Abstract
Polysialic acid (PSA) is a carbohydrate polymer of repeating α-2,8 sialic acid residues that decorates multiple targets, including neural cell adhesion molecule (NCAM). PST and STX encode the two enzymes responsible for PSA modification of target proteins in mammalian cells, but despite widespread polysialylation in embryonic development, the majority of studies have focused strictly on the role of PSA in neurogenesis. Using human pluripotent stem cells (hPSCs), we have revisited the developmental role of PST and STX and show that early progenitors of the three embryonic germ layers are polysialylated on their cell surface. Changes in polysialylation can be attributed to lineage-specific expression of polysialyltransferase genes; PST is elevated in endoderm and mesoderm, while STX is elevated in ectoderm. In hPSCs, PST and STX genes are epigenetically marked by overlapping domains of H3K27 and H3K4 trimethylation, indicating that they are held in a ‘developmentally-primed’ state. Activation of PST transcription during early mesendoderm differentiation is under control of the T-Goosecoid transcription factor network, a key regulatory axis required for early cell fate decisions in the vertebrate embryo. This establishes polysialyltransferase genes as part of a developmental program associated with germ layer establishment. Finally, we show by shRNA knockdown and CRISPR-Cas9 genome editing that PST-dependent cell surface polysialylation is essential for endoderm specification. This is the first report to demonstrate a role for a glycosyltransferase in hPSC lineage specification.
Keywords: polysialic acid, neural cell adhesion molecule, glycosylation, pluripotent stem cells
Introduction
Glycosylation is a carbohydrate modification that increases structural diversity and function of proteins and lipids. The significance of glycosylation in mammalian cell biology is emphasized by its contribution to the glycocalyx- a dense, glycan-rich structure on the outer surface of cells. The glycocalyx is composed of a complex array of glycans that vary in a cell type-specific manner but deciphering specific roles for carbohydrates in the function of specific cell surface targets has been problematic. The broad impact of cell surface glycans on human health, however, has become increasingly important due to strong clinical links between cell surface glycosylation defects and human disease [1]. From a developmental perspective, glycosylation is an integral and dynamic process associated with cell migration, cell fate specification, proliferation, and organogenesis in the embryo [2]. Specific roles for glycans in these early developmental processes are, however, poorly understood.
One aspect of embryonic development that has been well documented in mice, but limited in humans, is the expression of polysialic acid (PSA) on the surface of developing cells [3, 4]. PSA is a linear glycan homopolymer composed of repeating N-acetylnueraminic acid (Neu5Ac or sialic acid) residues linked by α-2,8 glycosidic bonds [4]. Two polysialyltransferases, ST8SIA2 (STX) and ST8SIA4 (PST), reside in the Golgi and add polysialic acid chains to target proteins or to themselves through autopolysialylation [5]. Polysialylated epitopes are elevated during the development of many tissues in mice including heart, kidney, pancreas and brain with cell type-specific expression of PST and STX [6-9] . Little is known, however, about the function and regulation of polysialylation during early stages of development.
Although neuropilin-2 and SynCAM1 have been reported to be polysialylated [10, 11], neural cell adhesion molecule (NCAM) is the most abundant and best characterized substrate for PSA modification [4]. NCAM is an immunoglobulin (Ig) adhesion protein residing on the cell surface and plays roles in the formation of intercellular contacts through homophilic and heterophilic binding, in addition to intracellular signaling [4]. NCAM is polysialylated by either PST or STX on the fifth Ig domain, forming a polymer chain typically between 60-90 sialic acid residues [12, 13]. The resulting modification of NCAM (PSA-NCAM) is a large, highly-negatively charged side chain that disrupts binding interactions between neighboring PSA-NCAM molecules to enhance cellular motility [14-16]. This property is utilized in the developing mouse nervous system in which PSA-NCAM is instrumental in neuronal migration, neurite outgrowth and synaptic plasticity [17, 18]. Evidence also suggests that PSA impacts NCAM-directed signaling events [19-21]. PSA is widely expressed on the surface of metastatic cancer cells including neuroblastoma, pancreatic ductal adenocarcinomas, small cell lung carcinoma and Wilms’ tumor [22-25], highlighting its clinical significance. Consistent with this, the two polysialyltransferases (PST and STX) are expressed at elevated levels in tumor cells and the resulting polysialylation is an adverse prognosis factor [26].
Understanding the role of cell surface glycosylation and how it is regulated during human development is very limited. In this report, we address this by identifying broad roles for polysialylation in cell fate specification of human pluripotent stem cells and provide the first report describing a role for a specific glycosyltransferase in hPSC lineage specification.
Materials and Methods
Stem Cell Culture and Differentiations
WA09, WA01, WA07, and TE03 hESCs (http://grants.nih.gov/stem_cells/registry/current.htm) were cultured on Geltrex (Life Technologies) coated plates and sustained using complete defined medium (CDM) containing recombinant Heregulin β-1 (10 ng/mL), Activin A (10 ng/mL), bFGF2 (8 ng/mL), and IGF-1 (200 ng/mL) as described previously [27]. Cells were plated at a density of 50,000 cells/cm2 and grown to 90% confluency before passaging with Accutase (ICT). Mesoderm cells were generated by culturing WA09 cells for 4 days in CDM supplemented with WNT3a (25 ng/mL) and BMP4 (100 ng/mL). Endoderm and ectoderm differentiations followed previously established protocols [28, 29].
qRT-PCR
RNA was collected using the E.Z.N.A. RNA isolation kit (Omega) and cDNA made from 1 μg RNA using the Iscript cDNA synthesis kit (Bio-Rad). Genes were assayed using Taqman primers (Life Technologies) on a ViiA 7 Real-Time PCR System (Life Technologies). Assays were performed in triplicate and normalized to GAPDH expression. Error bars indicate ± SEM.
Western blots
Protein was prepared using RIPA lysis buffer and Western blots done using 30 μg of protein lysate loaded into NuSep Tris-Glycine Gels. Protein was then transferred onto a nitrocellulose membrane and probed using antibodies shown in Table S1 with detection via HRP.
