Abstract
Glycoproteomics involves the study of the glycosylation events on protein sequences ranging from purified proteins to whole proteome scales. Understanding these complex post-translational modification (PTM) events requires elucidation of the glycan moieties (monosaccharide sequences and glycosidic linkages between residues), protein sequences, as well as site-specific attachment of glycan moieties onto protein sequences, in a spatial and temporal manner in a variety of biological contexts. Compared with proteomics, bioinformatics for glycoproteomics is immature and many researchers still rely on tedious manual interpretation of glycoproteomics data. As sample preparation protocols and analysis techniques have matured, the number of publications on glycoproteomics bioinformatics has increased substantially; however, the lack of consensus on tool development and code reuse limits the dissemination of bioinformatics tools because it requires significant effort to migrate a computational tool tailored for one method design to alternative methods. This review discusses algorithms and methods in glycoproteomics, and refers to the general proteomics field for potential solutions. It also introduces general strategies for tool integration and pipeline construction in order to better serve the glycoproteomics community.
I. INTRODUCTION
Protein glycosylation is the most common and complex form of post-translational modification (PTM) covering a large portion of the entire protein repertoire (Apweiler et al. 1999, Khoury et al. 2011). Glycosylation modulates the biophysical properties of the carrier proteins and strongly influences interactions with binding partners. Because glycosylation is essential to all physiological systems, characterizing glycoproteomes (Thaysen-Andersen & Packer 2014) is a critical step to understanding the functions of individual proteins and dynamically regulated protein networks in both normal and pathogenic conditions (Ohtsubo & Marth 2006, Freeze 2013).
Glycan biosynthesis occurs with strict enzymatic specificity; glycans are appended to specific attachment sites (sequons) on the protein sequences and then subjected to a series of enzymatic modifications. In eukaryotes, glycosylation is usually divided into two main categories: N-glycosylation and O-glycosylation; other types exist but are rare. N-Glycosylation sites consist of the characteristic sequon Asn-X-Ser/Thr, where X represents any amino acid except proline. Asparagine residues not located in canonical sequons occur in rare circumstances (Zielinska et al. 2010). All N-glycans share a common pentasaccharide core sequence, and are classified into three categories depending on the topology of the glycan residues attached to the core: high mannose, complex and hybrid type. By contrast, O-glycosylation shows more diverse forms in terms of the attachment site and linker monosaccharide. Mucin-type O-glycans are covalently attached to a hydroxyl group of Ser/Thr through N-acetylgalactosamine (GalNAc) and involved in host defense by trapping bacterial pathogens. Many nuclear, cytoplasmic and mitochondrial proteins are dynamically modified at Ser/Thr residues by β-N-acetylglucosamine (GlcNAc) or phosphorylation, and responsible for cellular signaling events. Proteoglycans have Ser residues modified by glycosaminoglycan (GAG) chains via a xylosyl linker, and participate into protein binding events via sulfated GAG chains. O-mannosylation and O-fucosylation are important modifications found only on a subset of animal proteins.
In contrast to classical information carrier macromolecules (DNA, RNA and protein), the synthesis of glycans is non-template driven, and therefore results in heterogeneous mature structures. Thus, spatial and temporal variability of glycosylation increases the diversity of the protein sequences by several orders of magnitude. Taken as a population, mature glycoproteins exist as a set of glycosylated forms distinct in structures and functions. Thus, within a protein population, some glycosylated forms will bind a given partner and have an associated biological function and others will not bind. In order to determine structures of functionally relevant glycoprotein forms, multiple techniques have been used to study the combinative structures of proteins and glycans (Mariño et al. 2010).
Mass spectrometry has emerged as an essential technique in glycoprotein and glycoproteomics analysis due to its high throughput, high sensitivity and capability of analyzing complex samples. Over the past few years, mass spectral sample throughput, sensitivity and data quality have increased rapidly. The consensus is that the best quality data result from the combination of collisional (including collision-induced dissociation (CID) / collisionally activated dissociation (CAD)), high-energy collision dissociation (HCD)) and activated electron dissociation (including electron capture dissociation (ECD) and electron-transfer dissociation (ETD)) to provide complementary sequence information of the glycopeptides, and allow the identification of glycopeptide from the tandem mass spectra in a single step (Mayampurath, Yu, et al. 2014).
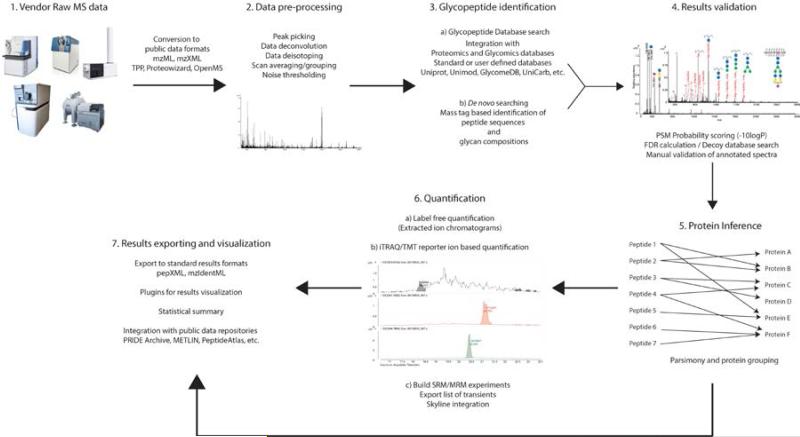
Compared with the rapid advance of instruments and experimental workflows, the development of tools supporting (semi-) automated glycoproteomics analysis remains immature compared to that for computational proteomics (Perez-Riverol et al. 2014). Most glycoproteomics software packages are tailored to the needs of individual glycoproteomics laboratories with little attention to code reuse. It would be beneficial to have integrated and automated frameworks for glycoproteomics, similar to those in proteomics (Sturm et al. 2008, Deutsch et al. 2010, McIlwain et al. 2014, Vaudel et al. 2015). These frameworks should in principle consist of modules for file conversion, spectra pre-processing, sequence identification, quantification and, favorably, visualization and interaction with online databases (Figure 1). However, many state-of-the-art tools in proteomics remain unassessed or incompatible (either on format or algorithmic principle) regarding their validity in glycoprotein/glycoproteomics study. As a result, researchers in glycoproteomics often find themselves in a dilemma whether to modify existing proteomics tools or to develop tailored tools from scratch.
Figure 1. Basic workflow for automated analysis of glycoproteins.
A comprehensive glycoproteomics pipeline should be able to either handle vendor specific multidimensional data formats or convert them to public data formats. Signal processing capabilities are an essential component of a mass spectrometry data analysis pipeline to ensure proper deconvolution, deisotoping, noise reduction and feature grouping. The pre-processed data must be fed into a search engine which uses database search or de novo sequencing algorithms to identify peptides/glycopeptides and then validates and scores matches. Glycopeptide identification and validation would be followed by glycoprotein inference and grouping. An optional but important quantification module would provide absolute or relative quantification capabilities at both glycopeptide and glycoprotein levels. Finally, the results from glycopeptide and glycoprotein identification and quantification must be exported appropriate data structures. It is also important to have a built-in results visualization tool for users, along with basic sorting and filtering capabilities.
In this review, we discuss current problems and computational solutions in glycoproteomics and explore the options of migrating comparable approaches in the larger proteomics field. While the overall workflow for glycoproteomics as shown in Figure 1 bears similarity to those used in proteomics, major differences exist in the strategies for glycopeptide identification, validation of results and subsequent quantification of the identified species. The data pre-processing and output visualization steps are integral to mass spectrometry based analytics, regardless of the analyte. Mature strategies from proteomics can therefore be directly applied to glycoproteomics data for data preprocessing, protein inference and results visualization. To that end, we discuss several strategies towards code reuse and pipeline deployment. Readers should refer to recent literatures (Li et al. 2013, Desaire 2013, Dallas et al. 2013, Woodin et al. 2013) for comprehensive overviews of specific software tools in glycoproteomics analysis.
II. IDENTIFICATION METHODS FOR GLYCOPEPTIDES
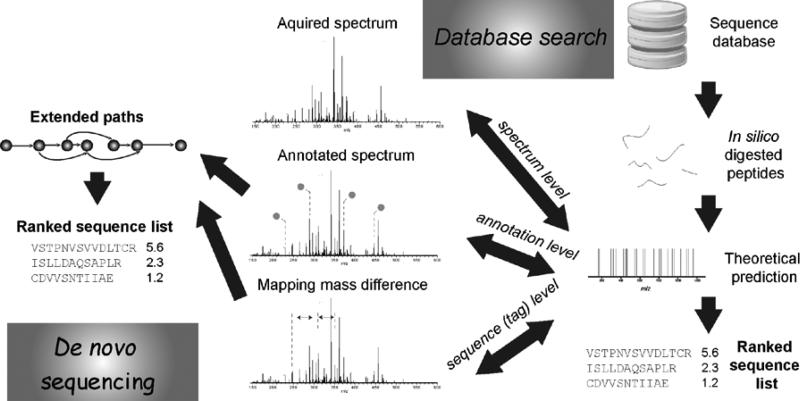
In a typical shotgun proteomics study, protein sequences are digested into peptides during proteolysis where the cleavage sites depend on the specificities of the enzymes. Trypsin, the most commonly used enzyme, specifically cleaves the sequences at the carboxyl side of arginine and lysine, unless preceded by proline. The peptide mixture undergoes chromatographic separation and mass spectrometric analysis. Designated peptide ions (precursor ions) are further dissociated into fragments (product ions) with their m/z values and abundances recorded as tandem mass spectra. The procedure of sequencing, or structure identification, involves proposing candidate sequences for the spectra that meet the instrumental, analytical and biological constraints. These constraints include: (1) precursor ion: the theoretical mass values, isotopic distribution and other characteristic features from the predicted sequence should match the observation; (2) product ion: the predicted product ion mass values and their expected relative ion abundances (if applicable) and neutral loss should match the actual tandem mass spectra; (3) spectra: structurally related sequences, e.g. alternative splicing variants and proteoforms (Smith et al. 2013), should produce similar spectra; and (4) sequence pattern: the predicted sequences should largely follow the enzymatic cleavage rules and biosynthetic rules posed on the molecule species. Depending on whether a prior sequence database is involved, identification methods are broadly divided into database search and de novo sequencing (Figure 2). Database search method looks for candidate peptides from in silico digested protein sequences, while de novo sequencing usually explores the relationship among peaks and constructs paths to represent candidate peptides. Once proposed, the candidate sequences are scrutinized and weighted according to their fitness to the constraints listed above, and top candidate(s) kept as the identified sequence(s) for the spectrum. Alternative methods also remain important in tandem MS-based sequence identification. These include hybrid methods, which combines the features of database searching and de novo sequencing, and spectral libraries, which store annotated tandem MS spectra for searching.
Figure 2. Illustration of different identification methods.
The connection between experimental tandem mass spectra and candidate sequences can be established in multiple styles. The experimental spectra can also be further converted to annotated peaks or sequence tags. The experimental data (spectra, peaks, or sequence tags) can either be used to build graph in a bottom-up way (de novo sequencing) or match against the theoretical data derived from database (database search).
In addition to generating candidate sequences, algorithms should also try to capture the characteristics of the generated tandem mass spectra. This step strongly depends on the mechanism of the fragmentation, and is affected simultaneously by multiple factors, including instrument types, collision energies, and experimental design (e.g. MSn). Using vibrational dissociation methods (including CID, CAD, HCD, and IRMPD), product ion abundance scales directly with bond lability. Thus, cleavage of glycosidic bonds is favored using vibrational dissociation. Nonetheless, at higher energies, dissociation of the amide/peptide bonds occurs and therefore yields peptide backbone information. As a complement, activated electron dissociation (ExD) methods, such as ECD and ETD, maintain the glycan moiety on peptide fragments, which is extremely useful in identifying the exact attachment site of the PTM. Sequential MS/MS (MSn) can also be used with either CID or ETD for multi-stage fragmentation of glycopeptides to yield the desired information on the glycan or peptide backbone. Different fragmentation methods and their corresponding patterns are summarized in Table 1. Identification of the peptides, glycan structures as well as their attachment sites all benefit from such complementary structural information. The study of fragmentation methods to generate informative fragments (Vékey et al. 2013) is a fast-moving field in glycoproteomics, as new instruments and dissociation techniques consistently emerge and couple with new fragmentation patterns, which requires frequent update of scoring models.
Table 1. Structural information provided by different fragmentation methods.
Analysis of de-glycosylated peptides may resolve to some extent the site(s) of glycan attachment to peptide but the information on site-specific glycan compositions is lost. Concomitantly, released glycans can be analyzed to identify the total pool of glycans on a glycoprotein but where multiple glycosylation sites exist, information linking glycan compositions to individual sites, is lost. It is therefore desirable to analyze intact glycopeptides. Glycopeptide analysis using mass spectrometry is challenging due to the large glycan post-translational modification attached to the peptide, which renders conventional peptide analysis methods inefficient. The macro and micro-heterogeneity of glycans leads to a non-linear increase in the theoretical search space or the number of possible compositions of glycopeptides. As a result, mass profiling (MS1) of glycopeptides yields composition assignments with a great deal of ambiguity and high FDR (Khatri et al. 2014), even when high resolution mass spectrometers are used (Desaire & Hua 2009). Therefore, tandem MS (MS2) becomes imperative for increasing confidence and decreasing FDR in glycopeptide assignment. Tandem MS is a technique where ionized analytes are fragmented in the mass spectrometer and the masses of the resulting fragments are then measured to further resolve any ambiguities in intact mass based assignments. Tandem MS of glycopeptides is typically performed using either of two major fragmentation modes involving vibrational/collisional activation (CID, HCD) and electron/radical based activation of the analyte (ETD, ECD). In addition to these methods, photodissociation mass spectrometry has recently made its way into glycopeptide analysis (Madsen et al. 2013), while other methods like SID (Surface Induced Dissociation) and Infrared multiphoton dissociation (IRMPD) (Seipert et al. 2008) are being explored. Each method is somewhat unique, in terms of accessibility and the kind of information generated.
| Fragmentation Method | Instrument type | Type of fragment ions generated |
|---|---|---|
| Trap CID | IT | • Stub-glycopeptide ions (intact peptide ions with small glycan fragments) • Abundant ions from loss of monosaccharide units from the precursor • Oxonium ions are observed based on acquisition range and precursor m/z • Peptide backbone ions (b and y) may be observed in sequential tandem MS. |
| Beam type CID | TOF-TOF, Q-TOF, Q-FTICR | • Mono, di or tri-saccharide oxonium ions.(Glycosidic bond cleavages) • Stub-glycopeptide ions • Ions from loss of monosaccharide units from the precursor • Peptide backbone ions. (b and y) • Peptide backbone (b and y) ions with the starting monosaccharide (HexNAc) |
| HCD | Orbitrap | • Mono, di or tri-saccharide oxonium ions. (Glycosidic bond cleavages) • Stub-glycopeptide ions. • Ions from loss of monosaccharides from the precursor • Peptide backbone ions. (b and y) |
| ECD | FTICR | • Peptide backbone ions (c and z ions with modification). • Labile PTMs like glycans and phosphate groups remain largely intact at the modification site. • Charge remote fragmentations, such as internal fragments and amino acid side chain losses can occur. • Better for shorter peptides than ETD due to additional fragmentation, which can be controlled by varying average electron energy.(Zhurov et al. 2013) |
| ETD | IT-FTICR, IT-Orbitrap | • Same as ECD (c and z ions with modifications intact). • Prone to steric effects and electron affinity depending on reagent used. • Fewer charge remote fragmentations and secondary cleavages than in ECD. (Li et al. 2010) |
| UVPD | IT-FTICR, IT-Orbitrap | • Peptide backbone ions with labile modification intact. (Predominantly a and x ions) • Diagnostic ions from glycosidic bond and cross-ring cleavages. |
A common strategy to simplify the glycopeptide identification complexity is to divide the task into known problems in proteomics and glycomics. This allows the software packages used in proteomics and glycomics to be integrated into the glycoproteomics workflow with little modification. From the perspective of the peptide, the attached glycan moieties correspond to PTM of large mass values, and may derive multiple PTM variants if the fragmentation causes loss of glycan residues. Each PTM variant expands the search space and therefore the computational time to generate the candidate sequences. Accompanied with deglycosylation or a genetic editing technique such as SimpleCell (Steentoft et al. 2011), glycopeptides are converted into peptides with homogeneous modifications, which is a routine PTM localization task supported by mainstream search engines (Chalkley & Clauser 2012). On the other hand, the glycan moieties can be detached and sequenced separately following traditional glycomics workflow (Leymarie & Zaia 2012), or built upon the peptide backbone in a de novo style through dynamic programming (DP) (Serang et al. 2013). We will discuss these strategies in the following subsections.
Analysis of glycoprotein site-specific glycosylation requires the identification of glycopeptides (component 3 in Figure 1), validation of the sequence-spectrum match (component 4 in Figure 1), and mapping of peptides into proteins (component 5 in Figure 1). Currently, most algorithms and tools focus on improving the performance in glycopeptide identification step (component 3 in Figure 1), which can further be divided into four smaller steps, including: (1) detection of glycopeptide spectra, (2) inference of peptide mass and selection of candidate sequences, (3) inference of the glycan structure information, (4) scoring the peptide/glycan-to-spectrum matches. The detection of glycopeptide spectra can be finished either on MS1 level based on mass defect (Froehlich et al. 2013) or MS2 using oxonium ions or other characteristic ions such as 0,2X ions (for low resolution CID on acidic glycopeptide (Irungu et al. 2007)). Intact peptide masses can be corrected from deglycosylated peptides or Y-ions of glycopeptides (Cheng et al. 2014, Lynn et al. 2015). In the case of one glycosylated site, the glycan mass can then be inferred by deducting the inferred peptide mass from the precursor ion mass. Alternatively, combinations of peptides and glycans can be enumerated and evaluated based on the matching between the spectrum and peptide/glycan.
