Abstract
Mild traumatic brain injury (mTBI) accounts for over one million emergency visits each year in the United States. The large-scale structural and functional network connectivity changes of mTBI are still unknown. This study was designed to determine the connectome-scale brain network connectivity changes in mTBI at both structural and functional levels. 40 mTBI patients at the acute stage and 50 healthy controls were recruited. A novel approach called Dense Individualized and Common Connectivity-based Cortical Landmarks (DICCCOLs) was applied for connectome-scale analysis of both diffusion tensor imaging and resting state functional MRI data. Among 358 networks identified on DICCCOL analysis, 41 networks were identified as structurally discrepant between patient and control groups. The involved major white matter tracts include the corpus callosum, and superior and inferior longitudinal fasciculi. Functional connectivity analysis identified 60 connectomic signatures that differentiate patients from controls with 93.75% sensitivity and 100% specificity. Analysis of functional domains showed decreased intra-network connectivity within the emotion network and among emotion-cognition interactions, and increased interactions among action-emotion and action-cognition as well as within perception networks. This work suggests that mTBI may result in changes of structural and functional connectivity on a connectome scale at the acute stage.
Keywords: Brain connectivity, Brain connectome, Magnetic resonance imaging, Neuroimaging, Traumatic brain injury
1. Introduction
Traumatic brain injury (TBI) is a significant public health care burden in the United States (US) and worldwide (Kay, 1993, National Institutes of Health, 1999). TBI has also gained national awareness because it has become the “signature wound” of soldiers in the antiterrorism wars in Iraq and Afghanistan (Zoroya, 2007) and because of the damage to both youngsters and adults caused by sports injuries. Most TBI patients have mild TBI (mTBI), with an incidence of over 1.2 million cases annually in the US (Kay, 1993, National Institutes of Health, 1999). Despite the term “mild,” mTBI causes a constellation of physical, cognitive, and emotional symptoms that significantly impact the patients' quality of life and cost the nation $16.7 billion each year (Bazarian et al., 2005, CDC, 2003, Ruff, 2005). In the emergency department (ED), the majority of mTBI patients have negative findings on clinical imaging, including computed tomography (CT) and conventional magnetic resonance imaging (MRI), such as at T1, T2*, and fluid attenuation inversion recovery (FLAIR) sequences, (Belanger et al., 2007, National Academy of Neuropsychology, 2002) despite a constellation of clinical presentations in their emergency stay.
To date, there are no properly sensitive tools to detect the underlying pathophysiological changes in the brain after mTBI at the acute stage. It has been reported that mTBI patients have both microstructural damage in major white matter tracts, detected by diffusion tensor imaging (DTI) (Kou and VandeVord, 2014, Kou et al., 2010, Niogi and Mukherjee, 2010, Niogi et al., 2008b), as well as functional network alterations, detected by functional MRI (fMRI) (Iraji et al., 2015, Johnson et al., 2012, Mayer et al., 2011, Stevens et al., 2012). For microstructural damage, the most susceptible white matter tracts include the corpus callosum (CC), superior coronal radiata, cingulate bundle, superior and inferior longitudinal fasciculi, and accurate fasciculus (Kou et al., 2012, Kou and VandeVord, 2014, Kou et al., 2010). The damage in these tracts, measured by fractional anisotropy (FA) on DTI, has been reported to be associated with mTBI patients' neurocognitive symptoms or post-concussive syndrome (PCS) scores. Irimia et al. used a connectogram approach to investigate structural connectivity among different functional parcellations of the brain in three moderate to severe TBI patients and suggested large scale structural network damages in TBI patients (Irimia et al., 2012). However, the study only investigated a handful of cases. To date, there is still a lack of data to investigate how brain injury affects the brain structural network connectivity at large scale or connectome-scale.
In addition to microstructural injury, alterations in functional activity may also occur after mTBI, either due to direct injury to the functional networks or the remodeling response to the microstructural injury. Therefore, an investigation of the brain's functional activity is also important for a further understanding of brain alterations. Task-based fMRI demonstrates functional alterations of mTBI patients' memory (Chen et al., 2012, McAllister et al., 1999) and language (Morgan et al., 2013, Tivarus et al., 2012) networks. Though task-based fMRI has been used to identify activity related to specific brain regions for years (Ogawa et al., 1992), functional brain activity is more complicated than is disclosed by investigating the brain during tasks. The brain includes many functional networks, such as memory, motor, attention, vision, and auditory networks, etc., that work together. It is impracticable to investigate all of these brain functional networks using multiple tasks, and in many scenarios, it is often not possible to perform task-based fMRI, due to subjects' health conditions or age. Furthermore, the output of task-based fMRI is vulnerable to subjects' willingness or ability to cooperate.
As an alternative, resting-state fMRI (rsfMRI) has been suggested to complement task-based fMRI (Kou and Iraji, 2014, Shimony et al., 2009). rsfMRI has the ability to simultaneously investigate several brain networks and to explore functional connectivity between brain regions (Damoiseaux et al., 2006, Iraji et al., 2016). Several studies investigating alterations in rsfMRI data in various brain disorders and mental conditions have shown its ability to differentiate patients from healthy subjects (Jafri et al., 2008, Sorg et al., 2007, Calhoun et al., 2008, Chen et al., 2015, Iraji et al., 2016). rsfMRI studies have reported several network alterations in mTBI, including the default mode network (DMN) (Iraji et al., 2015, Johnson et al., 2012, Mayer et al., 2011, Zhou et al., 2012), thalamus network (Tang et al., 2011) and others (Messe et al., 2013). Instead of looking at individual networks by using seed region based analysis, Stevens et al. (Stevens et al., 2012) used independent component analysis (ICA) to investigate mTBI data and reported several network alterations of the whole brain. In a longitudinal study, Messe et al. have shown that functional connectivity in temporal and thalamic regions was increased at the sub-acute stage in patient with post-concussion syndrome; however, it was decreased in frontal areas at six months after injury (Messe et al., 2013). Mayer et al. investigated the dynamic functional connectivity along with static functional connectivity in mTBI patients, and although their analysis did not show group differences after multiple comparisons correction, the static and dynamic functional connectivity shows trends of reduction in the DMN (Mayer et al., 2015), which is aligned with a previous study by the same group (Mayer et al., 2011) on static functional connectivity. Despite promising progress, these investigations of brain functional networks all focused on a limited number of brain networks and it is still unclear how mTBI changes the brain functional networks on a large-scale level, particularly at the connectome level.
The aforementioned evidence suggests that brain injury affects more than one or even several brain networks in both structural and functional connectivity. Instead, mTBI may cause large-scale network disruptions or alterations both structurally and functionally. A better assessment of the scale of structural disruptions and functional alterations of brain networks after mTBI may help physicians better diagnose brain injury and order appropriate rehabilitation. However, the field is still short of investigations on connectome-scale brain network changes, in both structural and functional connectivity, after mTBI.
When measuring the brain's structural and functional connectivity at large-scale network level, the choice of regions of interest (ROIs) plays a pivotal role in analysis. ROIs serve as a structural basis for measuring the connectivity between one brain region and others both structurally and functionally. The selected ROIs should not only be specific to individual brains but also be consistent and robust enough across the population for large-scale comparison (Zhu et al., 2013). Despite the promising abilities of rsfMRI, there are still several barriers and limitations that need to be resolved in the selection of functional nodes. The limitations can be divided in two general categories:
-
1)
Inability to identify an appropriate ROI due to unclear functional and/or cytoarchitectonic boundaries between cortical regions (Brett et al., 2002, Li et al., 2010a, Liu, 2011, Zhu et al., 2011). Considering the fact that a slight change in an ROI location can cause dramatic effects on results, the importance of identifying ROIs as precisely as possible has become clear (Li et al., 2012, Liu, 2011, Zhu et al., 2011). At the same time, each sulcus and gyrus can be included in several functions and be involved in several brain fiber bundles connections (Zoroya, 2007); therefore, current methods that use brain structure such as sulci and gyri are suboptimal to determine the most appropriate ROIs.
-
2)
Significant structural and functional variations among subjects (Zhu et al., 2011). Even if the complexity of cytoarchitecture and disagreement of the boundaries of cytoarchitectonic areas are addressed, due to notable brain morphological variations between subjects (for instance, in the prefrontal cortex), it is difficult to identify regions with similar structural and functional connectivity across individuals (Brett et al., 2002). The complex shape of the cortex makes it hard to find corresponding regions among individuals. Despite several magnificent efforts to identify common brain structures, we still lack reliable and consistent functional landmarks. All of this evidence of brain complexity brings up the question of reliability in using brain anatomy to identify corresponding meaningful functional regions among individuals.
To overcome the aforementioned problems, Zhu et al. identified a set of group-wise consistent brain landmarks based on common fiber connection profiles called Dense Individualized Common Connectivity-based Cortical Landmarks (DICCCOLs) (Zhu et al., 2014). In DICCCOL framework, the common cortical areas are defined based on fiber tracts derived from DTI fiber tractography – the common area carries common fiber tracks across individuals. Therefore, DICCCOL is a predictive framework for brain network regions. Instead of seeking one-to-one correspondence in brain image registration or whole-brain parcellation into sub-units, the DICCCOL system aims to identify the most common cortical landmarks based on the criteria of group-wise consistent DTI-derived fiber connections emanating from the corresponding landmarks. With a data driven approach, Zhu et al. explored and identified 358 consistent brain landmarks evenly distributed on the cortical surface. These landmarks have been shown to be highly reproducible across individuals (Zhu et al., 2014). Moreover, the functional role of each DICCCOL has been extensively examined (Yuan et al., 2013, Zhu et al., 2014). Based on the connectional fingerprint concept suggested by Passingham (Passingham et al., 2002), that each brain's cytoarchitectonic area has a unique set of extrinsic inputs and outputs that largely determines the functions that each brain area performs, and because each DICCCOL preserves consistent structural connectivity, its functional role in brain networks can also be determined. In previous extensive studies, it has been shown that DICCCOLs serve as a better landmark system than the traditionally used Brodmann map to investigate structural and functional connectomic abnormalities on a large scale (Li et al., 2013, Morgan et al., 2013, Zhu et al., 2014). The DICCCOL system can effectively address the aforementioned limitations. Unclear functional and/or cytoarchitectural boundaries and the nonlinear properties of cortex are appropriately satisfied by the search procedure, and group-wise consistency is used as a constraint of optimization. Structural and functional brain variations among individuals were addressed by maximizing group-wise consistency of fiber connections. Additionally, this method has the important advantage of the prevention of errors caused from image registration.