Immunohistochemistry
Cells were grown on Lab-Tek 4-well chamber slides, fixed using 4% paraformaldehyde treatment for 10 minutes, and permeabilized using 0.2% Triton in PBS. Antibodies and concentrations used for staining are shown in Table S1. Images were obtained using a Leica DM6000 B microscope and a Zeiss LSM 710 confocal microscope.
Flow Cytometry
Cells were harvested using Accutase and 1 million cells were incubated with antibodies or isotype control in 20 μL PBS containing 0.2% BSA. Antibodies and concentrations used are shown in Table S1. Results were obtained using a Beckman Coulter Cyan ADP analyzer with data analysis using FlowJo software. Cell sorting was done using a Beckman Coulter MoFlo XDP and a Bio-Rad S3.
ChIP-PCR assays
10 million cells were cross-linked using 2% formaldehyde and quenched using 2.5M glycine. Lysis was performed using the Agilent Mammalian ChIP protocol and DNA sonicated to ~500 bp using a Covaris S220 Focused-ultrasonicator. Immunoprecipitation was done by overnight incubation of cell lysate at 4°C with Dynabeads Protein G magnetic beads (Life Technologies) conjugated with 10 μg of antibody (Table S1). qRT-PCR of ChIP lysate was done using genomic primers on a ViiA 7 Real-Time PCR System with comparison to whole cell extract.
Luciferase assays
Reporter plasmids were constructed by cloning the 5kb upstream genomic region of PST and STX amplified using Expand Long Range dNTPack (Roche) with ligation into a pGL4.1 Luciferase plasmid (Promega). Cells were plated onto 24-well plates and transfected in triplicate with 1 μg reporter plasmid and 100 ng Renilla control plasmid (Promega) using the Dual-Luciferase Reporter Assay System (Promega). Cells were collected and analyzed 48h after transfection and analyzed for luminescence by a Biotek Synergy 2 plate reader. Data are shown in triplicate with error bars indicating ± SD.
shRNA knockdowns and CRISPR genome editing
shRNA knockdowns were done using TRC lentiviral shRNA plasmids (Dharmacon) targeting human PST (Cat. RHS4533-EG7903), NCAM (RHS4533-EG4684), and GSC (Cat. RHS4533-EG145258). Lentiviral particles were generated using a Trans-Lentiviral Packaging Kit (Thermo Scientific). Virus was concentrated upon collection using Lenti-X Concentrator (Clontech) and titer determined via qPCR titer kit (Mellgen Labs). Cells were plated in 24-well plates, transduced at MOI=1-5 in the presence of 6 μg/uL polybrene, and selected using 3 μg/mL Puromycin. CRISPR sgRNAs were designed using CRISPR Design software [30] and cells were transfected with sgRNAs, hCas9 [31], and repair plasmid. Cells were selected using Zeocin (100μg/mL) and grown as single cell clones in 96-well flat bottom plates with addition of 10μM Y-27632 (Tocris). PST−/− cells were verified via genomic PCR.
Results
Early PSC-derived progenitors are polysialylated on their cell surface
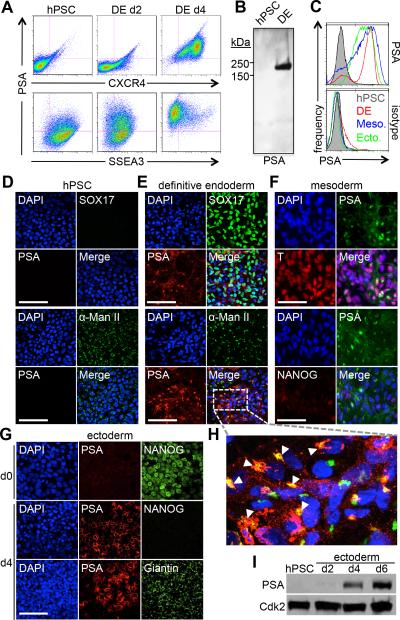
hPSCs are known to express characteristic glycans on their cell surface such as SSEA3 but do not present PSA (Fig. 1A). However, when differentiated towards early progenitor cells of the three embryonic germ layers, WA09 hPSCs transition to a PSA+ state (Fig. 1). This observation is apparent in multiple hPSC lines (WA01, WA07, TE03) as well as mESCs (R1) (Fig. S1). By immuno-blotting of definitive endoderm (DE) lysates, PSA-associated immuno-reactive bands are observed in the ~150-250 kDa range (Fig. 1B), characteristic of polysialylation. Within 2-4 days of Activin A-directed differentiation, CXCR4+ DE acquires cell surface (Fig. 1A, C, E, H and Fig. S1B,C) and Golgi-associated, α-mannosidase II co-localized PSA+ epitopes (Fig. 1H). T+/Isl1+ early mesoderm progenitors generated by BMP4 treatment also acquire cell surface and intracellular polysialylated epitopes within 4 days (Fig. 1C, F). Finally, early AP2+ ectoderm progenitors also become cell surface polysialylated but in contrast to mesoderm and endoderm, Golgi-associated (Giantin+) reactivity is less obvious (Fig. 1C, G, I). In all three germ layers, cell surface polysialylation is acquired as cells lose pluripotency markers, such as NANOG and SSEA3 (Fig. 1A, F, G).
Figure 1.
Polysialic acid is expressed upon differentiation to all three germ layers. (A) Flow cytometry of hPSC differentiation to definitive endoderm showing surface expression of PSA along with endoderm marker CXCR4 (Top) and pluripotency marker SSEA3 (Bottom). (B) Immunoblot of PSA expression in hPSC and DE. (C) Flow cytometry of PSA expression (Top) in hPSC (Shaded gray), DE (Red), mesoderm (Blue), and NCC (Green). PSA+ fractions in each cell type are 2.9%, 79.7%, 72.4% and 81.1%, respectively. Isotype control is shown for comparison (Bottom). (D-H) Immunostaining of PSA expression in hPSC differentiation. Scale bar, 100μm. (D) hPSC: PSA and DE marker SOX17 (Top), PSA and Golgi marker α-mannosidase II (Bottom). (E) DE: PSA and SOX17 (Top), PSA and α-mannosidase II (Bottom). (F) Mesoderm: PSA and T (Top), PSA and NANOG (Bottom). (G) NCC: PSA and NANOG (Top), PSA and Golgi marker Giantin (Bottom). (H) Zoom of PSA and α-mannosidase II in DE. Arrows point to overlap of PSA and α-mannosidase II. (I) Time course immunoblot of PSA expression during hPSC differentiation to NCC (Top). CDK2 shown as loading control (Bottom). Abbreviations: hPSC, human pluripotent stem cell; DE, definitive endoderm; NCC, neural crest cell.
