Abstract
Aquifoliaceae is the largest family in the campanulid order Aquifoliales. It consists of a single genus, Ilex, the hollies, which is the largest woody dioecious genus in the angiosperms. Most species are in East Asia or South America. The taxonomy and evolutionary history remain unclear due to the lack of a robust species-level phylogeny. We produced the first complete chloroplast genomes in this family, including seven Ilex species, by Illumina sequencing of long-range PCR products and subsequent reference-guided de novo assembly. These genomes have a typical bicyclic structure with a conserved genome arrangement and moderate divergence. The total length is 157,741 bp and there is one large single-copy region (LSC) with 87,109 bp, one small single-copy with 18,436 bp, and a pair of inverted repeat regions (IR) with 52,196 bp. A total of 144 genes were identified, including 96 protein-coding genes, 40 tRNA and 8 rRNA. Thirty-four repetitive sequences were identified in Ilex pubescens, with lengths >14 bp and identity >90%, and 11 divergence hotspot regions that could be targeted for phylogenetic markers. This study will contribute to improved resolution of deep branches of the Ilex phylogeny and facilitate identification of Ilex species.
Ilex, in the monogeneric family Aquifoliaceae, is the largest woody dioecious genus in the angiosperms with approximately 600 species1. Flowers and fruits are fairly uniform but Ilex species differ greatly in leaf characters, including size, texture, and margins. The genus is widespread in mesic habitats but the global diversity centers are in East Asia and South America, with a single species in tropical Africa, two in northern Australia, four in Europe1, and 17 in North America (http://plants.usda.gov/java/nameSearch). The four other families in the Campanulidae order Aquifoliales, Cardiopteridaceae, Stemonuraceae, Helwingiaceae and Phyllonomaceae, are all small2,3. Ilex species are economically important sources of teas and medicines. Ilex paraguariensis, yerba mate, is planted on 326,000 ha in Argentina, Brazil and Paraguay, with a total annual production of more than a million tonnes4. In China, several species of Ilex are used to produce a popular medicinal tea, kuding cha5,6. Ilex species are also widely grown as ornamental plants because of their persistent red fruits and often distinctive leaves.
Ilex has a good fossil record dating to the Eocene and Ilex-like pollen has been reported from the Cretaceous, although molecular evidence suggests that the extant crown clade diverged only in the middle Miocene, 13 million years ago1,7. Phylogenetic relationships within Ilex are still unclear. Cuénoud8 used the chloroplast markers atpB-rbcL spacer and rbcL to construct a phylogeny of 116 species, while Manen9 combined plastid (atpB-rbcL spacer, rbcL and trnL-trnF) and nuclear (ribosomal internal transcribed spacer and the 5S RNA spacer) markers for 105 species. Manen1 later increased the phylogenetic resolution by using nuclear markers ITS and ncpGS for 108 species, including species from America, Europe, Africa, and islands in the Atlantic and Pacific. However, this study included only 33 species of the 204 known Chinese species, of which 149 are endemic2. These studies show a striking incongruence between plastid and nuclear phylogenies1. Similar incongruence has been reported in recent studies of the grass tribe Arundinarieae10, and the genera Osmorhiza11, Hedyosmum12, and Medicago13, and has been attributed to incomplete lineage sorting, plastid capture, or hybridization.
Chloroplasts contain a circular genome ranging from 21 kb in Sciaphila densiflora14 to 217 kb in Pelargonium × hortorum15, with two copies of a large inverted repeat (IR), one large single-copy region (LSC), and one small single-copy region (SSC)16,17. Despite the previous problems with using chloroplast sequences to construct a species-level phylogeny for Ilex, chloroplast sequences have advantages for species identification as a result of their small size, uniparental inheritance, haploid nature, and highly conserved genomic structure16. Species identification is a particular problem in Ilex, where there are many similar species. Moreover, since numerous copies are present in each cell, useable fragments of the chloroplast genome are more likely to persist in dried herbarium specimens18,19, which is an important consideration for Ilex in China where many species are only known from the type collection. In addition, chloroplast genomes have been used to improve the resolution of the backbone of phylogenies built with nuclear markers10.
Here, we present the complete chloroplast genomes of seven Ilex species through Illumina sequencing and reference-guided assembly of de novo contigs. We then test the feasibility of phylogeny reconstruction using chloroplast genomes in Ilex. Topologies of the phylogenies constructed from different molecular datasets are compared, including the whole cp genome, the coding regions, and LSC, SSC, IR, and introns and spacers. A new chloroplast genome for Helwingia himalaica (Yao et al., submitted), from the most closely related family, Helwingiaceae, is used as the outgroup.
