Abstract
AIM: To investigate the relationship between ARID1A expression and clinicopathologic parameters, as well as its prognostic value, for patients with intrahepatic cholangiocarcinoma (IHCC).
METHODS: We assessed ARID1A protein and mRNA expression in IHCC tissues and paracarcinomatous (PC) tissues from 57 patients with IHCC using western blot and quantitative real-time reverse transcription polymerase chain reaction, respectively. We used Fisher’s exact and χ2 tests to analyze relationships between clinicopathological parameters and ARID1A expression. The Kaplan-Meier method and Cox regression were used to analyze survival.
RESULTS: The mean ARID1A protein level in IHCC tissues was 1.16 ± 0.36 relative units (RU), which was significantly lower than that in PC tissues (1.26 ± 0.21 RU, P < 0.01) and NL tissues (1.11 ± 0.31, P < 0.001). The mean ARID1A mRNA level in IHCC tissues (1.20 ± 0.18) was also lower than that in PC tissues (1.27 ± 0.15, P < 0.001) and normal liver tissues (1.15 ± 0.34, P < 0.001). Low ARID1A expression was significantly associated with tumor nodules, vein invasion, and recurrence. Median overall survival (OS) and disease-free survival (DFS) for the low ARID1A expression group was 15.0 and 7.0 mo, respectively, which were significantly shorter than those for the high ARID1A expression group at 25.0 and 22.0 mo (OS: P < 0.01; DFS: P < 0.001), respectively. Low ARID1A expression was significantly associated with worse OS (HR = 3.967, 95%CI: 1.299-12.118, P = 0.016) in multivariate analyses.
CONCLUSION: Low expression of ARID1A is associated with poor prognosis in patients with IHCC, and thus may be a potential prognostic biomarker candidate in IHCC.
Keywords: ARID1A, Intrahepatic cholangiocarcinoma, Progression, Prognosis, Biomarker
Core tip: We investigated the relationship between ARID1A expression and clinicopathologic parameters, as well as its prognostic value, for patients with intrahepatic cholangiocarcinoma (IHCC). We examined ARID1A protein and mRNA expression in IHCC and paracarcinomatous (PC) tissue from 57 patients with IHCC. The mean ARID1A protein and mRNA expression levels in IHCC tissues were significantly lower than those in PC tissues and normal liver tissues. Low ARID1A expression was significantly associated with tumor nodules, vein invasion, and recurrence. Low ARID1A expression was significantly associated with worse overall survival in multivariate analyses. ARID1A may be a potential prognostic biomarker candidate in IHCC.
INTRODUCTION
Primary liver cancer (PLC) is the fifth most common cancer worldwide, causing around 600000 deaths annually[1,2]. Intrahepatic cholangiocarcinoma (IHCC) accounts for 5%-10% of all PLCs in Western countries[3,4], with hepatocellular carcinoma (HCC) accounting for about 90%[5]. IHCC, a rare malignant tumor arising from the biliary tract, is aggressive and related to a very poor prognosis[6]. The incidence of IHCC is much higher in Asian countries than it is worldwide[7]. Although the incidence of IHCC is relatively low, it has been progressively and significantly increasing over the last 30 years[8]. Presently, surgical resection is the only option for treating IHCC. Despite surgery improving median survival when compared with conservative therapy alone (1.8 mo), the outcome is still poor, with a post-operative median survival of 12.2 mo[9,10]. Thus, in order to help develop diagnostic methods for enhanced therapeutic outcomes, information on somatic mutations that contribute to the oncogenesis of IHCC is an important first step.
The AT-rich interactive domain 1A (ARID1A) protein (BAF250) is a member of the switching defective/sucrose non-fermenting (SWI/SNF) complexes, which function as ATP-dependent chromatin remodelers[11,12]. The ARID1A gene is located at chromosome 1p36, which is related to the regulation of many cellular processes, including proliferation, DNA repair, development, differentiation, and tumor suppression[13]. Absence of ARID1A protein or gene expression has been found in the precursor stage of clear cell carcinoma of the ovary, ovarian endometrioid adenocarcinoma, breast cancer, and colorectal cancer, and correlates with tumor progression in these cancers[14-17]. These findings indicate that ARID1A may be a tumor-suppressor gene.
