Abstract
Introduction
Obestatin is a controversial gastrointestinal peptide purported to have metabolic actions.
Objectives
This study investigated whether treatment with a stable obestatin analogue (PEG-OB(Cys10, Cys13)) changed plasma metabolite levels firstly in lean and subsequently in diet-induced obesity (DIO) C57BL6/J mice.
Methods
Untargeted LC-HRMS metabolomics experiments were carried out in ESI + mode with plasma extracts from both groups of animals. Data were normalised, multivariate and univariate statistical analysis performed and metabolites of interest putatively identified.
Results
In lean mice, 39 metabolites were significantly changed by obestatin treatment and the majority of these were increased, including various C16 and C18 moieties of phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine and monoacylglycerol, along with vitamin A, vitamin D3, tyrosine, acetylcarnitine and 2α-(hydroxymethyl)-5α-androstane-3β,17β-diol. Decreased concentrations of glycolithocholic acid, 3-dehydroteasterone and various phospholipids were observed. In DIO mice, 25 metabolites were significantly affected and strikingly, the magnitudes of changes here were generally much greater in DIO mice than in lean mice, and in contrast, the majority of metabolite changes were decreases. Four metabolites affected in both groups included glycolithocholic acid, and three different long-chain (C18) phospholipid molecules (phosphatidylethanolamine, platelet activating factor (PAF), and monoacylglycerol). Metabolites exclusively affected in DIO mice included various phosphatidylcholines, lysophosphatidylcholines and fatty acyls, as well as creatine and oxidised glutathione.
Conclusion
This investigation demonstrates that obestatin treatment affects phospholipid turnover and influences lipid homeostasis, whilst providing convincing evidence that obestatin may be acting to ameliorate diet-induced impairments in lipid metabolism, and it may influence steroid, bile acid, PAF and glutathione metabolism.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-016-1063-0) contains supplementary material, which is available to authorized users.
Keywords: Obestatin, Nutrition, Obesity, Diabetes, UPLC–MS, Metabolomics
Introduction
Obestatin is a metabolic hormone recently discovered in rat stomach and initially described to inhibit food intake, decrease intestinal motility and curb body weight gain via the GPR39 G-protein coupled receptor. Its name is derived from two Latin words “obedere” meaning to devour and “statin” meaning to suppress. This 23 amino acid peptide hormone was originally thought to be a direct opponent of ghrelin, a peptide well characterised as an orexigenic hormone which negatively impacts glucose homeostasis; indeed, both obestatin and ghrelin are produced from post-translational modification of the same preproghrelin peptide encoded from the ghrelin gene (Dezaki 2013; Zhang et al. 2005). Primarily produced in the stomach, obestatin is also expressed in pancreatic and adipose tissue, skeletal muscle, liver, lung, thyroid, mammary glands and testes, suggesting it may be multifaceted in its functions and play autocrine/paracrine roles (Gesmundo et al. 2013; Seim et al. 2011). It has been found to act both centrally and peripherally with documented central effects most often relating to influence on food intake. The peptide has also been shown to inhibit thirst, regulate sleep patterns, and to influence anxiety levels and memory function actions which are thought to occur via stimulation of the vagal afferent pathway (Ataka et al. 2008; Carlini et al. 2007; Samson et al. 2007; Szentirmai and Krueger 2006). Peripheral effects have frequently been reported in the pancreas and adipose tissue, with additional actions also documented in the cardiovascular system and skeletal muscle (Agnew et al. 2012; Trovato et al. 2014).
Among the many gut hormones, obestatin is particularly controversial due to a lack of experimental reproducibility in its reported actions, which may in part be attributed to its short biological half-life in vivo (Vergote et al. 2008). Thus far, effects of obestatin on feeding behaviour, body weight and gastrointestinal activity have both been confirmed and refuted in numerous studies. Furthermore, any interaction with the GPR39 receptor has been refuted to the extent that obestatin is now widely considered to be an orphan ligand. Nevertheless, there is growing interest in obestatin’s apparent actions on glucose and lipid metabolism (Gargantini et al. 2013) as well as its ability to promote proliferation and to inhibit apoptosis in pancreatic β-cells and adipocytes (Granata et al. 2008; Tang et al. 2014). For example, there is evidence that obestatin modulates the expression of adipogenic and glucoregulatory genes and that it modulates insulin secretion, adipocyte GLUT-4 translocation, free fatty acid uptake, and glucose uptake and may inhibit lipolysis in white adipose tissue (Favaro et al. 2012; Granata et al. 2008, 2012; Tang et al. 2014). Although some findings are contradictory (Pruszynska-Oszmalek et al. 2013; Ren et al. 2013), it is becoming clear that plasma obestatin levels are disturbed following the development of obesity and diabetes (Seim et al. 2011; Trovato et al. 2014). Indeed, a growing body of evidence suggests that obestatin could play a positive role in obesity and obesity-induced diabetes.
Metabolomics is a systems biology technique used to investigate changes in the abundance of low molecular weight metabolites in tissues and biofluids and has been used to interrogate the mechanisms underlying many diseases and therapies (Bain et al. 2009). Indeed, a number of targeted and untargeted metabolomics studies have been conducted to examine the effect of obesity/diabetes on the metabolome, as well as the influence of clinically approved anti-diabetic therapies (Bao et al. 2009; Duggan et al. 2011; Park et al. 2015). This has provided new insights into the aetiology of obesity and diabetes and has pinpointed specific disturbances in amino acid, lipid, carbohydrate and nucleotide metabolism (Park et al. 2015).
Here, we examined the effects of obestatin treatment on the plasma metabolome. Our first aim was to assess the controversial status of obestatin as a hormone by establishing if 6 weeks of daily treatment affects metabolite levels (thereby indicating if there are modulatory effects on metabolism) in normal mice. Once this was verified the second aim was to then determine whether the observed obestatin-mediated effects on metabolism were substantially different in mice subjected to diet-induced obesity (DIO). Since the reported actions of obestatin are extremely diverse, potentially with more actions remaining undiscovered, this investigation focused on peripheral blood metabolites and employed an untargeted UPLC-MS metabolomics methodology. Importantly, since obestatin is physiologically degraded the treatment involved the use of a novel stable obestatin analogue resistant to degradation.
Materials and methods
Animals and sampling
C57BL6/J male mice were bred in-house, maintained under constant climatic conditions at 21 °C with 12:12 h light–dark cycles, housed either individually or in pairs and given free access to food and water. All experiments were performed in accordance with the Guidance on the Operation of the Animals (Scientific Procedures) Act, 1986 and approved by the Queen’s University Belfast Animal Welfare and Ethical Review Body. Lean mice (n = 12) received a standard laboratory diet, composed of 16.4 % protein, 4.0 % fat and 48.5 % carbohydrate (3.0 kcal/g; Teklad global rodent diet, Envigo, UK) for 23 weeks from birth. For the final 6 weeks animals were injected with either saline (n = 6) or a modified obestatin peptide N-PEGylated(Cys10, Cys13)obestatin (n = 6; PEG-OB(Cys10, Cys13)) (GL Biochem, China) at 50 nmol/kg/day. Injections were administered subcutaneously in the back of the neck at a volume of 10 ml/kg and the final weight range of the animals in each group was 25–33 g (saline) and 26–33 g (PEG-OB(Cys10, Cys13)).
In a separate experiment the effects of PEG-OB(Cys10, Cys13) were investigated in DIO mice (n = 11) which received a standard laboratory diet until 4 weeks of age, thereafter they received a high fat diet (20.5 % protein, 36.0 % fat and 35.7 % carbohydrate (5.49 kcal/g; Mouse Diet, 60 % Fat Calories, BioServ, USA) for 19 weeks. For the final 6 weeks, mice were separated into two groups and injected with either saline (n = 5) or PEG-OB(Cys10, Cys13) (n = 6) at 50 nmol/kg/day as for lean mice. The final weight range of the DIO animals in each group was 39–55 g (saline) and 42–56 g (PEG-OB(Cys10, Cys13)). At the end of the 23 week period, mice were fasted for 5 h and sacrificed by CO2 asphyxiation 3 h after the onset of the light cycle and exactly 1 h after injection on the final day of treatment. Blood obtained by cardiac puncture was collected into pre-chilled heparinised tubes, centrifuged at 3000×g for 10 min (4 °C) and the resulting plasma transferred to W/PTFE lined vials (Supelco, USA) prior to storage at −20 °C for metabolomic analysis.
Sample preparation
Frozen plasma aliquots were thawed on ice and 100 µl was added to 300 µl of ice cold methanol (100 %) in a 2 ml sterile Eppendorf tube. Samples were mixed for 30 s and the protein removed by centrifugation at 13,000×g for 15 min (4 °C). Supernatants were evaporated to dryness, reconstituted in 100 µl of ultra-pure water (Millipore) and filtered by centrifugation using a 0.22 µm Costar spin-X centrifuge tube filter (8000×g at 4 °C for 5 min; Corning Incorporated, Corning, NY 14831, USA).