Results
Output of genome sequencing and assembly
For the seven Ilex species, 239,377 to 748,662 paired-end reads (90bp in average reads length) were produced by Illumina sequencing. 233,558 to 692,191 reads were mapped to the reference genome Camellia yunnanensis (GenBank accession number KF156838), after screening these paired-end reads by aligning them to the reference genome, on average reaching over 100 × coverage of the cp genome. After de novo and reference-guided assembly, complete cp genomes of seven Ilex species were obtained. The four junction regions in each genome were validated using PCR-based sequencing (see Supplementary Table S1).
Genome features and sequence divergence
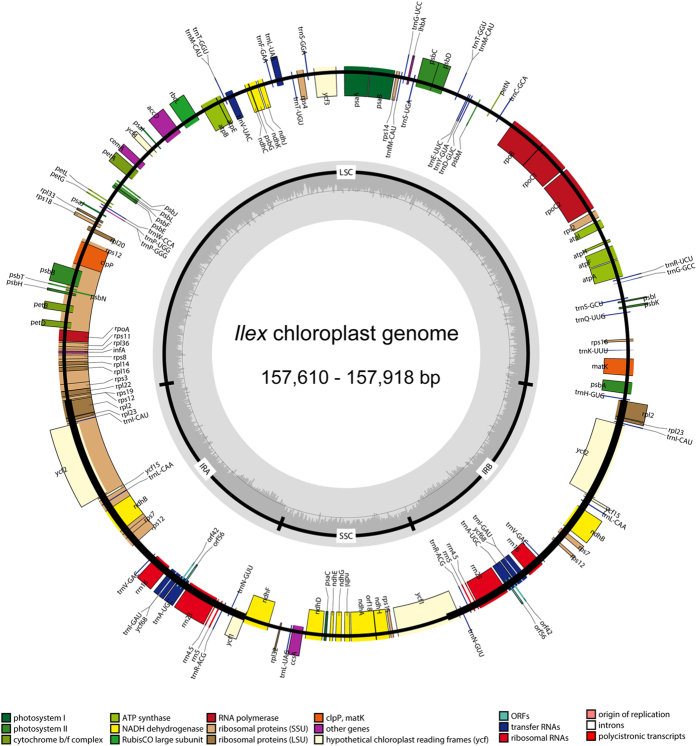
Chloroplast genomes of the seven species were assembled into single circular, double-stranded DNA sequences, presenting a typical quadripartite structure including one large single-copy region (LSC with 86,948–87,266 bp), one small single-copy region (SSC with 18,427–18,513 bp), and a pair of inverted repeat regions (IR with 52,122–52,235 bp) (Table 1). The full length ranged from 157,610 in I. latifolia to 157,918 bp in I. wilsonii (Table 1). In I. pubescens, which was investigated in detail as an example, the chloroplast encoded a set of 144 genes of which 100 are unique genes and 16 are duplicated in the IRs regions (Fig. 1). The same gene order and clusters were found in all seven species. The 100 unique genes were composed of 74 protein-coding genes and 26 tRNA genes (Table 2). All of the eight rRNA genes were duplicated in the IR regions (Table 1). Nine distinct genes (atpF, rpoC1, trnL-UAA, trnV-UAC, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA) contain one intron and three genes (ycf3, clpP and rps12) have two introns. Gene ycf1 in the junction region between SSC and IRb was the only pseudogene found because of the incomplete duplication of the normal copy in the junction region (Fig. 1).
Table 1. Comparison of plastid genomic characteristics in seven Ilex species and Helwingia himalaica.
