Abstract
The spatial and temporal evolution of concentration boundaries in sedimentation velocity analytical ultracentrifugation reports on the size distribution of particles with high hydrodynamic resolution. For large particles such as large protein complexes, fibrils, viral particles, or nanoparticles, sedimentation conditions usually allow migration from diffusion to be neglected relative to sedimentation. In this case, the shape of the sedimentation boundaries of polydisperse mixtures relates directly to the underlying size-distributions. Integral and derivative methods for calculating sedimentation coefficient distributions g*(s) of large particles from experimental boundary profiles have been developed previously, and are recapitulated here in a common theoretical framework. This leads to a previously unrecognized relationship between g*(s) and the time-derivative of concentration profiles. Of closed analytical form, it is analogous to the well-known Bridgman relationship for the radial derivative. It provides a quantitative description of the effect of substituting the time-derivative by scan differences with finite time intervals, which appears as a skewed box average of the true distribution. This helps to theoretically clarify the differences between results from time-derivative method and the approach of directly fitting the integral definition of g*(s) to the entirety of experimental boundary data.
Graphical Abstract
1 Introduction
The determination of the size and polydispersity of large macromolecules and nanoparticles is a traditional application of analytical ultracentrifugation (AUC). 1,2 It is an absolute method based on first principles and combines the virtues of an exquisite hydrodynamic resolution, afforded by the strongly size-dependent migration in the centrifugal field, with a size range spanning three orders of magnitude in Stokes radius. In the present work we recapitulate the theoretical relationships underpinning the different differential and integral approaches for determining sedimentation coefficient distributions of particles with negligible diffusion from experimental sedimentation data. These are of interest for the characterization by AUC of large particles including protein complexes, 3,4 fibrils, 5,6 viral particles, 7,8 entire organisms, 9, nanoparticles such as carbon nanotubes, 10–12 nanocrystals, 13 gold nanoparticles and their conjugates, 14 quantum dots, 15 and other colloids, 16 both in research and in regulatory environment. 17,18 Furthermore, sedimentation coefficient distributions have been the basis for determining molar mass distributions of such particles, 19,20 and apparent sedimentation coefficient distributions of diffusing species have been used for estimating molar mass21–23 and assess protein conformational changes. 24
Over the history of AUC several strategies to determine sedimentation coefficient distributions of non-diffusing particles have been described for different detector configurations. In 1942, Bridgman has presented a transformation of radial signal profiles a(r) into a sedimentation coefficient distribution g*(s) by virtue of the radial derivative da/dr, 25 which was naturally suited for Schlieren measurement of concentration gradients. Later, Scholtan & Lange developed an approach for determining the sedimentation coefficient distribution of latex particles from the time-course of signal obtained at a fixed-radius detector. 26 This could be combined with an increase of the rotor speed with time, providing an efficient characterization of particles over a large size range, which was adopted for industrial applications in several laboratories. 26–29 With computer-based analysis becoming routinely available in the early 1990s, Stafford has introduced a time-derivative method “dcdt” to calculate g*(s) from consecutively acquired radial concentration profiles across the solution column.30–32 Coinciding with the introduction of a new generation of commercial AUC instrumentation equipped with Rayleigh interference optics capable to produce digital fringe profiles at a high rate, it became widely popular for the study of apparent sedimentation coefficient distributions of medium-sized proteins and their interactions. 23,24 In particular, the dcdt-method has the virtue of being independent of time-invariant, radial-dependent baseline noise that is dominant in interference data at low sample concentrations. Most recently, the introduction of algebraic noise elimination techniques 33,34 and application of modern mathematical strategies for solving integral equations 35,36 made it possible to carry out least-squares fit to the entire temporal evolution of concentration profiles a(r, t) with explicit distribution models, ls-g*(s), 37 as a special case for large particles of a more general diffusion-deconvoluted particle sedimentation coefficient distribution c(s). 38
Even though these approaches share the same definition of the sedimentation coefficient distribution, different results may be obtained due to their differences in the strategies to accommodate experimental noise, and due to different approximations required in their practical application. In the present work we recapitulate the theoretical basis, and establish new relationships between the different approaches that help to understand their advantages and disadvantages.
2 Theory
Throughout we consider particles that are sufficiently large so that contributions of diffusion to migration in the centrifugal field can be neglected. Traditionally this approximation finds application also for diffusing species that form boundaries indistinguishable from distributions of non-diffusing species, in which case the resulting sedimentation coefficient distributions are considered ‘apparent’, and may be subject to secondary analysis to unravel the effects of finite diffusion 23,24. Practical application of the sedimentation coefficient distribution analysis of large particles may require the consideration of size-dependent signal increments, for example, from scattering, dependent on the optical detection used. This question is excluded from the present work, as it can be accounted for separately, as needed, by a renormalization of the distribution, which can be calculated at first entirely in signal units as concentration units.
