Abstract
Epithelial–mesenchymal transition (EMT) is a key event that is involved in the invasion and dissemination of cancer cells. Although typically considered as having tumour-suppressive properties, transforming growth factor (TGF)-β signalling is altered during cancer and has been associated with the invasion of cancer cells and metastasis. In this study, we report a previously unknown role for the cytoplasmic promyelocytic leukaemia (cPML) tumour suppressor in TGF-β signalling-induced regulation of prostate cancer-associated EMT and invasion. We demonstrate that cPML promotes a mesenchymal phenotype and increases the invasiveness of prostate cancer cells. This event is associated with activation of TGF-β canonical signalling pathway through the induction of Sma and Mad related family 2 and 3 (SMAD2 and SMAD3) phosphorylation. Furthermore, the cytoplasmic localization of promyelocytic leukaemia (PML) is mediated by its nuclear export in a chromosomal maintenance 1 (CRM1)-dependent manner. This was clinically tested in prostate cancer tissue and shown that cytoplasmic PML and CRM1 co-expression correlates with reduced disease-specific survival. In summary, we provide evidence of dysfunctional TGF-β signalling occurring at an early stage in prostate cancer. We show that this disease pathway is mediated by cPML and CRM1 and results in a more aggressive cancer cell phenotype. We propose that the targeting of this pathway could be therapeutically exploited for clinical benefit.
Introduction
The gene encoding the promyelocytic leukaemia (PML) tumour suppressor is a member of the Ring-B box-Coiled Coil family, originally identified at the breakpoint of the t(15;17) translocation found in acute promyelocytic leukaemia (APL).1, 2, 3 PML is involved in the regulation of many cellular pathways such as apoptosis, senescence, response to DNA damage and resistance to viral infection.4, 5, 6, 7 Although the function of PML has been extensively investigated because of its essential role in the formation of PML-nuclear bodies (PML-NBs), the subcellular localization of PML is not restricted to the PML-NBs and is observed in other cellular compartments including the nucleoplasm, the nucleolus, the nuclear envelope and the cytoplasm.5, 8 The roles of PML in these cellular compartments remain unclear, and may involve mechanisms that are independent of PML-NBs. There are seven main isoforms of PML (I–VIIb), which appear to have cellular functions that are related to their expression pattern in physiological and pathological cellular contexts. Most of the isoforms contain the nuclear localization signal (NLS), which enables them to localize in the nucleus (PMLI to PMLVI), whereas PMLVIIb lacks the NLS and is found in the cytoplasm.9 The PMLI isoform also possesses a nuclear export signal (NES), which allows its nuclear export in a CRM1-dependent manner.10
Recent studies suggest the involvement of cytoplasmic PML in tumorigenesis, glycolysis, anti-viral responses, laminopathies and cell cycle regulation.8 In a physiological context, cytoplasmic PML has been shown to activate transforming growth factor (TGF)-β signalling, a cellular pathway involved in tumour suppression, apoptosis and senescence.11, 12, 13 Furthermore, defects in TGF-β signalling in Pml−/− mouse embryonic fibroblasts have also been reported.14 TGF-β binding to its receptors results in the phosphorylation of the SMAD proteins, a cellular event that is facilitated by the TGF-β-receptor adapter SARA (SMAD Anchor for Receptor Activation). This process results in the translocation of SMADs to the nucleus and the activation of target genes.15 In this context, cytoplasmic PML appears to favour the interaction of SARA and the SMAD protein members, SMAD2 and SMAD3.14 Although TGF-β signalling is a potent inhibitor of cell proliferation, this function seems to be altered in tumour cells, in that it acts as an oncogene.16, 17, 18, 19, 20, 21 In particular, defects in normal TGF-β signalling have been shown to trigger tumorigenic events including cancer-associated epithelial–mesenchymal transition (EMT), an important process that is involved in the progression of advanced tumours.22, 23 Based on this background, we hypothesized that cytoplasmic PML promotes cancer-associated EMT and local tumour invasion via the induction of TGF-β signalling. Furthermore, we reveal a new role for cytoplasmic PML (cPML) in the progression of prostate cancer. To better understand the role of cPML in tumour progression, we investigated its influence on EMT and cell invasion using prostate cancer cell lines and mutant constructs. Furthermore, we showed the translational importance of this proposal in a clinical setting by showing that cPML expression correlates with the expression of the nuclear export factor CRM1 (Exportin-1), and is associated with a worse prognosis. These studies demonstrate that cPML induces cell invasion by promoting the transdifferentiation of epithelial cells into mesenchymal cells involving TGF-β signalling, via SMAD2 and SMAD3 phosphorylation. Collectively, these results support the concept that cPML promotes TGFβ-induced EMT promoting a more aggressive cancer cell phenotype resulting in increased cell invasion and reduced survival for patients with prostate cancer.