In this paper, we hypothesize that mTBI results in large-scale brain network connectivity changes. This study was designed to determine the large scale, or connectome scale, brain network connectivity changes in mTBI at both structural and functional levels. On the one hand, TBI is heterogeneous and complex, and each TBI patient is unique in terms of injury severity, location, biomechanical impact scenario and pathophysiology; on the other hand, the reported imaging investigations did report the common patterns of certain brain regions or major white matter tracts susceptible to injury, and cognitive neuroscience investigations also suggest several common functional domains that are susceptible to alterations. This study focuses on the common patterns of structural and functional connectivity changes after injury that are shared among patients to evaluate the utility of the DICCCOL analytical approach to brain injury research at the group level. The summary of processing steps (schematic of the pipeline) can be found in Fig. 1. Briefly, we first estimated the initial location of DICCCOL nodes by co-registering individual subjects onto a DICCCOL template to find initial DICCCOL node locations, then optimized the individual node locations to maximize the group consistency for each node. Next, we identified two sets of cortical landmarks in mTBI: common DICCCOLs and discrepant DICCCOLs. The common DICCCOLs have the consistent and similar fiber bundle patterns emanating from their corresponding DICCCOLs among healthy individuals and mTBI patients. In contrast, discrepant DICCCOLs have significant group difference between patients and controls in their extracted fiber bundle patterns emanating from their corresponding DICCCOLs. In other words, discrepant DICCCOLs refers to those DICCCOLs for which we could find group-consistent fiber bundle structures in control group but not in patient group, after exhaustive search for the optimal location within 5 mm radius range in our search algorithm. Therefore, the discrepant DICCCOLs could serve as a potential surrogate biomarker to identify the pathophysiological or structural abnormality in those affected white matter tracts. We also extracted and verified robust functional connectomic signatures that identify the cortical areas that have altered functional connectivity after brain injury despite having normal structural connectivity. These functional connectomic signatures can distinctively characterize mTBI patients from healthy subjects.
Fig. 1.
Schematic of the data analysis pipeline. The olive green boxes indicate general steps, and blue boxes provide details and particular procedures of each step. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2. Methods
2.1. Participants
This study was approved by both the Human Investigation Committee of Wayne State University and the Institutional Review Board of the Detroit Medical Center. Written informed consent was obtained from each subject before enrollment. A cohort of 40 mTBI patients were prospectively recruited from the ED of Detroit Receiving Hospital, a Level-1 trauma center which is an affiliated hospital of the Detroit Medical Center. Patient eligibility was based on the mTBI definition by the American Congress of Rehabilitation Medicine (American Congress of Rehabilitation Medicine, 1993) with the following inclusion criteria: patients aged 18 or older with an initial Glasgow Coma Scale (GCS) score of 13–15 in the ED with any loss of consciousness < 30 min or any post traumatic amnesia < 24 h, or recorded change of mental status (confused, disoriented or dazed). All patients required a CT scan as part of their clinical evaluation. All of them were able to speak English. The exclusion criteria included patients under the age of 18 years, pregnant woman, and patients with any medically documented history of brain injury, neurological disorders or psychoactive medications, history of substance abuse, computed tomography (CT) indication of any metal in the brain and body, known contraindication to MRI such as a pacemaker or other non-MR compatible implanted device as defined by metal screening procedure, and patients without a clear history of trauma as their primary event (e.g., seizure and epilepsy). In the acute stage, a patient's mental status change or amnesia may lead to errors in the report of their medical history, thus the patient's record was retrospectively screened as well to exclude any patient who does not fit our inclusion criteria. Additionally, we also recruited a cohort of 50 healthy controls without history of head injury or antecedents of central nervous system disease from the local community and patients' family members and relatives to match their demographics. The patients and healthy controls were not matched on a subject-to-subject basis; that is, a healthy control recruited from a specific patient's family was not chosen based on a close match to that specific patient, but rather to attempt to keep the groups relatively similar in terms of socioeconomic status, education, and other factors.
Our dataset includes 50 healthy subjects (age: 29.88/10.75 mean/std, male/female: 38/12) and 40 patients (age: 38.03/13.69 mean/std, male/female: 24/16) with mTBI, recruited at the acute stage. The rsfMRI data was not acquired for 8 patients; therefore, functional connectivity analysis was performed with 32 patients. Demographic information has been shown in Table 1. There was no significant gender difference between mTBI patients and healthy subjects; however, race and age are significantly different between two groups. The median time between injury and MRI scan is 20 h (Table 1).
Table 1.
Demographic characteristics of patients and healthy controls.
| Characteristic | Patients with mTBI (n = 40) |
Control subjects (n = 50) |
|---|---|---|
| Gender | ||
| Female | 16 | 12 |
| Male | 24 | 38 |
| Age | ||
| Mean ± SD | 38.03 ± 13.69 | 29.88 ± 10.75 |
| Median/range | 35/(73 ~ 19) | 26/(66 ~ 19) |
| Race | ||
| African American | 29 | 5 |
| White | 8 | 30 |
| Asian | 0 | 5 |
| Others | 3 | 10 |
| Time since injury (hours) | ||
| Mean ± SD | 74.43 ± 103.37 | – |
| Median/range | 20/(446 ~ 2) | – |
| Glasgow Coma Scale | ||
| Mean ± SD, range | 14.95 ± 0.22, (14–15) | – |
| MRI diagnosis | ||
| Traumatic bleeding and lesions | 3 | 0 |
| Non-specific hyperintensities on FLAIR | 10 | 11 |
2.2. Image acquisition
MRI data were collected on a 3-Tesla Siemens Verio scanner with a 32-channel radiofrequency head-only coil. Both DTI and rsfMRI images were acquired, in addition to baseline T1, T2, fluid attenuated inversion recovery (FLAIR), and susceptibility weighted imaging (SWI). The total data acquisition protocol was about 40 min. Specifically, diffusion imaging was acquired using a gradient echo EPI sequence with b = 0/1000 s/mm2 in 30 diffusion gradients directions with the following parameters: TR (repetition time) = 13.300 ms, TE (echo time) = 124 ms, slice thickness = 2 mm, pixel spacing size = 1.333 × 1.333 mm, matrix size = 192 × 192, flip angle = 90°, and number of averages (NEX) = 2. Resting state functional imaging was performed using a gradient echo EPI sequence with the following imaging parameters: TR/TE = 2000/30 ms, slice thickness = 3.5 mm, slice gap = 0.595 mm, pixel spacing size = 3.125 × 3.125 mm, matrix size = 64 × 64, flip angle = 90°, 240 volumes for whole-brain coverage, NEX = 1, acquisition time of 8 min. During resting state scans, subjects were instructed to keep their eyes closed, stay awake and not focus on anything in particular.
All structural MRI data, from both patients and controls, were reviewed by our board certified neuroradiologist (CZ) to identify any abnormalities. The neuroradiologist was blinded to all subjects' clinical information. Structural MRI findings are summarized in Table 1. Among 40 mTBI patients, twenty-seven of them experienced loss of consciousness for < 30 min. Three patients had positive structural imaging findings: one patient had a left temporal lobe hemorrhagic contusion and left ventricular hemorrhage on MRI, but only skull fracture and no parenchymal hemorrhage on CT; the second patient had small hemorrhagic foci in the ventricle and left lingual gyrus on MRI, but negative CT; and the third patient had small petechial hemorrahges on the right parietal cortical surface on MRI and bleeding and scalp edema on CT. This adds some level of heterogeneity to our data, which could potentially impact the group-level analysis. However, the third subject was excluded from the rsfMRI analysis as they were one of the eight subjects lacking fMRI data discussed above. Ten patients and 11 controls also had non-specific white matter hyper-intensities on structural MRI. Structural MRI and its role in mTBI detection and outcome prediction at the acute stage will be addressed in other works. This paper focuses on the large-scale network connectivity changes of the brain after head injury.
2.3. Image preprocessing
Preprocessing for both diffusion data and resting-state data was performed using the FSL software (Jenkinson et al., 2012) (http://www.fmrib.ox.ac.uk/fsl/). Diffusion data preprocessing included brain extraction, motion correction, and eddy current correction. The cortical surface was reconstructed based on the segmented fractional anisotropy (FA) image of white matter (Liu et al., 2007, Liu et al., 2008) (see Fig. 2). Since the surface is reconstructed upon an FA map of white matter, it marks the boundary between the white and gray matter. This reduces the impact of the cortical characteristics (i.e. thickness or shape) on the optimization process and identification of the optimized locations for DICCCOLs. This approach has been validated as effective in the DICCCOL modeling paper (Zhu et al., 2012). Fiber tracking was performed via MedINRIA (http://med.inria.fr/). For resting-state data, the first five volumes were excluded due to magnetization equilibrium. Brain extraction, motion correction, slice-time correction, spatial smoothing (FWHM = 5 mm), temporal prewhitening, grand mean removal, and temporal high-pass filtering were then applied on rsfMRI data accordingly. The preprocessed data were used to predict DICCCOLs as described below.
Fig. 2.

Examples of reconstructed cortical surfaces of (a) a healthy control subject and (b) an mTBI patient subject using fractional anisotropy (FA) data.
2.4. Prediction of DICCCOLs
The DICCCOL system (Zhu et al., 2014) is a cortical landmark prediction framework that contains 358 landmarks with consistent structural/functional roles across individuals. In this approach, all DICCCOLs were identified on the cortical surface based on their consistent white matter fiber connection profiles derived from DTI tract fibers. Prediction of DICCCOLs in a group of subjects is a process of searching optimal cortical regions in each subject from which the emanating fiber bundles share similar connectivity profiles within the group. In this approach, one key step is to quantitatively compare the fiber bundle connectivity profiles with each other. To do this, a fiber bundle shape descriptor called trace-map algorithm has been developed and utilized (Zhu et al., 2012). Briefly, the fiber bundle originating from an ROI is extracted by using the deterministic tractography algorithm (Fig. 3a). Next, the connection orientation profile for each fiber streamline is calculated (Fig. 3b), and projected onto a unit sphere (Fig. 3c). The result is the distribution of orientation vectors on the sphere (Fig. 3d). Next, the unit sphere is divided to 144 regions and the number of projected points was counted for each region. The result is a vector with 144 values represented the direction profile of fiber bundle connected to the ROI. The advantage of the trace-map algorithm is that it can be used to compare overall shape of different fiber bundles with tolerance of small variations among individuals.
Fig. 3.
Illustration of trace-map calculation and DICCCOL prediction. (a)–(d): trace-map calculation. (a) The fiber bundle emanating from a selected ROI; (b) visualization of two single streamlines of the fiber bundle and calculation of the connection orientation profiles; (c) projection of directions onto unit spheres; and (d) visualization of distribution of orientation vectors on the sphere (trace-map). (e)–(g): DICCCOL prediction. (e) DICCCOL templates with DICCCOL locations. (f) Subject surface and the initial location of a single DICCCOL. (g) Optimization process: Minimizing the dissimilarity of fibers passing through the DICCCOL between a subject and the templates.