Sedimentation boundaries of individual non-diffusing species approach step-functions. The comprehensive mathematical framework of generalized functions (mathematical distributions) was only developed in the 1950s 39 — decades after the development of analytical ultracentrifugation and the analysis of sedimentation coefficient distributions. However, Heaviside step-functions and Dirac δ -functions provide very efficient tools for the mathematical analysis of sedimentation and hydrodynamics. 31,37,40–45 Specifically, step-functions naturally lend themselves to the description of boundaries of non-diffusing, non-interacting 31,37 or interacting 45 species in the context of size-distributions, and can be easily numerically implemented. 37
2.1 g*(s) as a Sedimentation Coefficient Distribution of Polydisperse Mixtures of Large, Non-Diffusing Particles
We assume particles are initially distributed uniformly at unit concentration in a solution column with meniscus m. They are then subjected to the centrifugation at angular velocity ω. This causes particles of a single class of sedimenting species to migrate with the sedimentation coefficient s, such that after time t a boundary at a radius rb(t) is formed. Simultaneously, the plateau concentration decreases due to the dilution caused by increasing separation of radially moving particles. In more detail, the concentration distribution of this single class of particles χ1,nd(r, t) is
| (1) |
where rb(t) = mesω2t (Figure 1A). It can be easily shown that this function satisfies the Lamm equation 46 —the master equation for sedimentation in the gravitational field based on fluxes in radial geometry — for the special case of D = 0. The middle identity introduces the Heaviside step-function H(x), which has a discontinuous step from 0 to 1 at the origin, and is constant 0 for negative and constant 1 for positive values. 39
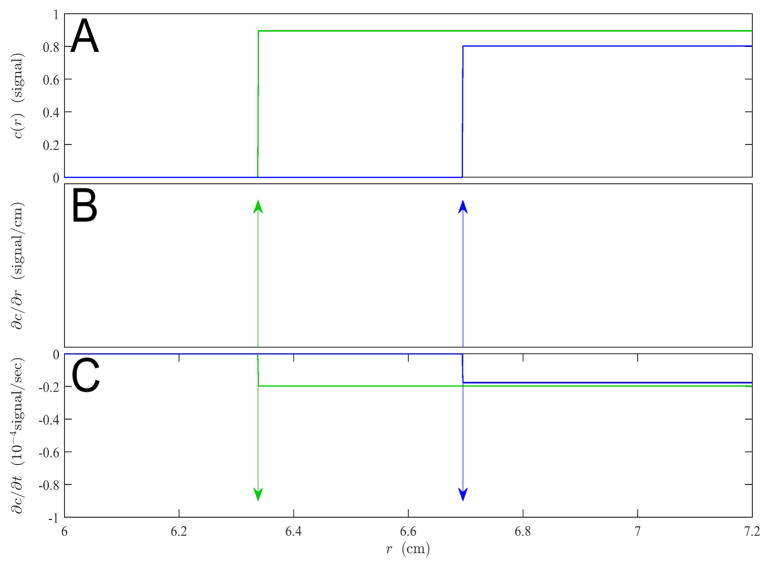
Fig. 1.
Sedimentation profiles (A), and their spatial derivatives (B) and temporal derivatives (C) of a single, non-diffusing species at a unit signal concentration with sedimentation coefficient of 100 S in a solution column with meniscus at 6.0 cm from the center of rotation, spinning at 10,000 rpm after a time of 5,000 sec (green) and 10,000 sec (blue). Both spatial and time derivatives involve δ -functions, which are depicted as arrows.
Based on the sedimentation of a single, non-diffusing species, in the following, it will be useful to highlight the general relationship between a radial position r* and the sedimentation coefficient s* of a non-diffusing species whose boundary arrives at that position at time t:
| (2) |
The asterisk distinguishes these as variables that relate the radial and sedimentation coefficient dimension, rather than coordinates of a boundary.
A polydisperse distribution of species with different s-values then sediments with the temporal evolution of total concentration
| (3) |
with g*(s)ds representing the total loading concentration of species with s-values between s and s+ds. smax is an upper limit of what could possibly be described, usually imposed by the time point and radial range of observation. As is custom, we apply an asterisk to the distribution as a reminder of the neglect of diffusion. The total concentration c(r, t) is what would be measured in an ideal experiment at radius r and time t; for clarity we assume this not to be subject to any time-invariant or radial-invariant baseline offsets that usually are superimposed to experimental data. 47 These are not the subject of the present work; for strategies how to address these noise contributions see, e.g., refs 32–34,47.
2.2 Direct Solution of the Integral Equation for g*(s)
It is possible to directly numerically solve the integral equation (3) in a least-squares fit to the experimental data, termed ls-g *(s). 37 This is accomplished by discretizing the s-values into a grid si from i = 1. . .N and determining the corresponding distribution values g*(si) such that the sum of their boundaries matches as closely as possible all available data points at radii rm and times tn:
| (4) |
If the experimental data are from the observation of an ideal sedimentation process of non-diffusing particles, then the residuals of the fit are solely random noise of data acquisition, which may be verified with bitmap representations or residual histograms. 47,48 Eq. (4) leads to a linear equation system, and with N usually being in the range of 100–500, the computational effort is minor and takes only seconds on office computers. 49 To avoid noise amplification it is essential that the solution of (4) be combined with regularization, usually Tikhonov regularization 37 or Bayesian approaches if other prior knowledge is available. 50 Time-invariant and radial-invariant noise can both be simultaneously included in the calculation. 33,34. One of the major virtues of the integral approach is that all experimental data can be incorporated, and as an explicit model for the sedimentation process, ls-g*(s) can be compared transparently with the experimental data in the original data space to examine the quality of fit. The approach is very general, and without any further complications refinements on χ1,nd are possible to account for solvent compressibility,51 time-varying centrifugal fields, 52, the finite scanning speed of the scanning systems, 3 optical signal magnification gradients, 53 or spatiotemporal modulation of signal increments in fluorescence detection. 54
2.3 The Spatial Derivative and the Bridgman Equation for g*(s)
To the extent that the distribution is additive in the species contributions, the relationships between g*(s) and derivatives of c(r, t) can be established by considering derivatives of χ1,nd(r, t). First we consider the radial derivative at a particular time t1:
| (5) |
The derivative of the Heaviside function is the Dirac δ -function, which can be imagined as a spike with the property
| (6) |
39 and for dχnd,1/dr we have
| (7) |
(Figure 1B). The coordinate transformation based on (2) allows us to express the radial coordinate r in terms of the sedimentation coefficient s*. With the chain rule for δ -functions 39 δ (ϕ (x)) = δ (x)×|dϕ (c)/dx|−1 at ϕ (c) = 0 we can re-write (7) as
| (8) |
and obtain after insertion into (5)
| (9) |
Executing the integral using the property (6) we arrive at the familiar Bridgman relationship 25
| (10) |
with the radial coordinate linked to the s*-value, r(s*), as in (2). It provides a recipe to determine the sedimentation coefficient distribution g*(s) on the basis of the spatial derivative of the total concentration profile, followed by transformation of the radial to the s* coordinate.