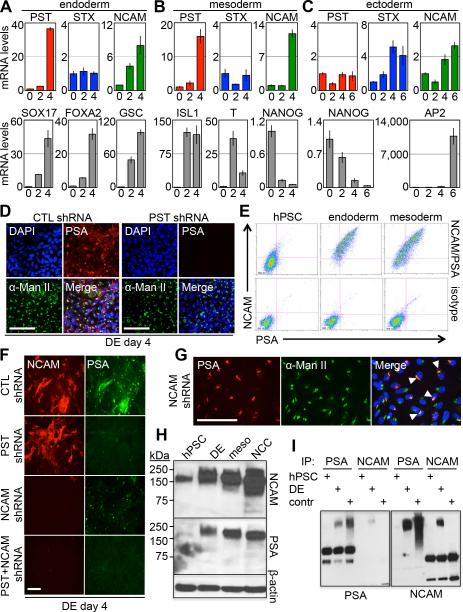
Polysialic acid can be attached to target proteins by two enzymes, PST and STX. To establish if polysialylation is regulated at the transcriptional level and whether PST or STX is involved in polysialylation during germ layer induction, transcript levels for these two genes were determined. This analysis showed germ layer specific differences in PST/STX transcript accumulation at the time when polysialylation increased (Fig. 2A-C). During early endoderm and mesoderm differentiation, PST transcript levels increase significantly while STX mRNA levels remain low (Fig. 2A,B and Fig. S1D). In contrast, STX transcript levels increase during early ectoderm differentiation, while PST mRNA remains low (Fig. 2C). NCAM transcript levels increase during the early stages of differentiation in all three germ layers (Fig. 2A-C). To confirm that PST was responsible for polysialylation in DE, shRNA was used to target PST transcripts. This resulted in ~85-90% reduction of PST transcript levels and loss of cell surface and Golgi-associated polysialylation (Fig. 2D and Fig. S2).
Figure 2.
Expression of polysialyltransferases is lineage specific and NCAM is the substrate of polysialylation. (A-C) qPCR expression of PST, STX, and NCAM (Top) and lineage markers (Bottom) in hPSC differentiation to DE (A), mesoderm (B), and NCC (C). (D) Immunostaining of PSA expression in DE day 4 cells transduced with shRNA targeting PST (Right) compared to control shRNA (Left). Scale bar, 100μm. (E) Flow cytometry of PSA and NCAM surface expression in hPSC, DE, and mesoderm. (F) Immunostaining of shRNA knockdowns of PST and NCAM in DE day 4. Scale bar, 100μm. (G) Immunostaining of PSA and α-mannosidase II expression in DE cells transduced with NCAM shRNA. Arrows point to overlap in expression. Scale bar, 100μm. (H) Immunoblot of NCAM (Top) and PSA (Middle) in all three germ layers. β-Actin shown as loading control (Bottom). (I) Immunoprecipitation of NCAM and PSA in hPSC and DE. IP lysate was then immunoblotted for PSA (Left) and NCAM (Right). Lysate from COS-1 cells overexpressing PST, STX, and NCAM was used as positive control. Abbreviations: hPSC, human pluripotent stem cell; DE, definitive endoderm; CTL, control.
We next addressed the issue of whether NCAM is a major target of polysialylation in endoderm and mesoderm differentiation. This is suggested by flow cytometry showing that PSA+ cells are also NCAM+ (Fig. 2E). This possibility is supported by a series of shRNA knockdown experiments showing that loss of NCAM eliminates cell surface polysialylation, but not Golgi-associated PSA reactivity (Fig. 2F,G). Next, NCAM was shown to be polysialylated in multiple PSC-derived cell types of endoderm, mesoderm and ectoderm origin by immunoprecipitation-immunoblot experiments using whole cell lysates (Fig. 2H,I). Finally, although ectopic PST expression in PSCs increased Golgi-dependent polysialylation, presumably through autocatalysis (see ref [5]), its co-expression with NCAM was required for decorating the cell surface with polysialylated substrates (Fig. S3). Taken together, these data indicate that NCAM is the major acceptor of polysialic acid on the surface of hPSC-derived progenitor cells.
Developmentally-regulated transcription factors control PST transcription and polysialylation
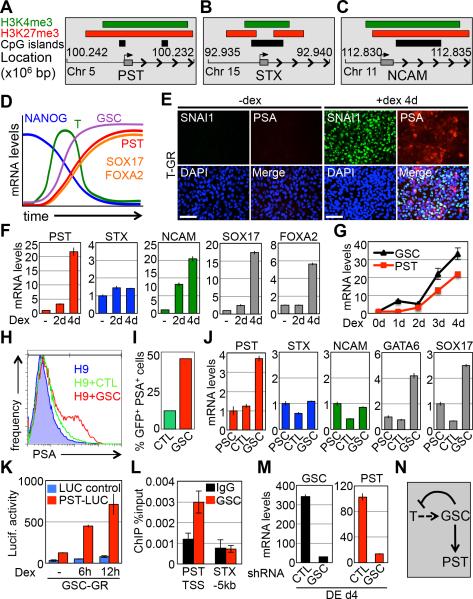
Overlapping domains of activating (H3K4me3) and repressive (H3K27me3) histone modifications mark developmental genes in pluripotent cells [32]. These epigenetic marks hold developmental genes in a ‘poised’ state, so they can be rapidly activated under differentiation conditions. Genes encoding components of the glycomics network have not been implicated in this developmental pathway before; however, based on our previous observations, it seemed reasonable that PST, STX, and NCAM could be part of this broad developmental program. ChIP-seq data from the UCSC genome database indicate bivalent regions of H3K4me3 and H3K27me3 marks overlapping the transcription start sites of PST, STX, and NCAM genes in WA01 hPSCs (Fig. 3A-C) [33]. To confirm this in WA09 hPSCs and to determine whether these histone marks change during differentiation, quantitative chromatin immunoprecipitation (qChIP) assays were performed. In agreement with ChIP-seq data, qChIP of the PST, STX, and NCAM promoters shows significant enrichment for both bivalent histone marks (Fig. S4), confirming them to be bivalent genes in hPSCs. During early germ layer formation, bivalent domains resolve in a pattern consistent with the transcription patterns for each gene as cells differentiate into endoderm, mesoderm and ectoderm (Fig. S4). These data indicate that genes implicated in cell surface polysialylation during germ layer formation are regulated by epigenetic mechanisms known to control a broader group of developmental regulators.
