SUMMARY
KDM2B (also known as FBXL10) controls stem cell self-renewal, somatic cell reprogramming and senescence, and tumorigenesis. KDM2B contains multiple functional domains, including a JmjC domain that catalyzes H3K36 demethylation and a CxxC zing finger that recognizes CpG islands and recruits the polycomb repressive complex 1 (PRC1). Here, we report that KDM2B, via its F-box domain, functions as a subunit of the CUL1-RING ubiquitin ligase (CRL1/SCFKDM2B) complex. KDM2B targets c-Fos for polyubiquitylation and regulates c-Fos protein levels. Unlike the phosphorylation of other SCF/CRL1 substrates that promotes substrates binding to F-box, EGF-induced c-Fos S374 phosphorylation dissociates c-Fos from KDM2B and stabilizes c-Fos protein. Non-phosphorylatable and phosphomimetic mutations at S374 result in c-Fos protein which cannot be induced by EGF and accumulates constitutively and lead to decreased or increased cell proliferation, respectively. Multiple tumor-derived KDM2B mutations impaired the function of KDM2B to target c-Fos degradation and to suppress cell proliferation. These results reveal a novel function of KDM2B in the negative regulation of cell proliferation by assembling an E3 ligase to targeting c-Fos protein degradation that is antagonized by mitogenic stimulations.
Keywords: KDM2B, SCF E3 ligase, c-Fos, EGF, ubiquitylation
INTRODUCTION
The activator protein-1 (AP-1) transcriptional factor complex is central to many biological processes (2, 3). AP-1 complexes perform such diverse functions by potentially forming a large number of homo- or hetero-dimeric complexes through combinatorial interaction between members of Fos, Jun, ATF (activating transcription factor) and MAF (musculoaponeurotic fibrosarcoma) protein families. Proto-oncogene c-Fos is one of the first genes identified to be induced by mitogenic stimulation (4). c-Fos forms a dimeric complex with c-Jun as the first identified AP-1 (1, 5-7). The regulation of c-Fos has been extensively studied and serves as paradigm for the tight, dynamic and multiple level regulation of stress and growth factor response (8). c-Fos is typically expressed at a very low level in both cells cultured in vitro and in vivo. Mitogenic stimulation, particularly the extracellular signal-regulated kinases 1/2 (ERK1/2) pathway, rapidly induces c-Fos mRNA (9, 10). The c-Fos protein is intrinsically unstable due to degradation by the 26S proteasome and is protected by phosphorylation (11, 12). Multiple putative phosphorylation sites, mostly in the C-terminal region of the c-Fos protein, have been reported to regulate c-Fos protein stability. In particular, two residues—Ser362 and Ser374—were found to be phosphorylated by RSK1/2 and ERK1/2, respectively (13) and their phosphorylation stabilizes c-Fos protein (11). Genetic studies using knock-in mutation demonstrated that phosphorylation on these two residues plays important roles for cell differentiation, cytokine response and tumorigenesis in vitro (14). In contrast, the identity of the E3 ubiquitin ligase that targets c-Fos degradation and is antagonized by ERK1/2-mediated phosphorylation has not been established. UBR1, a member of the N-end rule family E3 ligase, has been linked to c-Fos degradation in the cytoplasm, which is protected by ERK5-mediated phosphorylation at two separate sites, Thr232 which blocks c-Fos nuclear export and Ser32 which disrupts the interaction between c-Fos and UBR1 (15). The physiological significance of ERK5-mediated protection of c-Fos from UBR1-promoted degradation is currently unclear (16).
KDM2B (also known as FBXL10, NDY1, JHDM1B and CxxC2) controls stem cell self-renewal (17), somatic cell reprogramming (18), cell senescence (19, 20), and tumorigenesis (21-23). KDM2B/FBXL10 is a protein of multi-domains, including a JmjC domain situated at the N-terminal region of the protein, followed by a CxxC domain, a PHD domain, a F-box motif and seven leucine-rich repeats (LRRs, see Figure S2A). The JmjC domain catalyzes H3K36 demethylation (24) and the CxxC zing finger domain recognizes CpG islands and recruits polycomb repressive complex 1 (PRC1) to target genes (17, 25-28). KDM2B was also found to interact with SKP1 via its F-box domain (27, 28), a linker protein involved in the assembly of the SKP1-CUL1-F-box (SCF) E3 ubiquitin ligase complex, raising the possibility that KDM2B could additionally contain an E3 ligase function. The substrate of this putative KDM2B E3 ligase, however, has not been identified. Intriguingly, KDM2B has been reported to repress the transcription mediated by either c-Jun or c-Fos through a mechanism not completely understood (14, 28). In this study, we explore the possibility that a KDM2B-containing E3 ligase targets c-Fos for ubiquitination and degradation.
RESULTS
KDM2B/FBXL10 and CUL1 destabilize c-Fos protein
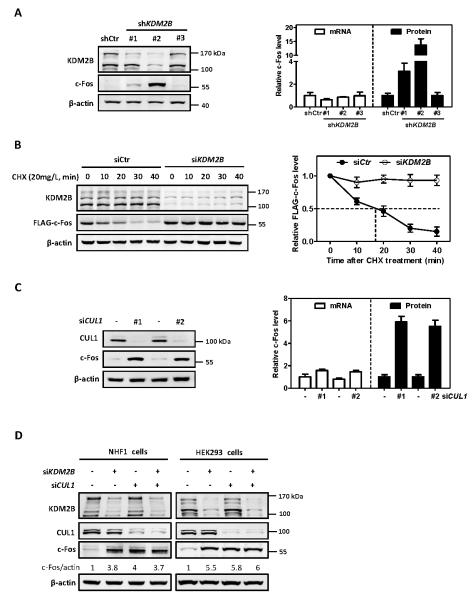
To determine whether KDM2B regulates c-Fos protein level, as well as its transcription, we first generated HEK293 cells with stable knock down of KDM2B. KDM2B expresses multiple isoforms produced by alternative splicing and differential transcriptional initiation. KDM2B-1 (NM_032590) is the longest and likely the full-length isoform and carries the histone demethylase JmjC domain, while KDM2B-2 (NM_013910) is translated from an mRNA initiated at an internal promoter of the KDM2B-1 transcript and lacks the JmjC domain and (Figure S1A). We first determined the subcellular localization and found that both long and short isoforms of KDM2B localized in the nucleus (Figure S1B). We then characterized three short-hairpin RNAs (shRNA) targeting different regions of KDM2B. We found that shKDM2B#1 targeted KDM2B-1 (upper-migrating) only and shKDM2B#2 targeted both of KDM2B-1 and KDM2B-2 effectively (Figure 1A). Notably, we found an increase of c-Fos protein levels in cells stably expressing shKDM2B#1 and an even more dramatic increase of c-Fos protein levels in cells stably expressing shKDM2B#2. This result suggested that KDM2B negatively regulated c-Fos protein level and this function of KDM2B did not require its JmjC domain-encoded histone demethylase activity. We next examined whether KDM2B controls the stability of c-Fos protein. Due to the extremely low basal level expression of c-Fos protein in un-stimulated cells, we established HEK293 cells stably expressing FLAG-tagged c-Fos (Figure S1C). FLAG-c-Fos has a half-life (t1/2) of approximately 18 minutes (Figure S1D), confirming that c-Fos is a very short-lived protein undergoing rapid turnover. Treatment of cells with MG132, a 26S proteasome inhibitor, effectively blocked c-Fos degradation (Figure S1E, S1F), confirming previous reports that c-Fos is degraded by the proteasome pathway (11, 12). Importantly, knockdown of KDM2B effectively stabilized FLAG-c-Fos, extending its half-life from 18 minutes beyond 40 minutes of experimental duration (Figure 1B).