Although correlations between ARID1A gene mutation and ARID1A protein expression with clinicopathologic parameters and prognosis in HCC have been recently reported[18], no study has examined its correlation with IHCC. In the current study, we investigated ARID1A gene and protein expression in surgically resected IHCC tumors in order to observe whether its expression status could be a prognostic biomarker for IHCC.
MATERIALS AND METHODS
Patients and tissue specimens
IHCC and adjacent paracarcinomatous (PC) liver specimens from 57 IHCC patients and normal liver tissues from 19 hepatic hemangioma patients (controls) were collected in the operation room. Samples were instantly frozen in liquid nitrogen and stored at -80 °C until testing. PC tissues were hepatic tissue collected 2-5 cm away from the tumor edge. All patients underwent surgery in the Department of Hepatobiliary Surgery, Yantaishan Hospital, from January 2012 to June 2013. Diagnoses were defined by pathological examination. The study protocol was approved by the Ethics Committee of Yantaishan Hospital. Informed written consent was obtained from each patient.
We collected clinicopathologic parameters, including age, gender, liver function, tumor size, tumor number, histopathological classification, vessel invasion, recurrence, and patient survival time. Vessel invasion was observed during pathological examination, which indicated tumor infiltration in the portal and/or hepatic veins. We monitored recurrence via ultrasound, computed tomography scan, and magnetic resonance imaging.
Western blot analysis
IHCC and PC tissues were homogenized and treated with RIPA lysis buffer (Dingguo, Beijing, China). Protein samples were resolved on a 4%-12% acrylamide gradient gel. Samples were transferred to a polyvinylidene fluoride membrane using iBlot fast electric transfer (Invitrogen, IL, United States). Membranes were blocked at room temperature for 1 h in 5% milk and incubated with primary antibodies against ARID1A or GAPDH (1:1000, Abcam, MA, United States) at 4 °C overnight. Membranes were then washed with TBST three times, followed by incubation with appropriate secondary antibodies (1:8000, Abcam, MA, United States) at room temperature for 2 h. After a further three washes in TBST, the membranes were exposed to film using the ECL kit (Pierce, CA, United States). ARID1A-specific signals were quantified from X-ray films using a scanner with BandScan 4.30 densitometry software and expressed as integrated intensity units relative to GAPDH signals[19]. The results were analyzed by physicians in a blinded manner.
Quantitative real-time reverse transcription-polymerase chain reaction
Total RNA was extracted from IHCC and PC tissues with TRIzol. The retroviral reverse transcriptase kit (Takara, Tokyo, Japan) was used to synthesize cDNA with the reaction conditions of 37 °C for 60 min and 95 °C for 3 min. Primers were sense: 5′- TTAACTCCAGCCACCAAAATGAAC-3′ and antisense: 5′- ATAGAGGCGATAGAGGTCCAGAGG-3′ for the ARID1A gene, and sense: 5′-GAAGGTGAAGGTCGGAGTC-3′ and antisense: 5′-GAAGATGGTGATGGGATTTC-3′ for GAPDH. Real-time polymerase chain reaction (PCR) was performed with the 7500 real-time quantitative PCR instrument (Applied Biosystems, CA, United States) using the following conditions: 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s for 40 cycles. Data were normalized using the GAPDH housekeeping gene and expressed as 2-ΔCt.
Patient follow-up
We obtained follow-up data after discharge for all 57 IHCC patients by direct communication with the patients or their relatives, or by reviewing hospital records. Disease-free survival (DFS) was measured from the date of hepatectomy until tumor recurrence. Overall survival (OS) was measured from the date of hepatectomy until death or the last follow-up point. The last follow-up evaluation was set as August 31, 2015, or up to the time of death.
