Phosphatidylethanolamine, an essential phospholipid for mitochondrial functions, is synthesized at the mitochondrial inner membrane. Miyata et al. demonstrate that Ups2–Mdm35, a protein complex in the mitochondrial intermembrane space, mediates phosphatidylserine transport for phosphatidylethanolamine synthesis in respiration-active mitochondria of Saccharomyces cerevisiae.
Abstract
Phosphatidylethanolamine (PE) is an essential phospholipid for mitochondrial functions and is synthesized mainly by phosphatidylserine (PS) decarboxylase at the mitochondrial inner membrane. In Saccharomyces cerevisiae, PS is synthesized in the endoplasmic reticulum (ER), such that mitochondrial PE synthesis requires PS transport from the ER to the mitochondrial inner membrane. Here, we provide evidence that Ups2–Mdm35, a protein complex localized at the mitochondrial intermembrane space, mediates PS transport for PE synthesis in respiration-active mitochondria. UPS2- and MDM35-null mutations greatly attenuated conversion of PS to PE in yeast cells growing logarithmically under nonfermentable conditions, but not fermentable conditions. A recombinant Ups2–Mdm35 fusion protein exhibited phospholipid-transfer activity between liposomes in vitro. Furthermore, UPS2 expression was elevated under nonfermentable conditions and at the diauxic shift, the metabolic transition from glycolysis to oxidative phosphorylation. These results demonstrate that Ups2–Mdm35 functions as a PS transfer protein and enhances mitochondrial PE synthesis in response to the cellular metabolic state.
Introduction
In eukaryotic cells, organelle membranes and plasma membranes are composed of specific sets of proteins and lipids. Most proteins are synthesized by ribosomes in the cytosol and specifically delivered to each cellular compartment. On the other hand, most membrane phospholipids are synthesized in the ER and transported to other compartments (Daum et al., 1998). Among phospholipids, cardiolipin (CL) and the majority of phosphatidylethanolamines (PEs) are synthesized in the mitochondrial inner membrane (MIM; Clancey et al., 1993; Trotter et al., 1993; Jiang et al., 1997; Chang et al., 1998; Tuller et al., 1998). PE and CL are required for mitochondrial functions and morphogenesis (Osman et al., 2011; Böttinger et al., 2012). Although PE is also synthesized in the ER and endosomes (Trotter and Voelker, 1995; Gibellini and Smith, 2010; Gulshan et al., 2010), the functions of PE synthesized in mitochondria cannot be performed by PE formed extramitochondrially under several conditions. For example, concomitant loss of CL and PE synthesis in mitochondria leads to lethality in yeast (Gohil et al., 2005), and loss of mitochondrial PE synthesis in mice causes embryonic lethality (Steenbergen et al., 2005).
Because phosphatidic acid (PA) and phosphatidylserine (PS), which are the starting material for CL biosynthesis and a precursor of PE, respectively, are synthesized in the ER, PA and PS should be transported from the ER to the MIM to synthesize CL and PE. Inversely, PE is transported from the MIM to the ER followed by conversion to phosphatidylcholine (PC) by phospholipid methylation enzymes (Kodaki and Yamashita, 1987). Accordingly, phospholipid traffic between the ER and MIM across the mitochondrial outer membrane (MOM) is a crucial process for cellular phospholipid homeostasis (Tamura et al., 2014). Recently, it was reported that phospholipid transport between the ER and MOM is mediated via organelle contact sites formed by membrane tethering complexes (Lang et al., 2015). The ER–mitochondria encounter structure (ERMES) was first identified in yeast as an ER–mitochondria contact site, and is thought to mediate phospholipid transport between the ER and MOM (Kornmann et al., 2009). A conserved ER membrane protein complex forms another ER–mitochondria contact site and functions cooperatively with ERMES in phospholipid transport (Lahiri et al., 2014). Recently, the vacuole-mitochondrial patch (vCLAMP) was identified as a novel contact site between yeast vacuoles and mitochondria and was implicated in phospholipid transfer (Elbaz-Alon et al., 2014; Hönscher et al., 2014). Interestingly, vCLAMP is formed only when cells are grown in glucose and disassembled upon a shift to a nonfermentable carbon source such as glycerol (Hönscher et al., 2014). Thus, phospholipid transport pathways into mitochondria could change depending on the metabolic state.
Phospholipid transport between the MOM and MIM is less understood than that between mitochondria and the ER or vacuoles. The conserved intermembrane space proteins Ups1 and Ups2, both of which form a complex with Mdm35, are reported to regulate phospholipid transport between the MOM and MIM (Tamura et al., 2012a). Ups1–Mdm35 functions as a PA transfer protein for CL synthesis (Connerth et al., 2012; Watanabe et al., 2015), and deletion of UPS1 (ups1Δ) causes a striking decrease in the CL level (Osman et al., 2009; Tamura et al., 2009). Ups2 is reported to function antagonistically with Ups1; deletion of UPS2 (ups2Δ) partially rescues the decrease in the CL level and the growth defect of ups1Δ cells (Osman et al., 2009; Tamura et al., 2009). Importantly, ups2Δ causes a decrease in the PE level in mitochondria (Osman et al., 2009; Tamura et al., 2009). From this observation and its similarity to Ups1 in amino acid sequence, Ups2–Mdm35 is a strong candidate for a PS transfer protein for PE synthesis. However, Ups2-dependent PE synthesis has not been observed, as far as we know. According to previous studies, the reason why loss of Ups2 leads to a decrease in the mitochondrial PE level is decreased stability of newly synthesized PE (Osman et al., 2009) or acceleration of PE export to the ER, followed by conversion to PC through phospholipid methylations (Tamura et al., 2012a,b). Thus, the function of Ups2 in PE metabolism is controversial. To address this issue, we examined the effect of loss of Ups2 on phospholipid metabolism of yeast cells in various genetic backgrounds and under various culture conditions, and we performed an in vitro phospholipid transport assay involving a recombinant Ups2–Mdm35 fusion protein. The results of in vivo and in vitro analyses demonstrated that Ups2–Mdm35 transports PS from the MOM to the MIM for PE synthesis in respiration-active mitochondria.
Results
Ups2 is involved in mitochondrial PE synthesis
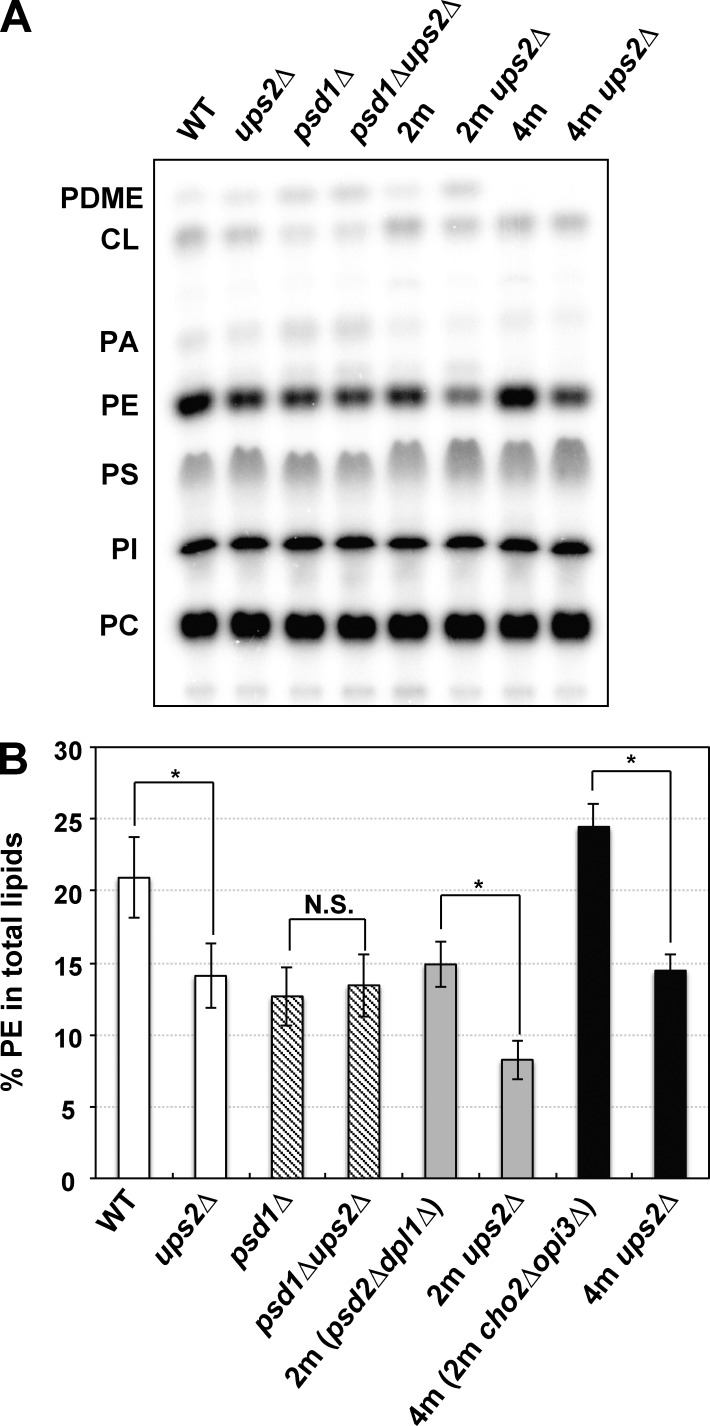
The cellular PE level is mainly defined by the balance between the synthesis and conversion to PC (Wu and Voelker, 2002). In the yeast Saccharomyces cerevisiae, PE is synthesized through decarboxylation of PS, catalyzed by PS decarboxylase 1 (Psd1) and 2 (Psd2), and the Kennedy pathway, in which ethanolamine (Etn) is converted sequentially to Etn phosphate, CDP-Etn, and then PE. Conversion of PE to PC is performed through three steps of methylation catalyzed by Cho2 and Opi3 (Kodaki and Yamashita, 1987). To facilitate investigation into the Ups2 function in PE metabolism, we constructed ups2Δ strains with several genetic backgrounds, including psd1Δ, psd2Δdpl1Δ (designated as 2m), and psd2Δdpl1Δcho2Δopi3Δ (designated as 4m). Dpl1 is a dihydrosphingosine lyase that generates Etn phosphate, which is incorporated into PE through the Kennedy pathway (Gottlieb et al., 1999). We examined the cellular PE levels in these strains as well as wild-type and ups2Δ cells. To label total cellular phospholipids, cells (initial OD600, 0.05) were incubated in YPD medium containing a fermentable carbon source in the presence of [32P]Pi for 15 h. As shown in Fig. 1, the PE level in ups2Δ cells decreased as compared with that in wild-type cells, being consistent with previous studies (Osman et al., 2009; Tamura et al., 2009). Loss of Ups2 no longer affected the PE level in psd1Δ cells. In contrast, loss of Ups2 decreased the PE level in 2m cells, in which endosomal Psd2 was deficient. These results suggest that loss of Ups2 affects only the level of PE synthesized in mitochondria. In 4m cells, the PE level was elevated as compared with that in 2m cells, probably because of a lack of conversion of PE to PC through phospholipid methylations. Even in 4m cells, the PE level was decreased by loss of Ups2 to the same extent in the wild-type and 2m cells, implying that the decrease in PE level of ups2Δ and 2m ups2Δ cells is not due to acceleration of conversion of PE to PC through phospholipid methylations.