| Ilex latifolia | Ilex szechwanensis | Ilex pubescens | Ilex polyneura | Ilexnew sp. | Ilex delavayi | Ilex wilsonii | Helwingia himalaica | |
|---|---|---|---|---|---|---|---|---|
| Total paired-end reads | 1,057,844 | 1,292,586 | 1,135,472 | 467,116 | 1,338,864 | 1,201,978 | 1,459,906 | 914,296 |
| Aligned paired-end reads | 1,045,069 | 1,257,399 | 1,131,722 | 465,040 | 1,327,085 | 1,198,537 | 1,448,390 | 907,908 |
| Mean coverage | 143.8 | 523.2 | 504 | 523.2 | 1006.0 | 562.3 | 342.6 | 188.2 |
| Number of contigs | 230 | 84 | 68 | 47 | 44 | 76 | 106 | 142 |
| Mean length (bp) | 1,649 | 2,419 | 2,834 | 3,505 | 3,654 | 2,564 | 1,982 | 1,820 |
| N50 (bp) | 2,806 | 8,762 | 9,089 | 9,503 | 9,506 | 8,343 | 8,346 | 4,534 |
| Sum contigs length (bp) | 379,388 | 203,271 | 192,740 | 164,767 | 160,803 | 194,865 | 210,138 | 258,537 |
| Size (bp) | 157,610 | 157,900 | 157,741 | 157,621 | 157,611 | 157,671 | 157,918 | 158,362 |
| LSC length (bp) | 86,952 | 87,204 | 87,109 | 87,064 | 86,948 | 87,000 | 87,266 | 87,810 |
| SSC length (bp) | 18,429 | 18,513 | 18,436 | 18,435 | 18,434 | 18,436 | 18,432 | 18,560 |
| IRs length (bp) | 52,229 | 52,183 | 52,196 | 52,122 | 52,229 | 52,235 | 52,220 | 51,992 |
| Protein Genes [unique] | 96[74] | 96[74] | 96[74] | 96[74] | 96[74] | 96[74] | 96[74] | 94[76] |
| tRNA [unique] | 40[26] | 40[26] | 40[26] | 40[26] | 40[26] | 40[26] | 40[26] | 40[26] |
| rRNA [unique] | 8[0] | 8[0] | 8[0] | 8[0] | 8[0] | 8[0] | 8[0] | 8[0] |
| GC content (%) | 37.60 | 37.70 | 37.60 | 37.60 | 37.60 | 37.60 | 37.60 | 37.70 |
Figure 1. Gene map of the Ilex chloroplast genome.
Genes shown outside the outer circle are transcribed clockwise and those inside are transcribed counterclockwise. Gray arrows indicate the direction of sequence coding. Genes belonging to different functional groups are color-coded. Dashed area in the inner circle indicates the GC content of the chloroplast genome.
Table 2. List of genes in the chloroplast genome of Ilex.
| Category | Groups of gene | Name of genes |
|---|---|---|
| Protein synthesis and DNA-replication | Transfer RNAs | trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, trnH-GUG, trnK-UUU, trnL-UAA, trnM-CAU, trnQ-UUG, trnP-GGG, trnP-UGG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-UAC, trnW-CCA, trnY-GUA, trnA-UGC(×2), trnI-CAU(×2), trnI-GAU(×2), trnL-CAA(×2), trnL-UAG, trnN-GUU(×2), trnR-ACG(×2), trnV-GAC(×2) |
| Ribosomal RNAs | rrn16(×2), rrn23(×2), rrn4.5(×2), rrn5(×2) | |
| Ribosomal protein small subunit | rps16, rps2, rps14, rps4, rps18, rps12(×2), rps11, rps8, rps3, rps19, rps7(×2), rps15 | |
| Ribosomal protein large subunit | rpl33, rpl20, rpl36, rpl14, rpl16, rpl22, rpl2(×2), rpl23(×2), rpl32 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Photosynthesis | photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbG, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, lhbA | |
| Cythochrome b/f complex | petA, petB, petD, petG, petL petN | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| NADH-dehydrogenase | ndhA, ndhB(×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Large subunit Rubisco | rbcL | |
| Miscellaneous group | Translation initiation factor | infA |
| Acetyl-CoA carboxylase | accD | |
| Cytochrome c biogenesis | ccsA | |
| Maturase | matK | |
| ATP-dependent protease | clpP | |
| Inner membrane protein | cemA | |
| Pseudogene unknown function | Conserved hypothetical chloroplast ORF | ycf3, ycf4, ycf2(×2), ycf15(×2), ycf68(×2), orf42(×2), orf56(×2), ycf1(×2), orf188 |
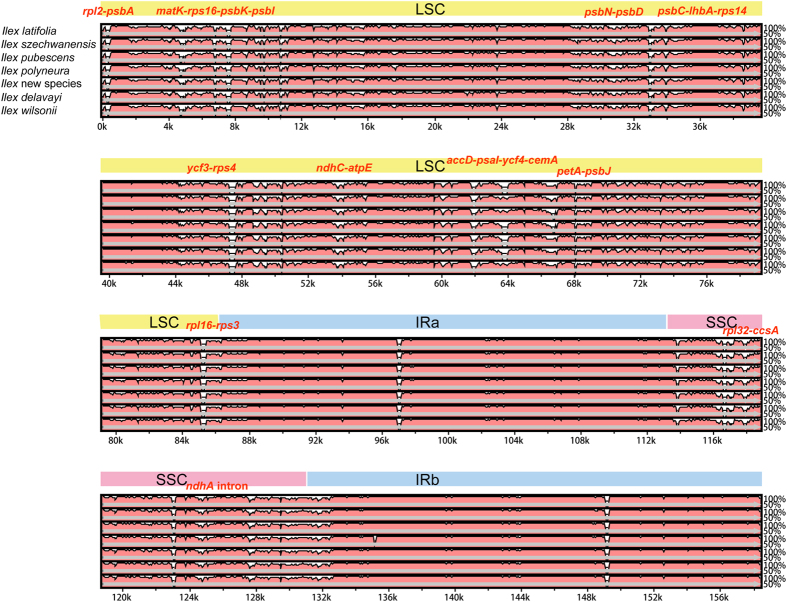
AT content is rich (62.3–62.4%) and sequence identity among the seven species was 97.9%. The whole aligned sequences disclosed moderate divergence with 11 regions containing sequence similarities below 50%, especially in intergenic regions. Eleven divergent hotspot regions were identified (Fig. 2). The p-distances between Ilex and Helwingia, and among Ilex species, were 0.03988 and 0.00288, respectively, both indicating moderate genetic divergences. The p-distance between the two most closely related species, I. latifolia and I. delavayi in subgenus Ilex section Ilex, was 0.00185.