A. DATABASE SEARCH-BASED METHODS
In 1994, Mann and Wilm introduced PeptideSearch (Mann & Wilm 1994) to extract sequence tags from tandem mass spectra and search them against the sequence database. In the same year, Eng et al. (Eng et al. 1994) implemented SEQUEST, an automated database search method based on a cross-correlation function. Since then, several other database search methods have been developed, including Mascot (Perkins et al. 1999), X!Tandem (Craig & Beavis 2004), OMSSA (Geer et al. 2004), MyriMatch (Tabb et al. 2007), ProteinProspector (Clauser et al. 1999, Chalkley et al. 2008), Andromeda (Cox et al. 2011), Morpheus (Wenger & Coon 2013), Comet (Eng et al. 2013), and MS Amanda (Dorfer et al. 2014). Database search methods are widely used in peptide identification (Steen & Mann 2004, Sadygov et al. 2004), and remain the standard procedure in large-scale proteome analysis workflow (Kim et al. 2014, Wilhelm et al. 2014). Despite their distinct features, all search engines follow the general procedure of matching the experimental tandem mass spectra against a sequence database.
There are multiple stages to associate a tandem mass spectrum to a candidate sequence in the database (Figure 2). Each protein sequence is digested to peptides in silico based on the cleavage specificity of the enzyme specified. These peptides may be further fragmented to generate theoretical tandem mass spectra according to the favored ion types of the selected fragmentation method. The similarity between experimental data and expected data may be measured either on the spectral level (spectrum-spectrum similarity, such as SEQUEST (Eng et al. 1994)), fragment level (ion count or scoring function based on matched ions, the method adopted by most database search engines) or peptide level (homology search, such as PEAKS DB (Zhang, Xin, et al. 2012)). Sadygov et al. categorized the scoring function of peptide-spectrum match (PSM) into four types: descriptive, interpretative, stochastic and probability-based (Sadygov et al. 2004). Despite its specific form, the actual performance of a scoring function relies on how its assumption fits the actual data. As suggested by Wenger & Coon, when high accuracy data are used, a simple counting strategy (used by Morpheus) beats the complex probabilistic models used by other search engines (Wenger & Coon 2013). When the searching performance is a concern, sequence tags (short amino acid sequences) derived from the spectrum can be used to filter the candidate sequences and therefore reduce the search space (Tabb et al. 2003).
Glycopeptide identification based on database searches usually requires separate identification of the peptides and glycans. GlycoPep Evaluator (Zhu et al. 2014) generates peptide candidates containing the characteristic sequon, and glycan list from GlycoMod (Cooper et al. 2001). GlycoPeptideSearch (Chandler et al. 2013) provides a comprehensive solution for glycopeptide identification, and implements functions such as glycopeptide spectra detection (oxonium ions recognition), intact-peptide fragment ion matching, glycan structure retrieval, evaluation of spectrum to peptide/glycan matches and FDR estimation. GlycoMaster DB (He et al. 2014) searches protein sequence database and glycan sequence database for the best pair, and uses the complementary information from HCD/ETD pairs to derive the glycopeptide sequences. The separation of peptide and glycan also means current search engines can be adapted to work on the glycopeptide data. GlyDB (Ren et al. 2007) represents the glycan structure in multiple linear sequences, and utilized SEQUEST (Eng et al. 1994) to search glycoforms from a theoretical glycan database. MAGIC (Lynn et al. 2015) generates in silico peptide tandem mass spectra in MGF format from the original glycopeptide spectra, and identified the corresponding peptide sequences using Mascot and X!Tandem. We expect that more software packages will emerge and gear the mature search engines towards the complex glycopeptide data.
B. DE NOVO SEQUENCING-BASED METHODS
Compared to database search methods, de novo sequencing requires no prior database. It is commonly treated as a subsidiary solution to support identification results from database searches due to the lack of statistical validation and concerns regarding its search performance (Allmer 2011). However, it becomes indispensable when the homologous sequence database is unavailable (e.g. snake venom proteomes (Fox & Serrano 2008)), the species of organism is unknown, single amino acid mutation (Su et al. 2014) or the target proteins undergo rapid mutation (e.g. viral pathogens) or recombination (antibody variable regions).
The definition of de novo sequencing is very broad. Allmer categorizes current de novo sequencing methods in proteomics into naïve approaches, spectrum graph models, probabilistic and combinatorics models (Allmer 2011). Naïve approaches follow a general procedure of enumerating sequences and filtering/optimizing sequences. PEAKS (Ma et al. 2003), which is the most popular commercial software for de novo sequencing, generates 105 sequences and merges them into a consensus sequence with local confidence on residues. Heredia-Langner et al. (Heredia-Langner et al. 2004) implemented a genetic algorithm to optimize the candidate peptides efficiently from a very large search space. In glycomics, STAT (Gaucher et al. 2000) enumerated all possible combinations of monosaccharide residues matching the mass values of precursor ions and product ions, and produces the most likely glycan structures that meet the monosaccharide connectivity constraint.
The principle of naïve approaches in essence is no different than searching in a database. For this reason many researchers view de novo sequencing as a generalization of database searching (Allmer 2011, Na & Paek 2014). However, this doesn't necessarily mean de novo sequencing has to rely on search. Given an unmodified peptide with perfect fragmentation (high coverage and few spurious peaks), the peptide sequence can be assembled immediately by connecting the product ion pairs whose mass difference matches the mass value of certain amino acid residue, which is actually independent of the size of the overall sequence space. As the consistent improvement of instrument accuracy and experimental design, researchers are gradually approaching the ideal data quality required for de novo sequencing.
The major class of de novo sequencing explores the peak relationships and builds a graph from the tandem mass spectrum with the vertex representing the observed peak and the edge representing the amino acid residue mapped from the mass difference between vertexes. The optimal sequence corresponds to the longest path in the graph, and dynamic programming is usually used to solve the problem (Dancik et al. 1999, Chen et al. 2001, Mo et al. 2007). However, dynamic programming usually gives only the optimal solution (Lu & Chen 2003). Due to the ambiguity and unknown fragmentation mechanisms, even the optimal sequence may not be the correct one. Therefore, algorithms looking for suboptimal solutions were also developed (Lu & Chen 2003). Modern de novo sequencing algorithms tend to integrate complementary information and take advantage of high accuracy instruments. pNovo+ (Chi et al. 2012) integrated complementary HCD/ETD spectra pairs and provided an efficient algorithm to identify the k-longest paths in a directed cyclic graph. PepNovo+ (Frank 2009a) evaluated the path scores from spectrum graph as well as features from peak rank prediction (Frank 2009b), peak annotation, peak offset and sequence composition, and achieved great accuracy improvement of peptide sequence tags. UniNovo (Jeong et al. 2013) offered an automatic learning procedure to determine the ion type of the spectra, and are applicable to different spectra types and spectra pairs.
In glycomics, similar strategies were used to identify glycan structure. The candidate structures can be represented by subgraphs or trees in data structure through dynamic programming. StrOligo built a relationship tree representing loss of monosaccharide compositions between peaks, which was used to determine the most likely composition and infer the candidate structures based on biosynthetic rules (Ethier et al. 2002, 2003). GlyCH predicted the most probable glycan sequence while taking the cross-ring ions and double fragmentation ions into account (Tang et al. 2005). Shan et al. proved that finding the optimal glycan structure is an NP-hard (Non-deterministic Polynomial-time hard) problem (Shan et al. 2008). In glycan de novo sequencing, repetitive peak counting is a potential problem affecting the scoring performance of algorithms. Compared to peptide sequence, glycans have larger chance to generate isomeric ions (ions with the same theoretical mass values) due to the similar ring structures of monosaccharide residues and high occurrence of a small set of residues. GlyCH simply allowed the repeated usage of the same peaks in candidate scoring (Tang et al. 2005). Shan et al. proposed a heuristic approach and enforced that the peak should be used only once (Shan et al. 2008). Böcker et al. also proposed an exact algorithm to efficiently generate candidate sequences while require the most k intensive peaks should be counted only once (Böcker et al. 2011). All the studies mentioned above work for N- and/or O-glycan sequences. For glycosaminoglycan identification, where single peaks also correspond to multiple fragment interpretations, we developed the first sequencing algorithm HS-SEQ using divide-and-conquer strategy and graphic model built from peak interpretations, and achieved highly accurate performance in heparan sulfate sequencing (Hu et al. 2014). HS-SEQ avoided the arbitrary decision of re-using the same peaks, and focused on selecting the most confident fragment interpretations.
Although the peptide part and the glycan part can be identified by de novo sequencing separately, few algorithms work on the glycopeptide as a whole. Serang et al. designed a dynamic programming tool SweetSEQer to sequentially build the longest path for the peptide sequence and the largest directed subgraph for the glycan part (Serang et al. 2013). However, when multiple glycan sequences are present on the peptide, it may be difficult for dynamic programming to find the right peaks to extend the monosaccharide residues due to multiple interpretations of peaks.
C. SPECTRAL LIBRARIES
Introduced by Yates et al. in 1998 (Yates et al. 1998), the spectral library approach has become widely used for peptide identification (Lam 2011). It utilizes assigned reference spectra to identify new experimental spectra. Compared to traditional database search methods, it limits the search space size to improve computational speed, and maintains complete spectral information to improve identification sensitivity (Zhang et al. 2011). Recently, it has been applied to glycoproteomics (Toghi Eshghi et al. 2015).
The spectral library framework is built upon the observation that for a given peptide sequence, tandem mass spectra are reproducible when instrument condition are controlled precisely (Yates et al. 1998). The spectral patterns usually include information that is not captured by traditional database search methods, including peak intensities, neutral losses, and non-canonical or unknown ions (Zhang et al. 2011). Once a spectrum has been assigned to a sequence, any future spectra showing high similarity with the reference spectrum will be linked to the sequence assignment. Due to the reduced sequence space, the spectral library approach can reduce computation time by a hundred fold or more compared to traditional approaches, with better sensitivity and accuracy (Zhang et al. 2011).
The spectral library approach typically contains library construction and library matching procedures (Lam 2011), and multiple algorithms have been developed for such identification. These include Bibliospec (Frewen et al. 2006), X! Hunter (Craig et al. 2006) and SpectraST (Lam et al. 2007). Library construction usually requires high-quality spectra which have been assigned with high confidence. The original identification information can be acquired through traditional database searches (e.g. SEQUEST), de novo sequencing, or even manual interpretation. One limitation of using the spectral library approach is that the search space is usually limited to spectra that have been identified previously. Sequences with no spectra in the library cannot be identified in subsequent experiments. Sequences with un-specified PTM types are also likely to be missing in the identification results. This places high importance on maximizing the coverage of the spectral libraries. As a remedy, simulation of peptide spectra using MassAnalyzer (Zhang 2004, 2005) has been introduced to the preparation of the library (Yen et al. 2009) in order to increase the coverage. A combination of a simulated spectral library and a public reference library (Cho et al. 2015) was also reported to improve the ability to detect proteins that are not identified using traditional database searches.
The presence of PTMs can affect fragmentation pathways of peptides, resulting in mass shifts and intensity variations of the fragment ions. Additional ions (resulting from neutral losses or internal fragmentation) may also be present in the experimental tandem mass spectra and affect the spectrum-spectrum matching performance. To address the issue, significant effort has been made to build comprehensive spectral libraries for peptides with site-specific phosphorylation (Hu & Lam 2013). In that study, the authors used database searching followed by site-localization tools to assign phosphopeptide sequences, and introduced mass shifts of fragment ions to predict the spectra of phosphopeptides from the dephosphorylated counterparts. Recently, an improved simulation strategy was proposed utilizing the intensity change of fragment ions (between the phosphopeptide spectrum and dephosphorylated counterparts) caused by site-specific phosphorylation (Suni et al. 2015). Efforts to develop spectral libraries for identification of unanticipated PTMs were also reported (Ye et al. 2010, Ahrné et al. 2011, Ma & Lam 2014).
In glycoproteomics, practical limitations impede the use of the spectral library approach. First, it is necessary to construct high-quality reference libraries. This requires confidently assigned sequences and clean tandem mass spectra with low noise levels. For many glycoproteomics studies focusing on improving experimental workflows and profiling glycosylation events, the spectral patterns are likely not to have been identified previously and therefore present in a spectral library. Second, glycopeptide product ions result from dissociation of the glycan to form oxonium ions and neutral losses from the precursor ion. Peptide backbone dissociation may also be observed, resulting in peptide fragment ions with or without glycans attached. The abundances and reproducibility of each ion type depends on specific instrument conditions and needs to be controlled and evaluated closely. Moreover, there lacks a consensus, among researchers studying glycosylation, on the analytical methods used for studying this modification, which limits the availability of glycoconjugate spectral libraries. Nevertheless, there are clear benefits to applying spectral libraries to glycoproteomics.
Deglycosylation of glycopeptides is an important step in many glycoproteomics studies, which reduces the complexity of the spectra, and can directly serve to the construction of spectral libraries. Moreover, modern spectral library approaches (Ye et al. 2010, Ahrné et al. 2011, Ma & Lam 2014) can identify peptides with unanticipated PTMs. This suggests that experimental tandem mass spectra (intact glycopeptide) can be linked to reference spectra (deglycosylated peptide) regardless of the glycan moieties attached. If so, researchers will be able to interpret the peptide backbone and the glycan compositions as separate entities in the tandem mass spectra. In case of ExD methods where the glycan remains intact during peptide fragmentation, the glycan information can be deduced from precursor mass and product ion mass shifts in the experimental spectra. On the other hand, for collisional dissociation where the glycan dissociates during the peptide fragmentation process, glycan information can be construed only from the masses of the glycopeptide precursor ion and the unmodified peptide mass extracted from the product ion pattern. Glycomics experiments or glycan database searches may follow to increase the level of glycan structural detail.
The spectral library approach has been used in a few glycomics and glycoproteomics studies. Kameyama et al. built a library of MSn CID spectra for N-glycans and further proposed a strategy to simulate the CID spectra based on the extracted fragmentation patterns (Kameyama et al. 2005, 2006). Aebersold et al. used deglycosylated peptides to build a spectral library, and conducted further analysis of abundances of inferred glycoproteins using SWATH-MS (Liu, Chen, et al. 2014). Toghi et al. examined the reproducibility of fragmentation patterns between the deglycosylated peptides and peptides containing glycan residues, and designed the algorithm GPQuest to support the site-specific glycopeptide identification (Toghi Eshghi et al. 2015). In their study, the spectral library approach was divided into following steps: 1) library construction using deglycosylated peptides; 2) classification of glycopeptide spectra by examining oxonium ions and other characteristic ions; 3) library search and 4) candidate refinement.
As the prevalence of the SWATH technique and the needs for larger coverage of glycoproteomes have grown, spectral libraries represent one of the future trends in glycopeptide identification. However, researchers should be cautious of the scenarios where the library matching performance might be affected, such as fragmentation patterns of ions carrying glycan residues, overlapping isotopic clusters of fragment ions, and multiple glycosylated sites. In addition, a large portion of peaks in the glycopeptide spectrum may be missing in the reference spectrum, so it is necessary to remove product ions resulting from glycan dissociation from the glycopeptide tandem mass spectrum (Lynn et al. 2015) or adjust the scoring function to achieve high accuracy. For ion types where the assumption of reproducibility does not hold, the matching procedure deteriorates into a normal database search, where no abundance information is assumed. In the next few years, we expect more studies that focus on exploration and automated extraction of spectra patterns in order to design robust scoring functions. Integrated software tools and accessible public spectral libraries will also benefit the glycoproteomics community.
D. INSIGHT FROM PTM PROTEOMICS
PTMs expand the functional diversity of linear protein sequences, and increase the number of structural variants exponentially. Almost all database search tools allow users to specify fixed (static) and flexible modifications. A fixed modification, such as phosphorylation, has the same effect as updating the mass of the modified residue, which causes no extra burden in searching. For sites with flexible modifications, the search engines must consider the situation of either modified or unmodified status for each such site, which causes the search space to grow exponentially (×2x, where x is the number of sites with modified/unmodified flexible modifications). Andromeda (Cox et al. 2011), the search engine of the quantification software MaxQuant (Cox & Mann 2008), exhaustively listed all the possible combinations of PTMs on the protein sequence to improve the identification rates. To speed up the analysis, it built multiple levels of indexing so that the data can be processed even on a laptop (Cox et al. 2011). From the perspective of the peptide, glycan attached to a peptide essentially represents a PTM of large mass value. When the redundant variation of mass shift on the glycosylation sites is removed, the searching procedure is identical to normal database search with flexible modifications. This also serves as theoretical basis for the SimpleCell technique (Steentoft et al. 2011).
Algorithms working on complicated PTM analysis may also shed light to the glycopeptide identification. MODa (Na et al. 2012) combines the advantages of sequence tags and spectral alignment to provide fast identification of multiple unknown modifications on a peptide. In glycoproteomics studies, ETD spectra offer such sequence tags, while high energy CID/HCD spectra can provide the scaffold of an “unmodified” peptide backbone. This should allow the spectra (sequence tags) using ETD to align to the paired spectra (sequence) using high energy CID/HCD. To the best of our knowledge, there is no such kind of tool tailored for glycopeptide identification.
