Abstract
K+ is an essential macronutrient for plants. It is acquired by specific uptake systems located in roots. Although the concentrations of K+ in the soil solution are widely variable, K+ nutrition is secured by uptake systems that exhibit different affinities for K+. Two main systems have been described for root K+ uptake in several species: the high-affinity HAK5-like transporter and the inward-rectifier AKT1-like channel. Other unidentified systems may be also involved in root K+ uptake, although they only seem to operate when K+ is not limiting. The use of knock-out lines has allowed demonstrating their role in root K+ uptake in Arabidopsis and rice. Plant adaptation to the different K+ supplies relies on the finely tuned regulation of these systems. Low K+-induced transcriptional up-regulation of the genes encoding HAK5-like transporters occurs through a signal cascade that includes changes in the membrane potential of root cells and increases in ethylene and reactive oxygen species concentrations. Activation of AKT1 channels occurs through phosphorylation by the CIPK23/CBL1 complex. Recently, activation of the Arabidopsis HAK5 by the same complex has been reported, pointing to CIPK23/CBL as a central regulator of the plant’s adaptation to low K+. Na+ is not an essential plant nutrient but it may be beneficial for some plants. At low concentrations, Na+ improves growth, especially under K+ deficiency. Thus, high-affinity Na+ uptake systems have been described that belong to the HKT and HAK families of transporters. At high concentrations, typical of saline environments, Na+ accumulates in plant tissues at high concentrations, producing alterations that include toxicity, water deficit and K+ deficiency. Data concerning pathways for Na+ uptake into roots under saline conditions are still scarce, although several possibilities have been proposed. The apoplast is a significant pathway for Na+ uptake in rice grown under salinity conditions, but in other plant species different mechanisms involving non-selective cation channels or transporters are under discussion.
Keywords: potassium, sodium, uptake, roots, Arabidopsis, rice
Introduction
Given the constant increase in world population, high-yield crop production has become a necessity for agriculture. As that the nutrient sources of the land are limited, the input of nutrients by the addition of fertilizers ensures a continuous supply for plants, circumventing reductions in plant yield. The use of fertilizers has raised crop yield considerably, for example, from 50 to 80% of wheat and corn grain yield is attributable to nutrient fertilization (Stewart et al., 2005). However, this practice comes with high economic and environmental costs.
Potassium (K+) is an essential macronutrient that is required by plants to complete their life cycle (Taiz and Zeiger, 1991). K+ can make up to 10% of the total plant dry weight (Leigh and Wyn Jones, 1984) and fulfills important functions for metabolism, growth, and stress adaptation. Specifically, it acts as an enzyme activator, protein synthesis stabilizer, neutralization of protein negative charges, and it participates in cytoplasmic pH homeostasis as well (Marschner, 2012). An optimal K+ concentration in the cytosol of around 100 mM is required for the performing of the functions mentioned above (Wyn Jones and Pollard, 1983), and plant cells maintain the cytosolic K+ concentration around this value (Walker et al., 1996).
K+ constitutes about 2.9% of the earth’s crust but the concentration of K+ in the soil solution is highly variable, in the 10-5 to 10-3 M range (Barraclough, 1989; Marschner, 2012). Since roots are able to take up K+ at a higher rate than this cation can diffuse from the bulk soil solution, a K+ depletion zone near the root surface can be formed with K+ concentrations of just a few micromolar (Baldwin et al., 1973; Claassen and Jungk, 1982). More importantly, increasing areas of the world are currently described as being K+ deficient for agricultural practices (Mengel et al., 2001; Moody and Bell, 2006; Römheld and Kirkby, 2010; Kirkby and Schubert, 2013).
K+ deficiency has a negative impact on plant growth since cellular expansion and photosynthesis are severely affected under these conditions (Bednarz and Oosterhuis, 1999; Hafsi et al., 2014). This deficiency also correlates with a decrease in protein synthesis and subsequent decline in growth (Walker et al., 1996, 1998). K+ deficiency has been shown to inhibit lateral root development in Arabidopsis (Armengaud et al., 2004; Shin and Schachtman, 2004; Kellermeier et al., 2014) and in barley (Drew, 1975) and in the up-regulation of genes involved in K+ uptake (Ashley et al., 2006; Nieves-Cordones et al., 2014). In addition, K+-deficient plants are more sensitive to abiotic and biotic stresses such as drought, cold, salinity, or fungal attacks (Marschner, 2012; Zörb et al., 2014).
Sodium (Na+) is not an essential element for plants but, for some species it can be a beneficial element that stimulates growth (Wakeel et al., 2010, 2011; Kronzucker et al., 2013). In these cases, Na+ can be regarded as a functional nutrient (Subbarao et al., 2003), that can partly replace K+ in some functions such as osmotic adjustment of the large central vacuole, cell turgor regulation leading to cell enlargement, or long-distance transport of anions (Subbarao et al., 2003; Horie et al., 2007; Gattward et al., 2012; Battie-Laclau et al., 2013).
On the other hand, Na+ has been extensively associated to its negative impact on crop yield. Excess of Na+ salts in the soil results in both reduced soil water availability (due to the decrease in water potential) and ionic toxicity. When accumulated at high concentrations in the cytoplasm, Na+ results in deleterious effects on cell biology, e.g., on photosynthetic activity or on membrane integrity (due to displacement of membrane-bound Ca2+ ions) (Cramer et al., 1985). Thus, Na+ is usually compartmentalized outside the cytoplasm (Morgan et al., 2014), in vesicles such as the vacuole, where it is used as an osmoticum. Estimates of the area of salt-affected soils vary widely, ranging from 6 to 10% of the earth’s land area (Eynard et al., 2005; Munns and Tester, 2008). Importantly, 20% of irrigated lands are affected by secondary salinization, limiting agriculture worldwide.
In the present review, we summarize recent advances in the field of K+ and Na+ uptake in the plant root, with special attention to the transport systems and their regulation mechanisms. We believe that the studies performed on the model plant Arabidopsis and the results of recent research in crops such as rice suggest that the results obtained with model species cannot be fully extended to other plant species.
K+ and Na+ Uptake By Roots: Kinetic Features and Sensitivity to Other Cations
K+ and Na+ can enter the root apoplast and diffuse toward inner cell layers (Sattelmacher et al., 1998). However, this pathway is interrupted by the endodermis, where the Casparian strip, which is impermeable to water and ions, is located (Schreiber et al., 1999; Tester and Leigh, 2001; Marschner, 2012; Geldner, 2013; Barberon and Geldner, 2014). To cross this impermeable barrier, nutrient ions enter the cytosol of a root peripheral cell either from the epidermis, cortex or endodermis and move from cell to cell (symplastic pathway) through plasmodesmata (Burch-Smith and Zambryski, 2012). Diffusion within the symplasm beyond the endodermic barrier allows nutrient ions to reach the stele, where they will initiate their travel toward the aerial parts within the xylem vessels (Lauchli, 1972).
It is worth noting that the Casparian strip may be absent in some places (Maathuis, 2014) allowing ions to reach the root stele and xylem vessels through the apoplastic pathway via bypass flow (Kronzucker and Britto, 2011). Since this flow is relatively low, most of the ions that reach the root xylem vessels are probably taken up across the plasma membrane of a root peripheral cell (Tester and Leigh, 2001). Thus, their entry into the root symplasm would have been mediated by membrane transport systems, channels, transporters or cotransporters. It should be noted that the bypass flow was observed in rice at Na+ external concentrations as low as 25 mM (Yeo et al., 1987) and it may contribute to salt stressing effects under high salinity by having an effect in shoot Na+ content (Yeo et al., 1987; Faiyue et al., 2012; Maathuis, 2014). Na+ bypass flow has been described in other species, besides rice, such as mangroves (Krishnamurthy et al., 2014), maize, and broad bean (Peterson et al., 1981), but not in Arabidopsis (Essah et al., 2003).
More than 60 years ago, through the application of the concept of enzyme kinetics for the study of root K+ absorption (Epstein and Hagen, 1952), Epstein et al. (1963) suggested that at least two transport systems were involved in root K+ uptake: a high-affinity system that operates at low external concentrations and a low-affinity system at higher concentrations. A similar scheme was also described for Na+ uptake (Rains and Epstein, 1967a). This biphasic behavior has since been observed in many plant species, with some exceptions. Maize, for example, shows a linear, non-saturating response to K+ in the low-affinity range (Kochian and Lucas, 1982). More recently, it has been shown that this linear response is dominated by the apoplastic movement of K+, and the “true” transmembrane flux saturates at modest rates (Coskun et al., 2016). In the high-affinity range of concentrations, K+ uptake is an active process that takes place against the K+ electrochemical potential, most likely by a K+/H+ symport, whilst the low-affinity uptake can take place by passive transport through inwardly rectifying K+ channels (Maathuis and Sanders, 1996a; Maathuis et al., 1997; Rodríguez-Navarro, 2000). It should also be noted that the limits in K+ concentrations for the operation of a symporter or a channel depend on the plasma membrane potential and the cytoplasmic pH and K+ concentrations. Thus, assuming, for example, that a cytoplasmic K+ concentration of 100 mM and a membrane potential of -240 mV exists, K+ uptake could take place through a channel from an external K+ concentration as low as 10 μM, which falls within the high-affinity system described by Epstein et al. (1963), Hirsch et al. (1998) and Spalding et al. (1999). High-affinity K+ uptake becomes apparent when K+ tissue concentrations decrease due to K+ starvation (Glass, 1976; Kochian and Lucas, 1982; Siddiqi and Glass, 1986; Martínez-Cordero et al., 2005). Providing NH4+ to the nutrient solution used to grow the plants, has a large influence on the NH4+ sensitivity of high-affinity K+ uptake. In some species such as barley (Santa-María et al., 2000), pepper (Martínez-Cordero et al., 2005), or Arabidopsis (Rubio et al., 2008), the presence of NH4+ in the growth solution induced an NH4+-insensitive high-affinity K+ uptake component. In others, such as tomato (Nieves-Cordones et al., 2007), high-affinity K+ uptake was dominated by an NH4+-sensitive component, irrespectively of the presence or the absence of NH4+ in the growth solution. By contrast, both Na+ (Martínez-Cordero et al., 2005; Kronzucker et al., 2006, 2008; Nieves-Cordones et al., 2007, 2010; Alemán et al., 2009; Cheng et al., 2015) and Cs+ (Rubio et al., 2000; White and Broadley, 2000; Qi et al., 2008) usually inhibit high-affinity K+ uptake.
Regarding low-affinity K+ transport, the role of TEA+, Cs+, and Ba2+, in blocking animal and plant K+ channels is well known, as these inhibit low-affinity K+ uptake, supporting the idea of the channel-mediated nature of this transport (Ketchum and Poole, 1991; Blatt, 1992; Hille, 1992; Very and Sentenac, 2002; Hoopen et al., 2010; Coskun et al., 2013). Unlike high-affinity K+ uptake, low-affinity K+ uptake is not down-regulated at high external K+ (Maathuis and Sanders, 1996b; Szczerba et al., 2006) and it is NH4+-insensitive (Spalding et al., 1999; Santa-María et al., 2000; Kronzucker et al., 2003; Szczerba et al., 2006). Na+ suppresses K+ uptake both in the low- and the high-affinity ranges, with low-affinity K+ uptake being more sensitive to this inhibition (Epstein et al., 1963; Rains and Epstein, 1967a; Kronzucker et al., 2006, 2008). It is worth noting that K+ uptake seems to be insensitive to Ca2+ in barley and Arabidopsis (Cramer et al., 1989; Caballero et al., 2012; Coskun et al., 2013).
