Abstract
Objective: Streptococcus mitis is a predominant oral colonizer, but difficulties in genetic manipulation of this species have hampered our understanding of the mechanisms it uses for colonization of oral surfaces. The aim of this study was to reveal optimal conditions for natural genetic transformation in S. mitis and illustrate its application in direct genome editing.
Methods: Luciferase reporter assays were used to assess gene expression of the alternative sigma factor (σX) in combination with natural transformation experiments to evaluate the efficiency by which S. mitis activates the competence system and incorporates exogenous DNA. Optimal amounts and sources of donor DNA (chromosomal, amplicon, or replicative plasmid), concentrations of synthetic competence-stimulating peptide, and transformation media were assessed.
Results: A semi-defined medium showed much improved results for response to the competence stimulating peptide when compared to rich media. The use of a donor amplicon with large homology flanking regions also provided higher transformation rates. Overall, an increase of transformation efficiencies from 0.001% or less to over 30% was achieved with the developed protocol. We further describe the construction of a markerless mutant based on this high efficiency strategy.
Conclusion: We optimized competence development in S. mitis, by use of semi-defined medium and appropriate concentrations of synthetic competence factor. Combined with the use of a large amplicon of donor DNA, this method allowed easy and direct editing of the S. mitis genome, broadening the spectrum of possible downstream applications of natural transformation in this species.
Keywords: pheromone, competence, natural transformation, mutagenesis, oral cavity, CSP, streptococcus
Introduction
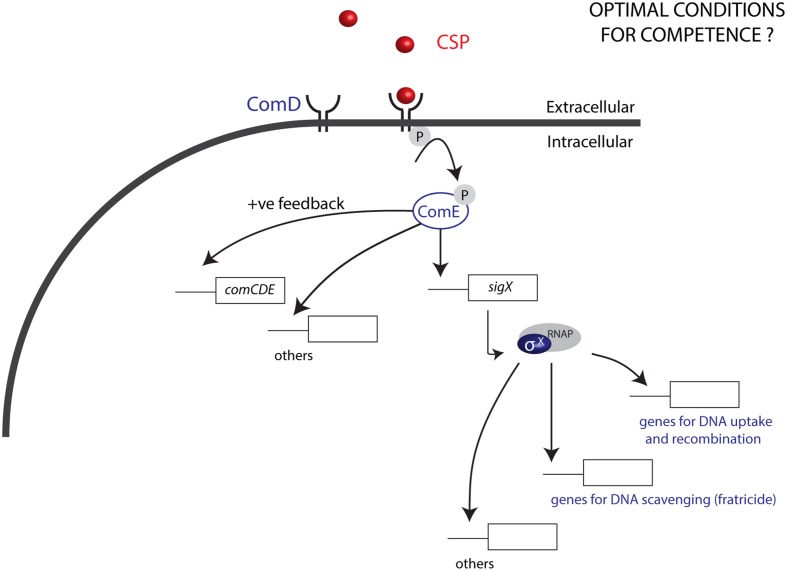
Natural transformation, a dominant force in the evolution of bacteria, is responsible for gene exchange and the acquisition of new DNA sequences, and is an important tool in genetic engineering. Most members of the Mitis group of streptococci are naturally competent for genetic transformation and produce competence pheromones, also known as competence stimulating peptides (CSPs), recognized by cognate surface receptors (Håvarstein et al., 1996). Competence development is most studied in S. pneumoniae, also a member of the Mitis group. In this model organism, competence is characterized by two main processes: (1) quorum-sensing (QS) and (2) expression of effectors that allow DNA binding, uptake, and integration into the bacterial genome (Claverys and Håvarstein, 2002; Luo and Morrison, 2003). The QS pathway launches the competence development cascade: first, comC is activated, and its activation leads to the transcription and translation of the gene into a pro-peptide before it is cleaved and exported as the CSP by an ABC transporter, ComAB (Håvarstein et al., 1996). Once CSP reaches a threshold concentration in the extracellular environment, it binds to its membrane receptor histidine kinase ComD, resulting in the phosphorylation of the response regulator ComE. ComE phosphorylation activates expression of the alternative sigma factor σX (Lee and Morrison, 1999; Claverys and Håvarstein, 2002) in addition to ∼20 early genes (Figure 1). The σX regulon includes in total 27 to 30 pan-streptococcal core genes (Khan et al., 2016), involved in exogenous DNA uptake, processing, and recombination. Environmental factors are also known to have an important impact on the development of competence and in the magnitude of the response to CSPs. However, in contrast to the molecular mechanisms involved in the CSP response and production that have been described in great detail, such environmental factors are still poorly defined (Johnsborg and Håvarstein, 2009). Notably, while conditions defined for some closely related species can be used as a start point for transformation, optimal levels of competence often require a series of optimization strategies, with an outcome that is not always predictable. In Streptococcus pyogenes, for instance, despite a series of optimization approaches, competence has only been demonstrated in the presence of epithelial cells and at levels that do not surpass 0.0001% (Marks et al., 2014).
FIGURE 1.
Competence in the Mitis group of streptococci. The CSP pheromone produced by streptococci, upon reaching a threshold concentration binds to the histidine kinase ComD, leading to phosphorylation of the response regulator ComE. ComE-P activates a positive feedback loop by inducing the expression of comCDE, and also induces the expression of sigX, which encodes the alternative sigma factor σX. σX binds to the RNA polymerase (RNAP), to activate the expression of 27 to 30 panstreptococcal competence genes responsible among other functions for DNA uptake and recombination, as well as DNA scavenging via fratricide. Optimization of conditions for transformation of the Mitis group has been mostly studied in S. pneumoniae. For other species in this group, conditions for optimal competence are generally unknown.
The human oral commensal Streptococcus mitis, a member of the Mitis group of streptococci, colonizes all surfaces of the oral cavity including teeth, tongue and mucosal surfaces, in addition to the tonsils and nasopharynx (Pearce et al., 1995). S. mitis and S. pneumoniae – one of the most important human pathogens –, share >80% of their genes. Being genetically very similar, their differences in pathogenic potential are striking (Kilian et al., 2014). In comparison to S. pneumoniae and their common ancestor, evolutionary analyses suggest that loss of virulence determinants in S. mitis may be the reason for the decrease in its pathogenicity (Kilian et al., 2008). In contrast with S. pneumoniae, in which most of the strains use one of two pheromones, in S. mitis almost each strain has its own CSP (Kilian et al., 2008). Despite the high interest in understanding S. mitis behavior, low transformation yields and challenges in genetic manipulation of this species, reflected in the literature for three decades, have probably hampered advances in the field (Gaustad, 1985; Potgieter and Chalkley, 1991; Bensing et al., 2001; Mitchell, 2011; Duran-Pinedo et al., 2014; Rukke et al., 2014; Xie et al., 2015). Prior to the introduction of synthetic CSPs for transformation, Gaustad (Gaustad, 1985) first reported S. mitis transformation efficiencies of around 0.0001%. This level was further shown to increase to 0.001% with the use of synthetic CSP by Bensing et al. (2001). Difficulties in transforming S. mitis were later also reported for B6, the first genome sequenced strain (Denapaite et al., 2010). For the type strain, Duran-Pinedo et al. (2014) reported failures in obtaining any transformants at all. We have been able to transform the type strain previously (Rukke et al., 2014), but only at low efficiency. Considering the multiple fields in which S. mitis has been studied, the development of a highly efficient method for S. mitis genetic manipulation through natural transformation is of great interest and value.
We recently described a markerless method for genome editing without introduction of selective markers that is applicable for streptococci with high transformation efficiencies (Morrison et al., 2015). Overcoming the challenges of low transformation efficiencies in S. mitis would thus not only facilitate the genetic manipulation of this organism using marker selection methods, but it would also allow the use of direct markerless genome editing approaches in S. mitis functional investigations. The aim of this study was to identify optimal conditions for natural genetic transformation in Streptococcus mitis, and to illustrate its application as a genome editing resource. We now report the development of a method for high efficiency transformation of the type strain, which is also applicable for strain SK321. Further, we demonstrate its use in direct genome editing.