In addition, many PTM proteomics studies assume the presence of unmodified peptides. In unrestrictive database searches, this assumption limits the PTM search only upon the unmodified peptides identified in the first round (Tharakan et al. 2010). In glycoproteomics studies, sample enrichment steps may invalidate this assumption unless unenriched samples are also used for identification.
E. STRATEGIES FOR IMPROVING SEQUENCING PERFORMANCES
Common complaints regarding de novo sequencing methods include time-consuming performance, poor identification rate and coverage, and lack of validation methods. For spectrum graph methods, the missing fragments and ambiguous paths may prevent an algorithm from finding the optimal sequence. For naïve methods, the enumeration of search space undergoes combinatorial explosion, which becomes intractable as the sequence length grows. Significant effort has been made to improve the covered sequence length and accuracy, including: combination of spectra pairs from multiple fragmentation mode (CID/ETD/HCD) and multiple enzymes (Jeong et al. 2013); combination of spectra from top-down and bottom-up proteomics experiments (Liu, Dekker, et al. 2014); and appending de novo sequencing with homologous database search (Ma & Johnson 2012). The emergence of high-resolution MS/MS data also contributed to the identification quality and proteomics-grade sequencing results (Chi et al. 2013).
As discussed above, the peptide part and glycan part built from the spectra can be further coupled to database searches for structure refinement. This hybrid approach searches partially sequenced structures to significantly improve the search performance, and has been adopted by several tools, includine Byonic (Bern et al. 2007, 2012), and PEAKS DB (Zhang, Xin, et al. 2012). Regardless of the final strategy (the integral approach or proteomics + glycomics approach) used, the de novo nature of these methods should allow sensitive identification of highly mutable glycoproteins, such as influenza hemagglutinin.
F. MISCELLANEOUS ISSUES
Glycopeptide identification involves a special problem regarding the discernment of mass spectra corresponding to glycopeptides from those of unmodified peptides. Froehlich et al. reported a MS1-based glycopeptide classifier utilizing the relationship between precursor mass and mass defect for peptides and glycopeptides respectively (Froehlich et al. 2013). On the MS2 level, oxonium ions and other characteristic product ions from HCD were used to flag to the identity of the glycopeptide sequence and triggered the subsequent ETD analysis of the precursor ion (Singh et al. 2012).
In the past few years, effort has been reported on integrating orthogonal, or complementary identification results. In the proteomics field, identification results generated from multiple search engines has been merged under the target-decoy approach (Shteynberg et al. 2011). The logic behind this strategy is that each search engine has a unique preference for the features in tandem mass spectra, and covers distinct sets of peptides that others strategies miss. In addition, it's also worthwhile to study the complementarity of different mainstream search engines in order to balance the identification performance and computation time (Shteynberg et al. 2011). Integration of information can also occur on the spectra level. For example, in proteomics, Guthals et al. combined the CID, ETD and HCD spectra to fill the gap of missing peptide fragments and improve the sequencing accuracy and resolution (Guthals et al. 2013). In glycoproteomics, Mayampurath et al. also integrated the CID/ETD/HCD spectra for complete glycopeptides identification in their GlycoFragWork program (Mayampurath, Song, et al. 2014). There was also effort reported on data integration in top-down and bottom-up experiments, as indicated by Liu et al. in whole protein sequencing (Liu et al. 2014).
Glycopeptides with more than one glycosylation site become more challenging to analyze, due to the aforementioned heterogeneity. Detailed information on site-specific glycoforms requires optimization of sample preparation, chromatography/separation and MS acquisition. Depending on the protein sequence and glycan heterogeneity, one of the following strategies may be used. If the most commonly used proteases do not yield peptides with a single glycosylation site, an alternative enzyme or a combination of proteolytic enzymes may be used. The most efficient strategy for analyzing glycopeptides with multiple glycosylation sites is the use of ExD, which cleaves peptide bonds while leaving the PTMs intact. Collisional dissociation on the other hand, may provide peptide backbone information along with a composite mass of all the glycans present on the peptide. ETD is performed on ion-trap instruments or hybrid instruments. Many such instruments also enable MSn, where a collisional dissociation may be performed on an isolated peptide/glycopeptide fragment to yield more information on glycan structure/topology.
The rapid improvements in the capabilities of high accuracy and high resolution instruments present a game changer in the identification algorithm design. Similar to the advance of database search methods, de novo sequencing approaches also benefit from high accuracy instruments and improvements in fragmentation coverage. Modern de novo sequencing approaches such as pNovo+ (Chi et al. 2012) have reported the achievement of results comparable to the state-of-the-art database search methods when high accurate instrument was used. At present, the current approaches in glycopeptide identification tend to follow classical solutions in proteomics; there remains a clear need to develop novel algorithms that address the unique needs of glycopeptides as hybrid molecules with both genetically templated and metabolically modified components.
III. VALIDATION STRATEGIES FOR GLYCOPEPTIDES
Glycoproteomics is a rapidly developing field. With the emergence of new instruments and experimental protocols, bioinformatics tools and workflows must undergo stringent validation procedures to demonstrate their effectiveness and limitations. In proteomics, there have been too many cases where unreliable conclusions were drawn from improper operation of the state-of-the-art software packages (Gupta et al. 2011). In this section, we start with the widely used target-decoy model and its assumptions in the context of glycopeptide validation, and then summarize several validation strategies reported in current studies.
A. FALSE DISCOVERY RATE AND TARGET-DECOY APPROACH
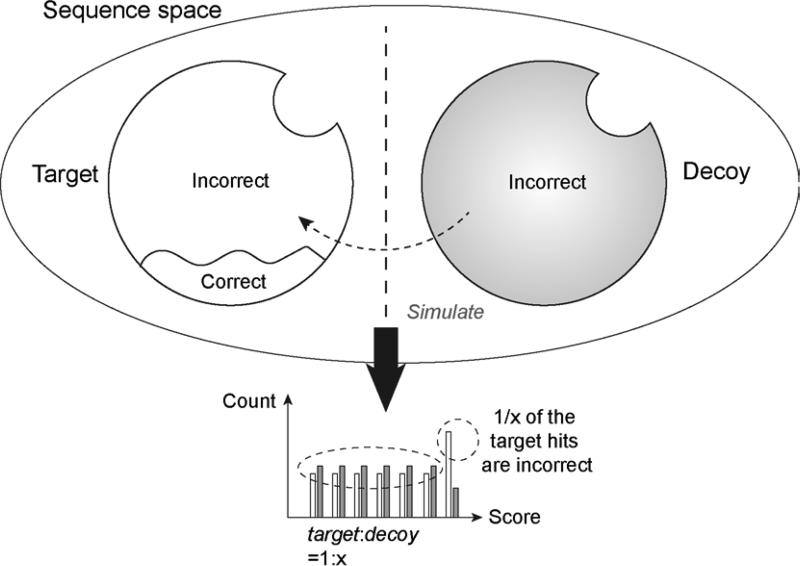
To date, the target-decoy approach (TDA) (Elias & Gygi 2007) has become the de facto standard for validating proteomics identification results. Its wide acceptance and application results from its simple concept and proven performance in estimating false-discovery rate (FDR). Figure 3 shows the basic framework of TDA strategy in estimating FDR.
Figure 3. Principle of target-decoy model in peptide identification.
The FP target hits and decoy hits (assumed to be FPs) share similar data structure (maintain a consistent ratio across the matching scores). Therefore, the FP target hits can be estimated from decoy hits and FDR can be calculated. This model requires the similarity and the non-overlapping feature (orthogonality) between target data and decoy data.
By assuming that the decoy hits simulate the distribution of incorrect target hits across scores (similarity) and the two share few common sequences (orthogonality), the false positives in the target hits can be estimated by replicating the hit number in the decoy peptides. This strategy appears to be universal regardless of the implementation of search engines and scoring functions, and thus enables the comparison across multiple searching tools: with fixed FDR (e.g. 1% or 5%), the tool with the largest number of hits performs best.
The TDA approach facilitates peptide identification in that once the FDR threshold is controlled, the more peptides a search tool identifies, the better the performance it provides. This also allows the strategy of combining multiple search engine results to improve the overall identification rate. The rationale is that each tool may be superior in detecting peptides of some features, but deteriorates in others. A set of tools were developed to support the combination approach, including MSBlender (Kwon et al. 2011), PepArML (Edwards et al. 2009), ConsensusID (Nahnsen et al. 2011), iProphet (Shteynberg et al. 2011), and PeptideShaker (Vaudel et al. 2015). Shteynberg et al. (Shteynberg et al. 2013) reviewed the identification results produced from different combinations of search engines, and suggested that it is important to consider complementarity and similarity between tools to balance the computational time and sensitivity. Similarly, with a controlled FDR, more strategies may be considered to improve the sensitivity, including combinations of multiple fragmentation modes (Guthals & Bandeira 2012), other omics data (Sheynkman et al. 2013), and digestion with multiple enzymes (Choudhary et al. 2003).
The uncertainty from peptide identification amplifies in the protein list. A true positive (TP) protein identification is usually considered as one with at least one TP PSM and a false positive (FP) protein identification as one with all FP PSMs. ProteinProphet (Nesvizhskii et al. 2003) calculated the protein identification error based on the probability of individual PSMs to be false. TDA-based FDR calculation has also been introduced to protein identification. Reiter et al. implemented a hypergeometric model in the MAYU program (Reiter et al. 2009) to calculate protein identification FDR in large-scale dataset, and showed a large gap between their protein identification FDR and PSM FDR.
B. STRATEGIES FOR IMPROVING VALIDATION
This simplification of TDA has caused concern regarding its validity and limitation in several scenarios, including datasets and databases with small sizes, biased sequence distributions in multi-pass searches, occurrence of homeometric peptides (different sequences producing similar spectra) (Gupta et al. 2011), and decoy generation strategies (Elias & Gygi 2010). While these concerns reflect extreme cases in routine proteomics studies, it is more likely for problems to occur in glycoproteomics data analysis (Zhu et al. 2014, Cheng et al. 2014). This is largely attributed to the diverse fragmentation patterns of glycopeptides resulting from different dissociation methods (Mayampurath, Yu, et al. 2014), the small size of identified glycopeptides in each run (Zhu et al. 2014), and different causes of false positives.
The first challenge in validating glycopeptide identification results is that these studies often produce relatively small set of glycopeptides. In contrast to proteomics experiments, where tens of thousands of spectra can be assigned to peptides, only hundreds of spectra are assigned to glycopeptides in typical glycoproteomics studies. This reduces the TDA accuracy in the context of 1:1 target-decoy ratio. For 100 identified glycopeptides with 1% FDR, one more/less incorrectly identified glycopeptide can cause the predicted FDR to deviate significantly from the true one. It is also likely that no decoy hits are above the score threshold. As a remedy, GlycoPep Evaluator introduces a 1:20 target-to-decoy ratio for validating small glycopeptide dataset, and corrects the FDR estimation by the multiplication factor (Zhu et al. 2014). The authors reported improved FDR accuracy for ETD tandem MS data using the software. A related question may arise as to determining the proper target-to-decoy ratio to produce confident FDR estimation. One may consider iteratively increasing the ratio until certain confidence interval threshold is achieved.
The second challenge in estimating glycopeptide FDR is that both vibrational (CID, HCD) and ExD are in widespread use for glycopeptide analysis, both of which have to unique characteristics of the glycopeptide structure: vibrational methods favor dissociation of the glycan moiety but produce peptide backbone dissociation at higher energies; ETD favors peptide backbone dissociation and can therefore produce the finer detail on the peptide sequence and glycan attachment site. In proteomics, by contrast, vibrational dissociation versus ExD both dissociate peptide bonds but through different mechanisms and bond propensities (Zubarev et al. 2008). In glycoproteomics, however, the content of true positives (TPs) and false positives (FPs) may change according to the experimental design. When the goal is to study the site-specific occupancy using deglycosylated peptides, a potential FP match may have a different amino acid sequence as well as different site occupancies although the sequon is not necessarily maintained. By forcing the decoy sequences to contain a sequon, many FPs in the decoy database are filtered implicitly; however, the similarity assumption of the target-decoy model may also be violated. When the experiment concerns the topology of the glycans attached to the peptides, a FP match will be a peptide with complex glycosylation from a large glycopeptide candidate space (different combinations of peptide backbones and glycan sequences). In this scenario, generating a decoy database containing unlikely glycan structures remains unverified in terms of the two basic assumptions in TDA, and searching in large database is also computationally inefficient. A potential solution is to treat the FDRs of peptide and glycan identification separately. Similar to the definition of TP and FP protein identifications (Nesvizhskii et al. 2003, Reiter et al. 2009), a TP glycopeptide identification can be defined as a glycopeptide with both TP peptide and TP glycans, while a FP glycopeptide identification can be defined as one with either FP peptide or FP glycan (or both). We expect that there will be more studies on the integration of peptide confidence and glycan confidence. In addition, other statistical measures such as q value may also be worth considering (Granholm & Käll 2011).
IV. QUANTIFICATION
Changes in protein expression level and PTMs are often linked to disease states. The changes are usually regulated at the gene expression level and by post-synthesis processing machinery. Measurement of gene expression at the mRNA level does not always correlate well with the actual levels of functional forms of a protein (Wang et al. 2014). It is therefore important to accurately determine protein levels in a given biological sample as part of projects to study altered biological states (Anderson & Seilhamer 1997, Anderson et al. 2009).
The advantage of using mass spectral methods for proteomics is that they enable both identification and quantification of proteins and their various functional forms in a mixture, in a single workflow. As a surrogate for proteins, peptides are used in bottom-up proteomics workflows, which can be easily adapted for quantitative analyses (Eng et al. 1994, Gygi et al. 1999, Zhou et al. 2002, Aebersold 2003). Mass spectrometry-based protein quantification can be divided into two groups: relative and absolute quantification. Of these, relative quantification can be performed using label-free or isotopic label-based methods. Absolute quantification, on the other hand, requires use of isotopically labeled internal standards. These strategies have been frequently presented and reviewed in literature (Aebersold 2003, Elliott et al. 2009, Wasinger et al. 2013). However, the methods may not be directly applicable to quantitative glycoproteomics due to the multiplicity of molecular forms or proteoforms (Smith et al. 2013) of a glycoprotein resulting from glycan macro- and micro-heterogeneity. In this section, we discuss current methods and strategies that show potential application in quantitative glycoproteomics analysis.
A. QUANTIFICATION OF DEGLYCOSYLATED PEPTIDES AS GLYCOPEPTIDE SURROGATES
While a surrogate peptide or a set of peptides can be used to quantify a non-glycosylated protein; multiple proteoforms of a particular glycoprotein exist, each of which has a different glycan composition and structure. This complexity increases exponentially with the number of glycosylation sites on a protein. As a result, it is almost impossible to quantify all proteoforms of a given glycoprotein given the existing capabilities of mass spectrometers; however, it is possible to quantify populations of molecular forms that differ based on their functional or physiochemical properties and can be separated based on these properties prior to MS analysis. For example, glycoprotein populations can be separated based on their glycan compositions using lectins and then quantified (Wei & Li 2009, Pan et al. 2011). Alternatively, glycan epitopes can be isolated using antibody pulldowns prior to quantification. Populations of glycoproteins or glycopeptides may be resolved using different chromatographic separation methods. The separation may be based on physiochemical properties like charge, hydrophobicity, size, etc., which will change the order of elution and chromatographic peak shapes. Consequently, data analysis methodologies and algorithms need to be designed to be compatible with the sample preparation and data acquisition workflows. Even in the recent literature, a majority of researchers prefer deglycosylating the glycoprotein/glycopeptides, after enrichment of modified/functional forms of interest, before proceeding to identification and quantification steps (Tian et al. 2007, Hill et al. 2009, Shakey et al. 2010, Liu et al. 2010, Kim et al. 2012, Pan et al. 2012, Zhang et al. 2014). Another technique that has become widely popular is enzymatic deglycosylation using PNGase F, in presence of H218O, which incorporates the heavy oxygen at the glycosylation site and allows facile N-glycosylation site identification and site-occupancy analysis (Hägglund et al. 2007, Liu et al. 2010, Zhang, Liu, et al. 2012). 18O Labeling also integrates easily with existing proteomics workflows for identification of the modified/labeled site and quantification of the labeled, deglycosylated peptides. Such studies allow efficient quantification of the population of interest with site occupancy information but detailed information on site-specific glycosylation is lost.
The advantage of using peptides as surrogates for glycopeptides is that the label-based and label-free approaches in quantitative proteomics can be used directly for data analysis. Label-based techniques like SILAC and iTRAQ allow multiplexing to increase throughput and minimize bias in analyses. Label/tag based quantification methods are well established and integrated into proteomics workflows and software for data analysis but add significant cost and effort to sample preparation. Label-free quantification (LFQ) on the other hand are easier to perform and more cost-effective. The challenges lay in selection and use of appropriate tools for data analysis and quantification, which include data pre- and post-processing and statistical analysis methods (Vaudel et al. 2010). In addition, differences in data acquisition methods and instrumentation may add complexity to LFQ methods. Comparisons between different quantitative proteomics strategies have already been reported in literature (Grossmann et al. 2010, Arike et al. 2012, Fabre et al. 2014).