In the process of predicting DICCCOLs, cortical landmarks were identified based on reconstructed cortical surface and fiber tracks were reconstructed from diffusion images with publicly available tools (http://dicccol.cs.uga.edu/). DICCCOL tools contain DICCCOL templates with DICCCOLs' locations (Fig. 3e) and trace-map feature vectors. The b0 image of diffusion data was aligned with DICCCOL templates using a linear registration with 12 degrees of freedom (DOF), and the initial location of DICCCOLs on the surface was obtained by alignment as illustrated in Fig. 3f. In the next step, for each DICCCOL, the optimized location has been identified by searching the neighborhood of up to 10 mm cortical distance (Zhang et al., 2012, Zhu et al., 2014) from the initial location to minimize the dissimilarity of fibers passing through the DICCCOL between a subject and the templates (Fig. 3g). Our previous comparison between DICCCOL prediction of functional nodes and task-activated functional locations demonstrated that the optimized locations are mostly within 5 mm of the predicted locations (Zhang et al., 2012, Zhu et al., 2014). Therefore, a radius of 10 mm search range will be a safe zone to identify optimal DICCCOLs. The processing pipeline is detailed in (Zhu et al., 2011).
2.5. Structural abnormality and discrepant DICCCOLs
In this study, “structural abnormality” does not mean gross structural damage, but instead refers to the situation in which the DTI deterministic tractography of the DICCCOL analysis fails. This could be caused by any source of structural abnormality in white matter connectivity that disables the tractography step of the DICCCOL analysis to identity the proper location for these DICCCOLs. The failure of the white matter fiber extraction step could be caused by several pathophysiological mechanisms after brain injury incident. Furthermore, most disruptions in white matter after mTBI might resolve during the recovery process. Thus, the structural abnormality does not indicate permanent structural damage. Therefore, discrepant DICCCOL is used to refer to those DICCCOLs for which the shape of connected fiber bundles is significantly different in patients as compared to healthy controls.
2.6. Classification of discrepant and common DICCCOLs
The DICCCOL system was designed to identify those regions with group-consistent structural connectivity patterns. After brain injury, water diffusion properties of white matter tracts can be changed due to various pathophysiological conditions. The DTI deterministic tractography of these white matter tracts will result in disrupted fibers. Despite the fact that these white matter tracts may not be physically disrupted or damaged, the “disrupted” fiber tractography certainly serves as a surrogate biomarker of the pathophysiological or structural abnormality in these affected white matter tracts. By comparing the fiber connectivity profiles between patients and controls, we could classify the DICCCOLs into two groups: commons vs. discrepant DICCCOLs. A common DICCCOL shares similar fiber connectivity patterns between patients and controls. However, in a discrepant DICCCOL, the fiber connectivity patterns reach statistical difference between patient and control groups. Since the trace-map represents the shape of fiber bundles connected to a DICCCOL, it can be used to find the DICCCOLs that have alterations in connected fibers in the patient group. The trace-map distance parameter measured by Euclidean distance can be used to measure similarity of structural connectivity patterns between two trace-maps (Eq. (1)). A lower value for the trace-map distance parameter represents greater similarity between two trace-maps.
| (1) |
where T1 and T2 are two trace-map vectors, i is the index of a 144 dimension feature vector which describe the trace-map.
For each DICCCOL, the trace-map distance parameter has been measured compared to the average trace-map vector of healthy subject group. A two-sample t-test (p-value = 0.01) has been applied to the trace-map distance parameters to compare structural connectivity characteristics between healthy subject and patient groups. The DICCCOLs that show significant differences between the two groups are considered discrepant DICCCOLs, i.e. structurally discrepant between patient and healthy control groups. The rest of the DICCCOLs are identified as common DICCCOLs, which means they have similar fiber connections patterns in patient and healthy subject groups. Identifying corresponding anatomical locations among subjects of both groups in common DICCCOLs gives us ability to assess functional connectivity patterns, which could still be affected in common DICCCOLs.
2.7. fMRI analysis
Only common DICCCOLs have been used for functional connectivity analysis. Our rationale is that patients could have variations in functional connectivity in spite of intact structural connectivity. Therefore, we expected to see alterations on functional connectivity among DICCCOLs that have similar structural connectivity in healthy subject and patient groups. In analysis, rsfMRI data was directly registered to the b0 image of diffusion data using a 6 DOF affine transformation. Both fMRI and diffusion data were collected in EPI sequence, and they distort in the same way (Li et al., 2010b). The 6 DOF affine transformation will be adequate for image registration as reported in (Li et al., 2010b, Penny et al., 2011). Since DICCCOLs are on the surface that was reconstructed using diffusion data, they are located on the interface between white matter and gray matter rather than on the surface of the gray matter. Therefore, the time series of the closest gray matter voxel has been assigned to the DICCCOL for functional analysis. Spatial smoothing has already been applied on functional data to improve signal-to-noise ratio. Our preliminary data demonstrated that this is an effective method for prediction of functional nodes (Zhang et al., 2012, Zhu et al., 2014). Functional connectivity has been calculated by measuring the Pearson correlation value between each pair of common DICCCOLs.
In order to find the most discriminate functional connectivities, which will be referred to as features, we have applied a two-step feature selection procedure. First, a two sample t-test (p-value = 0.01) was performed to exclude those features that could not reveal a statistically significant difference between the mTBI patient group and the healthy subject group (remove the true negative). In the second step, the correlation-based feature selection (CFS) was used to find the most distinctive features by minimizing the degree of redundancy among features (Zhu et al., 2011). The selected functional connectivities are the most distinctive and discriminative characteristic features to distinguish between healthy subjects and patients in our dataset.
In order to assess the ability of these functional connectivities to differentiate between two groups, supervised and unsupervised learning procedures were used. Specifically, a supervised learning procedure was performed using a support vector machine (SVM) classifier with 10-fold cross-validation to measure the specificity and sensitivity of the selected functional connectivities. At the same time, in order to measure the similarity within each group and dissimilarity between two groups, we used an unsupervised learning method. In other words, we are interested in seeing how well the data of each group belongs to the same category. XMeans clustering has been used to evaluate this similarity (Pelleg and Moore, 2000).
2.8. Meta-analysis
BrainMap (http://www.nitrc.org/projects/brainmap/) (Laird et al., 2009) is an accessible database of published neuroimaging literature, which allows us to identify the function of a specific brain region and compare the result of our study with reported literature. Use of this database can significantly enhance the reliability of neuroimaging studies. BrainMap includes the coordinate and the associated metadata of brain regions which are identified across a collection of studies (Laird et al., 2009). In our previous work, we have used it to identify the possible functional roles for each DICCCOL using meta-analysis (Yuan et al., 2013).
Briefly, the locations of DICCCOLs from our templates were registered to the MNI atlas, then a neighborhood with a radius of 3 mm around each DICCCOL was selected in order to assign a Brodmann area and determine a related functional role using the BrainMap software. 110 fMRI publications and their reported activation regions in the BrainMap database were examined to identify related functional roles for each DICCCOL (Yuan et al., 2013). Therefore, all selected DICCCOLs were categorized in five general classes, based on published fMRI data set in Brain Map Database: “Action,” “Perception,” “Cognition,” “Interoception,” and “Emotion”.
At the same time, the strength of functional connectivity between two DICCCOLs can represent the strength of functional connectivity between their networks. For instance, if DICCCOL A was identified as “Cognition”, and DICCCOL B was identified as “Action” then we can interpret that the strength of functional connectivity between DICCCOLs A and B is related to the strength of functional connectivity between “Cognition” and “Action”. Therefore, if the functional connectivity between two categories of DICCCOLs changes, we can conclude that the interaction between their networks has been affected. For example, if the majority of the strengths of functional connectivity between DICCCOLs belonging to “Cognition” with DICCCOLs belong to “Action” is changed due to a specific health condition, we can conclude that the interaction between “Cognition” and “Action” has been affected.
The interaction between brain networks was measured using five general functional categories: “Action,” “Perception,” “Cognition,” “Interoception,” and “Emotion”. Since “Interoception” is not significantly involved in connectomic signatures, it has been eliminated from this analysis step.
Two general conditions occur to the functional connectivity between each pair of DICCCOLs:
-
1.
Each DICCCOL designates to only one network. In this case, the correlation value between two DICCCOLs indicates the connectivity between two functional roles. For example, if DICCCOL A and DICCCOL B were respectively labeled as “Action” and “Cognition” functions, the correlation value between DICCCOL A and DICCCOL B associates to a “Action-Cognition” network interaction.
-
2.
One or both DICCCOLs designate to two or more networks. In this situation, the functional connectivity between the DICCCOL pair is associated to each pair of networks. For instance, if DICCCOL A was identified as “Action”, and DICCCOL B was identified as “Action” and “Cognition”, the correlation value between DICCCOL A and DICCCOL B associates to both “Action-Action” and “Action-Cognition” network interactions.
The interaction between DICCCOLs can also be divided in two categories:
-
1.
Within network interactions in which the same network is assigned to interactive DICCCOLs, like a “Perception-Perception” interaction.
-
2.
Between networks interactions in which two different networks interact with each other, like “Perception-Action”.
3. Results
3.1. DICCCOL prediction
DICCCOLs have been predicted on the cortical surfaces for all 50 healthy subjects and 40 mTBI patients. An example of DTI-derived axonal fiber bundles connected to a randomly selected common DICCCOL for 20 healthy subjects' brain and 20 patients' brain are shown in Fig. 4a and Fig. 4b. The corresponding trace-map distances are shown in Fig. 4c and Fig. 4d accordingly. By visual inspection, the shape patterns of fiber bundles are relatively consistent across individuals for both patients and healthy subjects. The trace-map distance similarity between patients and healthy subjects can also be observed in the trace-map distance obtained for the same common DICCCOL (Fig. 4c and Fig. 4d).
Fig. 4.