Results
PML cytoplasmic localization is mediated by CRM1-dependent nuclear export
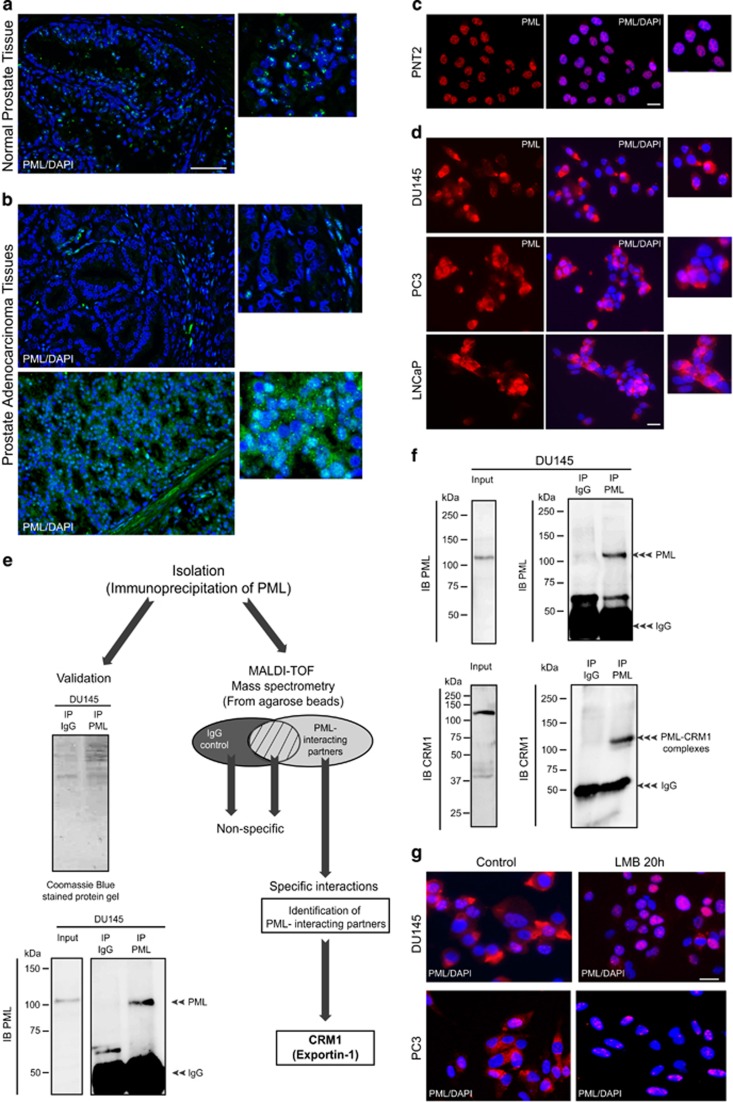
PML expression is altered in cancers of various histological origins.24 To investigate the expression status of PML, we performed immunostaining on prostate adenocarcinoma tissues using an antibody against PML. In normal prostate tissue, PML was expressed in the nucleus of cells within the prostate gland. The expression was typical of that of PML-NBs (Figure 1a). In contrast, PML expression was decreased (Figure 1b, upper panel) or exhibited nucleo-cytoplasmic and/or cytoplasmic expression patterns (Figure 1b, lower panel) in prostate adenocarcinoma cells. Similar results were obtained by immunofluorescence (IF), in that PML is expressed in the nucleus of the normal prostate cell line PNT2, and in the nucleo-cytoplasm and/or cytoplasm of the prostate cancer cell lines DU145, PC3 and LNCaP (Figures 1c and d). Although the decrease of PML expression in cancer tissues has been previously reported, PML localization in the cytoplasm and its role in that cellular compartment are not understood. To investigate this, we performed immunoprecipitation (IP) experiments using a PML antibody and whole-cell extracts from DU145 human prostate cancer cells. The efficiency of the IP was determined by investigating the presence of PML in PML-immunoprecipitates and using immunoblotting (IB) with PML antibody. PML immunoprecipitates were also used to identify PML-interacting partners using MALDI-TOF mass spectrometry. PML and several molecules were identified as PML-interacting partners (Figure 1e). Interestingly, one of the interacting proteins identified was CRM1 (Exportin-1), a molecule which is involved in nuclear export. Therefore, the cytoplasmic localization of PML within prostate cancer cells might be a consequence of its nuclear export in a CRM1-dependent manner. To confirm the interaction of PML with CRM1, IP of DU145 extracts with a PML antibody was performed (Figure 1f). In addition to PML, CRM1 was also pulled down as demonstrated by IB of the immunoprecipitates with PML and CRM1 antibodies. Finally, the nuclear export of endogenous PML in DU145 and PC3 cells was markedly decreased after treatment with Leptomycin B, an inhibitor of nuclear export mediated by CRM1 (Figure 1g). Taken together, these results demonstrate that the cytoplasmic localization of PML in prostate cancer cells involves, at least in part, a CRM1-dependent nuclear export.
Figure 1.
PML expression in normal and cancer prostate cells. (a and b) Immunohistochemistry/fluorescence micrographs showing PML expression (green) in normal prostate tissue and prostate adenocarcinoma. The nucleus is stained in blue (4,6-diamidino-2-phenylindole (DAPI)). Scale bar=100 μm. (c and d) Immunofluorescence micrograph showing PML expression (red) in normal prostate cell line (PNT2) and in prostate cancer cell lines (DU145, PC3 and LNCaP). Scale bar=20 μm. (e) Schematic representation of the methodology used to identify PML-interacting partners from DU145 cell extracts using immunoprecipitation (IP) with PML antibody and MALDI-TOF mass spectrometry. (f) Immunoprecipition of DU145 cell extract with PML antibody and immunoblotting (IB) with PML and CRM1 antibodies. (g) Immunofluorescence micrograph showing PML expression in DU145 and PC3 treated with or without Leptomycin B (15 ng/ml). Scale bar=20 μm.
Clinical significance of cytoplasmic PML and CRM1 expression: association with poor prognosis
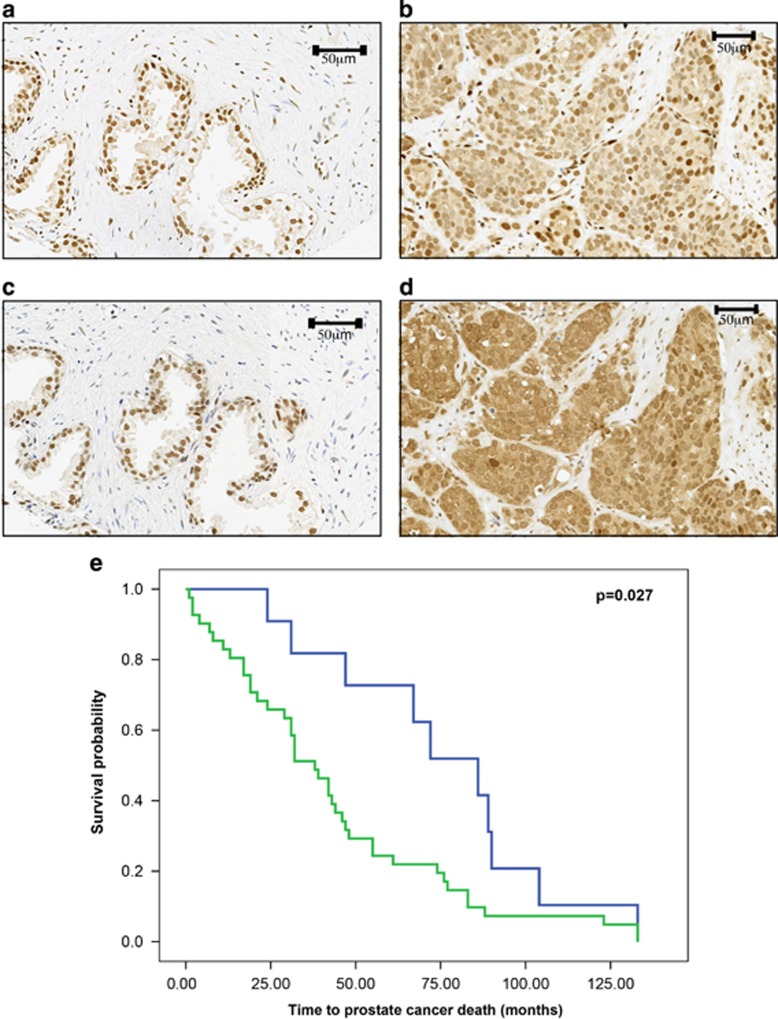
Although previous studies have shown a correlation between the loss of PML expression and prostate cancer progression,24, 25 the role of cytoplasmic PML in this process is still unclear. To examine this, the association of CRM1 and PML with cancer-specific mortality and metastasis in a cohort of patients with prostate cancer was investigated by immunohistochemical staining of prostate cancer tissue microarray (TMA). Both proteins were variously located within the nuclei of benign prostate tissue (Figures 2a and c) and in the nuclei and cytoplasm of prostate cancer tissue and were more abundant in the cytoplasm of tumours compared with normal prostate tissue (Figures 2b, d and Supplementary Table 2). Neither nuclear or cytoplasmic expression of PML, nor the expression of cytoplasmic CRM1 protein alone was found to be predictive of reduced disease-specific survival or metastasis (Supplementary Table 2). To test the hypothesis that PML is transported by CRM1, the capacity of PML/CRM1 expression to predict clinical outcome was assessed. Eighty percent (40/50) of patients exhibited either complete loss of PML and CRM1 in tumours or simultaneous expression of cytoplasmic PML and CRM1. This patient group showed reduced cancer-specific survival (χ2=6.47, P=0.011) over a 10-year term (Figure 2e), but not significantly increased metastasis over the same term (χ2=3.108, P=0.078). A Cox proportional hazards regression model demonstrated that simultaneous expression of cytoplasmic PML/CRM1 predicted reduced disease-specific survival that was independent of prostate-specific antigen levels (>4 ng/ml) and the Gleason score (>7; hazard ratio=0.264, P=0.009, 95% confidence interval=0.097–0.720).