Statistical analysis
Quantitative values are presented as mean ± SD or median (range). Student’s t-test was used to evaluate differences in ARID1A protein and mRNA expression between IHCC and PC tissues. Fisher’s exact and χ2 tests were used to analyze the correlation between ARID1A expression level and clinicopathologic parameters in patients with IHCC. Survival curves were plotted by the Kaplan-Meier method and compared using the log-rank test. Survival data were evaluated using the Cox proportional hazards model. All tests were two-tailed; P < 0.05 was considered significant. SPSS package 17.0 (SPSS Inc., Chicago, IL, United States) was used for all analyses. The statistical methods of this study were reviewed by Prof. Xiaoqing Liu from Peking Union Medical College Hospital.
RESULTS
Patient characteristics
The mean age of the IHCC patients was 55.1 ± 9.3 years and 68.4% (39/57) were male. The mean age of the controls was 51.2 ± 6.6 years and 57.9% (11/19) were male. Forty patients with IHCC had at least one tumor nodule larger than 3 cm. Tumors were well-differentiated in 22 patients, moderately-differentiated in 17 patients, and poorly-differentiated in 18 patients. The median follow-up time was 20 mo (range: 2-28 mo). IHCC recurred in 33 patients over a median recurrence time of 15.0 mo. During the follow-up, 30 patients died, with a mean survival time of 19.0 ± 6.4 mo.
ARID1A protein and mRNA expression in IHCC and PC tissues
The mean ARID1A protein level in IHCC tissues was 1.16 ± 0.36 relative units (RU). This was significantly lower than that in PC (1.26 ± 0.21 RU, P < 0.01) and normal liver (NL) tissues (1.11 ± 0.31, P < 0.001) (Figure 1). The mean ARID1A mRNA level in IHCC tissues (1.20 ± 0.18) was also lower than that in PC (1.27 ± 0.15, P < 0.001) and NL tissues (1.15 ± 0.34, P < 0.001) (Figure 2).
Figure 1.
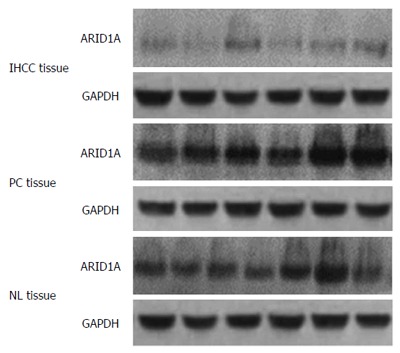
ARID1A protein expression in intrahepatic cholangiocarcinoma patients. Western blotting for ARID1A expression in intrahepatic cholangiocarcinoma tissues, paracarcinomatous tissues, and normal liver tissues. GAPDH was used as the internal loading control. IHCC: Intrahepatic cholangiocarcinoma; PC: Paracarcinomatous; NL: Normal liver.
Figure 2.
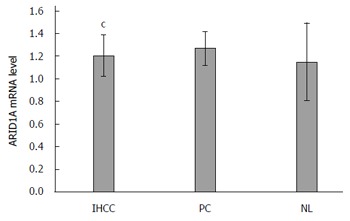
ARID1A mRNA level in intrahepatic cholangiocarcinoma patients. IHCC: Intrahepatic cholangiocarcinoma; PC: Paracarcinomatous; NL: Normal liver. cP < 0.001 vs PC and NL.
Correlations between ARID1A expression and clinicopathological features
IHCC tissues were then divided according to ARID1A expression. IHCC tissues with ARID1A protein expression lower than that in NL tissues were defined as low expression tumors. Of the 57 total IHCC cases, 19 cases showed low ARID1A expression and 38 cases showed high ARID1A expression. The correlations of ARID1A mRNA and protein expression with clinicopathological parameters are shown in Table 1. Low ARID1A expression was significantly associated with tumor nodules, vein invasion, and recurrence.
Table 1.