Materials and Methods
Bacterial Strains and Media
All strains of S. mitis used are listed in Table 1. Streptococci were stored at -80°C in Todd Hewitt Broth (THB, Becton Dickinson and Company, Le Pont de Claix, France) or Tryptic Soy Broth (TSB, Soybean-Casein Digest medium, BactoTM) supplemented with 30% glycerol. Pre-cultures used in the experiments described below were made from fresh liquid cultures incubated at 37°C in a 5% CO2-supplemented atmosphere and grown until the cultures reached an absorbance at 600 nm (optical density at 600 nm [OD600]; Biophotometer; Eppendorf) of 0.5, before storage at -80°C in 15% glycerol. For transformation assays, Tryptic Soy Broth (TSB, Soybean-Casein Digest medium, BactoTM), THB supplemented with 5% of horse serum (THB-HS) (Petersen and Scheie, 2010), semi-defined media C+YY B (Stevens et al., 2011) or C+Y (Martin et al., 1995) were used. For plating, blood agar base No. 2 (Oxoid, Hampshire, England) supplemented with 5% defibrinated sheep blood (TCS Biosciences Ltd., Buckingham, UK) was used. Final concentrations of selective levels of antibiotics were 500 μg mL-1 kanamycin, 10 μg mL-1 erythromycin, and 200 μg mL-1 spectinomycin.
Table 1.
Strains, synthetic peptides and primers used in this study.
| Strains | Descriptiona | Source or characteristics |
|---|---|---|
| Streptococcus mitis CCUG 31611T | S. mitis biovar 1 type strain, corresponds to NCTC 12261T; encapsulated, transformable strain | CCUG |
| S. mitis NCTC 8033 (SK321) | S. mitis strain isolated from the oral cavity | NCTC; (Kilian et al., 2008) |
| S. mitis CCUG 62644 (SK575) | S. mitis strain isolated from blood; ErmR | CCUG; (Kilian et al., 2008) |
| S. mitis CCUG 62641 (SK579) | S. mitis strain isolated from blood | CCUG |
| S. mitis CCUG 62642 (SK616) | S. mitis strain isolated from blood | CCUG; (Kilian et al., 2008) |
| MI009 | CCUG 31611, but ΔsrtA::erm; ErmR | This study |
| MI055 | CCUG 31611, but psigX luc::spc; SpcR | This study |
| MI074 | CCUG 31611, but ΔSM12261_0092::kan; KanR | This study |
| MI091 | CCUG 31611, but Δshp | This study |
| MI092 | SK321, but psigX luc::spc; SpcR | This study |
| Competence Stimulating Peptide (CSP) | Sequence | |
| S. mitis CCUG 31611 | EIRQTHNIFFNFFKRR | GenScript |
| S. mitis SK321 | ESRLPKIRFDFIFPRKK | GenScript |
| S. mitis SK575 | EMRKMNEKSFNFF | GenScript |
| S. mitis SK579 | EIRKSNSALVNFFK | GenScript |
| S. mitis SK616 | EMRRIDKIFINFLKRR | GenScript |
| Primers | Sequence (5′ to 3′) | |
| FP001b | ggcgcgccGTTTGATTTTTAATG | KanR cassette |
| FP015b | ggcgcgccCCGGGCCCAAAATTTGTTTGAT | ErmR cassette |
| FP016c | ggccggccAGTCGGCAGCGACTCATAGAAT | ErmR cassette |
| FP068c | aggccggccTAGGTACTAAAACAATTCATCCAGTA | KmR cassette |
| FP411 | AGCCGTTCGTGGTATGAGTC | MI009 mutant construction |
| FP412b | ggcgcgccTTTGATACCTGGCTGACTTGG | |
| FP413c | ggccggccAGATCGCGTTGATGAGATTGA | |
| FP414 | CTCAGCCGTTTTTCTCCAAG | |
| FP770d | aaagctagcAGCTGCTTTAGTCGCTGCTC | pFW5-luc |
| FP771e | aaaggatccCAATCCCCTGGACTTCTTCA | pFW5-luc |
| FP829c | ggccggccTTTTTGCTGTAAACTAAGGAGTAGAGA | MI074 mutant construction |
| FP830 | CGACTTCGATCTTTTAGTTGATG | |
| FP831 | CCCGTTTTCCCAACAAAAA | |
| FP832b | ggcgcgccTTTTCTGTACACCTCATTTCCTTT | |
| FP914 | TCTCGCGGAAGATAAAGTCG | MI009 mutant amplification |
| FP915 | GGGTTAGCAGCAACAAGGAA | |
| FP1163 | CATCTTGATAGCGTGGCTCA | |
| FP1164 | GAAGTCTAAATATGAAGAAGGTGTACACCTAAGTCT | MI091mutant construction |
| FP1165 | AGACTTAGGTGTACACCTTCTTCATATTTAGACTTC | |
| FP1166 | TTGAATTGAGACGGATTGGA | |
| FP1167 | GCTGAGTTCCTCCAAGCTGT | MI009 mutant amplification |
| FP1168 | ATCCAGAAACGACCGACAAG | |
| FP1169 | TCTTTCATCGTGTCCTCCAA | |
| FP1170 | TGATTCTCCCCGTTCAAATC | |
| FP1171 | AAACATTTTATCCACCGACAA | |
| FP1172 | TTGCACCAAGTTAGCGAATG | |
| FP1242 | CTTGAGCTGGGCTTCGTAGT | |
| FP1243 | ACAGGGGATGTCATGGGTAA | |
| FP1244 | CTGCTACGTTTGCAAGGTCA | |
| FP1245 | ATGAACGTGAACCCTTGGTC | |
| FP1248 | CAGCATGCAGTTGCAAAGTT | |
| FP1249 | CCCGAACGTGTTTCTCAAAT | |
| FP1250 | CTCAGCCGTTTTTCTCCAAG | |
| FP1251 | AGTATGGCCGTATTGCCAAG | |
| FP1280 | GCACCTGTATCACGAAGCAA | |
| FP1281 | TGCTGAAGCACAAGCTGAGT |
aKan, kanamycin; Erm, erythromycin; Spc, spectinomycin.bAscI restriction site underlined.cFseI restriction site underlined.dBamHI restriction site underlined.eNheI restriction site underlined.
Competence-Stimulating Peptide
Stock solutions of the lyophilized CSPs (Table 1) were prepared by re-suspending the material in distilled water to a concentration of 10 mM, and storing it at -20°C. Working solutions of 100 μM were aliquoted and stored at -20°C.
Luciferase Reporter Assays
Culture stocks of the PsigX-luciferase strain MI055 and MI092 at OD600 0.5 were diluted in the presence or absence of CSP, and luminescence was monitored as described previously (Morrison et al., 2015).
Construction of Mutants with Antibiotic Markers
All primers used for marker constructions are listed in Table 1. For construction of the S. mitis ΔsrtA (MI009) and ΔSM12261_0092 (MI074) mutants, the standard PCR ligation mutagenesis strategy was employed (Lau et al., 2002), with minor modifications (Rukke et al., 2014). The srtA flanking regions were amplified using primer pairs FP411–FP412 and FP413–FP414. The ΔSM12261_0092 flanking regions were amplified with the primer pairs FP829–FP830 and FP831–FP832. The erythromycin and kanamycin resistance cassettes (ErmR, KmR) (Claverys et al., 1995; Sung et al., 2001) were amplified using the primer pairs FP015–FP016 and FP001–FP068, respectively. Ligation and purification of the PCR products were performed using T4 DNA ligase (Fermentas) and the QIAquick PCR purification kit (Qiagen), respectively. The resultant amplicons were used for transformation of S. mitis as described below. Correct insertion of erythromycin or kanamycin markers in the deleted region was verified by PCR amplification of the chromosomal DNA extracted from each mutant, using primer pairs complementary to sequences within the antibiotic cassette sequence in combination with primers in regions up and downstream the insertion of the antibiotic cassette. To construct MI055, the luciferase reporter for sigX, the primer pair FP770–FP771 was used to amplify the specific promoter region from the S. mitis type strain. Following restriction with BamHI and NheI, the amplicon was ligated to pFW5-luc (SpecR) and cloned into Escherichia coli as previously described (Khan et al., 2012). A plasmid with the correct insert was further purified and transformed into the S. mitis type strain, where it was incorporated in the chromosome by single cross-over recombination.
Transformation Assays
For construction of marker insertions, the following protocol was used. Pre-cultures of the S. mitis type strain at OD600 0.5 were diluted 1:10 in TSB and liquid cultures were grown at 37°C, 5% CO2 for 15 min. Then, 250 nM CSP was added, together with the PCR constructs or donor plasmids. Cultures were incubated under aerobic conditions at 37°C for 4 h. Transformants were selected on blood agar plates supplemented with the appropriate antibiotics by 24 h of incubation at 37°C, 5% CO2.