UPLC-MS analysis
All solvents were purchased from Fisher Scientific (Pittsburg, USA) and were LC-MS grade or equivalent. Chromatography was performed on a Dionex Ultimate 3000 UHPLC system (Dionex, Softron GmbH, Germany) coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). 5 µl of extracted plasma was injected (n = 3 injections for each sample) onto an Acquity UPLC CSH C18 column (2.1 × 100 mm, 1.7 µm, Waters, Wexford, Ireland) operating at 50 °C and applying a binary mobile phase system. The sample manager temperature was maintained at 4 °C and the order in which the samples were injected was randomised throughout the experiment. The gradient elution buffers were A (water with 0.1 % formic acid (vol/vol)) and B (methanol with 0.1 % formic acid (vol/vol)). Solvent B was varied as follows: 0 min 1 %, 2.5 min 1 %, 16 min 99 %, 18 min 99 %, 18.1 min 1 % and 20 min 1 % with a flow rate of 0.4 ml min−1. Positive ionisation mode was employed with these conditions; source heater temperature at 400 °C, sheath gas at 60 (AU), aux gas at 45 (AU) and sweep gas at 1 (AU), capillary temp was maintained at 325 °C and source voltage at 3.5 kV. Mass spectra data were acquired in profile mode over the 50–1200 m/z range with a mass resolution of 60,000 at mass 400 (FWHM) and a scan time of 0.5 s. In further experiments, the samples were subjected to mass fragmentation analysis (FT HCD (10, 30 and 70 NCE), MS2) with an isolation width of 1 Da and 60,000 FWHM at 400 m/z.
Initially the mass spectrometer was calibrated using LTQ Velos ESI Positive Ion Calibration Solution (Thermo Scientific) and a mixture of metabolites (100 µg/ml uridine, nicotinic acid, tryptophan, hippuric acid and phenylalanine; ACROS organics) in water. Prior to sample analysis 10 pooled conditioning samples were injected. To determine chromatographic reproducibility of retention times and peak intensities, pooled samples were injected after every 10 sample injections throughout the experiment (Graham et al. 2013; Want et al. 2010). These pooled samples comprised of plasma from this study and a separate similar sized pharmacological study (C57BL6/J male mice).
Data analysis
UPLC-MS acquired data were analysed using Progenesis QI software (Waters Corporation, Milford, MA) for peak alignment, peak picking and data normalisation. A peak threshold filter of 2.5 AU was applied and peak picking thresholds were set between 0.5 and 20 min. Data were normalised to all compounds by correcting for multiple features to determine a global scaling factor. An output table was subsequently generated to include paired m/z retention times and raw and normalised peak intensities for pools and individual samples. These metabolic features were exported to Simca P v.14 (Umetrics, Umea, Sweden) for multivariate analysis by principal component analysis (PCA) and data quality was assessed via visualisation of clustering of pools and sample replicates. PCA observations were indicative of good platform stability (Supplementary Figure 1). UPLC-MS acquired data for all samples (lean and DIO) were subsequently reanalysed using Progenesis QI as previously described.
On the basis of normalised peak intensities, metabolic features were reduced by exclusion of those observations showing no change in magnitude (fold change = 1) and no significant statistical difference (P > 0.05, students t test). Metabolic features were reduced from 3819 to 736 and 3819 to 615 when PEG-OB(Cys10, Cys13) and saline treated mice were compared within lean and DIO groups, respectively. This was carried out to avoid over-fitting and to improve the model’s predictive ability. Using the average value for each biological replicate the filtered metabolic features were exported to Simca 14 Simca P v.14 (Umetrics, Umea, Sweden) for multivariate analysis. Data were mean centred, pareto scaled and grouped into PEG-OB(Cys10, Cys13) and saline treated groups for analysis using PCA and orthogonal projection to latent structures via partial least squares discriminant analysis (OPLS-DA). The S-plots were used to highlight the ions with the greatest influence on the separation between PEG-OB(Cys10, Cys13) and saline treated animals.
Overall model performance was assessed by R2 indicative of variation described by all components in the model and by Q2, measuring the models ability to predict class membership. The latter is made possible by the sevenfold cross validation default method of the SIMCA software whereby 1/7th of the data from the model is omitted and then predicted for class membership. Further to this, in each case a predictive model was built with the original data from 2/3 of the animals (training set) and used to blindly predict the remaining 1/3 (test set). Overall, the test set observations were assigned to the correct group in 87.5 % of cases. Permutation testing (n = 999) was also carried out to determine if the models were accurate i.e. the model fits the training set well and predicts Y accurately for new observations (Graham et al. 2016).
Discriminatory metabolites as highlighted in the S plot were investigated by using exact mass when searching HMDB (Wishart et al. 2009, 2013; Wishart et al. 2007) (http://www.hmdb.ca/) and the Metlin library (https://metlin.scripps.edu/) online databases. Putative identifications were further examined to increase confidence in identification by mass fragmentation analyses whereby ms/ms spectra were used to search spectral libraries via additional online databases m/z cloud (https://www.mzcloud.org/; http://www.highchem.com/) and MetFrag (http://msbi.ipb-halle.de/MetFrag/).
Results
Multivariate analysis
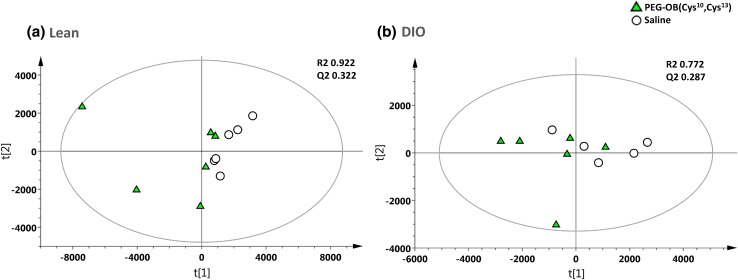
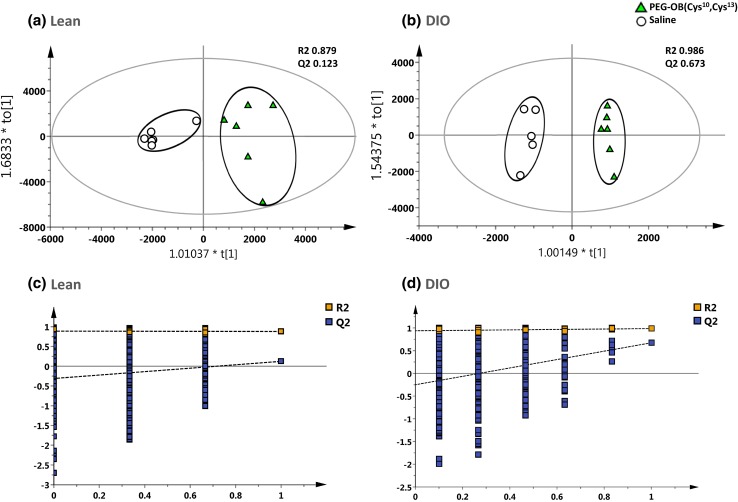
Figure 1 displays the PCA analysis of the raw mass spectral data acquired from PEG-OB(Cys10, Cys13) and saline treated plasma extracts using selected ion features (P < 0.05 and fold change ≠ 1). Figure 1a displays the PCA scores plot for lean mice and Fig. 1b displays the PCA scores plot for DIO mice. Both scores plots represent each group treated daily with PEG-OB(Cys10,Cys13) (triangles) or with saline (circles) for a period of 6 weeks. The PCA scores plots show subtle to moderate separation between respective classes. Subsequently OPLS-DA was used to build predictive models capable of discriminating between PEG-OB(Cys10, Cys13) (triangles) and saline (circles) treated animals (n = 3 replicates per sample) with increased power. Figure 2a and 2b display the OPLS-DA scores plots for PEG-OB(Cys10, Cys13) and saline treated animals in both lean and DIO groups, respectively. The scores plot for each model demonstrates clear separation between the two groups. R2 and Q2 values reflecting the goodness of fit and the predictive abilities of the models were 0.879 and 0.123 (1 orthogonal and 2 latent components) for the comparison in lean animals and 0.986 and 0.673 (1 orthogonal and 3 latent components) for the comparison in DIO animals, respectively. These values signify minimal model variance for both groups, and low to moderate predictive ability for lean and DIO groups respectively. These observations are further highlighted in Fig. 2c and 2d where the permutation plots (n = 999) of the OPLS-DA models are displayed for both sets of data respectively.
Fig. 1.
PCA scores plot displaying the separation for lean mice (a) and DIO mice (b) treated daily with PEG-OB(Cys10,Cys13) (triangles) or with saline (circles) for 6 weeks. Explained variances (R2) were 0.922 and 0.772 and predictive abilities (Q2) were 0.322 and 0.287 for lean and DIO mice respectively
Fig. 2.
