Abstract
Objective
The goal of this study is to devise a machine learning framework to assist care coordination programs in prognostic stratification to design and deliver personalized care plans and to allocate financial and medical resources effectively.
Materials and Methods
This study is based on a de-identified cohort of 2,521 hypertension patients from a chronic care coordination program at the Vanderbilt University Medical Center. Patients were modeled as vectors of features derived from electronic health records (EHRs) over a six-year period. We applied a stepwise regression to identify risk factors associated with a decrease in mean arterial pressure of at least 2 mmHg after program enrollment. The resulting features were subsequently validated via a logistic regression classifier. Finally, risk factors were applied to group the patients through model-based clustering.
Results
We identified a set of predictive features that consisted of a mix of demographic, medication, and diagnostic concepts. Logistic regression over these features yielded an area under the ROC curve (AUC) of 0.71 (95% CI: [0.67, 0.76]). Based on these features, four clinically meaningful groups are identified through clustering - two of which represented patients with more severe disease profiles, while the remaining represented patients with mild disease profiles.
Discussion
Patients with hypertension can exhibit significant variation in their blood pressure control status and responsiveness to therapy. Yet this work shows that a clustering analysis can generate more homogeneous patient groups, which may aid clinicians in designing and implementing customized care programs.
Conclusion
The study shows that predictive modeling and clustering using EHR data can be beneficial for providing a systematic, generalized approach for care providers to tailor their management approach based upon patient-level factors.
Index Terms: Electronic health records, secondary use, predictive modeling, patient stratification, chronic disease management
I. Introduction
Current models of ambulatory care are generally neither cost-effective nor adequately patient-centric. Chronic diseases are expensive and require careful management in the ambulatory care setting to achieve the best possible patient outcomes [1], [2]. Presently, the management of chronic diseases consumes over 90% of Medicare expenditure [3] and amounts to over $1.5 trillion per year [4]. The most prevalent chronic disease is hypertension, which affects over 30% of American adults and accounts for almost $70 billion in direct costs annually [5], [6].
Hypertension is the primary risk factor for stroke and a major risk factor for other debilitating diseases including coronary heart disease, renal failure, and heart failure [6]–[10]. Relatively short durations of uncontrolled hypertension, as brief as several months, have been associated with adverse clinical outcomes [11]. A reduction in systolic blood pressure (SBP) as small as 2 mmHg has been shown to reduce the risk of adverse clinical outcomes at a population-based level [11]–[13]. In addition, demographic factors such as race, gender, and age are important factors in the development and progression of hypertension [14].
Due to the complex interplay of multiple disease and patient-level factors, successful management of hypertension is rarely a “one size fits all” situation and is often more effective when tailored to individual patient needs, local culture, and available resources [15]. Chronic care management plans are more likely to succeed when they incorporate personalized care coordination [16], [17]. Yet, to ensure that coordination is personalized, patient information must be collected, shared, analyzed and leveraged to inform actions within and among a well-organized care team (which can include providers and staff from primary care and specialty medicine, nursing, pharmacy, social work, administration, family and the patient) [18], [19].
Identification of patient-level factors that enable the tailoring of hypertension management plans has proven difficult. We believe, however, that such factors may be uncovered through the analysis of existing data in electronic health records (EHRs). As such, the primary goal of our investigation is to devise a generalizable machine learning approach for EHR data to accomplish this task. To do so, we worked with data from a pilot care coordination program instituted at the Vanderbilt University Medical Center (VUMC). This program was specifically designed to improve the care of patients with chronic disease. It focuses on the management of patients with hypertension and various comorbidities. Since the pilot program places an emphasis on comprehensive data collection, the solicited data carries great potential for informing personalized, targeted therapy. Although prior work has shown that EHR data can be leveraged for the predictive modeling of diseases [20], to the best of our knowledge there are currently no standardized methods for automated stratification of patients with minimal human supervision.
We propose and implement a machine learning framework for identifying risk factors for targeted outcomes and for stratifying patients based upon those risk factors. We apply this framework on a cohort of 2,521 patients enrolled in the pilot program. Our approach consists of two main components:
Risk factor identification: We identify risk factors in the patient cohort that are associated with lowering the patients’ median blood pressure (BP) by at least 2 mmHg (a change shown to be associated with reduction of mortality in large populations) [11]–[13]. Based on these risk factors, we develop a logistic regression classifier for predicting changes in BP.
Patient stratification: We illustrate that clustering the patients can segment the cohort into four distinct groups, each of which exhibits a different disease subtype. We anticipate that the definition of such groups may lead to the design of more specific treatments and care management plans.
II. Background and Significance
Hypertension is a chronic disease whose management is influenced by a variety of factors including age, diet, exercise, drug use, body habitus, genetic factors, and the presence of comorbidities. Current approaches to hypertension management commonly treat all patients using similar management plans based upon standard treatment guidelines [11]. However, to achieve best possible patient outcomes, it is important to develop approaches to care that address unique aspects of specific patient subgroups, or clusters. Recent initiatives such as the Strategic Health IT Advanced Research Projects (SHARP) Area 4 Consortium [21], [22] have helped to facilitate this process by establishing standardized data formats for secondary uses. Our study aims to leverage EHR data specifically to identify and analyze subgroups of hypertension patients with distinct individual characteristics.
A. EHR-driven phenotyping for hypertension control and outcome prediction
Machine learning-based predictive models using data from medical records and clinical databases have been achieved for various chronic diseases, including type 2 diabetes [23], asthma [24], chronic kidney disease [25], and rheumatoid arthritis [26]. However, the prediction of hypertension control patterns has not been studied widely. The present study builds on prior work, which introduced a predictive model for detecting changes in hypertension control status using an earlier version of the same pilot dataset [20]. In that study, EHR records for medications, labs and ICD-9 codes were used as features to predict whether or not patients’ blood pressures will change (between the status of “in control” or “out of control”, or vice versa, as determined by a clinician). One element of the present study employs a similar method, but is aimed to predict specific magnitudes of BP changes. Furthermore, our current study goes one step further by applying clustering to automatically segment the patients into subgroups with distinct characteristics that reflect disease subtypes.
One challenging aspect in characterizing and managing hypertension is the fact that patients with similar clinical presentations may demonstrate significant variation in BP changes among each other. Gaining insight about which subgroups of these patients might benefit from more frequent monitoring and intervention would be helpful to clinicians as they participate in and allocate care coordination resources. Furthermore, clinicians could benefit from a more accurate predictive model for individual patients when designing treatment plans.
B. EHR-driven phenotyping for detection of disease subtypes
Current research on clinical phenotyping aims to develop methods for segmenting cohorts of patients into subgroups with distinct characteristics using EHR data. However, many of these approaches often involve expert specification and are not fully automated [27]–[30]. Other methods take a feedback-based approach with active learning [31]. Furthermore, some studies have applied machine learning for feature selection [32], [33].