Figure 3.
Polysialylation is bivalently regulated with PST under the control of the T/GSC network. (A-C) Graphical representation of H3K4me3, H3K27me3, and CpG islands at the PST (A), STX (B), and NCAM (C) genomic loci in WA01 hPSCs (Data from the UCSC genome browser database [33]). (D) Graphical illustration of gene kinetics during DE differentiation. (E) Immunostaining comparison of T-GR expression of PSA and SNAI1 after 4 day addition of Dex compared to −Dex. Scale bar, 100μm. (F) qPCR of genes in T-GR cells −Dex, +Dex 2d, and +Dex 4d. (G) Transcript analysis of GSC and PST over 4 day time course addition of Dex in T-GR cells. (H) Flow cytometry for surface PSA expression in hPSCs, hPSCs+GFP, and hPSCs+GSC-GFP 24h post electroporation. (I) Analysis of the percentage of GFP+/PSA+ cells from Fig. 4H. (J) qPCR of samples from Fig. 4H. (K) Luciferase assay of hPSCs transfected with GSC-GR, PST-LUC, and LUC-Control after Dex addition for 0, 6, and 12h. (L) GSC ChIP assay in hPSCs transfected with GSC-GFP. Genomic primers used probe the TSS of the PST promoter and the STX −5kb upstream region shown as negative control. (M) qPCR of DE d4 cells transduced with shRNA targeting GSC compared to control shRNA. (N) Diagram of proposed interaction of T, GSC, and PST. Abbreviations: hPSC, human pluripotent stem cell; Dex, dexamethasone; GR, glucocorticoid receptor; DE, definitive endoderm; LUC, luciferase; TSS, transcription start site; CTL, control.
The kinetics of PST transcript accumulation in endoderm differentiation follows that of Brachyury (T) and Goosecoid (GSC) and coincides with the endoderm markers SOX17 and FOXA2 (Fig. 2A,B and Fig. 3D). Since the polysialyltransferase genes are bivalently marked, they are likely to be part of a broader developmental program associated with lineage specification. Therefore, we hypothesized potential roles for T and GSC in PST/STX regulation because they are developmentally regulated transcription factors known to control early cell fate decisions, including the epithelial to mesenchymal transition (EMT) associated with early PSC differentiation [34-36]. Developmental transcription factors have not previously been implicated in regulation of cell surface glycans, making this a potentially important possibility. To investigate this, we first tested the possibility that T directly regulates the PST gene during endoderm differentiation. Ectopic activation of T using a T-glucocorticoid receptor (T-GR) fusion protein [37] elevates cell surface polysialylation in hPSCs within 4 days treatment with dexamethasone (Dex) (Fig. 3E). Transcript analysis of cells expressing T-GR shows an increase in PST and NCAM expression following Dex induction with no change in STX expression (Fig. 3F). This is consistent with potential roles for T and PST in mesoderm/endoderm differentiation, but not ectoderm formation, which is more closely associated with STX expression (Fig. 2A-C). Dex treatment also increased endoderm markers SOX17 and FOXA2 (Fig. 3F), mesoderm markers ISL1 and GATA6 (Fig. S5A), the EMT marker SNAI1 (Fig. 3E), and PSA (Fig. 3E) within 4 days. Although T-GR increased PST and NCAM transcript levels, indicating that T lies upstream of these genes, there was a significant delay between Dex addition and increased cell surface polysialylation. This points towards the effects of T-GR induction on PST transcription and polysialylation as being indirect or perhaps, requiring additional other factor(s).
Interestingly, we noticed that transcript levels for the developmental transcription factor GSC peaked slightly earlier than that for PST following T-GR induction (Fig. 3G). This is consistent with the respective kinetics of T, GSC and PST during Activin A-induced endoderm differentiation (Fig. S5B). This indicates that GSC is downstream of T and that GSC may act directly on PST. To investigate this possibility, GSC was ectopically expressed in hPSCs to establish if it could activate the PST gene and increase cell surface polysialylation. Within 24 hours of GSC expression, cell surface polysialylation was significantly elevated (Fig. 3H,I). Furthermore, GSC transfected cells showed increased PST, GATA6 and SOX17 mRNA levels (Fig. 3J), and an inducible GSC-GR construct activated a PST-luciferase reporter (PST-LUC) (Fig. S5C) within 6 hours (Fig. 3K) of Dex addition, whereas T-GR (+Dex) had no effect on the reporter (Fig. S5D). This reporter faithfully reproduces induction kinetics of the endogenous PST gene during differentiation (Fig. 2A-B and Fig. S5E), confirming it to be a suitable tool for these studies. Other luciferase reporter assays showed that GSC-GR strongly activated the PST promoter, whereas T-GR had a weaker effect and neither GSC-GR nor T-GR activated an STX-luciferase reporter (Fig. S5F). GSC was shown to bind the PST promoter, but not the STX promoter, by ChIP assays (Fig. 3L), confirming that it directly regulates PST transcription. Finally, shRNA knockdown of GSC blocks the up-regulation of PST during endoderm differentiation and suppresses the down-regulation of T (Fig. 3M and Fig. S5G). This latter observation is consistent with a previous report showing that GSC is a repressor of T transcription [38]. Taken together, these data indicate that T promotes expression of GSC, which then represses T and directly activates transcription of PST resulting in cell surface polysialylation (Fig. 3N).