Clinicopathological features of IHCC patients and the correlation between ARID1A protein expression and clinicopathological parameters
| Characteristics |
ARID1A |
P value | |
| Low expression | High expression | ||
| n = 19 | n = 38 | ||
| Age (yr) | 0.703 | ||
| < 50 | 7 | 16 | |
| ≥ 50 | 12 | 22 | |
| Sex | 0.546 | ||
| Female | 7 | 11 | |
| Male | 12 | 27 | |
| Child-classification | 0.500 | ||
| A | 15 | 34 | |
| B | 4 | 4 | |
| Differentiation | 0.546 | ||
| Well | 10 | 12 | |
| Moderately | 4 | 13 | |
| Poorly | 5 | 13 | |
| Tumor size (cm) | 0.306 | ||
| > 3 | 15 | 25 | |
| ≤ 3 | 4 | 13 | |
| Tumor nodule | < 0.001 | ||
| Solitary | 7 | 32 | |
| Multiple | 12 | 6 | |
| Vein invasion | 0.004 | ||
| Positive | 13 | 11 | |
| Negative | 6 | 27 | |
| Recurrence status | 0.004 | ||
| Yes | 16 | 17 | |
| No | 3 | 21 | |
IHCC: Intrahepatic cholangiocarcinoma.
ARID1A protein expression in IHCC tissues with multiple tumor nodules was significantly lower (n = 18, 0.93 ± 0.34 RU) than in those with solitary tumor nodules (n = 39, 1.27 ± 0.32, P < 0.001). ARID1A mRNA expression in IHCC tissues with multiple tumor nodules (n = 18, 1.13 ± 0.17) was also lower than in those with solitary tumor nodules (1.23 ± 0.16, P < 0.05).
ARID1A protein expression in IHCC tissues with vein invasion was significantly lower (n = 24, 0.93 ± 0.33 RU) than in those without vein invasion (n = 33, 1.33 ± 0.28, P < 0.001). ARID1A mRNA expression in IHCC tissues with vein invasion (1.18 ± 0.18) was also lower than in those without vein invasion (1.21 ± 0.17), although without statistical significance.
ARID1A protein levels in IHCC tissues from patients who experienced recurrence during follow-up (n = 33, 1.00 ± 0.33) were significantly lower than in those without recurrence (n = 24, 1.39 ± 0.27, P < 0.001). ARID1A mRNA levels in IHCC tissues with recurrence (1.20 ± 0.17) were also lower than in those without recurrence (1.25 ± 0.21), although without statistical significance.
Association of ARID1A expression with prognosis
Kaplan-Meier survival curves and log-rank tests showed that low ARID1A protein expression in IHCC tissues was associated with poor prognosis. The median OS for the low ARID1A expression group was 15.0 mo, which was significantly shorter than that of the high ARID1A expression group at 25.0 mo (P < 0.01; Figure 3A). The median DFS rates for the low and high ARID1A expression groups were 7.0 mo and 22.0 mo, respectively (P < 0.001; Figure 3B). Furthermore, multivariate analysis using the Cox proportional hazards model indicated that ARID1A expression levels, tumor nodules, and vein invasion were independent predictors of DFS in patients with IHCC (Table 2). However, ARID1A expression level was the only independent predictor of OS in a multivariate analysis. Low ARID1A expression was significantly associated with worse OS in patients with IHCC when compared with the results from high ARID1A expression (HR = 3.967, 95%CI: 1.299-12.118, P = 0.016, Table 2).
Figure 3.