Regardless of the method, most implementations of phenotyping are leveraged to detect patients with certain diseases rather than subgroups of patients within a disease (e.g., finding patients in a cohort who have hypertension, rather than finding patients in a hypertension cohort with different characteristics) [34], [35]. For example, the NIH-sponsored Electronic Medical Records and Genomics (eMERGE) network devised a phenotyping algorithm for resistant hypertension based upon EHR data from patients at the Marshfield Clinic [35]. Ho and colleagues [36] showed that phenotyping for disease subtypes can be accomplished with higher order tensor factorization. However, this approach may be limited in its scalability because it is exponential in complexity with respect to the dimensionality of the feature space, thus leading to challenges in analyzing a large number of features per patient. Furthermore, Ho et al. utilized only medications and diagnosis codes as features. In this work we extend the feature set to include vital signs and demographic information, with which we aim to enhance the stratification of patients into subgroups. We believe that identifying refined subgroups can be informative for important tasks in patient management, such as the identification of patients who are more costly to manage, or who may be more responsive to certain treatment protocols. Thus, while the use of phenotyping for identifying disease subtypes remains largely an unsolved problem, phenotyping could provide a valuable contribution towards hypertension management. Our current study provides a first step towards this goal.
III. Materials and Methods
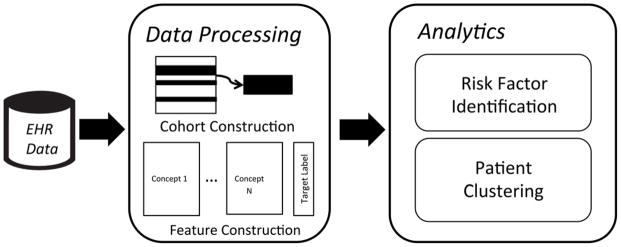
This section describes the data used, as well as details regarding the analytic pipeline, for the study. As shown in Figure 1, de-identified data from the VUMC EHR system are processed in cohort construction (where the study cohort is identified) and feature construction (where the feature variables are computed for all patients in the study cohort). The resulting data are fed into an analytics module for analysis comprised of risk factor identification and patient clustering. Next we describe the details in the analytic pipeline.
Fig. 1.
An illustration of the data processing and analytic pipeline utilized in this study.
A. Data description
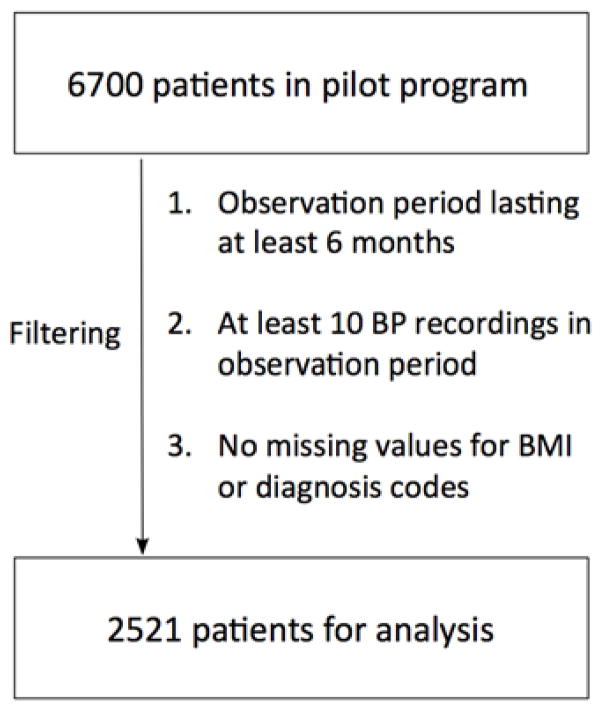
All data was collected from the de-identified copy of the VUMC EHR [37], in which all date information is randomly shifted between −1 and −356 days on a per patient basis to preserve the relative time between events. Data were collected from a group of 6,700 primary care patients with hypertension enrolled in the pilot program, which was filtered down to 2,521 patients (see Data Processing subsection). All enrolled patients were diagnosed with hypertension, as determined by an initial screening by ICD-9 codes and confirmed with manual chart review. Patients were followed for a median of 5.9 years before and 0.9 years after joining the program.
Table 1 summarizes the characteristics of the pilot program cohort. For each patient, the following data were collected longitudinally: demographics, BP, body mass index (BMI), clinic and hospital medications orders, medications extracted from clinical documents [38], and ICD-9 codes. We analyzed the patients according to several groupings: 1) all patients, 2) patients with a positive change in mean arterial pressure (MAP), indicated by reduction of median MAP by at least 2 mmHg after enrollment in the pilot program, and 3) patients without such a reduction in MAP (i.e., either no change in MAP or an increase in MAP). Our rationale for 2 mmHg reduction as a filtering criteria is based on various epidemiological studies that show such a decrease is associated with reduced rates of stroke and coronary heart disease, two conditions which lead to increased mortality [11]–[13]. Specifically, a 2 mmHg decrease in systolic blood pressure can result in approximately 10% reduction in mortality from stroke and 7% reduction in mortality from ischemic heart disease [39].
Table 1.
A summary of the characteristics of patient records used in this study. Features are reported for all patients, as well as subsets of patients whose mean arterial pressure decreased by 2mmHg after pilot program enrollment, and patients whose mean arterial pressure did not decrease by at least 2mmHg.
| n = | All Patients | MAP decreased by at least 2mmHg | No decrease in MAP of least 2mmHg | |
|---|---|---|---|---|
| 2521 | 1198 (47.5%) | 1323 (52.5%) | ||
| Demographics | Age, years (mean) | 67.6 | 67.3 | 67.9 |
| Gender (% male) | 45.00% | 44.90% | 45.10% | |
| Race (% white) | 82.80% | 81.50% | 84.00% | |
| Race (% black) | 15.90% | 17.20% | 14.80% | |
|
| ||||
| Disease Conditions | Diabetes | 35.40% | 36.00% | 34.80% |
| Heart Failure | 8.40% | 8.30% | 8.50% | |
|
| ||||
| Vital Signs | BMI (median) | 30.5 | 30.9 | 30.2 |
B. Data processing
1) Cohort Construction
Figure 2 describes the criteria for filtering patients to construct the study cohort. The observation period is the period of two years before the pilot program engagement date; engagement period is the time after the pilot program engagement date. We use patient data in observation periods to construct feature vectors and use the BP change between the observation and engagement periods to define the outcome variable.
Fig. 2.
The data filtering process with inclusion criteria for generating cohorts for analysis.
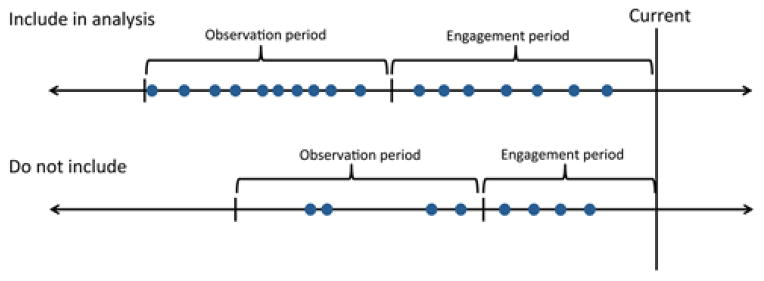
Figure 3 illustrates the inclusion criteria for patients based upon the observation and engagement periods. Patients were included in the analysis if BP recordings were available on at least 10 distinct days over a window of at least 6 months during the observation period. This step aims to filter out patients with sparse data whose samples may skew the results. Patients were filtered from further consideration if demographic information was missing and vital signs other than blood pressure (i.e., BMI) were not available during either period. We selected these filters to enhance the effectiveness of the predictive models. The filters were based on phenotype validation techniques from the eMERGE network, which illustrate that the confirmation of patients with certain a phenotype is enhanced when their inpatient visits span a specified period of time and their status is confirmed over multiple clinic visits [35], [40].