Figure 1. KDM2B/FBXL10 and CUL1 destabilize c-Fos protein.
(A) Stable knock down of KDM2B increases endogenous c-Fos protein level. Two distinct bands of KDM2B (170 KDa and 120 KDa, respectively) were observed in HEK293 cells, representing the two isoforms. The protein and mRNA levels of c-Fos were determined by western blot and qRT-PCR, respectively, and normalized against β-actin. Error bars represent ±SD for triplicate experiments.
(B) Ectopically expressed c-Fos is stabilized by knock down of KDM2B. HEK293 cells stably expressing FLAG-c-Fos were transfected with siRNA oligonucleotides targeting KDM2B or control siRNA. 72 hours after siRNA transfection, the half-life of FLAG-c-Fos protein levels was determined by cycloheximide (CHX, 20 mg/L) chase assay and normalized against β-actin. Error bars represent ±SD for triplicate experiments.
(C) Knock down of CUL1 increases c-Fos protein level. HEK293 cells were transfected with two different siRNAs targeting CUL1 or a control siRNA. 72 h after transfection, the protein and mRNA levels of c-Fos were determined by western blot and qRT-PCR respectively and normalized against β-actin. Error bars represent ±SD for duplicate experiments.
(D) Knock down of either KDM2B or CUL1 or combination increases c-Fos protein to a similar level. NHF1 cells and HEK293 cells were transfected with siRNA oligonucleotides targeting either KDM2B or CUL1 individually or in combination. The protein and mRNA levels of c-Fos were determined by western blot and qRT-PCR, respectively, and normalized against β-actin. Error bars represent ±SD for triplicate experiments.
To determine the mechanism by which KDM2B negatively regulates c-Fos, we tested the possibility that KDM2B promotes c-Fos degradation by binding to SKP1 and CUL1 through its F-box. We first determined whether CUL1 affects c-Fos protein level. We found that knockdown of CUL1 in HEK293 cells with two different siRNAs resulted in an approximate six-fold increase of c-Fos protein levels without significant change in c-Fos mRNA levels (Figure 1C). Likewise depletion of KDM2B or CUL1 in NHF1 normal human fibroblasts also resulted in the accumulation of c-Fos (Figure 1D). Furthermore, co-depletion of both KDM2B and CUL1 did not cause any additional increase of c-Fos protein level (Figure 1D), suggesting that KDM2B and CUL1 act in the same pathway for controlling c-Fos. Together, these results demonstrate that KDM2B plays a major role to target c-Fos protein degradation by a CUL1-based E3 ligase. Following commonly accepted nomenclature, we refer to the KDM2B-CUL1 E3 ligase as SCF/CRL1KDM2B where the substrate recruiter F-box protein KDM2B is in superscript.
c-Fos is a substrate of SCFKDM2B/FBXL10 E3 ubiquitin ligase
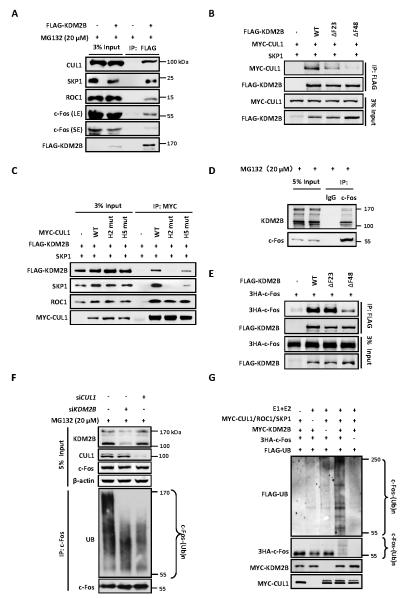
To demonstrate that KDM2B assembles the SCFKDM2B ligase complex to target c-Fos ubiquitylation, we examined its interaction with endogenous SCF E3 ligase complex components. Co-immunoprecipitation (Co-IP) assays demonstrated that FLAG-tagged KDM2B interacted with endogenous SKP1, CUL1 and ROC1 (Figure 2A). Meanwhile, we also detected the interaction between FLAG-KDM2B and endogenous c-Fos (Figure 2A). The F-box domain in F-box proteins is essential for its binding to SKP1 and thus, the assembly of SCF complex (29-31). To determine the CUL1/SKP1-binding region in KDM2B, we constructed various deletion mutants of KDM2B (Figure S2A). Co-IP assays revealed that deletion of both leucine-rich repeats (LRRs) and F-box domains from KDM2B (ΔLRF), but not the LRRs only (ΔLRR), completely disrupted its binding to CUL1 (Figure S2B). Consistently, the C-terminal of KDM2B (C300) containing the F-box and LRRs strongly interacted with CUL1 (Figure S2B), suggesting that the F-box domain of KDM2B is essential for its binding to CUL1. Structural analysis has previously shown that the N-terminal region of F-box domain in SKP2 is essential for its binding to SKP1 (32). Similar to SKP2, deletion of 23 amino acids in the N-terminal half of the F-box domain from KDM2B (residues 1056 to 1078, referred to as ΔF23) substantially reduced its binding to CUL1, and deletion of 48 residues containing the entire F-box domain (residues 1057 -1105, ΔF48) completely abolished its binding to CUL1 (Figure 2B). Two hydrophobic helical surfaces in the N-terminal tip of CUL1, H2 and H5, pack with hydrophobic and polar residues from SKP1 to form a large interface (32). Mutation of either helix in CUL1 significantly disrupted its binding to KDM2B (Figure 2C). Together, these results demonstrate that KDM2B, via its F-box domain, assembles a bona fide SCF-type E3 ubiquitin ligase complex.
Figure 2. c-Fos is a substrate of SCFKDM2B/FBXL10 E3 ubiquitin ligase.
(A) Ectopically expressed KDM2B binds to endogenous CUL1/SKP1/ROC1 E3 ligase. HEK293T cells were transfected with plasmid expressing FLAG-tagged KDM2B and treated with 20 μM MG132 for 6 h. The interactions between KDM2B and endogenous CUL1, SKP1, ROC1 and c-Fos proteins were determined by Co-IP assay. LE and SE refer to longer exposure and short exposure, respectively.
(B) The F-box domain of KDM2B is essential for its binding to CUL1. HEK293T cells were transfected with plasmids expressing indicated proteins. The interactions between WT or mutant KDM2B and CUL1 were determined by Co-IP assay.
(C) The H2 and H5 helixes of CUL1 are essential for its binding to KDM2B. HEK293T cells were transfected with plasmids expressing indicated proteins. The interactions between KDM2B and WT or mutant CUL1 were determined by Co-IP assay.
(D) Endogenous interaction between c-Fos and KDM2B two isoforms determined by Co-IP. HEK293 cells were treated with 20 μM MG132 for 6 h and cell lysates were used for IP with IgG or c-Fos antibody.