The use of data-independent acquisition (DIA) strategies such as SWATH-MS (Gillet et al. 2012) and multiplexed DIA (Egertson et al. 2013) increases the number of ions that can be quantified and their limits of detection and quantification. Such methods make an excellent alternative to targeted measurements while eliminating the need for the use of targeted precursor lists and triple-quadrupole (QQQ) mass spectrometers for quantitative analyses. Huang et al. recently showed that SWATH was able to offer outstanding LFQ performance using complex mouse-cell lysate samples (Huang et al. 2015). A few studies have employed SWATH-MS for site-specific glycosylation quantification from deglycosylated peptides (Liu et al. 2013, Liu, Chen, et al. 2014, Xu et al. 2015). Open-source data analysis tools like OpenSWATH (Röst et al. 2014) have already made their way into the field thereby minimizing the gap between analytics and informatics. DIA-Umpire (Tsou et al. 2015) combined untargeted peptide identification and targeted re-extraction/quantification, and achieved better sensitivity as well as consistent quantification performance. The authors showed that DIA-Umpire can significantly improve the identification rate of deamidated peptides compared to OpenSWATH (Röst et al. 2014) on standard glycoproteomics dataset from prostate cancer tissues (Liu, Chen, et al. 2014). However, the applicability of DIA to intact glycopeptides is still debatable because upon fragmentation glycopeptides with subtle differences in glycan compositions/topologies produce very similar fragment ion spectra using collisional-dissociation methods, which are best suited for the rapid SWATH acquisitions.
B. QUANTIFICATION OF SITE-SPECIFIC GLYCOFORMS
There has been a lot of interest in intact glycopeptide quantification to preserve information on site-specific glycosylation (Rebecchi et al. 2009). One of the major challenges with LFQ arises from the differences in ionization efficiencies of glycopeptides. Methods as simple as comparing LC-MS extracted ion chromatogram peak areas with mass spectral response may be used (Ding et al. 2009, Nilsson et al. 2009, Khatri et al. 2014) for LFQ, while keeping in mind the effects of sugar moieties leading to differences in ionization of the precursor. In order to compare abundances across multiple samples, isotopic labeling methodologies can be used just like in standard quantitative proteomics workflows (Wollscheid et al. 2009, Kurogochi & Amano 2014). Another strategy for minimizing difference in ionization efficiencies of glycopeptides is the use of isobaric tandem MS labeling strategies like iTRAQ and TMT (tandem mass tags) for quantification as described by Viner et al (Viner et al. 2009). Others chemical derivatization methods selectively enhance the ionization of glycopeptides in a complex sample and minimize bias in quantification (Amano et al. 2010).
For LFQ of site-specific glycoforms, targeted acquisition methods including selected reaction monitoring (SRM) or multiple reaction monitoring (MRM) have become the state-of-the-art workflows (Song et al. 2012, Hong et al. 2013). These methods make use of diagnostic oxonium ions resulting from glycan fragmentation as MRM transitions. A survey scan can be used to identify all precursors that generate oxonium ions to create a list of precursors for the MRM experiment. The use of the most common oxonium ions for quantification minimizes effects from differences in ionization efficiencies and eliminates the need to select specific transitions for individual glycopeptide precursors. The use of internal standards or spiked in synthetic peptides is recommended for minimizing bias from sample handling and preparation steps. There are several factors that can affect quantification using MRM, including number of transitions, duty cycle and collision energies, which must be optimized before data acquisition; this is now possible using software tools like Skyline (MacLean et al. 2010, Maclean et al. 2010). While MRM assays are typically performed using QQQ mass spectrometers, some studies have shown that hybrid instruments including quadrupole-time of flight (Q-TOF) can be used for glycopeptide quantification using this method (Sanda et al. 2013). Quadrupole-Orbitrap hybrid instruments have been used for parallel reaction monitoring of peptide abundances (Peterson et al. 2012, Gallien et al. 2014) and are likely to be useful for glycoproteomics. Skyline not only allows designing experiments for quantitative proteomics and glycoproteomics but also assists with efficient data analysis (Schilling et al. 2012). The open-source nature of the Skyline framework allows easy integration of external tools to suit the needs of various researchers (Broudy et al. 2014).
Other quantification methods rely on efficient analytical strategies, such as the Glyco-AMP workflow described by Hua and colleagues (Hua et al. 2013) for LFQ using multiple enzymes and comparing ion abundances for identified glycopeptides. This allows cross-validation of glycoform quantification based on different carrier peptides, while minimizing the bias from differences in ionization efficiencies. Recently, Mayampurath and coworkers (Mayampurath, Song, et al. 2014) presented a new ANOVA-based mixed effects model for LFQ of site-specific glycosylation on glycopeptides. The model was applied to study differences in glycosylation between sera from normal and cancer patients, in order to identify potential glycoprotein biomarkers.
Analysis and quantification of intact glycopeptides is more challenging than using deglycosylated peptides. Glycopeptides come with the aforementioned macro- and micro-heterogeneity, which must be accounted for while applying any quantification methodologies. The approach selected for quantification may depend on the goals of a study. Surrogate peptides may be used when overall glycoprotein expression or site-occupancy information are desired; however, when relative or absolute quantitation of the different glycoforms needs to be performed, experimental conditions and physiochemical factors need to be considered. The glycoform present on a glycopeptide affects its liquid chromatography retention time and mass spectrometric ionization efficiency, which will affect quantification. Therefore, LFQ may not be applicable, as it exists for proteomics, in quantification of intact glycopeptides and there is a need to modify existing LFQ strategies for application to glycopeptides.
There are several algorithms in use for LFQ that can be divided into two categories: feature intensity-based and spectral counting-based, as reviewed by Nahnsen et al (Nahnsen et al. 2013). T3PQ (Grossmann et al. 2010) and iBAQ (Schwanhäusser et al. 2011) are the most popular MS1 precursor-feature-based algorithms for quantification. T3PQ is based on Top3 (Silva et al. 2006), a method that that uses the peak intensities of the top 3 most abundant peptides identified for a specific protein. T3PQ is, therefore, only suitable for glycoproteomics level quantification, which can provide information on total protein abundance. iBAQ (intensity-based quantification), uses the sum of all peak intensities, for peptides matching to a particular protein and then normalizes using the total length or number of theoretical peptides of the protein. Such information can be used to compare different functional forms of a glycoprotein. Glycosylation site-identification and site-occupancy analysis is possible using iBAQ, by performing 18O labeling of glycosylation sites as described in previous sections.
Spectral counting, on the other hand, uses all the tandem spectra matching a particular protein, for quantification. Spectral counting can theoretically be used for quantification of the different proteoforms (glycoforms) of a given glycoprotein. emPAI (Ishihama et al. 2005) (exponentially modified protein abundance index) is a spectral counting method that uses number of observed peptides divided by the number of observable peptides for a protein as a measure of protein concentration in a complex sample. The central idea being that the number of observed peptides is proportional to abundance of a protein in a complex sample. emPAI has been implemented in the MASCOT search engine (Perkins et al. 1999), which is commonly used for proteomics experiments. This method has been used for glycoprotein quantification, based on deglycosylated peptides in an omics experiment but not for intact glycopeptides (Toyama et al. 2011, Qin et al. 2014). A value similar to emPAI could be used to infer the abundance of a particular proteoform of a glycoprotein. This would require integration of intact glycopeptide identification and statistical analysis of the relative abundances of glycoforms at each glycosylation site to infer the abundances of different glycoproteoforms. Ideally, bottom-up and middle-down or top-down glycoproteomics data would have to be combined to maximize confidence in quantitative results from this approach.
APEX (absolute protein expression) (Braisted et al. 2008), is another spectral counting method that accounts for differences in peptide detection in mass spectrometry and predicts a correlation between protein quantities and the number of peptides detected, based on their physiochemical properties. It is conceivable to build a prediction model for glycopeptide quantitation based on this approach, while taking into account the changes in physiochemical properties and MS detection introduced by the glycan modification. Such a model would be most appropriate for LFQ of glycopeptide glycoforms and would provide information on both quantitative protein expression as well as glycan micro-heterogeneity.
Since intact glycopeptide analysis requires tailored analytical strategies, it is important that the data analysis tools be designed accordingly. For label-free quantification of glycoforms, it may be important that the differences in physiochemical properties introduced by the glycan be accounted for. For example, addition of neutral saccharides vs. acidic (sialylated or sulfated) sugars may have very different effects on chromatography and gas-phase ionization of a glycopeptide, thus biasing the quantification results. In order to build toward automated analysis, orthogonal measurements, such as UV absorbance, are necessary to identify adjustment factors for differences in molecular compositions. A robust model for optimizing glycopeptide collision energies needs to be integrated into SRM/MRM workflow facilitating tools such as Skyline. While peptide backbone ions generated by collisional dissociation of glycopeptides are typically of low abundance, glycan fragment ions can be generated very reliably and can be used for relative quantification using MS/MS. Thus, the transitions used for glycopeptide quantification may be very different from those used for non-glycosylated peptides. It is important to recognize glycoproteins as heterogeneous populations with different glycoforms at each glycosylation site. Bottom-up identification and quantification workflows are only suitable for comparing site-specific glycoform abundances but information relating glycoforms at different sites is lost. Therefore, it is not be possible to directly comment on the abundance of every glycoproteoform present, using existing methods; however, pre-fractionation or isolation can help quantitate functionally different populations. Overall, there are quantification strategies that may be borrowed or modified from their current state in proteomics workflows but there is a clear need for better integration of glycopeptide characterization/identification and quantification tools to make robust workflows.
V. SOFTWARE DESIGN AND DEPLOYMENT
Discussions on the need for automated glycoproteomics solutions have existed for years, despite the fact that new tools emerge every few months. This awkward situation was attributed to the lack of accessibility to the source code of current tools (Dallas et al. 2013). From the perspective of developers, there are a lot of problems to work directly on the source code from other groups unless the code is well documented and organized. In this section, we summarize several strategies for code re-use and tool development, most of which have been adopted in the bioinformatics field.
A. EXTENSION FROM CURRENT FRAMEWORKS
A typical glycoproteomics data analysis workflow can be divided into steps that are well-established and steps that are still open and updating. The former covers data preprocessing (raw file parsing, peak picking, spectral alignment, and deconvolution), sequence identification, quantification and post-processing (e.g. statistical summary, functional analysis and data visualization), which have been addressed intensively in proteomics (Nesvizhskii 2010) and glycomics (Campbell et al. 2014). The latter includes problems that are glycopeptide specific, such as site-specific localization and profiling of the glycosylation, integration of scores from the candidate peptide and glycan part, tailored target-decoy model and FDR calculation. Solving the problems may require concurrent progress from both computational and experimental side, which constitutes the fast evolving parts of the glycoproteomics workflow.
We here discuss several basic forms of extending new functions from current frameworks. The final solution depends on the architecture and support of the frameworks as well as the requirements of the users. Readers can refer to recent publication on comprehensive review of libraries and frameworks in proteomics (Perez-Riverol et al. 2014).
One of the options are plugins, which are addable components to extend current software's functions. Mass++ (Tanaka et al. 2014) provides a flexible plugin architecture that allows users to integrate external tools. Each plugin contains a compiled function implementation file (.dll) and xml files specifying the parameter configurations and layout in the interface panel. Users can easily add new utilities without knowing the source code of the framework. Nevertheless, not all frameworks were designed to support such kind of extension, and users of the plugin system are responsible for converting existing external tools into the format accepted by the plugin system, which sometimes are not feasible. An alternative option is to develop new applications based on provided API (Application Program Interface). ProteoWizard (Kessner et al. 2008) API (C++) contains a uniform interface called MSData for users to access raw data without known the vendor-specific configurations. This allows flexible control of memory usage and avoids the problem of working with large mzML files. The Pyteomics (Goloborodko et al. 2013) API provides a set of utility functions to access common proteomics data files and calculate basic biochemical properties of peptides. MzJava (http://mzjava.expasy.org/), the library reengineered from Java Proteomic Library, provides a set of functions to manipulate the tandem mass spectra (MSn) and associates them with corresponding peptide or glycan molecules. Open source Java library compomics-utilities (Barsnes et al. 2011) offers a large set of commonly used features for data parsing, analysis and visualization, and has been used in multiple computational tools. Users usually have the flexibility of developing new applications with given API, but may spend extra time on integrating existing external tools.
Occasionally, some tools may meet the users’ requirements for functionality, but the output format cannot be used directly as input for the downstream tool. Comprehensive suites such as TOPP/OpenMS (Sturm et al. 2008) and TPP (Deutsch et al. 2010) contain their own file formats describing intermediate identification and quantification results, and support conversion from/to common external file formats; however, this is not guaranteed, especially for legacy programs or programs focusing on specific functions. One example is the deconvolution tool DeconTools, which is an upgraded version of Decon2LS (Jaitly et al. 2009). DeconTools produces two CSV files where one file records metadata (e.g. base peak information, total ion chromatogram abundances and peaks deisotoped) associated with each scan in a LC/MS dataset, and the other file contains information of the monoisotopic peaks (M), the second isotopic peak (M+1) and their corresponding scan number. The output files from Decon2LS have been used by a few glycomics tools such as GlyReSoft (Maxwell et al. 2012) and MultiGlycan (Yu et al. 2013). However, this is incompatible with tools that take XML identification files. In this case, module developers should consider designing a specific adapter for the CSV files and wrapping it with the same interface as the accepted XML format. In this way, the functions designed to process the XML file will remain intact. Note that adapters may cause loss of information, which can be problematic for downstream tools. In our case, when LC-tandem MS data was used for deconvolution, the linkage between precursor ion and the corresponding product ions was missing. As a remedy, we have to access the original raw files to recover the scan relationship between the precursor and product ions. Developing adapters for commonly used tools is as significant for the community as designing new dedicated tools, but requires deliberate consideration of adding and removing information.
Users should be aware of the available options of extending functionalities based on current framework in order to improve productivity, and focus on the part where significant modification and innovation is needed.
B. STANDARD FORMATS FOR MODULE COMMUNICATION
Researchers frequently need to combine external and in-house developed tools in order to process the glycoproteomics data generated using the latest techniques and experimental designs. For such purposes, it is more practical to develop modules based on file formats than legacy code, since the former removes the issues of compatibility and code maintenance, and minimizes the responsibilities of the developers. In addition, outputs from intermediate steps (e.g. identification results) can be stored in files, and picked up directly for downstream processing without re-running the whole program. This is beneficial when the program crashes during running or parameters in the downstream steps need to be changed. To facilitate the communication between tools, an established file format protocol should be pre-determined and the corresponding validation method should be provided. Even for self-contained software suites, it is still important for developers to deliver the function of exporting intermediate outputs into popular formats, so that users will have the flexibility to continue the data analysis using alternative programs. Recently, The Human Proteome Organization – Proteomics Standards Initiative (HUPO-PSI) proposed several standards covering raw data format mzML (Martens et al. 2011), peptide identification format mzIdentML (Jones et al. 2012) and quantification format mzQuantML (Walzer et al. 2013). These standards allow tools from different groups to communicate with each other effectively. It is important for developers to follow the standard data formats, or at least provide the export of these formats as an option. A detailed description of these formats, access approaches and JAVA-based examples were given by Gonzalez-Galarza et al (Gonzalez-Galarza et al. 2014).
Despite the fact that standard formats and guidelines in proteomics (Taylor et al. 2007) and glycoproteomics (Kolarich et al. 2013, York et al. 2014) have been made and consistently improved, there is a need to restrict the output formats produced by tools dedicated to different stages. For example, multiple preprocessing steps may be applied to the raw data file, and algorithm-specific information may be produced (e.g. DeconTools). Considering the diversity of results representation from different processing tools and distinct features of peptide and glycan sequences, it seems impractical to force tools at all stages to follow the standard formats. However, it is recommended that developers at least generate output formats in XML style appended with a schema file for format checking, or simply produce plain text files such as CSV files.
Platform dependency is another issue which may impede the integration of multiple tools. For example, msconvert from ProteoWizard project (Kessner et al. 2008, Chambers et al. 2012) requires the Windows system in order to parse vendor-specific raw data. This will cause problems if the pipeline has been deployed on the Linux cluster. Developers may consider using simulation software such as WineHQ to simulate the windows environment, or use a dedicated Windows server for file conversion and a Linux server for heavy duty data processing, and let the two platforms share the same hard drive partition through local network (e.g. using SAMBA network protocol).
C. USER INTERFACE AND BATCH PROCESSING
Typical proteomics pipelines such as TPP (Deutsch et al. 2010) and OpenMS (Sturm et al. 2008) provide simple graphic user interfaces (GUIs) for users to configure parameters and monitor work progress. TPP projects are managed through its web-based GUI Petunia by default, and a local Apache server needs to be launched in the backend to manage the data analysis. Users can submit multiple raw files, or input files at intermediate stages, and the analysis tasks stop in the end of each stage. LabKey Server (Nelson et al. 2011) also supports such web access to search engines and different TPP components, but provides more post-processing functions. In contrast, OpenMS offers better flexibility and portability in terms of workflow customization. Users can specify the parameters and number of threads/jobs of each module through its TOPPAS GUI (Junker et al. 2012). A nice feature of the TOPPAS interface is that users can export the whole workflow as well as the complete parameters into a template file, which can be used as input for command-line pipeline under different operation systems, or imported into TOPP on another computer.