For optimization steps, different protocols were used as described in the results section.
Preparation of Plasmids
The replicative shuttle plasmid pVA838 (Macrina et al., 1982) and the non-replicative plasmid pFW5-luc (Podbielski et al., 1996) were isolated from E. coli using the QIAprep Spin Miniprep kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s recommendations.
Preparation of Genomic Donor DNA
Genomic donor DNA was extracted from strains according to the CTAB method previously described (Petersen et al., 2006).
Preparation of Donor DNA Amplicons
Primers used for amplification of donor DNA from existing mutants are listed in Table 1 and corresponding amplicons are described in Table 2. PCR conditions and primers can be found elsewhere (Morrison et al., 2015).
Table 2.
Donor DNA amplicons.
| Amplicon | Primers, amplicon size, marker, and template | Source |
|---|---|---|
| aRJ19 | FP1167/FP1168 – 5.2 kb – Markerless, from MI091 | This study |
| aRJ20 | FP914/FP915 – 6.9 kb – ErmR, from MI009 | This study |
| aRJ21 | FP1163/FP1166 – 6.9 kb – KanR, from MI074 | This study |
| aRJ22 | FP1250/FP1251 – 1.9 kb – ErmR, from MI009 | This study |
| aRJ23 | FP1248/FP1249 – 3.0 kb – ErmR, from MI009 | This study |
| aRJ24 | FP1280/FP1281 – 3.9 kb – ErmR, from MI009 | This study |
| aRJ25 | FP1244/FP1245 – 4.9 kb – ErmR, from MI009 | This study |
| aRJ26 | FP1242/FP1243 – 5.9 kb – ErmR, from MI009 | This study |
Markerless Mutant Construction
To construct the markerless mutant MI091, with deletion of a putative pheromone gene (shp) located between ΔSM12261_0093 and ΔSM12261_0094, two flanking fragments of 3 kb were amplified from S. mitis type strain with the primer pairs FP1163–FP1164 and FP1165–FP1166. Overlapping sequences were used for fusion of the two PCR products, in a ligase-free PCR reaction that used less than 50 ng of each of the two amplified sequences and the nested primers FP1167 and FP1168, to produce an amplicon of 5224 bp. After transformation of competent cells of the S. mitis type strain, as described in the optimized protocol, colonies from blood plates were screened by PCR, and re-screened to identify a clone containing the correct deletion.
Detection of Markerless Mutation through Colony Genotyping Assay
Colonies on non-selective plates were screened by PCR using primers specific, respectively, for the donor and recipient alleles at targeted loci, as previously described (Morrison et al., 2015).
Statistical Analysis
Two-way ANOVA with multiple measures followed by Tukey’s test was used to analyze differences between groups. Student’s t-test was used to compare two independent samples. A p < 0.05 was considered statistically significant.
Results and Discussion
Semi-Defined Medium Enriched with Bovine Serum Albumin Supports High CSP Signaling in S. mitis
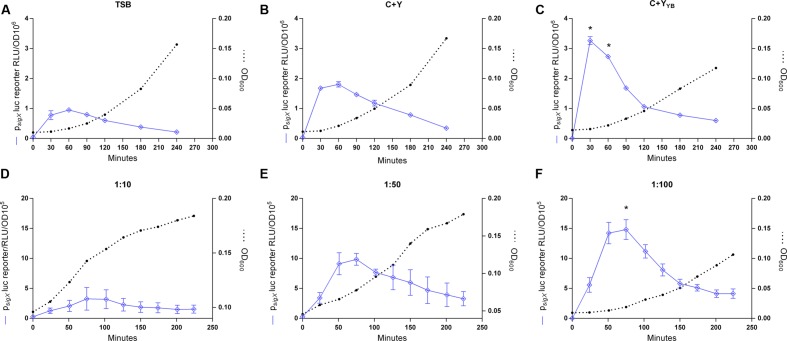
Previous protocols for transformation of S. mitis have suggested the use of a rich medium (Todd Hewitt Broth or Tryptic Soya Broth) for culture growth and transformation (Gaustad and Morrison, 1998; Petersen and Scheie, 2010). However, for some species of streptococci, such as Streptococcus thermophilus, transformation is mostly restricted to growth in defined media with restricted peptide contents (Gardan et al., 2013). Thus, when looking for alternatives to increase S. mitis low transformation efficiency, ranging from 0.001 to 0.5%, we thought to compare competence in TSB with two semi-defined media: C+Y, a medium often used for transformation of S. pneumoniae, and C+YY B, a medium derived from C+Y, with an increased concentration of yeast extract and additional bovine serum albumin, permissive for development of spontaneous competence in S. pneumoniae (Stevens et al., 2011). The sigX expression was significantly higher in the culture in C+YY B medium than in C+Y or TSB, suggesting that this medium might also allow increased rates of transformation in S. mitis (Figures 2A–C).
FIGURE 2.
Effect of growth medium and dilution ratios on CSP-induced sigX expression profiles. (A–C) The sigX reporter type strain (MI055; psigX luc) was grown in 200 μL of TSB, C+Y, or C+YY B in a 96-well plate in air at 37°C with 150 nM CSP. ∗ Significant statistical difference in C+YY B when compared to C+Y and TSB (p < 0.05). (D–F) Comparison of sigX expression profiles and culture growth in pre-culture dilution ratios of 1:10, 1:50, or 1:100. Strain MI055 (psigX luc) was grown in 200 μL volumes of C+YY B in a 96-well plate in air at 37°C with 150 nM CSP. ∗ Significant differences on sigX expression comparing 1:10 and 1:50 to 1:100 (p < 0.05). For media comparison, pre-cultures were prepared in TSB and on the day of experiment they were centrifuged and re-suspended 1:100 in fresh medium – TSB, C+Y or C+YY B. For all figures, sigX expression, presented as relative luminescence units (RLU), and culture optical density (OD) were measured periodically. The results are averages from 3 replicates and representative of three independent experiments. Error bars represent standard error of the mean. Differences in sigX expression among media and dilutions rates were analyzed by two-way ANOVA.
Dilution Rate of Cultures Influences the CSP Response
In S. pneumoniae, the basic in vitro requirements for high competence to appear include (1) a culture of cells that have not recently been competent since the refractory period, (2) a culture at the early exponential phase of growth, as planktonic cells close to the stationary phase respond poorly to CSP, and (3) exposure to a culture medium conducive to development of competence (Tomasz, 1965; Tomasz and Chambers, 1965; Morrison et al., 1984). The results in Figure 2 showing background levels of sigX expression at the start of the experiments indicate that the cells were not in a competent state, in line with the first requirement for competence in S. pneumoniae. As to the second and third requirements, we observed that expression of sigX in S. mitis is activated early in all three tested media and it is shut off after approximately 30 more minutes (Figure 2), and experiments with the addition of CSP to cultures entering stationary phase show a transformation efficiency below 0,001% (data not shown). Our next step was to investigate whether the dilution rate of the pre-cultures into the fresh medium would affect the S. mitis CSP response. Although the time to achieve optimal competence levels was not markedly affected, cells from cultures diluted in a 1:100 ratio presented a somewhat (50%) higher sigX signaling compared to cells diluted 1:50 or 1:10 (Figures 2D–F). Thus, this ratio was used in the further optimization steps described below.
S. mitis Competence for Genetic Transformation is Transient
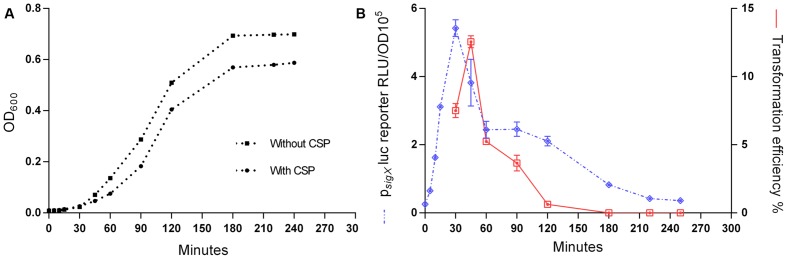
To explore the kinetics of transformation in S. mitis, we performed a time-course study evaluating sigX expression employing luciferase reporter assays and monitoring transformation efficiency throughout 240 min of incubation of an inoculum starting with an OD600 of 0.04. sigX expression increased rapidly after the addition of the synthetic peptide, with its peak occurring at 30 min and decreasing gradually until disappearing at 240 min (Figure 3). Following a similar pattern, transformation efficiency reached its highest value (12%) after 45 min of incubation with the competence factor, followed by a gradual decrease. Our results demonstrate that competence for genetic transformation in S. mitis type strain is transient, as it is in its close relative S. pneumoniae (Håvarstein et al., 1995; Bricker and Camilli, 1999).