(E) The C-terminal of F-box domain in KDM2B is required for its binding to c-Fos. HEK293T cells were transfected with plasmids expressing indicated proteins. The interactions between WT or mutant KDM2B and c-Fos were determined by Co-IP.
(F) Knock down of either KDM2B or CUL1 decreases c-Fos ubiquitylation in vivo. HEK293 cells were transfected with siRNA oligonucleotides targeting indicated genes and treated with 20 μM MG132 for 6 h. Endogenous c-Fos was immunoprecipitated and immunoblotted with an antibody specific against ubiquitin.
(G) In vitro ubiquitylation of c-Fos by SCFKDM2B E3 ubiquitin ligase. Purified c-Fos protein was incubated with immunopurified E3 CUL1 complex and KDM2B individually or in combination in the presence or absence of E1, E2, ATP and Flag-ubiquitin in vitro for 1 h. The reaction mixture was resolved by SDS-PAGE and blotted using antibodies (from top to bottom panels) recognizing FLAG, HA, and MYC.
To demonstrate that endogenously expressed c-Fos binds to KDM2B in vivo, HEK293 cells were treated with MG132 followed by immunoprecipitation using an antibody against c-Fos. The interactions between c-Fos and KDM2B two isoforms translated by different promoters were readily detected (Figure 2D, Figure S2C). A fragment containing the C-terminal 300 residues of KDM2B that includes the LRRs and F-box could bind c-Fos, and conversely, deletion of the LRRs substantially reduced the binding of KDM2B with c-Fos (Figure S2D). Deletion of the F-box from KDM2B (ΔF48) also reduced the interaction between KDM2B and c-Fos (Figure 2E). These results indicate that the LRRs domain in KDM2B is mainly responsible for the binding with the substrate c-Fos and that the F-box domain may also contribute to the binding.
To demonstrate the ubiquitylation of c-Fos by the SCFKDM2B E3 ligase, we carried out both in vivo and in vitro ubiquitylation assays. We first knocked down either CUL1 or KDM2B and found that knocking down either gene reduced the ubiquitylation of endogenous c-Fos (Figure 2F). An in vitro ubiquitylation assay demonstrated that the CUL1 E3 immunocomplexes efficiently ubiquitylated c-Fos, converting nearly all c-Fos into ubiquitylated form (Figure 2G). The ubiquitylation of c-Fos was dependent on the addition of E1 and E2, CUL1 E3 complex, substrate receptor KDM2B and substrate c-Fos. We therefore conclude that c-Fos is a substrate of SCFKDM2B E3 ubiquitin ligase.
EGF stabilizes c-Fos by dissociating c-Fos from KDM2B/FBXL10
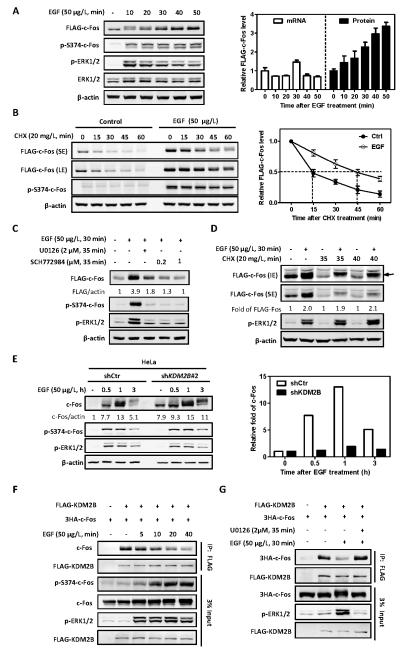
The MEK/ERK signaling pathway activates c-Fos transcription in response to extracellular growth factors such as EGF (9). Supporting this conclusion, EGF stimulation dramatically increased the endogenous c-Fos protein as early as within 20 minutes, which was compromised by the classical MEK1/2 (the ERK1/2 activating kinases) inhibitor U0126 (Figures S3A, S3B and S3C). In addition to transcriptional activation, MEK/ERK pathway has also been reported to increase c-Fos level by stabilizing c-Fos protein through an undefined mechanism that involves phosphorylation at Ser362 and Ser374 (11). We therefore examined whether EGF stabilizes c-Fos protein through regulating SCFKDM2B-mediated c-Fos ubiquitylation. To separate from the intrinsically transcriptional regulation of c-Fos, we took advantage of HEK293 cells that stably express ectopic FLAG-tagged c-Fos. EGF treatment induced FLAG-c-Fos protein accumulation as early as within 20 minutes and continuously in a time-dependent manner without significant change in FLAG-c-Fos mRNA levels (Figure 3A). Consistently, EGF treatment stabilized FLAG-c-Fos in HEK293 cells, extending its half-life from 15 minutes to 45 minutes (Figure 3B). Moreover, EGF-induced c-Fos accumulation was blocked by MEK1/2 inhibitor U0126 and an ERK1/2 inhibitor, SCH772984 (Figure 3C). Inhibition of translation by the treatment of cells with cycloheximide (CHX) reduced the steady state level of c-Fos protein, but this reduction was not seen in cells treated with EGF (Figure 3D), supporting a post-translational mechanism by which EGF stabilizes c-Fos. Notably, the c-Fos protein accumulated in EGF-treated cells was highly phosphorylated, as indicated by the appearance of a discrete slower migrating band (Figures 3A-3D), supporting a previous notion that the phosphorylated form c-Fos is resistant to the degradation.
Figure 3. EGF stabilizes c-Fos by dissociating c-Fos from KDM2B/FBXL10.
(A) EGF induces c-Fos protein accumulation in a time-dependent manner. HEK293 cells stably expressing FLAG-tagged c-Fos were deprived of serum for 8 h, and then treated with 50 μg/L EGF for the indicated length of time. The protein and mRNA levels of FLAG-c-Fos were determined by western blot and qRT-PCR, respectively, and normalized against β-actin. Error bars represent ±SD for triplicate experiments.
(B) EGF stabilizes c-Fos. HEK293 cells stably expressing FLAG-tagged c-Fos were pretreated with or without EGF (50 μg/L) for 30 min, followed by CHX treatment (20 mg/L) for the indicated time. The protein levels of c-Fos were determined by western blot and normalized against β-actin. Error bars represent ±SD for triplicate experiments.
(C) EGF-induced c-Fos accumulation is blocked by MEK1/2 inhibitor U0126 and an ERK1/2 inhibitor (SCH772984). HEK293 cells stably expressing FLAG-c-Fos were deprived of serum for 8 h, followed by treatment with U0126 (2 μM) or SCH772984 (0.2 or 1 μM) for 5 min. EGF (50 μg/L) was then added for additional 30 min. The protein levels of c-Fos were determined by western blot and normalized against β-actin.
(D) EGF-induced c-Fos accumulation is not affected by inhibition of protein synthesis. HEK293 cells stably expressing FLAG-tagged c-Fos were treated with CHX (20 mg/L) for 5 min or 10 min. Solvent or EGF (50 μg/L) was then added for further treatment for 30 min. The protein levels of FLAG-c-Fos were determined by western blot and normalized against β-actin. Data are shown as relative fold change over protein levels in cells without EGF treatment. The arrow represents highly phosphorylated c-Fos
(E) Knock down of KDM2B prolongs the high level of c-Fos protein following EGF stimulation. HeLa cells with stable knock down of KDM2B were deprived of serum for 8 h, followed by treatment with EGF (50 μg/L) for the indicated length of time. The protein levels of c-Fos were determined by western blot and normalized against β-actin. Data are shown as relative fold change over cells without EGF treatment (right panel).