Fig. 3.
An illustration of the inclusion criteria for patients in the analysis. Patients were included if there were at least 10 blood pressure recordings made on at least 10 distinct dates spanning a period of at least 6 months during the observation period.
2) Feature Construction
Features used were drawn from several different categories: i) demographics, ii) comorbid disease conditions, iii) vital signs, iv) medication status, and v) phenome-wide association study (PheWAS) phenotype code status [41].
Features in the demographics category include: a) age in years, b) gender, and c) self-reported race. Features in the vital signs category include systolic BP (SBP), diastolic BP (DBP), MAP, and BMI. Features in the medication category include any recordings for medications belonging to one of 13 classes of hypertension medications (see table 2 for details). We note that several of the medication classes correspond to combinations of medications (e.g., thiazide and beta blocker). These combinations are specified in classes distinct from the individual medications because they work synergistically and have been clinically shown to reduce blood pressure more than the separate medications [11]. Features in the PheWAS code status category correspond to PheWAS codes in the encoding described by Denny et al. [41], [42] PheWAS codes represent groupings of ICD-9 codes that are collectively indicative of similar medical conditions.
Table 2.
Example medications corresponding to each of the 13 classes of hypertension medications used in the study are listed.
| Medication Class | Example |
|---|---|
| ACE inhibitor or angiotensin receptor blocker | Lisinopril, losartan |
| Aldosterone antagonists | Spironolactone |
| Alpha antagonists | Prazosin |
| Beta blockers | Atenolol |
| Central alpha agonists | Clonidine |
| Dihydropyridine calcium channel blockers | Amlodipine |
| Diuretic combination | Aldactazide |
| Diuretics | Hydrochlorothiazide |
| Hydralazine | Hydralazine |
| Minoxidil | Minoxidil |
| Non-dihydropyridine calcium channel blockers | Diltiazem |
| Thiazide and ACE inhibitor or angiotensin receptor blocker | Hydrochlorothiazide and lisinopril or losartan |
| Thiazide and beta blocker | Hydrochlorothiazide and metropolol tartrate |
Features used in the analysis were generated by aggregating across all data points recorded during the observation period. Table 3 provides a summary of the aggregation methods used for each feature. The following features were generated by calculating the median values: i) SBP, DBP, MAP before initial engagement with the care program; ii) SBP, DBP, MAP after initial engagement with the program; and iii) BMI. Change in BMI was calculated by comparing mean values from the observation period before program initiation, with mean values from the observation after program initiation. For all of the above numerical features, we scaled the data by substituting the z-score for each observation.
Table 3.
A summary of the aggregation functions applied to the features in patient event sequences over their time windows. (BMI: body mass index; BP: blood pressure; PheWAS: phenome-wide association study).
| Category | Feature | Aggregation |
|---|---|---|
| Demographics | Age | Binary |
| Sex | Binary | |
| White | White | |
| Black | White | |
|
| ||
| BP | Systolic | Median per patient, mean across patients |
| Diastolic | Median per patient, mean across patients | |
| MAP | Median per patient, mean across patients | |
|
| ||
| Disease Conditions | Diabetes | Binary |
| Heart Failure | Binary | |
|
| ||
| Vital Signs | BMI | Median |
| Medications | Each of 13 classes of hypertension medications | Binary |
| PheWAS | PheWAS (phenotype) codes | Binary |
Demographic features pertaining to self-reported race and ethnicity were treated as binary variables: i) White, ii) Black, iii) Asian, and iv) Hispanic. Features pertaining to comorbidities, in the form of diabetes and heart failure (HF), were treated as binary variables indicating whether or not the patient was currently being treated for the disease in the pilot program.
The features pertaining to medication status were also denoted by binary variables. Each of the hypertension medication classes was treated as a separate feature. The appearance of at least one recording for a medication class in the observation period denoted a positive value for that feature.
The features pertaining to PheWAS phenotype status were obtained by counting occurrences of ICD-9 codes related to each PheWAS code. Features were generated from PheWAS codes at the disease category level (188 distinct categories). The presence of at least one recording for a PheWAS disease category during the observation period denotes a value of 1, otherwise a value of 0. In total, we constructed 213 features for each patient.
C. Analytics
The analytics module consists of two components: 1) Risk Factor Identification and 2) Patient Clustering. We define the target outcome as a binary indicator representing whether patients exhibit a decrease in MAP by at least 2 mmHg. A reduction of at least 2 mmHg was defined as the target outcome because such a decrease on the population level has been shown in epidemiological studies to be associated with major comorbidities (6% decrease in mortality due to stroke, 4% decrease in mortality due to coronary heart disease, and 3% decrease in all-cause mortality) [11]. Thus, such a reduction was considered a favorable outcome.
In Risk Factor Identification, we perform feature selection to identify the features deemed to be most predictive of the outcome, and classification to determine the accuracy of the predictions for the resulting model. In Patient Clustering, we use the most predictive features to cluster patients into coherent groups.
1) Risk Factor Identification
We investigated features that predicted subjects would achieve a minimum reduction in BP of 2 mmHg. We performed a forward stepwise logistic regression with all of the features. We performed 10-fold cross-validation to predict a binomial target using a logistic regression classifier. The targets for the classification model were denoted as follows:
1: decrease in MAP by at least 2 mmHg and
0: otherwise.
We use the Bayesian Information Criteria (BIC) statistic as the metric for measuring the goodness of the current model [43]. The BIC is formulated as the following:
where
refers to the value of the log likelihood loss function across all patients, such that β is a vector consisting of the parameters for the likelihood function,
yi is the target of patient i in the dataset, xi is a vector consisting of the values of the features for patient i, and β is the set of feature weights to be optimized. N refers to the number of patients used in a training set, and d refers to the number of features used in the model.
During each stage of the classification, the feature that minimizes BIC was added to the model until no additional feature reduced the BIC. Note that since the 10 experiment runs were conducted through 10-fold cross-validation, the selected features from fold to fold can vary. For the subsequent clustering task, we chose features that were consistently selected at least 8 times out of 10 runs as the predictive features.
To compare the performance of the stepwise logistic regression against other standard classification models, we also performed 10-fold cross-validation using decision tree, random forest, support vector machine (SVM) and artificial neural network classifiers. We used a J48 tree for decision tree and an RBF kernel with c = 1 for SVM.
2) Patient Clustering
Although risk factor identification can identify factors leading to disease, one would still need to manually stratify the patients into groups based upon the risk factors, a task that is cumbersome with a large number of features. Such a process would require one to differentiate groups in order to tailor treatment and to decide how to effectively allocate resources. Additionally, to aid in reducing waste and maximizing efficient usage of limited resources (e.g., time, labs and clinical specialists), there should be relative similarity among each group. To automate the process, we applied a clustering algorithm to stratify the cohort based on the features deemed to be predictive according to the stepwise regression. Specifically, we used the predictive features selected in the risk factor identification step as input to the clustering algorithm.