Figure 1.
PE levels in ups2Δ cells with various mutations. (A and B) Yeast cells, as indicated (OD600, 0.05), were cultured in YPD in the presence of [32P]Pi for 15 h. Total cellular phospholipids were extracted, separated by TLC, and then analyzed by an imaging analyzer. PDME, phosphatidyldimethylethanolamine; PI, phosphatidylinositol. (B) The percentages of PE relative to total phospholipids. Values are means ± SD (n = 3). *, P < 0.02. N.S., not significant.
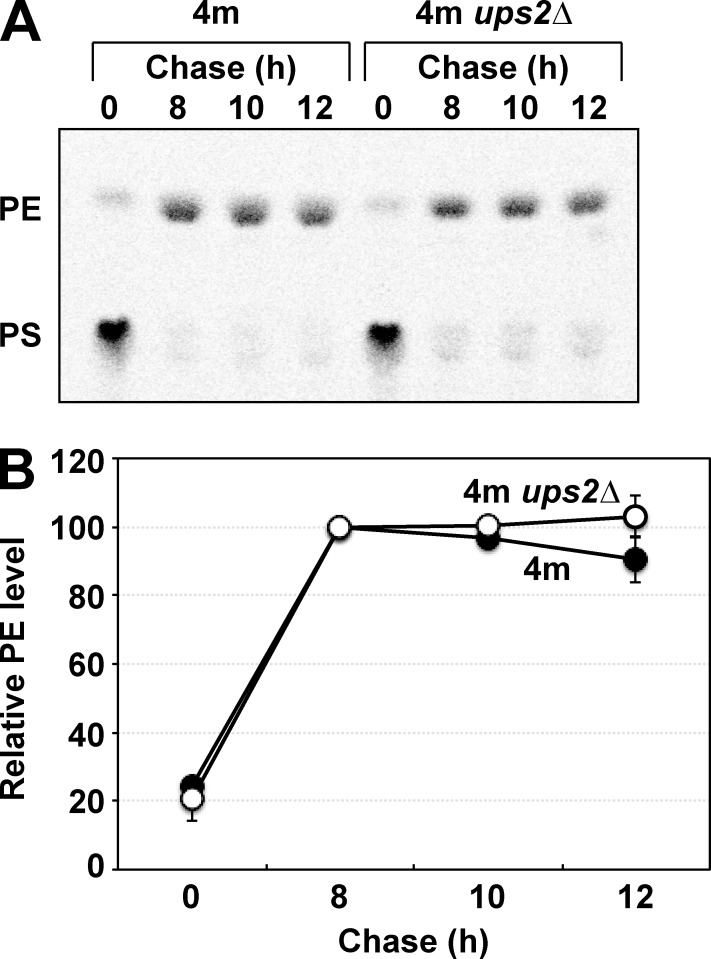
Next, we investigated whether loss of Ups2 affects the stability of mitochondrial PE using 4m and 4m ups2Δ cells. In 4m cells, newly synthesized PS is converted to PE exclusively by mitochondrial Psd1 because of a lack of endosomal Psd2. When 4m and 4m ups2Δ cells were pulse-labeled with l-[14C]serine for 15 min, followed by an 8-h incubation at 30°C in YPD, [14C]PS synthesized de novo was almost completely converted to [14C]PE in both strains (Fig. 2). Upon further incubation up to 12 h, the [14C]PE level was hardly reduced in 4m cells, suggesting that PE synthesized in mitochondria was stable unless it was converted to PC. Furthermore, loss of Ups2 did not affect PE stability in 4m cells (Fig. 2). These results suggest that the PE reduction in ups2Δ cells is caused by neither increased conversion of PE to PC through phospholipid methylations nor decreased PE stability. Thus, by means of elimination, the reason for PE reduction in ups2Δ cells was supposed to be a decrease in the rate of biosynthesis of PE in mitochondria.
Figure 2.
Loss of Ups2 does not affect PE stability. (A and B) 4m and 4m ups2Δ cells were pulse-labeled with l-[14C]serine for 15 min and then further cultured in YPD for the indicated periods. At each time point, total phospholipids were extracted, separated by TLC, and then analyzed by an imaging analyzer. (B) The amount of PE at 8 h was set to 100%, and the relative PE level at each time point is presented. The mean values for two independent experiments and deviations are shown. Closed circle, 4m cells; open circle, 4m ups2Δ cells.
Ups2 enhances PE accumulation in respiration-active mitochondria
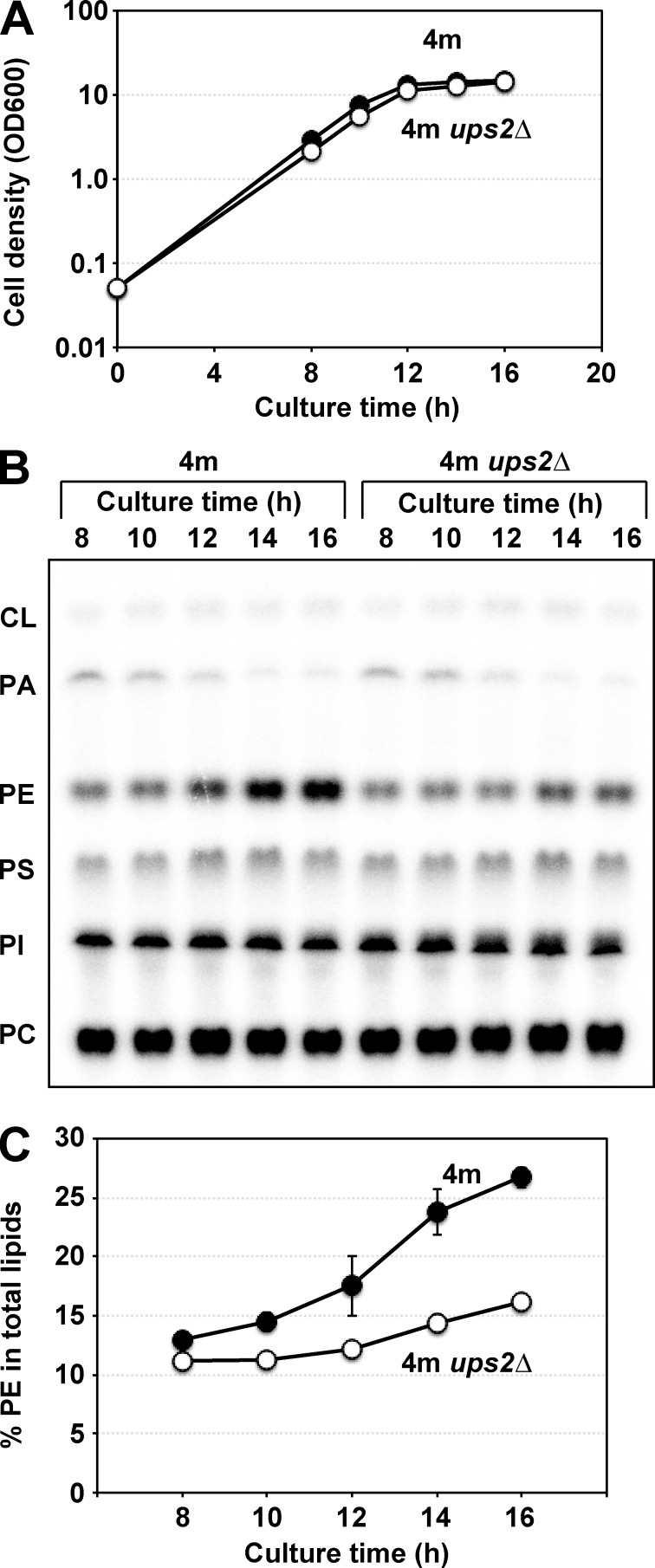
To further investigate the Ups2 function in PE metabolism, we examined the cellular PE levels in 4m and 4m ups2Δ cells at various time points during cultivation in YPD medium. At early time points in culture (8–10 h), when cells were growing logarithmically (Fig. 3 A), the cellular PE level was relatively low, and loss of Ups2 only slightly affected it (Fig. 3, B and C). In contrast, at later time points in culture (14–16 h) when cells had entered the post-log phase (Fig. 3 A), the PE level rapidly increased in 4m cells, and PE accumulation in 4m ups2Δ cells was much lower than that in 4m cells (Fig. 3 B, and C). Similar results were obtained when the cells were cultured in SCD medium containing 3 mM choline, in which PE synthesis via Kennedy pathway was absent for lack of Etn (Fig. S1). These results suggest that Ups2 is involved in PE synthesis during the post-log phase in YPD and SCD.
Figure 3.
Ups2 enhances PE accumulation in the post–log phase. (A) 4m and 4m ups2Δ cells (OD600, 0.05) were cultured in YPD for the indicated periods. At each time point, the cell density was analyzed by measuring OD600. Closed circle, 4m; open circle, 4m ups2Δ cells. (B and C) 4m and 4m ups2Δ cells (OD600, 0.05) were cultured in YPD in the presence of [32P]Pi for the indicated periods. At each time point, total cellular phospholipids were extracted, separated by TLC, and then analyzed by an imaging analyzer. (C) The percentages of PE relative to total phospholipids. The mean values for two independent experiments and deviations are shown. Closed circle, 4m cells; open circle, 4m ups2Δ cells.