(F) EGF treatment induces c-Fos S374 phosphorylation and concomitantly reduces the interaction between c-Fos and KDM2B. HEK293 cells were transfected with plasmids expressing indicated proteins and then treated with EGF (50 μg/L) for the indicated length of time. The interactions between c-Fos and KDM2B were determined by Co-IP.
(G) The reduction of c-Fos and KDM2B interaction by EGF is blocked by U0126. FLAG-KDM2B and 3HA-c-Fos were co-transfected into HEK293 cells. Cells were pretreated with U0126 (2 μM) for 5 min and then treated with EGF (50 μg/L) for another 30 min. The interactions between c-Fos and KDM2B were determined by Co-IP.
To determine whether KDM2B is involved in the regulation of c-Fos by EGF, we first examined the effect of KDM2B knockdown on EGF-induced c-Fos protein levels and found that knockdown of KDM2B prolonged the high level of c-Fos protein following EGF stimulation (Figure 3E). To determine how EGF stabilizes c-Fos, we next examined the association between c-Fos and KDM2B in EGF-treated cells and found that EGF treatment dissociated their binding in a time-dependent manner (Figure 3F). The reduction of c-Fos and KDM2B interaction by EGF was completely blocked by U0126 (Figure 3G), a highly selective inhibitor of both MEK1 and MEK2, suggesting the involvement of MEK/ERK signaling pathway in the regulation of c-Fos association with KDM2B. TPA is a potent tumor promoter that activates PKC signaling pathway (PKC-Ras-Raf-MEK-ERK), which causes phosphorylation of c-Fos and dissociates c-Fos from KDM2B. Consistently, we found that TPA treatment decreased the association of KDM2B with c-Fos (Figure S3F) and accumulated c-Fos protein (Figure S3D). Importantly, we found that knock down of KDM2B stabilized c-Fos protein by in the presence of TPA (Figure S3E). Together, these results demonstrate that c-Fos level is regulated at both transcriptional and posttranslational levels and that c-Fos protein is stabilized by EGF-promoted phosphorylation which dissociates c-Fos from its E3 ligase, KDM2B.
EGF-mediated phosphorylation at S374 stabilizes c-Fos and promotes cell proliferation
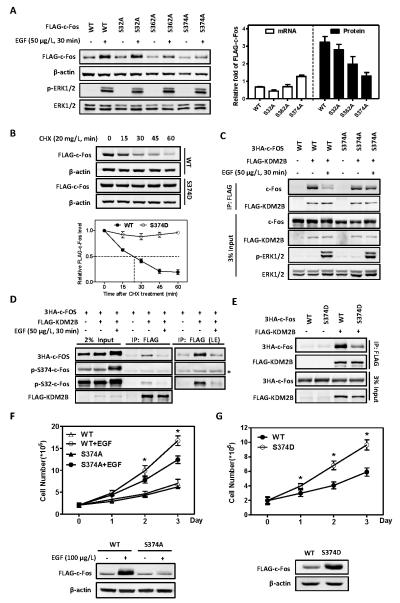
It has been previously reported that c-Fos can be stabilized by phosphorylation at several sites, including S32, S362, S374 sites (11, 15) that are stimulated by ERK5 and ERK1/2 pathways. To determine which of these sites is involved in the ubiquitylation of c-Fos by the SCFKDM2B E3 ligase, we established HEK293 cells stably expressing FLAG-c-Fos mutants targeting individual serine residues. We found that a non-phosphorylatable mutation at S374 (S374A) completely abolished EGF-induced c-Fos stabilization and protein accumulation, while S362A and S32A mutations had mild or no effect on EGF-induced increase of c-Fos stability and steady state c-Fos level (Figures 4A & S4A). In contrast to S374A mutation, a phosphomimetic mutation of S374 (S374D) dramatically extended the half-life of c-Fos from approximately 18 minutes to more than 1 hour of the experimental duration (Figure 4B), whereas neither S32D nor S362D mutation had significant effect on c-Fos stability (Figure S4B). Collectively, these results indicate that S374 is a major phosphorylation site that contributes to c-Fos stabilization in response to EGF stimulation.
Figure 4. EGF-mediated phosphorylation at S374 stabilizes c-Fos and promotes cell proliferation.
(A) Mutation of S374 site to alanine abolishes EGF-induced c-Fos protein accumulation. HEK293 cells stably expressing FLAG-tagged c-Fos were deprived of serum for 8 h, and then treated with EGF (50 μg/L) for 30 min. The protein and mRNA levels of FLAG-c-Fos were determined by western blot and qRT-PCR, respectively, and normalized against β-actin. Data are shown as relative fold change over cells without EGF treatment. Error bars represent ±SD for duplicate experiments.
(B) Mutation of S374 site to aspartic acid renders c-Fos resistant to degradation. HEK293 cells stably expressing FLAG-tagged c-Fos WT or S374D mutant were treated with 20mg/L CHX for the indicated length of time. The half-life of c-Fos protein levels were determined by western blot and normalized against β-actin (Bottom). Error bars represent ±SD for triplicate experiments.
(C) The reduction of c-Fos and KDM2B interaction by EGF is inhibited by c-Fos S374A mutant. HEK293 cells were co-transfected with plasmids expressing indicated proteins and then treated with or without EGF (50 μg/L) for 30 min. The interactions between c-Fos and KDM2B were determined by Co-IP.
(D) KDM2B preferentially interacts with S374 non-phosphorylatable c-Fos. HEK293 cells were co-transfected with plasmids expressing the indicated proteins and then treated with or without EGF (50 μg/L) for 30min. The FLAG-KDM2B was immunoprecipitated and western blot was performed to detect the co-precipitated c-Fos with indicated antibodies. * indicates the heavy chain background around 55 KDa.
(E) S374D mutation of c-Fos hinders its binding to KDM2B. HEK293 cells were co-transfected with plasmids expressing indicated proteins, and the interactions between c-Fos and KDM2B were determined by Co-IP.
(F) EGF-induced cell proliferation is compromised by S374A mutant of c-Fos. HEK293 cells stably expressing FLAG-c-Fos WT or S374A mutant were cultured in the absence of serum and treated with or without EGF (100 μg/L) for 0, 1, 2 or 3 days, as indicated. EGF was replenished every day. Cell numbers were counted each day. Western blot was performed to show FLAG-c-Fos protein levels on the 3rd day. * denotes the p < 0.05 for cells stably expressing S374A mutant versus wild-type c-Fos under EGF treatment. Error bars represent ±SD for triplicate experiments.
(G) c-Fos S374D mutant is more potent than wild-type in promoting cell proliferation. HEK293 cells stably expressing FLAG-c-Fos wild-type or S374D mutant were cultured in the absence of serum for 0, 1, 2 or 3 days, as indicated. Cell numbers were counted each day. Western blot was performed to show FLAG-c-Fos protein levels on the 3rd day. * denotes the p < 0.05 for cells stably expressing S374D mutant versus wild-type c-Fos. Error bars represent ±SD for triplicate experiments.