PST-dependent polysialylation is required for early cell fate specification
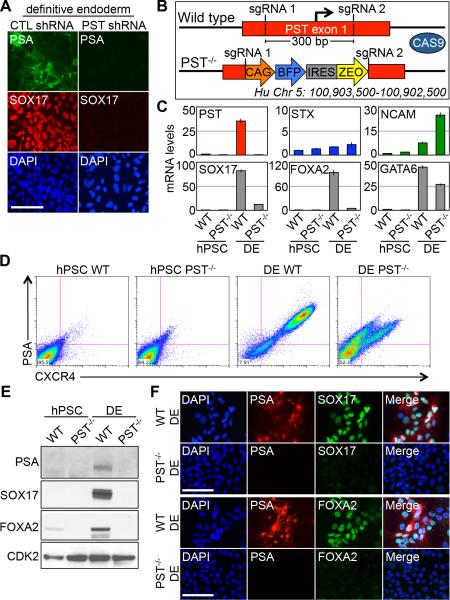
Earlier in this report, we showed that shRNA knockdown of PST activity significantly reduces polysialylation under endoderm differentiation conditions (Fig. 2D). qPCR and immunofluorescence staining analysis also showed that up-regulation of endoderm markers FOXA2 and SOX17 was blocked under these conditions (Fig. 4A and Fig. S6A-B), indicating that polysialylation is essential for normal differentiation. shRNA knockdown of NCAM, however, did not affect the ability to form DE despite a loss of PSA on the surface and its restriction to the Golgi (Fig. 2G and Fig. S6C-E).
Figure 4.
PST is required for efficient hPSC differentiation to endoderm. (A) Immunostaining of PSA and SOX17 expression in DE cells transduced with control shRNA (Left) and PST shRNA (Right). Scale bar, 100μm. (B) Schematic of CRISPR gene editing approach of the PST first exon. The 300 bp region containing the PST TSS was cleaved and replaced by a BFP-IRES-Zeo cassette via homology directed repair. (C) qPCR of genes in WT and PST−/− cells differentiated to DE. (D) Flow cytometry of CXCR4 and PSA expression in WT and PST−/− cells differentiated to DE. (E) Immunoblot comparisons of WT and PST−/− cells differentiated to DE. (F) Immunostaining of PSA with SOX17 (Top) and FOXA2 (Bottom) in WT and PST−/− cells differentiated to DE. Abbreviations: CTL, control; hPSC, human pluripotent stem cell; DE, definitive endoderm; TSS, transcription start site; sgRNA, single guide RNA.
To establish by an independent approach that PST is required for endoderm differentiation, we engineered hPSC lines carrying a homozygous deletion of the PST gene using CRISPR-Cas9 genome editing (Fig. 4B and Fig. S7). Culture of these cells with Activin A confirmed results obtained by shRNA knockdown of PST. After 4 days of Activin A-treatment of PST−/− cells, endoderm markers assayed by qPCR, Western blot, flow cytometry, and immunofluorescence are markedly reduced (Figure 4C-F) relative to hPSCs, indicating a major disruption to the differentiation program. Additionally, forced expression of either PST or STX restores cell surface polysialylation and rescues the DE differentiation defect of PST−/−cells (Fig. S8). These results suggest that polysialylation by PST is required for hPSC differentiation to DE and PSA plays an important role in hPSC cell fate determination.
In contrast to endoderm experiments, PST−/− cells differentiate to mesoderm and are comparable to WT cells (Fig. S9A-C). Flow cytometry indicates PST−/− mesoderm cells express a high level of cell surface PSA (Fig. S9A), but STX expression is elevated and appears to compensate for loss of PST activity (Fig. S9C). These results are surprising considering both endoderm and mesoderm express PST exclusively in WT cells, suggesting that the mechanisms controlling polysialylation are regulated differently. In addition, PST−/− cells were also able to generate NCCs and neural progenitors similar to WT cells (Fig. S9D-F); this result was expected given that STX is expressed in early neuroectoderm (Fig. 2A). Analysis of the bivalent marks on the STX promoter during WT mesoderm differentiation shows that there is a high level of activating H3K4me3 despite STX transcript not being expressed (Fig. S4). This is also true for PST−/− mesoderm (Fig. S9G) and may indicate that STX is unrepressed and capable of activation in the absence of PST.
To test if polysialylation is sufficient to promote exit from pluripotency and for transition towards endoderm, PST alone or PST with NCAM was over-expressed in hPSCs under self-renewal conditions. When polysialylated NCAM was presented on the cell surface following ectopic expression of PST and NCAM, hPSCs maintained the expression of pluripotency marker OCT4 (Fig. S10A,D), retained an epithelial morphology (Fig. S10B-D), and did not up-regulate mesendoderm markers such as T (Fig. S10C) or DE markers such as SOX17 and FOXA2 (Fig. S10A-B). Although cell surface polysialylation is a characteristic of early hPSC differentiation towards the three germ layers, ectopic expression of PST and ectopic cell surface polysialylation is not sufficient for exit from pluripotency.
Discussion
In this report, we show that as pluripotent cells commit towards the three germ layers, they become polysialylated on their cell surface. The major target for this polysialylation in endoderm, ectoderm and mesoderm differentiation appears to be the cell adhesion molecule NCAM. Polysialylation of cell surface NCAM is critical for endoderm differentiation but is not sufficient to trigger exit from pluripotency and is not directly involved in the EMT associated with transition through the mesendoderm state. This is surprising because polysialylation of NCAM has been reported to play role in cell migration by promoting an EMT [39]. Loss of NCAM eliminates surface polysialylation in our experiments, similar to that seen in PST loss of function experiments, but it does not impact endoderm differentiation. This may suggest that additional polysialylated targets control differentiation and that NCAM is only part of the general mechanism. Our preferred explanation however, relates to previous reports in mice where loss of NCAM rescues neurological defects seen in PST and STX mutants [40]. This has led to the idea that unpolysialylated NCAM inhibits neural development and that polysialylated NCAM is compatible with neurogenesis. The role of PST and STX in this context would be to suppress negative signaling inputs generated by NCAM. This model is consistent with our observations but the details of how unpolysisalylated NCAM blocks differentiation needs to be explored further.