ProteinProspector (Chalkley et al. 2005) provides all its tools through its online web server. Users are able to save and view the searched results. ProteinProspector also contains a utility program MS-Viewer (Baker & Chalkley 2014) that allows user to generate annotated spectra for publication purpose.
There are several advantages to deploy the computational tools in online web servers. The first is that the implementation is transparent to end users, so it is suitable for projects with copyright concerns, frequent update or complex configuration. The second is that users can choose to be notified only when the tasks are finished. This is advantageous especially for tasks that require long time processing, such as database searching. Developers may consider deploying the workflow on the cloud, where the computational resources can be acquired on demand. In addition, with the advance of modern web techniques such as HTML5 and D3 visualization (Bostock et al. 2011), it is expected that a lot of useful user-interactive features (PEPTAGRAM: http://boscoh.github.io/peptagram/) will be added to the web server, and users can even interact with the program, and involve in the algorithm decision-making process.
D. AVAILABLE TOOLS TOWARDS PIPELINE DEPLOYMENT
In addition to customizing modules under public proteomics framework, users may need to assemble their in house modules or legacy code into a pipeline. Shell script is commonly used to manage the computational tasks and input/output files at each stage; recent pipeline tools have emerged to facilitate module assembly and management that may be useful to the glycoproteomics community. Among these, Galaxy (Giardine et al. 2005, Blankenberg et al. 2010, Goecks et al. 2010) is the most widely used pipeline tool in life science, and has been successfully applied to proteomics and proteogenomics in the Galaxy-P platform (Sheynkman et al. 2014). Galaxy provides a user-interactive interface that allows users to manipulate their modules and data. It also contains an appstore-like service ToolShed (Blankenberg et al. 2014) so that tools deployed by other groups can be cloned with no extra efforts. In addition, the Galaxy framework also supports API access and cloud extension (Afgan et al. 2012), both of which can be manipulated easily through Python package bioblend (Sloggett et al. 2013).
For users who wish to have flexible control of a workflow in a cluster environment, a command-line based pipeline tool is a good option. Bpipe (Sadedin et al. 2012) focuses on the management of running tasks in a simple and flexible style. The Python package nestly (McCoy et al. 2013) allows nested combinations of parameters/inputs as well as aggregation of output results. Tools published recently also added support for parallelization and scaling of the pipeline (Köster & Rahmann 2012, Gafni et al. 2014, Cingolani et al. 2015), which provide users with the options to analyze large datasets using different high performance computing infrastructures such as traditional cluster or cloud.
In order to determine the proper pipeline tools, users may need to consider the support of features such as status monitoring and reporting, failure recovery, scalability to support large scale dataset; the platform/infrastructure needed, portability, and user interface. Pipeline tools supporting environment clone or portable configuration will be highly beneficial for the whole community to repeat or validate the published methods.
VI. CONCLUSIONS AND PROSPECTS
Protein glycosylation elaborates the functional roles of carrier proteins in a spatially and temporally regulated manner. Characterization of glycosylation in a specific biological context, as well as its crosstalk with other protein PTMs (Hart et al. 2011), is necessary for understanding the complete regulation network and drive the development of new therapeutics.
Glycoproteomics experiments benefit from technological advances in the larger proteomics field. Similarly, most algorithms in glycoproteomics analysis can be traced back to their respective prototypes in proteomics. In this review, we summarized current strategies used in identification, validation and quantification in glycoproteomics, and referred to the proteomics field for possible solutions. Nevertheless, the field of glycoproteomics is developing rapidly. Improvements in analytical methods and instrumentation now allow the production of high quality glycoproteomics data by non-expert proteomics laboratories. Each methodological improvement enables production of large datasets containing elaborate data structures and fragmentation patterns, the benefits of which can only be reaped with dedicated computational tools.
Concurrent efforts have been made on standardizing the formats for glycan structures (Lütteke & Frank 2015). Issues such as integrating protein reference and glycan identification information into a single file should concern the whole community. Progress and consensus on these issues will undoubtedly contribute to diminishing the inconsistency among tools and promote their reuse and further improvement. Instead of enforcing the open source for all scientific tools, we suggest that tools, regardless of their specific implementations, are best to be modularized to solve individual problems. This will maximize the reuse of the programs. Well-defined input/output formats and command-line interface are also critical to ensure the integration into new pipelines. All these only require minimal extra effort for developers in the beginning but will reward the whole community in the long run.
The paucity of computational tools in glycoproteomics analysis accessible to general users will likely last for years, as a corollary of consecutively emerged upgrades in experiment and instrument techniques. In order to meet this need, deployment of pipeline modules in framework with public accessibility will promote the popularization and integration of new tools. As consensus is achieved on standardizing the experimental procedure and analysis workflow, the tool scarcity situation will be changed in the foreseeable future.
Table 2.
Proteomics software
| Task | Category | Software | Features |
|---|---|---|---|
| Database search (peptide) | Comparison on spectra level | SEQUEST (Eng et al. 1994) | • Compares experimental spectra with theoretical spectra through cross-correlation function |
| Comet (Eng et al. 2013) | • Academic (open-source) version of SEQUEST • Included in TPP/SearchGUI |
||
| Tide | • Reimplementation of SEQUEST algorithm with speed optimization • Included in SearchGUI |
||
| Probability-based scoring | Mascot (Perkins et al. 1999) | • Most popular commercial database search engine • Integrates mass fingerprint search, sequence tag and normal database search • Probability based scoring • Widely supported |
|
| ProteinProspector (Clauser et al. 1999, Chalkley et al. 2008) | • Comprehensive analysis of proteomics data at increasing levels of depth • Searches user input modification types • Allows detection of novel modification types • Site localization in peptide (SLIP) scoring |
||
| X!Tandem (Craig & Beavis 2004) | • Multipass search for modified and non-tryptic peptides • Compatible with Proteome Discoverer, TPP, TOPP, LabKey, RforProteomics |
||
| Myrimatch (Tabb et al. 2007), | • Multivariate hypergeometric distribution to calculating random match • Stratification of peak intensities for better discrimination |
||
| OMSSA (Geer et al. 2004) | • Probability-based scoring (Poisson distribution) • No longer maintained |
||
| Andromeda (Cox et al. 2011) | • Probability-based scoring (binomial distribution) • Supports complex PTMs and large database • Standalone or included in MaxQuant |
||
| Optimized for high-resolution tandem mass spectra | Morpheus (Wenger & Coon 2013) | • Designed for high resolution tandem mass spectrometry • Simple scoring function • Fast speed |
|
| MS Amanda (Dorfer et al. 2014) | • Optimized for high resolution tandem mass spectra • Support multiple fragmentation types (HCD, ETD, and CID) • Probability-based scoring (cumulative binomial distribution) • Included in SearchGUI and node in Proteome Discover |
||
| Universal | MS-GF+ (Kim and Pevzner, 2014) | • Support for data from multiple fragmentation methods, multiple enzyme digestion, and phosphoproteomics • Bioconductor, SearchGUI |
|
| de novo sequencing | Naïve approach | PEAKS (Zhang, Xin, et al. 2012) | • Widely used commercial tool for de novo sequencing • Generates consensus sequences based on comparison of experimental spectrum with a large pool of randomly sampled sequences • Now a full- featured software suite for proteomics study |
| Spectrum graph (dynamic programming) | pNovo+ (Chi et al. 2012) | • Integrates complementary HCD and ETD spectral information. • pDAG algorithm looking for the top k longest paths • 0.018s for sequencing / spectrum |
|
| pepNovo+ (Frank 2009a) | • Boosting-based PSM ranking algorithm integrating multiple PSM features | ||
| UniNovo (Jeong et al 2013) | • Independent of specific spectral type • Estimates the probability that a constructed sequence is correct |
||
| Novor (Ma 2015) | • Currently the fastest sequencing tool for de novo searches • Contains two large decision trees built from NIST spectral library • Supported by DeNovoGUI |
||
| Hybrid approach | Byonic™ (Bern et al. 2012) | • Integration of database search with de novo sequencing • Supports the detection of glycosylation • Supports unexpected/novel PTMs • Supports multiple fragmentation methods • Standalone or node in Proteome Discoverer |
|
| PEAKS DB (Zhang et al. 2012) | • Integration of database search with de novo sequencing • Decoy fusion for result validation |
||
| Spectral library | Peptide | SpectraST (Lam et al. 2007) | • Open-source, extensible tandem MS spectral searching tool • Searches a spectral library; faster than database search engines • Included in TPP |
| BiblioSpec (Frewen et al. 2006) | • Stores tandem mass spectral libraries in an open-source data format • Builds peptide tandem MS libraries and removes redundant spectra • Searches library for matches using query spectra |
||
| X!Hunter (Craig et al. 2006) | • Based on annotated peptide tandem mass spectra in the Global Proteome Machine Database (http://gpmdb.thegpm.org/) • Different tandem mass spectra from the same peptide were averaged to facilitate comparison of experimental data to the library. |
||
| Validation | PSM validation | Percolator (Brosch et al 2009) | • A machine learning method for rescoring database search results • Outperforms Mascot scoring schemes • Provides significance measures |
| PeptideProphet (Ma et al 2012) | • Supports validation of PSMs from multiple search engines. • Uses the expectation-maximization algorithm to distinguish correctly assigned peptides to incorrectly assigned ones, and computes the probability of each PSM to be correct. • Included in TPP |
||
| Peptide sequence validation & integration of search engine | iProphet (Shteynberg et al 2011) | • tCalculates the probability of unique peptide sequences using PSM probabilities from PeptideProphet • combines results from multiple search engines • Included in TPP |
|
| ConsensusID (Nahnsen et al. 2011) | • Combines results from multiple search engines. • Converts each engine scores into probabilities, and generates consensus score for each sequence |
||
| MSBlender (Kwon et al. 2011) | • Converts raw proteomics database search scores into a probability score for every peptide-spectrum match. • Improves ability to identify and quantify peptides over sue of single search engines |
||
| Protein FDR | MAYU (Reiter et al. 2009) | • Uses a hypergeometric model to calculate FDR for protein identification for large datasets • Included in OpenMS/TOPP |
|
| Quantification | MS1 quantification | MaxQuant (Cox & Mann 2008) | • Supports quantification of label-free and labeling datasets (e.g. TMT, SILAC, iTRAQ) • Parameters optimized for general usage. • Integrates with Andromeda |
| MS1/MS2 quantification | ProteinQuantifier (Weisser et al. 2013) | • Top3 approach for quantification • Supports the import of protein inference results from outside software, e.g. ProteinProphet and Fido. |
|
| Targeted | Skyline (MacLean et al. 2010, Maclean et al. 2010) | • Open source windows client for targeted proteomics methods creation • Supports creation and use of tandem MS spectral libraries • Works with many MS vendor platforms • Allows integration of external tools |
|
| mProphet (Surinova et al 2013) | • Workflow for analysis of large quantitative selected reaction monitoring data sets • Designed for use with stable isotope labeled peptide internal standards • Included in TPP |
||
| Untargeted/data-independent analysis | OpenSWATH (Röst et al. 2014) | • Open source software for targeted analysis of DIA data sets • Compatible with multiple MS vendor data via open data formats • Provides retention time alignment, chromatogram extraction and statistical analysis |
|
| DIA-Umpire (Tsou et al. 2015) | • Detects precursor and production chromatographic features and assembles them into pseudo-tandem mass spectra • Pseudo tandem mass spectra can be identified with proteomics search engine • Combines untargeted peptide identification and targeted quantification |
||
| Integration | Identification | SearchGUI (Vaudel et al. 2011) | • Open-source graphical user interface for running OMSSA and X!Tandem search engines • Manages parameters and tasks for multiple search engines |
| DenovoGUI (Muth et al. 2014) | • Graphical user interface for running parallelized versions of the de novo sequencing software PepNovo+ • MS platform independent |
||
| Quantification | aLFQ (Rosenberger et al. 2014) | • Supports multiple absolute label-free protein quantification methods (TopN, iBAQ, APEX, NSAF and SCAMPI) • R package |
|
| Validation and protein inference | PeptideShaker (Vaudel et al. 2015) | • Utilizes the output from multiple search engines. • Calculates false discovery rate (FDR) and false negative rate (FNR) for PSMs, peptides, and proteins. • Provides user-friendly way of filtering and visualizing results. |
Table 3.
Glycoproteomics software
| Software | Features |
|---|---|
| Byonic™ (Bern et al. 2012) | • Commercial software • Supports the detection of glycosylation site • Supports unexpected/novo PTMs • Supports multiple fragmentation methods • Standalone and node in Proteome Discoverer |
| GlycoFragWork (Mayampurath, Song, et al. 2014) | • Performs label free quantification and glycopeptide identification • Uses CAD/CID/HCD data to identify gycan, ETD to identify peptide |
| GlycoMaster DB (He et al. 2014) | • Using a combination of glycopeptide HCD and ETD tandem mass spectra, searches a protein sequence database and a glycan database to identify the best glycan/peptide pair • With HCD only data, identifies the glycan |
| GlycoPep Detector (Zhu et al. 2013) | • Assigns glycopeptides from ETD tandem mass spectral data • User defines target protein and set of theoretical glycans • Algorithm scores ETD data against theoretical glycopeptides. |
| GlycoPep Evaluator (Zhu et al. 2014) | • Generates decoy database optimized for glycopeptides (target:decoy = 1:20) • Scores glycopeptide ETD data. |
| GlycoPep Grader (Woodin et al. 2012) | • Calculates theoretical compositions for glycopeptides from a user input target protein and a defined set of glycan compositions • Scores candidate glycopeptide compositions against tandem mass spectral data. • Generates false discovery rates using a set of decoy glycopeptides. |
| GlycoPeptideSearch (Chandler et al. 2013) | • Identifies glycopeptides from collisional tandem mass spectra using user defined peptide sequences and glycans from Glycome DB • Uses collisional dissociation tandem MS data • Identifies oxonium ions and peptide mass from peptide+saccharide ions • Does not match peptide backbone fragments |
| GlyDB (Ren et al. 2007) | • Linearizes a glycan structure database to allow searching of glycopeptide tandem mass spectra using Sequest |
| GlyPID (Wu et al 2010) | • Groups glycopeptides detected in reversed phase LC-MS data sets according to presence of ions differing by monosaccharide residue masses • Scores glycopeptides based on MS and tandem MS data |
| GP Finder (Strum et al. 2013) | • Uses deconvoluted deisotoped CID/CAD/HCD tandem mass spectra to identify glycopeptides produced using non-specific proteases • Filters for oxonium ions and self-consistency rules • Uses a decoy strategy to estimate false discovery rate |
| GPQuest (Toghi Eshghi et al. 2015) | • Spectral library matching algorithm for N-glycopeptides using HCD tandem MS • Constructs a library of deglycosylated peptides • Classifies glycopeptide tandem mass spectra based on presence of oxonium ions • Identifies glycopeptides using a search of intact glycopeptide tandem mass spectra against the library of deglycosylated peptides. |
| MAGIC (Lynn et al. 2015) | • Identification of intact glycopeptide from CID spectra • Extracts b/y ions for peptide database search • Uses mass shift, B/Y ions and look-up table (compiled from biosynthetic rules) to score glycan composition |
| Medicel N-gycopeptide library (Joenvaara et al. 2008) | • Assigns glycopeptides from deconvoluted tandem mass spectra • Calculates theoretical glycopeptides from the Uniprot database • Determines peptide mass and glycan composition from tandem mass spectra |
| Peptoonist (Goldberg et al. 2001) | • Identifies the glycopeptides in a proteolytic digest mixture using tandem MS data. • Expands the number of identified glycoforms at a given peptide sites using MS data and biosynthetic rules |
| SimGlycan™ (Apte et al 2010) | • Commercial software • Algorithm predicts glycan structure from tandem mass spectra using a database of theoretical dissociation • Applicable to glycopeptides |
| Sweet-Heart (Wu et al. 2013) | • A tool for application ion trap multistage tandem MS to glycopeptide analysis • Algorithm drives selection of MS3 stages to increase the abundances of peptide backbone product ions • HCD triggered CID or ETD |
| SweetSEQer (Serang et al. 2013) | • Identifies glycopeptide tandem mass spectra present in proteomics tandem mass spectrometric data sets • Automates assignment of glycan product ions in proteomics data |
| Sweet substitute (Clerens et al. 2004) | • Creates theoretical neutral mass glycopeptide collisional tandem mass spectra against which experimental data may be searched • Searches for glycan fragments from glycoconjugates |
ACKNOWLEDGEMENTS
The authors were supported by NIH grant P41RR10888, R01HL098950, and R21CA177479.