Joint visualization of a common DICCCOL (#178, yellow arrows) and a discrepant DICCCOL (#19, yellow arrows). (a) and (b) show the connected DTI-derived axonal fibers (colorful lines) connected to the common DICCCOL on the cortical surface for 20 patients and 20 control subjects, respectively. Visual inspection reveals the relatively consistent patterns of fiber bundles across individuals for both patients and healthy subjects. (c) and (d) respectively show the trace-map distance for 20 healthy controls and 20 patients selected in (a) and (b). As we can see, healthy subjects and patients have similar trace-map distance values for the common DICCCOL # 178. (e) and (f) show the connected DTI-derived axonal fibers (colorful lines) connected to the discrepant DICCCOL # 19 on the cortical surface for the same 20 patients and 20 control subjects as (a) and (b), respectively. Several patients have different fiber shapes (red outlines) in these randomly chosen 20 samples, which drive the group difference between patients and controls. At the same time, measuring the trace-map distances for DICCCOL #19 for same subjects shows that patients in general have a higher trace-map distance. The same scale has been used at (g) and (h) to make it visually easy to compare the trace-map distances of discrepant DICCCOLs across groups. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
On the other hand, Fig. 4e and Fig. 4f demonstrate the connected fibers to a randomly selected discrepant DICCCOL. By visual inspection, the connection pattern of this DICCCOL is still consistent in healthy subjects; however, as highlighted by the red boxes, it is obvious that the structural connectivity profile of the selected discrepant DICCCOL varies across patients' brains. This could be caused by any source of structural or pathophysiological abnormality in white matter tracts, which changes the water diffusion characteristics and consequently affect their fiber shapes in tractography reconstruction. Moreover, the trace-map distance for this discrepant DICCCOL has been demonstrated for the same 20 healthy subjects and 20 patients in the Fig. 4g and Fig. 4. Results show that patients have higher trace-map distances than that of controls. At the same time, the trace-map of fiber bundles for the common and discrepant DICCCOLs for these 20 healthy subjects and 20 patients also showed similar results.
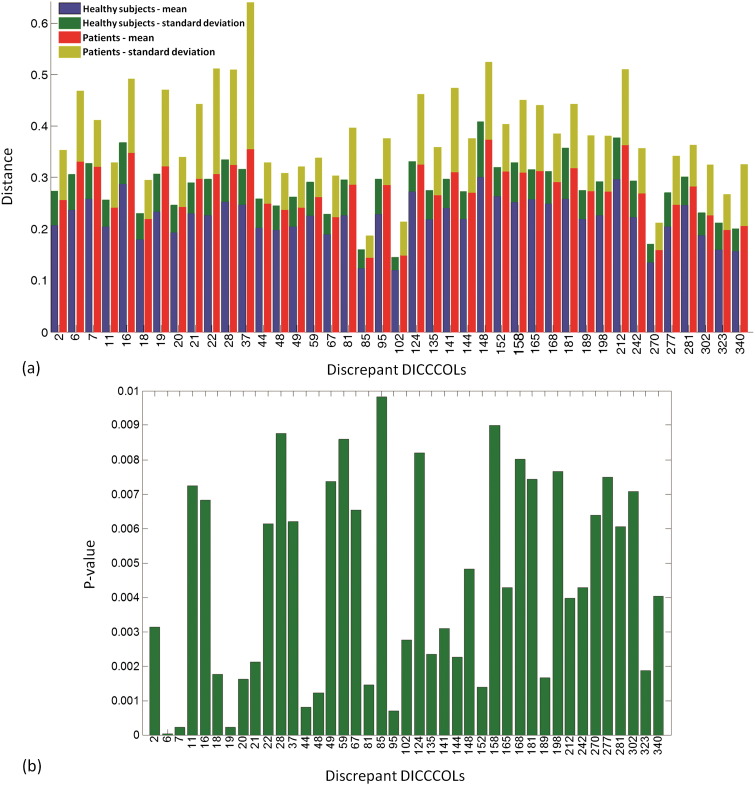
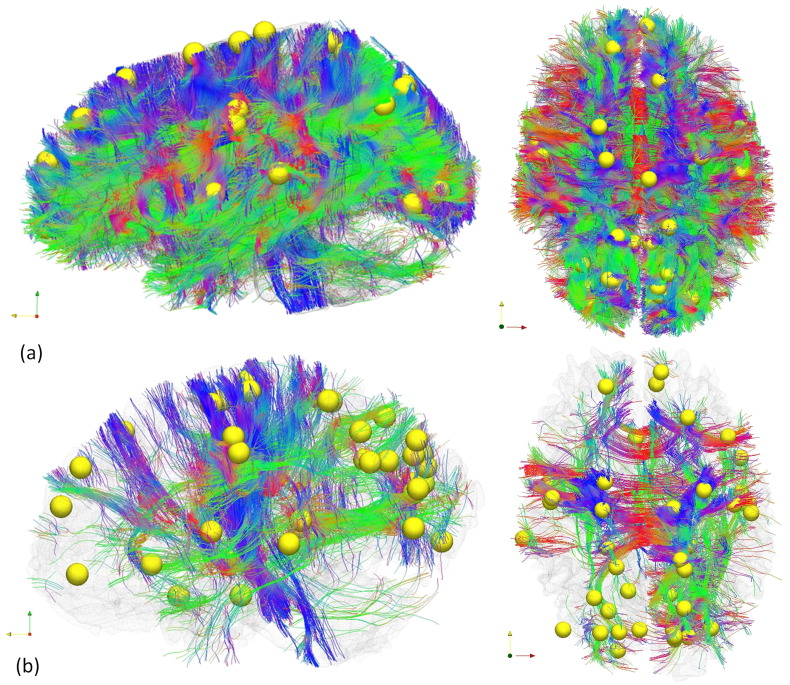
In total, 41 discrepant DICCCOLs were identified among total 358 DICCCOLs using statistical analysis on trace map distance. The quantitative measurement for trace-map distance is shown in Fig. 5. Fig. 5a demonstrates the average and standard deviation of the trace-map distance of all discrepant DICCCOLs for both groups. As we can see, patients in general have the higher trace-map distance. Their p-values indicate statistical difference between two groups in Fig. 5b. Examining discrepant DICCCOLs reveals that the somatosensory association cortex (Brodmann area 7) and visual cortex (Brodmann area 17, 18, 19) have been affected most, as determined by the number of discrepant DICCCOLs. The distribution of the discrepant DICCCOLs in the brain is shown in Fig. 6. Furthermore, by using these discrepant DICCCOLs as ROIs, fiber tractography can visualize the difference between a randomly chosen control and randomly chosen mTBI patient (see Fig. 7). In Fig. 7, the yellow spheres are the discrepant DICCCOL landmarks. Despite the negative findings on the mTBI patient's structural MRI, the patient's white matter structure shows significant difference in the white matter bundles emanating from these 41 discrepant DICCCOL landmarks. Of particular note, the sharp contrast of their fiber tractography between the mTBI patients and controls may not be due to physical disruption or loss of white matter tracts in the mTBI patient. Instead, it could be due to white matter abnormalities that change water diffusion properties in the affected tracts and consequently show “disrupted” fibers in the tractography representation of these 41 discrepant DICCCOL networks. Fiber tracking using these discrepant DICCCOLs would result in much sparser representation of the WM tracts. In comparison, the discrepant DICCCOLs in the healthy control were intact and widely distributed throughout the whole brain. Since each DICCCOL is involved one or more major WM tracts, fiber tracking using these 41 DICCCOLs in a healthy control brain would show the WM tracts across almost the whole brain.
Fig. 5.
(a) Trace-map distances of the discrepant DICCCOLs for the healthy control group (mean value in blue and standard deviation in green) and patient group (mean value in red and standard deviation in yellow). (b) p-values from two-sample t-tests, which show the difference between the two groups in discrepant DICCCOLs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Visualization of location of discrepant DICCCOLs (red sphere) and the rest DICCCOLs (green sphere) on cortical surface. ID numbers are shown for discrepant DICCCOLs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
White matter fiber tractography of a randomly chosen control subject (a) and a randomly chosen mTBI patient (b) by using the 41 discrepant DICCCOLs (yellow spheres) as seed points. Despite the negative findings on the mTBI patient's structural MRI, the patient's white matter structure shows significant differences in the 41 discrepant networks in comparison with controls. Of particular note, the differences in the major white matter tracts between the control and the patient are not because of the loss of white matter tracts in the patient; instead, it is that these discrepant networks fail the fiber tractography algorithm when using the selected DICCCOL landmark as seed regions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Our visual inspection of the connected white matter tracts of each discrepant DICCCOL also found white matter pathways that are structurally different between two groups in their fiber tractography presentations, which is confirmed in a group comparison using the trace-map algorithm. Table 2 demonstrates the locations of discrepant DICCCOLs in Brodmann areas and their connected major fibers. The superior and inferior longitudinal fasciculi, the corpus callosum, the arcuate fibers, and the cingulate bundle are the most commonly affected white matter tracts. This finding is consistent with the summary of published literature regarding the white matter fibers most susceptible to injury (Kou and VandeVord, 2014). Importantly, the DICCCOL-based analysis offers fine-granularity dissection and measurement of such coarse-granularity fiber pathways.
Table 2.
Locations of discrepant DICCCOLs and their Brodmann areas and connected major fiber tracts.
| DICCCOL ID | BA | CC | ILF | IFOF | SFOF | SLF | PF | AF | Cing | UF |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 18 | ✓ | ✓ | |||||||
| 6 | 18 | ✓ | ✓ | ✓ | ||||||
| 7 | 17 | ✓ | ✓ | ✓ | ||||||
| 11 | 18 | ✓ | ✓ | |||||||
| 16 | 19 | ✓ | ✓ | ✓ | ||||||
| 18 | 17 | ✓ | ✓ | |||||||
| 19 | 18 | ✓ | ✓ | ✓ | ||||||
| 20 | 18 | ✓ | ✓ | |||||||
| 21 | 18 | ✓ | ✓ | |||||||
| 22 | 18 | ✓ | ✓ | ✓ | ||||||
| 28 | 19 | ✓ | ✓ | |||||||
| 37 | 39 | ✓ | ✓ | |||||||
| 44 | 7 | ✓ | ✓ | |||||||
| 49 | 7 | ✓ | ✓ | ✓ | ||||||
| 59 | 7 | ✓ | ✓ | |||||||
| 67 | 7 | ✓ | ✓ | ✓ | ||||||
| 81 | 7 | ✓ | ✓ | ✓ | ||||||
| 85 | ✓ | ✓ | ||||||||
| 95 | 5 | ✓ | ✓ | ✓ | ||||||
| 102 | ✓ | ✓ | ||||||||
| 124 | 42 | ✓ | ||||||||
| 135 | 22 | ✓ | ||||||||
| 141 | 3 | ✓ | ||||||||
| 144 | 36 | ✓ | ||||||||
| 148 | 3 | ✓ | ✓ | |||||||
| 152 | 2 | ✓ | ||||||||
| 158 | 38 | ✓ | ✓ | |||||||
| 165 | 40 | ✓ | ||||||||
| 168 | 3 | ✓ | ✓ | ✓ | ||||||
| 181 | 4 | ✓ | ✓ | ✓ | ||||||
| 189 | 43 | ✓ | ||||||||
| 198 | 41 | ✓ | ||||||||
| 212 | 6 | ✓ | ||||||||
| 242 | 38 | ✓ | ✓ | |||||||
| 270 | ✓ | |||||||||
| 277 | 8 | ✓ | ✓ | |||||||
| 281 | 44 | ✓ | ||||||||
| 302 | 9 | ✓ | ✓ | ✓ | ||||||
| 323 | 10 | ✓ | ||||||||
| 340 | 10 | ✓ | ✓ | |||||||
| Area total | 25 | 18 | 10 | 6 | 6 | 6 | 5 | 3 | 1 |
BA: Brodmann Area; CC: corpus callosum; ILF: inferior longitudinal fasciculus; IFOF: inferior fronto-occipital fasciculus; SFOF: superior fronto-occipital fasciculus; SLF: superior longitudinal fasciculus; PF: projection fibers; AF: arcuate fasciculus; Cing: cingulum; UF: uncinate fasciculus.