Although cell surface polysialylation is synonymous with early differentiation, it is regulated differently in the three germ layers. In endoderm polysialylation is exclusively controlled by PST, while in ectoderm, STX is the primary enzyme for catalytic activity. Mesoderm is perhaps the most interesting case. Here PST is the principle polysialyltransferase under normal conditions, but this switches to STX when PST activity is eliminated. This raises some interesting questions about how the regulatory mechanisms of polysialyltransferase genes are connected and how under specific conditions they can compensate for one another.
Many reports have been published describing the role of polysialylation in mouse development, particularly during neurogenesis. However, there have been no reports describing roles for polysialylation as cells exit pluripotency and as they form the three embryonic germ layers. We found that genes encoding the polysialyltransferases (PST and STX) and the main PSA acceptor NCAM are epigeneticaly marked by bivalent domains in pluripotent cells, and under specific conditions, these marks become resolved depending on the lineage chosen. These epigenetic marks implicated PST, STX and NCAM as part of a broader developmental program required for germ layer formation. This was subsequently confirmed by shRNA and CRISPR-Cas9 genome editing based experiments. Despite showing an essential role for PST in early differentiation, its exact function has not yet been resolved. We know, however, that its function is required after the EMT associated with loss of pluripotency but before key lineage specification genes, such as SOX17 and FOXA2, are activated. We speculate that a signaling defect is associated with loss of PST activity and this will be the subject of further investigation.
In endoderm, PST activation is dependent on a well-characterized transcriptional regulatory network required for lineage specification. Most notably, T is a T-box transcription factor with downstream effectors, such as GSC, that are expressed during primitive streak formation. In our experiments, we find that T is required for GSC activation, and GSC directly binds and transcriptionally activates the PST gene. Regulation of polysialylation through PST is therefore part of the developmental program required for endoderm and probably mesoderm and ectoderm formation. The regulatory network required for STX activation in ectoderm still needs to be defined but is likely to involve suppression of Smad2,3 activity [41] and/or activation of Wnt activity [29].
T and GSC have been previously implicated as transcriptional drivers of metastasis in a range of cancers [35, 36]. It is intriguing to speculate that reactivation of the T/GSC transcriptional network in tumor progression could also involve activation of polysialyltransferase genes such as PST to facilitate migration and metastasis. This is particularly interesting because there are numerous reports describing cell surface polysialyation on metastatic, migratory cells [22-26]. Reactivation of the T/GSC-driven polysialyltransferase activity could therefore be an important component of T/GSC-driven tumor development.
Interestingly, our observations of PST requirement for endoderm specification in hPSCs contrast with previous reports showing that PST null mice are viable with only mild cognitive impairments [42]. Additional studies showed that mice lacking STX shared a similar phenotype [43] and that double knockout of PST and STX led to severe developmental defects and precocious death [40], indicating that polysialylation is required for development but that PST and STX are functionally redundant and capable of compensating for each other. The discrepancy between our findings and previous reports are intriguing as it could represent a differential requirement of individual polysialyltransferase genes between species. Furthermore, this observation of an essential human gene being inessential in mice has been documented previously [44]. Liao and Zhang demonstrated that null mutations of mouse genes orthologous to essential human genes often show mild or normal phenotypes and that this phenomenon is frequently observed in genes involved in glycosylation and carbohydrate metabolism [44]. Their work coupled with our observations implies that considerable differences exist between species in regard to single gene mutation and the subsequent phenotypic effect and that this gap may be particularly evident in genes involved in glycosylation.
Alternatively, this discrepancy may be the result of a non-cell autonomous rescue of early endoderm in vivo by neighboring cells utilizing STX in the absence of PST. Our results show that PST−/− cells form mesoderm and ectoderm similar to WT and it could be that these cells assist in the formation of endoderm in the embryo through mechanisms not present during in vitro differentiation.
The impact of glycosylation in embryonic development is not well understood and is an increasingly important topic. In addition to our findings there have been a number of recent reports documenting potential roles for sialylation and sialyltransferases in hPSC pluripotency and differentiation [45-49]. The full impact of these processes is to be determined but may point toward sialylated glycans playing key roles in early development.
Conclusion
Our observations indicate that the initial differentiation of hPSCs toward lineages of all three germ layers is marked by abundant cell surface polysialylation stemming from increases in polysialytransferase expression. This process is regulated epigenetically and transcriptionally via well-established developmental pathways and appears to be critical for cell fate specification.
Supplementary Material
Acknowledgements
We thank Dr. Karen J. Colley (University of Illinois at Chicago) for providing PST-v5 and STX-v5 plasmids and IP positive control lysate and Dr. Rita Gerardy-Schahn (Hannover Medical School) for providing mAb735 antibody. This work was supported by NIH grants P01 GM75334 and P41 GM103490.
Footnotes
Author Contributions: R.P.B.: conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing. Y.H.S., M.K., J.K.L., and A.V.N.: collection and assembly of data. A.V.N., K.W.M. and M.P.: conception and design. S.D.: conception and design, data analysis and interpretation, financial support, manuscript writing, and final approval of manuscript.
Disclosure of Potential Conflicts of Interest
We declare no potential conflict of interest at this time.