2.4 The Relationship Between g*(s) and the Time-Derivative
An alternative avenue to determine g*(s) is the time-derivative. We can proceed analogously for the temporal derivative of the total concentration distribution observed after a time t1 at the radius r, which is additive in the temporal derivative of each species:
| (11) |
The temporal derivative of a single non-diffusing species (1) has two terms: First, the derivative arising from the boundary can be found using the chain rule ∂H (r−rb(t))/∂ t = (∂H (r−rb(t))/∂ rb )×(∂ rb/∂ t ) = −(∂ rb/∂ t )δ (rb(t)−r). But in addition, all positions at higher radii experience a radially uniform concentration change due to the decreasing plateau concentration with time:
| (12) |
(Figure 1C). Inserted in (11) to expand the relationship for the whole distribution, the coordinate transform δ (rb(t1)−r*) = (ω2t1mes′ω2t1 )−1δ (s′ −s*) leads to
| (13) |
The δ -function resolves again the integral of the boundary term, whereas the step-function resolves into a limit for the integration range: Rather than integrating to the maximum considered value smax, non-zero contributions can only be made for species that have not migrated past r*(s*). Together, this leads to
| (14) |
With the abbreviation
| (15) |
the integral equation (14) simplifies to
| (16) |
and differentiation with regard to s* leads to a first-order linear differential equation
| (17) |
As shown in Appendix A, for t1 > 0 it has a solution
| (18) |
. Reinserting g* via (15) we find the compact expression
| (19) |
It is possible to resolve the double differentiation through integration by parts (making use that the time-derivative at the meniscus vanishes for t1 > 0, and species with s = 0 only contribute to the baseline):
| (20) |
For convenience of relating this result to changes of experimentally measured radial concentration profiles, we can back-transform to radial coordinates (using ds′ = (ω2t1r′)−1dr′ and e2s*ω2t1 = r*2/m2 ):
| (21) |
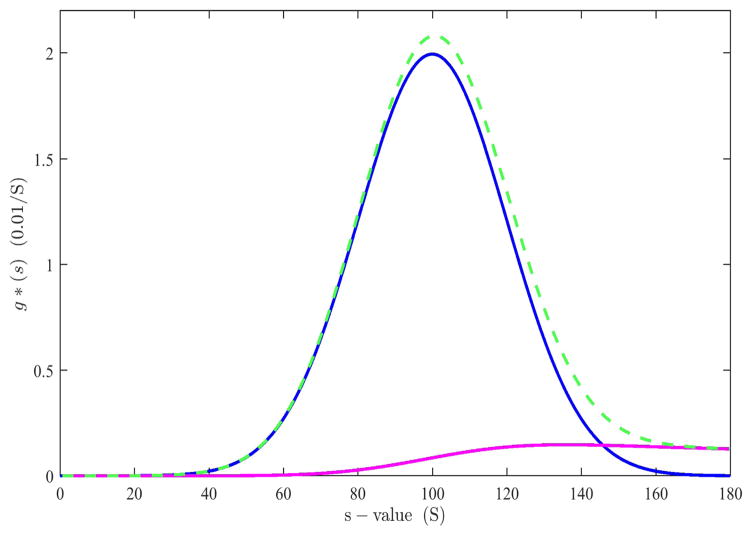
This is the analogue of the Bridgman equation (10) for the time-derivative. That it solves Eq. (14) is shown in Appendix B. The second term provides a correction for the radial dilution. It represents a squared radial-average of the time-derivative centripetal to r*. The relative magnitude of these terms can be discerned from Figure 2, for the example of a Gaussian sedimentation coefficient distribution g*(s) (blue): The first term generates an over-estimate at the high s-values (green dashed), and which is compensated by the radial dilution term (magenta) in (20).
Fig. 2.
Contributions to g*(s) from theoretical time-derivative data of an ensemble of non-diffusing particles with Gaussian sedimentation coefficient distribution centered at 100 S sedimenting at 10,000 rpm, shown at 9,000 sec (blue). Using only the first term of Eq. (21), corresponding to the simple transformation and normalization of the time-derivative into a s*-space (green dashed) leads to an overestimate of populations at high s-value, due to radial dilution of small species contributing to the time-derivative at high radii and high s-values. The second term of Eq. (21) corresponds to the magnitude of this radial dilution contribution (magenta), such that the difference of both terms produces the correct distribution (blue).
In the same framework, we can examine the response to sedimentation profiles from a single species with s=s1, which has the concentration profiles and time-derivative given in (1) and (12), respectively. This was carried out in Appendix C, arriving at the single-species distribution at unit concentration
| (22) |
as expected, showing consistency and correctness of the new expression Eq. (21).