OPLS-DA scores plot displaying the separation for lean mice (a) and DIO mice (b) treated daily with PEG-OB(Cys10, Cys13) (triangles) or with saline (circles) for 6 weeks. Explained variances (R2) were 0.879 and 0.986 and predictive abilities (Q2) were 0.123 and 0.673 for lean and DIO mice respectively. Corresponding validation plots for lean mice (c) and DIO mice (d) displaying 999 permutation tests for model built from PEG-OB(Cys10, Cys13) treated animals
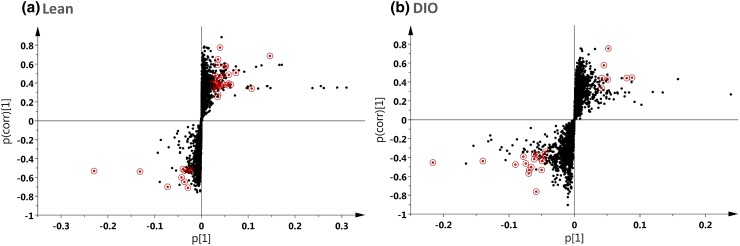
S-plots as demonstrated by Fig. 3a and b were generated to identify the metabolites contributing to the discrimination between PEG-OB(Cys10, Cys13) and saline treated animals for both lean and DIO groups respectively. Metabolites with both higher and lower values for pcorr and p(corr) (1) were those increased and decreased with PEG-OB(Cys10, Cys13) treatment respectively. These were the most significant for differentiating between PEG-OB(Cys10, Cys13) and saline treated animals and are highlighted by circling on the s-plots. Note, that some ions with high pcorr and p(corr) (1) values are not highlighted on the s-plots. These ions had large changes in magnitude when comparing PEG-OB(Cys10, Cys13) and saline treated groups but were not significantly different due to the presence of clear outliers.
Fig. 3.
S-Plot displaying ions of interest (circled) for lean (a) and DIO (b) mice. Ions of interest were selected from 736 and 615 of the original 3819 metabolic features for lean and DIO mice respectively. These 736 and 615 ions demonstrated differences in magnitude (fold change ≠ 1) and significance (P < 0.05) between PEG-OB(Cys10, Cys13) and saline treated animals
Multivariate analysis of technical replicates reported as Supplementary Figures 2 and 3 produced PCA and OPLS-DA scores plots for both data sets which showed good clustering of samples. Furthermore these data mirrored the scores plots acquired using the averaged biological replicates. The models produced using the larger technical replicate data set had greater R2, Q2 and better permutation results using the same ions of interest. These observations confirm the validity of the original model using biological replicates for ion selection and furthermore verify that using a larger sample size would produce better predictive models.
Metabolite profile changes with PEG-OB(Cys10, Cys13) obestatin treatment
Of the original n = 3819 metabolic features, 39 and 25 were tentatively assigned from comparison of PEG-OB(Cys10, Cys13) and saline treated animals in lean and DIO groups, respectively (Tables 1, 2). Data presented for each metabolic feature in Tables 1 and 2 include putative identifications, mass to charge ratio and retention time, empirical formula, adduct, deviation from exact mass, significance value, false discovery rate, percentage increase, and area under the curve (ROC analysis) values. Among the documented putatively identified metabolites, 29 were significantly increased (Table 1a) and 10 significantly decreased (Table 1b) with PEG-OB(Cys10, Cys13) treatment in lean mice, whilst 7 were significantly increased (Table 2a) and 18 were significantly decreased (Table 2b) in DIO mice. All of these metabolites also remained significantly altered after P value adjustment by false discovery rate (FDR), and furthermore, area under the curve values for all metabolites by ROC analysis were >0.6.
Table 1.
(a) Circulating metabolites increased in lean mice treated daily with PEG-OB(Cys10, Cys13) for 6 weeks, (b) Circulating metabolites decreased in lean mice treated daily with PEG-OB(Cys10, Cys13) for 6 weeks
| Putative ID | RT(min)_m/z | Formula | Adduct | Δ ppm | P value | FDR | % change | AUC (ROC) | Identifierd |
|---|---|---|---|---|---|---|---|---|---|
| (a) | |||||||||
| Phospholipids | |||||||||
| PC(16:0/20:4(5Z,8Z,10E,14Z)(12OH[S]))b | 16.75_820.5457 | C44H80NO9P | M + Na | 0 | 5.96E−03 | 1.55E−02 | 135.1 | 0.72 | MID82375 |
| PC(16:0/20:4(6E,8Z,11Z,14Z)(5OH[S]))b | 16.52_798.5638 | C44H80NO9P | M + H | 0 | 1.96E−02 | 3.29E−02 | 187.2 | 0.65 | MID82376 |
| PC(O-16:1(9Z)/2:0)[U]b | 14.91_544.3375 | C26H52NO7P | M + Na | 0 | 4.74E−03 | 1.32E−02 | 8.5 | 0.77 | MID40010 |
| PC(16:1(9Z)/2:0)b | 13.12_536.3323 | C26H50NO8P | M + H | 4 | 2.46E−02 | 3.43E−02 | 123.0 | 0.63 | MID39434 |
| PC(17:0/18:3(9Z,12Z,15Z))b | 16.94_802.5934 | C43H80NO8P | M + CH3OH +H | 2 | 2.78E−02 | 3.74E−02 | 83.5 | 0.71 | MID75787 |
| 1-Palmitoyl-2-epoxystearoylphosphatidylcholineb | 16.72_798.5625 | C42H82NO9P | M + Na | 0 | 1.69E−03 | 6.61E−03 | 78.9 | 0.77 | None |
| LysoPC(20:4(5Z,8Z,11Z,14Z))b | 14.77_544.3381 | C28H50NO7P | M + H | 3 | 2.19E−02 | 3.29E−02 | 10.3 | 0.71 | HMDB10395 |
| PE(20:2(11Z,14Z)/22:5(4Z,7Z,10Z,13Z,16Z))b | 16.99_850.5935 | C47H80NO8P | M + CH3OH +H | 2 | 8.60E−03 | 1.77E−02 | 139.8 | 0.72 | HMDB09307 |
| PE(18:2((9Z,11Z)/18:2(9Z,11Z))[U]b | 16.73_772.5472 | C41H74NO8P | M + CH3OH +H | 1 | 4.79E−02 | 4.79E−02 | 297.7 | 0.66 | MID40429 |
| PE(18:0/22:5(4Z,7Z,10Z,13Z,16Z))b | 17.06_826.5934 | C45H80NO8P | M + CH3OH +H | 2 | 7.07E−03 | 1.62E−02 | 117.2 | 0.76 | HMDB09010 |
| PE(18:0/20:4(5Z,8Z,11Z,14Z))b | 16.73_800.5777 | C43H78NO8P | M + CH3OH +H | 2 | 2.14E−02 | 3.29E−02 | 58.4 | 0.76 | HMDB09003 |
| PS(P-18:0/18:1(9Z))b | 16.68_774.5631 | C42H80NO9P | M + H | 1 | 1.03E−02 | 2.01E−02 | 62.1 | 0.73 | MID78831 |
| PS(18:0/22:4(7Z,10Z,13Z,16Z))b | 16.70_822.5625 | C46H82NO10P | M + H-H2O | 2 | 7.46E−03 | 1.62E−02 | 74.6 | 0.73 | MID78547 |
| Monoacylglycerols | |||||||||
| MG(18:1(9Z)/0:0/0:0)b | 15.80_339.2911 | C21H40O4 | M + H-H2O | 3 | 3.83E−03 | 1.24E−02 | 42.4 | 0.76 | HMDB11567 |
| MG(0:0/16:0/0:0)b | 15.68_313.2750 | C19H38O4 | M + H-H2O | 2 | 7.00E−03 | 1.62E−02 | 27.5 | 0.78 | HMDB11533 |
| Vitamins | |||||||||
| Vitamin D3b | 16.36_367.3364 | C27H44O | M + H-H2O | 0 | 3.93E−02 | 4.72E−02 | 142.3 | 0.64 | HMDB00876 |
| 25-Hydroxyvitamin-D3b | 16.46_401.3413 | C27H44O2 | M + H | 0 | 4.67E−02 | 4.79E−02 | 271.1 | 0.58 | HMDB05997 |
| Vitamin D3 derivativec | 16.04_521.4201 | C31H52O4 | M + CH3OH +H | 0 | 6.74E−04 | 3.76E−03 | 54.0 | 0.84 | Several |
| 9(cis)-retinal (Vitamin A)a | 14.33_285.2207 | C20H28O | M + H | 2 | 4.69E−02 | 4.79E−02 | 112.3 | 0.68 | HMDB06218 |
| Fatty acids | |||||||||
| α-Linolenic acidb | 14.05_279.2312 | C18H30O2 | M + H | 2 | 4.41E−02 | 4.79E−02 | 22.2 | 0.68 | HMDB01388 |
| 20-Hydroxyeicosatetraenoic acidb | 14.33_343.2243 | C20H32O3 | M + Na | 0 | 3.21E−02 | 4.18E−02 | 133.7 | 0.66 | HMDB05998 |
| 5,8,11-Eicosatriynoic acidb | 13.89_301.2156 | C20H28O2 | M + H | 1 | 3.76E−02 | 4.72E−02 | 144.5 | 0.67 | MID35294 |
| 5Z,8Z,11Z,14Z,17Z-Eicosapentaenoic acidb | 14.33_303.2314 | C20H30O2 | M + H | 1 | 4.24E−02 | 4.79E−02 | 108.8 | 0.67 | HMDB01999 |
| Amino acid, carnitine, steroid and unknowns | |||||||||
| Tyrosineb | 1.30_182.0809 | C9H11NO3 | M + H | 1 | 1.17E−08 | 4.55E−07 | 22.8 | 0.98 | HMDB00158 |
| Acetylcarnitinea | 0.95_204.1221 | C9H17NO4 | M + H | 4 | 4.49E−05 | 6.72E−04 | 15.6 | 0.85 | HMDB00201 |
| 2α-(Hydroxymethyl)-5α-androstane-3β,17β-diolb | 15.43_355.2843 | C20H34O3 | M + CH3OH +H | 0 | 4.49E−02 | 4.79E−02 | 34.9 | 0.75 | MID70469 |