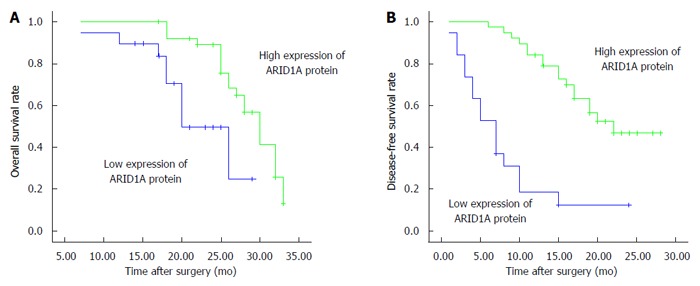
Overall survival (A) and disease-free survival (B) of patients with intrahepatic cholangiocarcinoma after surgical resection according to ARID1A protein expression in intrahepatic cholangiocarcinoma tissues (P < 0.01; log-rank test).
Table 2.
Multivariate analysis of overall survival and disease-free survival on ARID1A protein expression in patients with intrahepatic cholangiocarcinoma (Cox proportional hazards model)
| Prognostic factors |
Overall survival |
Disease-free survival |
||
| Hazard ration (95%CI) | P value | Hazard ration (95%CI) | P value | |
| Tumor nodule | ||||
| Solitary vs multiple | 0.292 (0.093-0.914) | 0.034 | 1.092 (0.328-3.639) | 0.886 |
| Vein invasion | ||||
| Positive vs negative | 0.181 (0.079-0.413) | < 0.001 | 1.207 (0.509-2.859) | 0.669 |
| ARID1A expression | ||||
| Low vs high | 7.240 (2.281-22.980) | < 0.001 | 3.967 (1.299-12.118) | 0.016 |
DISCUSSION
ARID1A/BAF250 is a component of the SWI/SNF family complexes and is extensively expressed in different human tissues[14,20,21]. The SWI/SNF chromatin remodeling complexes are recurrently mutated in various carcinomas[22]. Mutations in several subunits of these complexes have been identified, including BAF180, SNF5, BRM/SWI2-related genes, and ARID1A[22,23]. ARID1A/BAF250 provides SNF/SWI complex specificity and facilitates protein-protein or protein-DNA molecule interactions. Knockdown of ARID1A gene causes cell cycle arrest in osteoblasts cells, and previous studies have demonstrated a correlation of ARID1A loss and tumorigenesis[11]. Taken together, this supports the potential tumor suppressor function of ARID1A and suggests that ARID1A plays an important role in tumorigenesis and tumor progression.
Guichard et al[24] performed high-resolution copy-number analysis on 125 tumor tissues of patients with HCC, with whole-exome sequencing then being performed on 24 of these tumors. The author found new recurrent modifications in four genes (ARID1A, RPS6KA3, NFE2L2, and IRF2) that had not been formerly described in HCC. In a study by Fujimoto et al[25], the entire genomes of 27 HCCs were sequenced and analyzed. Twenty-five were associated with hepatitis B or C virus infections, including two groups of multicentric tumors. Statistical and functional analyses showed that ARID1A, ARID1B, ARID2, MLL, and MLL3 genes were mutated in about 50% of tumors. Huang et al[18] showed that ARID1A was mutated in 13% of HBV-associated HCC specimens. These studies suggested that ARID1A may be involved in advanced HCC.
IHCC is the second most common primary liver cancer after HCC, accounting for 5%-10% of all cholangiocarcinomas[26]. The origin of IHCC is normally demarcated as grade II intrahepatic bile duct epithelium[27,28]. The prognosis and mortality rate of IHCC is poor, as IHCC is usually diagnosed at terminal stages due to an absence of appropriate methods for early diagnosis, and surgical treatment only extends post-operative median survival to 12.2 mo[9,10]. A better understanding of the somatic mutations that contribute to the oncogenesis of IHCC is therefore critical for the development of diagnostic strategies. Through exome sequencing of 32 patients with IHCC, Jiao et al[29] found many inactivating mutations in various chromatin-remodeling genes (PBRM1, ARID1A, and BAP1), with activating mutations in one of these genes being observed in nearly half of all cancers sequenced. Zou et al[30] sequenced carcinoma and paracancerous tissues in a large cohort of 103 patients with IHCC in China and found that IHCC-specific somatic mutation was correlated with liver inflammation, fibrosis, and cirrhosis. The authors identified 25 mutated genes with eight possible driver genes, including KRAS, TP53, IDH1, ARID1A, PTEN, ECE2, EPPK1, and FYN. We observed that ARID1A may be a key point for the oncogenesis of IHCC; however, whether ARID1A status impacts clinical behavior has not been made clear. We performed this retrospective study in order to investigate the relationship between ARID1A expression and clinicopathologic parameters, as well as its predictive value for IHCC prognosis.