FIGURE 3.
Responses of the Streptococcus mitis sigX reporter in the type strain to CSP. A pre-culture of MI055 at OD600 0.5 was diluted 1:100 in C+YY B and grown until OD600 0.04 at 37°C in 5% CO2. At time0, the lag phase cell culture (OD600 0.039) was treated with 300 nM CSP and distributed into 200 μL aliquots that were further exposed to 150 ng mL-1 amplicon donor DNA of 7 kb (aRJ20) at different time points. 20 U mL-1 DNase I were added after 30 min of exposure to DNA and the culture was incubated in air at 37°C for additional 30 min. The % of CFU containing ErmR transformants was determined by plating on blood plates with or without 20 μg mL-1 erythromycin. (A) Growth curves with and without CSP, measured as OD600. (B) Induction of sigX and transformation. The results are average from 3 triplicates and representative of two independent experiments. Error bars represent the standard error of the mean.
Effect of CSP Concentration on Growth and Transformation
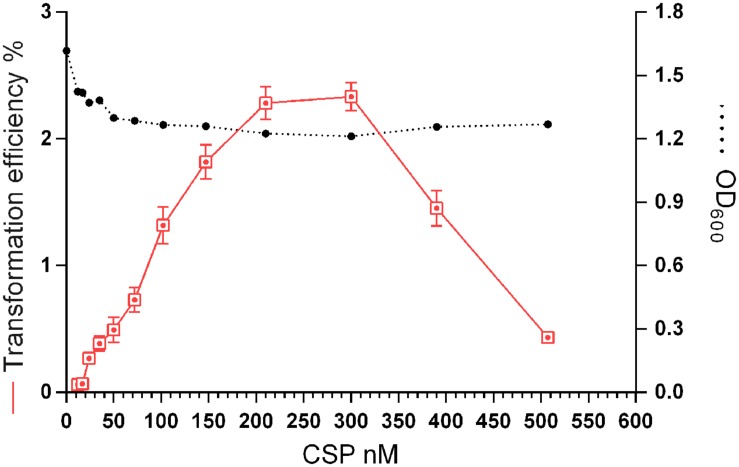
In S. pneumoniae, cells that are growing rapidly in a competence permissive medium, can be induced to competence by a low (45 nM CSP-1) level of the CSP (Håvarstein et al., 1995). However, a minority of the cells that do not develop competence are lysed by the competent cells, resulting in DNA release (Steinmoen et al., 2002). This fact, along with the previously described CSP-derived stress (Oggioni et al., 2004), may explain the detrimental effect of CSP on S. pneumoniae growth rates. Nonetheless, neither the concentration of synthetic CSP for optimal transformation of S. mitis nor the effects of the peptide on growth are known. We investigated the transformation efficiency and growth of the S. mitis type strain under CSP concentrations ranging from 10 to 510 nM (Figure 4). Transformation efficiency was stimulated by a concentration as low as 25 nM, reaching saturation at approximately 210 nM. The final culture density was only slightly affected in the presence of CSP.
FIGURE 4.
Effect of CSP concentration on transformation efficiency and culture density. S. mitis type strain pre-culture at OD600 0.5 was diluted 1:100 in C+YY B and grown until OD600 0.04 at 37°C in 5% CO2. Following, 500 μL aliquots were exposed to 150 ng mL-1of the 7 kb amplicon donor (aRJ21) and CSP concentrations varying from 0 to 507 nM. Cultures were further incubated in closed tubes for 4 h at 37°C in air atmosphere. Optical density of the cultures was measured at the time of plating. The % of CFU containing KanR transformants was determined by plating on blood plates with or without 500 μg mL-1 kanamycin. The results are averages from 3 replicates and representative of three independent experiments. Error bars represent standard error of the mean.
Transformation Efficiency Depends on Amount, Source, and Size of Donor DNA
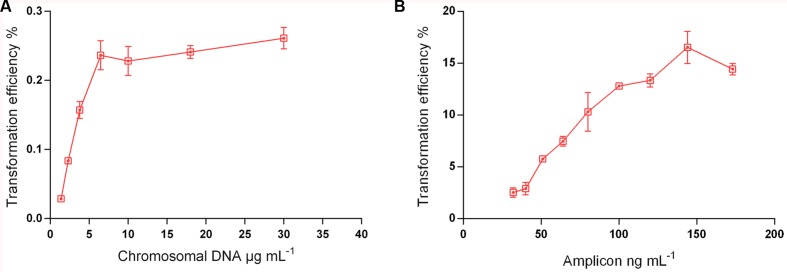
The complexity of the donor DNA is a crucial factor for the yield of recombinants (Morrison et al., 2015). A 7 kb PCR product is, for instance, ∼300-fold less complex than chromosomal DNA from the same genomic template. Thus, chromosomal DNA carries irrelevant genetic material that will compete for incorporation with the fragment of interest. To test this effect in S. mitis transformation, we compared chromosomal DNA and a large (7 kb) unique donor DNA amplicon (aRJ20), embodying the same antibiotic resistance insert. Figure 5 shows that maximum transformation yields were achieved with 6 μg mL-1 of chromosomal DNA and 150 ng mL-1 amplicon, respectively. These values were close to those previously found in other streptococci, in which saturated concentrations are in the range of 1 to 4 μg mL-1 of chromosomal DNA, and 75–100 ng mL-1 of amplicon donor DNA (Håvarstein et al., 1995; Bricker and Camilli, 1999; Johnston et al., 2013; Morrison et al., 2015). As expected, the two sources of DNA tested varied in efficiency, with transformation achieving 80-fold higher values with the amplicon than with the chromosomal DNA (Figure 5).
FIGURE 5.
Transformation as a function of donor DNA type and concentration. S. mitis type strain pre-culture at OD600 0.5 was diluted 1:100 in C+YY B and grown until OD600 0.04 at 37°C 5% in CO2. (A) 200 μL aliquots were exposed to 300 nM of CSP and chromosomal DNA concentrations (from strain MI009) ranging from 1,38 to 30 μg mL-1. Cells were further incubated in closed tubes for 3 h at 37°C in air atmosphere. (B) 200 μL aliquots were exposed to 300 nM of CSP and 7 kb amplicon (aRJ20) concentrations varying from 32 to 173 ng mL-1. Cells were further incubated in closed tubes for 3 h at 37°C in air atmosphere. The % of CFU containing ErmR transformants was determined by plating on blood plates with and without Erm. The results are averages from 3 replicates and representative of three independent experiments. Error bars represent standard error of the mean.