To cluster the patients, we applied a hybrid method that combined hierarchical and model-based clustering with expectation maximization (EM). We applied the clustering implementation from the Mclust package in R [44], [45].
We initialized clusters for patients using hierarchical clustering. Afterwards, to assign patients x1, …, xn into clusters, we maximized the mixture likelihood:
where xi is the feature vector for patient i, fk (xi|θk) is the density function representing the probability that a patient i belongs to cluster k, θk represents the corresponding parameters of cluster k, G represents the number of clusters in the mixture, and τk is the probability that a patient belongs to cluster . We assume that fk (xi|θk) is a multivariate Gaussian, where the parameters θk correspond to a mean vector μk and covariance matrix Σk, so that
where p is the number of features used in the model.
We fit the data to several Gaussian-mixture models with a different number of clusters and distribution shapes (e.g., spherical, diagonal, and ellipsoidal) as determined by the covariance matrix. To compare the models and associated number of clusters, the BIC was approximated for each model M as follows:
where ℓM(x, θ̂) is the maximized mixture likelihood for the model, and mM is the number of features in the model. We selected the combination of model shape and number of clusters that yielded the minimum BIC value.
IV. Results
In this section, we review the results for both parts of the analytics module, which includes risk factor identification and patient clustering.
A. Risk Factor Identification
All experiments were run on a MacBook Pro with a 2.4 GHz processor with 4 cores and 16 GB RAM. We identified all features that were added to the forward stepwise logistic regression model during each fold in cross validation. On average, the learning of a model required approximately 3 minutes. There were 9 features out of 213 total that were selected in the training models for at least 8 of 10 experiment runs. These were comprised of a mix of demographic features: i) age and ii) MAP before program engagement; medications: iii) aldosterone antagonists, iv) beta blockers, and v) central alpha agonists; and PheWAS codes: vi) non-ischemic and non-pulmonary heart disease, vii) disorders of female genital tract (other than inflammatory diseases of the pelvis), viii) symptoms involving the head and neck, and ix) disorders of pancreatic internal secretion (other than diabetes). The cross-validation for the feature selection step yielded an AUC (area under the receiver operating characteristic curve) of 0.71 (95% CI: [0.67,0.76]). The regression coefficients calculated in each fold of cross validation are reported in table A.1.
Additionally, we applied the standard classification methods (decision tree, random forest, support vector machine, artificial neural network) using the nine features identified with the stepwise logistic regression. The mean AUC values (across all folds in 10-folds) for these models were 0.67, 0.67, 0.51, and 0.70, respectively.
The demographic and vital signs features were both consistent with common knowledge regarding hypertension – older patients usually require more extensive efforts in controlling hypertension due to an increase in SBP with age and comorbid conditions; and improvement in BP may vary with a dependency on the initial severity of hypertension [46]–[49].
The medication status features are also meaningful from a clinical perspective. Beta blockers are used in both mild and severe forms of hypertension. On the other hand, aldosterone antagonists and central alpha agonists are usually given in later stages of hypertension treatment after other medications have been used in the regimen. Aldosterone antagonists such as spironolactone and eplenerone are commonly given in the setting of heart failure. Central alpha agonists such as clonidine may be prescribed after other antihypertensive drugs have failed [11]. Therefore, it is possible that these medications can be invoked as features to predict whether or not patients will exhibit a decrease in MAP by at least 2 mmHg.
Regarding the PheWAS codes, the feature “non-ischemic, non-pulmonary heart disease” has an association with hypertension status. Examples of such diseases include pericarditis, aortic stenosis, and aortic regurgitation. The other PheWAS codes do not necessarily indicate any obvious associations with hypertension. However, the feature “other disorders of female genital tract” could be a proxy for the feature of female gender (and associated conditions), which may play a factor in hypertension status. In addition, this feature may reflect differences in medication regimens between male and female patients [50]–[52].
B. Patient Clustering
We added the 9 features identified in the feature selection step as features in the clustering algorithm. The patients were subsequently grouped into four distinct clusters (C1, C2, C3, C4) based on the BIC criterion. For each cluster, we report BP statistics in Table 4 and summary statistics in Table 5.
Table 4.
A summary of blood pressure metrics (before program initiation, after program initiation, and the change in blood pressure). All values reported are means across the population subset, as well as across clusters identified through the clustering algorithm. Blood pressure values reflect the population mean of patient-level median blood pressures (mmHg). Reported P-values correspond to a Kolmogorov-Smirnov test that compares the distribution of blood pressure values before and after the care program was initiated.
| BEFORE | AFTER | CHANGE | p-value | ||
|---|---|---|---|---|---|
| All Patients (n = 2521) | SYSTOLIC | 131.6 | 130.6 | −1.0 | 1.3 × 10-3 * |
| DIASTOLIC | 74.2 | 72.8 | −1.4 | 2.9×10-6 * | |
| MAP | 93.4 | 92.1 | −1.3 | 2.7×10-9 * | |
|
| |||||
| Cluster 1 (n = 1697) | SYSTOLIC | 130.7 | 129.8 | −0.9 | 1.8 × 10-3 * |
| DIASTOLIC | 74.3 | 73.0 | −1.3 | 9.5 × 10-5 * | |
| MAP | 93.2 | 92.0 | −1.2 | 6.8 × 10-7 * | |
|
| |||||
| Cluster 2 (n = 392) | SYSTOLIC | 133.9 | 132.3 | −1.6 | 3.4 × 10-3 * |
| DIASTOLIC | 76.6 | 74.6 | −2.0 | 0.04 * | |
| MAP | 95.7 | 93.9 | −1.9 | 9.1 × 10-3 * | |
|
| |||||
| Cluster 3 (n = 204) | SYSTOLIC | 135.5 | 134.4 | −1.0 | 0.34 |
| DIASTOLIC | 72.0 | 70.4 | −1.6 | 0.12 | |
| MAP | 93.3 | 91.8 | −1.5 | 0.34 | |
|
| |||||
| Cluster 4 (n = 228) | SYSTOLIC | 130.6 | 130.3 | −0.4 | 0.63 |
| DIASTOLIC | 71.4 | 70.2 | −1.2 | 0.29 | |
| MAP | 91.4 | 90.3 | −1.1 | 0.34 | |
Table 5.