As for Na+ uptake, it has been shown that root high-affinity Na+ uptake is significant when plants are starved of K+ (Garciadeblas et al., 2003; Haro et al., 2010). Under these conditions, Na+ is able to partially replace K+ and support plant growth (Maathuis and Sanders, 1993; Garciadeblas et al., 2003; Horie et al., 2007; Wakeel et al., 2011; Wakeel, 2013). High-affinity Na+ uptake has been shown to be sensitive to K+ and Ca2+ in barley (Rains and Epstein, 1967b) and to K+ and Ba2+ in rice (Garciadeblas et al., 2003). By contrast, low-affinity Na+ uptake seems to be insensitive to Ca2+ and K+ in rice (Malagoli et al., 2008) while it is sensitive to Ca2+ in Arabidopsis, barley, and wheat (Cramer et al., 1987, 1989; Essah et al., 2003; D’Onofrio et al., 2005). Na+ uptake can be reduced by the application of K+, and such reduction can alleviate salt stress effects to some extent (Wakeel, 2013). For example, a reduction in tissue Na+ content due to increased K+ application was observed in strawberry or in Jatropha curcas (Khayyat et al., 2009; Rodrigues et al., 2012).
Identification of K+ Transport Systems in Plants
The molecular approaches developed in the last 25 years have led to the characterization of many K+ and Na+ transport systems in plants. The first K+ uptake system identified in plants, AKT1, was isolated by complementing a K+-uptake deficient yeast strain with an Arabidopsis cDNA library (Sentenac et al., 1992). Sequence analysis and heterologous expression in Sf9-insect cells showed that the AKT1 cDNA encoded an inward-rectifier K+ channel belonging to the Shaker family (Gaymard et al., 1996). The gene encoding this channel showed specific constitutive expression in epidermal root cells (Lagarde et al., 1996) and AKT1 was proposed as the system mediating low-affinity K+ uptake in the roots.
Later, a PCR-based approach led to the identification of a cDNA from barley, HvHAK1, that mediated high-affinity K+ uptake in yeast (Santa-María et al., 1997). Specific expression of its mRNA in K+-starved roots and its kinetic properties in yeast prompted researchers to propose it as the system mediating the high-affinity K+ uptake observed in barley roots (Epstein et al., 1963). Subsequent studies led to the identification of an Arabidopsis homolog of HvHAK1, that was named AtHAK5 (Rubio et al., 2000).
In addition to AKT1 and AtHAK5-like transporters, other K+ uptake systems could be involved in root K+ uptake and in trans-membrane K+ movements in other plant organs. Moreover, the sequence of whole genomes of plants evidenced the existence of large gene families encoding putative K+ transport systems (Grabov, 2007; Véry et al., 2014; Nieves-Cordones et al., 2016).
Initial Characterizations in Arabidopsis
The studies on heterologous systems and on gene expression patterns for AKT1 and AtHAK5 produced data suggesting that these two systems played important roles in K+ acquisition by the root. However, a demonstration for the proposed roles was only possible when Arabidopsis knock-out mutants for these two genes became available (Spalding et al., 1999; Gierth et al., 2005; Rubio et al., 2008, 2010; Pyo et al., 2010). The studies showed that while Arabidopsis wild-type (WT) and akt1 plants could deplete external K+ (Rb+) to values around 1 μM, the athak5 plants did not diminish its concentration below 30 μM. In agreement with this, reduced growth was observed in athak5 plants grown in the presence of 1 μM K+ (Qi et al., 2008) or 10 μM K+ (Pyo et al., 2010; Ragel et al., 2015). Moreover, K+ (Rb+) uptake in athak5 plants was completely inhibited by the presence of Ba2+ in the external solution. By contrast, a complete inhibition of K+ (Rb+) depletion was observed in the presence of NH4+ in the akt1 line. An athak5 akt1 double mutant did not show K+ (Rb+) uptake at external concentrations below 50 μM, and it could only promote K+ (Rb+) uptake at concentrations higher than 100 μM. All these results demonstrated that in Arabidopsis plants, AtHAK5 was the only system mediating K+ uptake at concentrations below 10 μM and that this system was inhibited by NH4+. At concentrations between 10 and 200 μM, both AtHAK5 and AKT1 contributed to K+ uptake, defining AKT1 as a Ba2+-sensitive component of K+ uptake. Above 500 μM AtHAK5 contribution was negligible as the AtHAK5 gene was repressed at this external K+ concentration, and AKT1 role became more relevant. Unidentified systems could compensate for the lack of AKT1 since the akt1 and the athak5 akt1 lines grew at similar rates than the WT line when the external K+ concentration was sufficiently high (∼10 mM).
The studies described for Arabidopsis allowed for depicting a model for the contribution of AtHAK5 and AKT1 to root K+ uptake (Alemán et al., 2011). They also allowed for extending the studies to species where knock-out mutants were not available, through the use of NH4+ and Ba2+ as specific inhibitors of HAK5 and AKT1, respectively. Thus, it was shown that in tomato and pepper plants grown in the absence of NH4+, an NH4+-sensitive component, probably mediated by SlHAK5 (formerly LeHAK5) and CaHAK1, respectively, dominated K+ uptake from concentrations corresponding to the high-affinity component (inhibition of K+ uptake by NH4+ was close to 80% in tomato, for example; Martínez-Cordero et al., 2005; Nieves-Cordones et al., 2007). These results contrast with those obtained for Arabidopsis, where both AtHAK5 and AKT1 contribute to K+ uptake within the range of the K+ concentration assigned to the high-affinity component, as demonstrated by the use of single (Rubio et al., 2008), and double KO mutants (Rubio et al., 2010) in AtHAK5 and AKT1. Therefore, it can be concluded that the Arabidopsis model cannot be completely extended to other plant species, crops included, and highlights the need for the characterization of knock-out lines in each particular species to address the relevance of the different K+ transport systems. In addition, the availability of whole genomes for many plant species revealed some differences with respect to Arabidopsis. In Arabidopsis, AtHAK5 is the only member that belongs to the cluster Ia of HAK transporters, which are involved in high-affinity K+ uptake in roots (Nieves-Cordones et al., 2016). In tomato, a highly homologous gene to SlHAK5 is located just 2580 bp downstream from it in the tomato genome (Fernandez-Pozo et al., 2015). In rice, the OsHAK21 gene, also belonging to cluster Ia is induced by salinity, something that has not been described in other species for members of this cluster. This transporter has been linked to rice tolerance to salinity through the maintenance of Na+/K+ homeostasis, although the physiological mechanism remains unclear (Shen et al., 2015).
Rice Transport Systems Contributing to Root K+ Uptake
The recent characterization of T-DNA insertion rice lines knocked-out for K+ uptake systems such as OsHAK1, OsHAK5, and OsAKT1, has importantly contributed to increase our understanding of the relative contribution of such systems to K+ uptake in a species different from Arabidopsis, which is of great importance in agriculture.
A T-DNA insertion mutant with OsAKT1 knocked-out (Golldack et al., 2003) showed reduced growth and decreased root and shoot K+ concentrations when grown in the presence of 1 and 0.1 mM K+ (Li et al., 2014). K+ flux experiments and electrophysiological approaches showed an impairment of K+ uptake in the osakt1 line. Strong expression of OsAKT1 was detected in epidermal root cells, but it was also found in cortex, endodermis, and vascular bundles, suggesting a direct or indirect role in K+ translocation. In addition, slight expression was detected in shoots.
Knock-out mutants of the gene encoding the high-affinity K+ transporter OsHAK1 (Bañuelos et al., 2002) were also characterized. The studies with oshak1 lines showed that OsHAK1 contributed about 50–55% of high-affinity K+ uptake in the range of 0.05–0.1 mM external K+ and about 30% of K+ uptake at 1 mM external K+ (Chen et al., 2015). Root and shoot growth, cell size and internal K+ concentrations were reduced in the oshak1 mutant at both 0.1 and 1 mM K+ and this deficient phenotype could not be rescued at high external K+ (5 mM K+). Transcripts of OsHAK1 are preferentially accumulated in roots of K+-starved plants, as it is observed with AtHAK5 in Arabidopsis. In addition, OsHAK1 is strongly expressed at the xylem parenchyma and phloem of root vascular tissues, shoot meristems and vascular bundles of leaf sheaths. Moreover, oshak1 plants show reduced K+ translocation from root to shoot. In addition, the osakt1 and oshak1 lines were inhibited throughout development, showing delayed grain filling and reduced grain yield, suggesting that they may play an important role in rice productivity.
OsHAK5 was isolated and characterized as a high-affinity transport system in heterologous systems (Horie et al., 2011). Expression studies showed that OsHAK5 localized to the plasma membrane and that under normal K+ supply its transcripts were detected in root, root–shoot junction and leaf sheath. K+ starvation enhanced its expression in root epidermis, parenchyma of stele tissue, primordials of lateral roots, mesophyll, and parenchyma cells of the vascular bundle. The expression pattern of OsHAK5 supported its role in K+ uptake, but also in K+ distribution between roots and shoots, especially at low external K+. The lower accumulation of K+ in roots of overexpressing lines and the higher K+ accumulation in the knock-out lines, grown in low K+, supported this idea (Yang et al., 2014). These authors suggested that OsHAK5 may mediate K+ accumulation in xylem parenchyma cells to enable K+ channels to release K+ efficiently into the xylem sap. A role for OsHAK5 in K+ signaling is also proposed, as K+ in the phloem may act as a signal to convey the shoot demand of K+ and K+ xylem loading (Engels and Marschner, 1992), and OsHAK5 is abundantly expressed in phloem tissue. A role for the AKT2 channel in K+ signaling by modulating K+ in the phloem has been also proposed (Deeken et al., 2002) and AKT2 has been recently suggested as a pathway for Na+ entry into the roots (Salvador-Recatala, 2016). Since AKT2-mediated K+ transport is Ca2+-sensitive (Latz et al., 2007), interesting interactions between salt stress, Ca2+, and AKT2 may emerge.
The differential abundance of OsHAK1, OsHAK5, and OsAKT1 transcripts suggests that they play non-redundant functions. OsHAK1 and OsAKT1 are highly expressed in all cell types of roots and at low levels in shoots. However, while the OsHAK1 gene was induced 8- to 12-fold by K+ starvation (Chen et al., 2015), OsAKT1 expression was not affected by the external K+ concentrations (Li et al., 2014). By contrast, OsHAK5 was less expressed in roots and strongly in shoots (Yang et al., 2014). It has been proposed that both OsHAK1 and OsAKT1 contribute to K+ acquisition at low and high external concentrations. At low K+, OsHAK1 dominates high-affinity K+ uptake over OsAKT1 and OsHAK5. By contrast, OsHAK5 dominates K+ transport from root to shoots (Yang et al., 2014).