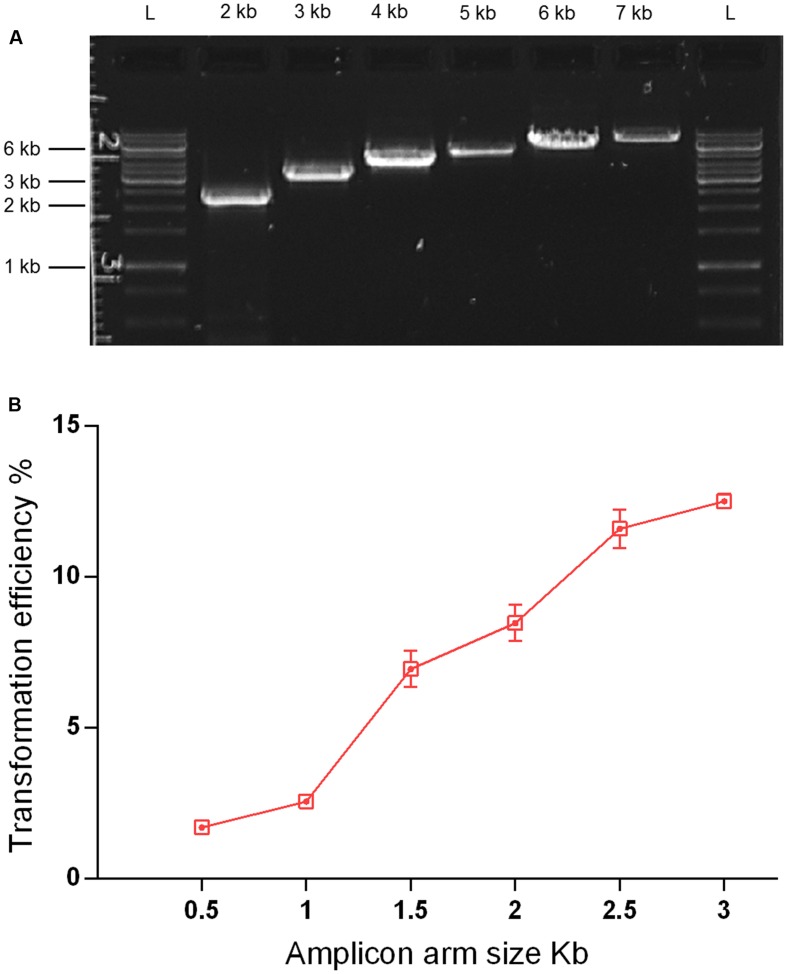
As streptococcal transformation efficiency is strongly dependent on donor DNA fragment size (Cato and Guild, 1968; Morrison et al., 2015), we investigated the effect of amplicon size in the S. mitis system. We used unique donor DNA amplicons with a final size from 2 to 7 kb (Figure 6A). The amplicons comprised two arms of ∼0.5 to ∼3 kb, flanking a central region with an erythromycin marker. The flanking homologous arms of 1.5 kb increased transformation efficiency by approximately threefold compared to 0.5 kb arms, while further enlargement to 3.5 kb achieved an approximately sixfold increase when compared to 1 kb arms (Figure 6B). This is similar to results recently obtained in S. mutans (Morrison et al., 2015).
FIGURE 6.
Effect of amplicon arm size on transformation. (A) Gel analysis of six amplicons of different sizes derived from the same genomic template (MI009). L, DNA ladder Gene Ruler 1kb (GenScript). (B) Transformation as a function of amplicon arm sizes. S. mitis type strain pre-culture at OD600 0.5 was diluted 1:100 in C+YY B and grown until OD600 0.04 at 37°C in 5% CO2. Following, 200 μL aliquots were exposed to 300 nM of CSP, 150 ng mL-1 donor DNA amplicon (aRJ20, aRJ22, aRJ23, aRJ24, aRJ25, and aRJ26) and further incubated in closed tubes for 4 h at 37°C in air atmosphere. The % of CFU containing ErmR transformants was determined by plating on blood plates with and without Erm. The results are averages from three replicates and representative of three independent experiments. Error bars represent standard error of the mean.
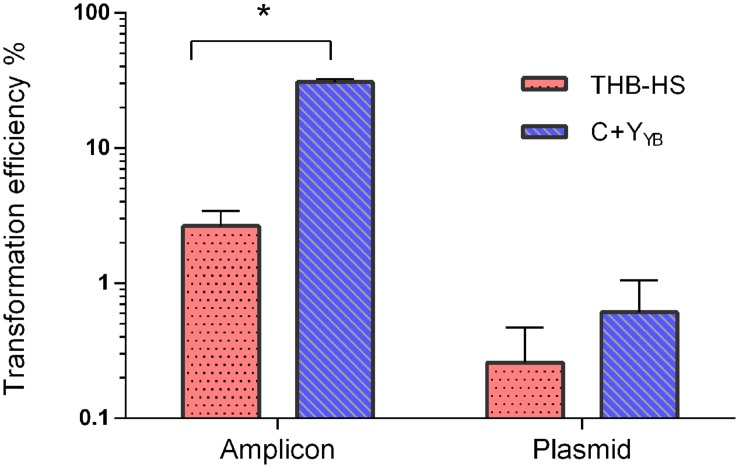
The Optimized Protocol Results in Transformation Efficiencies Significantly Higher Than the THB-HS Standard Protocol
We compared the optimized protocol in C+YY B with previously published protocols in THB-HS (Gaustad and Morrison, 1998; Petersen and Scheie, 2010). The protocol in THB-HS consists of 1:10 dilution of pre-cultures prepared in the same medium, followed by addition of 200 nM CSP and donor DNA, and incubation for 4 h. Figure 7 shows that the transformation efficiency with pVA838, a second order process, was slightly higher with the new protocol, while transformation with the aRJ20 amplicon as donor DNA resulted in 20-fold higher values, thus confirming the superiority of the optimized protocol in C+YY B.
FIGURE 7.
Transformation as a function of different protocols and donor DNA types. Following the optimized protocol, S. mitis type strain pre-culture at OD600 0.5 was diluted 1:100 in C+YY B and grown until OD600 0.04 at 37°C in 5% CO2. Then, 200 μL aliquots were exposed to 300 nM of CSP and 150 ng mL-1 amplicon (aRJ20) or 1 μg replicative plasmid pVA838. Cells were further incubated in closed tubes for 3 h at 37°C air atmosphere. The % of CFU containing ErmR transformants was determined by plating on blood plates with and without Erm. The results are averages from two independent experiments. Bars represent standard error of the mean. Differences in efficiency between media were analyzed by Student’s T-test (∗p < 0.05).
S. mitis SK321 Shows Similar Transformation Rates, but Higher sigX Expression than the Type Strain
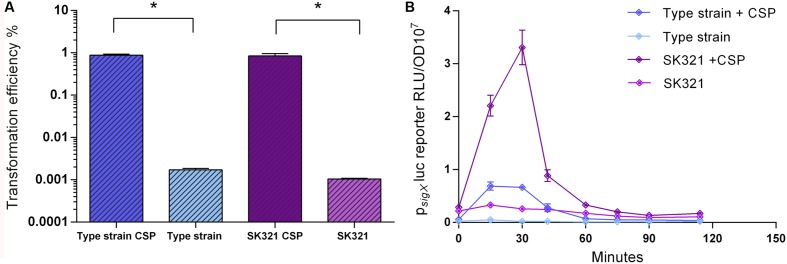
The competence apparatus in S. mitis is not always intact, as suggested by a recent study showing that 5 of 15 strains had defects in either sigX or other competence effector genes required for transformation (Kilian et al., 2014). This could be one reason why the transformation of S. mitis in the laboratory setting has been regarded as challenging. However, with 77% of the strains having apparently intact competence systems, it is expected that a significant range of strains will be amenable to transformation once optimal conditions are defined.
We used the improved transformation protocols for the S. mitis type strain presented above to test the transformation of 4 other S. mitis strains, using CSPs as deduced from their comC sequences. Among these, S. mitis SK321 was transformed with pVA838 at efficiencies similar to the type strain (Figure 8A), while the remaining strains were not amenable to transformation either with pVA838 or the 7 kb amplicon aRJ21 (percentage similarity of the corresponding region in the reference genomes was approximately 50%). We further constructed a sigX reporter in SK321 (MI092) to examine the kinetics of competence development in this strain compared to the type strain. Although sigX expression was significantly higher in SK321, the window of competence in the presence of CSP was similar in the two strains, with maximum levels of sigX expression between 15 and 30 min (Figure 8B).
FIGURE 8.
Comparison of transformation efficiency and sigX expression in S. mitis type strain and S. mitis strain SK321. (A) S. mitis type strain and SK321 pre-cultures at OD600 0.5 were diluted 1:100 in C+YY B and grown until OD600 0.04 at 37°C in 5% CO2. Following, 200 μL aliquots were exposed to 300 nM of their respective strain specific CSP or kept untreated. Cells were transformed with 1 μg mL-1 replicative plasmid pVA838 at 37°C in air atmosphere for 3 h. The % of CFU containing ErmR transformants was determined by plating on blood plates with and without Erm. The results are representative of three independent experiments. Bars represent standard error of the mean. Differences in efficiency between cultures with and without CSP were analyzed by Student’s T-test (∗p < 0.05). (B) The sigX reporter type strain (MI055) and SK321 (MI092) were grown in 200 μL of C+YY B in a 96-well plate in air at 37°C with 300 nM of their respective strain specific CSP or kept untreated. Culture optical density (OD600) and sigX expression (RLU/OD) were monitored periodically. The results are averages from three replicates and representative of three independent experiments. Error bars represent standard error of the mean.
Analyses of the genomes of these strains available at the Human Oral Microbiome Database1 revealed that they possess homologs of all genes known to be involved in development of competence for genetic transformation in S. pneumoniae, but with possible truncations or mutations involving the sigX gene (Table 3). This adds to the body of evidence indicating that some strains of S. mitis may not exhibit a fully functional competence apparatus (Kilian et al., 2014). Furthermore, all S. mitis strains tested present the constitutively expressed gene endA, which encodes for an indispensable endonuclease involved in DNA transport.