We found lower ARID1A protein in IHCC tumor tissues when compared with the PC tissues of patients with IHCC and the NL tissues of hepatic hemangioma patients. We also observed a significant difference between the expression level of ARID1A mRNA in IHCC samples and PC tissues.
An important result of the current study was that ARID1A protein expression was associated with tumor nodules, vein invasion, and tumor recurrence status. These factors are highly correlated with the invasion and metastasis of IHCC. Our results indicate that more invasive tumors have lower ARID1A protein expression, suggesting that ARID1A has a suppressive function in IHCC. ARID1A protein expression did not correlate with sex, age, liver function, tumor size, or tumor differentiation.
Many studies have examined the relevance of ARID1A mutation or protein loss to survival in several carcinomas, although the findings were varied. ARID1A mutation or protein loss was a predictor of poor prognosis in cervical carcinoma[31] and gastric cancer[21]. Other studies found no association between ARID1A mutation/protein loss and survival in endometrial clear-cell carcinoma[32] and ovarian clear cell adenocarcinoma[33]. Other reports suggested that ARID1A mutation or protein loss may be related to survival in endometrial carcinoma[20] and gastric cancer[21]. Our study is the first to explore the relationship between ARID1A expression and IHCC survival. OS and DFS were significantly shorted in the low ARID1A expression group when compared to that of the high ARID1A expression group. These associations need to be verified and the mechanisms clarified in future investigations. In univariate analyses, ARID1A expression, tumor nodules, vein invasion, and recurrence status were found to be significant prognostic factors. In multivariate analysis, all of the above factors were independent prognostic factors of DFS, while only ARID1A expression was an independent prognostic factor of OS. Taken together, our findings suggest that ARID1A may be a potential prognostic biomarker candidate in IHCC.
This is the first investigation into the correlation between ARID1A gene and protein expression with clinicopathologic features of IHCC. Expressions of ARID1A protein and mRNA of IHCC tissues were lower than in that of PC tissues. ARID1A protein levels in IHCC tissues from patients with recurrence during follow-up were significantly lower than in those without recurrence. IHCC tissues with vein invasions had significantly lower ARID1A protein levels than in those without vein invasions. ARID1A protein expression in IHCC tissues correlated with OS and DFS. Based on our results showing low ARID1A expression in IHCC, we speculate that the manipulation of ARID1A expression in IHCC patients might have therapeutic implications. However, the functions and mechanisms of ARID1A regulation in normal and IHCC tissues remain unclear and require further study.
COMMENTS
Background
Primary liver cancer (PLC) is the fifth most common cancer, with annually about 600000 deaths worldwide. Intrahepatic cholangiocarcinoma (IHCC) accounts for 5%-10% of all PLCs in western countries, while hepatocellular carcinoma (HCC) accounts for about 90%.
Research frontiers
The authors investigated ARID1A gene and protein expression in surgically resected IHCC tumors to observe whether its expression status could be a prognostic biomarker for IHCC.
Innovations and breakthroughs
This is the first investigation into correlations between ARID1A gene and protein expression with clinicopathologic features of IHCC. Expressions of ARID1A protein and mRNA of IHCC tissues were lower than in that of paracarcinomatous (PC) tissues.
Peer-review
In this very interesting study, the authors investigated the relationship between ARID1A expression and clinicopathologic parameters, as well as its prognostic value, for patients with IHCC. ARID1A protein and mRNA expression in IHCC and PC tissues from 57 patients with IHCC were assessed. The authors found that the low ARID1A expression was associated with a poor prognosis in patients with IHCC, and that ARID1A may be a potential prognostic biomarker candidate in IHCC.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Yantaishan Hospital Institutional Review Board.