2.5 Approximation of g*(s) from Time-Differences
Using the result (22) for a single species, it can be useful to rewrite the distribution as a linear combination of its normalized single-species response function g1*
| (23) |
. This is a reflection of the fact that g*(s*) in (20) is additive in the concentrations (and time-derivative, respectively) of different species. Eq. (23) seems redundant at first, but this consideration is useful for analyzing the effect of the substitution of finite time-differences for the time-derivative, as is required in practice when the data sets consist of a sequence of scans with finite time interval. Eq. (23) reduces the problem of what the impact of this practical imperfection is on the entire distribution to the question how a single-species response function is affected. We will label distributions arising from finite time-difference substitution with a tilde, and refer to the single-species response as , and the entire distribution as
| (24) |
If we take the time-interval between t1 and t2 (with t1 < t2 and Δt = t2 − t1) we have
| (25) |
. If this time difference is inserted in Eq. (21) in the place of the time-derivative we obtain
| (26) |
As shown in the Appendix D, this simplifies to
| (27) |
, i.e., a segment of a hyperbola 1/s* from s*1 =st1/t to s*2 =st2/t. Thus, substitution of the time-derivative with the analogous difference expression leads to an error that amounts to the convolution of the true g*(s) with a hyperbola segment. The width of this hyperbola segment is Δs = s*2−s*1 = sΔt/t1, and the relative width is Δs/s =Δt/t1 (Figure 3). As a visual guide for the relative broadening to be expected, in the rectangular approximation of the solution column we find Δs/s ≈ Δrb/(rb −m), i.e., the relative width of convolution in s is similar to the ratio of boundary migration during the time interval and the boundary distance from the meniscus.
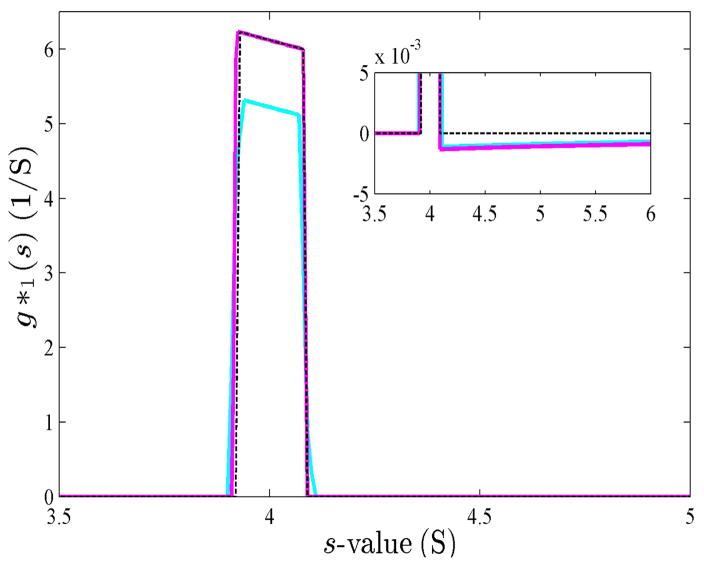
Fig. 3.
Response function for step-function boundaries of a single 4 S species sedimenting at 50,000 rpm, observed at 7,200 and 7,500 sec. Shown are the resulting g(s*) from DCDT (kindly provided by Dr. Walter Stafford) as solid cyan line, the result from DCDT+ (kindly provided by Dr. John Philo) as solid magenta line, and the theoretical prediction Eq. (27) as black dotted line.
3 Results and Discussion
In the present work we have applied a concise theoretical framework to comprehensively study different approaches for determining g*(s). A discretized form of the integral definition of g*(s) via step-functions can be fitted directly to the entirety of available data in the ls-g*(s) approach, and the well-known Bridgman equation for the radial-derivative approach arises naturally from the derivative properties of step-functions. We have found an analogous, previously unrecognized simple expression for the time-derivative.
A key feature of the time-derivative is the radial dilution term, which is superimposed to the boundary term. The radial dilution term makes significant contributions, as shown in Figures 1C and 2. For example, one can imagine that when using a fixed-radius detector recording a continuous time-trace of the macromolecular depletion, the signal will decrease already prior to any particle boundaries traversing the detector. If it would not be accounted for, then the steady radial dilution of all small particles would be mistaken for a broad population of large particles. Accordingly, for the interpretation of the signal with a fixed-radius detector, Scholtan & Lange26 have solved the problem exactly in a recursive approach by starting at the latest time points working backwards to earliest times, accounting for radial dilution at earlier times of the particles passing through the detector at later times.
In practice, however, the single fixed-position detector approach has the drawback of providing significantly lower information content and correspondingly lower statistical precision. Perhaps more importantly, the lack of radial scans monitoring the entire solution column prohibits diagnostics of systematic errors in the model or experimental problems, including significant contributions to particle migration from diffusion, convection, systematic noise contributions, and/or signal offsets from sedimenting small co-solute molecules. Fixed-radius detection has been applied recently in conjunction with a multiwavelength detector design for AUC55, and is used in disc centrifuges 56–58. However, in the latter approach, similar to band centrifugation configurations in AUC, samples are injected causing sedimenting lamella that will exhibit reduced, but more complex simultaneous detection and radial dilution effects.
For the time-derivative approach, which takes differences of entire scans, Stafford has initially neglected the radial dilution terms, 30 but later applied an iterative scheme to approximately account for radial dilution 31 via a numerical solution of the implicit integral equation Eq. (14). However, it stops short of an analytical solution. To still achieve the desired theoretical analogy to the Bridgman equation, Stafford has presented an equation postulating a single derivative term (dc/dt)corr, where a correction was conceptualized to account for the radial dilution. However, no expression could be provided for the conceptualized correction, and instead (dc/dt)corr was defined only indirectly after first approximately solving the implicit equation Eq. (14).