We next determined whether EGF-promoted c-Fos dissociation from KDM2B is mediated by S374 phosphorylation. Western blot showed that the S374-phosphorylated form is closely associated with the appearance of the slow-migrating form of c-Fos in EGF-treated cells that are resistant to KDM2B binding (Figures 3A, 3E and S3A). We then expressed wild-type and S374A mutant c-Fos and found that S374A mutation inhibited EGF-induced dissociation of c-Fos from KDM2B (Figure 4C). Notably, S374-phosphorylated c-Fos, while significantly induced by EGF, was not detected in the KDM2B immunocomplex (Figure 4D). Consistently, S374D mutation of c-Fos reduced its binding to KDM2B (Figure 4E). EGF-induced cell proliferation was partially compromised by the expression of S374A mutant of c-Fos and promoted by S374D mutation (Figure 4F, 4G). Consistently, c-Fos S374D mutant that is resistant to KDM2B-mediated degradation is more potent than wild-type in activating target genes (Figure S4C). Together, these results demonstrate that EGF-induced phosphorylation at S374 in c-Fos disassociates it from KDM2B, resulting in increased stability and level of c-Fos and contributing to cell proliferation.
KDM2B/FBXL10 inhibits cell proliferation via promoting c-Fos degradation
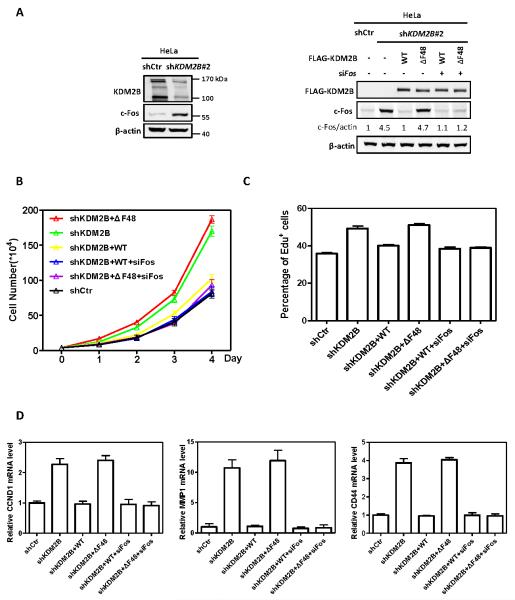
To determine the effect of KDM2B on cell proliferation, we first generated HeLa cells with stable knock down of KDM2B which resulted in increased c-Fos protein levels (Figure 5A, left panel). Re-introduction of wild-type, but not ΔF-box mutant (ΔF48), KDM2B reduced the c-Fos protein level back to that similar in the parental HeLa cells (Figure 5A, right panel). Consistent with increased level of c-Fos protein, ectopic expression of ΔF-box mutant, but not the wild-type KDM2B, increased the rate of cell proliferation, and this effect was not seen when c-Fos was knocked down (Figure 5B). EdU cell proliferation assay also showed that KDM2B ΔF-box mutant lost its ability to suppress cell proliferation and knockdown c-Fos reduced cell proliferation back to the rate similar to that seen in control cells (Figure 5C, S5). We also examined several target genes of c-Fos linked to cell proliferation and growth, including cyclin D1 (CCND1) involved in G1 cell cycle control, matrix metallopeptidase 1 (MMP1) linked to embryonic development and tissue remodeling and CD44 linked to cell-cell interactions, cell adhesion and migration. We found that the accumulation of c-Fos resulting from reduced expression or dysfunction of KDM2B increased the transcriptional level of these genes (Figure 5D). Together, these results demonstrated that KDM2B-promoted c-Fos degradation contributes to its function in the negative regulation of cell proliferation.
Figure 5. KDM2B/FBXL10 inhibits cell proliferation via promoting c-Fos degradation.
(A) KDM2B ΔF-box mutant abolished its function in promoting c-Fos degradation. FLAG-tagged wild-type or ΔF-box mutant of KDM2B was stably expressed in HeLa cells stably knocking down of KDM2B. Stable cells were transfected with siRNA targeting Fos or control siRNA as indicated. The protein levels of c-Fos were determined by western blot and normalized against β-actin.
(B) ΔF-box mutant lose the function of KDM2B in suppressing cell proliferation, and this effect is compromised by c-Fos knockdown. Stable HeLa cells identified as in Figure 5A were transfected with siRNAs targeting Fos. Cell numbers were counted each day. Error bars represent ±SD for triplicate experiments.
(C) ΔF-box mutant lose the function of KDM2B in inhibiting DNA synthesis, and this effect is compromised by c-Fos knockdown. Stable HeLa cells identified as in Figure 5A were transfected with siRNAs targeting Fos and labelled with 10 μM EdU for 1 h. The percentage of EdU-positive cells was determined by flow cytometry, indicating the percentage of S-phase cells in the population. Error bars represent ±SD for triplicate experiments.
(D) Knock down of KDM2B increases the transcriptional level of c-Fos-targeting genes, which is compromised by put-back of wild-type, but not ΔF-box mutant KDM2B. Stable HeLa cells identified as in Figure 5A were transfected with siRNAs targeting Fos. The mRNA levels of genes were determined by qRT-PCR respectively and normalized against β-actin. Error bars represent ±SD for triplicate experiments.
Tumor-derived mutations impair the function of KDM2B/FBXL10 to target c-Fos degradation and suppress cell proliferation
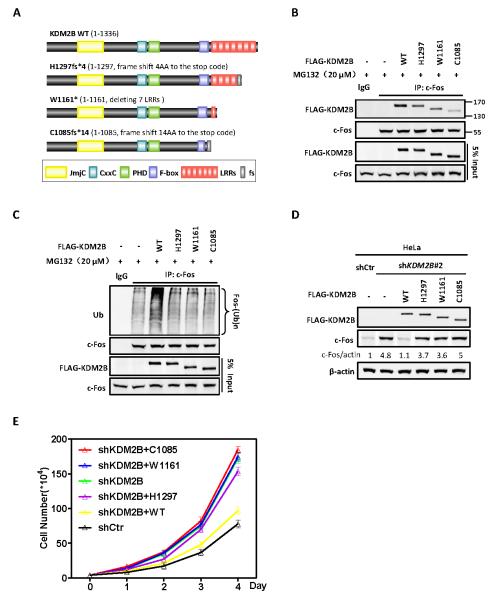
Recent cancer genome sequencing efforts have identified a number of mutations targeting KDM2B. For example, the COSMIC (Catalogue of Somatic Mutations in Cancer) database reports 16 deletions and 7 nonsense mutations of KDM2B in different type of human cancers (Figure S6). A loss of function of KDM2B could be viewed consistent with its function in suppressing cell proliferation. Among these reported mutations are three, C1085fs*14, W1161*, H1297fs*4, that predict to destroy the F-box or/and the leucine rich repeats (LRRs) domains of KDM2B (Figure 6A). We therefore set to determine whether these tumor-derived mutations affect the activity of KDM2B in promoting c-Fos degradation and suppressing cell proliferation. We recreated all these three mutations and first examined their effect on the binding of KDM2B with c-Fos. We found that all three mutations reduced the binding of KDM2B with c-Fos when compared with the wild-type KDM2B expressed at a similar level (Figure 6B). We next carried out in vivo ubiquitylation assay and demonstrated that all three tumor-derived mutations in KDM2B evidently reduced the activity of KDM2B in promoting c-Fos ubiquitylation (Figure 6C). Knockdown-replacement experiment demonstrates that these three mutations also abolished the ability to decrease c-Fos protein (Figure 6D). We further demonstrated that all three tumor-derived mutants lost the function of KDM2B in suppressing cell proliferation (Figure 6E). Together, these results identify the first functional consequence linked to the tumor mutations in KDM2B.