Figure 2. Visualization of the alignment of the seven Ilex chloroplast genomes.
VISTA-based identity plots showing sequence identity with the Helwingia himalaica chloroplast genome as a reference. LSC indicates long single copy region; SSC indicates short single copy region; IRa and IRb indicate two inverted regions. Locations of divergent hotspot regions are labeled above alignment.
Indels and repeated sequences
A total of 113 indels were detected in the Ilex species, 88 in spacers, 13 in introns of genes, and 12 in genes, with 89 in LSC, 08 in SSC, and 08 in IRb (see Supplementary Table S1). In I. pubescens we identified 34 dispersed repeats >14 bp with sequence identify >90%, ranging from 15 bp to 29 bp (Table 3). Most repeats were 16 bp (32.4%) or 17 bp (23.5%). A total of 23 repeats were located in intergenic regions, while 06 were in protein-coding genes and 05 in tRNA genes. Five repeats were identified in ycf2 which was the most in any gene.
Table 3. Repetitive sequences of Ilex pubescens calculated in REPuter.
| Repeat length (bp) | Repeat bases | Repeat location | Copy of repeat location |
|---|---|---|---|
| 15 | TCTTTCTTTTTTTTT | trnH-GUG psbA spacer | clpP |
| 16 | TTTTTTTTTTTTTTTT | trnH-GUG psbA spacer | trnM-CAU atpE spacer |
| 16 | TTGAAAAAAAAAAAAA | atpA-atpF spacer | trnS-UGA lhbA spacer |
| 16 | TGAAAAAAAAAAAAAA | atpA-atpF spacer | rps18-rpl20 spacer |
| 16 | ATTTCTTTTTTAGTTT | atpH-atpI spacer | ycf1 |
| 16 | TTTTTTGAAAAAAAAA | rps2-rpoC2 spacer | ndhF-rpl32 spacer |
| 16 | GAAAAAAAAAAAAAGA | lhbA trnG-UCC spacer | rps18-rpl20 spacer |
| 16 | AAAAAAAAAAAAAGAA | lhbA trnG-UCC spacer | rpl14-rpl16 spacer |
| 16 | ATTATTAATTTGTATG | trnF-GAA ndhJ spacer | ycf1 |
| 16 | TAGTCACTTCTTTTTT | psaJ-rpl33 spacer | ycf2 |
| 16 | CTTTCTTTTTTTTTTC | clpP | infA-rps8 spacer |
| 16 | ATTTTTATTTTGTTTT | rpl16-rps3 spacer | rps19-rpl2 spacer |
| 17 | TTTTTTTTTTTTTTATT | trnH-GUG psbA spacer | psbE-petL spacer |
| 17 | CTTTTTTGAAAAAAAAA | atpA-atpF spacer | rps2-rpoC2 |
| 17 | TTTTTTGAAAAAAAAAA | atpA-atpF spacer | ndhF-rpl32 |
| 17 | TAGTAAAAATAAAAAGA | trnM-CAU psbD spacer | accD-psaI spacer |
| 17 | AAGACGAAAAAAAAAAA | trnT-UGU trnL-UAA spacer | petA-psbJ spacer |
| 17 | CTATATATTTTTCCAGT | cemA-petA spacer | petD |
| 17 | GCTTTTGTTTATAAAAA | rpl16-rps3 spacer | rpl16-rps3 spacer |
| 17 | GATATTGATGCTAGTGA | ycf2 | ycf2 |
| 18 | TCCACTCAGCCATCTCTC | trnS-GCU | trnS-UGA |