Developing the theory further, in the present work we find that a simple closed-form expression can be given, which now allows to directly define Stafford’s (dc/dt)corr in a physically meaningful way as
| (28) |
The correction corresponds to the radial dilution contributions of all smaller species, in satisfying agreement with the corrections described earlier by Scholtan & Lange.
In principle, this should be useful to replace the iterative approach of current dcdt implementations by a more precise direct calculation. However, an added complication is the necessity to approximate the time-derivative in practice by a time difference expression, due to the finite time differences between scans. Dependent on the rotor speed, sample number, and scan settings the time between successive scans of the same cell may be between 1 and 5 min (or longer in some configurations). The theoretical analysis shows that this approximation of the time-derivative leads to an error in the resulting distribution corresponding to a convolution with a hyperbola segment. A similar prediction was made previously on the basis of the approximation of rectangular solution geometry37, and is shown in the current framework to hold strictly also for sector-shaped geometry. The effect of this is a convolution that will simultaneously broaden and skew the distribution; this corresponds well with experimental observations discussed previously. 21,37
Accounting for the finite time resolution, it is of interest to compare the new explicit expression (Eq. 21) with the performance of the iterative scheme. To this end, we calculated theoretical boundary profiles of a hypothetical non-diffusing 4 S species sedimenting at 50,000 rpm, observed at 7,200 and 7500 sec via Eq. (1) and created synthetic scan files. The resulting radial boundary profiles were analyzed with the programs DCDT and DCDT+. As may be discerned from Figure 3, there is a close correspondence between the iteratively calculated results using DCDT and DCDT+ and the theoretical expectation for this single-species case (Eq. 27), modulo an inconsequential small scale factor of unknown origin in DCDT. This agreement confirms both the software and the theoretical derivations for single species.
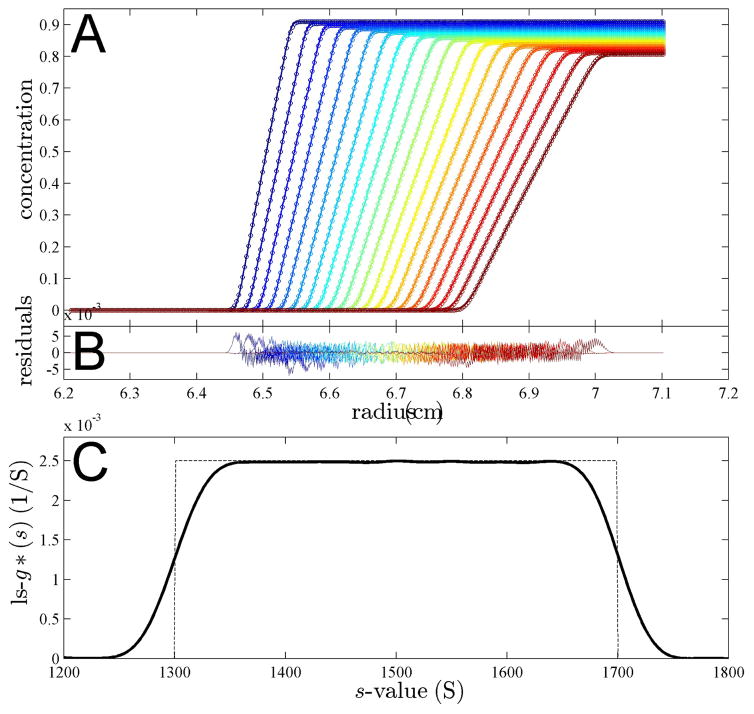
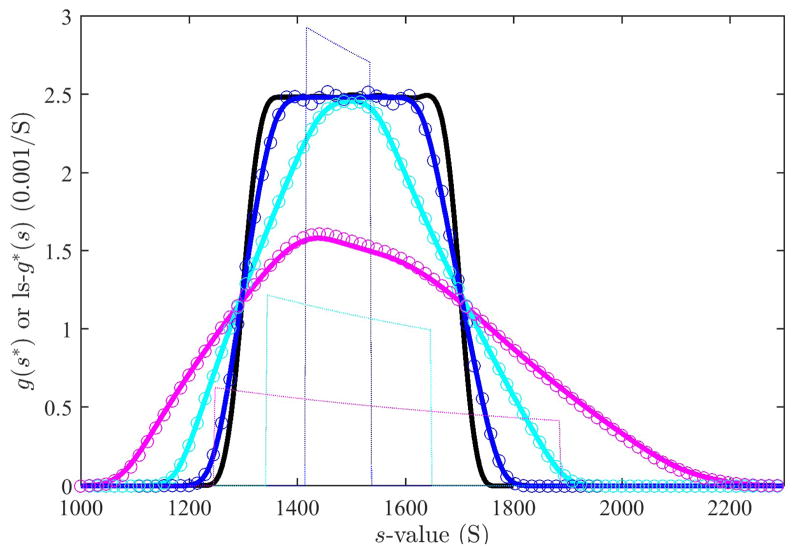
Next, we aimed at testing the predictions for extended distributions. To this end, we simulated sedimentation profiles of a very simple sedimentation coefficient distribution, only consisting of a constant value over a certain size range. Figure 4A shows sedimentation profiles for species between 1,300 S and 1,700 S with the constant distribution indicated as dotted line in Figure 4C. Such a rectangular distribution is unlikely to be found in a practical application, but the sharp features reveal most clearly the effects of convolution. The data were simulated in SEDFIT for close-to-spherical proteinaceous particles sedimenting at a rotor speed at 8,000 rpm. Under these conditions, diffusion effects are very minor and indistinguishable from the effects of a slight blurring of the sedimentation coefficient distribution. (This is advantageous for the simulation so as to remove effects from the discrete approximation of the continuous distribution simulated.) The effects of diffusion are reflected in the ls-g*(s) distribution calculated with SEDFIT, which fits the sedimentation profiles very well (shown for the time-interval from 420 sec to 1,020 sec), but results in an apparent sedimentation coefficient distribution with smoother transitions than the rectangular distribution underlying the simulation (Figure 4C bold solid line versus dotted line).