A summary of the results of the patient clustering process. The mean values are reported for mean arterial pressure (MAP) and median values are reported for body mass index (BMI).
| General Features | Cluster: | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|---|
| n = | 1697 | 392 | 204 | 228 | ||
| Demographics | Age, years | 67.8 | 63.2 | 71.9 | 69.9 | |
| Gender (% male) | 52.6% | 14.0% | 38.7% | 47.8% | ||
| Race (% white) | 85.7% | 76.8% | 75.0% | 78.5% | ||
| Race (% black) | 12.8% | 22.2% | 24.5% | 20.6% | ||
|
| ||||||
| Vital Signs | BMI (kg/m2) (median) | 30.3 | 30.7 | 30 | 31.1 | |
|
| ||||||
| Features Used in Clustering | ||||||
|
| ||||||
| Demographics | Age | 67.8 | 63.2 | 71.9 | 69.9 | |
|
| ||||||
| Vital Signs | MAP before pilot enrollment (mmHg) | 93.2 | 95.7 | 93.3 | 91.4 | |
|
| ||||||
| Medications | Aldosterone antagonists | 3.1% | 0.5% | 43.1% | 75.4% | |
| Beta blockers | 49.3% | 44.9% | 83.3% | 76.8% | ||
| Central alpha agonists | 0.0% | 38.8% | 90.2% | 29.8% | ||
|
| ||||||
| PheWAS Codes | Other forms of heart disease (non-ischemic, non-pulmonary heart disease) | 39.4% | 0.0% | 83.8% | 72.4% | |
| Other disorders of female genital tract (other than inflammatory diseases of pelvis) | 6.1% | 69.9% | 34.3% | 1.3% | ||
| Symptoms involving head and neck | 23.8% | 28.8% | 9.8% | 25.4% | ||
| Other disorders of pancreatic internal secretion (other than diabetes) | 0.4% | 2.0% | 0.0% | 12.7% | ||
With respect to demographics, comorbid disease conditions and vital signs, C3 and C4 appear to correspond to patients with more severe clinical makeups. Higher percentages of these clusters contain patients who also have diabetes (42.2% and 48.8%, respectively, compared to 33.5% and 32.1% for C1 and C2, respectively) and heart failure (27.0% and 24.1% for C3 and C4 respectively, compared to 5.8% and 1.3% for C1 and C2, respectively), compared to the other clusters. Furthermore, C3 contained the highest average SBP, which was 135.5 mmHg (25–75th percentile: 128–143) before and 134.4 mmHg (25–75th percentile: 126–144) after pilot program engagement, compared with 131.6 mmHg before and 130.6 mmHg after for the entire patient cohort.
With respect to features used specifically in the clustering, the patients in C3 and C4 were older (average age of 71.9 and 69.9 years, respectively) compared to C1 and C2 (average age of 67.8 and 63.2, respectively). Furthermore, a large proportion of the patients in C3 and C4 were treated with beta blockers, aldosterone antagonists, and central alpha agonists. In C3, 90.2% of the patients were treated with central alpha agonists, while 75.4% of patients in C4 were treated with aldosterone antagonists. These observations further support the notion that C3 and C4 consist of patients with more severe disease profiles that may benefit from a tailored care coordination strategy.
With respect to PheWAS codes, a much higher percentage of patients in C3 and C4 (83.8% and 72.4%, respectively) were diagnosed with non-ischemic, non-pulmonary heart disease, compared to C1 and C2 (39.4% and 0%, respectively). When comparing C1 and C2, which exhibit medication status characteristics that suggest lower disease severity, it should be noted that 39.4% of patients in C1 have non-ischemic, non-pulmonary heart disease compared to 0% for C2. Furthermore, 69.9% of patients in C2 were positive for other disorders of the female genital tract (other than inflammatory diseases of the pelvis). Since there is not an obvious biological explanation for female patients with genital tract disorders to cluster together, this result may simply reflect the 86% female gender in this cluster. Taken together, these results show that while C1 and C2 represent patients with less severe disease conditions, they can still be differentiated.
V. Discussion
A. Challenges in Distinguishing Disease Subtypes
There are several notable challenges regarding disease subtyping which apply to our study. Our study included patients with controlled and uncontrolled hypertension. However, it may be insightful to stratify patients for analysis based upon control status. Clinically, it is difficult to predict a patient’s future hypertension control status due to the complex nature of blood pressure, which has significant inherent variability. In the pilot, clinicians determined patient control based upon a combination of BP recordings taken at home and in the clinic. Hypertension is defined as clinic BP of 140/90 mmHg or higher (SBP/DBP, either value greater than or equal to the cutoff), or a home average over 135/85 mmHg [11]. However, the actual target blood pressure and control rates can be impacted by the patient’s age and comorbidities such as diabetes, kidney disease, and overall health.
Furthermore, our study measured the differences in aggregated BP values during the time periods before program engagement and during the program engagement. There may be changes due to trends over these periods which were not captured by our study.
Another limitation lies in the fact that patients were drawn from a specific, small group of patients and primary care providers, so the results may not reflect the population at large. Finally, our method aims to stratify patients based upon population-based target outcomes in an automated fashion. Our method does not provide specific decision support for individuals, although it should motivate future work in that direction. Nonetheless, our method was able to identify relevant risk factors and segment patients into clinically meaningful clusters. The clustering algorithm yielded realistic patient subgroups that can be adequately explained by differences in disease severity.
The challenge of distinguishing disease subtypes is rooted in the complex nature of hypertension, which may be primary (found in isolation) or secondary (caused by other complex conditions including endocrine diseases (e.g., diabetes, Cushing’s syndrome, congenital adrenal hyperplasia), renal disease (e.g., renal tubular defects, polycystic kidney disease), or tumors (e.g., multiple endocrine neoplasia). Table 1 shows that one may not be able to segregate patients effectively by using obvious risk factors for hypertension such as demographic features (sex, age, race), or the presence of common comorbidities, further supporting the fact that an automated method for identifying risk factors is needed in order to more effectively predict differences in patient outcomes. The nature of the disease may explain why the AUC of 0.71 found in the risk factor identification analysis is lower than in the application of logistic regression and feature selection towards more straightforward use cases such as the identification of rheumatoid arthritis or peripheral artery disease [32], [33], [53]. More importantly, this finding underscores the importance of identifying disease subtypes, each of which may exhibit hypertension with varying degrees of severity. Future research in predictive modeling and phenotyping of disease subtypes should address the aforementioned challenges.
B. Challenges Regarding the Use of EHR data
Although there is an opportunity to leverage existing data in EHRs to compose improved predictive models for patients in care coordination programs, this approach faces several notable challenges. First, EHR data is often limited to events observed upon a patient visit to a clinic or specific events (e.g., BP) which transpire outside of the clinic and which patients are instructed to record. Second, EHR data is often limited to the diagnoses, labs, and treatments prescribed to patients. While additional information may be documented in the history, physical exam, or the progress notes within an EHR, chronic diseases such as hypertension are influenced by patient behaviors outside of the clinic (e.g., diet and exercise) [11] which are not captured consistently in EHRs [54], [55]. Third, EHR data by its nature is often leveraged in a secondary fashion, which raises concerns about the reliability of the data [55]. In particular, BP measurements contain high variability depending on a variety of factors. BP measurements may vary with quality of the measurement techniques, for example patient positioning, cuff size, and an appropriate rest period. In addition, variation in BP occurs due to diurnal variation as well as medications, body habitus, smoking, exercise, and salt intake.
It should also be recognized that the size of the study carries certain limitations as well. Our patient clustering methods yielded groups of varying sizes – the largest cluster contained 1697 patients, while the smallest cluster contained 204 patients. While the clusters exhibited differences in BP change in response to the pilot program, it is difficult to compare outcomes in separate groups with vast differences in sample sizes because different magnitudes of change are required in order to meet significance thresholds. Furthermore, there may be other, less common subclasses of patients with hypertension that were not detected in our clustering analysis due to the small sample size. This issue may be ameliorated when clustering is performed on a larger scale. In addition, it is possible that the sample size may have prevented the set of features identified in the risk factor identification step from including less commonly seen risk factors. However, it is unclear that adding more features in the stepwise logistic regression model would improve the performance significantly. Finally, there may be a selection bias in the study as a result of the data filtering process, which decreased the size of the original cohort of 6700 patients down to 2521. It is possible that the patients with complete records may not be representative of the true underlying population because they may, for instance, be more compliant with follow-up clinic visits and blood pressure readings.