References
- 1.Varki A, RD C, JD E, et al. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.Haltiwanger RS, Lowe JB. Role of glycosylation in development. ANNUAL REVIEW OF BIOCHEMISTRY. 2004;73(1):491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 3.Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. NATURE REVIEWS NEUROSCIENCE. 2008;9(1):26–35. doi: 10.1038/nrn2285. [DOI] [PubMed] [Google Scholar]
- 4.Mühlenhoff M, Rollenhagen M, Werneburg S, et al. Polysialic acid: versatile modification of NCAM, SynCAM 1 and neuropilin-2. NEUROCHEMICAL RESEARCH. 2013;38(6):1134–1143. doi: 10.1007/s11064-013-0979-2. [DOI] [PubMed] [Google Scholar]
- 5.Close BE, Colley KJ. In vivo autopolysialylation and localization of the polysialyltransferases PST and STX. JOURNAL OF BIOLOGICAL CHEMISTRY. 1998;273(51):34586–34593. doi: 10.1074/jbc.273.51.34586. [DOI] [PubMed] [Google Scholar]
- 6.Finne J. Occurrence of unique polysialosyl carbohydrate units in glycoproteins of developing brain. JOURNAL OF BIOLOGICAL CHEMISTRY. 1982;257(20):11966–11970. [PubMed] [Google Scholar]
- 7.Phillips GR, Krushel LA, Crossin KL. Developmental expression of two rat sialyltransferases that modify the neural cell adhesion molecule, N-CAM. DEVELOPMENTAL BRAIN RESEARCH. 1997;102(2):143–155. doi: 10.1016/s0165-3806(97)00069-2. [DOI] [PubMed] [Google Scholar]
- 8.Hildebrandt H, Becker C, Mürau M, et al. Heterogeneous expression of the polysialyltransferases ST8Sia II and ST8Sia IV during postnatal rat brain development. JOURNAL OF NEUROCHEMISTRY. 1998;71(6):2339–2348. doi: 10.1046/j.1471-4159.1998.71062339.x. [DOI] [PubMed] [Google Scholar]
- 9.Lackie PM, Zuber C, Roth J. Polysialic acid of the neural cell adhesion molecule (N-CAM) is widely expressed during organogenesis in mesodermal and endodermal derivatives. DIFFERENTIATION. 1994;57(2):119–131. doi: 10.1046/j.1432-0436.1994.5720119.x. [DOI] [PubMed] [Google Scholar]
- 10.Curreli S, Arany Z, Gerardy-Schahn R, et al. Polysialylated Neuropilin-2 Is Expressed on the Surface of Human Dendritic Cells and Modulates Dendritic Cell-T Lymphocyte Interactions. JOURNAL OF BIOLOGICAL CHEMISTRY. 2007;282(42):30346–30356. doi: 10.1074/jbc.M702965200. [DOI] [PubMed] [Google Scholar]
- 11.Galuska SP, Rollenhagen M, Kaup M, et al. Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES. 2010;107(22):10250–10255. doi: 10.1073/pnas.0912103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galuska SP, Geyer R, Gerardy-Schahn R, et al. Enzyme-dependent Variations in the Polysialylation of the Neural Cell Adhesion Molecule (NCAM) in Vivo. JOURNAL OF BIOLOGICAL CHEMISTRY. 2007;283(1):17–28. doi: 10.1074/jbc.M707024200. [DOI] [PubMed] [Google Scholar]
- 13.Close BE, Mendiratta SS, geiger KM, et al. The Minimal Structural Domains Required for Neural Cell Adhesion Molecule Polysialylation by PST/ST8Sia IV and STX/ST8Sia II. JOURNAL OF BIOLOGICAL CHEMISTRY. 2003;278(33):30796–30805. doi: 10.1074/jbc.M305390200. [DOI] [PubMed] [Google Scholar]
- 14.Johnson CP, Fujimoto I, Rutishauser U, et al. Direct evidence that neural cell adhesion molecule (NCAM) polysialylation increases intermembrane repulsion and abrogates adhesion. JOURNAL OF BIOLOGICAL CHEMISTRY. 2005;280(1):137–145. doi: 10.1074/jbc.M410216200. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Dai G, Cheng YB, et al. Polysialylation promotes neural cell adhesion molecule-mediated cell migration in a fibroblast growth factor receptor-dependent manner, but independent of adhesion capability. GLYCOBIOLOGY. 2011;21(8):1010–1018. doi: 10.1093/glycob/cwr020. [DOI] [PubMed] [Google Scholar]
- 16.Rutishauser U, Watanabe M, Silver J, et al. Specific alteration of NCAM-mediated cell adhesion by an endoneuraminidase. THE JOURNAL OF CELL BIOLOGY. 1985;101(5):1842–1849. doi: 10.1083/jcb.101.5.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu H, Tomasiewicz H, Magnuson T, et al. The role of polysialic acid in migration of olfactory bulb interneuron precursors in the subventricular zone. NEURON. 1996;16(4):735–743. doi: 10.1016/s0896-6273(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 18.Landmesser L, Dahm L, Tang J, et al. Polysialic acid as a regulator of intramuscular nerve branching during embryonic development. NEURON. 1990;4(5):655–667. doi: 10.1016/0896-6273(90)90193-j. [DOI] [PubMed] [Google Scholar]
- 19.Ono S, Hane M, Kitajima K, et al. Novel Regulation of Fibroblast Growth Factor 2 (FGF2)-mediated Cell Growth by Polysialic Acid. JOURNAL OF BIOLOGICAL CHEMISTRY. 2012;287(6):3710–3722. doi: 10.1074/jbc.M111.276618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eggers K, Werneburg S, Schertzinger A, et al. Polysialic acid controls NCAM signals at cell-cell contacts to regulate focal adhesion independent from FGF receptor activity. JOURNAL OF CELL SCIENCE. 2011;124(19):3279–3291. doi: 10.1242/jcs.084863. [DOI] [PubMed] [Google Scholar]
- 21.Seidenfaden R, Krauter A, Hildebrandt H. The neural cell adhesion molecule NCAM regulates neuritogenesis by multiple mechanisms of interaction. NEUROCHEMISTRY INTERNATIONAL. 2006;49(1):1–11. doi: 10.1016/j.neuint.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Livingston BD, Jacobs JL, Glick MC, et al. Extended polysialic acid chains (n > 55) in glycoproteins from human neuroblastoma cells. JOURNAL OF BIOLOGICAL CHEMISTRY. 1988;263(19):9443–9448. [PubMed] [Google Scholar]
- 23.Schreiber SC, Giehl K, Kastilan C, et al. Polysialylated NCAM Represses E-Cadherin-Mediated Cell-Cell Adhesion in Pancreatic Tumor Cells. GASTROENTEROLOGY. 2008;134(5):1555–1566. doi: 10.1053/j.gastro.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka F, Otake Y, Nakagawa T, et al. Expression of polysialic acid and STX, a human polysialyltransferase, is correlated with tumor progression in non-small cell lung cancer. CANCER RESEARCH. 2000;60(11):3072–3080. [PubMed] [Google Scholar]