3.2. Functional connectivity analysis
Functional disruption has been reported in mTBI patients with normal looking structural MRI (Iraji et al., 2015, Johnson et al., 2012, Mayer et al., 2011, Stevens et al., 2012). Therefore, it is possible there can be alterations in brain activity as seen via changes in functional connectivity of common DICCCOLs despite their normal-looking structural connection patterns of white matter tracts. For this analysis, functional connectivity between each pair of common DICCCOLs was calculated to evaluate common functional alterations among mTBI patients. A symmetric 317 × 317 matrix of functional connectivities was created using the 317 common DICCCOLs for each individual. The 41 discrepant DICCCOLs have been excluded in functional connectivity analysis since the DICCCOL method could not identify their accurate anatomical locations. Despite searching within 10 mm radius scope, the optimized locations for these discrepant DICCCOLs still could not produce group-consistent fiber patterns. The inclusion of these DICCCOLs for functional connectivity analysis could induce false positives.
In the feature selection step, the feature reduction was performed using two-sample t-tests to exclude those features that could not reveal a statistically significant difference between two groups (p-value = 0.01). As a result, 385 connectivities out of the 50,086 features survived. To further control the false positives in these 385 features, in the second step, the CFS was utilized to select features (i.e. connectomic signatures) while minimizing the degree of redundancy among functional connectivities. CFS selected 60 out of 385 functional connectivities as the most distinctive and discriminative features of our data to differentiate patients from healthy control subjects. After controlling for the effect of age with an analysis of covariance (ANCOVA), 58 out these 60 connections were still significantly different between the two groups (p-value < 0.05). These were labeled as connectomic signatures for further analysis.
3.3. Sensitivity and specificity evaluation
The sensitivity, the probability of classifying a real patient correctly (a true positive), and the specificity, the probability of classifying a healthy subject correctly (a true negative), were calculated using a SVM classifier with 10 fold cross-validation. Classification using these 60 connectomic signatures correctly identified 80 out of 82 total subjects (50 controls and 32 patients with fMRI data), giving 97.56% classification accuracy. In fact, all healthy subjects were classified correctly (100% specificity), and only 2 mTBI patients were misclassified into the control subjects group (93.75% sensitivity). XMeans clustering has been used to evaluate the similarity between subjects in each group and the dissimilarity between subjects from different groups. Only one patient among 32 patients was incorrectly clustered in healthy control group; however, 5 healthy subjects among 50 healthy control subjects were incorrectly clustered in the patients' cluster (Incorrectly clustered instances = 7.32%). The clustering result demonstrates that connectomic signatures identified are truly different between groups. And together, they are a decent discriminant marker to categorize mTBI patients for this dataset.
3.4. Meta-analysis
The Brodmann areas of discrepant DICCCOLs are exhibited in Table 2. The results shows that the structural connectivity of the somatosensory association cortex (BA 7) and secondary visual cortex (BA 18) are disrupted the most. In our functional connectivity analysis using common DICCCOLs, 85 DICCCOLs involved in the 60 connectomic signatures have been identified.
Among these 85 DICCCOLs, the ones which have been affected the most are DICCCOL #73, which appears in 5 connectomic signatures and belongs to the angular gyrus (BA 39); DICCCOL #92, which appears in 5 connectomic signatures and belongs to the parahippocampal gyrus (BA 36); DICCCOL #93, which appears in 5 connectomic signatures and belongs to the fusiform gyrus (BA 37); and DICCCOL #271, which appears in 5 connectomic signatures and belongs to the caudate, (Supplementary Table 1).
At the same time, organizing the affected functional connectivity DICCCOLs based on their Brodmann areas reveals that the premotor area (BA 6) appears 10 times in 5 connectomic signatures, the temporopolar area (BA 38) appears 10 times in 5 connectomic signatures, and the angular gyrus (BA 39) appears 7 times in 5 connectomic signatures are the most commonly associated Brodmann areas (Supplementary Table 2). The affected connectivities for these regions include the following:
For Brodmann area 6, the affected connectivities are with Brodmann areas 18, 19, 37, 39, 44, and 47. For Brodmann area 38, the affected connectivities are with Brodmann areas 7, 11, 22, 32, and 47. For Brodmann area 39, the affected connectivities are with Brodmann areas 6, 8, 9, and 27. For a better understanding of the brain's response to injury, we should consider both structural and functional disruptions together. Therefore, by considering the results of common and discrepant DICCCOLs together, we can explain the alterations in the brain in a more appropriate way. Table 3 demonstrates the most affected Brodmann areas. Distribution of affected DICCCOLs has been shown in Fig. 8. The red spheres are discrepant DICCCOLs. The connectomic signatures are demonstrated by gray lines, and their related DICCCOLs are identified with blue spheres.
Table 3.
The most affected Brodmann areas identified by the combination of structural and functional DICCCOL analyses.
| Most effected Brodmann area (BA) | Common | Discrepant |
|---|---|---|
| BA 38 (temporopolar area, most rostral part of the superior and middle temporal gyri) | 10 | 2 |
| BA 6 (secondary motor cortex) | 10 | 1 |
| BA 18 (secondary visual cortex) | 4 | 7 |
| BA 39 (angular gyrus) | 7 | 1 |
| BA 7 (somatosensory association cortex) | 2 | 5 |
| BA 19 (associative visual cortex) | 5 | 2 |
| BA 36 (parahippocampal gyrus.) | 6 | 1 |
Fig. 8.
Visualization of discriminative functional connectivities (gray lines) between mTBI patients and healthy subjects and the location of related DICCCOLs. DICCCOLs were represented by color-coded spheres (blue: DICCCOLs related to discriminative functional connectivities, red: discrepant DICCCOLs, green: common DICCCOLs not related to discriminative functional connectivities). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Connectivity meta-analysis
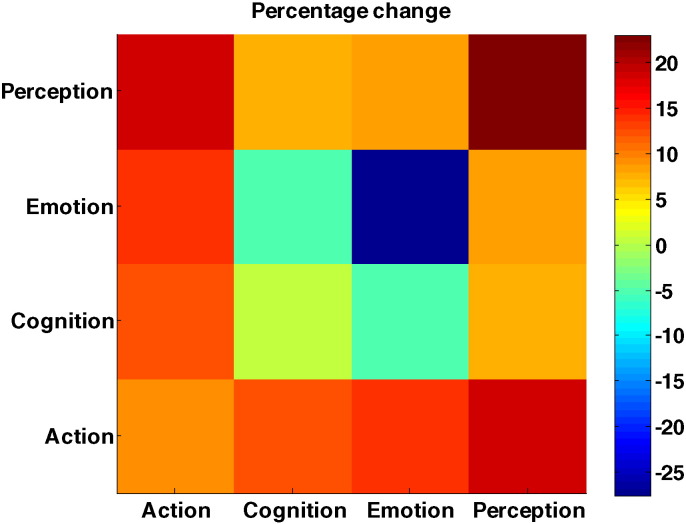
Results show that most of possible network interactions have been affected. Among them, the intra-emotion network interactions have been altered the most, with a significant decrease in the connectivity inside this network (by > 27%). The “Emotion-cognition” interaction also shows decreased connectivity (by 4.8%). On the other hand, intra-perception, “Action-Emotion,” and “Action-Cognition” interactions reveal increased functional connectivity, by 22.97%, 14.01%, and 12.11%, respectively. These results can explain a strong disruption in emotion and perception in patients during the acute stage (Fig. 9).
Fig. 9.
Changes in network interactions. Negative values indicate decreases in network interaction in patient group. Color-bar indicates the percent change. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
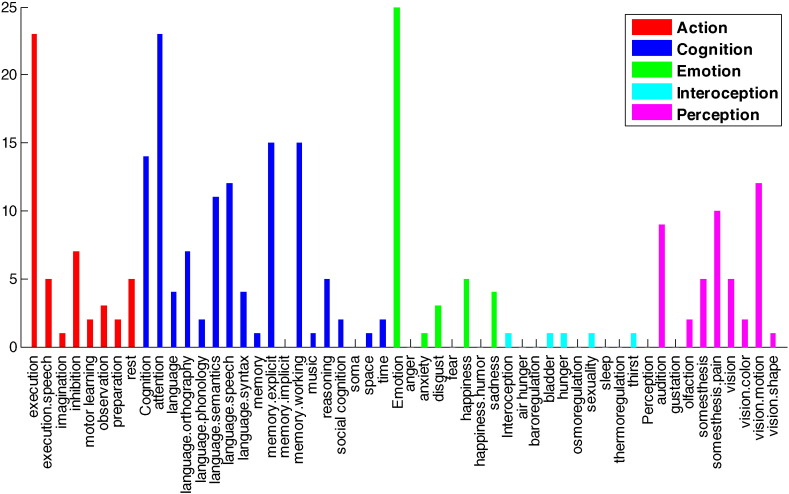
For further functional analysis, the roles of individual DICCCOLs were evaluated more specifically. In this step, the functional roles of DICCCOLs have been categorized in 53 classes using meta-analysis (Yuan et al., 2013). Among all functional roles “Emotion”, “Cognition/attention”, and “Action/execution” are involved more in connectomic signature than others, with 25, 23, and 23 DICCCOLs each, respectively (see Fig. 10 and Supplementary Table 3).
Fig. 10.
Functional roles of connectomic signatures. The affected DICCCOLs were divided in 5 categories with 53 sub-categories using meta-analysis. Meta-analysis reveals that the “Emotion”, “Cognition/attention”, and “Action/execution” categories are involved in more connectomic signature than others, with 25, 23, and 23 DICCCOLs each, respectively. The vertical axis is number of DICCCOLs associated with each functional role.