Figure 2.
Immunohistochemical localization of PML and CRM1 in prostate cancer patients' TMA. (a) Lack of cytoplasmic PML and (c) CRM1 in benign tissue. (b) Nuclear and cytoplasmic localization of PML and (d) CRM1 in cancer. (e) Kaplan–Meier plot. Scale bars as indicated.
The cytoplasmic fraction of PML promotes cell invasion and EMT
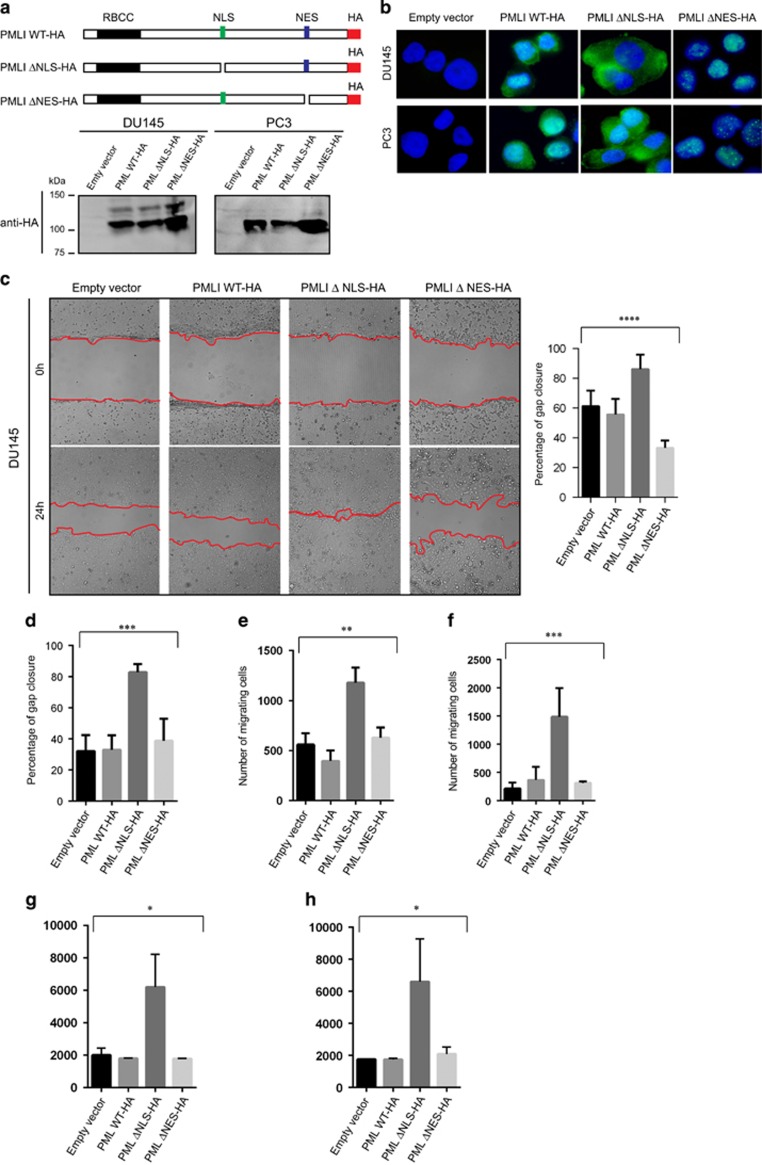
Although, PML and CRM1 do not appear to significantly correlate with metastasis in prostate cancer, the cytoplasmic localization of PML as a result of CRM1 nuclear export may be an early event, which has a role in tumour invasion of adjacent tissues. To investigate the role of cytoplasmic PML on cell migration and invasion, we generated three PMLI haemagglutinin (HA)-tagged mutant constructs: PMLI wild-type HA, PMLI ΔNLS-HA (without the NLS) and PMLI ΔNES-HA (without the NES). The constructs were stably expressed in DU145 and PC3 prostate cancer cell lines using a lentiviral expression system (Figure 3a). Their protein expression pattern was investigated by IB and IF (Figures 3a and b), and the effect of expression on cell migration and invasion determined using wound healing (Scratch assay), well cell migration and well cell invasion assays. Interestingly, DU145 and PC3 cells expressing PMLI ΔNLS exhibited an increased migration and invasive capacities in comparison to cells expressing the empty vector, PMLWT-HA and PMLΔNES constructs (Figures 3c–h and Supplementary Figures 2a and b). Furthermore, knockdown of endogenous PML in DU145 and PC3 resulted in a decreased cell migration. Treatment of these cells with Leptomycin B also affected cell migration (Supplementary Figure 2c). Finally, CRM1 knockdown in DU145-expressing PMLI ΔNLS did not significantly affect cell migration (Supplementary Figure 3d). The increased cell migration and invasion observed in the presence of PMLI ΔNLS was not caused by increased proliferation (Supplementary Figure 3e). These results suggest that the cytoplasmic fraction of PML promotes the migration and invasive capacities of prostate cancer cells.
Figure 3.