The studies described above suggest that OsHAK1, OsHAK5, and OsAKT1 are involved in K+ uptake at low and high concentrations as well as in K+ translocation from root to shoot. Contribution to K+ uptake over a wide range of K+ concentrations by a unique system has been demonstrated for the Arabidopsis AKT1 (Alemán et al., 2011). For transporters of the HAK family, the overexpression of AtKUP1 in Arabidopsis suspension cells produced enhanced K+ uptake at micromolar and millimolar K+ concentrations (Kim et al., 1998). The rice studies reviewed here suggest that OsHAK1 and OsHAK5 may contribute to K+ uptake at both low and high concentrations, which would explain why the growth of oshak1 and oshak5 lines is not rescued at high external K+. However, the idea of dual affinity for HAK transporters should be taken with caution. It is possible that, in addition to K+ uptake, the knock-out lines are affected in other processes. In fact, Yang et al. (2014) highlight the role of some HAK/KUP/KT transporters in auxin distribution, irrespective of K+ supply. At this stage, it cannot be ruled out that OsHAK1 and OsHAK5 play a part in this process.
Role of HAK Transporters in K+/Na+ Homeostasis under NaCl Stress
K+/Na+ homeostasis has been shown to be crucial for tolerance of plants to salinity (Maathuis and Amtmann, 1999). Maintaining K+ uptake rates at high external Na+ is crucial for K+/Na+ homeostasis and salt tolerance (Munns and Tester, 2008; Cuin et al., 2012; Cheng et al., 2015). However, in the presence of high Na+ concentrations, the low-K+ induction of genes encoding high-affinity K+ transporters is not observed (Nieves-Cordones et al., 2008, 2010). In addition, under salinity, K+ transport through high-affinity HAK transporters is competitively inhibited (Santa-María et al., 1997; Rubio et al., 2000). In fact, OsHAK1 has been defined as the Na+-sensitive high-affinity K+ pathway in rice (Chen et al., 2015). Nonetheless, studies in low-K+-grown Arabidopsis and rice plants showed that AtHAK5 and OsHAK1 function was still pivotal in maintaining K+ uptake and plant growth in the presence of high Na+ (Nieves-Cordones et al., 2010; Chen et al., 2015). In addition to OsHAK1, OsHAK5 may also play a role in salt tolerance, as high salt transiently enhances OsHAK5 expression (Yang et al., 2014) and the transporter mediates Na+-insensitive K+ uptake (Horie et al., 2011). In the presence of salinity, OsHAK5-overexpressing lines accumulated more K+ in shoots and showed enhanced growth as compared to WT. In contrast, oshak5 lines accumulated more Na+ in shoots and grew less than WT plants (Yang et al., 2014). The authors propose that the higher accumulation of Na+ in shoots in oshak5 may be due to a hyperpolarized membrane potential of mesophyll cells in knock-out mutants that would favor Na+ accumulation in shoots. Therefore, the K+ transport systems that contribute to maintaining a depolarized membrane potential of mesophyll cells to evade excessive Na+ accumulation under salinity may play a role in salt tolerance. The function of another rice HAK transporter, OsHAK21, seems to be important for salt tolerance (Shen et al., 2015). The gene encoding this transporter is enhanced by salinity and the protein is localized to the plasma membrane of xylem parenchyma and endodermal cells (putative passage cells). It has been shown that the knock-out oshak21 line is more salt sensitive because of a higher and a lower accumulation of Na+ and K+ respectively, which points to OsHAK21 as a key transporter needed for the maintenance of Na+/K+ homeostasis in rice under salt stress.
Other Systems That May Be Involved in Root K+ Uptake
Recently, a T-DNA insertion mutant in the Arabidopsis KUP7, which belongs to cluster V of the HAK family (Nieves-Cordones et al., 2016) has been characterized (Han et al., 2016). The results showed that the kup7 line was sensitive to low K+ (<100 μM), showing lower internal K+ concentrations. It could be rescued by higher K+ concentrations or by complementing the mutant with the WT gene. The KUP7 gene was ubiquitously expressed in many organs and the KUP7 protein was localized to the plasma membrane. KUP7 could complement a yeast strain deficient in K+ uptake. K+ transport studies showed that KUP7 was involved in root K+ uptake and K+ translocation to the shoot. It seemed to operate at higher concentrations than AtHAK5, and may be an alternative system involved in K+ uptake in the athak5 akt1 Arabidopsis line (Caballero et al., 2012). The observed effect on K+ translocation may be an indirect effect of a reduced uptake. However, Han et al. (2016) speculated that KUP7 may mediate K+ release into the xylem sap. It should be noted that besides K+ uptake, some HAK transporters have been shown to mediate K+ efflux (Bañuelos et al., 2002; Garciadeblas et al., 2002; Osakabe et al., 2013). Thus, K+ release into the xylem sap could take place through this type of transporters, if the electrochemical potential for K+ allows for this movement. It is well known that K+ loading of the xylem is mainly mediated by SKOR channels and the possible specific contribution of HAK transporters to K+ loading is yet to be determined.
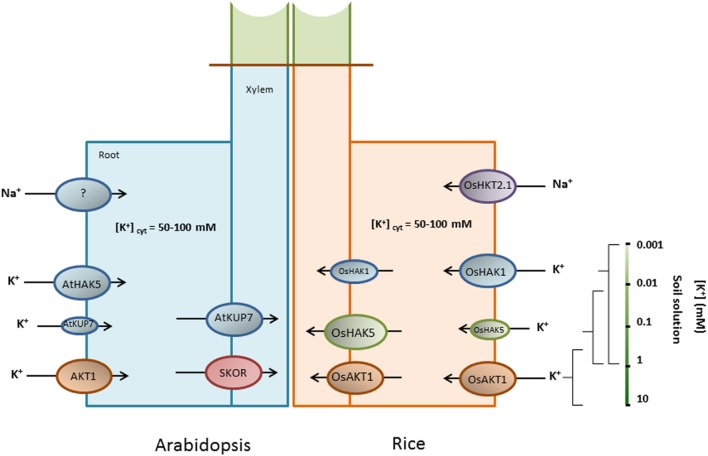
All of the above described results can be summarized into two different models for the K+ uptake systems in Arabidopsis and rice roots, shown in Figure 1 and Table 1.
FIGURE 1.
Schematic comparison of the systems involved in K+ and Na+ movements in Arabidopsis and rice roots. The availability of knock-out mutants in Arabidopsis and rice plants for specific transport systems has allowed for elucidating their roles in K+ transport. The figure shows the predicted function of each system for which knock-out mutants have been studied. AtHAK5 and AKT1 are the main systems for K+ uptake in Arabidopsis plants. In addition, a member of the KT/HAK/KUP family, AtKUP7, seems to also be involved in K+ uptake. K+ release into the xylem is mainly mediated by SKOR and also partially by AtKUP7. In rice, AtHAK5 and AKT1 functions are fulfilled by the rice homologs OsHAK1 and OsAKT1. An additional system, OsHAK5, partially contributes to high-affinity K+ uptake, but at higher concentrations than OsHAK1. These three rice systems may directly or indirectly facilitate K+ release into the xylem, with the contribution by OsHAK5 to K+ release into the xylem being more relevant. It is not clear if such contribution is a direct (by mediating K+ efflux into xylem vessels) or indirect (by favoring K+ accumulation in endodermal cells) outcome of the aforementioned transporters. Regarding Na+ uptake, the genetic identity of Na+ uptake systems in Arabidopsis remains to be elucidated. GLRs, CNGCs or other non-selective cation channels could be involved. In rice, OsHKT2;1 has been shown to mediate Na+ uptake during K+ deficiency. In the presence of K+ or high external Na+ concentrations other unknown systems should take part in Na+ uptake.
Table 1.
Summary for K+ and Na+ uptake features observed in Arabidopsis and rice roots.
| Species | Cation | Type of uptake† | Km (μM) | Sensitivity | Transport systems involved | Reference | |
|---|---|---|---|---|---|---|---|
| Arabidopsis | K+ | High-affinity | 24∗ | (-) NH4+, Ba+2, Cs+ (=) Ca+2 | AtHAK5, AKT1 | Spalding et al., 1999; Gierth et al., 2005; Rubio et al., 2008; Coskun et al., 2013 | |
| Low-affinity | 3,991∗ | (-) Ba+2, Cs+, TEA, La+3, Na+ (=) NH4+, Ca+2 | AKT1 | Spalding et al., 1999; Gierth et al., 2005; Caballero et al., 2012 | |||
| Na+ | High-affinity | n.d. | n.d. | – | – | ||
| Low-affinity | Linear response | (-) Ca+2, Ba+2, cyclic-nucleotides (+) La+3, GABA (=) TEA, Cs+ | – | Maathuis and Sanders, 2001; Essah et al., 2003 | |||
| Rice | K+ | High-affinity | 11–18 | (-) NH4+ | OsHAK1, OsHAK5, OsAKT1 | Bañuelos et al., 2002; Li et al., 2014; Yang et al., 2014; Chen et al., 2015 | |
| Low-affinity | n.d. | n.d. | OsHAK1, OsAKT1 | Li et al., 2014; Chen et al., 2015 | |||
| Na+ | High-affinity | 60 or 477–655¶ | (-) K+, Ba+2 | OsHKT2;1 | Garciadeblas et al., 2003; Horie et al., 2007; Haro et al., 2010 | ||
| Low-affinity | n.d. | (=) K+, Ca2+ | OsHKT2;1 | Horie et al., 2007; Malagoli et al., 2008 | |||
†High-affinity uptake takes into account cation uptake observed at external concentrations <0.5 mM while the low-affinity one does so at >0.5 mM. ∗Obtained with Rb+ as an analog for K+. (+), (-), and (=) stand for activation, inhibition, or no effect on cation uptake, respectively. n.d., not determined. ¶ Only one component was identified in Horie et al. (2007) for external Na+ concentrations up to 5 mM.
Regulation of K+ Uptake
In general terms, the genes encoding HAK transporters that mediate high-affinity K+ uptake in roots are strongly induced by K+ deprivation, whereas the genes encoding AKT1 channels are not. Several elements in the signal transduction cascade that results in the activation of HAK transcription have been identified. One of the first events when a root faces K+ deprivation is the hyperpolarization of the cell’s plasma membrane (Amtmann and Blatt, 2009). A positive correlation has been found between the membrane potential and the expression levels of SlHAK5 and AtHAK5, independent of the root’s K+ concentration (Nieves-Cordones et al., 2008; Rubio et al., 2014). Thus, it has been proposed that the hyperpolarization of the membrane potential may be the first element in the low-K+ signal cascade. The mechanisms linking membrane potential and gene expression are unknown but, changes in cytoplasmic Ca2+ derived from the activity of hyperpolarization-activated Ca2+ channels, could provide a connecting mechanism (Véry and Davies, 2000). Increases in ethylene (Jung et al., 2009) and reactive oxygen species (ROS; Shin and Schachtman, 2004; Hernandez et al., 2012) are also involved, probably acting following the hyperpolarization of the membrane potential. Other hormones such as jasmonic acid (Armengaud et al., 2004, 2010; Takehisa et al., 2013; Schachtman, 2015) and cytokinins (Nam et al., 2012) may also play a role in K+ signaling. At the end of the cascade, transcription factors such as DDF2, JLO, bHLH121, TFII_A for Arabidopsis AtHAK5 (Kim et al., 2012; Hong et al., 2013), bind the gene’s promoter to activate its expression.