| None | 10.19_394.2219 | N/A | N/A | N/A | 2.31E−02 | 3.34E−02 | 53.8 | 0.73 | N/A |
| None | 15.68_331.2864 | N/A | N/A | N/A | 1.35E−02 | 2.50E−02 | 19.3 | 0.73 | N/A |
| None | 15.80_357.3018 | N/A | N/A | N/A | 3.66E−03 | 1.24E−02 | 40.3 | 0.71 | N/A |
| (b) | |||||||||
| Phospholipids | |||||||||
| LysoPC(22:0)b | 16.70_580.4318 | C30H62NO7P | M + H | 3 | 1.07E−03 | 5.24E−03 | −22.3 | 0.79 | HMDB10398 |
| PC(18:1(9Z)/20:4(5Z,8Z,11Z,14Z))b | 17.29_830.5675 | C46H82NO8P | M + Na | 0 | 3.99E−02 | 4.72E−02 | −11.6 | 0.71 | HMDB08114 |
| PAF C-18b | 16.32_552.401 | C28H58NO7P | M + H | 2 | 2.20E−02 | 3.29E−02 | −18.9 | 0.71 | MID43410 |
| LysoPC or PCc | 17.03_608.4627 | C32H66NO7P | M + H | 3 | 4.51E−03 | 1.32E−02 | −15.7 | 0.81 | Several |
| LysoPE(0:0/20:3(5Z,8Z,11Z))b | 14.93_526.2902 | C25H46NO7P | M + Na | 0 | 1.38E−04 | 1.34E−03 | −34.4 | 0.77 | HMDB11485 |
| PE(18:2(9Z,12Z)/0:0)b | 14.54_500.2756 | C23H44NO7P | M + Na | 1 | 5.17E−05 | 6.72E−04 | −42.1 | 0.89 | MID7767 |
| PE(18:1(9Z)/0:0)b | 14.80_502.2914 | C23H46NO7P | M + Na | 2 | 2.00E−02 | 3.29E−02 | −22.4 | 0.65 | MID40778 |
| Sterol lipids and unknown | |||||||||
| Glycolithocholic acidb | 14.81_478.2915 | C26H43NO4 | M + 2Na-H | 2 | 3.89E−04 | 3.04E−03 | −25.6 | 0.83 | HMDB00698 |
| 3-Dehydroteasteroneb | 16.86_447.3469 | C28H46O4 | M + H | 0 | 6.58E−04 | 3.76E−03 | −17.9 | 0.82 | HMDB41527 |
| None | 11.82_616.1743 | N/A | N/A | N/A | 1.48E−03 | 6.42E−03 | −31.7 | 0.83 | N/A |
aSpectral library identification(m/z cloud)
bIn silico identification(MetFrag)
cPrecursor mass identification(hmdb/Metlin)
dhmdb(HMDB)/Metlin(MID) identifier code
Table 2.
(a) Circulating metabolites increased in DIO mice treated daily with PEG-OB(Cys10, Cys13) for 6 weeks, (b) circulating metabolites decreased in DIO mice treated daily with PEG-OB(Cys10, Cys13) for 6 weeks
| Putative ID | RT(min)_m/z | Formula | Adduct | Δ ppm | P value | FDR | % change | AUC (ROC) | Identifierd |
|---|---|---|---|---|---|---|---|---|---|
| (a) | |||||||||
| Amino acid and antioxidant related metabolites | |||||||||
| Creatinea | 0.65_132.0763 | C4H9N3O2 | M + H | 3 | 4.72E−06 | 1.18E−04 | 26.2 | 0.94 | HMDB00064 |
| Oxidised glutathioneb | 1.61_307.0828 | C20H32N6O12S2 | M + 2H | 1 | 4.59E−02 | 4.78E−02 | 622.3 | 0.73 | HMDB03337 |
| Unknowns | |||||||||
| None | 7.22_770.3435 | None | N/A | N/A | 1.67E−02 | 2.69E−02 | 761.8 | 0.86 | N/A |
| None | 6.17_599.6132 | None | N/A | N/A | 1.27E−02 | 2.29E−02 | 475.6 | 0.79 | N/A |
| None | 7.88_163.1329 | None | N/A | N/A | 9.83E−04 | 6.14E−03 | 39.1 | 0.74 | N/A |
| None | 7.08_820.8670 | None | N/A | N/A | 1.23E−02 | 2.29E−02 | 977.5 | 0.80 | N/A |
| None | 6.27_565.9308 | None | N/A | N/A | 1.22E−02 | 2.29E−02 | 427.2 | 0.75 | N/A |
| (b) | |||||||||
| Phospholipids | |||||||||
| LysoPC(20:2(11Z,14Z))b | 15.28_570.3528 | C28H54NO7P | M + Na | 0 | 2.96E−02 | 3.45E−02 | −17.5 | 0.75 | HMDB10392 |
| LysoPC, LysoPE, PC or PEc | 15.56_510.3558 | C25H52NO7P | M + H | 0 | 1.79E−02 | 2.69E−02 | −6.1 | 0.76 | Several |
| PAF C-18b | 16.32_552.401 | C28H58NO7P | M + H | 2 | 1.10E−02 | 2.29E−02 | −20.3 | 0.76 | MID43410 |
| PE(18:1(9Z)/0:0)b | 15.42_480.3072 | C23H46NO7P | M + H | 2 | 3.64E−02 | 3.96E−02 | −12.2 | 0.67 | MID40778 |
| LysoPE(0:0/20:2(11Z,14Z))b | 15.30_528.3059 | C25H48NO7P | M + Na | 0 | 1.25E−03 | 6.27E−03 | −32.1 | 0.82 | HMDB11483 |
| Fatty acid and fatty acyls | |||||||||
| 7,10,13,16- Docosatetraenoic acidb | 16.16_350.3062 | C22H36O2 | M + NH4 | 2 | 1.94E−02 | 2.69E−02 | −13.6 | 0.74 | HMDB02226 |
| 1-(O-α-d-glucopyranosyl)-29-keto-(1,3R,31R)-dotriacontanetriolb | 17.11_675.5415 | C38H74O9 | M + H | 1 | 1.28E−02 | 2.29E−02 | −15.2 | 0.73 | MID46604 |
| N-oleoyl threoninec | 15.98_348.2916 | C22H41NO4 | M + H-2H2O | 2 | 7.76E−04 | 6.14E−03 | −17.8 | 0.85 | MID75490 |
| Fatty acid, fatty acid ester, fatty alcohol or steroid esterc | 16.15_333.28 | C22H36O2 | M + H | 3 | 1.84E−02 | 2.69E−02 | −18.8 | 0.75 | Several |
| Monoacylglycerol, bile acid and unknowns | |||||||||
| MG(18:1(9Z)/0:0/0:0)b | 15.80_339.2911 | C21H40O4 | M + H-H2O | 3 | 2.55E−02 | 3.35E−02 | −15.2 | 0.74 | HMDB11567 |
| Glycolithocholic acidb | 14.81_478.2915 | C26H43NO4 | M + 2Na-H | 2 | 9.69E−03 | 2.29E−02 | −29.1 | 0.74 | HMDB00698 |
| None | 0.91_169.0355 | N/A | N/A | N/A | 8.53E−03 | 2.29E−02 | −12.4 | 0.78 | N/A |
| None | 11.64_244.1325 | N/A | N/A | N/A | 3.04E−02 | 3.45E−02 | −98.6 | 0.69 | N/A |
| None | 10.02_530.8273 | N/A | N/A | N/A | 3.86E−05 | 4.82E−04 | −33.9 | 0.89 | N/A |
| None | 15.80_357.3018 | N/A | N/A | N/A | 2.73E−02 | 3.42E−02 | −14.6 | 0.73 | N/A |
| None | 16.07_374.3067 | N/A | N/A | N/A | 2.29E−03 | 8.18E−03 | −28.6 | 0.82 | N/A |
| None | 7.58_449.2146 | N/A | N/A | N/A | 4.88E−02 | 4.88E−02 | −33.3 | 0.70 | N/A |
| None | 15.98_331.2653 | N/A | N/A | N/A | 2.13E−03 | 8.18E−03 | −23.9 | 0.81 | N/A |
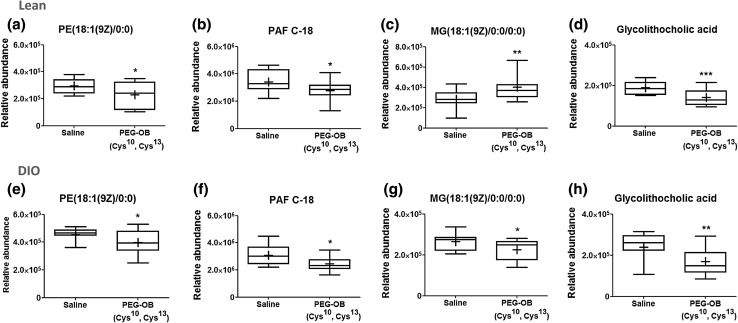
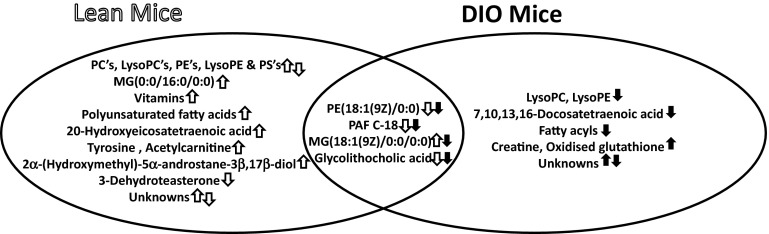
Across both groups, 3 metabolite identifications were assigned based on fragmentation pattern match with high resolution spectral library (m/z cloud) (available as online resources 4–6), and 40 metabolite identifications were putatively assigned based on in silico fragmentation match. Among those assigned, four were detected as altered between PEG-OB(Cys10, Cys13) and saline treated animals in both lean and DIO groups. Of these, two phospholipids (PE(18:1(9Z)/0:0) and PAF C-18), and a secondary bile acid (glycolithocholic acid) were decreased with treatment in both groups. A monoacylglycerol (MG(18:1(9Z)/0:0/0:0)) was also increased in lean mice and decreased in DIO mice (Fig. 4). When comparing PEG-OB(Cys10, Cys13) and saline treated lean mice, further metabolites were found to be present at increased levels and these comprised phospholipids (phosphatidylcholines (PC’s), a lysophosphatidylcholine (LysoPC), phosphatidylethanolamines(PE’s), and phosphatidylserines(PS’s)), monoacylglycerols, polyunsaturated fatty acids (α linolenic acid, arachidonic acid and metabolite) and vitamins (A, D and D derivatives). Moreover, tyrosine, acetylcarnitine and 2α-(hydroxymethyl)-5α-androstane-3β,17β-diol were also found to be elevated along with three unidentified features. Additional metabolites found to be at lower levels in lean mice were phospholipids (PC’s, PE’s, lysoPC’s and a lysoPE), 3-dehydroteasterone and one unidentified feature. PEG-OB(Cys10, Cys13) treated DIO animals showed increased levels of creatine, oxidised glutathione and five unidentified features when compared to saline treated controls. Conversely we identified more metabolites at lower levels in DIO treated animals. These consisted of phospholipids (PC’s, PE’s, lysoPC’s and lysoPE’s), as well as 1,7,10,13,16-docosatetraenoic acid, 1-(O-alpha-d-glucopyranosyl)-29-keto-(1,3R,31R)-dotriacontanetriol, N-oleoyl threonine, another metabolite of lipid class (a fatty acid, fatty acid ester, fatty alcohol or steroid ester by exact mass search in metlin/hmdb databases) and seven unidentified features (Fig. 5).
