094–5107. doi: 10.2741/3066. [DOI] [PubMed] [Google Scholar]
- 64.Sun M, Chen M, Dawood F, et al. Tumor necrosis factor-alpha mediates cardiac remodeling and ventricular dysfunction after pressure overload state. Circulation. 2007;115:1398–1407. doi: 10.1161/CIRCULATIONAHA.106.643585. [DOI] [PubMed] [Google Scholar]
- 65.Sarzi-Puttini P, Atzeni F, Shoenfeld Y, et al. TNF-α, rheumatoid arthritis, and heart failure: a rheumatological dilemma. Autoimmunity reviews. 2005;4:153–161. doi: 10.1016/j.autrev.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 67.Shaked Y, Ciarrocchi A, Franco M, et al. Therapy-induced acute recruitment of circulating endothelial progenitor cells to tumors. Science. 2006;313:1785–1787. doi: 10.1126/science.1127592. [DOI] [PubMed] [Google Scholar]
- 68.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 69.Sasi SP, Song J, Park D, et al. TNF-TNFR2/p75 signaling inhibits early and increases delayed nontargeted effects in bone marrow-derived endothelial progenitor cells. Journal of Biological Chemistry. 2014;289:14178–14193. doi: 10.1074/jbc.M114.567743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lerner DJ, Kannel WB. Patterns of coronary heart disease morbidity and mortality in the sexes: a 26-year follow-up of the Framingham population. American heart journal. 1986;111:383–390. doi: 10.1016/0002-8703(86)90155-9. [DOI] [PubMed] [Google Scholar]
- 71.Robbins EM, Krupp AJ, Perez de Arce K, et al. SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron. 2010;68:894–906. doi: 10.1016/j.neuron.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fogel AI, Stagi M, Perez de Arce K, et al. Lateral assembly of the immunoglobulin protein SynCAM 1 controls its adhesive function and instructs synapse formation. EMBO J. 2011;30:4728–4738. doi: 10.1038/emboj.2011.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas LA, Akins MR, Biederer T. Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. J Comp Neurol. 2008;510:47–67. doi: 10.1002/cne.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sara Y, Biederer T, Atasoy D, et al. Selective capability of SynCAM and neuroligin for functional synapse assembly. J Neurosci. 2005;25:260–270. doi: 10.1523/JNEUROSCI.3165-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biederer T, Sara Y, Mozhayeva M, et al. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 76.Koma Y-i, Ito A, Wakayama T, et al. Cloning of a soluble isoform of the SgIGSF adhesion molecule that binds the extracellular domain of the membrane-bound isoform. Oncogene. 2004;23:5687–5692. doi: 10.1038/sj.onc.1207761. [DOI] [PubMed] [Google Scholar]
- 77.Toshimori K. Dynamics of the Mammalian Sperm Head. Springer; 2009. Sperm-Head Formation and Factors Affecting It. pp. 17–25. [Google Scholar]
- 78.Wakayama T, Koami H, Ariga H, et al. Expression and functional characterization of the adhesion molecule spermatogenic immunoglobulin superfamily in the mouse testis. Biology of reproduction. 2003;68:1755–1763. doi: 10.1095/biolreprod.102.012344. [DOI] [PubMed] [Google Scholar]
- 79.Liu D, Feng X, Wu X, et al. Tumor suppressor in lung cancer 1 (TSLC1), a novel tumor suppressor gene, is implicated in the regulation of proliferation, invasion, cell cycle, apoptosis, and tumorigenicity in cutaneous squamous cell carcinoma. Tumour Biol. 2013;34:3773–3783. doi: 10.1007/s13277-013-0961-2. [DOI] [PubMed] [Google Scholar]