Table 3.
Transformability and putative competence genes in five S. mitis strains.
| Genes |
S. mitis strainsa |
|||||
|---|---|---|---|---|---|---|
| CCUG31611T | SK321 | SK575 | SK579 | SK616 | ||
| Transformability | + | + | - | - | - | |
| Early genes | comAB | + | + | + | + | + |
| comCDE | + | + | + | + | + | |
| sigX1 | + | + | ∗ | ∗∗ | ∗∗ | |
| sigX2 | + | + | - | ∗∗ | - | |
| Core SigX regulon | Recombination | |||||
| radA | + | + | + | + | + | |
| coiA | + | + | + | + | + | |
| dprA | + | + | + | + | + | |
| ssbB | + | + | + | + | + | |
| recA | + | + | + | + | + | |
| comFA-comFC | + | + | + | + | + | |
| cilC | + | + | + | + | + | |
| comEA-comEC | + | + | + | + | + | |
| comGA-comGG | + | + | + | + | + | |
| Fratricide | ||||||
| cbpD | + | + | + | + | + | |
| Unknown functions | ||||||
| dut | + | + | + | + | + | |
| rmuC, ccs50 | + | + | + | + | + | |
| yhaM, cbf1 | + | + | + | + | + | |
| yfiA | + | + | + | + | + | |
| pepF | + | + | + | + | + | |
| pilC | + | + | + | + | + | |
| radC | + | + | + | + | + | |
| cinA | + | + | + | + | + | |
aGenome data used for the identification of competence genes: NCTC12261 (CCUG 31611), accession AEDX00000000; SK321, accession AEDT00000000; SK575, accession AICU00000000; SK579, accession AJJL00000000; SK616, accession AICR00000000. ∗Substitution of two conserved N-terminal aminoacid residues (E4K; L5M). ∗∗Lack of the 5 to 8 C-terminal aminoacid residues possibly essential for proper function of σX. +Present, – Absent.
Application for Markerless Transformation of S. mitis
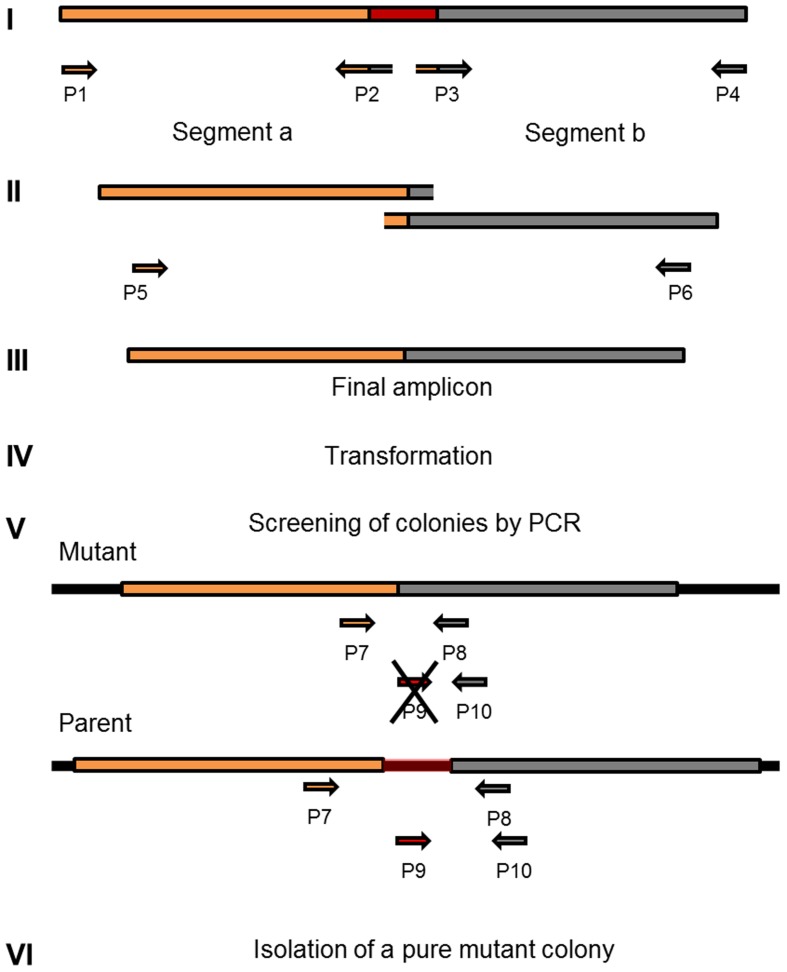
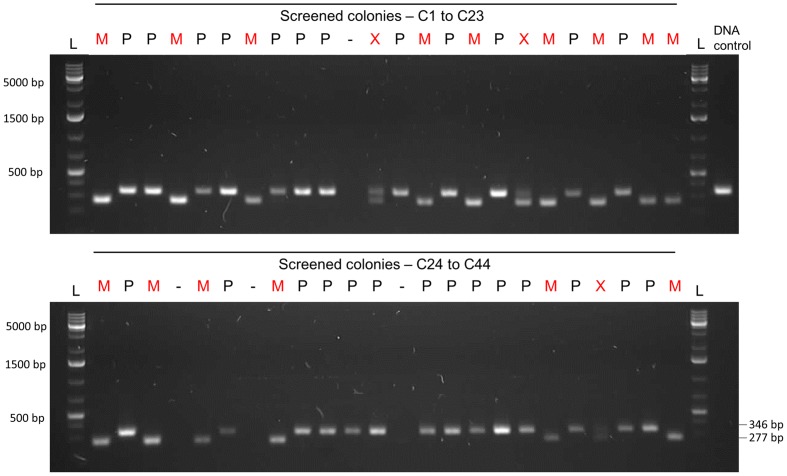
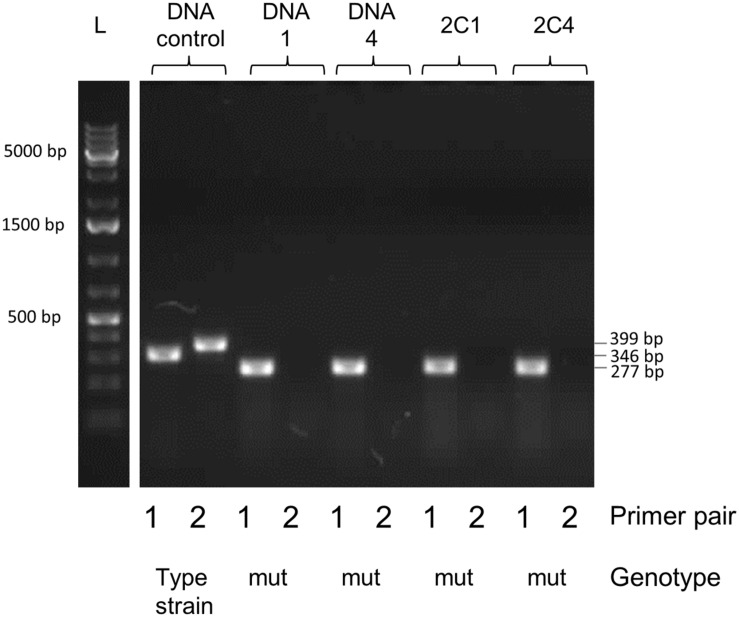
Selective markers are often used for microbial mutagenesis, especially when transformation efficiencies are low. Despite their value, markers carry unwanted information and might interfere with gene expression or subsequent analysis. The need for multiple mutations may also lead to an accumulation of antibiotic resistance genes. With markerless mutations, it is possible to make as many genome modifications as desired, as long as the mutation is tolerated. In addition, a markerless method based on highly competent cells can be an important tool considering the wide range of possible downstream applications, such as gene knockouts, direct base mutations, promoter editing, and missense mutations. Over the years different markerless mutations strategies have been developed and most of them are based in two-step counter-selection methods (Dailidiene et al., 2006; Xie et al., 2011; Farkas et al., 2012; Thiel et al., 2014). We recently described a one-step method applied in S. mutans (Morrison et al., 2015). The high efficiency of our optimized protocol opened the possibility to apply this methodology to create a markerless deletion (69 bp) in an ORF encoding a peptide in the S. mitis type strain. Fragments a and b were amplified with primers FP1163/FP1164 and FP1165/FP1166. Both sections were then fused and amplified with nested primers FP1167/FP1168 creating a final donor amplicon of 5.2 kb. This amplicon (aRJ19) carried the 69 bp deletion and ∼2.6 kb flanking regions of homology with the recipient chromosome. The aRJ19 product was used directly to transform S. mitis type strain. Forty-four colonies from plates without antibiotics were screened with specific flanking primers for the insertion of the mutation (Figures 9 and 10). Forty-one per cent of the colonies contained the mutation allele, among which only 3 colonies (6.8%) presented both the parent and the mutant alleles. Complete segregation was observed using DNA from C1 and C4 (DNA 1 and DNA 4), as well of derived colonies (2C1 and 2C4) that were grown for several generations (Figure 11). Thus, we can conclude that segregation took place already within the 3 h of DNA exposure used in the transformation assay. The summary of all variables tested in the optimization of the protocol is presented in Figure 12.