- 25.Roth J, Zuber C, Wagner P, et al. Presence of the long chain form of polysialic acid of the neural cell adhesion molecule in Wilms' tumor. Identification of a cell adhesion molecule as an oncodevelopmental antigen and implications for tumor histogenesis. THE AMERICAN JOURNAL OF PATHOLOGY. 1988;133(2):227. [PMC free article] [PubMed] [Google Scholar]
- 26.Amoureux M-C, Coulibaly B, Chinot O, et al. Polysialic Acid Neural Cell Adhesion Molecule (PSA-NCAM) is an adverse prognosis factor in glioblastoma, and regulates olig2 expression in glioma cell lines. BMC CANCER. 2010;10(1):91. doi: 10.1186/1471-2407-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Schulz TC, Sherrer ES, et al. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. BLOOD. 2007;110(12):4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLean AB, D'Amour KA, Jones KL, et al. Activin A Efficiently Specifies Definitive Endoderm from Human Embryonic Stem Cells Only When Phosphatidylinositol 3-Kinase Signaling Is Suppressed. STEM CELLS. 2007;25(1):29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- 29.Menendez L, Yatskievych TA, Antin PB, et al. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES. 2011;108(48):19240–19245. doi: 10.1073/pnas.1113746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. NATURE PROTOCOLS. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mali P, Yang L, Esvelt KM, et al. RNA-Guided Human Genome Engineering via Cas9. SCIENCE. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernstein BE, Mikkelsen TS, Xie X, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. CELL. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 33.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE Data in the UCSC Genome Browser: year 5 update. NUCLEIC ACIDS RESEARCH. 2012;41(D1):D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Showell C, Binder O, Conlon FL. T-box genes in early embryogenesis. DEVELOPMENTAL DYNAMICS. 2004;229(1):201–218. doi: 10.1002/dvdy.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernando RI, Litzinger M, Trono P, et al. The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. JOURNAL OF CLINICAL INVESTIGATION. 2010;120(2):533–544. doi: 10.1172/JCI38379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartwell KA, Muir B, Reinhardt F, et al. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES. 2006;103(50):18969–18974. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carey KL, Richards SA, Lounsbury KM, et al. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the Ran/TC4 GTPase mediates an essential function independent of nuclear protein import. THE JOURNAL OF CELL BIOLOGY. 1996;133(5):985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Artinger M, Blitz I, Inoue K, et al. Interaction of goosecoid and brachyury in Xenopus mesoderm patterning. MECHANISMS OF DEVELOPMENT. 1997;65:187–196. doi: 10.1016/s0925-4773(97)00073-7. [DOI] [PubMed] [Google Scholar]
- 39.Lehembre F, Yilmaz M, Wicki A, et al. NCAM-induced focal adhesion assembly: a functional switch upon loss of E-cadherin. EMBO JOURNAL. 2008;27(19):2603–2615. doi: 10.1038/emboj.2008.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinhold B, Seidenfaden R, Röckle I, et al. Genetic Ablation of Polysialic Acid Causes Severe Neurodevelopmental Defects Rescued by Deletion of the Neural Cell Adhesion Molecule. JOURNAL OF BIOLOGICAL CHEMISTRY. 2005;280(52):42971–42977. doi: 10.1074/jbc.M511097200. [DOI] [PubMed] [Google Scholar]
- 41.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. NATURE BIOTECHNOLOGY. 2009;27(3):275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckhardt M, Bukalo O, Chazal G, et al. Mice deficient in the polysialyltransferase ST8SiaIV/PST-1 allow discrimination of the roles of neural cell adhesion molecule protein and polysialic acid in neural development and synaptic plasticity. THE JOURNAL OF NEUROSCIENCE. 2000;20(14):5234–5244. doi: 10.1523/JNEUROSCI.20-14-05234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angata K, Long JM, Bukalo O, et al. Sialyltransferase ST8Sia-II assembles a subset of polysialic acid that directs hippocampal axonal targeting and promotes fear behavior. JOURNAL OF BIOLOGICAL CHEMISTRY. 2004;279(31):32603–32613. doi: 10.1074/jbc.M403429200. [DOI] [PubMed] [Google Scholar]
- 44.Liao B-Y, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES. 2008;105(19):6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y-C, Stein JW, Lynch CL, et al. Glycosyltransferase ST6GAL1 contributes to the regulation of pluripotency in human pluripotent stem cells. SCIENTIFIC REPORTS. 2015:1–13. doi: 10.1038/srep13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alisson-Silva F, de Carvalho Rodrigues D, Vairo L, et al. Evidences for the involvement of cell surface glycans in stem cell pluripotency and differentiation. GLYCOBIOLOGY. 2014;24(5):458–468. doi: 10.1093/glycob/cwu012. [DOI] [PubMed] [Google Scholar]
- 47.Swindall AF, Londono-Joshi AI, Schultz MJ, et al. ST6Gal-I Protein Expression Is Upregulated in Human Epithelial Tumors and Correlates with Stem Cell Markers in Normal Tissues and Colon Cancer Cell Lines. CANCER RESEARCH. 2013;73(7):2368–2378. doi: 10.1158/0008-5472.CAN-12-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y-C, Nakagawa M, Garitaonandia I, et al. Specific lectin biomarkers for isolation of human pluripotent stem cells identified through array-based glycomic analysis. CELL RESEARCH. 2011;21(11):1551–1563. doi: 10.1038/cr.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tateno H, Toyota M, Saito S, et al. Glycome Diagnosis of Human Induced Pluripotent Stem Cells Using Lectin Microarray. JOURNAL OF BIOLOGICAL CHEMISTRY. 2011;286(23):20345–20353. doi: 10.1074/jbc.M111.231274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.