Figure 6. Tumor-derived mutations impair the function of KDM2B/FBXL10 to target c-Fos degradation and suppress cell proliferation.
(A) Schematic representations of three tumor-derived mutants of KDM2B examined in this study. Additional tumor-derived mutations in KDM2B are shown in Figure S6.
(B) Tumor-derived mutations in KDM2B impair their binding to c-Fos. HEK293 cells were transfected with plasmids expressing indicated proteins. The interactions between WT or mutant KDM2B and c-Fos were determined by Co-IP.
(C) Tumor-derived mutations in KDM2B impair their ability to ubiquitylate c-fos in vivo. HEK293 cells were transfected with plasmids expressing indicated proteins and treated with 20 μM MG132 for 6 h. Endogenous c-Fos was immunoprecipitated and immunoblotted with an antibody specific against ubiquitin.
(D) Tumor-derived mutants of KDM2B abolish their function in promoting c-Fos degradation. HeLa cells with stable knockdown of KDM2B were established by lentivirus transduction. FLAG-tagged wild-type or tumor-derived mutants KDM2B was then stably expressed in these cells. The protein levels of c-Fos were determined by western blot and normalized against β-actin.
(E) Tumor-derived mutations lose the function of KDM2B in suppressing cell proliferation. Sable HeLa cells were identified as in Figure 6D. Cell numbers were counted each day. Error bars represent ±SD for triplicate experiments.
DISCUSSION
This paper reports two key findings. First, KDM2B/FBXL10 functions as a bona fide F-box protein and assembles into an activate SCF/CRL1-type E3 ubiquitin ligase. KDM2B contains a JmjC domain that catalyzes histone H3K36 demethylation. Our finding identifies a second enzymatic activity to this multi-domain protein. Of 32 human histone demethylases proteins, KDM2B, and its closest homologue, KDM2A/FBXL11, are the only two KDMs that contain an F-box domain. The F-box domains encode by KDM2B and KDM2A/FBXL11 are highly related (78%). Our results suggest that KDM2A/FBLX11, which, like KDM2B, also catalyzes H3K36 demethylation and recognizes CpG through its JmjC and CxxC domains (33), is likely to form a functional SCF E3 ligase. It remains to be determined whether and how these two enzymatic activities are mechanistically linked in gene regulation. It will also be interesting to determine whether KDM2B catalyzes ubiquitylation of any subunit of PRC1 complex or a histone.
Second, c-Fos is robustly ubiquitylated and degraded by SCFKDM2B E3 ligase. As an immediate early gene, c-Fos level is rapidly accumulated by both transcriptional activation and protein stabilization, thereby conferring cells the ability to rapidly regulate AP-1 target genes during stress and mitogenic responses. Our finding provides a mechanism—dissociating c-Fos from its E3 ligase by EGF-promoted phosphorylation at S374—for cells to stabilize c-Fos protein in response to mitogenic stimulation. Our observations also provide a plausible explanation for the previously observed inhibition of c-Jun- and c-Fos-mediated transcriptional activity by KDM2B (14, 28). Conversely, this mechanism allows cells to maintain the c-Fos at a very low after withdrawal of the mitogenic stimulation through turning off the c-Fos transcription and constitutive degradation of c-Fos protein once the S374 is dephosphorylated. Many other SCF substrates are also regulated by phosphorylation which targets a small motif, known as degron, and promotes the substrate to bind with an F-box protein and be degraded by the SCF ligase. Linking signal-dependent phosphorylation to ubiquitylation provides cells an efficient mechanism to regulate protein function through crosstalk between different posttranslational modifications. The finding reported here further expands the spectrum of this crosstalk.
EXERIMENTAL PROCEDURES
Plasmids and chemicals
Full-length human KDM2B cDNA was a kind gift from Michele Pagano of NYU. Wild-type and mutants of KDM2B were constructed into pRK7-N-FLAG vector for transient expression or pQCXIH-N-FLAG vector for stable transduction by retrovirus infection. Wild-type and mutants of CUL1 were constructed into pRK7-N-MYC vector for transient expression. c-Fos cDNA was amplified from a cDNA library of HEK293 cells. Wild-type and mutants of c-Fos were constructed into pCDNA3-N-3HA vector for transient expression or pQCXIH-N-FLAG vector for stable transduction by retrovirus infection. The lentivirus packaging plasmids containing shKDM2B were purchased from Shanghai Genechem Company. The target sequences of shKDM2B are below:
shKDM2B #1 TGAGCGTGAAAGGTTGTTT
shKDM2B #2 GCCTTTACAAGAAGACATT
shKDM2B #3 TTCTTCAAACGCTGTGGAA
MG132 (C2211, Sigma-Aldrich), CHX (94271, AMRESCO), EGF (AF-100-15, PeproTech), U0126 (S1102, Selleckchem) and SCH772984 (S7101, Selleckchem) were purchased commercially.
Cell culture, transfection and proliferation assay
HEK293, HEK293T and HeLa cells were purchased from the American Type Culture Collection. Normal human fibroblast strains 1 (NHF1) was a kind gift from Dr. William K. Kaufmann of UNC. NHF1 was derived from neonatal foreskins and established in secondary culture according to established methods (34). Mycoplasma contamination was not detected by PCR. HEK293, HEK293T and HeLa cells were cultured in Dulbecco's Modified Eagle's Medium (Life Technologies) supplemented with 10% Newborn Calf Serum, 8mM L-glutamine, 50 μg/mL penicillin and streptomycin (Gibco). NHF1 cells were cultured in DMEM supplemented with 10% Fetal Bovine Serum. HEK293 and HeLa cells with stable knockdown or over-expression were established by lentivirus or retrovirus transduction, respectively, selected and maintained in medium containing 1 μg/mL puromycin or 50 μg/mL hygromycin B (Amresco). Cells were cultured in a 37°C incubator with 5% CO2. Cell transfection was performed using Lipofectamine 2000 (Life Technologies) for plasmid DNA or Lipofectamine RNAi MAX (Life Technologies) for siRNAs following the manufacturer’s instructions. Cells were harvested at 48-72 hours post-transfection for protein analyses.
For cell proliferation assay, stable HEK293 cells were triply seeded in 6-well plates at a density of 2×105 cells per well and deprived of serum for indicated length of time after adhering for 12 h. Stable HeLa cells were triply seeded in 6-well plates at a density of 4×104 cells per well. Cell numbers were counted daily by using Countstar IC1000. For EdU cell proliferation assay, HeLa cells were labelled with 10 μM EdU (5-ethynyl-2′-deoxyuridine) for 1 h, then collected with 1% BSA in PBS and fixed in 4% PFA in PBS. After incubation for 20 min, cells were washed with 1% BSA in PBS and permeabilized in 0.5% TritonX-100 for 20 min. Cells were washed and resuspended in the cocktail of [250 μL for each sample: PBS 215 μL, CuSO4 10μL, Sodium Ascorbate 25 μL, Azide Alexa Fluor (A10266, Life technologies) 0.6 μL] for 30 min. Cells were washed and resuspended in 500 μL 1% BSA in PBS. The percentage of EdU-positive cells was determined by flow cytometry (BD ACCURI C6).