The migration of DU145 and PC3 cells is promoted by cytoplasmic PML. (a) Schematic representation of PMLI mutant constructs (PMLI WT-HA, PMLI ΔNLS-HA and PML ΔNES-HA) and their protein expression profiles as shown by immunoblotting from DU145 and PC3 whole-cell extracts and using an anti-HA antibody. (b) Immunofluorescence images demonstrating stable expression of PMLI mutants in DU145 and PC3 cells using an anti-HA antibody. Scale bar=10 μm. (c, d) Wound healing assay (Scratch assay) using DU145 and PC3 cells expressing PMLI mutant constructs and the corresponding data quantifying gap closure at time points 0 and 24 h following scratching. Analysis of variance (ANOVA): ****P<0.0001 and ***P=0.0001. Scale bar=200 μm. (e, f) Well cell migration assay using DU145 (left panel) and PC3 cells expressing PMLI mutant constructs and the data relating to the number of migrated cells after 24 h of culture. ANOVA: **P=0.0018 and ANOVA: ***P=0.0002. (g, h) Cell invasion assay using DU145 (left panel) and PC3 cells (right panel) expressing PMLI mutant constructs and the data relating to the number of migrated cells after 24 h of culture. ANOVA: *P=0.0108 and ANOVA: *P=0.0304.
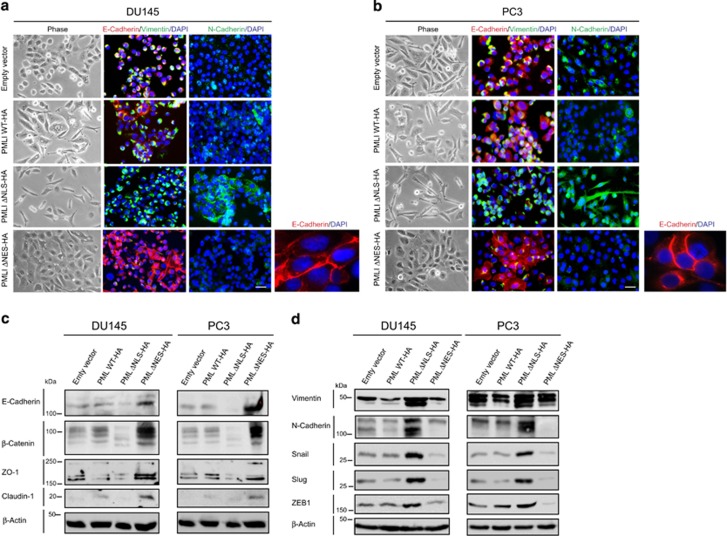
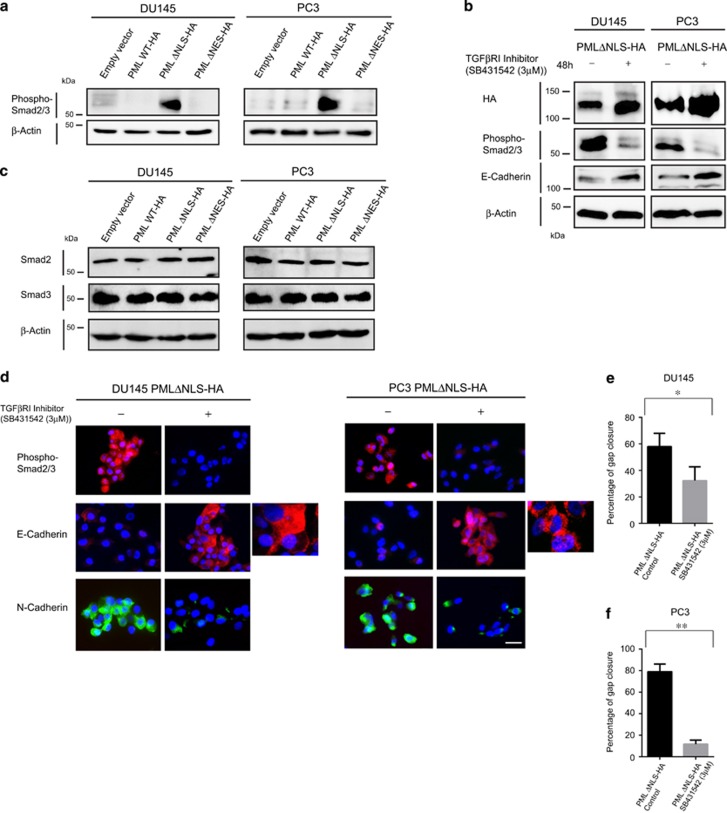
The expression of PML constructs also influenced the cellular morphology of DU145 and PC3 prostate cancer cells. Notably, cells expressing the PMLΔNLS-HA construct exhibited a mesenchymal phenotype, which was characterized by the presence of individual elongated cells. In contrast, DU145 and PC3 cells expressing PML ΔNES-HA exhibited a more epithelial phenotype. The cells expressing the wild-type construct presented an intermediate phenotype, which was probably a result of its dual nuclear and cytoplasmic expression (Figures 4a and b). To confirm these observations, we performed IF on these cells using antibodies that recognize key markers of epithelial cells (E-Cadherin) and mesenchymal cells (Vimentin and and N-Cadherin). Lower E-cadherin expression and increased expression of Vimentin and N-Cadherin were observed in cells expressing PMLΔNLS. On the contrary, cells expressing PML ΔNES-HA, expressed higher levels of E-Cadherin, but a lower expression of Vimentin and N-cadherin. Some of the cells that expressed PMLWT-HA exhibited a higher expression of E-Cadherin, whereas others expressed higher levels of Vimentin and N-Cadherin. To confirm these observations, we performed IB of lysates from cells expressing the empty vector, PML WT-HA, PML ΔNLS-HA and PML ΔNES using antibodies recognizing key markers of epithelial and mesenchymal phenotypes (Figures 4c and d). PMLΔNLS cells expressed lower levels of epithelial markers and higher levels of mesenchymal markers (Figures 4c and d). PMLΔNES expressed higher levels of epithelial markers and lower levels of mesenchymal markers. These results demonstrate that cytoplasmic PML promotes EMT, whereas nuclear PML promotes an epithelial phenotype.
Figure 4.
Epithelial–mesenchymal transition (EMT) is induced by cytoplasmic PML. (a, b) Micrographs of immunofluorescences on DU145 and PC3 cells expressing PML mutant constructs using antibodies to E-Cadherin (red), Vimentin (green) or N-Cadherin (green). Scale bar=20 μm. (c, d) Immunoblotting analysis of whole-cell extracts from DU145 and PC3 cells expressing PML mutant constructs using antibodies against molecules expressed by epithelial cells (E-Cadherin, β-Catenin, ZO-1 and Claudin1) and mesenchymal cells (Vimentin, N-Cadherin, SNAIL, SLUG and ZEB1). β-Actin represents the loading control.