Interestingly, some environmental conditions such as the lack of N, P, or S, that hyperpolarize root cell membrane potential (Rubio et al., 2014) and elevate ROS levels (Shin et al., 2005), also activate the transcription of AtHAK5-type genes. However, under such conditions, no HAK-mediated uptake is observed, suggesting a post-transcriptional regulation for these HAK transporters that is elicited specifically by K+ starvation (Rubio et al., 2014). Recently, it has been shown that the Arabidopsis AtHAK5 transporter is activated by complexes that contain the CIPK23 kinase and CBL1, CBL8, CBL9, or CBL10 Ca2+ sensors. AtHAK5 phosphorylation by CIPK23 leads to increases in the maximal rate of transport (Vmax) and the affinity for K+ transport (Ragel et al., 2015). It can be assumed that a specific low-K+-induced Ca2+ signal is registered by the CBL Ca2+ sensor, that in turn promotes phosphorylation of the transporter by CIPK23.
As a voltage-dependent inward-rectifier K+ channel, AKT1 activity is regulated by the membrane potential (Gaymard et al., 1996; Xu et al., 2006). In addition, its activity is modulated by interaction with other proteins. AKT1 forms tetrameric channels by interacting with the AtKC1 subunit (Daram et al., 1997; Pilot et al., 2003; Duby et al., 2008; Geiger et al., 2009). Upon interaction with AtKC1 in root cells, the activation potential for AKT1-containing channels becomes more negative, in comparison with AKT1 homotetramers (Reintanz et al., 2002; Wang et al., 2010). Moreover, AtKC1 interacts, in turn, with the SNARE proteins SYP121 (Honsbein et al., 2009) and VAMP721 (Zhang et al., 2015). SYP121 was shown to activate K+ uptake through AKT1/AtKC1 channels while VAMP721 negatively regulated this process. Phosphorylation also plays a role in AKT1 regulation. AKT1 activity is enhanced at low K+ by the same regulators as AtHAK5, i.e., the CIPK23/CBL1/9 complex, supporting the putative role of Ca2+ as a secondary messenger involved in low-K+ signaling (Li et al., 2006; Xu et al., 2006). Inactivation of the channel is achieved by the AIP1 phosphatase (Luan, 2009). It is worth to note that CBL proteins can also modify AKT1 activity in absence of CIPKs, as it is the case of CBL10 which is a negative regulator of AKT1 (Ren et al., 2013). Recently it has been shown that CIPK23 and AtKC1 act synergistically and balance K+ uptake/leakage to modulate AKT1-mediated responses of Arabidopsis to low K+ (Wang et al., 2016). Finally, nitric oxide has recently been shown to lower AKT1 activity by modulating vitamin B6 biosynthesis, constituting a new mechanism for the regulation of K+ uptake (Xia et al., 2014).
The current model for the regulation of these two main systems involved in K+ uptake, i.e., AtHAK5 and AKT1, is depicted in Figure 2.
FIGURE 2.
Main pathways for root K+ uptake and their regulatory mechanisms. HAK5-type transporters are high-affinity K+ transporters involved in K+ uptake at very low concentrations. When the external K+ concentration increases, the inward-rectifier K+ channel AKT1 together with HAK5, contributes to K+ uptake. At K+ concentrations above 200 μM, HAK5 is not present and AKT1 is the main system for low-affinity K+ uptake. At very high concentrations, other unknown systems can secure K+ supply if AKT1 is not functional. The HAK5 and AKT1 uptake systems are subjected to finely tuned regulation. At low K+ concentrations, a hyperpolarization of the plasma membrane induces HAK5 transcription. The signal cascade of HAK5 regulation is dependent on ethylene and ROS production. In addition, low external K+ likely produces a specific cytoplasmic Ca2+ signal that is registered by the Ca2+ sensor CBL1, which induces CIPK23 recruitment to the plasma membrane resulting in the phosphorylation and subsequent activation of HAK5 and AKT1. The channel activity is downregulated through dephosphorylation by the AIP1 phosphatase, and interaction with CBL10 and vitamin B6. Other subunits such as KC1 and the SNARE protein SYP121 also cooperate in AKT1 regulation. It can be concluded that whereas HAK5 is subjected to transcriptional and post-transcriptional regulation, K+ uptake through AKT1 is mainly regulated post-transcriptionally. The CIPK23/CBL1 complex emerges as a key regulator of K+ nutrition.
It is worth mentioning that CIPK23/CBL is also involved in the regulation of NO3- uptake (Ho et al., 2009; Tsay et al., 2011; Léran et al., 2015), and that K+ starvation significantly reduced the NO3- concentrations in tomato plants (Rubio et al., 2014) and induced several genes for NO3- uptake (Armengaud et al., 2004). This indicates that a cross-regulation between K+ and NO3- nutrition exists and that the CIPK23/CBL complex may constitute one of the key elements for such regulation.
Uptake of Na+ and Cell Mechanisms Involved
As for the identities of genes involved in Na+ uptake from external solutions, the scenario is far less clear than that for K+. With respect to high-affinity Na+ uptake, despite this activity being widely observed in roots from several species (Garciadeblas et al., 2003; Haro et al., 2010), only a few transport systems belonging to the HKT and HAK transporter families have been shown to take up Na+ within this range of concentrations. High-affinity K+ transporters (HKT) are related to fungal and bacterial K+ transporters from the Trk/Ktr families (Corratgé-Faillie et al., 2010). In plants, however, HKT transporters display varying Na+/K+ permeabilities. Phylogenetic and functional analyses have led to the identification of two HKT subfamilies (Platten et al., 2006): subfamily I, present in both monocotyledonous and dicotyledonous species, and subfamily II, identified only in monocotyledonous species so far. Subfamily II HKT transporters are expected to be all K+-permeable and can operate as Na+/K+ symporters (Rubio et al., 1995; Jabnoune et al., 2009; Yao et al., 2010; Oomen et al., 2012) or K+-selective uniporters (Horie et al., 2011; Sassi et al., 2012) when heterologously expressed in yeast and/or Xenopus oocytes. Subfamily I HKT transporters are Na+-selective in Arabidopsis and rice and are mostly involved in Na+ recirculation through vascular tissues (Maathuis, 2014; Véry et al., 2014), thus falling beyond the scope of the present review. In the rice cultivar Nipponbare, OsHKT2;1 provides a major pathway for root high-affinity Na+ uptake that supports plant growth under limiting K+ supply (Garciadeblas et al., 2003; Horie et al., 2007; Figure 1). Plants lacking a functional OsHKT2;1 gene have shown reduced growth and lower Na+ content when starved of K+ in the presence of 0.5 mM Na+, and under such conditions Na+ can partially compensate K+ demand (Horie et al., 2007). Besides Na+, OsHKT2;1 can also transport K+ when expressed in Xenopus oocytes (Jabnoune et al., 2009; Oomen et al., 2012), but K+ transport is not detected when it was expressed in yeast or in tobacco BY2 cells (Horie et al., 2001; Yao et al., 2010). In OsHKT2;1 expressing oocytes, the shifts in reversal potentials induced by K+ depended on the pre-treatment of oocytes. When the oocytes were pre-treated in low-Na+ (0.5 mM Na+) they showed smaller shifts that when pretreated with high-Na+ (96 mM Na+; Yao et al., 2010). Comparisons of Rb+ influx between WT and oshkt2;1 roots did not reveal significant differences between these genotypes (Horie et al., 2007). Thus, the possible involvement of OsHKT2;1 in root K+ uptake remains to be verified. Another subfamily II HKT transporter, OsHKT2;2, is absent in the Nipponbare (japonica) cultivar but present in the indica cultivar Pokkali. The transporter obtained from the latter cultivar is permeable to both Na+ and K+ at low external concentrations when expressed in tobacco BY2 cells, yeast and Xenopus oocytes (Horie et al., 2001; Yao et al., 2010; Oomen et al., 2012). It is important to note that a natural chimera OsHKT2;2/1 present in the Nona Bokra (indica) cultivar maintains high-affinity K+ uptake even at high Na+ concentrations, something that is also observed for Pokkali OsHKT2;2 but not for Nipponbare OsHKT2;1 (Oomen et al., 2012). Despite the lack of data concerning oshkt2;2 knock-out mutants, it is tempting to speculate that OsHKT2;2 contributes to both K+ and Na+ high-affinity uptake in rice roots. As for HAK transporters, two members, none of them from higher plants, have been shown to mediate high-affinity Na+ uptake: PpHAK13 from the moss Physcomitrella patens and YlHAK1 from the yeast Yarrowia lipolytica (Benito et al., 2012). PpHAK13, which belongs to cluster IV (Nieves-Cordones et al., 2016), transports Na+, but not K+, and high-affinity Na+ uptake is abolished in pphak13 mutants plants which evidences that PpHAK13 forms the major pathway for Na+ entry at low external concentrations in P. patens plants. On the other hand, YlHAK1 is able to transport Na+ and K+ when expressed in yeast, but the latter cation is only transported when Na+ is not added to the experimental solution. High-affinity Na+ transporters from the HAK family in higher plants are still to be identified.
Concerning low-affinity Na+ uptake, it is generally accepted that Na+ can enter the plant through ion channels (Maathuis, 2014). Na+-permeable channels include glutamate-like receptors (GLRs; Davenport, 2002) and cyclic nucleotide gated channels (CNGCs; Assmann, 1995; Bolwell, 1995; Trewavas, 1997; Newton and Smith, 2004) and possibly other, non-identified, non-selective cation channels (NSCCs; Maathuis and Sanders, 1993; Tyerman et al., 1997; Demidchik and Tester, 2002; Essah et al., 2003). Voltage-independent NSCCs (VI-NSCC) may constitute the main class of NSCCs involved in Na+ entry since they are highly sensitive to Ca2+ as observed for Na+ uptake in roots (Demidchik and Maathuis, 2007). Moreover, VI-NSCC blockers such as quinine or lanthanides also inhibited root Na+ influx (Essah et al., 2003; Wang et al., 2006). Despite these encouraging observations linking VI-NSCCs and root Na+ uptake, the molecular identity of these channels remains obscure at present. Several HAK and HKT transporters, which are expressed in roots, are also permeable at millimolar Na+ concentrations (Santa-María et al., 1997; Horie et al., 2001; Takahashi et al., 2007; Mian et al., 2011; Oomen et al., 2012). It is worth to note that OsHKT2;1 and HvHKT2;1 contribute to Na+ uptake in the millimolar range but they are downregulated in the presence of salt stress or K+. Thus, when taking into account other experimental conditions and plant species, it remains unclear which other type/family of transport systems constitute the major pathway for low-affinity Na+ uptake. It is likely that there is a large redundancy between the aforementioned channels and transporters. Insights into the identification of the contributing transport proteins would be of extraordinary biotechnological value since low-affinity Na+ uptake allows for the massive entry of Na+ within the plant that gives rise to toxicity. Interestingly, the secondary messengers cyclic AMP and GMP affect Na+ influx. Studies on Arabidopsis seedlings (Maathuis and Sanders, 2001; Essah et al., 2003) and on pepper plants (Rubio et al., 2003) have shown that unidirectional Na+ influx is reduced by cGMP addition. CNGCs have a cyclic-nucleotide binding domain and their activity is modulated by cyclic-nucleotides (Gao et al., 2014, 2016). It can be expected that the cyclic-nucleotide regulation of Na+ fluxes occurs through direct cGMP (or cAMP) binding to this domain as it is the case of animal CNGCs (Craven and Zagotta, 2006).