FIGURE 9.
Workflow for markerless genome editing. Amplicon design and construction (I to III) are prepared according to the desired mutation, exemplified in this diagram as a gene deletion (marked in red). This step is followed by a highly efficient (>10%) method for natural transformation (IV). Two dozen colonies recovered from the transformation step are screened with specific primers that amplify fragments with different sizes in the mutant when compared to the parent (V). Isolation of colonies with only the new recombined chromosomal DNA (VI).
FIGURE 10.
Gel analysis of the first PCR-screening for the markerless mutation. Forty-four colonies were screened with pair of primers FP1169/FP1170. Fifteen colonies presented only the mutant allele (M – 277 bp band), while three presented both parent and mutant alleles (X – two bands). Twenty-six colonies presented only the parent allele (P – 346 bp). Transformation efficiency was 41%. L, DNA ladder Gene Ruler 1kb plus (GenScript); DNA control, control with type strain DNA; C, colony followed by the number; M, mutant colony; P, parent strain colony; X. mixed colony presenting both parent and mutant alleles;-, DNA not detected.
FIGURE 11.
Gel analysis of segregants from candidate mutant-containing clones. Primer pair 1 (FP1169/FP1170) amplifies a 346 bp fragment in the parent and a 277 bp fragment in the mutant. Primer pair 2 (FP1171/FP1172) amplifies a 399 bp fragment in the parent but produces no band in the mutant, since primer 1171 is located in the deleted locus. DNA 1 and 4 were extracted from two pure mutant colonies retrieved from the first screening procedure (C1 and C4 in Figure 10). Colonies 1 and 4 (C1, C4) were isolated and grown from the first screening procedure for several generations in order to promote segregation of the mutant strain. L, DNA ladder Gene Ruler 1kb plus (GenScript); DNA control, control with type strain DNA; DNA, extracted DNA after the first screening followed by the colony number; 2C, tested colony after growing for several generations to allow segregation, followed by the colony number.
FIGURE 12.
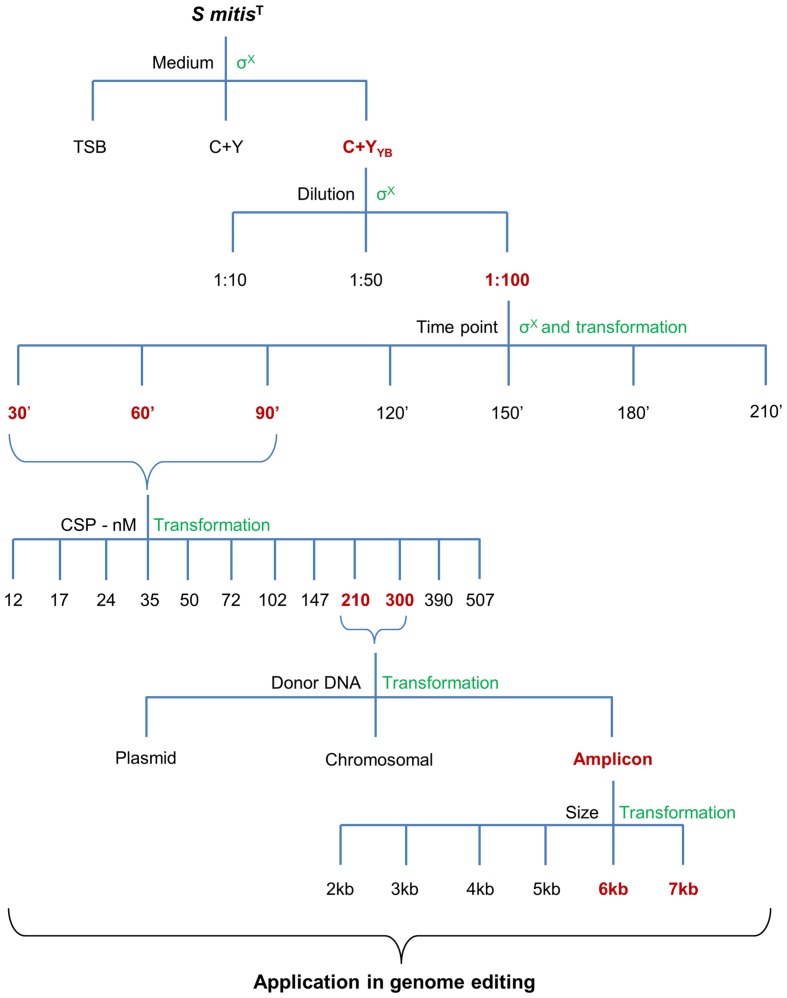
Summary of all variables tested in the optimization of the protocol. Different media, culture dilution, time points of transformation, CSP concentration, type of donor DNA, and amplicon sizes were used during the optimization steps. Chosen optimal conditions are in  , and the methods used to assess competence are in
, and the methods used to assess competence are in  .
.
Conclusion
Mechanisms of natural genetic transformation in S. mitis have not been thoroughly studied, leaving a gap in the knowledge about this species. As one plausible origin of this lacuna is the challenge in genetically transforming S. mitis in the laboratory setting. Thus, the optimized protocol we present here will facilitate future studies of this organism. Based upon the use of a semi-defined medium, specific dilution ratios, increased concentration of CSP, and a large amplicon, we increased S. mitis transformation efficiency by more than 3000-fold. This improvement in the protocol allows easy and direct genetic manipulation of the type strain, which also applied to strain SK321, broadening future prospects for the study of genetic transformation in the species as well as of other aspects of S. mitis biology.
Author Contributions
GS, RJ, DM, and FP participated in the conception and design of the work, as well as in the analysis and interpretation of data; GS and RJ conducted the experiments; all authors participated in drafting and revising the work for intellectual content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Andreas Podbielski for providing the plasmid pFW5-luc.
Funding. This work was supported by the Faculty of Dentistry, University of Oslo, by the Norwegian surveillance system for antibiotic resistance in microbes (NORM) and by the Norwegian Research Council (NFR) grant ES530425.