Cytoplasmic PML-induced EMT and cell invasion are mediated by TGF-β signalling
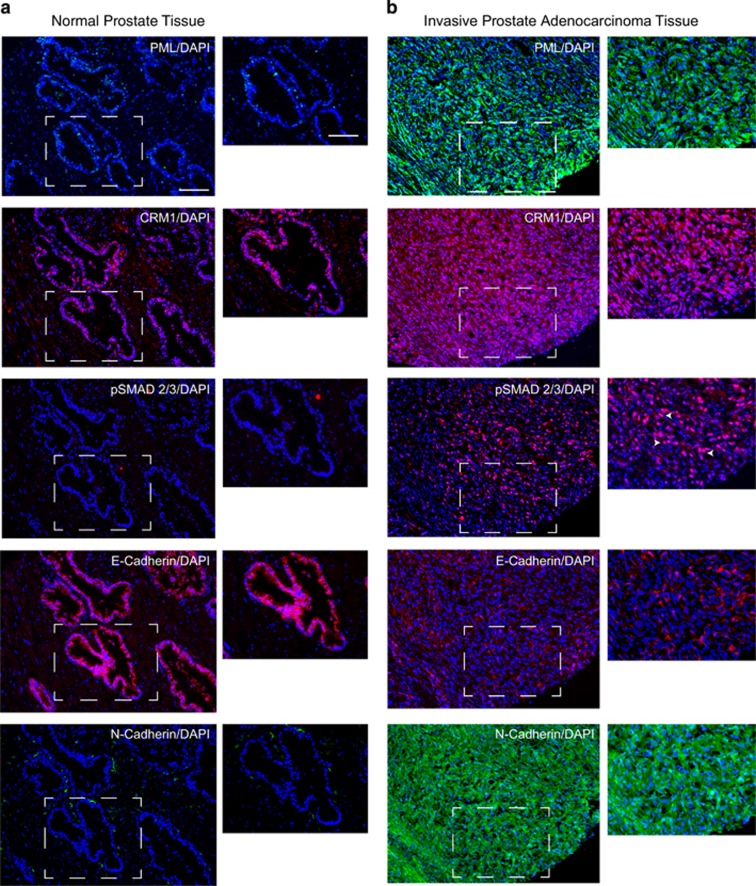
To investigate the potential involvement of TGF-β signalling pathway in PML-mediated EMT and cell migration and invasion, we performed IB of lysates from DU145 and PC3 cells expressing the empty vector, PMLWT-HA, PMLΔNLS-HA and PMLΔNES using an antibody against the phosphorylated forms of SMAD2 and 3. Interestingly, higher levels of phospho-SMAD2/3 were observed in PMLΔNLS-expressing cells, when compared with cells expressing the other constructs (Figure 5a). To confirm this result, we performed IB using the phospho-SMAD2/3 antibody on cell lysates from DU145 and PC3 cells expressing PMLΔNLS that had been treated with a specific TGFβRI inhibitor (untreated cells as controls). Treatment with the inhibitor significantly reduced the level of phospho-SMAD2/3 expression, but did not affect cPML expression (Figures 5b and c). E-Cadherin expression levels were increased following treatment with the inhibitor (Figures 5b and c), whereas the expression of N-Cadherin was decreased (Figure 5c). As the inhibition of TGF-β signalling in this experimental setting may also affect cell migration, we tested the influence of the TGFβRI inhibitor on DU145 and PC3 cells expressing PMLΔNLS using a wound healing assay. Remarkably, the inhibitor significantly reduced the migration-inducing capacity of PMLΔNLS (Figures 5d and e). This result was confirmed by knocking down TGF-β receptor I in DU145 cells expressing PMLΔNLS (Supplementary Figure 3a). Finally, in invasive prostate adenocarcinoma tissue, the cytoplasmic expression of PML and CRM1 correlated with the nuclear expression of phospho-SMAD2/3, an increased expression of N-Cadherin and a decreased expression of E-Cadherin (Figures 6a and b). Taken together, these results demonstrate a role of cytoplasmic PML in TGFβ-mediated EMT and invasion (Figure 7).
Figure 5.
Cytoplasmic PML induces SMAD2/3 phosphorylation, EMT and cell invasion in vitro. (a) Immunoblotting of whole-cell extracts from DU145 and PC3 cells expressing PML mutant constructs using antibodies against phosphorylated forms of SMAD2/3 and β-actin (loading control). (b) Immunoblotting analysis of whole-cell extracts from DU145 and PC3 cells expressing the PML ΔNLS construct, treated with DMSO (control) or TGFβ-RI inhibitor (3 μM for 48 h) using antibodies against HA, phosphorylated forms of SMAD2/3, E-Cadherin and β-actin (loading control). (c) Immunoblotting of whole-cell extracts from DU145 and PC3 cells expressing PML mutant constructs using antibodies against total SMAD2, SMAD3 and β-actin (loading control). (d) Immunofluorescence analysis of DU145 and PC3 cells expressing the PML ΔNLS construct, treated with DMSO (control) or TGFβ-RI inhibitor (3 μM for 48 h) using antibodies against phosphorylated forms of SMAD2/3, E-Cadherin and N-Cadherin. Scale bar=20 μm. (e, f) Wound healing assay (Scratch assay) using DU145 and PC3 cells expressing the PML ΔNLS construct, treated with DMSO (control) or TGFβ-RI inhibitor (3 μM for 48h) and the data relating to the percentages of gap closure at time points 0 and 24 h following scratching. Two-tailed paired test: *P=0.0451 and **P=0.0088.
Figure 6.
Expression of PML, CRM1, p-SMAD2/3, E-Cadherin and N-Cadherin in normal prostate and invasive adenocarcinoma tissues. (a, b) Immunohistochemistry/immunofluorescence using antibodies against PML, CRM1, p-SMAD2/3, E-Cadherin and N-Cadherin in normal prostate tissue (a) and invasive prostate adenocarcinoma tissue (b). Arrows indicates nuclear p-SMAD2/3. Scale bar=100 μm and 50 μm. DAPI, 4,6-diamidino-2-phenylindole.
Figure 7.