Recently, it has been shown that salt stress triggers the formation of endocytic vesicles via a clathrin-independent mechanism (Baral et al., 2015). Such vesicles lead to the formation of vacuole-like structures that may help plants to better cope with salt stress. Endocytosis can modify the transporter complement of the plasma membrane (Sutter et al., 2007), thus affecting the Na+ uptake pathways. But more interestingly, the endocytic process involves bulk-flow entry into root cells that may transport Na+ ions from the apoplast to the vacuole-like structures. If this were true, it would constitute a parallel pathway for Na+ uptake, independent from that mediated by transmembrane proteins. Moreover, this direct Na+ transport into vacuoles would prevent Na+ accumulation in the cytosol which leads to cell toxicity. Interestingly, vesicle trafficking has been recently suggested to play a role in plant adaptation to salt stress (Garcia de la Garma et al., 2015).
Concluding Remarks
K+ is an essential macronutrient for plants while Na+ may be beneficial or detrimental at low or high concentrations, respectively. Plant roots possess specific K+ transport systems that can function under a wide range of concentrations to secure K+ ions. The studies with knock-out mutants of the model plant Arabidopsis have led to the identification of two major pathways for K+ uptake: the high-affinity K+ transporter AtHAK5 and the inward-rectifying K+ channel AKT1. These systems operate at low (micromolar) and high (millimolar) external K+ concentrations, although an overlap in their operation is observed in the 10–200 μM K+ range. Different mechanisms that include transcriptional and post-transcriptional regulation modulate the activity of these two systems in response to K+ supply. Importantly, the CIPK23/CBL1 complex activates AtHAK5 as well as AKT1, pointing to its role as a central regulator of K+ nutrition. Homologs of AtHAK5 and AKT1 have been found in many plant species, and in some of them, paralog genes exist which suggest function redundancy. This precludes assigning a function by only using sequence homology or heterologous expression studies. The recent characterization of rice knock-out plants has shed light on this matter. OsHAK1, OsHAK5, and OsAKT1 seem to contribute to root K+ uptake as well as K+ release into xylem, and they probably play additional unknown functions in the shoot. However, they play non-redundant roles: (i) OsHAK1 is mainly involved in root K+ uptake at low concentrations, (ii) OsAKT1 mostly at high K+ concentrations, and (iii) OsHAK5 is more relevant for K+ translocation to the shoot. These systems may also play a role in salinity tolerance by maintaining the K+/Na+ homeostasis.
Regarding Na+ transport systems, the information is scarcer. High-affinity Na+ uptake and high-affinity Na+ transporters have been described in some species, but they are lacking in many others. The pathways for low-affinity Na+ uptake are not clearly identified yet, and several families of transporters contain members that could be good candidates. Recently, it has been described that salt stress induced a new endocytic pathway that is clathrin-independent, non-discriminatory in its choice of cargo, and that operates across all layers of the root. This new pathway may contribute to the bulk Na+ uptake and distribution across the root cells under saline stress conditions.
All of the above highlight the importance of characterizing the function of each transporter in each particular species. The studies with knock-out lines in Arabidopsis and rice evidence that the conclusions drawn in model species cannot be always fully extended to other, non-model species.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was funded by the Ministerio de Economía y Competitividad, Spain (grant number AGL2015-66434-R).
References
- Alemán F., Nieves-Cordones M., Martínez V., Rubio F. (2009). Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana. Environ. Exp. Bot. 65 263–269. 10.1016/j.envexpbot.2008.09.011 [DOI] [Google Scholar]
- Alemán F., Nieves-Cordones M., Martínez V., Rubio F. (2011). Root K+ acquisition in plants: the Arabidopsis thaliana model. Plant Cell Physiol. 52 1603–1612. 10.1093/pcp/pcr096 [DOI] [PubMed] [Google Scholar]
- Amtmann A., Blatt M. R. (2009). Regulation of macronutrient transport. New Phytol. 181 35–52. 10.1111/j.1469-8137.2008.02666.x [DOI] [PubMed] [Google Scholar]
- Armengaud P., Breitling R., Amtmann A. (2004). The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136 2556–2576. 10.1104/pp.104.046482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P., Breitling R., Amtmann A. (2010). Coronatine-insensitive 1 (COI1) mediates transcriptional responses of Arabidopsis thaliana to external potassium supply. Mol. Plant 3 390–405. 10.1093/mp/ssq012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley M. K., Grant M., Grabov A. (2006). Plant responses to potassium deficiencies: a role for potassium transport proteins. J. Exp. Bot. 57 425–436. 10.1093/jxb/erj034 [DOI] [PubMed] [Google Scholar]
- Assmann S. M. (1995). Cyclic AMP as a second messenger in higher plants (status and future prospects). Plant Physiol. 108 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin J. P., Nye P. H., Tinker P. B. (1973). Uptake of solutes by multiple root systems from soil. 3. Model for calculating solute uptake by a randomly dispersed root system developing in a finite volume of soil. Plant Soil 38 621–635. 10.1007/BF00010701 [DOI] [Google Scholar]
- Bañuelos M. A., Garciadeblas B., Cubero B., Rodríguez-Navarro A. (2002). Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 130 784–795. 10.1104/pp.007781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral A., Irani N. G., Fujimoto M., Nakano A., Mayor S., Mathew M. K. (2015). Salt-induced remodeling of spatially restricted clathrin-independent endocytic pathways in Arabidopsis root. Plant Cell 27 1297–1315. 10.1105/tpc.15.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M., Geldner N. (2014). Radial transport of nutrients: the plant root as a polarized epithelium. Plant Physiol. 166 528–537. 10.1104/pp.114.246124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough P. B. (1989). Root-growth, macro-nutrient uptake dynamics and soil fertility requirements of a high-yielding winter oilseed rape crop. Plant Soil 119 59–70. 10.1007/BF02370269 [DOI] [Google Scholar]
- Battie-Laclau P., Laclau J.-P., Piccolo M. D. C., Arenque B. C., Beri C., Mietton L., et al. (2013). Influence of potassium and sodium nutrition on leaf area components in Eucalyptus grandis trees. Plant Soil 371 19–35. 10.1007/s11104-013-1663-7 [DOI] [Google Scholar]
- Bednarz C. W., Oosterhuis D. M. (1999). Physiological changes associated with potassium deficiency in cotton. J. Plant Nutr. 22 303–313. 10.1016/j.jphotobiol.2012.02.002 [DOI] [Google Scholar]
- Benito B., Garciadeblas B., Rodriguez-Navarro A. (2012). HAK transporters from Physcomitrella patens and Yarrowia lipolytica mediate sodium uptake. Plant Cell Physiol. 53 1117–1123. 10.1093/pcp/pcs056 [DOI] [PubMed] [Google Scholar]
- Blatt M. R. (1992). K+ channels of stomatal guard cells. Characteristics of the inward rectifier and its control by pH. J. Gen. Physiol. 99 615–644. 10.1085/jgp.99.4.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell G. P. (1995). Cyclic AMP, the reluctant messenger in plants. Trends Biochem. Sci. 20 492–495. 10.1016/S0968-0004(00)89114-8 [DOI] [PubMed] [Google Scholar]
- Burch-Smith T. M., Zambryski P. C. (2012). Plasmodesmata paradigm shift: regulation from without versus within. Annu. Rev. Plant Biol. 63 239–260. 10.1146/annurev-arplant-042811-105453 [DOI] [PubMed] [Google Scholar]
- Caballero F., Botella M. A., Rubio L., Fernández J. A., Martínez V., Rubio F. (2012). A Ca2+-sensitive system mediates low-affinity K+ uptake in the absence of AKT1 in Arabidopsis plants. Plant Cell Physiol. 53 2047–2059. 10.1093/pcp/pcs140 [DOI] [PubMed] [Google Scholar]
- Chen G., Hu Q., Luo L., Yang T., Zhang S., Hu Y., et al. (2015). Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 38 2747–2765. 10.1111/pce.12585 [DOI] [PubMed] [Google Scholar]
- Cheng D., Wu G., Zheng Y. (2015). Positive correlation between potassium uptake and salt tolerance in wheat. Photosynthetica 53 447–454. 10.1007/s11099-015-0124-3 [DOI] [Google Scholar]
- Claassen N., Jungk A. (1982). Potassium dynamics at the soil-root interface in relation to the uptake of potassium by maize plants. Z. Pflanzenernahr. Bodenkd. 145 513–525. 10.1002/jpln.19821450603 [DOI] [Google Scholar]
- Corratgé-Faillie C., Jabnoune M., Zimmermann S., Véry A. A., Fizames C., Sentenac H. (2010). Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cell. Mol. Life Sci. 67 2511–2532. 10.1007/s00018-010-0317-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun D., Britto D. T., Kochian L. V., Kronzucker H. J. (2016). How high do ion fluxes go? A re-evaluation of the two-mechanism model of K(+) transport in plant roots. Plant Sci. 243 96–104. 10.1016/j.plantsci.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Coskun D., Britto D. T., Li M., Oh S., Kronzucker H. J. (2013). Capacity and plasticity of potassium channels and high-affinity transporters in roots of barley and Arabidopsis. Plant Physiol. 162 496–511. 10.1104/pp.113.215913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer G., Epstein E., Lauchli A. (1989). Na-Ca interactions in barley seedlings - relationship to ion-transport and growth. Plant Cell Environ. 12 551–558. 10.1111/j.1365-3040.1989.tb02128.x [DOI] [Google Scholar]
- Cramer G. R., Lauchli A., Polito V. S. (1985). Displacement of Ca2+ by Na+ from the plasmalemma of root-cells - A primary response to salt stress. Plant Physiol. 79 207–211. 10.1104/pp.79.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer G. R., Lynch J., Lauchli A., Epstein E. (1987). Influx of Na+, K+ and Ca2+ into roots of alt-stressed cotton seedlings. Plant Physiol. 79 207–211. 10.1104/pp.79.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven K. B., Zagotta W. N. (2006). CNG and HCN channels: two peas, one pod. Annu. Rev. Physiol. 68 375–401. 10.1146/annurev.physiol.68.040104.134728 [DOI] [PubMed] [Google Scholar]
- Cuin T. A., Zhou M., Parsons D., Shabala S. (2012). Genetic behaviour of physiological traits conferring cytosolic K+/Na+ homeostasis in wheat. Plant Biol. (Stuttg) 14 438–446. 10.1111/j.1438-8677.2011.00526.x [DOI] [PubMed] [Google Scholar]