4. Discussion
To the best of our knowledge, this is the first effort towards a connectome-scale assessment of structural and functional brain connectivity changes after mild TBI. Our work demonstrates that a) mTBI can render the structural abnormality in major white matter tracts that provide structural support of important functional networks; and b) connectivity alterations in functional networks happen across the whole brain, including both increased and decreased functional connectivity in large-scale networks. We identified 60 functional connectivity patterns that differentiate mTBI patients from controls in our data. This work confirms our hypothesis that brain connectivity changes, including both structural and functional changes, happen at the whole brain connectome level. This finding could have significant implications in mTBI diagnosis in the acute setting as well as in understanding of brain recovery after TBI. A better understanding of the extent of common structural abnormalities and functional alterations of brain networks in mTBI could help physicians order proper rehabilitation plans to address specific domains of brain functions for speedy recovery.
4.1. Structural abnormality
Our tractography inspection of the white matter tracts that give rise to discrepant DICCCOL nodes revealed that most major brain white matter tracts are involved, including corpus callosum, inferior/superior longitudinal fasciculi, and inferior/superior fronto-occipital fasciculi, projection fibers, arcuate fasciculus, cingulum, and uncinate fasciculus. Particularly, the discrepant DICCCOLs are those DICCCOLs for which a similar shape of connected fiber bundles could not be identified due to its structural abnormality, which does not necessarily mean gross structural damage or disruption. Among functional domains that the discrepant DICCCOL nodes belong to, “action” and “cognition” are the most affected. This finding is well consistent with the published literature on DTI structural analysis (Kou and VandeVord, 2014, Niogi and Mukherjee, 2010). Major white matter tracts, including the corpus callosum, the major tract that connects the two hemispheres; the cingulum; long association fibers, including superior and inferior longitudinal fasciculus; and white matter structure in the frontal and temporal lobes are well studied in an mTBI population.
Structural abnormality in many of these white matter tracts has been shown to be associated with symptoms. The corpus callosum, the largest white matter fiber bundle in the brain, is highly susceptible to injury during rotational motion of the brain in the sagittal and coronal planes, as it strikes the falx cerebri and tentorium cerebri (Blumbergs, 1997). Autopsy studies show that even mTBI patients could have disrupted axons in the corpus callosum, as evidenced by axonal retraction balls (Blumbergs et al., 1994, Blumbergs et al., 1995). Injury of the callosal fibers has been reported to be associated with post concussion symptom (PCS) scores in both adolescent (Wilde et al., 2008) and adult mTBI patients (Bazarian et al., 2007) at the acute stage. Similarly, Treble et al.'s (Treble et al., 2013) DTI study of pediatric TBI suggested that reduced microstructural integrity of the CC, particularly in the sub-regions connecting the parietal and temporal cortices, may act as a neuropathological mechanism contributing to long-term working memory deficits (Treble et al., 2013). Injury at the temporal white matter tracts or in the cingulum bundle has been reported being associated with memory problems. Niogi et al. (Niogi et al., 2008a) reported that reduced FA in the uncinate fascilulus correlated with memory performance. Wu et al. (Wu et al., 2010) reported that the fractional anisotropy of the left cingulum bundle correlated with delayed recall. Baek et al.'s (Baek et al., 2013) study of chronic TBI patients reported that the integrity of the basal forebrain and cingulum are associated with patients' overall cognition and memory. Regarding the long association fibers, Messé et al. (Messé et al., 2011) reported that mTBI patients with poor outcomes showed significantly higher mean diffusivity values than both controls and good-outcome mTBI patients in the corpus callosum, the right anterior thalamic radiations, the superior longitudinal fasciculus, the inferior longitudinal fasciculus and the fronto-occipital fasciculus. Fig. 7b demonstrates tractography of the white matter fibers by using these discrepant nodes as seed regions. It shows that the white matter structural abnormality happens across the brain instead of at just one or two tracts.
4.2. Alternations in functional network connectivity
Our data identified 60 functional networks that were altered in the mTBI group in comparison with controls. These networks span throughout the whole brain.
Using meta-analysis and comparing our finding with previous functional and structural studies reveal that the four major functional domains, perception, emotion, cognition and action, are involved in the connectomic signatures. Among these four functional domains, our data shows reduced connectivity between cognition and emotion networks as well as in intra-emotion networks. It further shows increased functional connectivity among other network domains. This result is consistent with the collective evidence from different groups who investigated different functional networks. Several groups who investigated the DMN of mTBI patients at the subacute and chronic stages have reported decreased connectivity among different regions of DMN and increased connectivity of DMN regions with other networks (Iraji et al., 2015, Johnson et al., 2012, Mayer et al., 2011, Stevens et al., 2012). In addition to in the DMN, hyper-connectivities in the thalamocortical network have also been reported in mTBI patients (Messe et al., 2013, Tang et al., 2011), including a more distributed thalamus correlation map as compared to the healthy controls (Tang et al., 2011). Our data further support the evidence reported by Stevens et al. that multiple resting state networks could have functional abnormalities after mTBI (Stevens et al., 2012), who reported diminished functional connectivity in the patient group in the DMN and in many other networks, as well as greater connectivity in the precuneus connectivity map. This suggests that the brain tends to respond to head injury by recruiting a cohort of networks in a global manner instead of just one or two networks.
4.3. Combining structural and functional information
Structural information has been used to identify corresponding anatomical locations across individuals to compare functional connectivity between two groups. Thus, for the discrepant DICCCOLs which did not have identifiable corresponding anatomical locations in the mTBI patient group, we cannot perform functional analysis. Therefore, we are unable to identify the functional alteration in structural abnormal DICCCOLs although it would be ideal to combine brain structural and functional alterations.
4.4. The uniqueness and advantage of DICCCOL approach
The determination of DICCCOL functional nodes is based on the concept of a “connectional fingerprint” which provides an anatomical basis of functional localization from basic neuroscience findings (Passingham et al., 2002). The approach consists of several key steps (Zhu et al., 2014): i) initial determination of DICCCOL nodes based on a template, ii) quantification of fiber tracts for each structural network using a trace-map algorithm, iii) optimization of individual node locations to make them group-wise consistent, and iv) determination of functional nodes at the cortex. As a result, each DICCCOL node is individually different in their anatomical locations but group-wise consistent in their connectional finger prints across the population (Zhu et al., 2014). Further validation of major functional nodes by using a task-oriented fMRI benchmark demonstrated a remarkable consistency between the prediction of a functional region and actual activation with the location error < 5 mm on average (Zhang et al., 2012). The 10 mm range for the search algorithm in our work is very likely to find the optimal node location. This would allow researchers to determine large-scale functional networks with high confidence. In comparison, a template-based approach does not account for individual difference, hand-drawn ROIs are susceptible to inter-rater variability, and task-oriented ROIs are not suitable for large-scale network determination. Similar to published data, most of our mTBI patients have normal structural imaging findings, making the DICCCOL approach particularly suitable for mTBI data analysis. Furthermore, the predicted functional nodes are located on the overlap of the reconstructed cortical surface and the real cortex, which makes it less likely to be affected by the susceptibility effect of any hemorrhages on the cortical surface or in the subdural space. However, traumatic hemorrhages at the junction of gray and white matter could induce strong susceptibility artifacts that potentially affect the determination of functional nodes at these locations.
4.5. Limitations and future work
To date, alterations in the brain's structural and functional connectivity after mTBI are still unclear considering the high level of complexity in several factors, such as the time between injury and scan, individual subject-specific features, different mechanisms of injury, and other sources of heterogeneity (Eierud et al., 2014, Ilvesmaki et al., 2014, Sharp and Jenkins, 2015). Moreover, the lack of investigations of large-scale connectivity analysis further hinders our understanding of brain injury phenomena. Therefore, our findings, particularly the identified discriminant features in this work, should be independently tested in separate mTBI datasets as a reproducibility study. Additionally, group studies like ours are made more difficult by the heterogeneity of mTBI, which is largely due to: 1) different mechanisms of injury (e.g. vehicular collision vs. sports) which results in different scenarios of biomechanical loading (strain and stress); 2) different injury pathologies (e.g. neuronal injury vs. axonal injury vs. vascular injury); and 3) different subject samples with different pre-existing conditions and demographic characteristics (age, gender, and education level) (Eierud et al., 2014). One important category of heterogeneity which confounds group-level connectome study is gross structural abnormalities, which can be seen in structural MRI and CT modalities. In our data, three mTBI patients had positive structural imaging findings, one of whom did not have rsfMRI data and therefore it was not included in functional connectivity analyses. The structural abnormalities of these two patients (positive CT and/or structural MRI) might be a confounding factor that drives the patient group (n = 32) apart from the control group (n = 50) in their functional connectivity. In future investigations, excluding these patients with structural abnormalities by focusing on the patients with normal structure findings might yield a more convincing conclusion. In our dataset, given the relative size of the number, these two patients might not play a significant role in the whole group of 32 patients. Moreover, by investigating only the common DICCCOLs in our functional connectivity analysis, we attempted to minimize the effect of structural damage.
In contrast with group analysis, an individual-level analysis is also required to properly identify specific brain connectivity alterations, especially personalized alterations. Unlike group-level analysis, which is related to the common alterations of brain functions among a group of individuals, individual-level analysis is personalized and related to the origin and location of the injury of each individual. Due to the heterogeneous characteristic of each individual patient and the close relationship between the alterations of structural and functional connectivity, combining the structural and functional connectivity analyses together could allow us to characterize the interplay between structural and functional connectivities after brain injury. A combined structural and functional analysis could have the potential to ameliorate the clinical management of mTBI patients in the future, which should be investigated in future studies with a large number of subjects.
Another potential confounding factor is the race difference between patients and controls in our data sample. To our best knowledge, there is no rsfMRI study specifically investigating or ever reporting the effect of race on functional connectivity. Instead, different studies across the world reported similar patterns of resting state brain networks, particularly in the DMN, though their recruited local populations are different in race (Biswal et al., 2010). Considering our limited number of patients, we did not include race as a control factor in our analysis to avoid overfitting. In future studies, race could also be included as a control factor in a large number of data sample.
The current work also has several other limitations to be addressed in future work. One is the relatively small number of subjects (40 patients and 50 controls) in comparison with a large number of networks. Moreover, the patients' emotional alterations at the acute stage, such as pain, stress, anxiety or sadness, could significantly affect their functional connectivity results. Inclusion of orthopedic controls in future studies could help to control for this potential problem. Another limitation of the current work is that it does not include a longitudinal analysis of patients' recovery. It is our future plan to analyze the connectome-scale network changes longitudinally to investigate patients' recovery and brain plasticity patterns. Finally, given the magnitude of large-scale functional network changes, correlation of imaging findings with patients' functional assessment and interpretation of the imaging findings with clinical data is still a challenge to be addressed. In our ongoing longitudinal analysis in collaboration with psychologists and statisticians, we are in the process of evaluating the relationship of mTBI patients' neuropsychological testing outcomes with DICCCOL parameters. The results will be reported in the near future.