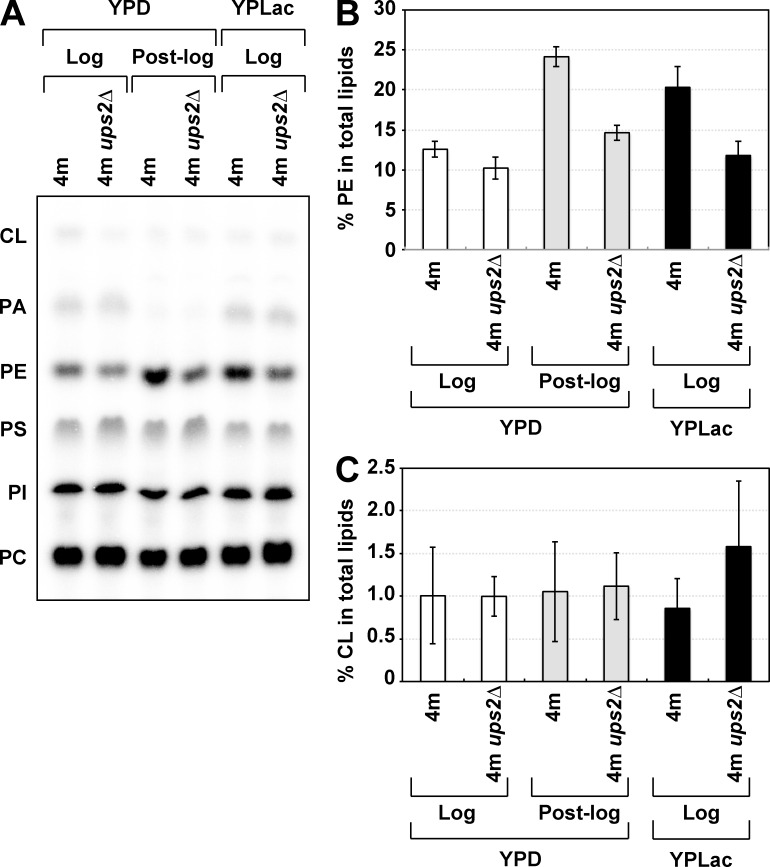
While growing logarithmically in glucose-containing medium, yeastcells produce ATP only by glycolysis, i.e., they do not use mitochondrial respiration (Entian and Barnett, 1992). When glucose is exhausted, cells undergo the diauxic shift and enter the post–log phase (post–diauxic phase). Through the diauxic shift, cells switch their energy metabolism from glycolysis to mitochondrial respiration; hence, mitochondria should be remodeled into the respiration-active state at the diauxic shift. Therefore, we assumed that Ups2 is required for PE synthesis in respiration-active mitochondria. To test this notion, 4m and 4m ups2Δ cells were cultured in a medium containing a nonfermentable carbon source, lactate (YPLac), in which mitochondrial respiration is required for cell growth. The PE level in 4m cells growing logarithmically in YPLac was higher than that in YPD (Fig. 4, A and B). Loss of Ups2 strikingly decreased the PE level of 4m cells growing in YPLac, as well as 4m cells in the post–log phase in YPD (Fig. 3 and Fig. 4, A and B). Collectively, these results suggest that Ups2 is involved in PE synthesis in respiration-active mitochondria.
Figure 4.
Ups2 enhances PE accumulation under nonfermentable conditions. (A–C) 4m and 4m ups2Δ cells (OD600, 0.05 in YPD or 0.1 in YPLac) were cultured for 8 h in YPD (YPD log phase) or 16 h in YPD (YPD post-log phase) or YPLac (YPLac log phase). Total phospholipids were extracted from the cells, separated by TLC, and then analyzed by an imaging analyzer. (B) The percentages of PE relative to total phospholipids. Values are means ± SD (n = 3). (C) The percentages of CL relative to total phospholipids. Values are means ± SD (n = 3).
Ups2–Mdm35 is involved in conversion of PS to PE in mitochondria
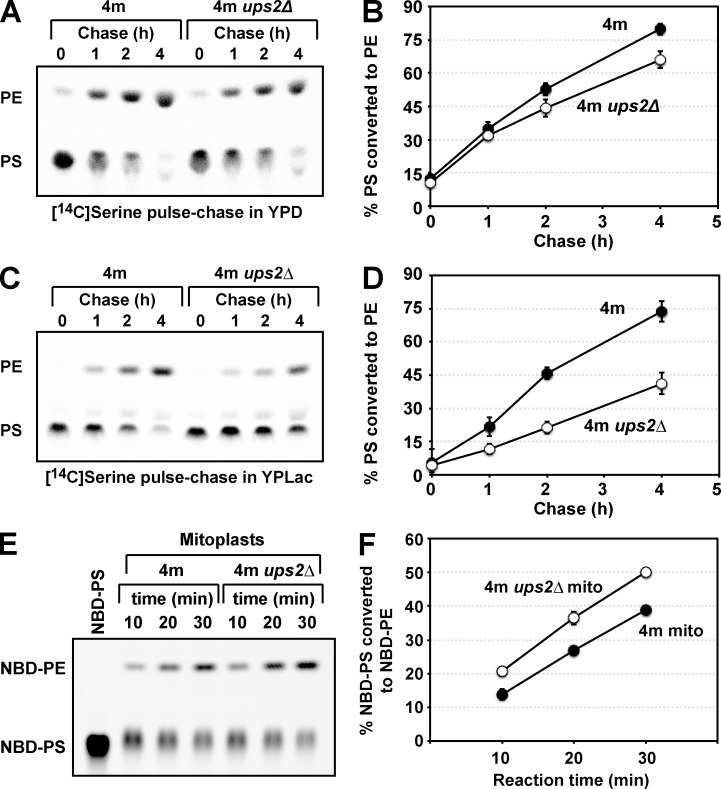
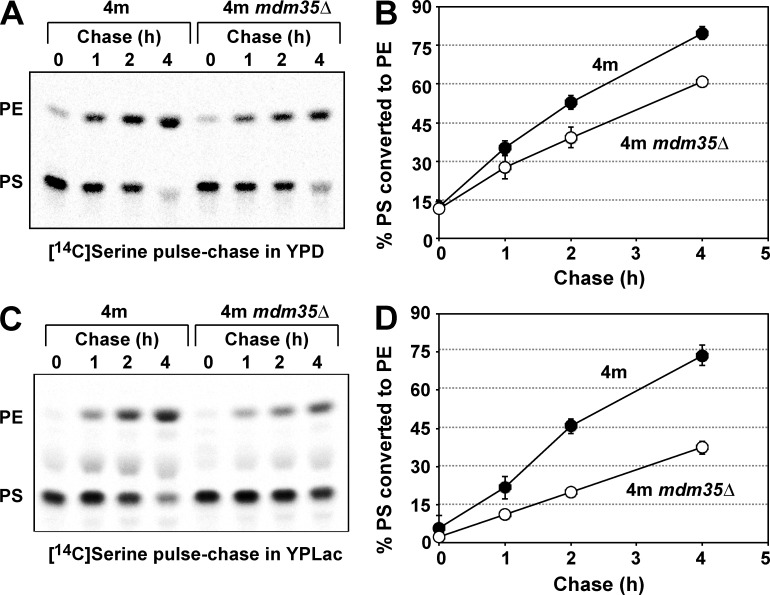
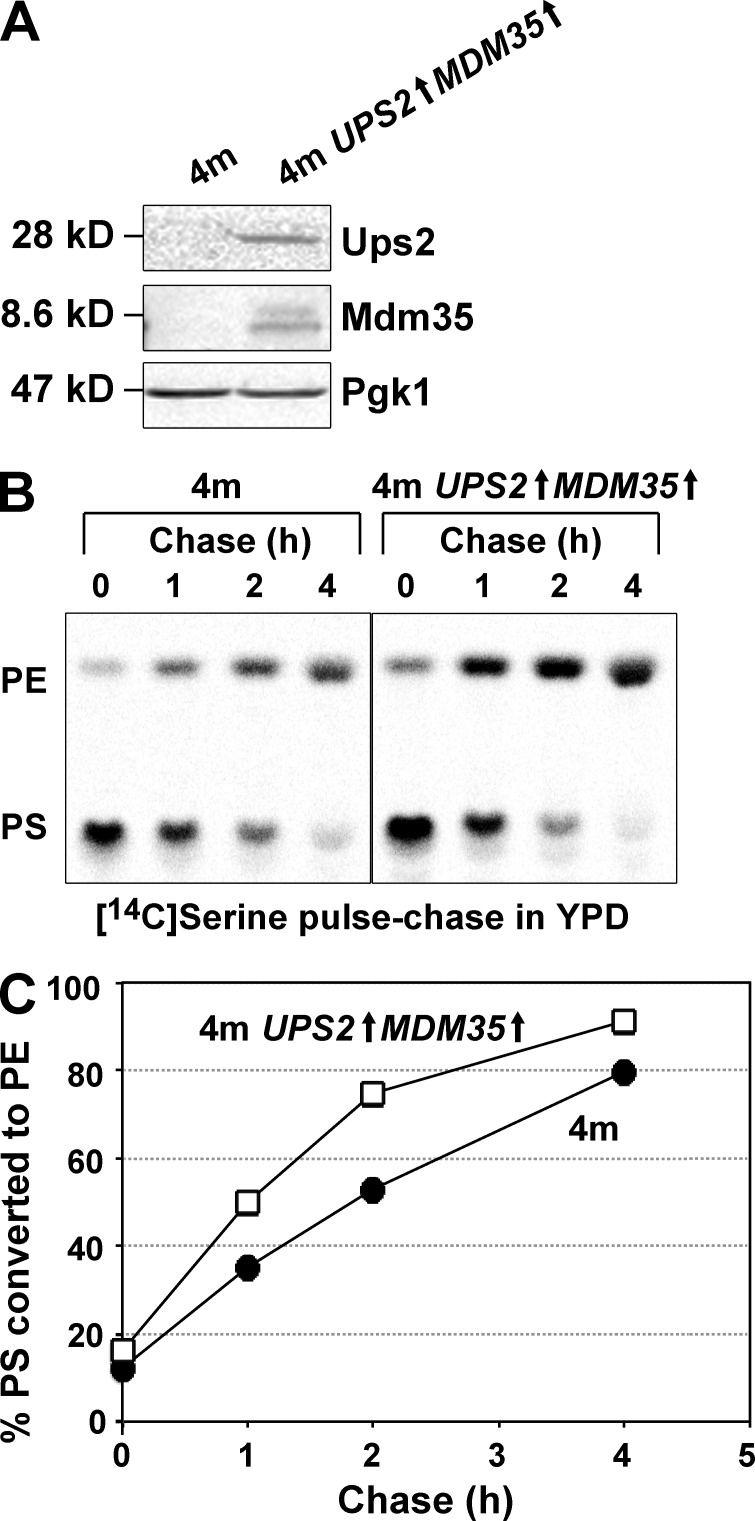
Next, we examined the function of Ups2 in PS conversion to PE by means of an in vivo pulse-chase experiment with l-[14C]serine. Because of a deficiency of Cho2 and Opi3, 4m and 4m ups2Δ cells are unable to incorporate the radiolabel from l-[14C]serine into PC through not only methylation of [14C]PE but also methylation of nonradiolabeled PE by [14C]S-adenosylmethionine synthesized from l-[14C]serine by one-carbon metabolism (Trotter and Voelker, 1995). Therefore, these strains would be suitable for monitoring conversion of PS to PE with the in vivo pulse-chase experiment. 4m and 4m ups2Δ cells were pulse-labeled with l-[14C]serine for 15 min and then further incubated for 1–4 h in either YPD or YPLac. In YPD, the rate of conversion of [14C]PS to [14C]PE in 4m ups2Δ cells was slightly lower than that in 4m cells (Fig. 5, A and B). However, in YPLac, ups2Δ caused striking retardation of conversion of [14C]PS to [14C]PE (Fig. 5 C, and D). This retardation did not seem to be caused by the pleiotropic effect of PSD2, DPL1, CHO2, and/or OPI3 deletion, because, in addition to 4m ups2Δ cells, ups2Δ and 2m ups2Δ cells also exhibited remarkable retardation of conversion of [14C]PS to [14C]PE in nonfermentable conditions (Fig. S2). To determine whether the retardation is relevant to the Psd1 enzymatic activity, we assessed PS decarboxylase activity. Mitochondria isolated from 4m and 4m ups2Δ cells grown in YPLac were osmotically swelled to disrupt the MOM and resulting mitoplasts were incubated with NBD-labeled PS (NBD-PS). The conversion of NBD-PS to NBD-PE by mitoplasts of 4m ups2Δ cells was similar to or slightly higher than that in the case of 4m cells (Fig. 5, E and F), suggesting that ups2Δ does not affect the Psd1 enzymatic activity. We also examined the effect of depletion of Mdm35, a binding partner of Ups2, on PS conversion to PE by an in vivo pulse-chase experiment with l-[14C]serine. The rate of conversion of [14C]PS to [14C]PE in 4m mdm35Δ cells was slightly lower than that in 4m cells (Fig. 6, A and B) in YPD and largely retarded in YPLac (Fig. 6, C and D). This phenotype of 4m mdm35Δ cells was very similar to that of 4m ups2Δ cells (Fig. 5, A–D). Furthermore, concomitant overexpression of UPS2 and MDM35 (UPS2↑MDM35↑) under the control of a strong glyceraldehyde-3-phosphate dehydrogenase promoter in 4m cells (Fig. 7 A) facilitated conversion of PS to PE in YPD (Fig. 7, B and C). These in vivo and in vitro observations suggest that Ups2–Mdm35 is involved in PS transport to the site of PE synthesis, namely, to the MIM in respiration-active mitochondria.