- Daram P., Urbach S., Gaymard F., Sentenac H., Cherel I. (1997). Tetramerization of the AKT1 plant potassium channel involves its C- terminal cytoplasmic domain. EMBO J. 16 3455–3463. 10.1093/emboj/16.12.3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport R. (2002). Glutamate receptors in plants. Ann. Bot. 90 549–557. 10.1093/aob/mcf228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R., Geiger D., Fromm J., Koroleva O., Ache P., Langenfeld-Heyser R., et al. (2002). Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 216 334–344. 10.1007/s00425-002-0895-1 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Maathuis F. (2007). Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol. 175 387–404. 10.1111/j.1469-8137.2007.02128.x [DOI] [PubMed] [Google Scholar]
- Demidchik V., Tester M. (2002). Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol. 128 379–387. 10.1104/pp.010524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Onofrio C., Kader A., Lindberg S. (2005). Uptake of sodium in quince, sugar beet, and wheat protoplasts determined by the fluorescent sodium-binding dye benzofuran isophthalate. J. Plant Physiol. 162 421–428. 10.1016/j.jplph.2004.07.017 [DOI] [PubMed] [Google Scholar]
- Drew M. C. (1975). Comparison of effects of a localized supply of phosphate, nitrate, ammonium and potassium on growth of seminal root system, and shoot, in barley. New Phytol. 75 479–490. 10.1111/j.1469-8137.1975.tb01409.x [DOI] [Google Scholar]
- Duby G., Hosy E., Fizames C., Alcon C., Costa A., Sentenac H., et al. (2008). AtKC1, a conditionally targeted Shaker-type subunit, regulates the activity of plant K+ channels. Plant J. 53 115–123. 10.1111/j.1365-313X.2007.03324.x [DOI] [PubMed] [Google Scholar]
- Engels C., Marschner H. (1992). Adaptation of potassium translocation into the shoot of maize (Zea mays) to shoot demand: evidence for xylem loading as a regulating step. Physiol. Plant. 86 263–268. 10.1034/j.1399-3054.1992.860211.x [DOI] [Google Scholar]
- Epstein E., Hagen C. E. (1952). A kinetic study of the absorption of alkali cations by barley roots. Plant Pshyiol. 27 457–474. 10.1104/pp.27.3.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E., Rains D. W., Elzam O. E. (1963). Resolution of dual mechanisms of potassium absorption by barley roots. Proc. Natl. Acad. Sci. U.S.A. 49 684–692. 10.1073/pnas.49.5.684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essah P. A., Davenport R., Tester M. (2003). Sodium influx and accumulation in Arabidopsis. Plant Physiol. 133 307–318. 10.1104/pp.103.022178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eynard A., Lal R., Wiebe K. (2005). Crop response in salt-affected soils. J. Sustain. Agric. 27 5–50. 10.1007/BF00223914 [DOI] [Google Scholar]
- Faiyue B., Al-Azzawi M. J., Flowers T. J. (2012). A new screening technique for salinity resistance in rice (Oryza sativa L.) seedlings using bypass flow. Plant Cell Environ. 35 1099–1108. 10.1111/j.1365-3040.2011.02475.x [DOI] [PubMed] [Google Scholar]
- Fernandez-Pozo N., Menda N., Edwards J. D., Saha S., Tecle I. Y., Strickler S. R., et al. (2015). The sol genomics network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res. 43 D1036–D1041. 10.1093/nar/gku1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q. F., Fei C. F., Dong J. Y., Gu L. L., Wang Y. F. (2014). Arabidopsis CNGC18 is a Ca2+-permeable channel. Mol. Plant 7 739–743. 10.1093/mp/sst174 [DOI] [PubMed] [Google Scholar]
- Gao Q. F., Gu L. L., Wang H. Q., Fei C. F., Fang X., Hussain J., et al. (2016). Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113 3096–3101. 10.1073/pnas.1524629113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia de la Garma J., Fernandez-Garcia N., Bardisi E., Pallol B., Asensio-Rubio J. S., Bru R., et al. (2015). New insights into plant salt acclimation: the roles of vesicle trafficking and reactive oxygen species signalling in mitochondria and the endomembrane system. New Phytol. 205 216–239. 10.1111/nph.12997 [DOI] [PubMed] [Google Scholar]
- Garciadeblas B., Benito B., Rodriguez-Navarro A. (2002). Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa. Plant Mol. Biol. 50 623–633. 10.1023/A:1019951023362 [DOI] [PubMed] [Google Scholar]
- Garciadeblas B., Senn M. E., Bañuelos M. A., Rodriguez-Navarro A. (2003). Sodium transport and HKT transporters: the rice model. Plant J. 34 788–801. 10.1046/j.1365-313X.2003.01764.x [DOI] [PubMed] [Google Scholar]
- Gattward J. N., Almeida A.-A. F., Souza J. O., Jr., Gomes F. P., Kronzucker H. J. (2012). Sodium-potassium synergism in Theobroma cacao: stimulation of photosynthesis, water-use efficiency and mineral nutrition. Physiol. Plant. 146 350–362. 10.1111/j.1399-3054.2012.01621.x [DOI] [PubMed] [Google Scholar]
- Gaymard F., Cerutti M., Horeau C., Lemaillet G., Urbach S., Ravallec M., et al. (1996). The baculovirus/insect cell system as an alternative to xenopus oocytes. J. Biol. Chem. 271 22863–22870. 10.1074/jbc.271.37.22863 [DOI] [PubMed] [Google Scholar]
- Geiger D., Becker D., Vosloh D., Gambale F., Palme K., Rehers M., et al. (2009). Heteromeric AtKC1/AKT1 channels in Arabidopsis roots facilitate growth under K+ limiting conditions. J. Biol. Chem. 284 21288–21295. 10.1074/jbc.M109.017574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N. (2013). The endodermis. Annu. Rev. Plant Biol. 64 531–558. 10.1146/annurev-arplant-050312-120050 [DOI] [PubMed] [Google Scholar]
- Gierth M., Maser P., Schroeder J. I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137 1105–1114. 10.1104/pp.104.057216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass A. (1976). Regulation of potassium absorption in barley roots. An allosteric model. Plant Physiol. 58 33–37. 10.1104/pp.58.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Quigley F., Michalowski C. B., Kamasani U. R., Bohnert H. J. (2003). Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Mol. Biol. 51 71–81. 10.1023/A:1020763218045 [DOI] [PubMed] [Google Scholar]
- Grabov A. (2007). Plant KT/KUP/HAK potassium transporters: single family - multiple functions. Anna. Bot. 99 1035–1041. 10.1093/aob/mcm066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafsi C., Debez A., Abdelly C. (2014). Potassium deficiency in plants: effects and signaling cascades. Acta Physiol. Plant. 36 1055–1070. 10.1007/s00709-015-0845-y [DOI] [Google Scholar]
- Han M., Wu W., Wu W.-H., Wang Y. (2016). Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol. Plant 9 437–446. 10.1016/j.molp.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Haro R., Banuelos M. A., Rodriguez-Navarro A. (2010). High-affinity sodium uptake in land plants. Plant Cell Physiol. 51 68–79. 10.1093/pcp/pcp168 [DOI] [PubMed] [Google Scholar]
- Hernandez M., Fernandez-Garcia N., Garcia-Garma J., Rubio-Asensio J. S., Rubio F., Olmos E. (2012). Potassium starvation induces oxidative stress in Solanum lycopersicum L. roots. J. Plant Physiol. 169 1366–1374. 10.1016/j.jplph.2012.05.015 [DOI] [PubMed] [Google Scholar]
- Hille B. (1992). Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Hirsch R. E., Lewis B. D., Spalding E. P., Sussman M. R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280 918–921. 10.1126/science.280.5365.918 [DOI] [PubMed] [Google Scholar]
- Ho C. H., Lin S. H., Hu H. C., Tsay Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138 1184–1194. 10.1016/j.cell.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Hong J. P., Takeshi Y., Kondou Y., Schachtman D. P., Matsui M., Shin R. (2013). Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol. 54 1478–1490. 10.1093/pcp/pct094 [DOI] [PubMed] [Google Scholar]
- Honsbein A., Sokolovski S., Grefen C., Campanoni P., Pratelli R., Paneque M., et al. (2009). A tripartite snare-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21 2859–2877. 10.1105/tpc.109.066118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopen F. T., Cuin T. A., Pedas P., Hegelund J. N., Shabala S., Schjoerring J. K., et al. (2010). Competition between uptake of ammonium and potassium in barley and Arabidopsis roots: molecular mechanisms and physiological consequences. J. Exp. Bot. 61 2303–2315. 10.1093/jxb/erq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Costa A., Kim T. H., Han M. J., Horie R., Leung H. Y., et al. (2007). Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 26 3003–3014. 10.1038/sj.emboj.7601732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Sugawara M., Okada T., Taira K., Kaothien-Nakayama P., Katsuhara M., et al. (2011). Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 111 346–356. 10.1016/j.jbiosc.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Horie T., Yoshida K., Nakayama H., Yamada K., Oiki S., Shinmyo A. (2001). Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 27 129–138. 10.1046/j.1365-313x.2001.01077.x [DOI] [PubMed] [Google Scholar]
- Jabnoune M., Espeout S., Mieulet D., Fizames C., Verdeil J. L., Conejero G., et al. (2009). Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 150 1955–1971. 10.1104/pp.109.138008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. Y., Shin R., Schachtman D. P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21 607–621. 10.1105/tpc.108.063099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F., Armengaud P., Seditas T. J., Danku J., Salt D. E., Amtmann A. (2014). Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26 1480–1496. 10.1105/tpc.113.122101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum K. A., Poole R. J. (1991). Cytosolic calcium regulates a potassium current in corn (Zea mays) protoplasts. J. Membr. Biol. 119 277–288. 10.1007/BF01868732 [DOI] [PubMed] [Google Scholar]
- Khayyat M., Tafazoli E., Rajaee S., Vazifeshenas M., Mahmoo- Dabadi M. R., Sajjadinia A. (2009). Effects of NaCl and supple- mentary potassium on gas exchange, ionic content, and growth of salt-stressed strawberry plants. J. Plant Nutr. 32 907–918. 10.1080/01904160902870689 [DOI] [Google Scholar]
- Kim E. J., Kwak J. M., Uozumi N., Schroeder J. I. (1998). AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10 51–62. 10.2307/3870628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J., Ruzicka D., Shin R., Schachtman D. P. (2012). The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol. Plant 5 1042–1057. 10.1093/mp/sss003 [DOI] [PubMed] [Google Scholar]
- Kirkby E., Schubert S. (2013). Management of potassium in plant and soil systems in China. J. Plant Nutr. Soil Sci. 176 303–304. 10.1002/jpln.201390018 [DOI] [Google Scholar]
- Kochian L. V., Lucas W. J. (1982). Potassium transport in corn roots: I. Resolution of kinetics into a saturable and linear component. Plant Physiol. 70 1723–1731. 10.1104/pp.70.6.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy P., Jyothi-Prakash P. A., Qin L., He J., Lin Q., Loh C.-S., et al. (2014). Role of root hydrophobic barriers in salt exclusion of a mangrove plant Avicennia officinalis. Plant Cell Environ. 37 1656–1671. 10.1111/pce.12272 [DOI] [PubMed] [Google Scholar]