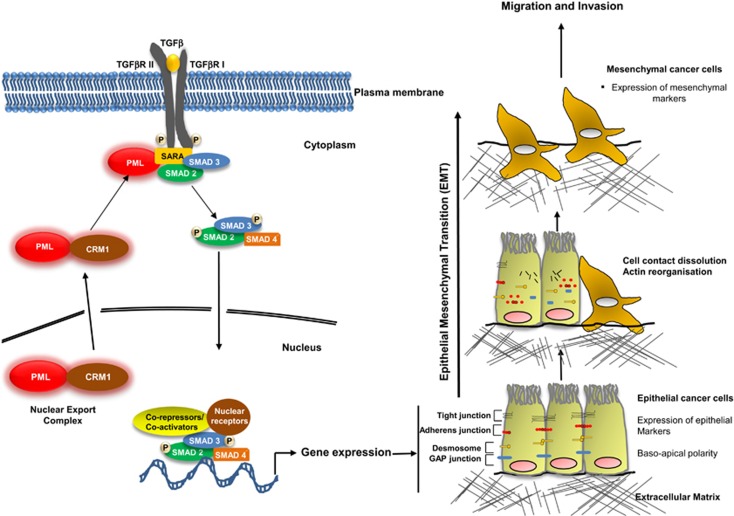
Schematic representation of the role played by cytoplasmic PML in promoting TGF-β signalling in tumour microenvironment. PML is exported from the nucleus to the cytoplasm in a CRM1-dependent manner. Cytoplasmic PML interacts and promotes the association of SARA with SMAD2 and SMAD3, which activates their phosphorylation. Phosphorylated SMAD2 and SMAD3 interact with SMAD4 and translocate to the nucleus where they regulate gene expression of factors involved in promoting EMT and invasion. During EMT, epithelial cells characterized by the expression of epithelial markers and a baso-apical polarity start to lose their cell–cell contacts and to express mesenchymal markers characteristic of mesenchymal cells. The later have a high migratory and invasive properties and have a key role in cancer progression.
Discussion
EMT is a key cellular event that is involved in embryonic development, wound healing and cancer progression.22, 26 The baso-apical polarity of epithelial cells is maintained through lateral cell–cell junctions that include desmosomes, adherens junctions, tight junctions and gap junctions. During EMT, epithelial cells, which begin their transdifferentiation via the loss of lateral junctions, start to express molecules associated with the mesenchymal phenotype and this results in the generation of mesenchymal cells. These later have an essential role in the spread of disease to adjacent tissues and other organs via a process called metastasis. Upon reaching the target organ, these cells begin a reverse cellular process (mesenchymal–epithelial transition), which results in the generation of cancer epithelial cells, which integrate into the structure of target tissue and therefore ‘ignite' its oncogenic transformation. EMT can be naturally induced by several cellular pathways such as growth factors, TGF-β, WNT, HH and Notch signalling. In cancer, these pathways can be altered and result in cancer-associated EMT. Notably, TGF-β signalling is a well-known tumour-suppressive pathway, and cancerous cells circumvent this function by inactivating the anti-proliferative arm of the pathway or by altering the function of downstream components of the pathway.27
EMT is induced as a consequence of TGF-β signalling dysfunction, to result in an increase in the invasion and dissemination of cancer cells. In this regard, cytoplasmic PML appears to activate the phosphorylation of SMAD2 and SMAD3, two main components of the canonical SMAD pathway in response to TGF-β. In this study, we observe a similar mechanism, in that cPML induces TGF-β signalling through cPML induction of SMAD2 and SMAD3 phosphorylation. However, and in comparison with Lin et al. study which used a physiological model (mouse embryonic fibroblasts), our work investigated the role of cPML in the regulation TGF-β signalling within a pathological setting, namely prostate cancer. In this context, we showed that cPML promotes TGF-β-induced prostate cancer-associated EMT and invasion, a mechanism that was previously unknown (Figure 7). This difference is not surprising given that the consequences of TGF-β signalling are dependent on whether the cellular context is physiological or pathological. Moreover, cPML is mainly found in the cytoplasm of tumour tissue for patients with prostate cancer that exhibited a poor clinical prognosis. In addition, several reports have reported on the oncogenic potential of cPML mutants.8 Two different PML mutations (1272delAG and IVS3–1G-A) have been identified in aggressive APL patients. The mutations generated a stop codon before the NLS domain leading to the generation of cPML mutants.24 Other studies have shown that cPML is upregulated in hepatocellular carcinoma.28, 29 In the present study, we showed increased cPML and cytoplasmic expression of Exportin-1 (CRM1) in tumour tissue of patients with prostate cancer correlates with a poor clinical prognosis, suggesting a functional link between the two molecules. Indeed, the PMLI isoform is the only known isoform that contains a NES, and thereby could be exported to the cytoplasm. This possibility has been demonstrated, in that we have shown that the mutant PMLI which lacked the NES promoted a more epithelial phenotype, which is associated with an increased expression of epithelial markers. This suggests that nuclear PML may prevent cancer-associated EMT and may have a role in promoting mesenchymal–epithelial transition. Taken together, these observations highlight the involvement of cPML in prostate cancer progression and provide further insights on the role of deregulated TGF-β signalling in pathogenesis of cancer invasion.
Materials and methods
Antibodies
For this study, we used antibodies to PML (1:250 for IB, 1:100 for IP, sc-966, Santa Cruz Biotechnology, Delaware Ave., Santa Cruz, CA, USA), PML (1:500 for immunohistochemistry (IHC), sc5621, Santa Cruz Biotechnology, Delaware Ave.), HA (1:1000 for IB, 1:200 for IF, A190-108 A, Bethyl Laboratories Inc., Montgomery, TX, USA), β-actin (1:5000 for IB, A5441, Sigma-Aldrich, St Louis, MO, USA), Exportin-1 (CRM1; 1:5000 for IB, 1:2000 for IHC, A300-469A, Bethyl Laboratories Inc.), E-Cadherin (1:500 for IF, 610181, BD Transduction Laboratories, San Jose, CA, USA), EMT antibody Sampler Kit (9782, Cell Signaling Technology, Danvers, MA, USA), which contains rabbit antibodies each used at 1:300 (IB), N-Cadherin and Vimentin antibodies were used at 1:500 for IF, Phospho-Smad2/Smad3 (1:300 for IB, 8828, Cell Signaling Technology), Phospho-Smad2/3 (1:100 for IHC, sc-11769, Santa Cruz Biotechnology), Anti-rabbit IgG, HRP-linked Antibody (1:1000 for IB, 7074, Cell Signaling Technology), Anti-TGFβ Receptor I (1:1000 for IB, 3712, Cell Signaling Technology), Anti-SMAD2 (1:1000 for IB, 3122, Cell Signaling Technology), Anti-SMAD3 (1:1000 for IB, 9513, Cell Signaling Technology), Anti-phospho-SMAD2 (1:500, SAB4504207, Sigma-Aldrich), Anti-phospho-SMAD3 (1:500, SAB4300253, Sigma-Aldrich), Anti-mouse IgG, HRP-linked antibody (1:1000 IB, 7076, Cell Signaling Technology).