| 18 | CGAAAATTCTTTTTTCTC | trnE-UUC trnM-CAU spacer | rpl32 trnL-UAG spacer |
| 18 | ATTGTATCCATTGAGCAA | psaB | psaA |
| 18 | ATGCAATAGCTAAATGAT | psaB | psaA |
| 18 | CTTTTCTGAGTGAACTAG | accD | accD |
| 18 | AGAACTACGAGATCACCC | trnI-GAU | trnA-UGC |
| 19 | TGCGGGTTCGATTCCCGCT | trnG-GCC | trnG-UCC |
| 21 | ATGCTGCTGCAGAATAAACCA | trnH-GUG psbA spacer | rpl22 |
| 21 | AAGAGAGGGATTCGAACCCTC | trnS-GCU | trnS-UGA |
| 21 | AGACAGGATTTGAACCCGTGA | trnfM-CAU | trnP-UGG |
| 23 | TCATTGTTCCACTCTTTGACAAC | rrn4.5-rrn5 spacer | rrn4.5-rrn5 spacer |
| 26 | GTGAGATTTTCATCTCATACGGCTCC | ycf3 | ndhA |
| 26 | TTATTTATTTTATATTCTATTTCAAT | rps4 trnT-UGU spacer | rps4 trnT-UGU spacer |
| 29 | TCGATATTGATGATAGTGACGATATTGAT | ycf2 | ycf2 |
IRs region
Extensions of the IR into the genes rps19 and ycf1 were identified (Fig. 1): a small part of the 5′-end of rps19 is in the IRb region and the 5′-end of ycf1 extended into the IRa region, resulting in its pseudogenization due to the incomplete duplication.
Genome divergence hotpot regions
Genome-wide comparative analyses among the seven Ilex species identified 11 hotspot regions for genome divergence that could be utilized as potential phylogenetic markers to reconstruct the phylogeny in this genus. These were rpl2-psbA, matK-rps16-psbK-psbI, psbN-psbD, psbC-IhbA-rps14, ycf3-rps4, ndhC-atpE, accD-psaI-ycf4-cemA, petA-psbJ, rpl16-rps3, rpl32-ccsA, and ndhA intron (Fig. 2). Character diversity of these hotspot regions was more than 4%.
Phylogeny construction
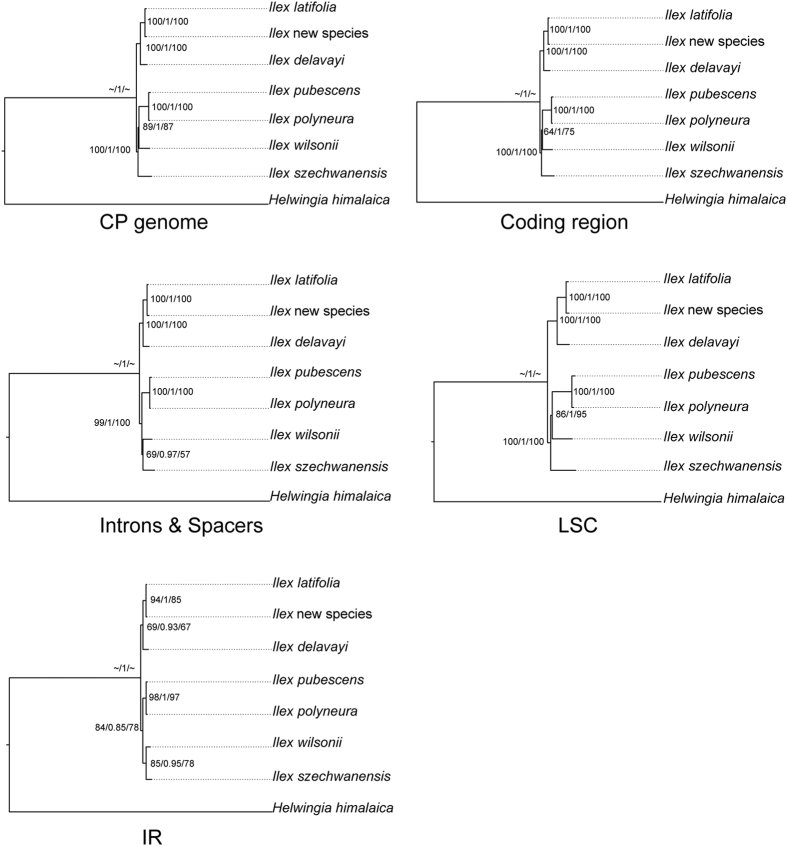
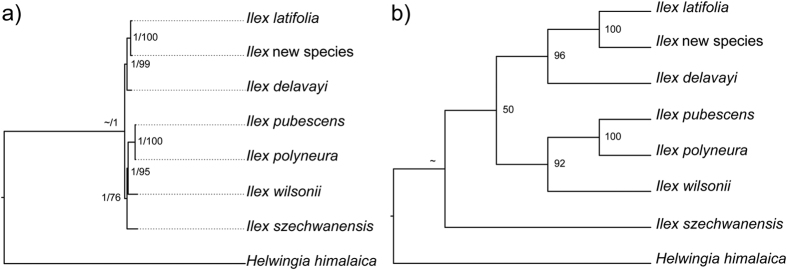
Among the cp genome sequences, protein-coding regions, LSC, SSC, IR, and introns and spacers, introns and spacers had the highest percentage variation at 1.7%, followed by LSC at 1.3%. The IR regions were least variable at 0.1%. The cp genome, SSC, and coding region, were 0.9%, 0.7% and 0.6%, respectively. Different methods of reconstructing phylogenies did not influence the topologic structure (Fig. 3) except with SSC, where maximum likelihood and Bayesian inference reconstructions differed in the position of I. szechwanensis from the one built by maximum parsimony (Fig. 4). However, in other respects all the phylogenies were the same, with I. latifolia, the new species, and I. delavayi forming one clade and the other species forming another. The cp genome and LSC phylogenies had higher bootstrap values and posterior probabilities than the others.