Fig. 4.
Panel A: Simulated sedimentation profiles of a population of species with uniform distribution between 1,300 S and 1,700 S, assuming a partial specific volume of 0.73 ml/g and a frictional ratio of 1.2, sedimenting at 8,000 rpm, shown in 300 sec intervals from 420 sec to 1,020 sec (circles). The solid line is the best-fit boundary of a ls-g*(s) model, neglecting diffusion. Panel B: Residuals of the fit. Panel C: The best-fit ls-g*(s) distribution (bold line) and the distribution underlying the simulation (thin dotted line).
Finally, the synthetic scan files from the data of Figure 4A were subjected to dcdt analysis in DCDT+ using the same 600 sec time-range (Figure 5 magenta line) and smaller time-intervals of 170 sec (cyan line) and for 90 sec (blue line). To test the prediction of Eq. (24), we took the ls-g*(s) distribution of Figure 4 as the correct distribution (Figure 5, black line), and convoluted it with the respective response functions as given for each time interval in Eq. (27), the latter shown in Figure 5 as thin dotted lines in the middle of the distribution. The resulting predictions of the convoluted sedimentation coefficient distributions g̃*(s*, t, Δt) (Figure 5, symbols) are in good agreement with the numerical results in DCDT+ (Figure 5, colored lines), corroborating the theoretical analysis and numerical computations.
Fig. 5.
Effect of the finite time interval in the dcdt method in the analysis of the uniform distribution of Figure 4. For reference the ls-g*(s) distribution from the analysis of the entire data set is shown in black. The results from the dcdt method, calculated with DCDT+ (version 2.4.3, kindly provided by Dr. John Philo) for the same time range (bold magenta line), for a smaller subset comprising 10 scans between 600 sec and 870 sec (bold cyan line), and for the a subset comprising only 4 scans between 690 sec and 780 sec (bold blue line). The circles are the theoretical dcdt results calculated on the basis of the convolution Eq. 24 with the response function from Eq. (27), using the ls-g*(s) distribution as an approximation of the true g*(s) distribution. In calculating , in analogy to the calculation in DCDT+, the reference time was taken as the harmonic mean of times of scan pairs, and the results averaged over all pairs of scans. The response functions in the center of the time range is shown as thin lines, reduced in scale by 1/3.
It is unfortunately not customary in the dcdt method to transform the distributions back into the original data space, although this is easily possible via insertion of g̃*(s*, t, Δt) into the definition Eq. (3), implemented as a tool in SEDFIT. As shown previously, 59 this would reveal a misfit of the original scan data in cases where the convolution from using the finite time-interval in dcdt creates significant artifacts, and thereby provide a rational quantitative guide for the scan selection in this method.
4 Conclusions
The uniform framework of integral and differential approaches for sedimentation coefficient distributions has led to a satisfying analogy between methods. We found a simple analogue of the Bridgman relationship for time-derivative (which was previously hypothesized to exist but not identified) provided an avenue to theoretically predict the impact of approximations of time-derivative by time-differences. This is practically relevant to interpret results from the application of the dcdt method, and provides simple approaches to rationally guide the selections of scans subsets. Good agreement between theory and numerical results was found, although the detailed results may further depend on the details of implementation, such as the averaging scheme adopted to condense sets of scans into an approximation of dcdt.
The effects of the convolution of g*(s) observed in dcdt does not depend on the size-range of particles, though it will be less apparent for smaller particles with diffusion-broadened apparent sedimentation coefficient distributions. It can be minimized by exclusion of most of the experimental data. The absence of such artifacts favors the application of the direct boundary modeling approach in ls-g*(s). This is true in particular for large particles where diffusion is negligible: In this case g*(s) is the correct model to describe the sedimentation process across the entire time of the experiment, and all data can be included in lsg *(s) to produce the best possible estimate of the particle sedimentation coefficient distribution. In general, the successful fit of the experimental data with ls-g*(s) can provide a simple and rational criterion for the application of the model of non-diffusing particles, and establish a rigorous approach for the determination of weight-average sedimentation coefficients via the transport method59 for further thermodynamic analysis.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health.
Appendix
A. Solution of the Integral Equation for g∘
For clarity we may rewrite the differential equation (17) in a general way as
identifying x = s*, and b = −2ω2t1. The ansatz
with any function p(x) has the derivative
. To satisfy our differential equation, we can identify
or
, leading to the solution
with the unknown integration constant q. Back-substitution leads to
Based on the definition of g∘ in (15) it is easy to see that g∘ (s = 0) = 0 will always be true, and therefore q = 0. Further, considering that r1(s*= 0) = m, and using the fact that for any time t1 > 0 the concentration of a species at the meniscus is zero, H (m−rb(s)) = 0, the second term in the brackets also disappears. With only the first term in the bracket remaining we arrive at
for t1 > 0.
B. Consistency of the Temporal Bridgman Equation
We want to examine whether the temporal Bridgman equation (21) is a solution of the integral equation (14). To this end, we rephrase Eq. (14) as
In comparison with Eq. (20), which arises from (21) through coordinate transformation, the only difference is in the second term on the right hand side. For (20) to be true, these must be identical:
This simplifies to
Differentiation with regard to s* resolves the left-hand-side integral, and the product rule applies on the right-hand side
which after simplification of terms reduces to Eq. (14), and is therefore true.