Fig. 4.
Box-and-whisker plots show the relative abundance of 4 circulating metabolites which were commonly affected by PEG-OB(Cys10,Cys13) treatment in lean mice (a–d) and DIO mice (e–h). Metabolites are a, e PE(18:1(9Z)/0:0), b, f PAF C-18, c, g MG(18:1(9Z)/0:0/0:0) and d, h glycolithocholic acid. Whiskers indicate min and max, bottom and top of boxes indicate 25th and 75th percentiles, and line through centre of each box and ‘+’ within box represent median and mean, respectively. P values were calculated by the Student’s t test with ***P < 0.001; **P < 0.01 and *P < 0.05 indicating significant differences between PEG-OB(Cys10,Cys13) treatment and saline
Fig. 5.
Venn diagram shows metabolites/metabolite classes increased and/or decreased in lean and DIO mice treated with PEG-OB(Cys10,Cys13). Directions of change are represented by opened and closed arrows for lean and DIO animals respectively
Discussion
A variety of previous studies have provided evidence suggesting that obestatin has effects on metabolism. Whilst many have focused on effects on food intake and body weight, actions on glucose and lipid metabolism have also been investigated both under normal physiological conditions and in experimental models of obesity or diabetes (Agnew et al. 2011; Egido et al. 2009; Granata et al. 2008, 2010, 2012; Green et al. 2007; Nagaraj et al. 2014). In order to build on these data and to provide a detailed assessment of the metabolic actions of obestatin, the current study employed untargeted metabolomic analysis to assess whether obestatin influences the levels of circulating small molecule metabolites. In lean mice, treatment with the obestatin analogue was found to significantly affect 39 different metabolites, a finding which served as the basis for a follow-on investigation in DIO mice to understand if this peptide’s activity was dependent upon nutritional status. Importantly, the experimental protocol adopted for these lean and DIO studies was identical with the exception that DIO mice received a high fat (HF) diet for 13 weeks before commencing obestatin treatment at which time they displayed a mean bodyweight of 46 % higher than lean mice. To our knowledge, this is the first metabolomic investigation focussing on the actions of obestatin. Importantly, we employed a peptide analogue of obestatin (PEG-OB(Cys10, Cys13)) which demonstrates enhanced stability and preserved biological activity (Burch et al. 2015; Cowan et al. 2015). The results clearly show that obestatin treatment alters metabolite profiles in both lean and DIO mice. OPLS-DA analysis demonstrated clear separation of mice treated with PEG-OB(Cys10,Cys13) from saline controls (Fig. 2a, b) and this led to the shortlisting and identification of metabolite ions which were significantly altered (Tables 1, 2). It was immediately evident that lean and DIO mice responded differently to obestatin treatment. Of the 39 and 25 metabolites significantly modulated in lean and DIO mice, respectively, there were only 4 which were commonly affected in both groups. Of particular note, the majority of affected metabolites were increased in lean mice, whilst in contrast in DIO mice the majority of metabolite changes were decreased. Furthermore, regardless of whether metabolites were increased or decreased the overall magnitude of the responses was clearly greater in DIO mice (≤978 %) compared with lean mice (≤298 %). Interestingly, a large proportion of metabolites altered in DIO mice could not be putatively identified potentially suggesting the involvement of lesser known or uncharacterised biochemical pathways.
The four metabolites commonly affected in both lean and DIO mice included a phosphatidylethanolamine moiety (PE(18:1(9Z)/0:0)), a platelet activating factor (PAF C-18), a monoacylglycerol (MG(18:1(9Z)/0:0/0:0)) and a secondary bile acid (glycolithocholic acid). This clearly demonstrates a role for obestatin in lipid metabolism and suggests that some actions may be independent of nutritional status. These 4 metabolites were all significantly decreased in DIO mice (12–29 %) by PEG-OB(Cys10,Cys13) treatment, whilst in lean mice three were also decreased although one (MG(18:1(9Z)/0:0/0:0)) was elevated. Three of the above metabolites are biochemically similar to one another all being phospholipid molecules with a single 18 carbon chain. Interestingly, long chain phospholipids are negatively associated with obesity and diabetes and PAF is an inflammatory mediator which acts in part to promote cytokine production (Giesbertz et al. 2015; Li and Mitra 1996; Menezes-Garcia et al. 2014). Indeed, PAF receptor deficient mice fed a high carbohydrate diet have been shown to display reduced adipose tissue inflammation and improved glucose homeostasis (Menezes-Garcia et al. 2014), whilst other studies have reported increases in bile acid levels with HF feeding for relatively short periods (Sayin et al. 2013; Sun et al. 2015). Therefore, a key function of obestatin may be to control inflammation and to preserve healthy lipid and cholesterol metabolism both under normal physiological circumstances and also under situations of DIO/diabetes, and such actions could be mediated via the observed reductions in these particular metabolites. In support of this suggestion, obestatin has been shown to reduce obesity-induced increases in inflammatory cytokines, to decrease insulin sensitivity and to decrease triglyceride levels. Furthermore, it has also been shown to decrease the expression of cholesterol transporter ATP-binding cassette A1 (ABCA1) in bovine white adipose tissue (A. Agnew et al. 2011; Grala et al. 2010; Granata et al. 2012; Hotamisligil 2006).
Notably, the majority of circulating metabolites affected by PEG-OB(Cys10,Cys13) treatment in lean mice were phospholipids. Most of these were increased and most were species of PC, LysoPC, PE or PS, although there were also some instances of decreased PC, LysoPC, PE or LysoPE phospholipid species. This suggests that obestatin influences the turnover of phospholipids. Indeed, many plasma phospholipids originate from lipoproteins (Miller et al. 2011), and therefore obestatin’s ability to regulate phospholipid levels could be linked to its ability to regulate HDL, LDL and VLDL levels (Catak et al. 2014; Nagaraj et al. 2008, 2009). Other metabolites which were increased with PEG-OB(Cys10,Cys13) treatment in lean mice included monoacylglycerols, polyunsaturated fatty acids, vitamin A and D forms, an amino acid (tyrosine), an acylcarnitine (acetylcarnitine), a steroid (2α-(hydroxymethyl)-5α-androstane-3β,17β-diol) and some unknown metabolites. As acylcarnitines facilitate transport of fatty acids into the mitochondria for β-oxidation, an increase in acetylcarnitine could reflect decreased mitochondrial utilisation (Jüllig et al. 2007). Obestatin may therefore promote glucose utilisation over fatty acid utilisation for energy production when fatty acids are present under normal physiological conditions. This assertion is based on the observed increases in the circulating levels of polyunsaturated fatty acids and monoacylglycerols in PEG-OB(Cys10,Cys13) treated animals. It may even reflect a tendency for obestatin to safeguard against potential hyperglycaemia. Interestingly, physiological levels of polyunsaturated fatty acids have been associated with decreased and increased proinflammatory and anti-inflammatory markers, respectively (Ferrucci et al. 2006). Therefore, slightly elevated levels observed in lean mice treated with PEG-OB(Cys10, Cys13) further corroborate a role for obestatin in limiting inflammation. In addition, the observed increases in 2α-(hydroxymethyl)-5α-androstane-3β,17β-diol, vitamin D3 and vitamin A metabolites could also be viewed as beneficial. 2α-(hydroxymethyl)-5α-androstane-3β,17β-diol is a metabolite of dehydroepiandrosterone (DHEA) and dihydrotestosterone (DHT) and both insulin resistance and obesity have been associated with low androgen and vitamin D levels (Navarro et al. 2015; Pereira-Santos et al. 2015). Furthermore, vitamin A is known to improve insulin sensitivity and has been suggested essential for maintenance of pancreatic β-cell function and mass (Jeyakumar et al. 2011; Trasino et al. 2015).