Abbreviations
- API
application program interface
- CID
collision-induced dissociation
- ECD
electron capture dissociation
- ETD
electron transfer dissociation
- DIA
data-independent acquisition
- FDR
false discovery rate
- FP
false positive
- FTMS
Fourier transform mass spectrometry
- HCD
higher energy dissociation
- LC-MS
liquid chromatography-mass spectrometry
- LFQ
label-free quantification
- MALDI
matrix-assisted laser desorption/ionization
- MRM
multiple reaction monitoring
- PSM
peptide-spectrum matching
- PTM
post-translational modification
- TDA
target-decoy approach
- TMT
tandem mass tags
- TOF
time-of-flight
- TP
true positive
- TPP
Trans-Proteomic Pipeline
- QTOF
quadrupole time-of-flight
REFERENCES
- Aebersold R. Quantitative Proteome Analysis: Methods and Applications. J Infect Dis. 2003;187(s2):S315–S320. doi: 10.1086/374756. [DOI] [PubMed] [Google Scholar]
- Afgan E, Chapman B, Taylor J. CloudMan as a platform for tool, data, and analysis distribution. BMC Bioinformatics. 2012;13(1):315. doi: 10.1186/1471-2105-13-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrné E, Nikitin F, Lisacek F, Müller M. QuickMod: A Tool for Open Modification Spectrum Library Searches. J Proteome Res. 2011;10(7):2913–2921. doi: 10.1021/pr200152g. [DOI] [PubMed] [Google Scholar]
- Allmer J. Algorithms for the de novo sequencing of peptides from tandem mass spectra. Expert Rev Proteomics. 2011;8(5):645–657. doi: 10.1586/epr.11.54. [DOI] [PubMed] [Google Scholar]
- Amano J, Nishikaze T, Tougasaki F, Jinmei H, Sugimoto I, Sugawara S, Fujita M, Osumi K, Mizuno M. Derivatization with 1-Pyrenyldiazomethane Enhances Ionization of Glycopeptides but Not Peptides in Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Anal Chem. 2010;82(20):8738–8743. doi: 10.1021/ac101555a. [DOI] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG, Pearson TW, Borchers CH, Paulovich AG, Patterson SD, Gillette M, Aebersold R, Carr SA. A human proteome detection and quantitation project. Mol Cell Proteomics MCP. 2009;8(5):883–886. doi: 10.1074/mcp.R800015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. ELECTROPHORESIS. 1997;18(3-4):533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta BBA - Gen Subj. 1999;1473(1):4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Apte A, Meitei NS. Bioinformatics in glycomics: glycan characterization with mass spectrometric data using SimGlycan. Methods Mol Biol. 2010;600:269–281. doi: 10.1007/978-1-60761-454-8_19. [DOI] [PubMed] [Google Scholar]
- Arike L, Valgepea K, Peil L, Nahku R, Adamberg K, Vilu R. Comparison and applications of label-free absolute proteome quantification methods on Escherichia coli. J Proteomics. 2012;75(17):5437–5448. doi: 10.1016/j.jprot.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Baker PR, Chalkley RJ. MS-Viewer: A Web-based Spectral Viewer for Proteomics Results. Mol Cell Proteomics. 2014;13(5):1392–1396. doi: 10.1074/mcp.O113.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsnes H, Vaudel M, Colaert N, Helsens K, Sickmann A, Berven FS, Martens L. compomics-utilities: an open-source Java library for computational proteomics. BMC Bioinformatics. 2011;12(1):70. doi: 10.1186/1471-2105-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern M, Cai Y, Goldberg D. Lookup Peaks: A Hybrid of de Novo Sequencing and Database Search for Protein Identification by Tandem Mass Spectrometry. Anal Chem. 2007;79(4):1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- Bern M, Kil YJ, Becker C. Current Protocols in Bioinformatics. John Wiley & Sons, Inc.; 2012. Byonic: Advanced Peptide and Protein Identification Software. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Kuster GV, Bouvier E, Baker D, Afgan E, Stoler N, Galaxy Team. Taylor J, Nekrutenko A. Dissemination of scientific software with Galaxy ToolShed. Genome Biol. 2014;15(2):403. doi: 10.1186/gb4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Ed Frederick M Ausubel Al. 2010;10:1–21. doi: 10.1002/0471142727.mb1910s89. Chapter 19:Unit 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcker S, Kehr B, Rasche F. Determination of Glycan Structure from Tandem Mass Spectra. IEEEACM Trans Comput Biol Bioinforma. 2011;8(4):976–986. doi: 10.1109/TCBB.2010.129. [DOI] [PubMed] [Google Scholar]
- Bostock M, Ogievetsky V, Heer J. D3 Data-Driven Documents. IEEE Trans Vis Comput Graph. 2011;17(12):2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- Braisted JC, Kuntumalla S, Vogel C, Marcotte EM, Rodrigues AR, Wang R, Huang S-T, Ferlanti ES, Saeed AI, Fleischmann RD, Peterson SN, Pieper R. The APEX Quantitative Proteomics Tool: Generating protein quantitation estimates from LC MS/MS proteomics results. BMC Bioinformatics. 2008;9(1):529. doi: 10.1186/1471-2105-9-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch M, Yu L, Hubbard T, Choudhary J. Accurate and sensitive peptide identification with Mascot Percolator. J Proteome Res. 2009;8(6):3176–3181. doi: 10.1021/pr800982s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broudy D, Killeen T, Choi M, Shulman N, Mani DR, Abbatiello SE, Mani D, Ahmad R, Sahu AK, Schilling B, Tamura K, Boss Y, Sharma V, Gibson BW, Carr SA, Vitek O, MacCoss MJ, MacLean B. A framework for installable external tools in Skyline. Bioinforma Oxf Engl. 2014;30(17):2521–2523. doi: 10.1093/bioinformatics/btu148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MP, Ranzinger R, Lütteke T, Mariethoz J, Hayes CA, Zhang J, Akune Y, Aoki-Kinoshita KF, Damerell D, Carta G, York WS, Haslam SM, Narimatsu H, Rudd PM, Karlsson NG, Packer NH, Lisacek F. Toolboxes for a standardised and systematic study of glycans. BMC Bioinformatics. 2014;15(1):1–11. doi: 10.1186/1471-2105-15-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans. J Proteome Res. 2008;7(4):1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- Chalkley RJ, Baker PR, Huang L, Hansen KC, Allen NP, Rexach M, Burlingame AL. Comprehensive Analysis of a Multidimensional Liquid Chromatography Mass Spectrometry Dataset Acquired on a Quadrupole Selecting, Quadrupole Collision Cell, Time-of-flight Mass Spectrometer II. New Developments in Protein Prospector Allow for Reliable and Comprehensive Automatic Analysis of Large Datasets. Mol Cell Proteomics. 2005;4(8):1194–1204. doi: 10.1074/mcp.D500002-MCP200. [DOI] [PubMed] [Google Scholar]
- Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. In-depth Analysis of Tandem Mass Spectrometry Data from Disparate Instrument Types. Mol Cell Proteomics. 2008;7(12):2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley RJ, Clauser KR. Modification Site Localization Scoring: Strategies and Performance. Mol Cell Proteomics. 2012;11(5):3–14. doi: 10.1074/mcp.R111.015305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. A Cross-platform Toolkit for Mass Spectrometry and Proteomics. Nat Biotechnol. 2012;30(10):918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler KB, Pompach P, Goldman R, Edwards N. Exploring Site-Specific N Glycosylation Microheterogeneity of Haptoglobin Using Glycopeptide CID Tandem Mass Spectra and Glycan Database Search. J Proteome Res. 2013;12(8):3652–3666. doi: 10.1021/pr400196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Chen R, Seebun D, Ye M, Figeys D, Zou H. Large-scale characterization of intact N-glycopeptides using an automated glycoproteomic method. J Proteomics. 2014;110:145–154. doi: 10.1016/j.jprot.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Chen T, Kao M-Y, Tepel M, Rush J, Church GM. A Dynamic Programming Approach to De Novo Peptide Sequencing via Tandem Mass Spectrometry. J Comput Biol. 2001;8(3):325–337. doi: 10.1089/10665270152530872. [DOI] [PubMed] [Google Scholar]
- Chi H, Chen H, He K, Wu L, Yang B, Sun R-X, Liu J, Zeng W-F, Song C-Q, He S-M, Dong M-Q. pNovo+: De Novo Peptide Sequencing Using Complementary HCD and ETD Tandem Mass Spectra. J Proteome Res. 2013;12(2):615–625. doi: 10.1021/pr3006843. [DOI] [PubMed] [Google Scholar]
- Chi H, Chen H, He K, Wu L, Yang B, Sun R-X, Liu J, Zeng W-F, Song C-Q, He S-M, others pNovo+: de novo peptide sequencing using complementary HCD and ETD tandem mass spectra. J Proteome Res. 2012;12(2):615–625. doi: 10.1021/pr3006843. [DOI] [PubMed] [Google Scholar]
- Cho J-Y, Lee H-J, Jeong S-K, Kim K-Y, Kwon K-H, Yoo JS, Omenn GS, Baker MS, Hancock WS, Paik Y-K. Combination of Multiple Spectral Libraries Improves the Current Search Methods Used to Identify Missing Proteins in the Chromosome-Centric Human Proteome Project. J Proteome Res. 2015 doi: 10.1021/acs.jproteome.5b00578. in press. [DOI] [PubMed] [Google Scholar]
- Choudhary G, Wu S-L, Shieh P, Hancock WS. Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J Proteome Res. 2003;2(1):59–67. doi: 10.1021/pr025557n. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Sladek R, Blanchette M. BigDataScript: a scripting language for data pipelines. Bioinformatics. 2015;31(1):10–16. doi: 10.1093/bioinformatics/btu595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauser KR, Baker P, Burlingame AL. Role of Accurate Mass Measurement (±10 ppm) in Protein Identification Strategies Employing MS or MS/MS and Database Searching. Anal Chem. 1999;71(14):2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- Cooper CA, Gasteiger E, Packer NH. GlycoMod –A software tool for determining glycosylation compositions from mass spectrometric data. PROTEOMICS. 2001;1(2):340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- Craig R, Cortens JC, Fenyo D, Beavis RC. Using Annotated Peptide Mass Spectrum Libraries for Protein Identification. J Proteome Res. 2006;5(8):1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- Dallas DC, Martin WF, Hua S, German JB. Automated glycopeptide analysis—review of current state and future directions. Brief Bioinform. 2013;14(3):361–374. doi: 10.1093/bib/bbs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancik V, Addona TA, Clauser KR, Vath JE, Pevzner PA. De novo peptide sequencing via tandem mass spectrometry. J Comput Biol. 1999;6(3-4):327–342. doi: 10.1089/106652799318300. [DOI] [PubMed] [Google Scholar]
- Desaire H. Glycopeptide Analysis, Recent Developments and Applications. Mol Cell Proteomics. 2013;12(4):893–901. doi: 10.1074/mcp.R112.026567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaire H, Hua D. When can glycopeptides be assigned based solely on high-resolution mass spectrometry data? Int J Mass Spectrom. 2009;287(1–3):21–26. [Google Scholar]
- Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10(6):1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Nothaft H, Szymanski CM, Kelly J. Identification and Quantification of Glycoproteins Using Ion-Pairing Normal-phase Liquid Chromatography and Mass Spectrometry. Mol Cell Proteomics. 2009;8(9):2170–2185. doi: 10.1074/mcp.M900088-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfer V, Pichler P, Stranzl T, Stadlmann J, Taus T, Winkler S, Mechtler K. MS Amanda, a Universal Identification Algorithm Optimized for High Accuracy Tandem Mass Spectra. J Proteome Res. 2014;13(8):3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards N, Wu X, Tseng C-W. An Unsupervised, Model-Free, Machine-Learning Combiner for Peptide Identifications from Tandem Mass Spectra. Clin Proteomics. 2009;5(1):23–36. [Google Scholar]
- Egertson JD, Kuehn A, Merrihew GE, Bateman NW, MacLean BX, Ting YS, Canterbury JD, Marsh DM, Kellmann M, Zabrouskov V, Wu CC, MacCoss MJ. Multiplexed MS/MS for improved data-independent acquisition. Nat Methods. 2013;10(8):744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large- scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-Decoy Search Strategy for Mass Spectrometry-Based Proteomics. Methods Mol Biol Clifton NJ. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. J Mass Spectrom. 2009;44(12):1637–1660. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- Eng JK, Jahan TA, Hoopmann MR. Comet: An open-source MS/MS sequence database search tool. PROTEOMICS. 2013;13(1):22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Ethier M, Saba JA, Ens W, Standing KG, Perreault H. Automated structural assignment of derivatized complex N-linked oligosaccharides from tandem mass spectra. Rapid Commun Mass Spectrom. 2002;16(18):1743–1754. doi: 10.1002/rcm.779. [DOI] [PubMed] [Google Scholar]
- Ethier M, Saba JA, Spearman M, Krokhin O, Butler M, Ens W, Standing KG, Perreault H. Application of the StrOligo algorithm for the automated structure assignment of complex N-linked glycans from glycoproteins using tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17(24):2713–2720. doi: 10.1002/rcm.1252. [DOI] [PubMed] [Google Scholar]
- Fabre B, Lambour T, Bouyssié D, Menneteau T, Monsarrat B, Burlet-Schiltz O, Bousquet-Dubouch M-P. Comparison of label-free quantification methods for the determination of protein complexes subunits stoichiometry. EuPA Open Proteomics. 2014;4:82–86. [Google Scholar]
- Fox JW, Serrano SMT. Exploring snake venom proteomes: multifaceted analyses for complex toxin mixtures. PROTEOMICS. 2008;8(4):909–920. doi: 10.1002/pmic.200700777. [DOI] [PubMed] [Google Scholar]
- Frank AM. A Ranking-Based Scoring Function for Peptide—Spectrum Matches. J Proteome Res. 2009a;8(5):2241–2252. doi: 10.1021/pr800678b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AM. Predicting Intensity Ranks of Peptide Fragment Ions. J Proteome Res. 2009b;8(5):2226–2240. doi: 10.1021/pr800677f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze HH. Understanding Human Glycosylation Disorders: Biochemistry Leads the Charge. J Biol Chem. 2013;288(10):6936–6945. doi: 10.1074/jbc.R112.429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frewen BE, Merrihew GE, Wu CC, Noble WS, MacCoss MJ. Analysis of Peptide MS/MS Spectra from Large-Scale Proteomics Experiments Using Spectrum Libraries. Anal Chem. 2006;78(16):5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
- Froehlich JW, Dodds ED, Wilhelm M, Serang O, Steen JA, Lee RS. A Classifier Based on Accurate Mass Measurements to Aid Large Scale, Unbiased Glycoproteomics. Mol Cell Proteomics. 2013;12(4):1017–1025. doi: 10.1074/mcp.M112.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni E, Luquette LJ, Lancaster AK, Hawkins JB, Jung J-Y, Souilmi Y, Wall DP, Tonellato PJ. COSMOS: Python library for massively parallel workflows. Bioinformatics. 2014;30(20):2956–2958. doi: 10.1093/bioinformatics/btu385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S, Bourmaud A, Kim SY, Domon B. Technical considerations for large- scale parallel reaction monitoring analysis. J Proteomics. 2014;100:147–159. doi: 10.1016/j.jprot.2013.10.029. [DOI] [PubMed] [Google Scholar]
- Gaucher SP, Morrow J, Leary JA. STAT: A Saccharide Topology Analysis Tool Used in Combination with Tandem Mass Spectrometry. Anal Chem. 2000;72(11):2331–2336. doi: 10.1021/ac000096f. [DOI] [PubMed] [Google Scholar]
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open Mass Spectrometry Search Algorithm. J Proteome Res. 2004;3(5):958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted Data Extraction of the MS/MS Spectra Generated by Data-independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Mol Cell Proteomics. 2012;11(6):O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg D, Bern M, Parry S, Sutton-Smith M, Panico M, Morris HR, Dell A. Automated N-glycopeptide identification using a combination of single- and tandem-MS. J Proteome Res. 2007;6:3995–4005. doi: 10.1021/pr070239f. [DOI] [PubMed] [Google Scholar]
- Goloborodko AA, Levitsky LI, Ivanov MV, Gorshkov MV. Pyteomics—a Python Framework for Exploratory Data Analysis and Rapid Software Prototyping in Proteomics. J Am Soc Mass Spectrom. 2013;24(2):301–304. doi: 10.1007/s13361-012-0516-6. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, Qi D, Fan J, Bessant C, Jones AR. A tutorial for software development in quantitative proteomics using PSI standard formats. Biochim Biophys Acta BBA - Proteins Proteomics. 2014;1844(1, Part A):88–97. doi: 10.1016/j.bbapap.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granholm V, Käll L. Quality assessments of peptide–spectrum matches in shotgun proteomics. PROTEOMICS. 2011;11(6):1086–1093. doi: 10.1002/pmic.201000432. [DOI] [PubMed] [Google Scholar]
- Grossmann J, Roschitzki B, Panse C, Fortes C, Barkow-Oesterreicher S, Rutishauser D, Schlapbach R. Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J Proteomics. 2010;73(9):1740–1746. doi: 10.1016/j.jprot.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Gupta N, Bandeira N, Keich U, Pevzner PA. Target-Decoy Approach and False Discovery Rate: When Things May Go Wrong. J Am Soc Mass Spectrom. 2011;22(7):1111–1120. doi: 10.1007/s13361-011-0139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthals A, Bandeira N. Peptide Identification by Tandem Mass Spectrometry with Alternate Fragmentation Modes. Mol Cell Proteomics. 2012;11(9):550–557. doi: 10.1074/mcp.R112.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthals A, Clauser KR, Frank AM, Bandeira N. Sequencing-Grade De novo Analysis of MS/MS Triplets (CID/HCD/ETD) From Overlapping Peptides. J Proteome Res. 2013;12(6):2846–2857. doi: 10.1021/pr400173d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Hägglund P, Matthiesen R, Elortza F, Højrup P, Roepstorff P, Jensen ON, Bunkenborg J. An Enzymatic Deglycosylation Scheme Enabling Identification of Core Fucosylated N-Glycans and O-Glycosylation Site Mapping of Human Plasma Proteins. J Proteome Res. 2007;6(8):3021–3031. doi: 10.1021/pr0700605. [DOI] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross Talk Between O GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease. Annu Rev Biochem. 2011;80(1):825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia-Langner A, Cannon WR, Jarman KD, Jarman KH. Sequence optimization as an alternative to de novo analysis of tandem mass spectrometry data. Bioinformatics. 2004;20(14):2296–2304. doi: 10.1093/bioinformatics/bth242. [DOI] [PubMed] [Google Scholar]