Figure 3. Phylogenetic relationships of the seven Ilex species and Helwingia himalaica constructed from chloroplast genes.
Numbers near nodes indicate the maximum parsimony bootstrap (left) values for each clade present in the 50% majority-rule consensus tree, Bayesian posterior probability (middle), and maximum likelihood bootstrap (right) values for each clade present in the 50% majority-rule consensus tree.
Figure 4. Phylogenetic relationships of the seven Ilex species and Helwingia himalaica based on the SSC region.
In plot ‘a’ numbers near nodes indicate the Bayesian posterior probability (left) and maximum likelihood bootstrap (right) values for each clade present in the 50% majority-rule consensus tree; in plot ‘b’ numbers near nodes indicate the maximum parsimony bootstrap of each clade present in the 50% majority-rule consensus tree.
Discussion
Modifications of chloroplast genome composition and gene order have been identified in many species in the asterid subclass Campanulidae, which includes the Aquifoliales, Asterales, Escalloniales, Bruniales, Apiales, Paracryphiales and Dipsacales. In this study, gene ycf68 was found in Ilex pubescens, but not Helwingia himalaica. The cp genome of Adenophora remotiflora (Asterales) (KF889213 in GenBank) does not have accD, clpP and infA, while these genes are present in Anethum graveolens (Apiales)20, Panax ginseng (Apiales)21, I. pubescens and H. himalaica (Aquifoliales). I. pubescens, H. himalaica and Panax ginseng have lhbA, but Adenophora remotiflora and Anethum graveolens do not. In Adenophora remotiflora (KF889213 in GenBank) and Trachelium caeruleum (Asterales)22 genes atpI, rps2, rpoC2, rpoC1 and rpoB are between ycf3 and rps12, while in Anethum graveolens (Apiales)20, Anthriscus cerefolium (Apiales)23, Tiedemannia filiformis (Apiales)23, Panax ginseng (Apiales)21, Schefflera delavayi (Apiales)24, Lonicera japonica (Dipsacales) (NC_026839 in GenBank), and I. pubescens (Aquifoliales) they are between atpH and trnC-GCA. The distances between the locations of these five genes in the two groups are about 82,000 bp and their order is also different. Consequently, the total length of the cp genome differs between lineages in the Campanulidae.
In addition, variability in the extent of the inverted repeat (IR) regions has been found, with the boundaries between IR and LSC or SSC very fluid. Gene rps19 is nearest to the LSC-IR boundary: in some species, like I. pubescens, H. himalaica, and Panax ginseng21, it spans the boundary, in others, like Millettia pinnata25 and Lupinus luteus26, it does not extend into the IR, while in others, like Phaseolus vulgaris27, Vigna radiata28 and Vigna unguiculata (JQ755301 in GenBank), the whole gene is inside the IR. Gene ycf1, nearest to the SSC-IR boundary, is similar.
I. pubescens had fewer and smaller dispersed repeats than reported for some other campanulids. The largest was 29 bp, while in the Apiales 9–29 repeats >30 bp were recorded in various species, with the largest 79 bp in length23.
Variable plastid regions have been used to design markers to investigate phylogenetic relationships, for instance rbcL, matK, and atpB, which have been widely used in phylogenic reconstruction from the genus level upwards. However, these genes are most divergent and informative among distantly related species, and are not suited for studying relationships between species in the same genus, like Curcuma29 and some genera in the Lauraceae30. According to the alignment of the cp genome of seven Ilex species studied here, rbcL, matK, and atpB were not appropriate for studies within Ilex because their divergences, which were 0.2%, 0.16% and 0.07%, respectively, were too low. However, 11 divergent regions were identified with sequence divergences around 4%.