Numerous studies have associated increased levels of the amino acid tyrosine with diabetes and obesity, although a few have reported decreased levels in this setting (Mihalik et al. 2012; Tranchida et al. 2015; X. Zhang et al. 2009). Furthermore, 3-dehydroteasterone is a sterol lipid of dietary source and a decreased level of this metabolite with PEG-OB(Cys10,Cys13) treatment in animals maintained on the same diet suggests that obestatin may inhibit its uptake from the gut.
Of the seven metabolites increased with PEG-OB(Cys10,Cys13) treatment in DIO mice only two could be putatively identified: creatine and oxidised glutathione (GSSG). GSSG is the oxidised form of the antioxidant reduced glutathione (GSH), and increased GSSG and GSSG/GSH ratio are considered to be indicative of oxidative stress and are associated with obesity and diabetes (Savini et al. 2013). However, it is not clear what effect PEG-OB(Cys10, Cys13) treatment has on the glutathione system, when the observed increase in GSSG may also be explained by sample storage conditions. Indeed, GSH is known to degrade at a much faster rate than GSSG, and is also susceptible to autoxidation (Lin et al. 2006); therefore, GSSG measured here may not reflect true plasma GSSG levels.
Increased plasma creatine could suggest decreased creatine uptake by skeletal muscle, and interestingly, skeletal muscle absorption of creatine is believed to be promoted by insulin (Steenge et al. 1998). The increased creatine levels observed here may thus reflect decreased insulin production with PEG-OB(Cys10, Cys13) treatment. Furthermore, hyperinsulinemia is one of the hallmarks of insulin resistance and thus obestatin could play a protective role against the development of insulin resistance. Indeed, supportive evidence for this suggestion is provided by a report that creatine levels decrease whilst glucose and insulin levels increase in HF fed mice (Duggan et al. 2011).
In addition to those already discussed, further metabolites found to decrease with PEG-OB(Cys10, Cys13) treatment in DIO mice included polyunsaturated fatty acids and fatty acyls. Significantly increased levels of fatty acids are well documented in obesity and diabetes, and so reduced polyunsaturated fatty acids and fatty acyls with PEG-OB(Cys10, Cys13) treatment in DIO animals suggests that obestatin has the potential to protect against lipid dysregulation under these conditions (Lopaschuk et al. 2010). Such protective effects of obestatin could involve decreasing adipose tissue lipolysis (Granata et al. 2012) and/or increasing fatty acid clearance by β-oxidation.
Altered levels of plasma lipoproteins (the source of phospholipids) in plasma are linked to inflammation, β-cell dysfunction, obesity and diabetes (Anderson et al. 2014; Miller et al. 2011; Roehrich et al. 2003). LysoPC’s are major components of oxidized low density lipoproteins (Ox-LDL), and not surprisingly increased levels have been correlated with endothelial dysfunction and atherogenesis, as well as inflammation and oxidative stress (Aiyar et al. 2007; Galili et al. 2007; Schmitz and Ruebsaamen 2010). Furthermore, phospholipids have been reported to be susceptible to glycoxidation and are suggested to be a potential source of oxidative damage to tissues (Miyazawa et al. 2008; Podrez 2002). The ability of PEG-OB(Cys10, Cys13) treatment to reduce high phospholipid levels in plasma of DIO animals therefore suggests that obestatin may be able to ameliorate lipoprotein/phospholipid mediated proinflammatory stress associated with complications of obesity and/or diabetes.
Interestingly, as previously discussed, longer rather than shorter chain phospholipids have been negatively associated with obesity and diabetes, and the hypoglycaemic effect of insulin has been shown to improve when phospholipids of reducing carbon numbers from C18 to C10 are involved (Giesbertz et al. 2015; Li and Mitra 1996). Therefore, reduction of phospholipids with carbon lengths of ≥18 as observed here in DIO mice treated with PEG-OB(Cys10, Cys13) suggests that obestatin may be promoting insulin absorption/sensitivity by reducing the ratio of longer to shorter carbon chain phospholipids.
Concluding remarks
In summary, this UPLC-MS metabolomic investigation has elucidated a number of plasma metabolite changes which occur as a consequence of obestatin treatment. In lean mice 39 metabolites were significantly affected, strongly suggesting that obestatin modulates metabolism and thereby appearing to justify its proposed status as a hormone.
The fact that there were 4 metabolites commonly affected in DIO and lean mice demonstrates that there is at least some consistency in their responses to obestatin. However, there were a further 21 unique metabolite responses in DIO mice, clearly showing that nutritional status has a bearing on obestatin’s mode of action. The metabolite changes observed here suggest that this peptide influences lipid metabolism and may also play an anti-inflammatory role. Furthermore, some of the observed metabolite changes may underlie the previously reported beneficial actions of obestatin on insulin resistance, hyperglycaemia and lipid dysregulation. Together, the findings of this study clearly highlight the potential value of metabolomics to delineate the metabolic actions of poorly characterised physiological hormones.
Electronic supplementary material
Below is the link to the electronic supplementary material.
PCA scores plot displaying pools (red triangles), samples from lean and DIO mice (black triangles) and samples from another study (grey triangles; data not presented)Supplementary material 1 (PDF 237 kb)
PCA scores plot displaying the separation for lean mice (a) and DIO mice (b) treated daily with PEG-OB(Cys10,Cys13) (triangles) or with saline (circles) for 6 weeks. Explained variances (R2) were 0.978 and 0.854 and predictive abilities (Q2) were 0.861 and 0.742 for lean and DIO mice, respectively (PDF 164 kb)
OPLS-DA scores plot displaying the separation for lean mice (a) and DIO mice (b) treated daily with PEG-OB(Cys10, Cys13) (triangles) or with saline (circles) for 6 weeks. Explained variances (R2) were 0.983 and 0.969 and predictive abilities (Q2) were 0.948 and 0.932 for lean and DIO mice respectively. Corresponding validation plots for lean mice (c) and DIO mice (d) displaying 999 permutation tests for model built from PEG-OB(Cys10,Cys13) treated animals (PDF 249 kb)
Detection and identification of creatine. MS2 mass spectrum obtained at 0.15–1.15 min (a), corresponding MS2 peak at 0.15–1.15 min (b) and high resolution spectral library match in m/z cloud (reference no 357)(c) (PDF 189 kb)
Detection and identification of acetylcarnitine. MS2 mass spectrum obtained at 0.45–1.44 min (a), corresponding MS2 peak at 0.45–1.44 min (b), and high resolution spectral library match in m/z cloud (reference no 879)(c) (PDF 187 kb)
Detection and identification of 9-cis retinal. MS2 mass spectrum obtained at 13.83–14.82 min (a), corresponding MS2 peak at 13.83–14.82 min (b), and high resolution spectral library match in m/z cloud (reference no 836)(c) (PDF 191 kb)
Acknowledgments
This research was supported by funding from the British Heart Foundation (BHF, FS/12/47/29703) and the Department for Employment and Learning Northern Ireland (DELNI), travel scholarships from the Translational Research Group (TRG) in Northern Ireland and Queens University, and specific funds allocated by Dr. Stewart Graham at Beaumont Health, Royal Oak, MI.
Compliance with ethical standards
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical standard
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
Footnotes
Brian D. Green and Stewart F. Graham have contributed equally to this work.