Figure 5.
Ups2 is involved in PS conversion to PE in mitochondria. (A and B) 4m and 4m ups2Δ cells were pulse-labeled with l-[14C]serine for 15 min and then further cultured in YPD for the indicated periods. At each time point, total phospholipids were extracted from the cells, separated by TLC, and then analyzed by an imaging analyzer. (B) The percentage of PS converted to PE. Values are means ± SD (n = 5 [4m ups2Δ cells] or 9 [4m cells]). Closed circle, 4m cells; open circle, 4m ups2Δ cells. (C and D) PS conversion to PE in 4m cells and 4m ups2Δ cells growing in YPLac was analyzed as in A and B. Values are means ± SD (n = 5 [4m ups2Δ cells] or 7 [4m cells]). Closed circle, 4m cells; open circle, 4m ups2Δ cells. (E and F) Mitoplasts of 4m and 4m ups2Δ cells grown in YPLac were incubated with NBD-PS. At indicated time points, phospholipids were extracted. Conversion of NBD-PS to NBD-PE was analyzed by TLC (E) and quantitated by an image analyzer (F). Values are means ± SD (n = 3). Closed circle, 4m cells; open circle, 4m ups2Δ cells.
Figure 6.
Mdm35 is involved in PS conversion to PE in mitochondria. (A and B) PS conversion to PE in 4m and 4m mdm35Δ cells growing in YPD was analyzed as in Fig. 5. The mean values for nine (4m cells) or two (4m mdm35Δ cells) independent experiments and deviations are shown. Closed circle, 4m cells; open circle, 4m mdm35Δ cells. (C and D) PS conversion to PE in 4m and 4m mdm35Δ cells growing in YPLac was analyzed as in Fig. 5. The mean values for seven (4m cells) or two (4m mdm35Δ cells) independent experiments and deviations are shown. Closed circle, 4m cells; open circle, 4m mdm35Δ cells.
Figure 7.
Ups2–Mdm35 facilitates PS conversion to PE in mitochondria. (A) 4m and 4m UPS2↑MDM35↑ cells were cultured in YPD. Logarithmically growing cells were harvested and analyzed by Western blotting with antibodies against Ups2, Mdm35, and Pgk1. Of note, anti-Ups2 and anti-Mdm35 antibodies could hardly detect endogenous Ups2 and Mdm35 in 4m cells, whereas clear Ups2 and Mdm35 signals were detected in a protein sample of 4m UPS2↑MDM35↑ cells with the respective antibodies. (B and C) PS conversion to PE in 4m and 4m UPS2↑MDM35↑ cells growing in YPD was analyzed as in Fig. 5. The mean values for nine (4m cells) or two (4m UPS2↑MDM35↑ cells) independent experiments and deviations are shown. Closed circle, 4m cells; open square, 4m UPS2↑MDM35↑ cells.
Deletion of UPS1 does not suppress the defect in conversion of PS to PE of ups2Δ cells
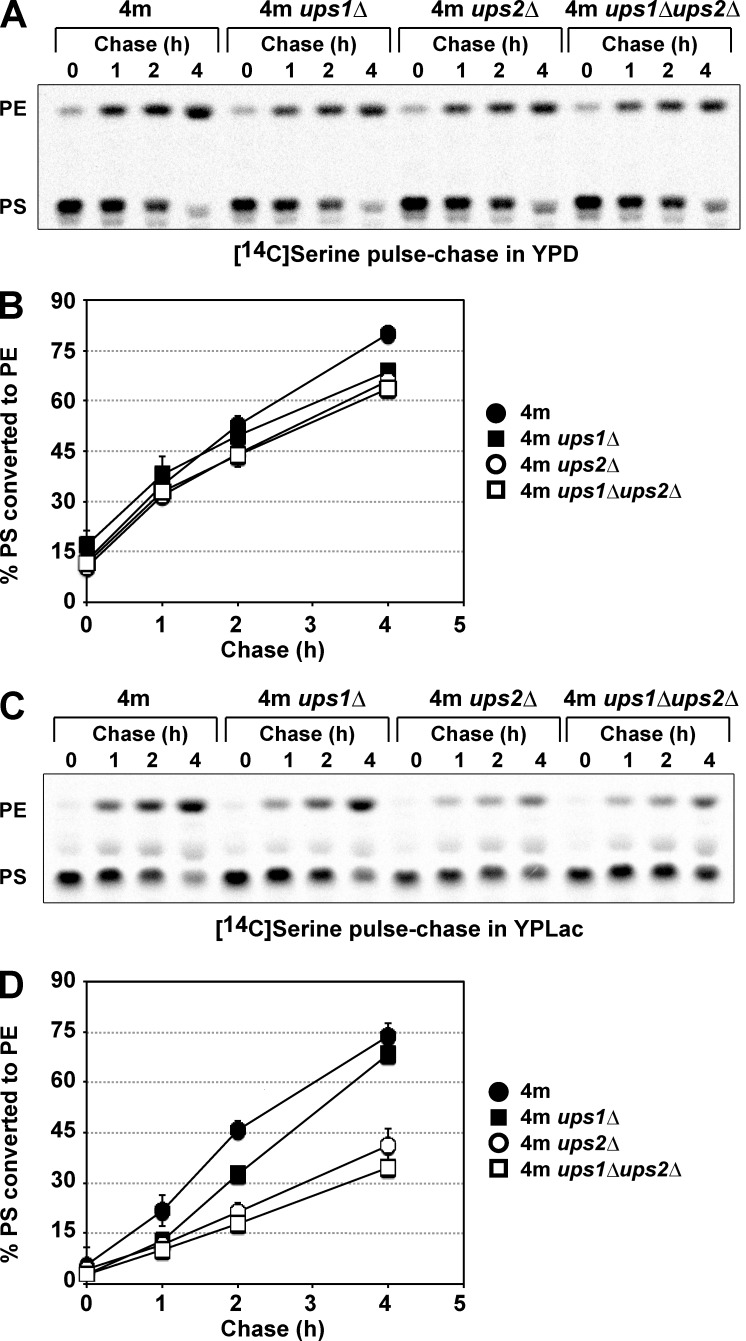
Deletion of UPS2 partially suppresses the defect in CL synthesis of ups1Δ cells (Osman et al., 2009; Tamura et al., 2009). Therefore, we next asked if deletion of UPS1 suppresses the defect in conversion of PS to PE of ups2Δ cells. The rates of conversion of PS to PE in 4m ups1Δ and 4m ups1Δups2Δ cells were determined by in vivo pulse-chase experiments in either YPD or YPLac, as described above, and compared with those in 4m and 4m ups2Δ cells. UPS1 deletion did not greatly affect the rate of conversion of [14C]PS to [14C]PE of cells cultivated in either YPD or YPLac (Fig. 8), suggesting that Ups1 is dispensable for intramitochondrial PS transport. In 4m ups1Δups2Δ cells, the rate of conversion of [14C]PS to [14C]PE was similar to that in 4m ups2Δ cells in both YPD and YPLac (Fig. 8). These results show that deletion of UPS1 does not suppress the defect in conversion of PS to PE of ups2Δ cells.
Figure 8.
Deletion of UPS1 does not suppress the defect in conversion of PS to PE of ups2Δ cells. 4m, 4m ups1Δ, 4m ups2Δ, and 4m ups1Δups2Δ cells were pulse labeled with l-[14C]serine for 15 min and then further cultured in either YPD (A and B) or YPLac (C and D) for the indicated periods. PS conversion to PE in cells was analyzed as in Fig. 5. (B) The percentage of PS converted to PE in YPD. The mean values for two (4m ups1Δ and 4m ups1Δups2Δ cells), five (4m ups2Δ cells), or nine (4m cells) independent experiments and deviations are shown. (D) The percentage of PS converted to PE in YPLac. The mean values for two (4m ups1Δ and 4m ups1Δups2Δ cells) or seven (4m and 4m ups2Δ cells) independent experiments and deviations are shown.