Informed consent statement: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest statement: All authors have no conflict of interest to disclosure.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Peer-review started: April 6, 2016
First decision: May 12, 2016
Article in press: June 2, 2016
P- Reviewer: Higuchi K, Ryan EM, Okada S S- Editor: Qi Y L- Editor: Rutherford A E- Editor: Ma S
References
- 1.Miao R, Luo H, Zhou H, Li G, Bu D, Yang X, Zhao X, Zhang H, Liu S, Zhong Y, et al. Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J Hepatol. 2014;61:840–849. doi: 10.1016/j.jhep.2014.05.025. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Zhu C, Zhao Y, Li M, Wu L, Yang X, Wan X, Wang A, Zhang MQ, Sang X, et al. Long non-coding RNA expression profiles of hepatitis C virus-related dysplasia and hepatocellular carcinoma. Oncotarget. 2015;6:43770–43778. doi: 10.18632/oncotarget.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aljiffry M, Abdulelah A, Walsh M, Peltekian K, Alwayn I, Molinari M. Evidence-based approach to cholangiocarcinoma: a systematic review of the current literature. J Am Coll Surg. 2009;208:134–147. doi: 10.1016/j.jamcollsurg.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Wan X, Zhao H. New avenues to treatment of liver cirrhosis. Sci China Life Sci. 2014;57:1049–1050. doi: 10.1007/s11427-014-4751-x. [DOI] [PubMed] [Google Scholar]
- 5.Chan SL, Johnson PJ, Mo F, Berhane S, Teng M, Chan AW, Poon MC, Lai PB, Yu S, Chan AT, et al. International validation of the Chinese university prognostic index for staging of hepatocellular carcinoma: a joint United Kingdom and Hong Kong study. Chin J Cancer. 2014;33:481–491. doi: 10.5732/cjc.014.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 7.Lo RC, Ng IO. Hepatocellular tumors: immunohistochemical analyses for classification and prognostication. Chin J Cancer Res. 2011;23:245–253. doi: 10.1007/s11670-011-0245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaib YH, Davila JA, McGlynn K, El-Serag HB. Rising incidence of intrahepatic cholangiocarcinoma in the United States: a true increase? J Hepatol. 2004;40:472–477. doi: 10.1016/j.jhep.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 9.Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004;24:115–125. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 10.Soares KC, Kamel I, Cosgrove DP, Herman JM, Pawlik TM. Hilar cholangiocarcinoma: diagnosis, treatment options, and management. Hepatobiliary Surg Nutr. 2014;3:18–34. doi: 10.3978/j.issn.2304-3881.2014.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagl NG, Patsialou A, Haines DS, Dallas PB, Beck GR, Moran E. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res. 2005;65:9236–9244. doi: 10.1158/0008-5472.CAN-05-1225. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Nagl NG, Wilsker D, Van Scoy M, Pacchione S, Yaciuk P, Dallas PB, Moran E. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem J. 2004;383:319–325. doi: 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayhan A, Mao TL, Seckin T, Wu CH, Guan B, Ogawa H, Futagami M, Mizukami H, Yokoyama Y, Kurman RJ, et al. Loss of ARID1A expression is an early molecular event in tumor progression from ovarian endometriotic cyst to clear cell and endometrioid carcinoma. Int J Gynecol Cancer. 2012;22:1310–1315. doi: 10.1097/IGC.0b013e31826b5dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samartzis EP, Samartzis N, Noske A, Fedier A, Caduff R, Dedes KJ, Fink D, Imesch P. Loss of ARID1A/BAF250a-expression in endometriosis: a biomarker for risk of carcinogenic transformation? Mod Pathol. 2012;25:885–892. doi: 10.1038/modpathol.2011.217. [DOI] [PubMed] [Google Scholar]