5. Conclusions
In summary, our work demonstrated that mTBI renders changes in brain network connectivity on a large scale despite normal clinical imaging findings in most cases. These changes include both structural network disruption, which involves major white matter tracts, and connectivity changes in functional networks at different domains. Further work is needed to determine their clinical correlates and functional recovery over time.
Acknowledgements
The authors would like to thank the following co-workers for their contribution: Mr. Zahid Latif, Mr. Yang Xuan and Mr. Yashwanth Katkuri from Wayne State University MR Research Facility (MRRF) for their help of data acquisition; Ms. Anamika Chaudhary and Ms. Judith Farah from MRRF for their coordination; Detroit Receiving Hospital Emergency Medicine research team for subject screening and recruitment; and other colleagues in the Cortical Architecture Imaging and Discovery Laboratory in the University of Georgia for their assistance in image analysis. This work was supported by a Seed Grant from the International Society for Magnetic Resonance in Medicine (PI: Zhifeng Kou) and a research grant from the US Department of Defense (award number W81XWH-11-1-0493). Research reported in this publication was also partially supported by the National Institute of Neurological Disorders And Stroke of the National Institutes of Health under Award Number R21NS090153 and the Eunice Kennedy Shriver National Institute of Child Health and Human Development under Award Number F30HD084144. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nicl.2016.06.012.
Contributor Information
Tianming Liu, Email: tliu@cs.uga.edu.
Zhifeng Kou, Email: zhifeng_kou@wayne.edu.
Appendix A. Supplementary data
Supplementary tables.
References
- American Congress of Rehabilitation Medicine Definition of mild traumatic brain injury. J. Head Trauma Rehabil. 1993;8:86–88. [Google Scholar]
- Baek S.O., Kim O.L., Kim S.H., Kim M.S., Son S.M., Cho Y.W., Byun W.M., Jang S.H. Relation between cingulum injury and cognition in chronic patients with traumatic brain injury; diffusion tensor tractography study. NeuroRehabilitation. 2013;33:465–471. doi: 10.3233/NRE-130979. [DOI] [PubMed] [Google Scholar]
- Bazarian J.J., McClung J., Shah M.N., Cheng Y.T., Flesher W., Kraus J. Mild traumatic brain injury in the United States, 1998–2000. Brain Inj. 2005;19:85–91. doi: 10.1080/02699050410001720158. [DOI] [PubMed] [Google Scholar]
- Bazarian J.J., Zhong J., Blyth B., Zhu T., Kavcic V., Peterson D. Diffusion tensor imaging detects clinically important axonal damage after mild traumatic brain injury: a pilot study. J. Neurotrauma. 2007;24:1447–1459. doi: 10.1089/neu.2007.0241. [DOI] [PubMed] [Google Scholar]
- Belanger H.G., Vanderploeg R.D., Curtiss G., Warden D.L. Recent neuroimaging techniques in mild traumatic brain injury. J. Neuropsychiatr. Clin. Neurosci. 2007;19:5–20. doi: 10.1176/jnp.2007.19.1.5. [DOI] [PubMed] [Google Scholar]
- Biswal B.B., Mennes M., Zuo X.N., Gohel S., Kelly C., Smith S.M., Beckmann C.F., Adelstein J.S., Buckner R.L., Colcombe S., Dogonowski A.M., Ernst M., Fair D., Hampson M., Hoptman M.J., Hyde J.S., Kiviniemi V.J., Kotter R., Li S.J., Lin C.P., Lowe M.J., Mackay C., Madden D.J., Madsen K.H., Margulies D.S., Mayberg H.S., McMahon K., Monk C.S., Mostofsky S.H., Nagel B.J., Pekar J.J., Peltier S.J., Petersen S.E., Riedl V., Rombouts S.A., Rypma B., Schlaggar B.L., Schmidt S., Seidler R.D., Siegle G.J., Sorg C., Teng G.J., Veijola J., Villringer A., Walter M., Wang L., Weng X.C., Whitfield-Gabrieli S., Williamson P., Windischberger C., Zang Y.F., Zhang H.Y., Castellanos F.X., Milham M.P. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumbergs P.C. Pathology. In: Reilly P., Bullock R., editors. Head Injury. Chapman & Hill; London: 1997. pp. 39–70. [Google Scholar]
- Blumbergs P.C., Scott G., Manavis J., Wainwright H., Simpson D.A., McLean A.J. Staining of amyloid precursor protein to study axonal damage in mild head injury. Lancet. 1994;344:1055–1056. doi: 10.1016/s0140-6736(94)91712-4. [DOI] [PubMed] [Google Scholar]
- Blumbergs P.C., Scott G., Manavis J., Wainwright H., Simpson D.A., McLean A.J. Topography of axonal injury as defined by amyloid precursor protein and the sector scoring method in mild and severe closed head injury. J. Neurotrauma. 1995;12:565–572. doi: 10.1089/neu.1995.12.565. [DOI] [PubMed] [Google Scholar]
- Brett M., Johnsrude I.S., Owen A.M. The problem of functional localization in the human brain. Nat. Rev. Neurosci. 2002;3:243–249. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- Calhoun V.D., Maciejewski P.K., Pearlson G.D., Kiehl K.A. Temporal lobe and “default” hemodynamic brain modes discriminate between schizophrenia and bipolar disorder. Hum. Brain Mapp. 2008;29:1265–1275. doi: 10.1002/hbm.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . National Center for Injury Prevention and Control; Atlanta (GA): 2003. Report to Congress on Mild Traumatic Brain Injury in the United States: Steps to Prevent a Serious Public Health Problem. Centers for Disease Control and Prevention. [Google Scholar]
- Chen H., Iraji A., Jiang X., Lv J., Kou Z., Liu T. Longitudinal analysis of brain recovery after mild traumatic brain injury based on groupwise consistent brain network clusters. In: Navab N., Hornegger J., Wells W.M., Frangi A.F., editors. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2015. Springer International Publishing; 2015. pp. 194–201. [Google Scholar]
- Chen C.J., Wu C.H., Liao Y.P., Hsu H.L., Tseng Y.C., Liu H.L., Chiu W.T. Working memory in patients with mild traumatic brain injury: functional MR imaging analysis. Radiology. 2012;264:844–851. doi: 10.1148/radiol.12112154. [DOI] [PubMed] [Google Scholar]
- Damoiseaux J.S., Rombouts S.A., Barkhof F., Scheltens P., Stam C.J., Smith S.M., Beckmann C.F. Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. U S A. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eierud C., Craddock R.C., Fletcher S., Aulakh M., King-Casas B., Kuehl D., LaConte S.M. Neuroimaging after mild traumatic brain injury: review and meta-analysis. Neuroimage Clin. 2014;4:283–294. doi: 10.1016/j.nicl.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilvesmaki T., Luoto T.M., Hakulinen U., Brander A., Ryymin P., Eskola H., Iverson G.L., Ohman J. Acute mild traumatic brain injury is not associated with white matter change on diffusion tensor imaging. Brain. 2014;137:1876–1882. doi: 10.1093/brain/awu095. [DOI] [PubMed] [Google Scholar]
- Iraji A., Benson R.R., Welch R.D., O'Neil B.J., Woodard J.L., Ayaz S.I., Kulek A., Mika V., Medado P., Soltanian-Zadeh H., Liu T., Haacke E.M., Kou Z. Resting state functional connectivity in mild traumatic brain injury at the acute stage: independent component and seed-based analyses. J. Neurotrauma. 2015;32:1031–1045. doi: 10.1089/neu.2014.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraji A., Calhoun V.D., Wiseman N.M., Davoodi-Bojd E., Avanaki M.R., Haacke E.M., Kou Z. The connectivity domain: Analyzing resting state fMRI data using feature-based data-driven and model-based methods. Neuroimage. 2016;134:494–507. doi: 10.1016/j.neuroimage.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraji A., Chen H., Wiseman N., Welch R.D., O'Neil B.J., Haacke E.M., Liu T., Kou Z. Compensation through functional hyperconnectivity: a longitudinal connectome assessment of mild traumatic brain injury. Neural. Plast. 2016;2016:4072402. doi: 10.1155/2016/4072402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia A., Wang B., Aylward S.R., Prastawa M.W., Pace D.F., Gerig G., Hovda D.A., Kikinis R., Vespa P.M., Van Horn J.D. Neuroimaging of structural pathology and connectomics in traumatic brain injury: toward personalized outcome prediction. Neuroimage Clin. 2012;1:1–17. doi: 10.1016/j.nicl.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri M.J., Pearlson G.D., Stevens M., Calhoun V.D. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. NeuroImage. 2008;39:1666–1681. doi: 10.1016/j.neuroimage.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E., Woolrich M.W., Smith S.M. Fsl. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Johnson B., Zhang K., Gay M., Horovitz S., Hallett M., Sebastianelli W., Slobounov S. Alteration of brain default network in subacute phase of injury in concussed individuals: resting-state fMRI study. NeuroImage. 2012;59:511–518. doi: 10.1016/j.neuroimage.2011.07.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay T. Neuropsychological treatment of mild traumatic brain injury. J. Head Trauma Rehabil. 1993;8:74–85. [Google Scholar]
- Kou Z., Iraji A. Imaging brain plasticity after trauma. Neural. Regen. Res. 2014;9:693–700. doi: 10.4103/1673-5374.131568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Z., VandeVord P.J. Traumatic white matter injury and glial activation: from basic science to clinics. Glia. 2014;62:1831–1855. doi: 10.1002/glia.22690. [DOI] [PubMed] [Google Scholar]
- Kou Z., Benson R.R., Haacke E.M. Magnetic resonance imaging biomarkers of mild traumatic brain injury. In: Dambinova S., editor. Biomarkers for Traumatic Brain Injury. Royal Society of Chemistry; 2012. [Google Scholar]
- Kou Z., Wu Z., Tong K.A., Holshouser B., Benson R.R., Hu J., Haacke E.M. The role of advanced MR imaging findings as biomarkers of traumatic brain injury. J. Head Trauma Rehabil. 2010;25:267–282. doi: 10.1097/HTR.0b013e3181e54793. [DOI] [PubMed] [Google Scholar]
- Laird A.R., Eickhoff S.B., Kurth F., Fox P.M., Uecker A.M., Turner J.A., Robinson J.L., Lancaster J.L., Fox P.T. ALE meta-analysis workflows via the brainmap database: progress towards a probabilistic functional brain atlas. Front. Neuroinform. 2009;3:23. doi: 10.3389/neuro.11.023.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Guo L., Faraco C., Zhu D., Deng F., Zhang T., Jiang X., Zhang D., Chen H., Hu X. Individualized ROI optimization via maximization of group-wise consistency of structural and functional profiles. Adv. Neural Inf. Proces. Syst. 2010:1369–1377. [Google Scholar]