- He L, Xin L, Shan B, Lajoie GA, Ma B. GlycoMaster DB: Software To Assist the Automated Identification of N-Linked Glycopeptides by Tandem Mass Spectrometry. J Proteome Res. 2014;13(9):3881–3895. doi: 10.1021/pr401115y. [DOI] [PubMed] [Google Scholar]
- Hill JJ, Moreno MJ, Lam JCY, Haqqani AS, Kelly JF. Identification of secreted proteins regulated by cAMP in glioblastoma cells using glycopeptide capture and label-free quantification. Proteomics. 2009;9(3):535–549. doi: 10.1002/pmic.200800257. [DOI] [PubMed] [Google Scholar]
- Hong Q, Lebrilla CB, Miyamoto S, Ruhaak LR. Absolute Quantitation of Immunoglobulin G and Its Glycoforms Using Multiple Reaction Monitoring. Anal Chem. 2013;85(18):8585–8593. doi: 10.1021/ac4009995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Hu CY, Kim BJ, Totten SM, Oh MJ, Yun N, Nwosu CC, Yoo JS, Lebrilla CB, An HJ. Glyco-Analytical Multispecific Proteolysis (Glyco-AMP): A Simple Method for Detailed and Quantitative Glycoproteomic Characterization. J Proteome Res. 2013;12(10):4414–4423. doi: 10.1021/pr400442y. [DOI] [PubMed] [Google Scholar]
- Huang Q, Yang L, Luo J, Guo L, Wang Z, Yang X, Jin W, Fang Y, Ye J, Shan B, Zhang Y. SWATH enables precise label-free quantification on proteome scale. Proteomics. 2015;15(7):1215–1223. doi: 10.1002/pmic.201400270. [DOI] [PubMed] [Google Scholar]
- Hu H, Huang Y, Mao Y, Yu X, Xu Y, Liu J, Zong C, Boons G-J, Lin C, Xia Y, Zaia J. A Computational Framework for Heparan Sulfate Sequencing Using High- resolution Tandem Mass Spectra. Mol Cell Proteomics. 2014;13(9):2490–2502. doi: 10.1074/mcp.M114.039560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lam H. Expanding Tandem Mass Spectral Libraries of Phosphorylated Peptides: Advances and Applications. J Proteome Res. 2013;12(12):5971–5977. doi: 10.1021/pr4007443. [DOI] [PubMed] [Google Scholar]
- Irungu J, Go EP, Dalpathado DS, Desaire H. Simplification of Mass Spectral Analysis of Acidic Glycopeptides Using GlycoPep ID. Anal Chem. 2007;79(8):3065–3074. doi: 10.1021/ac062100e. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially Modified Protein Abundance Index (emPAI) for Estimation of Absolute Protein Amount in Proteomics by the Number of Sequenced Peptides per Protein. Mol Cell Proteomics. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Decon2LS: An open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics. 2009;10(1):87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K, Kim S, Pevzner PA. UniNovo: a universal tool for de novo peptide sequencing. Bioinformatics. 2013;29(16):1953–1962. doi: 10.1093/bioinformatics/btt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joenvaara S, Ritamo I, Peltoniemi H, Renkonen R. N-glycoproteomics - an automated workflow approach. Glycobiology. 2008;18(4):339–349. doi: 10.1093/glycob/cwn013. [DOI] [PubMed] [Google Scholar]
- Jones AR, Eisenacher M, Mayer G, Kohlbacher O, Siepen J, Hubbard SJ, Selley JN, Searle BC, Shofstahl J, Seymour SL, Julian R, Binz P-A, Deutsch EW, Hermjakob H, Reisinger F, Griss J, Vizcaíno JA, Chambers M, Pizarro A, Creasy D. The mzIdentML Data Standard for Mass Spectrometry-Based Proteomics Results. Mol Cell Proteomics. 2012;11(7):M111.014381. doi: 10.1074/mcp.M111.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker J, Bielow C, Bertsch A, Sturm M, Reinert K, Kohlbacher O. TOPPAS: A Graphical Workflow Editor for the Analysis of High-Throughput Proteomics Data. J Proteome Res. 2012;11(7):3914–3920. doi: 10.1021/pr300187f. [DOI] [PubMed] [Google Scholar]
- Kameyama A, Kikuchi N, Nakaya S, Ito H, Sato T, Shikanai T, Takahashi Y, Takahashi K, Narimatsu H. A Strategy for Identification of Oligosaccharide Structures Using Observational Multistage Mass Spectral Library. Anal Chem. 2005;77(15):4719–4725. doi: 10.1021/ac048350h. [DOI] [PubMed] [Google Scholar]
- Kameyama A, Nakaya S, Ito H, Kikuchi N, Angata T, Nakamura M, Ishida H-K, Narimatsu H. Strategy for Simulation of CID Spectra of N-Linked Oligosaccharides toward Glycomics. J Proteome Res. 2006;5(4):808–814. doi: 10.1021/pr0503937. [DOI] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri K, Staples GO, Leymarie N, Leon DR, Turiák L, Huang Y, Yip S, Hu H, Heckendorf CF, Zaia J. Confident Assignment of Site-Specific Glycosylation in Complex Glycoproteins in a Single Step. J Proteome Res. 2014;13(10):4347–4355. doi: 10.1021/pr500506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep. 2011;1:90. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014:55277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-S, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang T-C, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TSK, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue CA, Gowda H, Pandey A. A draft map of the human proteome. Nature. 2014;509(7502):575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zaidi-Ainouch Z, Gallien S, Domon B. Mass spectrometry-based detection and quantification of plasma glycoproteins using selective reaction monitoring. Nat Protoc. 2012;7(5):859–871. doi: 10.1038/nprot.2012.023. [DOI] [PubMed] [Google Scholar]
- Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, Agravat S, Campbell MP, Kato M, Ranzinger R, Kettner C, York WS. The Minimum Information Required for a Glycomics Experiment (MIRAGE) Project: Improving the Standards for Reporting Mass-spectrometry-based Glycoanalytic Data. Mol Cell Proteomics. 2013;12(4):991–995. doi: 10.1074/mcp.O112.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012;28(19):2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- Kurogochi M, Amano J. Relative quantitation of glycopeptides based on stable isotope labeling using MALDI-TOF MS. Mol Basel Switz. 2014;19(7):9944–9961. doi: 10.3390/molecules19079944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Choi H, Vogel C, Nesvizhskii AI, Marcotte EM. MSblender: A Probabilistic Approach for Integrating Peptide Identifications from Multiple Database Search Engines. J Proteome Res. 2011;10(7):2949–2958. doi: 10.1021/pr2002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H. Building and Searching Tandem Mass Spectral Libraries for Peptide Identification. Mol Cell Proteomics. 2011;10(12):R111.008565. doi: 10.1074/mcp.R111.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Development and validation of a spectral library searching method for peptide identification from MS/MS. PROTEOMICS. 2007;7(5):655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- Leymarie N, Zaia J. Effective Use of Mass Spectrometry for Glycan and Glycopeptide Structural Analysis. Anal Chem. 2012;84(7):3040–3048. doi: 10.1021/ac3000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Glinskii OV, Glinsky VV. Glycobioinformatics: Current strategies and tools for data mining in MS-based glycoproteomics. Proteomics. 2013;13(2):341–354. doi: 10.1002/pmic.201200149. [DOI] [PubMed] [Google Scholar]
- Li X, Lin C, Han L, Costello CE, O'Connor PB. Charge Remote Fragmentation in Electron Capture and Electron Transfer Dissociation. J Am Soc Mass Spectrom. 2010;21(4):646–656. doi: 10.1016/j.jasms.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Cao J, He Y, Qiao L, Xu C, Lu H, Yang P. Tandem 18O Stable Isotope Labeling for Quantification of N-Glycoproteome. J Proteome Res. 2010;9(1):227–236. doi: 10.1021/pr900528j. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen J, Sethi A, Li QK, Chen L, Collins B, Gillet LCJ, Wollscheid B, Zhang H, Aebersold R. Glycoproteomic Analysis of Prostate Cancer Tissues by SWATH Mass Spectrometry Discovers N-acylethanolamine Acid Amidase and Protein Tyrosine Kinase 7 as Signatures for Tumor Aggressiveness. Mol Cell Proteomics. 2014;13(7):1753–1768. doi: 10.1074/mcp.M114.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Dekker LJM, Wu S, Vanduijn MM, Luider TM, Tolić N, Kou Q, Dvorkin M, Alexandrova S, Vyatkina K, Paša-Tolić L, Pevzner PA. De Novo Protein Sequencing by Combining Top-Down and Bottom-Up Tandem Mass Spectra. J Proteome Res. 2014;13(7):3241–3248. doi: 10.1021/pr401300m. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hüttenhain R, Surinova S, Gillet LC, Mouritsen J, Brunner R, Navarro P, Aebersold R. Quantitative measurements of N-linked glycoproteins in human plasma by SWATH-MS. PROTEOMICS. 2013;13(8):1247–1256. doi: 10.1002/pmic.201200417. [DOI] [PubMed] [Google Scholar]
- Lu B, Chen T. A Suboptimal Algorithm for De Novo Peptide Sequencing via Tandem Mass Spectrometry. J Comput Biol. 2003;10(1):1–12. doi: 10.1089/106652703763255633. [DOI] [PubMed] [Google Scholar]
- Lütteke T, Frank M, editors. Handling and Conversion of Carbohydrate Sequence Formats and Monosaccharide Notation - Springer. Springer; New York: 2015. [DOI] [PubMed] [Google Scholar]
- Lynn K-S, Chen C-C, Lih TM, Cheng C-W, Su W-C, Chang C-H, Cheng C-Y, Hsu W-L, Chen Y-J, Sung T-Y. MAGIC: An Automated N-Linked Glycoprotein Identification Tool Using a Y1-Ion Pattern Matching Algorithm and in Silico MS2 Approach. Anal Chem. 2015;87(4):2466–2473. doi: 10.1021/ac5044829. [DOI] [PubMed] [Google Scholar]
- Ma B. Novor: Real-Time Peptide de Novo Sequencing Software. J Am Soc Mass Spectrom. 2015;26(11):1885–1894. doi: 10.1007/s13361-015-1204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean B, Tomazela DM, Abbatiello SE, Zhang S, Whiteaker JR, Paulovich AG, Carr SA, Maccoss MJ. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal Chem. 2010;82(24):10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinforma Oxf Engl. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JA, Ko BJ, Xu H, Iwashkiw JA, Robotham SA, Shaw JB, Feldman MF, Brodbelt JS. Concurrent Automated Sequencing of the Glycan and Peptide Portions of O-Linked Glycopeptide Anions by Ultraviolet Photodissociation Mass Spectrometry. Anal Chem. 2013;85(19):9253–9261. doi: 10.1021/ac4021177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Johnson R. De Novo Sequencing and Homology Searching. Mol Cell Proteomics. 2012;11(2):O111.014902. doi: 10.1074/mcp.O111.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CWM, Lam H. Hunting for Unexpected Post-Translational Modifications by Spectral Library Searching with Tier-Wise Scoring. J Proteome Res. 2014;13(5):2262–2271. doi: 10.1021/pr401006g. [DOI] [PubMed] [Google Scholar]
- Ma K, Vitek O, Nesvizhskii AI. A statistical model-building perspective to identification of MS/MS spectra with PeptideProphet. BMC Bioinformatics. 2012;13(Suppl 16):S1. doi: 10.1186/1471-2105-13-S16-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Wilm M. Error-Tolerant Identification of Peptides in Sequence Databases by Peptide Sequence Tags. Anal Chem. 1994;66(24):4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- Mariño K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol. 2010;6(10):713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Römpp A, Neumann S, Pizarro AD, Montecchi-Palazzi L, Tasman N, Coleman M, Reisinger F, Souda P, Hermjakob H, Binz P-A, Deutsch EW. mzML—a Community Standard for Mass Spectrometry Data. Mol Cell Proteomics. 2011;10(1):R110.000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell E, Tan Y, Tan Y, Hu H, Benson G, Aizikov K, Conley S, Staples GO, Slysz GW, Smith RD, Zaia J. GlycReSoft: A Software Package for Automated Recognition of Glycans from LC/MS Data. PLoS ONE. 2012;7(9):e45474. doi: 10.1371/journal.pone.0045474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayampurath A, Song E, Mathur A, Yu C, Hammoud Z, Mechref Y, Tang H. Label-Free Glycopeptide Quantification for Biomarker Discovery in Human Sera. J Proteome Res. 2014;13(11):4821–4832. doi: 10.1021/pr500242m. [DOI] [PubMed] [Google Scholar]
- Mayampurath A, Yu C-Y, Song E, Balan J, Mechref Y, Tang H. Computational Framework for Identification of Intact Glycopeptides in Complex Samples. Anal Chem. 2014;86(1):453–463. doi: 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]
- Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17(20):2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- McCoy CO, Gallagher A, Hoffman NG, Matsen FA. nestly—a framework for running software with nested parameter choices and aggregating results. Bioinformatics. 2013;29(3):387–388. doi: 10.1093/bioinformatics/bts696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain S, Tamura K, Kertesz-Farkas A, Grant CE, Diament B, Frewen B, Howbert JJ, Hoopmann MR, Käll L, Eng JK, MacCoss MJ, Noble WS. Crux: Rapid Open Source Protein Tandem Mass Spectrometry Analysis. J Proteome Res. 2014;13(10):4488–4491. doi: 10.1021/pr500741y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo L, Dutta D, Wan Y, Chen T. MSNovo: A Dynamic Programming Algorithm for de Novo Peptide Sequencing via Tandem Mass Spectrometry. Anal Chem. 2007;79(13):4870–4878. doi: 10.1021/ac070039n. [DOI] [PubMed] [Google Scholar]
- Muth T, Weilnbock L, Rapp E, Huber CG, Martens L, Vaudel M, Barsnes H. DeNovoGUI: an open source graphical user interface for de novo sequencing of tandem mass spectra. J Proteome Res. 2014;13:1143–1146. doi: 10.1021/pr4008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na S, Bandeira N, Paek E. Fast Multi-blind Modification Search through Tandem Mass Spectrometry. Mol Cell Proteomics. 2012;11(4):M111.010199. doi: 10.1074/mcp.M111.010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahnsen S, Bertsch A, Rahnenführer J, Nordheim A, Kohlbacher O. Probabilistic Consensus Scoring Improves Tandem Mass Spectrometry Peptide Identification. J Proteome Res. 2011;10(8):3332–3343. doi: 10.1021/pr2002879. [DOI] [PubMed] [Google Scholar]
- Nahnsen S, Bielow C, Reinert K, Kohlbacher O. Tools for Label-free Peptide Quantification. Mol Cell Proteomics. 2013;12(3):549–556. doi: 10.1074/mcp.R112.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na S, Paek E. Software eyes for protein post-translational modifications. Mass Spectrom Rev. 2014 doi: 10.1002/mas.21425. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- Nelson EK, Piehler B, Eckels J, Rauch A, Bellew M, Hussey P, Ramsay S, Nathe C, Lum K, Krouse K, Stearns D, Connolly B, Skillman T, Igra M. LabKey Server: An open source platform for scientific data integration, analysis and collaboration. BMC Bioinformatics. 2011;12(1):71. doi: 10.1186/1471-2105-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73(11):2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Rüetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, Larson G. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Methods. 2009;6(11):809–811. doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. Glycosylation in Cellular Mechanisms of Health and Disease. Cell. 2006;126(5):855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Pan S, Chen R, Aebersold R, Brentnall TA. Mass spectrometry based glycoproteomics--from a proteomics perspective. Mol Cell Proteomics MCP. 2011;10(1):R110.003251. doi: 10.1074/mcp.R110.003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Tamura Y, Chen R, May D, McIntosh MW, Brentnall TA. Large-scale quantitative glycoproteomics analysis of site-specific glycosylation occupancy. Mol Biosyst. 2012;8(11):2850–2856. doi: 10.1039/c2mb25268f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Wang R, Hermjakob H, Müller M, Vesada V, Vizcaíno JA. Open source libraries and frameworks for mass spectrometry based proteomics: A developer's perspective. Biochim Biophys Acta BBA - Proteins Proteomics. 2014;1844(1, Part A):63–76. doi: 10.1016/j.bbapap.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel Reaction Monitoring for High Resolution and High Mass Accuracy Quantitative, Targeted Proteomics. Mol Cell Proteomics. 2012;11(11):1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhong Y, Yang G, Ma T, Jia L, Huang C, Li Z. Profiling of Concanavalin A-Binding Glycoproteins in Human Hepatic Stellate Cells Activated with Transforming Growth Factor-β1. Molecules. 2014;19(12):19845–19867. doi: 10.3390/molecules191219845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecchi KR, Wenke JL, Go EP, Desaire H. Label-free quantitation: A new glycoproteomics approach. J Am Soc Mass Spectrom. 2009;20(6):1048–1059. doi: 10.1016/j.jasms.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein Identification False Discovery Rates for Very Large Proteomics Data Sets Generated by Tandem Mass Spectrometry. Mol Cell Proteomics. 2009;8(11):2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren JM, Rejtar T, Li L, Karger BL. N-Glycan Structure Annotation of Glycopeptides Using a Linearized Glycan Structure Database (GlyDB). J Proteome Res. 2007;6(8):3162–3173. doi: 10.1021/pr070111y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger G, Ludwig C, Rost HL, Aebersold R, Malmstrom L. aLFQ: an R-package for estimating absolute protein quantities from label-free LC-MS/MS proteomics data. Bioinformatics. 2014;30(17):2511–2513. doi: 10.1093/bioinformatics/btu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röst HL, Rosenberger G, Navarro P, Gillet L, Miladinović SM, Schubert OT, Wolski W, Collins BC, Malmström J, Malmström L, Aebersold R. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32(3):219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- Sadedin SP, Pope B, Oshlack A. Bpipe: a tool for running and managing bioinformatics pipelines. Bioinformatics. 2012;28(11):1525–1526. doi: 10.1093/bioinformatics/bts167. [DOI] [PubMed] [Google Scholar]