Cell lines, growth conditions, cell fractionation and cell proliferation assay
DU145 and PC3 prostate cancer cell lines were purchased from American Type Culture Collection (Manassas, VA, USA) and cultured in Eagle's Minimum Essential Medium (BE12-6621, Lonza, Basel, Switzerland) and F-12k Nut Mix (1x; 21127-022, Gibco Life Technologies, Waltham, MA, USA) respectively. The media were supplemented with 10% (v/v) fetal calf serum and 1% (w/v) L-glutamine (Lonza). Cells were incubated at 37 °C in 5% (v/v) CO2 and 100% (v/v) humidity. For the relevant experiments, cells were treated for 48 h with 3 μM of TGFβR1 inhibitor SB431542 (1614, Tocris Bioscience, Moorend Farm Avenue, Bristol, UK) and for 20 h with 15 ng/ml of Leptomycin B. TGF-β1 was purchased from PeproTech (London, UK) (100-22) and used at 10 ng/ml. Cell proliferation was quantified using CyQUANT NF Cell Proliferation Assay Kit (C35006, Invitrogen, Waltham, MA, USA).
Retroviral expression of PML constructs
The PMLI gene fragment was obtained by PCR amplification from pcDNA3.1-PMLI (a kind gift from Dr Mounira Chelbi-Alix, University PARIS V) using forward primer SpeI/PML F′: 5′-CATCTAACTAGTATGGAGAGCCTGCACCCGCCC-3′ and reverse primer PML R′/BamHI 5′-CATCTAGGATCCTCAGCTCTGCTCGGAGGCC-3′ (Eurofins MWG Operon). The fragment obtained was cloned into the pLKOpuro vector using the SpeI and BamHI restriction sites (SHC001, Sigma, St Louis, MO, USA). An HA-tag was introduced in the C-terminal end of PMLI by PCR using QuikChange Site-Directed Mutagenesis Kit (200518, Stratagene, Cedar Creek, TX, USA) and the following primers: HA-tag-sense 5′-CCTCCCAGCAGAGCTACCCTATCGATGTTCCAGATTACGCTTGAGGATCCACCGG-3′ and HA-tag-antisense 5′-CCGGTGGATCCTCAAGCGTAATCTGGAACATCGTATGGGTAGCTCTGCTGGGAGG-3′. The deletions of PMLI NLS and PMLI NES were also performed by PCR using QuikChange Site-Directed Mutagenesis Kit using ΔNLS-sense 5′-GCCCCAGGAAGGTCGGGAAGGAGGCAAG-3′ ΔNLS-antisense 5′-CTTGCCTCCTTCCCGACCTTCCTGGGGC-3′ or ΔNES-sense 5′-ACATTAACAGGCTGTGGGAAGTGCCCGGGGC-3′ ΔNES-antisense 5′-GCCCCGGGCACTTCCCACAGCCTGTTAATGT-3′. The lentiviral particles used to infect DU145 and PC3 with pLKO-PML constructs were produced according to the manufacturer's recommendations. The lentiviral packaging mix was also purchased from Sigma (SHP001).
Stable and transient transfection of cell lines
The transient knockdown of CRM1 and TGFβRI was carried out using an CRM1- or TGFβRI-specific small interfering RNA molecule (CRM1 small interfering RNA (h), sc-35116, Santa Cruz Biotechnology, Inc; TGFβRI small interfering RNA (h), sc-40222, Santa Cruz Biotechnology, Inc) and Interferin transfection reagent (Polyplus, Woodley, Reading, UK) following the manufacturer's recommendations. PML lentiviral plasmids short hairpin RNA control and PML short hairpin RNA 1 and 2 were purchased from Sigma (TRCN0000003866, TRCN0000003869 and SHC001).
Immunoblotting and immunoprecipitation
The experimental procedures were performed as described previously.30 For IB, the cells lysed in 1 × solution containing 50 mM Tris-HCl (pH 6.8), 100 mM dithiothreitol, 2% (w/v) SDS, 0.1% (w/v) bromophenol blue and 10% (v/v) glycerol, and loaded onto Tris/glycine SDS–polyacrylamide gels for electrophoresis. The proteins were transferred onto Amersham Hybond-P PVDF membranes (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK). Membranes were blocked with 10% (w/v) Marvel milk powder/tris-buffered saline solution with 0.1% (v/v) Tween-20. Following washes in tris-buffered saline solution with 0.1% (v/v) Tween-20, membranes were incubated with primary antibodies (in blocking solution) overnight at 4 °C followed by washing and incubation with secondary antibodies for 1 h at room temperature before visualization using Rapid Step ECL reagent (Calbiochem, San Diego, CA, USA) and a CCD camera (Fujifilm, Martins Way, Bedford, UK). For IP, the cells were lysed in IP buffer (0.05 M Tris, pH 7.4, 0.15 M NaCl, 0.5% (v/v) Triton X-100 and 0.001 M EDTA). The extracts were pre-cleared for 1 h at 4 °C using Protein G Plus–Agarose beads (IP08, Calbiochem) and corresponding IgG. The PML antibody was incubated with agarose beads in IP buffer for 2 h before adding the pre-cleared extracts for overnight IP. The beads were washed four times in IP buffer and suspended in lysis buffer, after which samples were analysed by mass spectrometry and IB.
Immunohistochemistry (IHC) and immunofluorescence
Paraffin-embedded human prostate cancer TMAs were purchased from US Biomax (Rockville, MD, USA). The sections were treated and stained as previously reported.30 For IHC, 3 3' diaminobenzidine staining was performed using Dako Real Detection Systems LSAB+ (K5001, Agilent Technologies, Santa Clara, CA, USA).
IHC for PML and CRM1 expression in patient's prostate cancer samples
Patient selection and TMA construction
A TMA was constructed from formalin-fixed wax-embedded tumour and benign tissue samples collected from 192 patients diagnosed between 1999 and 2001 at the Nottingham City Hospital, UK. Two 0.6 mm tissue cores (per patient) of trans-urethral resection of prostate or radical prostatectomy specimens with a histological diagnosis of cancer were assembled using a Grand Master arrayer (3DHistech Ltd, Budapest, Konkoly-Thege Miklós út, Hungary). Multiple clinicopathological variables were recorded for each patient and the patient characteristics are shown in Supplementary Table 1.
Use of the tissue samples for this study was approved by the North West 7 Research Ethics Committee–Greater Manchester Central REC number 10/H1008/72.
IHC and analysis
Sections (4 μm) were subjected to microwave antigen retrieval in 10 mM sodium citrate, pH 6 and immunostained using the PML (1:300) and CRM1 (1:100) antibodies. Antibody localization and detection were achieved using Novolink reagents (Leica, Milton Keynes, UK), a diaminibenzidine chromogen and haematoxylin counterstain.
Stained TMA sections were microscopically assessed (DGP, GVS) to semi-quantify the level of protein expression in the nuclei and cytoplasm of malignant prostate tissue using a Histochemical scoring (H-score) technique resulting in a score between 0 and 300.31 Patients were dichotomously categorized for biomarker expression using the software programme X-tile to determine the optimal cutoff.32 The cutoff values used were PML: nuclear (230), cytoplasm (80); CRM: cytoplasm (110). Patients were classified according to PML and CRM1 expression patterns (Supplementary Table 2) and a Kaplan–Meier survival plot performed to compare the different classes according to disease-specific survival. Subsequently, a group of patients exhibiting a poor prognosis was defined on the basis of an absence of PML and CRM1 expression, or simultaneous cytoplasmic expression of PML and CRM1.