Antibodies and immunological procedures
Antibodies against FLAG (SG4110-16, Shanghai Genomics Technology), MYC (SG4110-18, Shanghai Genomics Technology), β-actin (A00702, GenScript), HA (sc-7392, Santa Cruz), KDM2B (09-864, Millipore), CUL1 (2436-1, Epitomics), SKP1 (2538-1, Epitomics), ROC1 (5296-1, Epitomics), Ubiquitin (Z045801-5, Dako), Phospho-ERK1/2(Thr202/Tyr204)(4370S, Cell Signaling Technology), ERK1/2(4695P, Cell Signaling Technology), c-Fos (3620-1, Epitomics and sc-52, Santa Cruz), Phospho-c-Fos (Ser32) (5348S, Cell Signaling Technology) and Phospho-c-Fos (Ser374) (sc-81485, Santa Cruz) were purchased commercially.
For immunoprecipitation experiments, cells were washed with cold phosphate buffered saline (PBS) once and lysed in a NP-40 lysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 0.5% NP-40 and supplemented with protease inhibitors and phosphatase inhibitors) for 1 h at 4°C with gentle shaking. 10% of cell lysates were mixed directly with 5x Laemmli sample buffer and used as input on western blot. The 90% cell lysates were incubated with specific antibody for 3 h at 4°C, followed by addition of protein A conjugated beads for 1 h at 4°C. Immunoprecipitates were washed three times with lysis buffer, and proteins were eluted from beads with 50 μL 1x Laemmli loading buffer.
For western blotting, cells were washed with cold PBS once and lysed in 1x Laemmli loading buffer directly or in NP-40 buffer if used for a subsequent immunoprecipitation. Lysates were heated at 99°C for 5 min, and resolved on 8-15% SDS-PAGE and transferred onto nitrocellulose membrane. Membrane was blocked in 5% milk in Tris-buffered saline and Tween 20 (TBST) for 1 h at room temperature, followed by incubation with a primary antibody overnight at 4°C, and a horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. The membrane was imaged either by a Typhoon Scanner (GE healthcare) or a LAS 4000 Imager system (GE healthcare).
In vivo and in vitro ubiquitination assay
The procedures for both assays were modified from our previously published report (35). Briefly, for in vivo ubiquitylation assay, at 36-48 h after transfection, cells were treated with proteasome inhibitor, MG132 (20μM) for 6 h to accumulate polyubiquinated c-Fos and thus increase the sensitivity of detection. Cells were collected and lysed in 1% SDS lysis buffer (50 mM Tris-HCl, pH 7.5, 0.5 mM EDTA and 1 mM DTT) and boiled for 10 min. For immunoprecipitation, the clarified SDS lysates were diluted 10-fold in 0.5% NP-40 lysis buffer, 10% of total lysates were mixed directly with 5x Laemmli sample buffer and used as input on western blot. The rest were incubated with anti-c-Fos antibody for 3 h at 4°C, followed by addition of protein A-agarose for 1 h at 4°C. Immunoprecipitates were washed 3 times with lysis buffer and boiled in 1x Laemmli loading buffer prior to SDS-PAGE. The ubiquitylation levels of c-Fos were determined by immunoblot with anti-Ub antibody.
For in vitro ubiquitylation assay, plasmids expressing different genes, as indicated in the figures, were transfected into HEK293T cells. 48 hours after transfection, cells were lysed in NP-40 lysis buffer supplemented with a cocktail of protease inhibitors and phosphatase inhibitors, followed by immunoprecipitation using anti-c-MYC agarose (A7470, Sigma-Aldrich) or anti-HA Sepharose (sc-7392 AC, Santa Cruz). Immunocomplexes were washed with the lysis buffer 3 times and eluted with MYC (EQKLISEEDL) or HA (YPYDVPDYA) antigen peptides, respectively. 3HA-c-Fos protein was immunopurified and used as the substrate. CUL1-based E3 ligase complex and substrate receptor KDM2B were derived from HEK293 cells co-transfected with plasmids expressing MYC-CUL1, ROC1 and SKP1 or singularly transfected with plasmid expressing MYC-KDM2B, respectively. In vitro ubiquitylation was initiated by mixing the substrate with immunopurified E3 complex and KDM2B in a ubiquitin ligation buffer [50 mM Tris-HCl at pH 7.4, 5 mM MgCl2, 2 mM NaF, 2 mM ATP, 10 nM okadaic acid (Beyotime Biotechnology), 0.6 mM DTT, 12 μg of bovine ubiquitin (Sigma-Aldrich), 1 μg of FLAG-tagged ubiquitin (Sigma-Aldrich), 60 ng of E1 (E301, Boston Biochem), 500 ng of E2 (human Ubc5c), final volume: 30 μL]. The reactions were incubated at 37°C for 1 h on a rotator with gentle shaking, then terminated with SDS sample buffer and boiled at 99°C for 5 min prior to SDS-PAGE. The ubiquitylation levels of c-Fos were determined by immunoblot with either anti-FLAG or anti-HA antibody.
RNA interference
RNAi-mediated down-regulation of human KDM2B, CUL1 was performed by transfecting small interference RNAs (siRNAs) into HEK293 cells in accordance with the manufacturer’s instructions of Lipofectamine RNAi MAX (Life Technologies). A non-targeting control siRNA duplex (sense 5′-UUCUCCGAACGUGUCACGUTT-3′) was included as a negative control. The knockdown efficiency was assessed 72 h after transfection by western blot.