The Ups2–Mdm35 complex transports PS in vitro
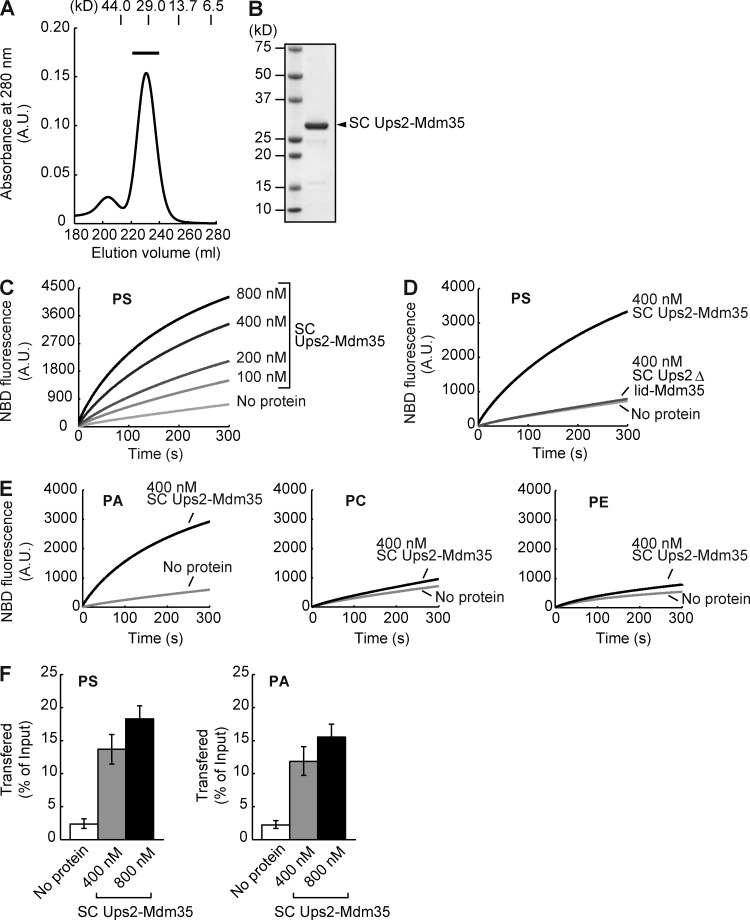
To substantiate the function of Ups2 in PS transport, we investigated whether or not Ups2 facilitates the PS transport between liposomes in vitro. Ups2 as well as Ups1 forms a complex with Mdm35 (Potting et al., 2010; Tamura et al., 2010). Mdm35 is required for the import of Ups1 and Ups2 into the mitochondrial intermembrane space (Potting et al., 2010; Tamura et al., 2010). Although we tried to bacterially coexpress Ups2 and Mdm35 from a thermotolerant yeast, Kluyveromyces marxianus (Km), in Escherichia coli cells, we failed in expression of the former (KmUps2). Because the crystal structure of the Ups1–Mdm35 complex showed that the C terminus of Ups1 is close to the N terminus of Mdm35 (Watanabe et al., 2015), we attempted to fuse KmUps2 to the N terminus of KmMdm35 to make single-chain (SC) KmUps2-KmMdm35 for expression in E. coli. To bring the C terminus of KmUps2 to the proximity of the N terminus of KmMdm35, the C-terminal region (residues 174–232) of KmUps2, which is not an essential region for the Ups2 function and is not present in Ups1 (Hall et al., 2011), was deleted (KmUps2(1–173)). SC KmUps2(1–173)-KmMdm35 was successfully expressed in E. coli and could be purified (Fig. 9, A and B). We hereafter call SC KmUps2(1−173)-KmMdm35 “SC Ups2–Mdm35.” The PS transport activity of the purified SC Ups2–Mdm35 was measured by means of a fluorescent-based assay similar to the PA transport assay for the Ups1–Mdm35 complex reported previously (Watanabe et al., 2015). Donor liposomes containing NBD-PS and Rhodamine-labeled PE (Rhod-PE) were incubated with SC Ups2–Mdm35 and acceptor liposomes without fluorescent lipids. When NBD-PS and Rhod-PE are on the same liposome, NBD fluorescence is quenched by Rhodamine. If SC Ups2–Mdm35 transports NBD-PS from donor liposomes to acceptor liposomes, the fluorescence of NBD-PS will be dequenched and increased in intensity. As shown in Fig. 9 C, SC Ups2–Mdm35 increased the NBD-fluorescence of PS in a time- and dose-dependent manner, indicating that SC Ups2–Mdm35 has PS transport activity. According to the crystal structure of the Ups1–Mdm35 complex with PA, the Ω1 loop domain forms a lid for the phospholipid-binding pocket. The Ω1 loop domain of Ups1 is a prerequisite for PA extraction from liposomes and PA transport between liposomes (Watanabe et al., 2015). Therefore, to examine the role of the Ω1 loop domain in the PS transport function of Ups2, we measured the PS transport activity of a Ups2 mutant lacking the region corresponding to the Ω1 loop domain (residues 62–71; Δlid). SC Ups2Δlid–Mdm35 showed no PS transport activity (Fig. 9 D), suggesting that the Ups2–Mdm35 complex transports PS in a manner similar to the Ups1-mediated PA transport. We also examined the substrate preference of SC Ups2–Mdm35. As shown in Fig. 9 E, SC Ups2–Mdm35 exhibited transport activity for NBD-PA in addition to NBD-PS but only little activity for transport of NBD-PE and NBD-PC.
Figure 9.
Phospholipid transfer activity of Ups2–Mdm35. (A) Size-exclusion chromatography of SC Ups2–Mdm35 using a HiLoad 26/60 Superdex 200 PG column. Positions of molecular mass standards (in kilodaltons) are shown. A fraction indicated with a black bar in the elution profile was used in the phospholipid transfer assay. (B) Purified SC Ups2–Mdm35 was analyzed by SDS-PAGE followed by CBB staining. (C) PS transfer activity of SC Ups2–Mdm35 was measured at 25°C by the fluorescent-based phospholipid transfer assay between liposomes (see Materials and methods for details). (D) PS transfer activity of SC Ups2ΔLid–Mdm35 was analyzed as in C. (E) PA, PC, and PE transfer activities of SC Ups2–Mdm35 were analyzed as in C. (F) PS and PA transfer activities of SC Ups2–Mdm35 were measured by the mass spectrometry–based phospholipid transfer assay. SC Ups2–Mdm35 was incubated with heavy donor liposomes containing DOPS or DOPA and light acceptor liposomes. Liposomes were separated by sucrose density-gradient ultracentrifugation. Amounts of DOPS (left) or DOPA (right) in acceptor liposomes relative to those in input donor liposomes were quantified by mass spectrometry. Values are means ± SD (n = 3).
Because NBD-PS and NBD-PA have an attached bulky fluorescent group as compared with authentic PS and PA, respectively, we attempted to confirm nonfluorescently labeled PS and PA transfer activities of Ups2 by the mass spectrometry–based phospholipid transfer assay. Heavy donor liposomes with nonlabeled PS or PA and light acceptor liposomes without PS and PA were incubated with SC Ups2–Mdm35 and separated by sucrose density-gradient centrifugation. Then, phospholipids in the acceptor liposomes were analyzed by mass spectroscopy (Figs. 9 F and S3). SC Ups2–Mdm35 increased the amount of both PS and PA transferred to acceptor liposomes in a dose-dependent manner (Figs. 9 F and S3). These results suggest that Ups2–Mdm35 is able to transport PS and PA between liposomes. However, in contrast to the loss of Ups1 (Osman et al., 2009; Tamura et al., 2009), the loss of Ups2 did not lead to a significant decrease in the level of CL in vivo (Fig. 4 C). This is consistent with previous studies (Osman et al., 2009; Tamura et al., 2009) and implies that Ups2 does not transport PA from the MOM to the MIM for CL synthesis in vivo (see also Discussion). Taking these results together, we conclude that Ups2–Mdm35 transports PS from the MOM to the MIM for PE synthesis in respiration-active mitochondria.
The expression of UPS2 is elevated under nonfermentable conditions and at the diauxic shift
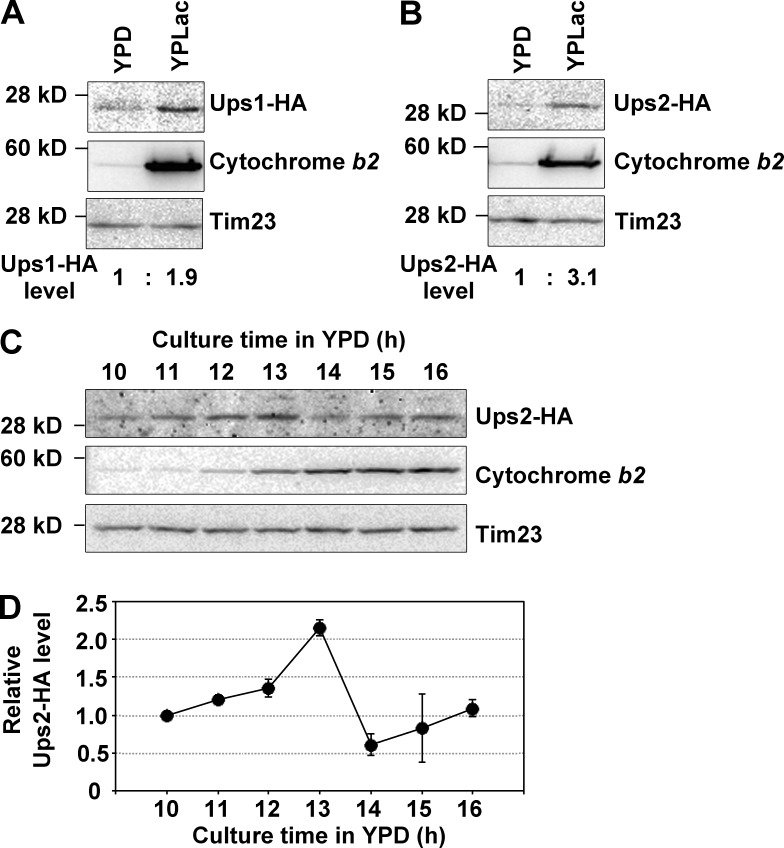
To examine the correlation of function with expression, the protein levels of Ups1 and Ups2 in the fermentable and nonfermentable conditions were analyzed by C-terminal tagging of the chromosomal UPS1 and UPS2 gene with an HA epitope, followed by Western blotting. Cells expressing Ups1-HA or Ups2-HA were cultured in YPD and YPLac to the log phase, and then total protein extracts were analyzed by SDS-PAGE followed by Western blotting. As shown in Fig. 10 (A and B), Ups1-HA and Ups2-HA levels were increased 1.9- and 3.1-fold, respectively, in YPLac culture as compared with those in YPD culture. Next, we investigated the detailed expression profiles of Ups2-HA during YPD culture. Cells expressing Ups2-HA were grown in YPD, harvested at various time points, and then analyzed by Western blotting. At an early time in culture (10–12 h), cytochrome b2, a subunit of respiratory chain complex III, was poorly expressed in cells, indicating that respiration activity was low. At 13 h in culture, when the growth phase shifted from the log phase to the post–log phase (Fig. 3), cytochrome b2 expression extensively increased (Fig. 10 C). These results demonstrated that cells underwent the diauxic shift at 13 h in the culture and changed their metabolism from glycolysis to respiration. Notably, expression of Ups2-HA was the highest at this time point (Fig. 10, C and D). At a late time in culture (15–16 h), Ups2-HA was quickly down-regulated, which is consistent with a previous finding that Ups2 is unstable and readily degraded by mitochondrial proteases (Potting et al., 2010). These results suggest that Ups2 has an important function under nonfermentable conditions and at the diauxic shift.
Figure 10.
The expression of UPS2 is elevated under nonfermentable conditions and at the diauxic shift. (A and B) Yeast cells expressing Ups1-HA (A) or Ups2-HA (B) (OD600, 0.05 in YPD or 0.1 in YPLac) were cultured for 8 h in YPD or 12 h in YPLac. After cultivation, cells were harvested and analyzed by Western blotting with antibodies against HA, cytochrome b2, and Tim23. Ups1-HA and Ups2-HA levels were quantified, normalized to Tim23 levels, and expressed as fold increase relative to those in YPD culture, respectively. (C and D) Yeast cells expressing Ups2-HA (OD600, 0.05) were cultured in YPD for the indicated periods. At each time point, cells were harvested and analyzed by Western blotting with antibodies against HA, cytochrome b2, and Tim23. (D) Ups2-HA levels were quantified, normalized to Tim23 levels, and expressed as fold increase relative to one at 10 h. The mean values for two independent experiments and deviations are shown.