C. Testing the Temporal Bridgman Equation with Single- Species Profiles
Let us consider the sedimentation profiles of a single non-diffusing species sedimenting with s1. It has the concentration profiles and time-derivative given in (1) and (12), respectively, with a boundary at rb = rb(s1, t1). Inserted in the temporal variant of the Bridgman equation (20), using r* short for r*(s*, t1), and after straightforward rearrangement including separation and cancelation of terms, as well as the insertion of e−s1ω2t1 = m/rb , we obtain
The two integrals can be resolved in the following way: The first integral over the δ -function can only produce a non-zero result for radii r* ≥ rb(t) (i.e., when the integration limits cover its argument across zero) which can be expressed conveniently by multiplying the Heaviside function H(r*−rb) to the executed integral, as in . In the second integral we can eliminate the Heaviside function by adjustment of integration limits (eliminating the range over which H = 0 and starting integration where H = 1). But, again, to express the fact that integral can be non-zero only for radii r* ≥ rb(t), the analytic result of the integration must be multiplied with a new Heaviside function H(r*−rb), as in . This leads to :
and an equation
where all terms cancel out except for the first
Finally, with the transformation of the δ -function into s*-space δ(rb – r*) = (ω2t1 rb)−1 δ(s1 – s*) we arrive at
where the second identity arises from the fact that a non-zero results is only obtained at s* = s1, in which case also r*= rb. Thus, we find the expected result for the distribution of a single species sedimenting with s1, which was underlying the concentration profile and its time-derivative.
D. The Single-Species Response with Finite Time-Difference
Instead of the time-derivative we use the difference between the signals at times t2 and t1, which are thought to be slightly larger and smaller than the time t, respectively:
(where Δt = t2 −t1, and r*= r*(s*)). If we insert this approximation into Eq. (21) in place of ∂ c(r*, t)/∂ t , after simplification, we have
The Heaviside functions in the integrals are equivalent to a change of the lower integration limit, but non-vanishing only if r*> rb(t2) and r*> rb(t1), respectively. As outlined above, the latter can be conveniently expressed by multiplication of the integration result with new Heaviside functions:
which, after integration and sorting of terms, leads to
or
This function is the hyperbola segment shown in Figure 3.
References
- 1.Svedberg T, Pedersen K. Die Ultrazentrifuge. Theodor Steinkopff; Dresden: 1940. [Google Scholar]
- 2.Svedberg T, Rinde H. J Am Chem Soc. 1924;46:2677– 2693. [Google Scholar]
- 3.Brown P, Balbo A, Schuck P. Eur Biophys J. 2009;38:1079–1099. doi: 10.1007/s00249-009-0514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthos J, Cicala C, Steenbeke TD, Van Ryk D, Dela Cruz C, Khazanie P, Selig SM, Hanback DB, Nam D, Schuck P, Chun TW, Chaikin M, Fauci AS. J Biol Chem. 2002;277:11456–11464. doi: 10.1074/jbc.M111191200. [DOI] [PubMed] [Google Scholar]
- 5.Hatters D, MacRaild C, Daniels R, Gosal W, Thomson N, Jones J, Davis J, MacPhee C, Dobson C, Howlett G. Biophys J. 2003;85:3979–3990. doi: 10.1016/S0006-3495(03)74812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mok YF, Ryan T, Yang S, Hatters D, Howlett G, Griffin MDW. Methods. 2011;54:67–75. doi: 10.1016/j.ymeth.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz S, Philo J. Anal Biochem. 2006;362:16–37. doi: 10.1016/j.ab.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Agarwala S, Ravindran S, Vellekamp G. J Pharm Sci. 2008;97:746–763. doi: 10.1002/jps.21008. [DOI] [PubMed] [Google Scholar]
- 9.Trachtenberg S, Schuck P, Phillips T, Andrews S, Leapman R. PLoS One. 2014;9:e87921. doi: 10.1371/journal.pone.0087921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silvera Batista C, Zheng M, Khripin C, Tu X, Fagan J. Langmuir. 2014;30:4895–4904. doi: 10.1021/la404892k. [DOI] [PubMed] [Google Scholar]
- 11.Fagan J, Zheng M, Rastogi V, Simpson J, Khripin C, Silvera Batista C, Hight Walker A. ACS Nano. 2013;7:3373– 3387. doi: 10.1021/nn4002165. [DOI] [PubMed] [Google Scholar]
- 12.Arnold M, Suntivich J, Stupp S, Hersam M. ACS Nano. 2008;2:2291–2300. doi: 10.1021/nn800512t. [DOI] [PubMed] [Google Scholar]
- 13.Calabretta M, Jamison J, Falkner JC, Liu Y, Yuhas BD, Matthews KS, Colvin V. Nano Lett. 2005;5:963–967. doi: 10.1021/nl047926f. [DOI] [PubMed] [Google Scholar]
- 14.Pease LF, Tsai DH, Zangmeister Ra, Zachariah MR, Tarlov MJ. J Phys Chem C. 2007;111:17155–17157. [Google Scholar]
- 15.Lees EE, Gunzburg MJ, Nguyen TLL, Howlett G, Rothacker J, Nice EC, Clayton AHA, Mulvaney P. Nano Lett. 2008;8:2883–2890. doi: 10.1021/nl801629f. [DOI] [PubMed] [Google Scholar]
- 16.Cölfen H, Völkel A. Prog Colloid Polym Sci. 2004;127:31–47. [Google Scholar]