C. Towards an Improved Model for Care Coordination
Care coordination programs have evolved over time to follow changes in standards of care and incorporate larger populations of patients. However, the evidence suggests that chronic disease management programs may have different outcomes for specific subgroups of patients, as illustrated by the differences in BP changes across the patient subgroups identified through clustering. The results of the clustering analysis show that clustering of patients in a care program can stratify patients by profile using a collection of several features identified via stepwise regression for feature selection. This suggests that it is worthwhile to conduct more detailed investigations regarding clustering of patients for purposes such as assignment to different treatment regimens and triaging of patients based upon risk assessment. In addition, our results suggest that the EHR may be a valuable resource in the future to identify patients with similar risk for poor clinical outcomes and determine when and how to spend limited healthcare resources. Results from studies such as ours could be used to explore differences in responsiveness of patients to care coordination programs. Such insights could be leveraged for making iterative improvements to care management programs and personalized treatment plans.
The improvement of care coordination programs could be accelerated by adoption of analytics methods for medical decision making. Although our results could have been obtained without analyzing EHR data, the fact that we have successfully obtained such results using machine learning approaches shows that secondary use of EHR data can be beneficial for the purposes of care coordination. Also, it is important to note that our unique combination of stepwise logistic regression and clustering showcases a framework for automated patient stratification requiring minimal human supervision, which may be helpful for organizations managing large care coordination programs. Future studies should further refine specific subareas of this framework.
VI. Conclusion
This study identified relevant features for predicting a patient’s BP response to a chronic care management program via stepwise regression and clustered the patients using these features. Our clustering algorithm stratified the patients based upon differences in characteristics relevant to treatment outcomes, illustrating the potential of using EHR data to inform the personalization of treatment plans for care coordination. This study underscores the potential usefulness of EHR data in performing automated risk factor identification and patient stratification. It provides a first step in the development of personalized hypertension treatment programs via a data-driven approach.
Acknowledgments
This work was supported in part by the National Center for Advancing Translational Sciences (UL1TR000445), the National Heart, Lung, and Blood Institute (HL 1K12HL109019), and the National Science Foundation (IIS 1418504, IIS 1418511).
Appendix
In the appendix, we show the detailed results for the risk factor identification step.
Table A.1.
Coefficients for forward stepwise logistic regression obtained in each fold of cross validation during the risk factor identification step. A value of “N/A” indicates that the feature was not selected during that particular fold.
| Fold of Cross Validation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Features Identified | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Mean | |
| Demographics | Age | 0.26 | 0.23 | 0.23 | 0.28 | 0.27 | 0.24 | 0.24 | 0.28 | 0.27 | 0.24 | 0.25 |
|
| ||||||||||||
| Vital Signs | MAP before pilot enrollment (mmHg) | 1.10 | 1.14 | 1.11 | 1.07 | 1.10 | 1.11 | 1.06 | 1.06 | 1.10 | 1.08 | 1.09 |
| Aldosterone antagonists | 0.38 | 0.36 | 0.49 | 0.38 | 0.36 | 0.47 | 0.43 | 0.48 | 0.45 | 0.40 | 0.42 | |
| Medications | Beta Blockers | −0.19 | −0.32 | −0.22 | −0.22 | −0.28 | −0.19 | N/A | −0.25 | −0.27 | −0.22 | −0.24 |
| Central alpha agonists | −0.28 | −0.39 | −0.39 | −0.27 | −0.33 | −0.34 | −0.30 | −0.36 | −0.39 | −0.31 | −0.34 | |
|
| ||||||||||||
| PheWAS Codes | Other disorders of pancreatic internal secretion (other than diabetes) | N/A | 0.96 | 0.80 | 0.78 | 0.68 | 0.93 | N/A | 0.71 | 0.89 | 0.76 | 0.81 |
| Other forms of heart disease (non-ischemic, non-pulmonary heart disease) | 0.27 | 0.29 | 0.31 | 0.26 | 0.25 | 0.26 | 0.27 | 0.31 | 0.29 | 0.25 | 0.28 | |
| Other disorders of female genital tract (other than inflammatory diseases of pelvis) | 0.30 | 0.34 | 0.48 | 0.40 | 0.37 | 0.35 | 0.30 | 0.32 | 0.34 | 0.41 | 0.36 | |
| Symptoms involving head and neck | −0.37 | −0.38 | −0.45 | −0.45 | −0.36 | −0.39 | −0.40 | −0.42 | −0.40 | −0.36 | −0.40 | |
Contributor Information
Robert Chen, Email: rchen87@gatech.edu, School of Computational Science and Engineering at the Georgia Institute of Technology, Atlanta, GA 30332 USA.
Jimeng Sun, School of Computational Science and Engineering at the Georgia Institute of Technology, Atlanta, GA 30332 USA.
Robert S. Dittus, Institute for Medicine and Public Health, Vanderbilt University, Nashville, TN, the Geriatric Research, Education, and Clinical Center, VA Tennessee Valley Healthcare System, Nashville, TN, and the Department of Medicine, School of Medicine, Vanderbilt University, Nashville, TN
Daniel Fabbri, Department of Biomedical Informatics, School of Medicine, Vanderbilt University, Nashville, TN, and the Department of Electrical Engineering and Computer Science, School of Engineering, Vanderbilt University, Nashville, TN.
Jacqueline Kirby, Vanderbilt Institute for Clinical and Translational Research, Vanderbilt University, Nashville, TN.
Cheryl L. Laffer, Department of Medicine, School of Medicine, Vanderbilt University, Nashville, TN
Candace D. McNaughton, Department of Emergency Medicine, School of Medicine, Vanderbilt University, Nashville, TN
Bradley Malin, Department of Biomedical Informatics, School of Medicine, Vanderbilt University, Nashville, TN, and the Department of Electrical Engineering and Computer Science, School of Engineering, Vanderbilt University, Nashville, TN.