Discussion
CL and PE are essential phospholipids for mitochondrial functions, including respiratory chain complex assembly (Böttinger et al., 2012), development of the cristae structure (Tasseva et al., 2013), and mitochondrial fusion (Joshi et al., 2012). Because mitochondria are dynamic organelles and change their function and morphology in response to cellular circumstances, the synthesis of CL and PE should be strictly regulated during mitochondrial remodeling. However, little is known about how the synthesis of these phospholipids is regulated.
In the present study, we tried to clarify the function of Ups2 in mitochondrial PE metabolism both in vivo and in vitro. For this purpose, we established a 4m yeast strain lacking Psd2, Dpl1, Cho2, and Opi3. By using this 4m strain, we could exclude the influence of endosomal PE production and PE methylation on the cellular PE level. Because PE production via the Kennedy pathway comprises a minor portion (∼30%) of cellular PE in budding yeast (Trotter and Voelker, 1995), the PE level in 4m yeast cells mostly reflects PE synthesis in mitochondria. Thus, the 4m yeast strain would be a suitable tool for monitoring mitochondrial PE synthesis. Loss of Ups2 decreased the PE level even in the absence of endosomal PE synthesis and PE methylation in the ER (Fig. 1). Furthermore, we found that PE generated in mitochondria is unexpectedly stable in cells in the absence of phospholipid methylation and that loss of Ups2 does not affect PE stability (Fig. 2). Collectively, these results suggest that Ups2 is involved in the synthesis of PE rather than the conversion of PE to other metabolites.
In the log phase in glucose-containing medium, when yeast cells produce ATP mainly through glycolysis, mitochondrial PE production was relatively low and loss of Ups2 did not significantly affect the PE level (Fig. 3). In contrast, in the post–log phase, when cells undergo the diauxic shift and produce ATP via mitochondrial respiration, the PE level dramatically increased in a Ups2-dependent manner (Fig. 3). In the medium containing a nonfermentable carbon source, lactate, loss of Ups2 also significantly affected the PE level (Fig. 4) and retarded conversion of PS to PE in mitochondria (Fig. 5). These results suggest that Ups2 enhances PE synthesis in respiration-active mitochondria. At the diauxic shift, because mitochondria would quickly develop respiratory chain complexes and the cristae structure, Ups2-dependent PE accumulation would contribute to such mitochondrial remodeling at the diauxic shift. Supporting this notion, Ups2 expression was the highest at the diauxic shift (Fig. 10).
From the similarity in amino acid sequences between Ups2 and Ups1 (Tamura et al., 2009), we supposed that Ups2 is involved in phospholipid transport. Indeed, we demonstrated the PS transfer activity of SC Ups2–Mdm35 in vitro (Fig. 9). Taking all the results together, we conclude that Ups2–Mdm35 functions as a PS transfer protein between the MOM and MIM in respiration-active mitochondria to enhance PE production.
SC Ups2–Mdm35 was able to transport PA and PS between liposomes, although loss of Ups2 did not lead to a significant decrease in the level of CL in vivo (Fig. 4 C), which is consistent with previous studies (Osman et al., 2009; Tamura et al., 2009). Interestingly, it has been shown that Ups2 overexpression decreases the CL level (Osman et al., 2009) and that loss of Ups2 partially rescues CL synthesis in Ups1-defective cells (Osman et al., 2009; Tamura et al., 2009). Therefore, Ups2 seems to give a negative effect on CL synthesis. Given that Ups2–Mdm35 can transport PA between liposomes, one possible explanation for this negative effect is that there is an unknown factor inhibiting the release of PA from the PA–Ups2–Mdm35 complex in vivo, and the inhibition results in retardation of PA transport through authentic physiological pathways. The second explanation is that the rate of PS and PA transport reconstituted in vitro with SC Ups2–Mdm35 is insufficient for physiological transports and that there exists a factor accelerating transport of PS, but not PA, by Ups2–Mdm35 in vivo. In this case, the formation of the PA–Ups2–Mdm35 complex also seems to retard PA transport through authentic physiological pathways. The third explanation is that in mitochondria Ups2–Mdm35 transports PA backward from the MIM, where PA is converted to CL, to the MOM. It is noteworthy that the PS transfer proteins oxysterol-binding protein–related protein (ORP) 5 and 8 and oxysterol-biding homology (Osh) protein 6 exchange PS for PI 4-phosphate between the ER and the plasma membrane (Chung et al., 2015; Moser von Filseck et al., 2015). Similarly, Ups2–Mdm35 might mediate PS–PA exchange between the mitochondrial membranes, thereby transporting PA backward from the MIM to the MOM. In future, the study addressing these possibilities should be performed to clarify the mechanism of the negative effect of Ups2 on CL biosynthesis.
PS conversion to PE was only weakly dependent on Ups2 under fermentable conditions (Fig. 5). This is likely the reason why Ups2-dependent PE synthesis was not observed in the previous studies (Osman et al., 2009; Tamura et al., 2012a). In addition, loss of Ups2 did not completely abolish PS conversion to PE under nonfermentable conditions (Fig. 5). These results imply that another factor in addition to Ups2 is involved in PS transport within mitochondria. The unidentified factor would mainly act as PS transport machinery under fermentable conditions, whereas Ups2–Msm35 mainly acts under nonfermentable conditions. Furthermore ERMES and vCLAMP, which are ER–mitochondria and vacuole–mitochondria contact sites, respectively, are reciprocally regulated in response to cellular metabolism. Under fermentable conditions, ERMES and vCLAMP cooperatively mediate phospholipid transport. In contrast, under nonfermentable conditions, Vps39, a constituent of vCLAMP, is inactivated through phosphorylation, whereas ERMES is up-regulated (Hönscher et al., 2014). Therefore, phospholipid transport pathways into and within mitochondria seem to change depending on the cellular metabolic state.
Ups1 and Ups2 are highly conserved among species. PRELI, the mammalian homologue of Ups1 is reported to function in PA transport and CL synthesis in mammalian cells (Potting et al., 2013). Therefore, the Ups2 function in PS transport may be also conserved among species. In mammals, mitochondrial energy metabolism is intimately linked to cell differentiation. The energy production by pluripotent stem cells (PSCs) depends on glycolysis even in aerobic circumstances (Folmes et al., 2011; Zhang et al., 2011), and energy production shifts to mitochondrial respiration during PSC differentiation (Varum et al., 2011). This metabolic transition during cell differentiation is very similar to that during the diauxic shift in yeast. Thus, Prelid2, the mammalian homologue of Ups2 (Gao et al., 2009), might be involved in PE production in differentiating cells, contributing to mitochondrial remodeling. Furthermore, recent studies implied that mitochondrial energy metabolism could regulate the differentiation rate in stem cells (Chung et al., 2007; Mandal et al., 2011; Carey et al., 2015). Accordingly, functional analysis of Ups2 homologues in mammalian PSCs would contribute to improvement of stem cell technology and regenerative medicine.
In conclusion, the present work shows that a protein complex consisting of Ups2, a yeast member of a highly conserved family of intermembrane space proteins, and Mdm35 functions as a phospholipid transfer protein that can transport PS from the MOM to the MIM for PE synthesis and that the PE synthesis depending on Ups2–Mdm35 is enhanced in respiration-active mitochondria. This enhancement may be required for the remodeling of the mitochondrial membrane in response to the cellular metabolic state to meet the demand of cellular functions.
Materials and methods
Yeast strains
The yeast strains used in this study are listed in Table S1. Complete disruption, promoter replacement, and tagging of the yeast gene were accomplished by PCR-mediated gene replacement (Lorenz et al., 1995) with a pair of primers and a template plasmid, as listed in Table S2.
Plasmids
Plasmids pCgLEU2 (National Bio-Resource Project [NBRP] ID BYP1419), pCgHIS3 (NBRP ID BYP1804), pCgTRP1 (NBRP ID BYP1805), and pCgURA3 (NBRP ID BYP1806), which carry Candida glabrata LEU2, HIS3, TRP1, and URA3, respectively, were provided by the NBRP of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
pCgLEU2-NT1, which was used for disruption of the yeast gene with a LEU2 marker, was constructed as follows. A DNA fragment of the LEU2 gene was amplified from pCgLEU2 using a forward primer containing an NcoI site, 5′-ATACCATGGCTGTGACCAAGAC-3′, and a reverse primer containing a XhoI site, 5′-ATACTCGAGCTAAGCTAATAGTTCCCTG-3′, digested with NcoI and XhoI, and then ligated with a large DNA fragment of pFA6a-hphNT1 (Janke et al., 2004) digested with NcoI and XhoI.
pCgHIS3-TB, which was used for disruption of the yeast gene with a HIS3 marker, was constructed as follows. A DNA fragment of the HIS3 gene was amplified from pCgHIS3 using a forward primer containing a BamHI site, 5′-ATAGGATCCAATTCTGGCCCGAGTCAC-3′, and a reverse primer containing an EcoRI site, 5′-GATGAATTCGAGCTCGAGCTTGGGTCTTCTGGAG-3′, digested with BamHI and EcoRI, and then ligated with a large DNA fragment of pFA6a-kanMX4 (Wach et al., 1994) digested with BamHI and EcoRI.
pCgURA3-NT2, which was used for disruption of the yeast gene with a URA3 marker, was constructed as follows. A DNA fragment of the URA3 gene was amplified from pCgURA3 using a forward primer, 5′-TCCAGTGCCTCATATTTAC-3′, and a reverse primer containing a XhoI site, 5′-ATACTCGAGTCAGTTTCCTATTCTTTTCAAG-3′, digested with XhoI, and then ligated with a large DNA fragment obtained by means of XhoI-digestion of pFA6a-natNT2 (Janke et al., 2004) pretreated with NcoI, followed by blunt-ending with DNA polymerase I (Klenow fragment).
pCgTRP1-TB, which was used for disruption of the yeast gene with a TRP1 marker, was constructed as follows. A DNA fragment of the TRP1 gene was amplified from pCgTRP1 using a forward primer containing a BamHI site, 5′-ATAGGATCCGGTACCGTCGCTTTGAGAG-3′, and a reverse primer containing a SacI site, 5′-TTCGAGCTCGGTACCCGGAAGTTGGCTG-3′, digested with BamHI and SacI, and then ligated with a large DNA fragment of pFA6a-kanMX4 (Wach et al., 1994) digested with BamHI and SacI.