References
- Bensing B. A., Rubens C. E., Sullam P. M. (2001). Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect. Immun. 69 1373–1380. 10.1128/IAI.69.3.1373-1380.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker A. L., Camilli A. (1999). Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172 131–135. 10.1111/j.1574-6968.1999.tb13460.x [DOI] [PubMed] [Google Scholar]
- Cato A., Jr., Guild W. R. (1968). Transformation and DNA size. i. activity of fragments of defined size and a fit to a random double cross-over model. J. Mol. Biol. 37 157–178. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Dintilhac A., Pestova E. V., Martin B., Morrison D. A. (1995). Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164 123–128. 10.1016/0378-1119(95)00485-O [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Håvarstein L. S. (2002). Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798–d1814. [DOI] [PubMed] [Google Scholar]
- Dailidiene D., Dailide G., Kersulyte D., Berg D. E. (2006). Contraselectable streptomycin susceptibility determinant for genetic manipulation and analysis of Helicobacter pylori. Appl. Environ. Microbiol. 72 5908–5914. 10.1128/AEM.01135-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denapaite D., Bruckner R., Nuhn M., Reichmann P., Henrich B., Maurer P., et al. (2010). The genome of Streptococcus mitis B6–what is a commensal? PLoS One 5:e9426 10.1371/journal.pone.0009426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo A. E., Baker V. D., Frias-Lopez J. (2014). The periodontal pathogen Porphyromonas gingivalis induces expression of transposases and cell death of Streptococcus mitis in a biofilm model. Infect. Immun. 82 3374–3382. 10.1128/IAI.01976-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas J., Stirrett K., Lipscomb G. L., Nixon W., Scott R. A., Adams M. W., et al. (2012). Recombinogenic properties of Pyrococcus furiosus strain COM1 enable rapid selection of targeted mutants. Appl. Environ. Microbiol. 78 4669–4676. 10.1128/AEM.00936-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardan R., Besset C., Gitton C., Guillot A., Fontaine L., Hols P., et al. (2013). Extracellular life cycle of ComS, the competence-stimulating peptide of Streptococcus thermophilus. J. Bacteriol. 195 1845–1855. 10.1128/JB.02196-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaustad P. (1985). Genetic transformation in Streptococcus sanguis. Effects on genetic transformation by culture filtrates of Streptococcus sanguis (serogroups H and W) and streptococcus mitis (mitior) with reference to identification. Acta Pathol. Microbiol. Immunol. Scand. B. 93 283–287. [PubMed] [Google Scholar]
- Gaustad P., Morrison D. A. (1998). Induction of transformation in streptococci by synthetic competence stimulating peptides. Methods Cell Sci. 20 65–70. 10.1023/A:1009882608636 [DOI] [Google Scholar]
- Håvarstein L. S., Coomaraswamy G., Morrison D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92 11140–11144. 10.1073/pnas.92.24.11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein L. S., Gaustad P., Nes I. F., Morrison D. A. (1996). Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21 863–869. 10.1046/j.1365-2958.1996.521416.x [DOI] [PubMed] [Google Scholar]
- Johnsborg O., Håvarstein L. S. (2009). Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol. Rev. 33 627–642. 10.1111/j.1574-6976.2009.00167.x [DOI] [PubMed] [Google Scholar]
- Johnston C., Caymaris S., Zomer A., Bootsma H. J., Prudhomme M., Granadel C., et al. (2013). Natural genetic transformation generates a population of merodiploids in Streptococcus pneumoniae. PLoS Genet. 9:e1003819 10.1371/journal.pgen.1003819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R., Rukke H. V., Høvik H., Åmdal H. A., Chen T., Morrison D. A., et al. (2016). Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core Streptococcal competence genes. mSystems 1 1–27. 10.1128/mSystems.00038-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R., Rukke H. V., Ricomini Filho A. P., Fimland G., Arntzen M. O., Thiede B., et al. (2012). Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J. Bacteriol. 194 3781–3788. 10.1128/JB.00624-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian M., Poulsen K., Blomqvist T., Håvarstein L. S., Bek-Thomsen M., Tettelin H., et al. (2008). Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683 10.1371/journal.pone.0002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian M., Riley D. R., Jensen A., Bruggemann H., Tettelin H. (2014). Parallel evolution of Streptococcus pneumoniae and Streptococcus mitis to pathogenic and mutualistic lifestyles. MBio 5:e1490-14. 10.1128/mBio.01490-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. C., Sung C. K., Lee J. H., Morrison D. A., Cvitkovitch D. G. (2002). PCR ligation mutagenesis in transformable Streptococci: application and efficiency. J. Microbiol. Methods 49 193–205. 10.1016/S0167-7012(01)00369-4 [DOI] [PubMed] [Google Scholar]
- Lee M. S., Morrison D. A. (1999). Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181 5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Morrison D. A. (2003). Transient association of an alternative sigma factor. ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Tobian J. A., Jones K. R., Evans R. P., Clewell D. B. (1982). A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19 345–353. 10.1016/0378-1119(82)90025-7 [DOI] [PubMed] [Google Scholar]
- Marks L. R., Mashburn-Warren L., Federle M. J., Hakansson A. P. (2014). Streptococcus pyogenes biofilm growth in vitro and in vivo and its role in colonization, virulence, and genetic exchange. J. Infect. Dis. 210 25–34. 10.1093/infdis/jiu058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B., Garcia P., Castanie M. P., Glise B., Claverys J. P. (1995). The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls an SOS regulon. Dev. Biol. Stand. 85 293–300. [PubMed] [Google Scholar]
- Mitchell J. (2011). Streptococcus mitis: walking the line between commensalism and pathogenesis. Mol. Oral. Microbiol. 26 89–98. 10.1111/j.2041-1014.2010.00601.x [DOI] [PubMed] [Google Scholar]
- Morrison D. A., Khan R., Junges R., Amdal H. A., Petersen F. C. (2015). Genome editing by natural genetic transformation in Streptococcus mutans. J. Microbiol. Methods 119 134–141. 10.1016/j.mimet.2015.09.023 [DOI] [PubMed] [Google Scholar]
- Morrison D. A., Trombe M. C., Hayden M. K., Waszak G. A., Chen J. D. (1984). Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAM beta 1. J. Bacteriol. 159 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oggioni M. R., Iannelli F., Ricci S., Chiavolini D., Parigi R., Trappetti C., et al. (2004). Antibacterial activity of a competence-stimulating peptide in experimental sepsis caused by Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48 4725–4732. 10.1128/AAC.48.12.4725-4732.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce C., Bowden G. H., Evans M., Fitzsimmons S. P., Johnson J., Sheridan M. J., et al. (1995). Identification of pioneer viridans Streptococci in the oral cavity of human neonates. J. Med. Microbiol. 42 67–72. 10.1099/00222615-42-1-67 [DOI] [PubMed] [Google Scholar]
- Petersen F. C., Assev S., Scheie A. A. (2006). Combined effects of NaF and SLS on acid- and polysaccharide-formation of biofilm and planktonic cells. Arch. Oral Biol. 51 665–671. 10.1016/j.archoralbio.2006.02.003 [DOI] [PubMed] [Google Scholar]
- Petersen F. C., Scheie A. A. (2010). Natural transformation of oral Streptococci. Methods Mol. Biol. 666 167–180. 10.1007/978-1-60761-820-1_12 [DOI] [PubMed] [Google Scholar]
- Podbielski A., Spellerberg B., Woischnik M., Pohl B., Lutticken R. (1996). Novel series of plasmid vectors for gene inactivation and expression analysis in group a Streptococci (GAS). Gene 177 137–147. 10.1016/0378-1119(96)84178-3 [DOI] [PubMed] [Google Scholar]
- Potgieter E., Chalkley L. J. (1991). Reciprocal transfer of penicillin resistance genes between Streptococcus pneumoniae Streptococcus mitior and Streptococcus sanguis. J. Antimicrob. Chemother. 28 463–465. 10.1093/jac/28.3.463 [DOI] [PubMed] [Google Scholar]
- Rukke H. V., Kalluru R. S., Repnik U., Gerlini A., Jose R. J., Periselneris J., et al. (2014). Protective role of the capsule and impact of serotype 4 switching on Streptococcus mitis. Infect. Immun. 82 3790–3801. 10.1128/IAI.01840-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmoen H., Knutsen E., Håvarstein L. S. (2002). Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. U. S. A. 99 7681–7686. 10.1073/pnas.112464599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens K. E., Chang D., Zwack E. E., Sebert M. E. (2011). Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. MBio 2:e00071-11 10.1128/mBio.00071-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C. K., Li H., Claverys J. P., Morrison D. A. (2001). An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67 5190–5196. 10.1128/AEM.67.11.5190-5196.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel A., Michoud G., Moalic Y., Flament D., Jebbar M. (2014). Genetic manipulations of the hyperthermophilic piezophilic archaeon Thermococcus barophilus. Appl. Environ. Microbiol. 80 2299–2306. 10.1128/AEM.00084-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A. (1965). Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208 155–159. 10.1038/208155a0 [DOI] [PubMed] [Google Scholar]
- Tomasz M., Chambers R. W. (1965). Periodate oxidation of pseudouridine. Biochim. Biophys. Acta 108 510–512. 10.1016/0005-2787(65)90044-4 [DOI] [PubMed] [Google Scholar]
- Xie E., Kotha A., Biaco T., Sedani N., Zou J., Stashenko P., et al. (2015). Oral delivery of a novel recombinant Streptococcus mitis vector elicits robust vaccine antigen-specific Oral Mucosal and systemic antibody responses and T Cell tolerance. PLoS ONE 10:e0143422 10.1371/journal.pone.0143422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Okinaga T., Qi F., Zhang Z., Merritt J. (2011). Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl. Environ. Microbiol. 77 8025–8033. 10.1128/AEM.06362-11 [DOI] [PMC free article] [PubMed] [Google Scholar]