References
- Agnew A, Calderwood D, Chevallier OP, Greer B, Grieve DJ, Green BD. Chronic treatment with a stable obestatin analog significantly alters plasma triglyceride levels but fails to influence food intake; fluid intake; body weight; or body composition in rats. Peptides. 2011;32(4):755–762. doi: 10.1016/j.peptides.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Agnew AJ, Robinson E, McVicar CM, Harvey AP, Ali IH, Lindsay JE, et al. The gastrointestinal peptide obestatin induces vascular relaxation via specific activation of endothelium-dependent NO signalling. British Journal of Pharmacology. 2012;166(1):327–338. doi: 10.1111/j.1476-5381.2011.01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar N, Disa J, Ao Z, Ju H, Nerurkar S, Willette RN, et al. Lysophosphatidylcholine induces inflammatory activation of human coronary artery smooth muscle cells. Molecular and Cellular Biochemistry. 2007;295(1–2):113–120. doi: 10.1007/s11010-006-9280-x. [DOI] [PubMed] [Google Scholar]
- Anderson SG, Dunn WB, Banerjee M, Brown M, Broadhurst DI, Goodacre R, et al. Evidence that multiple defects in lipid regulation occur before hyperglycemia during the prodrome of type-2 diabetes. PLoS ONE. 2014;9(9):e103217. doi: 10.1371/journal.pone.0103217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataka K, Inui A, Asakawa A, Kato I, Fujimiya M. Obestatin inhibits motor activity in the antrum and duodenum in the fed state of conscious rats. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2008;294(5):G1210–G1218. doi: 10.1152/ajpgi.00549.2007. [DOI] [PubMed] [Google Scholar]
- Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research: Moving from information to knowledge. Diabetes. 2009;58(11):2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Zhao T, Wang X, Qiu Y, Su M, Jia W. Metabonomic variations in the drug-treated type 2 diabetes mellitus patients and healthy volunteers research articles. Journal of Proteome Research. 2009;8:1623–1630. doi: 10.1021/pr800643w. [DOI] [PubMed] [Google Scholar]
- Burch KJ, Cowan E, Green BD, Grieve DJ. A novel obestatin analogue improves metabolic and endothelial function in a diet-induced model of pre-diabetes. Journal of Vascular Research. 2015;52(Suppl 1):1–88. [Google Scholar]
- Carlini VP, Schiöth HB, Debarioglio SR. Obestatin improves memory performance and causes anxiolytic effects in rats. Biochemical and Biophysical Research Communications. 2007;352(4):907–912. doi: 10.1016/j.bbrc.2006.11.112. [DOI] [PubMed] [Google Scholar]
- Catak Z, Aydin S, Sahin İ, Kuloglu T, Aksoy A, Dagli AF. Regulatory neuropeptides (ghrelin, obestatin and nesfatin-1) levels in serum and reproductive tissues of female and male rats with fructose-induced metabolic syndrome. Neuropeptides. 2014;48(3):167–177. doi: 10.1016/j.npep.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Cowan E, Burch KJ, Greer B, McClune D, Scantlebury M, Grieve DJ, Green BD. Chronic treatment with a stable obestatin analogue curbs insulin desensitisation in high fat fed mice. Diabetic Medicine. 2015;32(Suppl. 1):30–206. [Google Scholar]
- Dezaki K. Ghrelin function in insulin release and glucose metabolism. Endocrine Development. 2013;25:135–143. doi: 10.1159/000346064. [DOI] [PubMed] [Google Scholar]
- Duggan GE, Hittel DS, Sensen CW, Weljie AM, Vogel HJ, Shearer J. Metabolomic response to exercise training in lean and diet-induced obese mice. Journal of Applied Physiology. 2011;110(5):1311–1318. doi: 10.1152/japplphysiol.00701.2010. [DOI] [PubMed] [Google Scholar]
- Egido EM, Hernández R, Marco J, Silvestre RA. Effect of obestatin on insulin, glucagon and somatostatin secretion in the perfused rat pancreas. Regulatory Peptides. 2009;152(1–3):61–66. doi: 10.1016/j.regpep.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Favaro E, Granata R, Miceli I, Baragli A, Settanni F, Cavallo Perin P, et al. The ghrelin gene products and exendin-4 promote survival of human pancreatic islet endothelial cells in hyperglycaemic conditions, through phosphoinositide 3-kinase/Akt, extracellular signal-related kinase (ERK)1/2 and cAMP/protein kinase A (PKA) signalling pathways. Diabetologia. 2012;55(4):1058–1070. doi: 10.1007/s00125-011-2423-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L, Cherubini A, Bandinelli S, Bartali B, Corsi A, Lauretani F, et al. Relationship of plasma polyunsaturated fatty acids to circulating inflammatory markers. The Journal of Clinical Endocrinology & Metabolism. 2006;91(2):439–446. doi: 10.1210/jc.2005-1303. [DOI] [PubMed] [Google Scholar]
- Galili O, Versari D, Sattler KJK, Olson ML, Mannheim D, McConnell JP, et al. Early experimental obesity is associated with coronary endothelial dysfunction and oxidative stress. American Journal of Physiology-Heart and circulatory physiology. 2007;292:H904–H911. doi: 10.1152/ajpheart.00628.2006. [DOI] [PubMed] [Google Scholar]
- Gargantini E, Grande C, Trovato L, Ghigo E, Granata R. The role of obestatin in glucose and lipid metabolism. Hormone and Metabolic Research. 2013;45(13):1002–1008. doi: 10.1055/s-0033-1351325. [DOI] [PubMed] [Google Scholar]
- Gesmundo I, Gallo D, Favaro E, Ghigo E, Granata R. Obestatin: A new metabolic player in the pancreas and white adipose tissue. IUBMB Life. 2013;65(12):976–982. doi: 10.1002/iub.1226. [DOI] [PubMed] [Google Scholar]
- Giesbertz P, Padberg I, Rein D, Ecker J, Höfle AS, Spanier B, Daniel H. Metabolite profiling in plasma and tissues of ob/ob and db/db mice identifies novel markers of obesity and type 2 diabetes. Diabetologia. 2015 doi: 10.1007/s00125-015-3656-y. [DOI] [PubMed] [Google Scholar]
- Graham SF, Chevallier OP, Kumar P, Türkoğlu O, Bahado-Singh RO. High resolution metabolomic analysis of ASD human brain uncovers novel biomarkers of disease. Metabolomics. 2016;12(4):62. doi: 10.1007/s11306-016-0986-9. [DOI] [Google Scholar]
- Graham SF, Chevallier OP, Roberts D, Ho C, Elliott CT, Green BD. Investigation of the human brain metabolome to identify potential markers for early diagnosis and therapeutic targets of Alzheimer’s disease. Analytical Chemistry. 2013;85(1):1803–1811. doi: 10.1021/ac303163f. [DOI] [PubMed] [Google Scholar]
- Grala TM, Kay JK, Walker CG, Sheahan AJ, Littlejohn MD, Lucy MC, Roche JR. Expression analysis of key somatotropic axis and liporegulatory genes in ghrelin- and obestatin-infused dairy cows. Domestic Animal Endocrinology. 2010;39(1):76–83. doi: 10.1016/j.domaniend.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Granata R, Gallo D, Luque RM, Baragli A, Scarlatti F, Grande C, et al. Obestatin regulates adipocyte function and protects against diet-induced insulin resistance and inflammation. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2012;26(8):3393–3411. doi: 10.1096/fj.11-201343. [DOI] [PubMed] [Google Scholar]
- Granata R, Settanni F, Gallo D, Trovato L, Biancone L, Cantaluppi V, et al. Obestatin promotes survival of pancreatic β-cells and human islets and induces expression of genes involved in the regulation of β-cell mass and function. Diabetes. 2008;57(April):967–979. doi: 10.2337/db07-1104. [DOI] [PubMed] [Google Scholar]
- Granata R, Volante M, Settanni F, Gauna C, Ghé C, Annunziata M, et al. Unacylated ghrelin and obestatin increase islet cell mass and prevent diabetes in streptozotocin-treated newborn rats. Journal of Molecular Endocrinology. 2010;45(1):9–17. doi: 10.1677/JME-09-0141. [DOI] [PubMed] [Google Scholar]
- Green BD, Irwin N, Flatt PR. Direct and indirect effects of obestatin peptides on food intake and the regulation of glucose homeostasis and insulin secretion in mice. Peptides. 2007;28(5):981–987. doi: 10.1016/j.peptides.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(1476–4687):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Jeyakumar SM, Vijaya Kumar P, Giridharan NV, Vajreswari A. Vitamin A improves insulin sensitivity by increasing insulin receptor phosphorylation through protein tyrosine phosphatase 1B regulation at early age in obese rats of WNIN/Ob strain. Diabetes, Obesity & Metabolism. 2011;13(10):955–958. doi: 10.1111/j.1463-1326.2011.01407.x. [DOI] [PubMed] [Google Scholar]
- Jüllig M, Chen X, Hickey AJ, Crossman DJ, Xu A, Wang Y, et al. Reversal of diabetes-evoked changes in mitochondrial protein expression of cardiac left ventricle by treatment with a copper(II)-selective chelator. Proteomics-Clinical Applications. 2007;1(4):387–399. doi: 10.1002/prca.200600770. [DOI] [PubMed] [Google Scholar]
- Li Y, Mitra AK. Effects of phospholipid chain length, concentration, charge, and vesicle size on pulmonary insulin absorption. Pharmaceutical Research. 1996;13(1):76–79. doi: 10.1023/A:1016029317299. [DOI] [PubMed] [Google Scholar]
- Lin S, Tsai S, Lee S, Wu S, Wu S, Lin J, Tsai L. Effects of storage time and temperature on the stability of glutathione in deproteinized blood sample. Journal of Food and Drug Analysis. 2006;14(2):141–146. [Google Scholar]
- Lopaschuk GD, Ussher JR, Folmes CDL, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiological Reviews. 2010;90(1):207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- Menezes-Garcia Z, Oliveira MC, Lima RL, Soriani FM, Cisalpino D, Botion LM, et al. Lack of platelet-activating factor receptor protects mice against diet-induced adipose inflammation and insulin-resistance despite fat pad expansion. Obesity. 2014;22(3):663–672. doi: 10.1002/oby.20142. [DOI] [PubMed] [Google Scholar]