An E. coli expression plasmid for N-terminally hexahistidine (His6)-tagged SC KmUps2(1–173)–KmMdm35 was constructed as follows. A DNA fragment of KmUps2(1–173) was amplified by PCR using a forward primer containing an EcoRI site, 5′-CGGAATTCGATGAGACTCTTTGAGAACCAA-3′, and a reverse primer containing a NotI site, 5′-ATAAGAATGCGGCCGCTTAGCTTTCATCAAGAAGTTGTAA-3′, and digested with EcoRI and NotI. A DNA fragment of KmMdm35 was amplified by PCR using a forward primer containing an NdeI site, 5′-GGAATTCCATATGGGTAACGTGATGAGTGC-3′, and a reverse primer containing a XhoI site, 5′-CCGCTCGAGTTACTCGTTTACGGGTTTGCC-3′, and digested with NdeI and XhoI. Then, the DNA fragments of KmUps2(1–173) and KmMdm35 were inserted into the EcoRI–NotI site and the NdeI–XhoI site in pETDuet-1 (EMD Millipore), respectively. The open reading flames of the N-terminally His6-tagged KmUps2(1–173) and KmMdm35 in pETDuet-1 were fused by inverse PCR using a forward primer, 5′-ATGGGTAACGTGATGAGTGCTAGTTTCGCG-3′, and a reverse primer, 5′-GCTTTCATCAAGAAGTTGTAAAACTGAATC-3′. The expression plasmid for the N-terminally His6-tagged SC KmUps2(1–173)Δlid–KmMdm35 was constructed by inverse PCR using a forward primer, 5′-GGATCCAATGTGAGTTATATCCGTGAGGTG-3′, and a reverse primer, 5′-CGACTGTTTACATGTGATCAAACGTTCGCT-3′. The resulting constructs were sequenced to confirm their identities. We refer to His6-tagged single-chain KmUps2(1–173)–KmMdm35 as SC Ups2–Mdm35.
Cell culture
Yeast cells were grown in YPD (1% yeast extract, 2% peptone, 0.008% adenine, and 2% glucose), YPLac (1% yeast extract, 2% peptone, 0.008% adenine, and 3% lactic acid, pH 6.0), or SCD (0.67% yeast nitrogen base without amino acids, 0.2% drop out mix, 0.008% adenine, and 2% glucose, pH 6.0). For metabolic labeling with l-[14C]serine, SCD-serine (0.67% yeast nitrogen base without amino acids, 0.2% drop out mix lacking serine, 0.008% adenine, and 2% glucose, pH 6.0) or SCLac-serine (0.67% yeast nitrogen base without amino acids, 0.2% drop out mix lacking serine, 0.008% adenine, and 2% lactic acid, pH 6.0), both supplemented with 2 mM ethanolamine and 2 mM choline, were used.
Pulse-chase experiment
Cells growing logarithmically were resuspended in SCD-serine supplemented with 2 mM choline and 2 mM ethanolamine and then incubated with 2.5 µCi/ml l-[14C]serine at 30°C for 15 min. After washing twice with YPD, cells were further incubated in YPD at 30°C for different periods. After incubation, cells were harvested, resuspended in 150 µl 80% ethanol, and kept at −80°C until all samples had been collected. The samples were heated at 95°C for 15 min, mixed with 800 µl chloroform/methanol (1:1, vol/vol), and then vortexed. 330 µl of 0.1 M HCl/0.1 M KCl was then added to the samples. The organic phase was separated by centrifugation at 3,000 g for 2 min, dried in a centrifugal evaporator, and resuspended in chloroform/methanol (1:2, vol/vol). The samples were then subjected to TLC (silica gel 60 TLC plate; Merck) with the solvent system chloroform/methylacetate/1-propanol/methanol/0.25% KCl (25:25:25:10:9). 14C-labeled phospholipids were detected and quantitated with an imaging analyzer (FLA-5000; Fujifilm) and MultiGauge software (Fujifilm). In the pulse-chase experiment with a nonfermentable carbon source, YPLac and SCLac-serine were used instead of YPD and SCD-serine, respectively.
Analysis of cellular phospholipid compositions
Yeast cells were grown at 30°C to saturation in either YPD or YPLac. The cells were then diluted to an OD600 of 0.05 (in YPD) or 0.1 (in YPLac) and further incubated at 30°C for different periods in the presence of 1 µCi/ml [32P]Pi. After incubation, cells were harvested, resuspended in 150 µl of 80% ethanol, and kept at −80°C until all samples had been collected. The samples were heated at 95°C for 15 min, mixed with 800 µl of chloroform/methanol (1:1, vol/vol), and then vortexed. 330 µl of 0.1 M HCl/0.1 M KCl was then added to the samples. The organic phase was separated by centrifugation at 3,000 g for 2 min. The equivalent radioactivity of each sample was collected and dried in a centrifugal evaporator and resuspended in chloroform/methanol (1:2, vol/vol). The samples were then subjected to TLC on a TLC plate (LK5 silica gel 150A TLC plate; Whatman), which had been pretreated with 1.8% boric acid, with the solvent system chloroform/ethanol/water/triethylamine (30:35:7:35, vol/vol; Vaden et al., 2005). 32P-labeled phospholipids were detected and quantitated with an imaging analyzer, FLA-5000 (Fujifilm) and MultiGauge software (Fujifilm).
Protein extraction from yeast cells for Western blotting
Yeast cells growing in YPD were harvested, washed with H2O, and then resuspended in 1 ml H2O. 150 µl 2 M NaOH/8% 2-mercaptoethanol was added to the cell suspensions; the mixtures were incubated on ice for 10 min, 75 µl 100% trichloroacetic acid was added to the samples, and the samples were further incubated on ice for 10 min. After incubation, the proteins were precipitated by centrifugation at 20,000 g for 2 min. The precipitates were resuspended in 1 ml acetone, followed by centrifugation at 20,000 g for 2 min. The precipitates were then resuspended with SDS sample buffer and subjected to Western blotting. Band intensities of Tim23, Ups1-HA, and Ups2-HA were quantified with ImageJ software.
Determination of Psd1 activity
Mitochondria from 4m or 4m ups2Δ cells grown in YPLac were resuspended in assay buffer (0.1 M Tris-HCl, pH 7.4, 10 mM EDTA, and 1 µM PS-C6-NBD; Avanti Polar Lipids, Inc.) to a final concentration of 1 mg/ml and then incubated at 30°C. At the time points indicated, mitochondria (100 µg) were removed from the reaction mixture. Phospholipids were extracted and subjected to TLC (Silica gel 60 TLC plate; Merck) with the solvent system chloroform/methylacetate/1-propanol/methanol/0.25% KCl (25:25:25:10:9). NBD signals were detected and quantified with an imaging analyzer (FLA-5000; Fujifilm) and MultiGauge software (Fujifilm).
Protein expression and purification
The target proteins were expressed in E. coli SHuffle T7 cells (New England Biolabs, Inc.) cultured in LB medium. After addition of 0.1 mM isopropyl-d-thiogalactoside, the cells were cultured at 20°C for 20 h, and were disrupted by sonication. The His6-tagged SC Ups2–Mdm35 was affinity-purified by a Ni-NTA Superflow column (QIAGEN). The obtained protein was purified by a HiLoad 26/60 Superdex 200 PG column (GE Healthcare) with elution buffer (20 mM Tris-HCl, pH 7.5, and 150 mM NaCl).
Liposomes
The phospholipids DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine, 850375C), DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 850725C), DOPS (1,2-dioleoyl-sn-glycero-3-phospho-l-serine [sodium salt], 840035C), DOPA (1,2-dioleoyl-sn-glycero-3-phosphate [sodium salt], 840875C), 18:1 CL (1’,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho-]-sn-glycerol, 710355C), 18:1-12:0 NBD-PA (1-oleoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphate, 810176C), 18:1-12:0 NBD-PS (1-oleoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoserine, 810195C), 18:1-12:0 NBD-PC (1-oleoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphocholine, 810133C), 18:1-12:0 NBD-PE (1-oleoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoethanolamine, 810156C), Egg Liss-Rhod-PE (L-α-phosphatidylethanolamine-N-(lissamine rhodamine B sulfonyl), 810146C), and 18:1 Liss-Rhod-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) [ammonium salt], 810150C) were obtained from Avanti Polar Lipids, Inc. Lipids in stock solutions in chloroform were mixed at the desired molar ratio, and the solvent was evaporated. The lipid film was hydrated in appropriate buffer. The lipid suspension was incubated at room temperature for 30 min and extruded 20 times through a polycarbonate 0.1-µm filter using a mini-extruder (Avanti Polar Lipids, Inc.).
Phospholipid transfer assay
NBD-phospholipid transfer activities of SC Ups2–Mdm35 and SC Ups2ΔLid–Mdm35 were measured by the fluorescent dequenching assay as described previously (Watanabe et al., 2015), with several modifications. Donor liposomes (12.5 µM; DOPC/DOPE/Egg Liss-Rhod-PE/18:1–12:0 NBD-phospholipids = 50:40:2:8) were incubated with acceptor liposomes (50 µM; DOPC/DOPE/18:1 CL = 50:40:10) in the presence or absence of the recombinant proteins in 2 ml of assay buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2 mM EDTA) at 25°C. NBD fluorescence was monitored by a FP-8500 spectrofluorometer (Jasco).
The mass spectrometry–based phospholipid transfer assay was performed as described previously, with several modifications (Connerth et al., 2012). SC Ups2–Mdm35 was incubated with donor liposomes (200 µM; DOPC/DOPE/DOPS/18:1 Liss-Rhod-PE or DOPC/DOPE/DOPA/18:1 Liss-Rhod-PE = 50:29.9:20:0.1) containing 10% sucrose and acceptor liposomes (200 µM; DOPC/DOPE/18:1 CL = 50:40:10) in 600 µl assay buffer (20 mM Tris-HCl, pH 7.5, and 150 mM NaCl) at 25°C. After 10-min incubation at 25°C, the sample was mixed with 200 µl assay buffer containing 30% sucrose and incubated for 10 min on ice. The sample was placed on an ultracentrifuge tube and overlaid with 1,200, 800, and 150 µl of assay buffer containing 5%, 2.5%, and 0% sucrose, respectively, and subjected to ultracentrifugation at 217,000 g for 2.5 h. The top fraction (1.5 ml) containing the floated acceptor liposomes was collected, and 200 µl out of 1.5 ml was used for extraction of phospholipids. The absence of donor liposomes in the top fraction was confirmed by measurement of Rhod-PE fluorescence. 200 µl of the fraction was mixed with 800 µl chloroform/methanol (2:1, vol/vol) and then vortexed. 200 µl of 0.1 M HCl and 0.1 M KCl was then added to the sample and vortexed. The organic phase was separated by centrifugation at 800 g for 2 min, collected, and dried under N2 gas. The resulting lipid film was dissolved in 1 ml of 2-propanol containing 5 mM ammonium formate and 0.1% formate. Lipid samples were injected into a Turbo V electrospray ion source by using a syringe pump with a flow rate of 10 µl/min and analyzed by direct infusion mass spectrometry using a 3200 QTrap system (SCIEX). DOPS and DOPA were analyzed by a multiple reaction monitoring (MRM) method. MRM analyses were performed in negative ion mode. For quantification, the singly charged precursors (DOPS, m/z 786.6; DOPA, m/z 699.5) and the fragments corresponding to the 18:1 fatty acids (m/z 281.1) were selected as MRM transitions.