- 17.Wohlleben W. J Nanopart Res. 2012;14:1300. doi: 10.1007/s11051-012-1300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berkowitz S, Engen JR, Mazzeo JR, Jones GB. Nat Rev Drug Discov. 2012;11:527–40. doi: 10.1038/nrd3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding S, Schuck P, Abdelhameed AS, Adams G, Kök MS, Morris G. Methods. 2011;54:136–44. doi: 10.1016/j.ymeth.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita H. Mathematical Theory of Sedimentation Analysis. Academic Press; New York: 1962. [Google Scholar]
- 21.Philo J. Anal Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 22.Philo J. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 23.Stafford W. Curr Opin Biotechnol. 1997;8:14–24. doi: 10.1016/s0958-1669(97)80152-8. [DOI] [PubMed] [Google Scholar]
- 24.Errington N, Byron O, Rowe A. Biophys Chem. 1999;80:189–197. doi: 10.1016/s0301-4622(99)00077-0. [DOI] [PubMed] [Google Scholar]
- 25.Bridgman W. J Am Chem Soc. 1942;64:2349–2356. [Google Scholar]
- 26.Scholtan W, Lange H. Kolloid-Z u Z Polym. 1972;250:782–796. [Google Scholar]
- 27.Ortlepp B, Panke D. Prog Coll Polym Sci. 1991;86:57–61. [Google Scholar]
- 28.Mächtle W. Biophys J. 1999;76:1080–1091. doi: 10.1016/S0006-3495(99)77273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller H. Colloid Polym Sci. 1989;267:1113–1116. [Google Scholar]
- 30.Stafford W. Anal Ultracentrifugation Biochem Polym Sci. The Royal Society of Chemistry; Cambridge, U.K: 1992. pp. 359–393. [Google Scholar]
- 31.Stafford W. Anal Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 32.Stafford W. Methods Enzym. 1994;240:478–501. doi: 10.1016/s0076-6879(94)40061-x. [DOI] [PubMed] [Google Scholar]
- 33.Schuck P, Demeler B. Biophys J. 1999;76:2288–2296. doi: 10.1016/S0006-3495(99)77384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuck P. Anal Biochem. 2010;401:280–287. doi: 10.1016/j.ab.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Provencher S. Makromol Chem. 1979;180:201–209. [Google Scholar]
- 36.Schuck P. 26th South. Biomed. Eng. Conf. SBEC 2010; April 30 – May 2, 2010; Coll. Park. Maryland, USA. Berlin Heidelberg: Springer; 2010. pp. 340–343. [Google Scholar]
- 37.Schuck P, Rossmanith P. Biopolymers. 2000;54:328– 341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 38.Schuck P. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoskins R. Delta Functions. Introduction to Generalized Functions. 2. Horwood Publishing; Coll House, U.K: 1999. [Google Scholar]
- 40.Batchelor G. J Fluid Mech. 1972;52:245–268. [Google Scholar]
- 41.Fujita H. Foundations of Ultracentrifugal Analysis. John Wiley & Sons; New York: 1975. [Google Scholar]
- 42.Ramaswamy S. Adv Phys. 2001;50:297–341. [Google Scholar]
- 43.Berres S, Bürger R, Tory EM. Chem Eng J. 2005;111:105–117. [Google Scholar]
- 44.Koch DL, Shaqfeh ESG. J Fluid Mech. 2006;224:275. [Google Scholar]
- 45.Schuck P. Biophys J. 2010;98:2005–2013. doi: 10.1016/j.bpj.2009.12.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamm O. Ark Mat Astr Fys. 1929;21B(2):1–4. [Google Scholar]
- 47.Schuck P, Zhao H, Brautigam C, Ghirlando R. Basic Principles of Analytical Ultracentrifugation. CRC Press; Boca Raton, FL: 2015. [Google Scholar]
- 48.Ma J, Zhao H, Schuck P. Anal Biochem. 2015;483:1–3. doi: 10.1016/j.ab.2015.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuck P. Eur Biophys J. 2010;39:1261–1275. doi: 10.1007/s00249-009-0545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown P, Balbo A, Schuck P. Biomacromolecules. 2007;8:2011–2024. doi: 10.1021/bm070193j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuck P. Biophys Chem. 2004;108:201–214. doi: 10.1016/j.bpc.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 52.Ma J, Zhao H, Sandmaier J, Liddle J, Schuck P. Biophys J. 2016;110:103–112. doi: 10.1016/j.bpj.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao H, Casillas E, Shroff H, Patterson G, Schuck P. PLoS One. 2013;8:e77245. doi: 10.1371/journal.pone.0077245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao H, Ma J, Ingaramo M, Andrade E, MacDonald J, Ramsay G, Piszczek G, Patterson G, Schuck P. Anal Chem. 2014;86:9286–9292. doi: 10.1021/ac502478a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter J, Peukert W. Nanoscale. 2016:7484–7495. doi: 10.1039/c5nr08547k. [DOI] [PubMed] [Google Scholar]
- 56.Kamiti M, Boldridge D, Ndoping LM, Remsen EE. Anal Chem. 2012;84:10526–10530. doi: 10.1021/ac3022086. [DOI] [PubMed] [Google Scholar]
- 57.Neumann A, Hoyer W, Wolff MW, Reichl U, Pfitzner A, Roth B. Colloids Surfaces B Biointerfaces. 2013;104:27–31. doi: 10.1016/j.colsurfb.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 58.Arosio P, Cedervall T, Knowles TP, Linse S. Anal Biochem. 2016 doi: 10.1016/j.ab.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 59.Schuck P. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]