Divergent hotspot regions in the chloroplasts are particularly useful for species-level identification in Ilex, which has many similar species represented by few collections. For example, several Chinese species, including Ilex chengkouensis C. J. Tseng, Ilex euryoides C. J. Tseng, Ilex synpyrena C. J. Tseng, and Ilex ningdeensis C. J. Tseng, have only been collected once. Moreover, some species currently recognized, such as Ilex huoshanensis Y. H. He, Ilex dabieshanensis K. Yao & M. P. Deng, Ilex urceolatus C. B. Shang, K. S. Tang & D. Q. Du, and Ilex wugonshanensis C. J. Tseng ex S. K. Chen & Y. X. Feng, are not clearly distinct in morphology and distribution from their nearest relatives, and may not deserve species status. A study of randomly sampled herbarium specimens in the National Herbarium in Beijing found that, although the DNA was usually highly degraded and most fragment <300 bp, it was still possible to extract usable genetic material from around a third of specimens18. This suggests that similar techniques could be used to clarify the diversity and status of the rare and little-known Ilex species in China.
For the phylogenetic reconstructions presented here, most informative characters occurred in intergenic regions, with some of these identified as divergent hotspot regions, as was also shown in Camellia16 and the Bambusoideae31. Among the phylogenies built by six different subsets of the genomic data, trees based on the complete cp genome and LSC displayed the highest support, although most nodes had high support in all. The topologies of different phylogenies were very similar, as also shown in Camellia and the Bambusoideae. These studies also showed that the methods used to build the phylogeny (MP, ML or BI) had a relatively minor influence.
The results of our phylogenetic analyses agree in part with the traditional classification system used in the Flora of China2. I. delavayi and I. latifolia in section Aquifolium form one clade with a new, large-fruited species, which is similar to but distinct from I. latifolia (Tan et al., submitted). However, in the other clade, the evergreen species I. pubescens and I. wilsonii are in section Pseudoaquifolium, while the deciduous I. polyneura is in Micrococca. In the classification used in the FOC all the deciduous species form a separate clade. Data from nuclear genes will be needed to resolve these differences, as shown previously for Ilex by Manen1. The chloroplast genome is also expected to be useful in helping to resolve the deeper branches of the phylogeny as more whole-genome sequences become available.
Conclusions
The chloroplast genomes of seven Ilex species are reported for the first time in this study and their organization is described and compared with that of other campanulids. Eleven divergent regions were identified, which can be used to develop phylogenetic markers. Phylogenies were constructed using the entire genomes and various subsets and their topologies and resolutions compared. Our results will be useful for identification at the species level and for helping to resolve the deeper branches of the phylogeny.
Methods
Plant materials
Plant materials used in this study were intact, fresh, young leaves collected in Yunnan. Species were identified with the Flora of China2 and specimens were deposited in the herbarium of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (HITBC) (Table 4).
Table 4. Sampled species and their voucher specimens used in this study according to the taxonomic treatment in the Flora of China.
| Species | Subgenus | Section | Geographic origin | Voucher | Accession number in GenBank |
|---|---|---|---|---|---|
| Ilex latifolia | Ilex | Aquifolium | XTBG | YX1303 | KX426465 |
| Ilex szechwanensis | Ilex | Paltoria | Dali, Yunnan | YX1418 | KX426466 |
| Ilex pubescens | Ilex | Pseudoaquifolium | Xishuangbanna, Yunnan | YX1676 | KX426467 |
| Ilex polyneura | Prinos | Micrococca | Xishuangbanna, Yunnan | YX1680 | KX426468 |
| Ilex new species | – | – | Xishuangbanna, Yunnan | YX1681 | KX426469 |
| Ilex delavayi | Ilex | Aquifolium | Dali, Yunnan | YX1723 | KX426470 |
| Ilex wilsonii | Ilex | Pseudoaquifolium | Yichun, Jiangxi | YX1748 | KX426471 |
| Helwingia himalaica | – | – | Gongshan, Nujiang | YX1678 | KX434807 |
Chloroplast genome sequencing and assembly
About 100 mg of fresh leaf material of each species was used to extract total DNA by a modified CTAB method17,32, with 4% CTAB with 0.2% DL-dithiothreitol (DTT) replacing 2% CTAB, and adding approximately 1% polyvinyl polypyrrolidone (PVP) while milling the materials. Long-range PCR was used for DNA amplification of the plastome using nine universal primers developed by Yang17. Each amplification was performed in 25 μL of a reaction mixture containing 1 × PrimeSTAR GXL buffer (10 mM Tris-HCl (pH 8.2), 1 mM MgCl2, 20 mM NaCl, 0.02 mM EDTA, 0.02 mM DTT; 0.02% Tween 20, 0.02% Nonidet P-40, and 10% glycerol); 1.6 mM of dNTPs, 0.5 μM of each primer; 1.25 U of Prime-STAR GXL DNA polymerase (TAKARA BIO INC., Dalian, China), and 30–100 ng of DNA template. The amplification was conducted using 94 °C for 1 min, 30 cycles of 98 °C for 10 s and 68 °C for 15 min, followed by a final extension step at 72 °C for 10 min.