- Li K., Guo L., Li G., Nie J., Faraco C., Zhao Q., Miller L.S., Liu T. Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on. IEEE. 2010. Cortical surface based identification of brain networks using high spatial resolution resting state fMRI data; pp. 656–659. [Google Scholar]
- Li K., Guo L., Zhu D., Hu X., Han J., Liu T. Individual functional ROI optimization via maximization of group-wise consistency of structural and functional profiles. Neuroinformatics. 2012;10:225–242. doi: 10.1007/s12021-012-9142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Zhu D., Guo L., Li Z., Lynch M.E., Coles C., Hu X., Liu T. Connectomics signatures of prenatal cocaine exposure affected adolescent brains. Hum. Brain Mapp. 2013;34:2494–2510. doi: 10.1002/hbm.22082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. A few thoughts on brain ROIs. Brain Imaging Behav. 2011;5:189–202. doi: 10.1007/s11682-011-9123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Li H., Wong K., Tarokh A., Guo L., Wong S.T. Brain tissue segmentation based on DTI data. NeuroImage. 2007;38:114–123. doi: 10.1016/j.neuroimage.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Nie J., Tarokh A., Guo L., Wong S.T. Reconstruction of central cortical surface from brain MRI images: method and application. NeuroImage. 2008;40:991–1002. doi: 10.1016/j.neuroimage.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A.R., Ling J.M., Allen E.A., Klimaj S.D., Yeo R.A., Hanlon F.M. Static and dynamic intrinsic connectivity following mild traumatic brain injury. J. Neurotrauma. 2015;32:1046–1055. doi: 10.1089/neu.2014.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A.R., Mannell M.V., Ling J., Gasparovic C., Yeo R.A. Functional connectivity in mild traumatic brain injury. Hum. Brain Mapp. 2011;32:1825–1835. doi: 10.1002/hbm.21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister T.W., Saykin A.J., Flashman L.A., Sparling M.B., Johnson S.C., Guerin S.J., Mamourian A.C., Weaver J.B., Yanofsky N. Brain activation during working memory 1 month after mild traumatic brain injury: a functional MRI study. Neurology. 1999;53:1300–1308. doi: 10.1212/wnl.53.6.1300. [DOI] [PubMed] [Google Scholar]
- Messé A., Caplain S., Paradot G., Garrigue D., Mineo J.F., Soto Ares G., Ducreux D., Vignaud F., Rozec G., Desal H. Diffusion tensor imaging and white matter lesions at the subacute stage in mild traumatic brain injury with persistent neurobehavioral impairment. Hum. Brain Mapp. 2011;32:999–1011. doi: 10.1002/hbm.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messe A., Caplain S., Pelegrini-Issac M., Blancho S., Levy R., Aghakhani N., Montreuil M., Benali H., Lehericy S. Specific and evolving resting-state network alterations in post-concussion syndrome following mild traumatic brain injury. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A.T., Masterton R., Pigdon L., Connelly A., Liegeois F.J. Functional magnetic resonance imaging of chronic dysarthric speech after childhood brain injury: reliance on a left-hemisphere compensatory network. Brain. 2013;136:646–657. doi: 10.1093/brain/aws355. [DOI] [PubMed] [Google Scholar]
- National Academy of Neuropsychology . National Academy of Neuropsychology; Denver, Co: 2002. Mild Traumatic Brain Injury—An Online Course. [Google Scholar]
- National Institutes of Health NIH consensus development panel on rehabilitation of persons with traumatic brain injury. J. Am. Med. Assoc. 1999;282:974–983. [PubMed] [Google Scholar]
- Niogi S.N., Mukherjee P. Diffusion tensor imaging of mild traumatic brain injury. J. Head Trauma Rehabil. 2010;25:241–255. doi: 10.1097/HTR.0b013e3181e52c2a. [DOI] [PubMed] [Google Scholar]
- Niogi S.N., Mukherjee P., Ghajar J., Johnson C.E., Kolster R., Lee H., Suh M., Zimmerman R.D., Manley G.T., McCandliss B.D. Structural dissociation of attentional control and memory in adults with and without mild traumatic brain injury. Brain. 2008;131:3209–3221. doi: 10.1093/brain/awn247. [DOI] [PubMed] [Google Scholar]
- Niogi S.N., Mukherjee P., Ghajar J., Johnson C., Kolster R.A., Sarkar R., Lee H., Meeker M., Zimmerman R.D., Manley G.T., McCandliss B.D. Extent of microstructural white matter injury in postconcussive syndrome correlates with impaired cognitive reaction time: a 3T diffusion tensor imaging study of mild traumatic brain injury. AJNR Am. J. Neuroradiol. 2008;29:967–973. doi: 10.3174/ajnr.A0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S., Tank D.W., Menon R., Ellermann J.M., Kim S.G., Merkle H., Ugurbil K. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc. Natl. Acad. Sci. U. S. A. 1992;89:5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passingham R.E., Stephan K.E., Kotter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3:606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Pelleg D., Moore W. X-means: extending K-means with efficient estimation of the number of clusters. ICML. 2000:727–734. [Google Scholar]
- Penny D.W., Friston J.K., Ashburner T.J., Kiebel J.S., Nichols E.T. Academic press; 2011. Statistical Parametric Mapping: The Analysis of Functional Brain Images: The Analysis of Functional Brain Images. [Google Scholar]
- Ruff R. Two decades of advances in understanding of mild traumatic brain injury. J. Head Trauma Rehabil. 2005;20:5–18. doi: 10.1097/00001199-200501000-00003. [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Jenkins P.O. Concussion is confusing us all. Pract. Neurol. 2015;15:172–186. doi: 10.1136/practneurol-2015-001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimony J.S., Zhang D., Johnston J.M., Fox M.D., Roy A., Leuthardt E.C. Resting-state spontaneous fluctuations in brain activity: a new paradigm for presurgical planning using fMRI. Acad. Radiol. 2009;16:578–583. doi: 10.1016/j.acra.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg C., Riedl V., Muhlau M., Calhoun V.D., Eichele T., Laer L., Drzezga A., Forstl H., Kurz A., Zimmer C., Wohlschlager A.M. Selective changes of resting-state networks in individuals at risk for Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M.C., Lovejoy D., Kim J., Oakes H., Kureshi I., Witt S.T. Multiple resting state network functional connectivity abnormalities in mild traumatic brain injury. Brain Imaging Behav. 2012;6:293–318. doi: 10.1007/s11682-012-9157-4. [DOI] [PubMed] [Google Scholar]
- Tang L., Ge Y., Sodickson D.K., Miles L., Zhou Y., Reaume J., Grossman R.I. Thalamic resting-state functional networks: disruption in patients with mild traumatic brain injury. Radiology. 2011;260:831–840. doi: 10.1148/radiol.11110014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivarus M.E., Starling S.J., Newport E.L., Langfitt J.T. Homotopic language reorganization in the right hemisphere after early left hemisphere injury. Brain Lang. 2012;123:1–10. doi: 10.1016/j.bandl.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treble A., Hasan K.M., Iftikhar A., Stuebing K.K., Kramer L.A., Cox C.S., Jr., Swank P.R., Ewing-Cobbs L. Working memory and corpus callosum microstructural integrity after pediatric traumatic brain injury: a diffusion tensor tractography study. J. Neurotrauma. 2013;30:1609–1619. doi: 10.1089/neu.2013.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde E.A., McCauley S.R., Hunter J.V., Bigler E.D., Chu Z., Wang Z.J., Hanten G.R., Troyanskaya M., Yallampalli R., Li X., Chia J., Levin H.S. Diffusion tensor imaging of acute mild traumatic brain injury in adolescents. Neurology. 2008;70:948–955. doi: 10.1212/01.wnl.0000305961.68029.54. [DOI] [PubMed] [Google Scholar]
- Wu T.C., Wilde E.A., Bigler E.D., Yallampalli R., McCauley S.R., Troyanskaya M., Chu Z., Li X., Hanten G., Hunter J.V., Levin H.S. Evaluating the relationship between memory functioning and cingulum bundles in acute mild traumatic brain injury using diffusion tensor imaging. J. Neurotrauma. 2010;27:303–307. doi: 10.1089/neu.2009.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Jiang X., Zhu D., Chen H., Li K., Lv P., Yu X., Li X., Zhang S., Zhang T., Hu X., Han J., Guo L., Liu T. Meta-analysis of functional roles of DICCCOLs. Neuroinformatics. 2013;11:47–63. doi: 10.1007/s12021-012-9165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Guo L., Li K., Jing C., Yin Y., Zhu D., Cui G., Li L., Liu T. Predicting functional cortical ROIs via DTI-derived fiber shape models. Cereb. Cortex. 2012;22:854–864. doi: 10.1093/cercor/bhr152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Milham M.P., Lui Y.W., Miles L., Reaume J., Sodickson D.K., Grossman R.I., Ge Y. Default-mode network disruption in mild traumatic brain injury. Radiology. 2012;265:882–892. doi: 10.1148/radiol.12120748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Li K., Faraco C.C., Deng F., Zhang D., Guo L., Miller L.S., Liu T. Optimization of functional brain ROIs via maximization of consistency of structural connectivity profiles. NeuroImage. 2012;59:1382–1393. doi: 10.1016/j.neuroimage.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Li K., Guo L., Jiang X., Zhang T., Zhang D., Chen H., Deng F., Faraco C., Jin C., Wee C.Y., Yuan Y., Lv P., Yin Y., Hu X., Duan L., Hu X., Han J., Wang L., Shen D., Miller L.S., Li L., Liu T. DICCCOL: dense individualized and common connectivity-based cortical landmarks. Cereb. Cortex. 2013;23:786–800. doi: 10.1093/cercor/bhs072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Li K., Terry D.P., Puente A.N., Wang L., Shen D., Miller L.S., Liu T. Connectome-scale assessments of structural and functional connectivity in MCI. Hum. Brain Mapp. 2014;35:2911–2923. doi: 10.1002/hbm.22373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Zhang D., Faraco C., Li K., Deng F., Chen H., Jiang X., Guo L., Miller L.S., Liu T. Information Processing in Medical Imaging. Springer; 2011. Discovering dense and consistent landmarks in the brain; pp. 97–110. [DOI] [PubMed] [Google Scholar]
- Zoroya G. 2007. Scientists: Brain Injuries From War Worse Than Thought. (USA Today) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.