- 17.Mao TL, Ardighieri L, Ayhan A, Kuo KT, Wu CH, Wang TL, Shih IeM. Loss of ARID1A expression correlates with stages of tumor progression in uterine endometrioid carcinoma. Am J Surg Pathol. 2013;37:1342–1348. doi: 10.1097/PAS.0b013e3182889dc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, Zhu ZD, Zhou B, Liu XY, Liu RF, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Yang H, Mao Y, Xu H, Zhang J, Li G, Lu X, Sang X, Zhao H, Zhong S, et al. Increased Golgi protein 73 expression in hepatocellular carcinoma tissue correlates with tumor aggression but not survival. J Gastroenterol Hepatol. 2011;26:1207–1212. doi: 10.1111/j.1440-1746.2011.06733.x. [DOI] [PubMed] [Google Scholar]
- 20.Allo G, Bernardini MQ, Wu RC, Shih IeM, Kalloger S, Pollett A, Gilks CB, Clarke BA. ARID1A loss correlates with mismatch repair deficiency and intact p53 expression in high-grade endometrial carcinomas. Mod Pathol. 2014;27:255–261. doi: 10.1038/modpathol.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang DD, Chen YB, Pan K, Wang W, Chen SP, Chen JG, Zhao JJ, Lv L, Pan QZ, Li YQ, et al. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS One. 2012;7:e40364. doi: 10.1371/journal.pone.0040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Li M, Liu Q, Zhang Y, Qian J, Wan X, Wang A, Zhang H, Zhu C, Lu X, et al. Dr.VIS v2.0: an updated database of human disease-related viral integration sites in the era of high-throughput deep sequencing. Nucleic Acids Res. 2015;43:D887–D892. doi: 10.1093/nar/gku1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 26.Cardinale V, Bragazzi MC, Carpino G, Torrice A, Fraveto A, Gentile R, Pasqualino V, Melandro F, Aliberti C, Bastianelli C, et al. Cholangiocarcinoma: increasing burden of classifications. Hepatobiliary Surg Nutr. 2013;2:272–280. doi: 10.3978/j.issn.2304-3881.2013.10.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi SB, Kim KS, Choi JY, Park SW, Choi JS, Lee WJ, Chung JB. The prognosis and survival outcome of intrahepatic cholangiocarcinoma following surgical resection: association of lymph node metastasis and lymph node dissection with survival. Ann Surg Oncol. 2009;16:3048–3056. doi: 10.1245/s10434-009-0631-1. [DOI] [PubMed] [Google Scholar]
- 28.Miwa S, Miyagawa S, Kobayashi A, Akahane Y, Nakata T, Mihara M, Kusama K, Soeda J, Ogawa S. Predictive factors for intrahepatic cholangiocarcinoma recurrence in the liver following surgery. J Gastroenterol. 2006;41:893–900. doi: 10.1007/s00535-006-1877-z. [DOI] [PubMed] [Google Scholar]
- 29.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 31.Cho H, Kim JS, Chung H, Perry C, Lee H, Kim JH. Loss of ARID1A/BAF250a expression is linked to tumor progression and adverse prognosis in cervical cancer. Hum Pathol. 2013;44:1365–1374. doi: 10.1016/j.humpath.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Fadare O, Gwin K, Desouki MM, Crispens MA, Jones HW, Khabele D, Liang SX, Zheng W, Mohammed K, Hecht JL, et al. The clinicopathologic significance of p53 and BAF-250a (ARID1A) expression in clear cell carcinoma of the endometrium. Mod Pathol. 2013;26:1101–1110. doi: 10.1038/modpathol.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto S, Tsuda H, Takano M, Tamai S, Matsubara O. PIK3CA mutations and loss of ARID1A protein expression are early events in the development of cystic ovarian clear cell adenocarcinoma. Virchows Arch. 2012;460:77–87. doi: 10.1007/s00428-011-1169-8. [DOI] [PubMed] [Google Scholar]