- Mihalik SJ, Michaliszyn SF, Heras J, de las Bacha F, Lee S, Chace DH, et al. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: Evidence for enhanced mitochondrial oxidation. Diabetes Care. 2012;35(3):605–611. doi: 10.2337/DC11-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Stone NJ, Ballantyne C, Bittner V, Criqui MH, Ginsberg HN, et al. Triglycerides and cardiovascular disease: A scientific statement from the American Heart Association. Circulation. 2011;123(20):2292–2333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- Miyazawa T, Ibusuki D, Yamashita S, Nakagawa K. Analysis of Amadori-glycated phosphatidylethanolamine in the plasma of healthy subjects and diabetic patients by liquid chromatography–tandem mass spectrometry. Annals of the New York Academy of Sciences. 2008;1126:291–294. doi: 10.1196/annals.1433.033. [DOI] [PubMed] [Google Scholar]
- Nagaraj S, Peddha MS, Manjappara UV. Fragments of obestatin as modulators of feed intake, circulating lipids, and stored fat. Biochemical and Biophysical Research Communications. 2008;366(3):731–737. doi: 10.1016/j.bbrc.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Nagaraj S, Peddha MS, Manjappara UV. Fragment analogs as better mimics of obestatin. Regulatory Peptides. 2009;158(1–3):143–148. doi: 10.1016/j.regpep.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Nagaraj S, Raghavan AV, Rao SN, Manjappara UV. Obestatin and Nt8U influence glycerolipid metabolism and PPAR gamma signaling in mice. The International Journal of Biochemistry & Cell Biology. 2014;53:414–422. doi: 10.1016/j.biocel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Navarro G, Allard C, Xu W, Mauvais-Jarvis F. The role of androgens in metabolism, obesity, and diabetes in males and females. Obesity (Silver Spring, Md.) 2015;23(4):713–719. doi: 10.1002/oby.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Sadanala KC, Kim E. A metabolomic approach to understanding the metabolic link between obesity and diabetes. Molecules and Cells. 2015;38(7):587–596. doi: 10.14348/molcells.2015.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Santos M, Costa PRF, Assis AMO, Santos CAST, Santos DB. Obesity and vitamin D deficiency: A systematic review and meta-analysis. Obesity Reviews. 2015;16(April):341–349. doi: 10.1111/obr.12239. [DOI] [PubMed] [Google Scholar]
- Podrez EA. A novel family of atherogenic oxidized phospholipids promotes macrophage foam cell formation via the scavenger receptor CD36 and is enriched in atherosclerotic lesions. Journal of Biological Chemistry. 2002;277(41):38517–38523. doi: 10.1074/jbc.M205924200. [DOI] [PubMed] [Google Scholar]
- Pruszynska-Oszmalek E, Szczepankiewicz D, Hertig I, Skrzypski M, Sassek M, Kaczmarek P, et al. Obestatin inhibits lipogenesis and glucose uptake in isolated primary rat adipocytes. Journal of Biological Regulators and Homeostatic Agents. 2013;27(1):23–33. [PubMed] [Google Scholar]
- Ren G, He Z, Cong P, Yu J, Qin Y, Chen Y, Liu X. Effect of TAT-obestatin on proliferation, differentiation, apoptosis and lipolysis in 3T3-L1 preadipocytes. Journal of Peptide Science. 2013;19(11):684–691. doi: 10.1002/psc.2550. [DOI] [PubMed] [Google Scholar]
- Roehrich M, Mooser V, Lenain V, Herz J, Azhar S, Bideau M, et al. Insulin-secreting β-cell dysfunction induced by human lipoproteins. The Journal of Biological Chemistry. 2003;278(20):18368–18375. doi: 10.1074/jbc.M300102200. [DOI] [PubMed] [Google Scholar]
- Samson WK, White MM, Price C, Ferguson AV. Obestatin acts in brain to inhibit thirst. American Journal of Physiology: Regulatory, Integrative and Comparative Physiology. 2007;292(1):R637–R643. doi: 10.1152/ajpregu.00395.2006. [DOI] [PubMed] [Google Scholar]
- Savini I, Catani M, Evangelista D, Gasperi V, Avigliano L. Obesity-associated oxidative stress: Strategies finalized to improve redox state. International Journal of Molecular Sciences. 2013;14(5):10497–10538. doi: 10.3390/ijms140510497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metabolism. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Ruebsaamen K. Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis. 2010;208(1):10–18. doi: 10.1016/j.atherosclerosis.2009.05.029. [DOI] [PubMed] [Google Scholar]
- Seim I, Walpole C, Amorim L, Josh P, Herington A, Chopin L. The expanding roles of the ghrelin-gene derived peptide obestatin in health and disease. Molecular and Cellular Endocrinology. 2011;340(1):111–117. doi: 10.1016/j.mce.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Steenge GR, Lambourne J, Casey A, Macdonald IA, Greenhaff PL. Stimulatory effect of insulin on creatine accumulation in human skeletal muscle. The American Journal of Physiology. 1998;275(6 Pt 1):E974–E979. doi: 10.1152/ajpendo.1998.275.6.E974. [DOI] [PubMed] [Google Scholar]
- Sun J, Monagas M, Jang S, Molokin A, Harnly JM, Urban JF, et al. A high fat, high cholesterol diet leads to changes in metabolite patterns in pigs—A metabolomic study. Food Chemistry. 2015;173:171–178. doi: 10.1016/j.foodchem.2014.09.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentirmai E, Krueger JM. Obestatin alters sleep in rats. Neuroscience Letters. 2006;404(1–2):222–226. doi: 10.1016/j.neulet.2006.05.053. [DOI] [PubMed] [Google Scholar]
- Tang S, Dong X, Zhang W. Obestatin changes proliferation, differentiation and apoptosis of porcine preadipocytes. Annales d’endocrinologie. 2014;75(1):1–9. doi: 10.1016/j.ando.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Tranchida F, Shintu L, Rakotoniaina Z, Tchiakpe L, Deyris V, Hiol A, Caldarelli S. Metabolomic and lipidomic analysis of serum samples following curcuma longa extract supplementation in high-fructose and saturated fat fed rats. PLoS ONE. 2015;10(8):e0135948. doi: 10.1371/journal.pone.0135948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasino SE, Benoit YD, Gudas LJ. Vitamin A deficiency causes hyperglycemia and loss of pancreatic β-cell mass. Journal of Biological Chemistry. 2015;290(3):1456–1473. doi: 10.1074/jbc.M114.616763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovato L, Gallo D, Settanni F, Gesmundo I, Ghigo E, Granata R. Obestatin: Is it really doing something? Frontiers of Hormone Research. 2014;42:175–185. doi: 10.1159/000358346. [DOI] [PubMed] [Google Scholar]
- Vergote V, Van Dorpe S, Peremans K, Burvenich C, De Spiegeleer B. In vitro metabolic stability of obestatin: Kinetics and identification of cleavage products. Peptides. 2008;29(10):1740–1748. doi: 10.1016/j.peptides.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J, et al. Global metabolic profiling procedures for urine using UPLC-MS. Nature Protocols. 2010;5(6):1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Research. 2013;41(D1):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Research. 2009;37(Database):D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: The human metabolome database. Nucleic acids research. 2007;35(Database issue):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JV, Ren P-G, Avsian-Kretchmer O, Luo C-W, Rauch R, Klein C, Hsueh AJW. Obestatin, a peptide encoded by the ghrelin gene, opposes ghrelin’s effects on food intake. Science (New York, N.Y.) 2005;310(5750):996–999. doi: 10.1126/science.1117255. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang Y, Hao F, Zhou X, Han X, Tang H, Ji L. Human serum metabonomic analysis reveals progression axes for glucose intolerance and insulin resistance statuses. Journal of Proteome Research. 2009;8(11):5188–5195. doi: 10.1021/pr900524z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCA scores plot displaying pools (red triangles), samples from lean and DIO mice (black triangles) and samples from another study (grey triangles; data not presented)Supplementary material 1 (PDF 237 kb)
PCA scores plot displaying the separation for lean mice (a) and DIO mice (b) treated daily with PEG-OB(Cys10,Cys13) (triangles) or with saline (circles) for 6 weeks. Explained variances (R2) were 0.978 and 0.854 and predictive abilities (Q2) were 0.861 and 0.742 for lean and DIO mice, respectively (PDF 164 kb)
OPLS-DA scores plot displaying the separation for lean mice (a) and DIO mice (b) treated daily with PEG-OB(Cys10, Cys13) (triangles) or with saline (circles) for 6 weeks. Explained variances (R2) were 0.983 and 0.969 and predictive abilities (Q2) were 0.948 and 0.932 for lean and DIO mice respectively. Corresponding validation plots for lean mice (c) and DIO mice (d) displaying 999 permutation tests for model built from PEG-OB(Cys10,Cys13) treated animals (PDF 249 kb)
Detection and identification of creatine. MS2 mass spectrum obtained at 0.15–1.15 min (a), corresponding MS2 peak at 0.15–1.15 min (b) and high resolution spectral library match in m/z cloud (reference no 357)(c) (PDF 189 kb)
Detection and identification of acetylcarnitine. MS2 mass spectrum obtained at 0.45–1.44 min (a), corresponding MS2 peak at 0.45–1.44 min (b), and high resolution spectral library match in m/z cloud (reference no 879)(c) (PDF 187 kb)
Detection and identification of 9-cis retinal. MS2 mass spectrum obtained at 13.83–14.82 min (a), corresponding MS2 peak at 13.83–14.82 min (b), and high resolution spectral library match in m/z cloud (reference no 836)(c) (PDF 191 kb)