- Kronzucker H. J., Britto D. T. (2011). Sodium transport in plants: a critical review. New Phytol. 189 54–81. 10.1111/j.1469-8137.2010.03540.x [DOI] [PubMed] [Google Scholar]
- Kronzucker H. J., Coskun D., Schulze L. M., Wong J. R., Britto D. T. (2013). Sodium as nutrient and toxicant. Plant Soil 369 1–23. 10.1007/s11104-013-1801-2 [DOI] [Google Scholar]
- Kronzucker H. J., Szczerba M. W., Britto D. T. (2003). Cytosolic potassium homeostasis revisited: K-42-tracer analysis in Hordeum vulgare L. reveals set-point variations in [K+]. Planta 217 540–546. 10.1007/s00425-003-1032-5 [DOI] [PubMed] [Google Scholar]
- Kronzucker H. J., Szczerba M. W., Moazami-Goudarzi M., Britto D. T. (2006). The cytosolic Na+:K+ ratio does not explain salinity-induced growth impairment in barley: a dual-tracer study using 42K+ and 24Na+. Plant Cell Environ. 29 2228–2237. 10.1111/j.1365-3040.2006.01597.x [DOI] [PubMed] [Google Scholar]
- Kronzucker H. J., Szczerba M. W., Schulze L. M., Britto D. T. (2008). Non-reciprocal interactions between K+ and Na+ ions in barley (Hordeum vulgare L.). J. Exp. Bot. 59 2793–2801. 10.1093/jxb/ern139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde D., Basset M., Lepetit M., Conejero G., Gaymard F., Astruc S., et al. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9 195–203. 10.1046/j.1365-313X.1996.09020195.x [DOI] [PubMed] [Google Scholar]
- Latz A., Ivashikina N., Fischer S., Ache P., Sano T., Becker D., et al. (2007). In planta AKT2 subunits constitute a pH- and Ca2+-sensitive inward rectifying K+ channel. Planta 225 1179–1191. 10.1007/s00425-006-0428-4 [DOI] [PubMed] [Google Scholar]
- Lauchli A. (1972). Translocation of inorganic solutes. Ann. Rev. Plant Physiol. 23 197–218. 10.1146/annurev.pp.23.060172.001213 [DOI] [Google Scholar]
- Leigh R. A., Wyn Jones R. G. (1984). A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant-cell. New Phytol. 97 1–13. 10.1111/j.1469-8137.1984.tb04103.x [DOI] [Google Scholar]
- Léran S., Edel K. H., Pervent M., Hashimoto K., Corratgé-Faillie C., Offenborn J. N., et al. (2015). Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 8:ra43 10.1126/scisignal.aaa4829 [DOI] [PubMed] [Google Scholar]
- Li J., Long Y., Qi G.-N., Li J., Xu Z.-J., Wu W.-H., et al. (2014). The os-akt1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26 3387–3402. 10.1105/tpc.114.123455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kim B. G., Cheong Y. H., Pandey G. K., Luan S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K+ response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103 12625–12630. 10.1073/pnas.0605129103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14 37–42. 10.1016/j.tplants.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Maathuis F. J., Ichida A. M., Sanders D., Schroeder J. I. (1997). Roles of higher plant K+ channels. Plant Physiol. 114 1141–1149. 10.1104/pp.114.4.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis F. J., Sanders D. (2001). Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 127 1617–1625. 10.1104/pp.010502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis F. J. M. (2014). Sodium in plants: perception, signalling, and regulation of sodium fluxes. J. Exp. Bot. 65 849–858. 10.1093/jxb/ert326 [DOI] [PubMed] [Google Scholar]
- Maathuis F. J. M., Amtmann A. (1999). K+ Nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann. Bot. 84 123–133. 10.1006/anbo.1999.0912 [DOI] [Google Scholar]
- Maathuis F. J. M., Sanders D. (1993). Energization of potassium uptake in Arabidopsis thaliana. Planta 191 302–307. 10.1007/BF00195686 [DOI] [Google Scholar]
- Maathuis F. J. M., Sanders D. (1996a). Characterization of csi52, a Cs+ resistant mutant of Arabidopsis thaliana altered in K+ transport. Plant J. 10 579–589. 10.1046/j.1365-313X.1996.10040579.x [DOI] [PubMed] [Google Scholar]
- Maathuis F. J. M., Sanders D. (1996b). Mechanisms of potassium absorption by higher plant roots. Physiol. Plant. 96 158–168. 10.1111/j.1399-3054.1996.tb00197.x [DOI] [Google Scholar]
- Malagoli P., Britto D. T., Schulze L. M., Kronzucker H. J. (2008). Futile Na+ cycling at the root plasma membrane in rice (Oryza sativa L.): kinetics, energetics, and relationship to salinity tolerance. J. Exp. Bot. 59 4109–4117. 10.1093/jxb/ern249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P. (2012). Marschner’s Mineral Nutrition of Higher Plants 3rd Edn San Diego, CA: Academic Press. [Google Scholar]
- Martínez-Cordero M. A., Martinez V., Rubio F. (2005). High-affinity K+ uptake in pepper plants. J. Exp. Bot. 56 1553–1562. 10.1093/jxb/eri150 [DOI] [PubMed] [Google Scholar]
- Mengel K., Kirkby E. A., Kosegarten H., Appel T. (2001). Principles on Plant Nutrition. Dordrecht: Kluwer. [Google Scholar]
- Mian A., Oomen R. J., Isayenkov S., Sentenac H., Maathuis F. J. M., Very A.-A. (2011). Over-expression of an Na+- and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 68 468–479. 10.1111/j.1365-313X.2011.04701.x [DOI] [PubMed] [Google Scholar]
- Moody P. W., Bell M. J. (2006). Availability of soil potassium and diagnostic soil tests. Aust. J. Soil Res. 44 265–275. 10.1071/SR05154 [DOI] [Google Scholar]
- Morgan S. H., Maity P. J., Geilfus C. M., Lindberg S., Muehling K. H. (2014). Leaf ion homeostasis and plasma membrane H+-ATPase activity in Vicia faba change after extra calcium and potassium supply under salinity. Plant Physiol. Biochem. 82 244–253. 10.1016/j.plaphy.2014.06.010 [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Ann. Rev. Plant Biol. 59 651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Nam Y.-J., Tran L.-S. P., Kojima M., Sakakibara H., Nishiyama R., Shin R. (2012). Regulatory roles of cytokinins and cytokinin signaling in response to potassium deficiency in Arabidopsis. PLoS ONE 7:e47797 10.1371/journal.pone.0047797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R. P., Smith C. J. (2004). Cyclic nucleotides. Phytochemistry 65 2423–2437. 10.1016/j.phytochem.2004.07.026 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M., Aleman F., Martinez V., Rubio F. (2010). The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol. Plant 3 326–333. 10.1093/mp/ssp102 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M., Alemán F., Martínez V., Rubio F. (2014). K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 171 688–695. 10.1016/j.jplph.2013.09.021 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M., Martinez-Cordero M. A., Martinez V., Rubio F. (2007). An NH4+-sensitive component dominates high-affinity K+ uptake in tomato plants. Plant Sci. 172 273–280. 10.1016/j.plantsci.2006.09.003 [DOI] [Google Scholar]
- Nieves-Cordones M., Miller A., Alemán F., Martínez V., Rubio F. (2008). A putative role for the plasma membrane potential in the control of the expression of the gene encoding the tomato high-affinity potassium transporter HAK5. Plant Mol. Biol. 68 521–532. 10.1007/s11103-008-9388-3 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M., Rodenas R., Chavanieu A., Rivero R. M., Martinez V., Gaillard I., et al. (2016). Uneven HAK/KUP/KT protein diversity among angiosperms: species distribution and perspectives. Front. Plant Sci. 7:127 10.3389/fpls.2016.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen R. J., Benito B., Sentenac H., Rodriguez-Navarro A., Talon M., Very A.-A., et al. (2012). HKT2;2/1, a K+-permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism. Plant J. 71 750–762. 10.1111/j.1365-313X.2012.05031.x [DOI] [PubMed] [Google Scholar]
- Osakabe Y., Arinaga N., Umezawa T., Katsura S., Nagamachi K., Tanaka H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25 609–624. 10.1105/tpc.112.105700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. A., Emanuel M. E., Humphreys G. B. (1981). Pathway of movement of apoplastic fluorescent dye tracers through the endodermis at the site of secondary root-formation in corn (Zea mays) and broad bean (Vicia faba). Can. J. Bot. 59:618 10.1139/b81-087 [DOI] [Google Scholar]
- Pilot G., Gaymard F., Mouline K., Cherel I., Sentenac H. (2003). Regulated expression of Arabidopsis Shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 51 773–787. 10.1023/A:1022597102282 [DOI] [PubMed] [Google Scholar]
- Platten J. D., Cotsaftis O., Berthomieu P., Bohnert H., Davenport R. J., Fairbairn D. J., et al. (2006). Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 11 372–374. 10.1016/j.tplants.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Pyo Y. J., Gierth M., Schroeder J. I., Cho M. H. (2010). High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol. 153 863–875. 10.1104/pp.110.154369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Hampton C. R., Shin R., Barkla B. J., White P. J., Schachtman D. P. (2008). The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 59 595–607. 10.1093/jxb/erm330 [DOI] [PubMed] [Google Scholar]
- Ragel P., Rodenas R., Garcia-Martin E., Andres Z., Villalta I., Nieves-Cordones M., et al. (2015). The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 169 2863–2873. 10.1104/pp.15.01401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rains D. W., Epstein E. (1967a). Sodium absorption by barley roots: its mediation by mechanism 2 of alkali cation transport. Plant Physiol. 42 319–323. 10.1104/pp.42.3.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rains D. W., Epstein E. (1967b). Sodium absorption by barley roots: role of the dual mechanisms of alkali cation transport. Plant Physiol. 42 314–318. 10.1104/pp.42.3.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz B., Szyroki A., Ivashikina N., Ache P., Godde M., Becker D., et al. (2002). AtKC1, a silent Arabidopsis potassium channel alpha -subunit modulates root hair K+ influx. Proc. Natl. Acad. Sci. U.S.A. 99 4079–4084. 10.1073/pnas.052677799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X.-L., Qi G.-N., Feng H.-Q., Zhao S., Zhao S.-S., Wang Y., et al. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 74 258–266. 10.1111/tpj.12123 [DOI] [PubMed] [Google Scholar]
- Rodrigues C. R. F., Silveira J. A. G., Silva E. N., Dutra A. T. B., Viegas R. A. (2012). Potassium transport and partitioning alle- viates toxic effects of sodium on young physic nut plants. Rev. Bras. Cien. Solo 36 223–232. 10.1590/S0100-06832012000100023 [DOI] [Google Scholar]
- Rodríguez-Navarro A. (2000). Potassium transport in fungi and plants. Biochim. Biophys. Acta 1469 1–30. 10.1016/S0304-4157(99)00013-1 [DOI] [PubMed] [Google Scholar]
- Römheld V., Kirkby E. (2010). Research on potassium in agriculture: needs and prospects. Plant Soil 335 155–180. 10.1007/s11104-010-0520-1 [DOI] [Google Scholar]