REMARK guidelines for reporting on prognostic biomarkers were followed and a flow diagram showing the final number of cases that were analysed as a proportion of the whole cohort is shown in Supplementary Figure 1.33 Following immunostaining, the number of samples available for analysis was reduced for several reasons: the amount of available malignant tissue was considered too small to be representative of the tumour, tissue cores were detached during processing, cases had data for only one biomarker/case or clinical data (survival or metastasis) were not available for follow-up.
Liquid chromatography-matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of PML-interacting partners
Following IP, identification of the potential interacting partner proteins was carried out by LC-MALDI mass spectrometry. Samples were tryptically digested and fractionated by nano-LC (C18 PepMap100, 150 mm × 75 μm; 5 μm particle size) before spotting in 10 s fractions over a 384-spot target plate. Analysis was carried out on a Bruker UltrafleXtreme MALDI-TOFTOF mass spectrometer (Bruker Daltonics, Banner La, Coventry, UK) in reflectron-positive mode over a m/z range of 650–3500. Tandem mass spectrometry fragmentation of the top 10 precursors per spot was carried out and results exported using Biotools 3.1 software (Bruker Daltonics) for searching via Mascot 2.1 against the human Swissprot database.
Cell migration and invasion assays
For wound healing assay (Scratch assay), DU145 and PC3 cells expressing the empty vector, PMLI WT-HA, PMLI ΔNLS-HA and PMLI ΔNES-HA were cultured to 80% confluence and serum starved for 24 h, after which a scratch was made in the middle of each well using a 10-μl pipette tip. Images from triplicate experiments at 0 and 24 h were taken and the distances between the edges of the scratch were measured at three different points using Carl Zeiss AxioVision software (Carl Zeiss, Oberkochen, Germany). The measurements were expressed as percentages of gap closure. For the well cell migration and invasion assay, we used Cultrex 96-Well Cell Migration Assay (3465-96-K, Cultrex) and Cultrex 96-Well BME Cell Invasion Assay following the manufacturer's recommendations (Trevigen, Gaithersburg, MD, USA). The cells in the bottom chamber were labelled by Calcein AM (Trevigen) and the quantification of the number of cells was performed using the plate reader Infinite M200 Pro TECAN (Tecan Group Ltd, Männedorf, Switzerland) at 585 nm excitation and 520 nm emission.
Acknowledgments
This work was funded by the John and Lucille van Geest Foundation and Cancer and Polio Research Fund. We thank Mrs Anne Schneider, Mr Stephen Reeder, Mr Chris Nolan and Dr Claire Hawkes for their assistance in this study.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- de Thé H, Chomienne C, Lanotte M, Degos L, Dejean A. The t (15; 17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor α gene to a novel transcribed locus. Nature 1990; 347: 558–561. [DOI] [PubMed] [Google Scholar]
- Borrow J, Goddard AD, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science 1990; 249: 1577–1580. [DOI] [PubMed] [Google Scholar]
- Kakizuka A, Miller Jr WH, Umesono K, Warrell Jr RP, Frankel SR, Murty VV VS et al. Chromosomal translocation t (15; 17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell 1991; 66: 663–674. [DOI] [PubMed] [Google Scholar]
- Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell 2002; 108: 165–170. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V. PML nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2: a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 2001; 20: 7274–7286. [DOI] [PubMed] [Google Scholar]
- Mazza M, Pelicci PG. Is PML a tumor suppressor? Front Oncol 2013; 3: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, Wang YJ, Lin HK. Emerging cellular functions of cytoplasmic PML. Front Oncol 2013; 3: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. Differential roles of PML isoforms. Front Oncol 2013; 3: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson BR, Eleftheriou AA. comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 2000; 256: 213–224. [DOI] [PubMed] [Google Scholar]
- Katakura Y, Nakata E, Miura T, Shirahata S. Transforming growth factor β triggers two independent-senescence programs in cancer cells. Biochem Biophys Res Commun 1999; 255: 110–115. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-β signaling in tumor suppression and cancer progression. Nat Genet 2001; 29: 117–129. [DOI] [PubMed] [Google Scholar]
- Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat Rev Cancer 2003; 3: 807–820. [DOI] [PubMed] [Google Scholar]
- Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-β signalling. Nature 2004; 431: 205–211. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol 2012; 13: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2: 442–454. [DOI] [PubMed] [Google Scholar]
- Grünert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol 2003; 4: 657–665. [DOI] [PubMed] [Google Scholar]
- Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res 2006; 66: 8319–8326. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFβ in cancer. FEBS Lett 2012; 586: 1959–1970. [DOI] [PubMed] [Google Scholar]
- Morrison CD, Parvani JG, Schiemann WP. The relevance of the TGF-β paradox to EMT-MET programs. Cancer Lett 2013; 341: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principe DR, Doll JA, Bauer J, Jung B, Munshi HG, Bartholin L et al. TGF-β: duality of function between tumor prevention and carcinogenesis. J Natl Cancer Inst 2014; 106: djt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H, Moon A. Epithelial-mesenchymal transition and cell invasion. Toxicol Res 2010; 26: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiczkova L, Sedlarikova L, Hajek R, Sevcikova S. TGF-beta-an excellent servant but a bad master. J Transl Med 2013; 10: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ et al. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst 2004; 96: 269–279. [DOI] [PubMed] [Google Scholar]
- Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC et al. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell 2011; 20: 214–228. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. TGFβ in cancer. Cell 2008; 134: 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Hénin D et al. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res 1995; 55: 1590–1597. [PubMed] [Google Scholar]
- Chan JYH, Chin W, Liew CT, Chang KS, Johnson PJ. Altered expression of the growth and transformation suppressor PML gene in human hepatocellular carcinomas and in hepatitis tissues. Eur J Cancer 1998; 34: 1015–1022. [DOI] [PubMed] [Google Scholar]
- Mathieu MG, Miles AK, Ahmad M, Buczek ME, Pockley AG, Rees RC et al. The helicase HAGE prevents interferon-α-induced PML expression in ABCB5+ malignant melanoma-initiating cells by promoting the expression of SOCS1. Cell Death Dis 2014; 5: e1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty Jr KS, Miller LS, Cox EB, Konrath J, McCarty KS Sr. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 1985; 109: 716–721. [PubMed] [Google Scholar]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004; 10: 7252–7259. [DOI] [PubMed] [Google Scholar]
- Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med 2012; 10: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.