- Sadygov RG, Cociorva D, Yates JR. Large-scale database searching using tandem mass spectra: Looking up the answer in the back of the book. Nat Methods. 2004;1(3):195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R. Quantitative Liquid Chromatography-Mass Spectrometry-Multiple Reaction Monitoring (LC MS-MRM) Analysis of Site-specific Glycoforms of Haptoglobin in Liver Disease. Mol Cell Proteomics MCP. 2013;12(5):1294–1305. doi: 10.1074/mcp.M112.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, Verdin E, Kahn CR, Maccoss MJ, Gibson BW. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics MCP. 2012;11(5):202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10(12):M111 007690. doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipert RR, Dodds ED, Clowers BH, Beecroft SM, German JB, Lebrilla CB. Factors That Influence Fragmentation Behavior of N-Linked Glycopeptide Ions. Anal Chem. 2008;80(10):3684–3692. doi: 10.1021/ac800067y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serang O, Froehlich JW, Muntel J, McDowell G, Steen H, Lee RS, Steen JA. SweetSEQer, Simple de Novo Filtering and Annotation of Glycoconjugate Mass Spectra. Mol Cell Proteomics. 2013;12(6):1735–1740. doi: 10.1074/mcp.O112.025940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakey Q, Bates B, Wu J. An Approach to Quantifying N-Linked Glycoproteins by Enzyme-Catalyzed 18O3-Labeling of Solid-Phase Enriched Glycopeptides. Anal Chem. 2010;82(18):7722–7728. doi: 10.1021/ac101564t. [DOI] [PubMed] [Google Scholar]
- Shan B, Ma B, Zhang K, Lajoie G. Complexities and algorithms for glycan sequencing using tandem mass spectrometry. J Bioinform Comput Biol. 2008;06(01):77–91. doi: 10.1142/s0219720008003291. [DOI] [PubMed] [Google Scholar]
- Sheynkman GM, Johnson JE, Jagtap PD, Shortreed MR, Onsongo G, Frey BL, Griffin TJ, Smith LM. Using Galaxy-P to leverage RNA-Seq for the discovery of novel protein variations. BMC Genomics. 2014;15(1):703. doi: 10.1186/1471-2164-15-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheynkman GM, Shortreed MR, Frey BL, Smith LM. Discovery and Mass Spectrometric Analysis of Novel Splice-junction Peptides Using RNA-Seq. Mol Cell Proteomics. 2013;12(8):2341–2353. doi: 10.1074/mcp.O113.028142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: Multi-level Integrative Analysis of Shotgun Proteomic Data Improves Peptide and Protein Identification Rates and Error Estimates. Mol Cell Proteomics. 2011;10(12):M111.007690. doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shteynberg D, Nesvizhskii AI, Moritz RL, Deutsch EW. Combining Results of Multiple Search Engines in Proteomics. Mol Cell Proteomics. 2013;12(9):2383–2393. doi: 10.1074/mcp.R113.027797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C, Zampronio CG, Creese AJ, Cooper HJ. Higher Energy Collision Dissociation (HCD) Product Ion-Triggered Electron Transfer Dissociation (ETD) Mass Spectrometry for the Analysis of N-Linked Glycoproteins. J Proteome Res. 2012;11(9):4517–4525. doi: 10.1021/pr300257c. [DOI] [PubMed] [Google Scholar]
- Sloggett C, Goonasekera N, Afgan E. BioBlend: automating pipeline analyses within Galaxy and CloudMan. Bioinformatics. 2013;29(13):1685–1686. doi: 10.1093/bioinformatics/btt199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Kelleher NL, Proteomics TC for TD. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10(3):186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Pyreddy S, Mechref Y. Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2012;26(17):1941–1954. doi: 10.1002/rcm.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen H, Mann M. The abc's (and xyz's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5(9):699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KT-BG, Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H. Mining the O- glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8(11):977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- Strum JS, Nwosu CC, Hua S, Kronewitter SR, Seipert RR, Bachelor RJ, An HJ, Lebrilla CB. Automated Assignments of N- and O-Site Specific Glycosylation with Extensive Glycan Heterogeneity of Glycoprotein Mixtures. Anal Chem. 2013;85:5666–5675. doi: 10.1021/ac4006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm M, Bertsch A, Gröpl C, Hildebrandt A, Hussong R, Lange E, Pfeifer N, Schulz-Trieglaff O, Zerck A, Reinert K, Kohlbacher O. OpenMS –An open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9(1):163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suni V, Imanishi SY, Maiolica A, Aebersold R, Corthals GL. Confident Site Localization Using a Simulated Phosphopeptide Spectral Library. J Proteome Res. 2015;14(5):2348–2359. doi: 10.1021/acs.jproteome.5b00050. [DOI] [PubMed] [Google Scholar]
- Su Z-D, Sheng Q-H, Li Q-R, Chi H, Jiang X, Yan Z, Fu N, He S-M, Khaitovich P, Wu J R, Zeng R. De novo identification and quantification of single amino-acid variants in human brain. J Mol Cell Biol. 2014;6(5):421–433. doi: 10.1093/jmcb/mju031. [DOI] [PubMed] [Google Scholar]
- Tabb DL, Fernando CG, Chambers MC. MyriMatch: Highly Accurate Tandem Mass Spectral Peptide Identification by Multivariate Hypergeometric Analysis. J Proteome Res. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, Saraf A, Yates JR. GutenTag: High-Throughput Sequence Tagging via an Empirically Derived Fragmentation Model. Anal Chem. 2003;75(23):6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Fujita Y, Parry HE, Yoshizawa AC, Morimoto K, Murase M, Yamada Y, Yao J, Utsunomiya S, Kajihara S, Fukuda M, Ikawa M, Tabata T, Takahashi K, Aoshima K, Nihei Y, Nishioka T, Oda Y, Tanaka K. Mass++: A Visualization and Analysis Tool for Mass Spectrometry. J Proteome Res. 2014;13(8):3846–3853. doi: 10.1021/pr500155z. [DOI] [PubMed] [Google Scholar]
- Tang H, Mechref Y, Novotny MV. Automated interpretation of MS/MS spectra of oligosaccharides. Bioinformatics. 2005;21(suppl 1):i431–i439. doi: 10.1093/bioinformatics/bti1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz P-A, Julian RK, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, Dunn MJ, Heck AJR, Leitner A, Macht M, Mann M, Martens L, Neubert TA, Patterson SD, Ping P, Seymour SL, Souda P, Tsugita A, Vandekerckhove J, Vondriska TM, Whitelegge JP, Wilkins MR, Xenarios I, Yates JR, Hermjakob H. The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol. 2007;25(8):887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- Tharakan R, Edwards N, Graham DRM. Data maximization by multipass analysis of protein mass spectra. PROTEOMICS. 2010;10(6):1160–1171. doi: 10.1002/pmic.200900433. [DOI] [PubMed] [Google Scholar]
- Thaysen-Andersen M, Packer NH. Advances in LC–MS/MS-based glycoproteomics: Getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim Biophys Acta BBA - Proteins Proteomics. 2014;1844(9):1437–1452. doi: 10.1016/j.bbapap.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N- linked glycopeptides. Nat Protoc. 2007;2(2):334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toghi Eshghi S, Shah P, Yang W, Li X, Zhang H. GPQuest: A Spectral Library Matching Algorithm for Site-Specific Assignment of Tandem Mass Spectra to Intact N-glycopeptides. Anal Chem. 2015;87(10):5181–5188. doi: 10.1021/acs.analchem.5b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama A, Nakagawa H, Matsuda K, Ishikawa N, Kohno N, Daigo Y, Sato T-A, Nakamura Y, Ueda K. Deglycosylation and label-free quantitative LC MALDI MS applied to efficient serum biomarker discovery of lung cancer. Proteome Sci. 2011;9(1):1–12. doi: 10.1186/1477-5956-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou C-C, Avtonomov D, Larsen B, Tucholska M, Choi H, Gingras A-C, Nesvizhskii AI. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods. 2015 doi: 10.1038/nmeth.3255. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudel M, Burkhart JM, Zahedi RP, Oveland E, Berven FS, Sickmann A, Martens L, Barsnes H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat Biotechnol. 2015;33(1):22–24. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]
- Vaudel M, Sickmann A, Martens L. Peptide and protein quantification: A map of the minefield. Proteomics. 2010;10(4):650–670. doi: 10.1002/pmic.200900481. [DOI] [PubMed] [Google Scholar]
- Vaudel M, Barsnes H, Berven FS, Sickmann A, Martens L. SearchGUI: An open- source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics. 2011;11(5):996–999. doi: 10.1002/pmic.201000595. [DOI] [PubMed] [Google Scholar]
- Vékey K, Ozohanics O, Tóth E, Jekő A, Révész Á , Krenyácz J, Drahos L. Fragmentation characteristics of glycopeptides. Int J Mass Spectrom. 2013;345–347:71–79. [Google Scholar]
- Viner RI, Zhang T, Second T, Zabrouskov V. Quantification of post-translationally modified peptides of bovine α-crystallin using tandem mass tags and electron transfer dissociation. J Proteomics. 2009;72(5):874–885. doi: 10.1016/j.jprot.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Walzer M, Qi D, Mayer G, Uszkoreit J, Eisenacher M, Sachsenberg T, Gonzalez-Galarza FF, Fan J, Bessant C, Deutsch EW, Reisinger F, Vizcaíno JA, Medina-Aunon JA, Albar JP, Kohlbacher O, Jones AR. The mzQuantML Data Standard for Mass Spectrometry–based Quantitative Studies in Proteomics. Mol Cell Proteomics. 2013;12(8):2332–2340. doi: 10.1074/mcp.O113.028506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu Q, Zhang B. Leveraging the complementary nature of RNA-Seq and shotgun proteomics data. PROTEOMICS. 2014;14(23-24):2676–2687. doi: 10.1002/pmic.201400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasinger VC, Zeng M, Yau Y. Current Status and Advances in Quantitative Proteomic Mass Spectrometry. Int J Proteomics. 2013;2013:e180605. doi: 10.1155/2013/180605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Li L. Comparative glycoproteomics: approaches and applications. Brief Funct Genomic Proteomic. 2009;8(2):104–113. doi: 10.1093/bfgp/eln053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser H, Nahnsen S, Grossmann J, Nilse L, Quandt A, Brauer H, Sturm M, Kenar E, Kohlbacher O, Aebersold R, Malmstrom L. An automated pipeline for high- throughput label-free quantitative proteomics. J Proteome Res. 2013;12(4):1628–1644. doi: 10.1021/pr300992u. [DOI] [PubMed] [Google Scholar]
- Wenger CD, Coon JJ. A Proteomics Search Algorithm Specifically Designed for High-Resolution Tandem Mass Spectra. J Proteome Res. 2013;12(3):1377–1386. doi: 10.1021/pr301024c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese J-H, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Wollscheid B, Bausch-Fluck D, Henderson C, O'Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27(4):378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodin CL, Hua D, Maxon M, Rebecchi KR, Go EP, Desaire H. GlycoPep grader: a web-based utility for assigning the composition of N-linked glycopeptides. Anal Chem. 2012;84(11):4821–4829. doi: 10.1021/ac300393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodin CL, Maxon M, Desaire H. Software for Automated Interpretation of Mass Spectrometry Data from Glycans and Glycopeptides. The Analyst. 2013;138(10):2793–2803. doi: 10.1039/c2an36042j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SW, Liang SY, Pu TH, Chang FY, Khoo KH. Sweet-Heart - an integrated suite of enabling computational tools for automated MS2/MS3 sequencing and identification of glycopeptides. J Proteomics. 2013;84:1–16. doi: 10.1016/j.jprot.2013.03.026. [DOI] [PubMed] [Google Scholar]
- Xu Y, Bailey U-M, Schulz BL. Automated measurement of site-specific N- glycosylation occupancy with SWATH-MS. Proteomics. 2015;15(13):2177–2186. doi: 10.1002/pmic.201400465. [DOI] [PubMed] [Google Scholar]
- Yates JR, Morgan SF, Gatlin CL, Griffin PR, Eng JK. Method To Compare Collision-Induced Dissociation Spectra of Peptides: Potential for Library Searching and Subtractive Analysis. Anal Chem. 1998;70(17):3557–3565. doi: 10.1021/ac980122y. [DOI] [PubMed] [Google Scholar]
- Ye D, Fu Y, Sun R-X, Wang H-P, Yuan Z-F, Chi H, He S-M. Open MS/MS spectral library search to identify unanticipated post-translational modifications and increase spectral identification rate. Bioinformatics. 2010;26(12):i399–i406. doi: 10.1093/bioinformatics/btq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C-Y, Meyer-Arendt K, Eichelberger B, Sun S, Houel S, Old WM, Knight R, Ahn NG, Hunter LE, Resing KA. A Simulated MS/MS Library for Spectrum-to-spectrum Searching in Large Scale Identification of Proteins. Mol Cell Proteomics. 2009;8(4):857–869. doi: 10.1074/mcp.M800384-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Agravat S, Aoki-Kinoshita KF, McBride R, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson N, Khoo K-H, Kolarich D, Liu Y, Novotny M, Packer NH, Paulson JC, Rapp E, Ranzinger R, Rudd PM, Smith DF, Struwe WB, Tiemeyer M, Wells L, Zaia J, Kettner C. MIRAGE: The minimum information required for a glycomics experiment. Glycobiology. 2014;24(5):402–406. doi: 10.1093/glycob/cwu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-Y, Mayampurath A, Hu Y, Zhou S, Mechref Y, Tang H. Automated annotation and quantification of glycans using liquid chromatography–mass spectrometry. Bioinformatics. 2013;29(13):1706–1707. doi: 10.1093/bioinformatics/btt190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Prediction of Low-Energy Collision-Induced Dissociation Spectra of Peptides. Anal Chem. 2004;76(14):3908–3922. doi: 10.1021/ac049951b. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Prediction of Low-Energy Collision-Induced Dissociation Spectra of Peptides with Three or More Charges. Anal Chem. 2005;77(19):6364–6373. doi: 10.1021/ac050857k. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jiao J, Yang P, Lu H. Mass spectrometry-based N-glycoproteomics for cancer biomarker discovery. Clin Proteomics. 2014;11(1):18. doi: 10.1186/1559-0275-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li Y, Shao W, Lam H. Understanding the improved sensitivity of spectral library searching over sequence database searching in proteomics data analysis. PROTEOMICS. 2011;11(6):1075–1085. doi: 10.1002/pmic.201000492. [DOI] [PubMed] [Google Scholar]
- Zhang S, Liu X, Kang X, Sun C, Lu H, Yang P, Liu Y. iTRAQ plus 18O: A new technique for target glycoprotein analysis. Talanta. 2012;91:122–127. doi: 10.1016/j.talanta.2012.01.033. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, Ma B. PEAKS DB: De Novo Sequencing Assisted Database Search for Sensitive and Accurate Peptide Identification. Mol Cell Proteomics. 2012;11(4):M111.010587. doi: 10.1074/mcp.M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Ranish JA, Watts JD, Aebersold R. Quantitative proteome analysis by solid-phase isotope tagging and mass spectrometry. Nat Biotechnol. 2002;20(5):512–515. doi: 10.1038/nbt0502-512. [DOI] [PubMed] [Google Scholar]
- Zhurov KO, Fornelli L, Wodrich MD, Laskay ÜA, Tsybin YO. Principles of electron capture and transfer dissociation mass spectrometry applied to peptide and protein structure analysis. Chem Soc Rev. 2013;42(12):5014. doi: 10.1039/c3cs35477f. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Hua D, Clark DF, Go EP, Desaire H. GlycoPep Detector: a tool for assigning mass spectrometry data of N-linked glycopeptides on the basis of their electron transfer dissociation spectra. Anal Chem. 2013;85:5023–5032. doi: 10.1021/ac400287n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Su X, Go EP, Desaire H. New Glycoproteomics Software, GlycoPep Evaluator, Generates Decoy Glycopeptides de Novo and Enables Accurate False Discovery Rate Analysis for Small Data Sets. Anal Chem. 2014;86(18):9212–9219. doi: 10.1021/ac502176n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wiśniewski JR, Mann M. Precision Mapping of an In Vivo N-Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell. 2010;141(5):897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Zubarev RA, Zubarev AR, Savitski MM. Electron Capture/Transfer versus Collisionally Activated/Induced Dissociations: Solo or Duet? J Am Soc Mass Spectrom. 2008;19(6):753–761. doi: 10.1016/j.jasms.2008.03.007. [DOI] [PubMed] [Google Scholar]