- Rubio F., Alemán F., Nieves-Cordones M., Martínez V. (2010). Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5 AtAKT1 and unknown systems mediate K+ uptake. Physiol. Plant. 139 220–228. 10.1111/j.1399-3054.2010.01354.x [DOI] [PubMed] [Google Scholar]
- Rubio F., Flores P., Navarro J. M., Martinez V. (2003). Effects of Ca2+, K+ and cGMP on Na+ uptake in pepper plants. Plant Sci. 165 1043–1049. 10.1016/S0168-9452(03)00297-8 [DOI] [Google Scholar]
- Rubio F., Fon M., Rodenas R., Nieves-Cordones M., Aleman F., Rivero R. M., et al. (2014). A low K+ signal is required for functional high-affinity K+ uptake through HAK5 transporters. Physiol. Plant. 152 558–570. 10.1111/ppl.12205 [DOI] [PubMed] [Google Scholar]
- Rubio F., Gassmann W., Schroeder J. I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270 1660–1663. 10.1126/science.270.5242.1660 [DOI] [PubMed] [Google Scholar]
- Rubio F., Nieves-Cordones M., Alemán F., Martinez V. (2008). Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol. Plant. 134 598–608. 10.1111/j.1399-3054.2008.01168.x [DOI] [PubMed] [Google Scholar]
- Rubio F., Santa-Maria G. E., Rodríguez-Navarro A. (2000). Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol. Plant. 109 34–43. 10.1034/j.1399-3054.2000.100106.x [DOI] [Google Scholar]
- Salvador-Recatala V. (2016). The AKT2 potassium channel mediates NaCl induced depolarization in the root of Arabidopsis thaliana. Plant Signal. Behav. 11:e1165381 10.1080/15592324.2016.1165381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-María G. E., Danna C. H., Czibener C. (2000). High-affinity potassium transport in barley roots. Ammonium-sensitive and -insensitive pathways. Plant Physiol. 123 297–306. 10.1104/pp.123.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-María G. E., Rubio F., Dubcovsky J., Rodriguez-Navarro A. (1997). The hak1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9 2281–2289. 10.2307/3870585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi A., Mieulet D., Khan I., Moreau B., Gaillard I., Sentenac H., et al. (2012). The rice monovalent cation transporter OsHKT2;4: revisited ionic selectivity. Plant Physiol. 160 498–510. 10.1104/pp.112.194936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattelmacher B., Muhling K. H., Pennewiss K. (1998). The apoplast - its significance for the nutrition of higher plants. Z. Pflanzen. Boden. 161 485–498. 10.1002/jpln.1998.3581610502 [DOI] [Google Scholar]
- Schachtman D. P. (2015). The role of ethylene in plant response to K+ deficiency. Front. Plant Sci. 6:1153 10.3389/fpls.2015.01153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber L., Hartmann K., Skrabs M., Zeier J. (1999). Apoplastic barriers in roots: chemical composition of endodermal and hypodermal cell walls. J. Exp. Bot. 50 1267–1280. 10.1093/jexbot/50.337.1267 [DOI] [Google Scholar]
- Sentenac H., Bonneaud N., Minet M., Lacroute F., Salmon J. M., Gaymard F., et al. (1992). Cloning and expression in yeast of a plant potassium ion transport system. Science 256 663–665. 10.1126/science.1585180 [DOI] [PubMed] [Google Scholar]
- Shen Y., Shen L., Shen Z., Jing W., Ge H., Zhao J., et al. (2015). The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 38 2766–2779. 10.1111/pce.12586 [DOI] [PubMed] [Google Scholar]
- Shin R., Berg R. H., Schachtman D. P. (2005). Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 46 1350–1357. 10.1093/pcp/pci145 [DOI] [PubMed] [Google Scholar]
- Shin R., Schachtman D. P. (2004). Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. U.S.A. 101 8827–8832. 10.1073/pnas.0401707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi M. Y., Glass A. D. (1986). A model for the regulation of k influx, and tissue potassium concentrations by negative feedback effects upon plasmalemma influx. Plant Physiol. 81 1–7. 10.1104/pp.81.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding E. P., Hirsch R. E., Lewis D. R., Qi Z., Sussman M. R., Lewis B. D. (1999). Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J. Gen. Physiol. 113 909–918. 10.1085/jgp.113.6.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart W. M., Dibb D. W., Johnston A. E., Smyth T. J. (2005). The contribution of commercial fertilizer nutrients to food production. Agron. J. 97 1–6. 10.2134/agronj2005.0001 [DOI] [Google Scholar]
- Subbarao G. V., Ito O., Berry W. L., Wheeler R. M. (2003). Sodium - A functional plant nutrient. Crit. Rev. Plant Sci. 22 391–416. 10.1080/07352680390243495 [DOI] [Google Scholar]
- Sutter J.-U., Sieben C., Hartel A., Eisenach C., Thiel G., Blatt M. R. (2007). Abscisic acid triggers the endocytosis of the Arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr. Biol. 17 1396–1402. 10.1016/j.cub.2007.07.020 [DOI] [PubMed] [Google Scholar]
- Szczerba M. W., Britto D. T., Kronzucker H. J. (2006). Rapid, futile K+ cycling and pool-size dynamics define low-affinity potassium transport in barley. Plant Physiol. 141 1494–1507. 10.1104/pp.106.082701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L., Zeiger E. (1991). Plant Physiology, Chapters 8, 9, 10, 2nd Edn Sunderland, MA: Sinauer. [Google Scholar]
- Takahashi R., Liu S., Takano T. (2007). Cloning and functional comparison of a high-affinity K+ transporter gene PhaHKT1 of salt-tolerant and salt-sensitive reed plants. J. Exp. Bot. 58 4387–4395. 10.1093/jxb/erm306 [DOI] [PubMed] [Google Scholar]
- Takehisa H., Sato Y., Antonio B., Nagamura Y. (2013). Global transcriptome profile of rice root in response to essential macronutrient deficiency. Plant Signal. Behav. 8:e24409 10.4161/psb.24409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M., Leigh R. A. (2001). Partitioning of nutrient transport processes in roots. J. Exp. Bot. 52 445–457. 10.1093/jexbot/52.356.653 [DOI] [PubMed] [Google Scholar]
- Trewavas A. J. (1997). Plant cyclic AMP comes in from the cold. Nature 390 657–658. 10.1038/37720 [DOI] [PubMed] [Google Scholar]
- Tsay Y.-F., Ho C.-H., Chen H.-Y., Lin S.-H. (2011). Integration of nitrogen and potassium signaling. Ann. Rev. Plant Biol 62 207–226. 10.1146/annurev-arplant-042110-103837 [DOI] [PubMed] [Google Scholar]
- Tyerman S. D., Skerrett M., Garrill A., Findlay G. P., Leigh R. A. (1997). Pathways for the permeation of Na+ and Cl- into protoplasts derived from the cortex of wheat roots. J. Exp. Bot. 48 459–480. 10.1093/jxb/48.Special_Issue.459 [DOI] [PubMed] [Google Scholar]
- Véry A.-A., Davies J. M. (2000). Hyperpolarization-activated calcium channels at the tip of Arabidopsis root hairs. Proc. Natl. Acad. Sci. U.S.A. 97 9801–9806. 10.1073/pnas.160250397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry A.-A., Nieves-Cordones M., Daly M., Khan I., Fizames C., Sentenac H. (2014). Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171 748–769. 10.1016/j.jplph.2014.01.011 [DOI] [PubMed] [Google Scholar]
- Very A. A., Sentenac H. (2002). Cation channels in the Arabidopsis plasma membrane. Trends Plant Sci. 7 168–175. 10.1016/S1360-1385(02)02262-8 [DOI] [PubMed] [Google Scholar]
- Wakeel A. (2013). Potassium-sodium interactions in soil and plant under saline-sodic conditions. J. Plant Nutr. Soil Sci. 176 344–354. 10.1002/jpln.201200417 [DOI] [Google Scholar]
- Wakeel A., Farooq M., Qadir M., Schubert S. (2011). Potassium substitution by sodium in plants. Crit. Rev. Plant Sci. 30 401–413. 10.1080/07352689.2011.587728 [DOI] [Google Scholar]
- Wakeel A., Steffens D., Schubert S. (2010). Potassium substitution by sodium in sugar beet (Beta vulgaris) nutrition on K-fixing soils. J. Plant Nutr. Soil Sci. 173 127–134. 10.1002/jpln.200900270 [DOI] [Google Scholar]
- Walker D. J., Black C. R., Miller A. J. (1998). The role of cytosolic potassium and ph in the growth of barley roots. Plant Physiol. 118 957–964. 10.1104/pp.118.3.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. J., Leigh R. A., Miller A. J. (1996). Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. U.S.A. 93 10510–10514. 10.1073/pnas.93.19.10510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Davenport R. J., Volkov V., Amtmann A. (2006). Low unidirectional sodium influx into root cells restricts net sodium accumulation in Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana. J. Exp. Bot. 57 1161–1170. 10.1093/jxb/erj116 [DOI] [PubMed] [Google Scholar]
- Wang X. P., Chen L. M., Liu W. X., Shen L., Wang F. L., Zhou Y., et al. (2016). AtKC1 and CIPK23 synergistically modulate AKT1-mediated low potassium stress responses in Arabidopsis. Plant Physiol. 170 2264–2277. 10.1104/pp.15.01493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., He L., Li H. D., Xu J. A., Wu W. H. (2010). Potassium channel alpha-subunit AtKC1 negatively regulates AKT1-mediated K+ uptake in Arabidopsis roots under low-K+ stress. Cell Res. 20 826–837. 10.1038/cr.2010.74 [DOI] [PubMed] [Google Scholar]
- White P. J., Broadley M. R. (2000). Mechanisms of caesium uptake by plants. New Phytol. 147 241–256. 10.1046/j.1469-8137.2000.00704.x [DOI] [Google Scholar]
- Wyn Jones R. G., Pollard A. (1983). “Proteins, enzymes and inorganic ions,” in Inorganic Plant Nutrition-Encyclopedia of Plant Physiology New Series 15, eds Lauchli A., Bieleski R. L. (Berlin: Springer-Verlag; ), 528–562. [Google Scholar]
- Xia J., Kong D., Xue S., Tian W., Li N., Bao F., et al. (2014). Nitric oxide negatively regulates AKT1-mediated potassium uptake through modulating vitamin B6 homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 16196–16201. 10.1073/pnas.1417473111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li H. D., Chen L. Q., Wang Y., Liu L. L., He L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 1347–1360. 10.1016/j.cell.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Yang T., Zhang S., Hu Y., Wu F., Hu Q., Chen G., et al. (2014). The role of a potassium transporter oshak5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 166 945–959. 10.1104/pp.114.246520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Horie T., Xue S., Leung H. Y., Katsuhara M., Brodsky D. E., et al. (2010). Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol. 152 341–355. 10.1104/pp.109.145722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo A. R., Yeo M. E., Flowers T. J. (1987). The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J. Exp. Bot. 38 1141–1153. 10.1093/jxb/38.7.1141 [DOI] [Google Scholar]
- Zhang B., Karnik R., Wang Y., Wallmeroth N., Blatt M. R., Grefen C. (2015). The Arabidopsis R-SNARE VAMP721 Interacts with KAT1 and KC1 K+ channels to moderate k+ current at the plasma membrane. Plant Cell 27 1697–1717. 10.1105/tpc.15.00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zörb C., Senbayram M., Peiter E. (2014). Potassium in agriculture – Status and perspectives. J. Plant Physiol. 171 656–669. 10.1016/j.jplph.2013.08.008 [DOI] [PubMed] [Google Scholar]