Statistical analysis
The results of all quantitative experiments except for cell density measurements in Fig. 3 A and Fig. S1 A are shown as means of independent experiments performed multiple times as indicated. The statistical significance of mean differences in Fig. 1 B was assessed by Student’s t test.
Online supplemental material
Fig. S1 shows a defect in PE accumulation of 4m ups2Δ cells cultivated in SCD medium containing 3 mM choline, in which PE synthesis via Kennedy pathway is absent. Fig. S2 documents the effect of UPS2 deletion in wild-type and 2m cells on the conversion of PS to PE. Fig. S3 is supplemental data for mass spectrometry–based phospholipid transfer assay. Table S1 lists yeast strains used in this study. Table S2 lists PCR templates and primers used for gene manipulation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201601082/DC1.
Supplementary Material
Acknowledgments
We thank Junko Suzuki for making plasmids and Tadashi Ogishima and Motohiro Tani for discussions.
We acknowledge support of this work by Japan Society for the Promotion of Science KAKENHI (grants 15H05705 and 22227003 to T. Endo, 15H05595 and 25870311 to Y. Tamura, and 16K07354 to O. Kuge), a Core Research for Evolutional Science and Technology Grant from Japan Science and Technology Agency (T. Endo), and a grant from Kyushu University Interdisciplinary Programs in Education and Projects in Research development (N. Miyata). Y. Watanabe is a Research Fellow of the Japan Society for the Promotion of Science.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CL
- cardiolipin
- ERMES
- ER–mitochondria encounter structure
- Etn
- ethanolamine
- Km
- Kluyveromyces marxianus
- MIM
- mitochondrial inner membrane
- MOM
- mitochondrial outer membrane
- MRM
- multiple reaction monitoring
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- SC
- single chain
- vCLAMP
- vacuole-mitochondrial patch
References
- Böttinger L., Horvath S.E., Kleinschroth T., Hunte C., Daum G., Pfanner N., and Becker T.. 2012. Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J. Mol. Biol. 423:677–686. 10.1016/j.jmb.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Finley L.W., Cross J.R., Allis C.D., and Thompson C.B.. 2015. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 518:413–416. 10.1038/nature13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.C., Heacock P.N., Mileykovskaya E., Voelker D.R., and Dowhan W.. 1998. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J. Biol. Chem. 273:14933–14941. 10.1074/jbc.273.24.14933 [DOI] [PubMed] [Google Scholar]
- Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L.B., Narayanaswamy P., Wenk M.R., Nakatsu F., and De Camilli P.. 2015. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER–plasma membrane contacts. Science. 349:428–432. 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Dzeja P.P., Faustino R.S., Perez-Terzic C., Behfar A., and Terzic A.. 2007. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat. Clin. Pract. Cardiovasc. Med. 4(Suppl 1):S60–S67. 10.1038/ncpcardio0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancey C.J., Chang S.C., and Dowhan W.. 1993. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J. Biol. Chem. 268:24580–24590. [PubMed] [Google Scholar]
- Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., and Langer T.. 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 338:815–818. 10.1126/science.1225625 [DOI] [PubMed] [Google Scholar]
- Daum G., Lees N.D., Bard M., and Dickson R.. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 14:1471–1510. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y., Rosenfeld-Gur E., Shinder V., Futerman A.H., Geiger T., and Schuldiner M.. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell. 30:95–102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Entian K.D., and Barnett J.A.. 1992. Regulation of sugar utilization by Saccharomyces cerevisiae. Trends Biochem. Sci. 17:506–510. 10.1016/0968-0004(92)90341-6 [DOI] [PubMed] [Google Scholar]
- Folmes C.D., Nelson T.J., Martinez-Fernandez A., Arrell D.K., Lindor J.Z., Dzeja P.P., Ikeda Y., Perez-Terzic C., and Terzic A.. 2011. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 14:264–271. 10.1016/j.cmet.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu Q., Zhang F., Han Z., Gu T., Tian W., Chen Y., and Wu Q.. 2009. Conserved expression of the PRELI domain containing 2 gene (Prelid2) during mid-later-gestation mouse embryogenesis. J. Mol. Histol. 40:227–233. 10.1007/s10735-009-9234-1 [DOI] [PubMed] [Google Scholar]
- Gibellini F., and Smith T.K.. 2010. The Kennedy pathway: de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 62:414–428. 10.1002/iub.354 [DOI] [PubMed] [Google Scholar]
- Gohil V.M., Thompson M.N., and Greenberg M.L.. 2005. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280:35410–35416. 10.1074/jbc.M505478200 [DOI] [PubMed] [Google Scholar]
- Gottlieb D., Heideman W., and Saba J.D.. 1999. The DPL1 gene is involved in mediating the response to nutrient deprivation in Saccharomyces cerevisiae. Mol. Cell Biol. Res. Commun. 1:66–71. 10.1006/mcbr.1999.0109 [DOI] [PubMed] [Google Scholar]
- Gulshan K., Shahi P., and Moye-Rowley W.S.. 2010. Compartment-specific synthesis of phosphatidylethanolamine is required for normal heavy metal resistance. Mol. Biol. Cell. 21:443–455. 10.1091/mbc.E09-06-0519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B.M., Owens K.M., and Singh K.K.. 2011. Distinct functions of evolutionary conserved MSF1 and late embryogenesis abundant (LEA)-like domains in mitochondria. J. Biol. Chem. 286:39141–39152. 10.1074/jbc.M111.259853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönscher C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., and Ungermann C.. 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell. 30:86–94. 10.1016/j.devcel.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jiang F., Rizavi H.S., and Greenberg M.L.. 1997. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol. Microbiol. 26:481–491. 10.1046/j.1365-2958.1997.5841950.x [DOI] [PubMed] [Google Scholar]
- Joshi A.S., Thompson M.N., Fei N., Hüttemann M., and Greenberg M.L.. 2012. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 287:17589–17597. 10.1074/jbc.M111.330167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T., and Yamashita S.. 1987. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J. Biol. Chem. 262:15428–15435. [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S., Chao J.T., Tavassoli S., Wong A.K., Choudhary V., Young B.P., Loewen C.J., and Prinz W.A.. 2014. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12:e1001969 10.1371/journal.pbio.1001969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A., John Peter A.T., and Kornmann B.. 2015. ER-mitochondria contact sites in yeast: beyond the myths of ERMES. Curr. Opin. Cell Biol. 35:7–12. 10.1016/j.ceb.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Muir R.S., Lim E., McElver J., Weber S.C., and Heitman J.. 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 158:113–117. 10.1016/0378-1119(95)00144-U [DOI] [PubMed] [Google Scholar]
- Mandal S., Lindgren A.G., Srivastava A.S., Clark A.T., and Banerjee U.. 2011. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells. 29:486–495. 10.1002/stem.590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser von Filseck J., Čopič A., Delfosse V., Vanni S., Jackson C.L., Bourguet W., and Drin G.. 2015. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 349:432–436. 10.1126/science.aab1346 [DOI] [PubMed] [Google Scholar]
- Osman C., Haag M., Potting C., Rodenfels J., Dip P.V., Wieland F.T., Brügger B., Westermann B., and Langer T.. 2009. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184:583–596. 10.1083/jcb.200810189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Voelker D.R., and Langer T.. 2011. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192:7–16. 10.1083/jcb.201006159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potting C., Wilmes C., Engmann T., Osman C., and Langer T.. 2010. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 29:2888–2898. 10.1038/emboj.2010.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potting C., Tatsuta T., König T., Haag M., Wai T., Aaltonen M.J., and Langer T.. 2013. TRIAP1/PRELI complexes prevent apoptosis by mediating intramitochondrial transport of phosphatidic acid. Cell Metab. 18:287–295. 10.1016/j.cmet.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Steenbergen R., Nanowski T.S., Beigneux A., Kulinski A., Young S.G., and Vance J.E.. 2005. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J. Biol. Chem. 280:40032–40040. 10.1074/jbc.M506510200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Endo T., Iijima M., and Sesaki H.. 2009. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185:1029–1045. 10.1083/jcb.200812018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Iijima M., and Sesaki H.. 2010. Mdm35p imports Ups proteins into the mitochondrial intermembrane space by functional complex formation. EMBO J. 29:2875–2887. 10.1038/emboj.2010.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Onguka O., Hobbs A.E., Jensen R.E., Iijima M., Claypool S.M., and Sesaki H.. 2012a Role for two conserved intermembrane space proteins, Ups1p and Ups2p, [corrected] in intra-mitochondrial phospholipid trafficking. J. Biol. Chem. 287:15205–15218. 10.1074/jbc.M111.338665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Onguka O., Itoh K., Endo T., Iijima M., Claypool S.M., and Sesaki H.. 2012b Phosphatidylethanolamine biosynthesis in mitochondria: phosphatidylserine (PS) trafficking is independent of a PS decarboxylase and intermembrane space proteins UPS1P and UPS2P. J. Biol. Chem. 287:43961–43971. 10.1074/jbc.M112.390997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Sesaki H., and Endo T.. 2014. Phospholipid transport via mitochondria. Traffic. 15:933–945. 10.1111/tra.12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasseva G., Bai H.D., Davidescu M., Haromy A., Michelakis E., and Vance J.E.. 2013. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J. Biol. Chem. 288:4158–4173. 10.1074/jbc.M112.434183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter P.J., and Voelker D.R.. 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270:6062–6070. 10.1074/jbc.270.11.6062 [DOI] [PubMed] [Google Scholar]
- Trotter P.J., Pedretti J., and Voelker D.R.. 1993. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268:21416–21424. [PubMed] [Google Scholar]
- Tuller G., Hrastnik C., Achleitner G., Schiefthaler U., Klein F., and Daum G.. 1998. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 421:15–18. 10.1016/S0014-5793(97)01525-1 [DOI] [PubMed] [Google Scholar]
- Vaden D.L., Gohil V.M., Gu Z., and Greenberg M.L.. 2005. Separation of yeast phospholipids using one-dimensional thin-layer chromatography. Anal. Biochem. 338:162–164. 10.1016/j.ab.2004.11.020 [DOI] [PubMed] [Google Scholar]
- Varum S., Rodrigues A.S., Moura M.B., Momcilovic O., Easley C.A. IV, Ramalho-Santos J., Van Houten B., and Schatten G.. 2011. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 6:e20914 10.1371/journal.pone.0020914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pöhlmann R., and Philippsen P.. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10:1793–1808. 10.1002/yea.320101310 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Tamura Y., Kawano S., and Endo T.. 2015. Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria. Nat. Commun. 6:7922 10.1038/ncomms8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.I., and Voelker D.R.. 2002. Biochemistry and genetics of interorganelle aminoglycerophospholipid transport. Semin. Cell Dev. Biol. 13:185–195. 10.1016/S1084-9521(02)00047-2 [DOI] [PubMed] [Google Scholar]
- Zhang J., Khvorostov I., Hong J.S., Oktay Y., Vergnes L., Nuebel E., Wahjudi P.N., Setoguchi K., Wang G., Do A., et al. 2011. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 30:4860–4873. 10.1038/emboj.2011.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.