References
- 1.Bodenheimer T, Fernandez A. High and Rising Health Care Costs. Part 4: Can Costs Be Controlled While Preserving Quality? Ann Intern Med. 2005 Jul;143(1):26–31. doi: 10.7326/0003-4819-143-1-200507050-00007. [DOI] [PubMed] [Google Scholar]
- 2.Trivedi AN, Moloo H, Mor V. Increased Ambulatory Care Copayments and Hospitalizations among the Elderly. N Engl J Med. 2010;362(4):320–328. doi: 10.1056/NEJMsa0904533. [DOI] [PubMed] [Google Scholar]
- 3.Braunstein ML. Health Informatics in the Cloud. Chapter 1. New York: Springer New York; 2013. Healthcare Delivery in the US; pp. 1–8. [Google Scholar]
- 4.Thrall JH. Prevalence and Costs of Chronic Disease in a Health Care System Structured for Treatment of Acute Illness. Radiology. 2005 Apr;235(1):9–12. doi: 10.1148/radiol.2351041768. [DOI] [PubMed] [Google Scholar]
- 5.Heidenreich PA, Trogdon JG, Khavjou OA, Butler J, Dracup K, Ezekowitz MD, Finkelstein EA, Hong Y, Johnston SC, Khera A, Lloyd-Jones DM, Nelson SA, Nichol G, Orenstein D, Wilson PWF, Woo YJ. Forecasting the Future of Cardiovascular Disease in the United States A Policy Statement From the American Heart Association. Circulation. 2011 Mar;123(8):933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 6.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart Disease and Stroke Statistics—2013 Update A Report From the American Heart Association. Circulation. 2013 Jan;127(1):e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacMahon S, Peto R, Collins R, Godwin J, MacMahon S, Cutler J, Sorlie P, Abbott R, Collins R, Neaton J, Abbott R, Dyer A, Stamler J. Blood pressure, stroke, and coronary heart disease: Part 1, prolonged differences in blood pressure: prospective observational studies corrected for the regression dilution bias. The Lancet. 1990 Mar;335(8692):765–774. doi: 10.1016/0140-6736(90)90878-9. [DOI] [PubMed] [Google Scholar]
- 8.Levy D, Larson MG, Vasan RS, Kannel WB, Ho KL. THe progression from hypertension to congestive heart failure. JAMA. 1996 May;275(20):1557–1562. [PubMed] [Google Scholar]
- 9.Egan BM, Zhao Y, Axon R. US trends in prevalence, awareness, treatment, and control of hypertension, 1988–2008. JAMA. 2010 May;303(20):2043–2050. doi: 10.1001/jama.2010.650. [DOI] [PubMed] [Google Scholar]
- 10.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart Disease and Stroke Statistics—2012 Update A Report From the American Heart Association. Circulation. 2012 Jan;125(1):e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jones DW, Materson BJ, Oparil S, Wright JT, Roccella EJ. Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension. 2003 Dec;42(6):1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 12.Whelton PK, He J, Appel LJ, et al. Primary prevention of hypertension: Clinical and public health advisory from the national high blood pressure education program. JAMA. 2002 Oct;288(15):1882–1888. doi: 10.1001/jama.288.15.1882. [DOI] [PubMed] [Google Scholar]
- 13.James PA, Oparil S, Carter BL, et al. 2014 Evidence-based guideline for the management of high blood pressure in adults: Report from the panel members appointed to the eighth joint national committee (JNC 8) JAMA. 2014 Feb;311(5):507–520. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 14.Ashley EA, Hershberger RE, Caleshu C, Ellinor PT, Garcia JGN, Herrington DM, Ho CY, Johnson JA, Kittner SJ, MacRae CA, Mudd-Martin G, Rader DJ, Roden DM, Scholes D, Sellke FW, Towbin JA, Eyk JV, Worrall BB. Genetics and Cardiovascular Disease A Policy Statement From the American Heart Association. Circulation. 2012 Jul;126(1):142–157. doi: 10.1161/CIR.0b013e31825b07f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas KL, Shah BR, Elliot-Bynum S, Thomas KD, Damon K, LaPointe NMA, Calhoun S, Thomas L, Breathett K, Mathews R, Anderson M, Califf RM, Peterson ED. Check It, Change It A Community-Based, Multifaceted Intervention to Improve Blood Pressure Control. Circ Cardiovasc Qual Outcomes. 2014 Nov;7(6):828–834. doi: 10.1161/CIRCOUTCOMES.114.001039. [DOI] [PubMed] [Google Scholar]
- 16.Katon WJ, Lin EHB, Von Korff M, Ciechanowski P, Ludman EJ, Young B, Peterson D, Rutter CM, McGregor M, McCulloch D. Collaborative Care for Patients with Depression and Chronic Illnesses. N Engl J Med. 2010;363(27):2611–2620. doi: 10.1056/NEJMoa1003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starfield B, Shi L, Macinko J. Contribution of Primary Care to Health Systems and Health. Milbank Q. 2005 Sep;83(3):457–502. doi: 10.1111/j.1468-0009.2005.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peikes D, Chen A, Schore J, Brown R. Effects of care coordination on hospitalization, quality of care, and health care expenditures among medicare beneficiaries: 15 randomized trials. JAMA. 2009 Feb;301(6):603–618. doi: 10.1001/jama.2009.126. [DOI] [PubMed] [Google Scholar]
- 19.Sochalski J, Jaarsma T, Krumholz HM, Laramee A, McMurray JJV, Naylor MD, Rich MW, Riegel B, Stewart S. What Works In Chronic Care Management: The Case Of Heart Failure. Health Aff (Millwood) 2009 Jan;28(1):179–189. doi: 10.1377/hlthaff.28.1.179. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, McNaughton CD, Zhang P, Perer A, Gkoulalas-Divanis A, Denny JC, Kirby J, Lasko T, Saip A, Malin BA. Predicting changes in hypertension control using electronic health records from a chronic disease management program. J Am Med Inform Assoc. 2013 Sep; doi: 10.1136/amiajnl-2013-002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pathak J, Bailey KR, Beebe CE, Bethard S, Carrell DS, Chen PJ, Dligach D, Endle CM, Hart LA, Haug PJ, Huff SM, Kaggal VC, Li D, Liu H, Marchant K, Masanz J, Miller T, Oniki TA, Palmer M, Peterson KJ, Rea S, Savova GK, Stancl CR, Sohn S, Solbrig HR, Suesse DB, Tao C, Taylor DP, Westberg L, Wu S, Zhuo N, Chute CG. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. J Am Med Inform Assoc. 2013 Dec;20(e2):e341–e348. doi: 10.1136/amiajnl-2013-001939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rea S, Pathak J, Savova G, Oniki TA, Westberg L, Beebe CE, Tao C, Parker CG, Haug PJ, Huff SM, Chute CG. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: The SHARPn project. J Biomed Inform. 2012 Aug;45(4):763–771. doi: 10.1016/j.jbi.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins GS, Mallett S, Omar O, Yu LM. Developing risk prediction models for type 2 diabetes: a systematic review of methodology and reporting. BMC Med. 2011 Sep;9(1):103. doi: 10.1186/1741-7015-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CH, Chen JCY, Tseng VS. A novel data mining mechanism considering bio-signal and environmental data with applications on asthma monitoring. Comput Methods Programs Biomed. 2011 Jan;101(1):44–61. doi: 10.1016/j.cmpb.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Drawz PE, Archdeacon P, McDonald CJ, Powe NR, Smith KA, Norton J, Williams DE, Patel UD, Narva A. CKD as a Model for Improving Chronic Disease Care through Electronic Health Records. Clin J Am Soc Nephrol. 2015 Jun;:CJN.00940115. doi: 10.2215/CJN.00940115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chin CY, Weng MY, Lin TC, Cheng SY, Yang YHK, Tseng VS. Mining Disease Risk Patterns from Nationwide Clinical Databases for the Assessment of Early Rheumatoid Arthritis Risk. PLoS ONE. 2015 Apr;10(4):e0122508. doi: 10.1371/journal.pone.0122508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denny JC. Chapter 13: Mining Electronic Health Records in the Genomics Era. PLoS Comput Biol. 2012 Dec;8(12):e1002823. doi: 10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conway M, Berg RL, Carrell D, Denny JC, Kho AN, Kullo IJ, Linneman JG, Pacheco JA, Peissig P, Rasmussen L, Weston N, Chute CG, Pathak J. Analyzing the Heterogeneity and Complexity of Electronic Health Record Oriented Phenotyping Algorithms. AMIA Annu Symp Proc. 2011;2011:274–283. [PMC free article] [PubMed] [Google Scholar]