The siRNAs targeting KDM2B (siGENOME SMARTpool M-014930-01-0005, Dharmacon, GE Healthcare) were purchased commercially, which was a mixture of 4 siKDM2B oligonucleotides. The siRNAs targeting CUL1 (Stealth siRNAs HSS112311, HSS112310) were purchased from Life Technologies. The sequences of all siRNAs used in this study are shown below:
siKDM2B #1: CAGCAUAGACGGCUUCUCU
siKDM2B #2: GGGAGUCGAUGCUUAUUGA
siKDM2B #3: GACCUCAGCUGGACCAAUA
siKDM2B #4: GCAAUAAGGUCACUGAUCA
siCUL1 #1: GGCCACUGAAUAAACAGGUAACAAA
siCUL1 #2: GGAGCUCAGUUUGUUGGCCUGGAAU
siFos GGGAUAGCCUCUCUUACUA
RNA isolation and quantitative RT-PCR (qRT-PCR) analysis
Total RNA was isolated from cultured cells using Trizol reagent (Life Technologies) following the manufacturer’s instructions. 2-5 μg total RNA was reversely transcribed with oligo-dT primers and preceded to qRT-PCR with gene-specific primers in the presence of SYBR Premix Ex Taq (TaKaRa). All data was performed in triplicate and β-actin (ACTB) was used as a housekeeping control. Relative abundance of mRNA was determined in 7500 real-time PCR system (Applied Biosystems). Primers (FLAG-c-FOS-F/R) were designed to specifically detect the ectopic FLAG-tagged c-Fos, but not endogenous c-Fos FLAG-c-FOS. Primer sequences are below:
ACTB-Forward: GCACAGAGCCTCGCCTT
ACTB-Reverse: GTTGTCGACGACGAGCG
c-Fos-F: CTACCACTCACCCGCAGACT
c-Fos-R: GTGGGAATGAAGTTGGCACT
FLAG-c-FOS-F: GGTAGGCCTCGTACGCTTAAT
FLAG-c-FOS-R: AGGATGACGCCTCGTAGTCT
CCND1-F: GCTGCGAAGTGGAAACCATC
CCND1-R: CCTCCTTCTGCACACATTTGAA
MMP1-F: TTCGGGGAGAAGTGATGTTC
MMP1-R: TCTCTGTCGGCAAATTCGTA
MMP9-F: TGTACCGCTATGGTTACACTCG
MMP9-R: GGCAGGGACAGTTGCTTCT
MMP13-F: TCCTGATGTGGGTGAATACAATG
MMP13-R: GCCATCGTGAAGTCTGGTAAAAT
CD44-F: GACAAGTTTTGGTGGCACG
CD44-R: CACGTGGAATACACCTGCAA
PTGS2-F: GGCGCTCAGCCATACAG
PTGS2-R: CCGGGTACAATCGCACTTAT
Statistical analysis
Statistical analyses were performed with a paired, two-tailed Student's t-test. All data shown represent the results obtained from triplicated independent experiments with standard errors of the mean (MEAN ± S.D.). The values of p<0.05 were considered statistically significant. No sample was excluded from the analysis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Fudan MCB laboratory for discussions and support throughout this study and Michele Pagano of NYU for providing plasmids expressing full-length human KDM2B cDNA. This work was supported by Chinese Ministry of Sciences and Technology 973 (Grant No. 2015CB910401), NSFC (Grant No. 81225016, 81430057), Shanghai Key basic research program (12JC1401100), Shanghai Outstanding Academic Leader (Grant No.13XD1400600) and the Youth Science and Technology Leading Talent by MOST (to Q.Y.L), NIH grants EY022611 and CA132809 (to K.L.G.) and GM067113 (to Y.X.).
Footnotes
CONFLICT OF INTEREST
The authors declare that there is no conflict of interests regarding the publication of this paper.
REFERENCES
- 1.Rauscher FJ, 3rd, Sambucetti LC, Curran T, Distel RJ, Spiegelman BM. Common DNA binding site for Fos protein complexes and transcription factor AP-1. Cell. 1988 Feb 12;52(3):471–80. doi: 10.1016/s0092-8674(88)80039-4. [DOI] [PubMed] [Google Scholar]
- 2.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003 Nov;3(11):859–68. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 3.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002 May;4(5):E131–6. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4-10;311(5985):433–8. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 5.Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 1988 Aug 12;54(4):541–52. doi: 10.1016/0092-8674(88)90076-1. [DOI] [PubMed] [Google Scholar]
- 6.Sassone-Corsi P, Ransone LJ, Lamph WW, Verma IM. Direct interaction between fos and jun nuclear oncoproteins: role of the 'leucine zipper' domain. Nature. 1988 Dec 15;336(6200):692–5. doi: 10.1038/336692a0. [DOI] [PubMed] [Google Scholar]
- 7.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995 Jan 19;373(6511):257–61. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 8.Nakakuki T, Birtwistle MR, Saeki Y, Yumoto N, Ide K, Nagashima T, et al. Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell. 2010 May 28;141(5):884–96. doi: 10.1016/j.cell.2010.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995 Oct 16;14(20):4905–13. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gille H, Sharrocks AD, Shaw PE. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992 Jul 30;358(6385):414–7. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 11.Okazaki K, Sagata N. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J. 1995 Oct 16;14(20):5048–59. doi: 10.1002/j.1460-2075.1995.tb00187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stancovski I, Gonen H, Orian A, Schwartz AL, Ciechanover A. Degradation of the proto-oncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol Cell Biol. 1995 Dec;15(12):7106–16. doi: 10.1128/mcb.15.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen RH, Abate C, Blenis J. Phosphorylation of the c-Fos transrepression domain by mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):10952–6. doi: 10.1073/pnas.90.23.10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakiri L, Reschke MO, Gefroh HA, Idarraga MH, Polzer K, Zenz R, et al. Functions of Fos phosphorylation in bone homeostasis, cytokine response and tumourigenesis. Oncogene. 2011 Mar 31;30(13):1506–17. doi: 10.1038/onc.2010.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki T, Kojima H, Kishimoto R, Ikeda A, Kunimoto H, Nakajima K. Spatiotemporal regulation of c-Fos by ERK5 and the E3 ubiquitin ligase UBR1, and its biological role. Mol Cell. 2006 Oct 6;24(1):63–75. doi: 10.1016/j.molcel.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Gilley R, March HN, Cook SJ. ERK1/2, but not ERK5, is necessary and sufficient for phosphorylation and activation of c-Fos. Cellular Signalling. 2009;21(6):969–77. doi: 10.1016/j.cellsig.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 17.He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nature cell biology. 2013;15(4):373–84. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nature cell biology. 2012 May;14(5):457–66. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008 Nov;15(11):1169–75. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfau R, Tzatsos A, Kampranis SC, Serebrennikova OB, Bear SE, Tsichlis PN. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):1907–12. doi: 10.1073/pnas.0711865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kottakis F, Foltopoulou P, Sanidas I, Keller P, Wronski A, Dake BT, et al. NDY1/KDM2B functions as a master regulator of polycomb complexes and controls self-renewal of breast cancer stem cells. Cancer Res. 2014 Jul 15;74(14):3935–46. doi: 10.1158/0008-5472.CAN-13-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood. 2011 Apr 7;117(14):3869–80. doi: 10.1182/blood-2010-10-312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Invest. 2013 Feb;123(2):727–39. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006 Feb 16;439(7078):811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 25.Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Johansen Jens V, Helin K. Fbxl10/Kdm2b Recruits Polycomb Repressive Complex 1 to CpG Islands and Regulates H2A Ubiquitylation. Molecular Cell. 2013;49(6):1134–46. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006 Sep;26(18):6880–9. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koyama-Nasu R, David G, Tanese N. The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nature cell biology. 2007;9(9):1074–80. doi: 10.1038/ncb1628. [DOI] [PubMed] [Google Scholar]
- 29.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996 Jul 26;86(2):263–74. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 30.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997 Oct 17;91(2):209–19. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 31.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997 Oct 17;91(2):221–30. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 32.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002 Apr 18;416(6882):703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 33.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010 Oct 29;143(3):470–84. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maher VM, Heflich RH, McCormick JJ. Repair of DNA damage induced in human fibroblasts by N-substituted aryl compounds. Natl Cancer Inst Monogr. 1981 Dec;58:217–22. [PubMed] [Google Scholar]
- 35.Furukawa M, Andrews PS, Xiong Y. Assays for RING family ubiquitin ligases. Methods in molecular biology. 2005;301:37–46. doi: 10.1385/1-59259-895-1:037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.