- 80.Lei W, Liu HB, Wang SB, et al. Tumor suppressor in lung cancer-1 (TSLC1) mediated by dual-regulated oncolytic adenovirus exerts specific antitumor actions in a mouse model. Acta Pharmacol Sin. 2013;34:531–540. doi: 10.1038/aps.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang QL, Wang BR, Li ZY, et al. Effect of TSLC1 gene on growth and apoptosis in human esophageal carcinoma Eca109 cells. Arch Med Sci. 2012;8:987–992. doi: 10.5114/aoms.2012.31251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu B, Di W, Wang H, et al. Tumor suppressor TSLC1 is implicated in cell proliferation, invasion and apoptosis in laryngeal squamous cell carcinoma by regulating Akt signaling pathway. Tumour Biol. 2012;33:2007–2017. doi: 10.1007/s13277-012-0460-x. [DOI] [PubMed] [Google Scholar]
- 83.You Y, Wang SH, Zhang JF, et al. TSLC1 expression discriminates cutaneous melanomas from dysplastic nevi. Melanoma Res. 2012;22:430–435. doi: 10.1097/CMR.0b013e328358d9a2. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J, Ning J, Geng J, et al. Down-regulation of tumor suppressor in lung cancer 1 (TSLC1) expression correlates with poor prognosis in patients with colon cancer. J Mol Histol. 2012;43:715–721. doi: 10.1007/s10735-012-9438-7. [DOI] [PubMed] [Google Scholar]
- 85.Nakahata S, Morishita K. CADM1/TSLC1 is a novel cell surface marker for adult T-cell leukemia/lymphoma. J Clin Exp Hematop. 2012;52:17–22. doi: 10.3960/jslrt.52.17. [DOI] [PubMed] [Google Scholar]
- 86.Nakahata S, Saito Y, Marutsuka K, et al. Clinical significance of CADM1/TSLC1/IgSF4 expression in adult T-cell leukemia/lymphoma. Leukemia. 2012;26:1238–1246. doi: 10.1038/leu.2011.379. [DOI] [PubMed] [Google Scholar]
- 87.He G, Lei W, Wang S, et al. Overexpression of tumor suppressor TSLC1 by a survivin-regulated oncolytic adenovirus significantly inhibits hepatocellular carcinoma growth. J Cancer Res Clin Oncol. 2012;138:657–670. doi: 10.1007/s00432-011-1138-2. [DOI] [PubMed] [Google Scholar]
- 88.Tatsumi K, Taatjes DJ, Wadsworth MP, et al. Cell adhesion molecule 1 (CADM1) is ubiquitously present in the endothelium and smooth muscle cells of the human macro- and micro-vasculature. Histochem Cell Biol. 2012;138:815–820. doi: 10.1007/s00418-012-1024-2. [DOI] [PubMed] [Google Scholar]
- 89.Hasstedt SJ, Bezemer ID, Callas PW, et al. Cell adhesion molecule 1: a novel risk factor for venous thrombosis. Blood. 2009;114:3084–3091. doi: 10.1182/blood-2009-05-219485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moh MC, Shen S. The roles of cell adhesion molecules in tumor suppression and cell migration: a new paradox. Cell adhesion & migration. 2009;3:334. doi: 10.4161/cam.3.4.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hasstedt S, Scott B, Callas P, et al. Genome scan of venous thrombosis in a pedigree with protein C deficiency. Journal of Thrombosis and Haemostasis. 2004;2:868–873. doi: 10.1111/j.1538-7836.2004.00663.x. [DOI] [PubMed] [Google Scholar]
- 92.Scott BT, Bovill EG, Callas PW, et al. Genetic screening of candidate genes for a prothrombotic interaction with type I protein C deficiency in a large kindred. THROMBOSIS AND HAEMOSTASIS-STUTTGART- 2001;85:82–87. [PubMed] [Google Scholar]
- 93.Chee YL, Culligan DJ, Watson HG. Inferior vena cava malformation as a risk factor for deep venous thrombosis in the young. British journal of haematology. 2001;114:878–880. doi: 10.1046/j.1365-2141.2001.03025.x. [DOI] [PubMed] [Google Scholar]
- 94.Cowley AW, Jr., Liang M, Roman RJ, et al. Consomic rat model systems for physiological genomics. Acta Physiol Scand. 2004;181:585–592. doi: 10.1111/j.1365-201X.2004.01334.x. [DOI] [PubMed] [Google Scholar]
- 95.Cowley AW, Jr., Roman RJ, Kaldunski ML, et al. Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension. 2001;37:456–461. doi: 10.1161/01.hyp.37.2.456. [DOI] [PubMed] [Google Scholar]