- 29.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2012 Sep; doi: 10.1136/amiajnl-2012-001145. amiajnl–2012–001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overby CL, Pathak J, Gottesman O, Haerian K, Perotte A, Murphy S, Bruce K, Johnson S, Talwalkar J, Shen Y, Ellis S, Kullo I, Chute C, Friedman C, Bottinger E, Hripcsak G, Weng C. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc. 2013 Dec;20(e2):e243–e252. doi: 10.1136/amiajnl-2013-001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Carroll RJ, Hinz ERM, Shah A, Eyler AE, Denny JC, Xu H. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. J Am Med Inform Assoc. 2013 Dec;20(e2):e253–e259. doi: 10.1136/amiajnl-2013-001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll RJ, Eyler AE, Denny JC. Naive Electronic Health Record Phenotype Identification for Rheumatoid Arthritis. AMIA Annu Symp Proc. 2011;2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, Pacheco JA, Boomershine CS, Lasko TA, Xu H, Karlson EW, Perez RG, Gainer VS, Murphy SN, Ruderman EM, Pope RM, Plenge RM, Kho AN, Liao KP, Denny JC. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012 Jun;19(e1):e162–e169. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei WQ, Leibson CL, Ransom JE, Kho AN, Chute CG. The Absence of Longitudinal Data Limits the Accuracy of High-Throughput Clinical Phenotyping for Identifying Type 2 Diabetes Mellitus Subjects. Int J Med Inf. 2013 Apr;82(4):239–247. doi: 10.1016/j.ijmedinf.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, Basford M, Chute CG, Kullo IJ, Li R, Pacheco JA, Rasmussen LV, Spangler L, Denny JC. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc JAMIA. 2013 Jun;20(e1):e147–e154. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho JC, Ghosh J, Steinhubl SR, Stewart WF, Denny JC, Malin BA, Sun J. Limestone: High-throughput candidate phenotype generation via tensor factorization. J Biomed Inform. 2014 Dec;52:199–211. doi: 10.1016/j.jbi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roden D, Pulley J, Basford M, Bernard G, Clayton E, Balser J, Masys D. Development of a Large-Scale De-Identified DNA Biobank to Enable Personalized Medicine. Clin Pharmacol Ther. 2008 Sep;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010 Jan;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prospective Studies Collaboration. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–13. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 40.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, Crane PK, Pathak J, Chute CG, Bielinski SJ, Kullo IJ, Li R, Manolio TA, Chisholm RL, Denny JC. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011 Apr;3(79):79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010 May;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, Kullo IJ, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013 Dec;31(12):1102–1111. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz G. Estimating the Dimension of a Model. Ann Stat. 1978 Mar;6(2):461–464. [Google Scholar]
- 44.Fraley C, Raftery AE. MCLUST Version 3: An R Package for Normal Mixture Modeling and Model-Based Clustering. Technical report no 597. 2006 Sep; [Google Scholar]
- 45.Fraley C, Raftery AE. MCLUST: Software for Model-Based Cluster Analysis. J Classif. 1999 Jul;16(2):297–306. [Google Scholar]
- 46.Bulpitt CJ, Beckett NS, Cooke J, Dumitrascu DL, Gil-Extremera B, Nachev C, Nunes M, Peters R, Staessen JA, Thijs L and on behalf of the Hypertension in the Very Elderly Trial (HYVET) Working Group. Results of the pilot study for the Hypertension in the Very Elderly Trial. J Hypertens. 2003 Dec;21(12):2409–2417. doi: 10.1097/00004872-200312000-00030. [DOI] [PubMed] [Google Scholar]
- 47.Beckett NS, Peters R, Fletcher AE, Staessen JA, Liu L, Dumitrascu D, Stoyanovsky V, Antikainen RL, Nikitin Y, Anderson C, Belhani A, Forette F, Rajkumar C, Thijs L, Banya W, Bulpitt CJ. Treatment of Hypertension in Patients 80 Years of Age or Older. N Engl J Med. 2008;358(18):1887–1898. doi: 10.1056/NEJMoa0801369. [DOI] [PubMed] [Google Scholar]
- 48.Mattila R, Malmivaara A, Kastarinen M, Kivelä SL, Nissinen A. Effectiveness of multidisciplinary lifestyle intervention for hypertension: a randomised controlled trial. J Hum Hypertens. 2003;17(3):199–205. doi: 10.1038/sj.jhh.1001531. [DOI] [PubMed] [Google Scholar]
- 49.Svetkey LP, Pollak KI, Yancy WS, Dolor RJ, Batch BC, Samsa G, Matchar DB, Lin PH. Hypertension improvement project: randomized trial of quality improvement for physicians and lifestyle modification for patients. Hypertension. 2009 Dec;54(6):1226–1233. doi: 10.1161/HYPERTENSIONAHA.109.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Niepen P, Verbeelen D. Gender and hypertension management: a sub-analysis of the I-inSYST survey. Blood Press. 2011 Apr;20(2):69–76. doi: 10.3109/08037051.2010.532304. [DOI] [PubMed] [Google Scholar]
- 51.Journath G, Hellénius M-L, Petersson U, Theobald H, Nilsson PM and Hyper-Q Study Group Sweden. Sex differences in risk factor control of treated hypertensives: a national primary healthcare-based study in Sweden. Eur J Cardiovasc Prev Rehabil Off J Eur Soc Cardiol Work Groups Epidemiol Prev Card Rehabil Exerc Physiol. 2008 Jun;15(3):258–262. doi: 10.1097/HJR.0b013e3282f37a45. [DOI] [PubMed] [Google Scholar]
- 52.Jones CA, Nagpal S. An update: women, hypertension and therapeutic efficacy. Can J Cardiol. 2001 Dec;17(12):1283–1289. [PubMed] [Google Scholar]
- 53.Fan J, Arruda-Olson AM, Leibson CL, Smith C, Liu G, Bailey KR, Kullo IJ. Billing code algorithms to identify cases of peripheral artery disease from administrative data. J Am Med Inform Assoc. 2013 Dec;20(e2):e349–e354. doi: 10.1136/amiajnl-2013-001827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Estabrooks PA, Boyle M, Emmons KM, Glasgow RE, Hesse BW, Kaplan RM, Krist AH, Moser RP, Taylor MV. Harmonized patient-reported data elements in the electronic health record: supporting meaningful use by primary care action on health behaviors and key psychosocial factors. J Am Med Inform Assoc. 2012;19(4):575–582. doi: 10.1136/amiajnl-2011-000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. J Am Med Inform Assoc. 2013 Dec;20(e2):e206–e211. doi: 10.1136/amiajnl-2013-002428. [DOI] [PMC free article] [PubMed] [Google Scholar]