The 6 μg PCR product was fragmented for constructing short-insert (500 bp) libraries according to the Illumina manual, using the Illumina Nextera XT library (Illumina, San Diego, CA, USA). DNA from each individual was indexed using tags and pooled together in one lane of a Illumina Hiseq 2000 for sequencing at the Germplasm Bank of Wild Species in Southwest China, Kunming Institution of Botany, Chinese Academy of Sciences.
Raw reads were filtered by quality control software NGSQCToolkit v2.3.333 to obtain high quality Illumina data (cut-off value for percentage of read length = 80, cut-off value for PHRED quality score = 30) and vector- and adaptor-free reads. Filtered reads were then assembled into contigs in the software CLC Genomics Workbench 8 (http://www.clcbio.com), by de novo method using a k-mer of 63 and a minimum contig length of 1 kb. Outputted contigs were aligned with a reference Camellia yunnanensis chloroplast genome (Genbank accession number KF156838), which was the most similar genome identified via BLAST (http://blast.ncbi.nlm.nih.gov/), and ordered according to the reference genome. Contigs were aligned with the reference genome for assembly of the chloroplast genome of each species in Geneious 4.834. Lastly, junctions between LSC/IRs and SSC/IRs were validated by Sanger sequencing of PCR-based products using newly designed primers (see Supplementary Table S2).
Genome annotation and repeat analysis
Assembled genomes were annotated using the Dual Organellar GenoMe Annotator (DOGMA) database35, then manually edited for start and stop codons. All annotated cp genomes will be deposited in GenBank. Genome maps were drawn in OGDraw 1.236. Multiple sequence alignment was done with MAFFT 537 and manually edited where necessary. A comparative plot of full alignment with annotation of these eight genomes was produced by mVISTA38,39, using Helwingia himalaica as a reference. REPuter was used to detect and assess repeats40 in I. pubescens. Average genetic divergences of these eight species were estimated using p-distances. The genetic divergence between the two most closely related species, I. latifolia and I. delavayi, was also estimated.
Molecular marker identification
In order to explore the divergence of chloroplast genes in Ilex and its utilization in identification, all coding genes, introns and spacers were extracted. Every homologous region was aligned by MUSCLE41 and manually edited where necessary.
Phylogenetic analyses
Sequences of the seven Ilex species and Helwingia himalaica were aligned using MAFFT37 and manually edited where necessary. Unambiguously aligned DNA sequences were used for phylogeny construction. Phylogenies were constructed by maximum parsimony (MP), maximum likelihood (ML) and Bayesian Inference analyses (BI) using the entire cp genome and also using exons of protein-coding regions, introns and spacers, LSC, SSC, and IR. Lengths of all alignment matrices of these datasets are shown in Supplementary Table S3. In all phylogenetic analyses, Helwingia himalaica was used as outgroup.
MP and ML analyses were conducted in PAUP 4.0b1042. For MP analysis, heuristic searches were conducted with tree bisection-reconnection (TBR) branch swapping, with the ‘Multrees’ option in effect. Bootstrap analysis was conducted with 1,000 replicates with TBR branch swapping. For ML analysis, the best substitution model was tested according to the Akaike information criterion (AIC) by jModeltest version 243,44 (see Supplementary Table S3). BI analysis was conducted using MrBayes version 3.2.245 and the best substitution model tested by AIC. Two independent Markov Chain Monte Carlo chains were calculated simultaneously for 10,000,000 generations and sampled every 1,000 generations. Potential Scale Reduction Factor (PSRF) values were used to determine convergence in Bayesian Inference using MrBayes version 3.2.245. All PSRF values were 1, indicating that these analyses converged. The first 25% of calculated trees was discarded as burn-in and a consensus tree constructed using the remaining trees.
Additional Information
How to cite this article: Yao, X. et al. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci. Rep. 6, 28559; doi: 10.1038/srep28559 (2016).
Supplementary Material
Acknowledgments
The authors would like to acknowledge Hong-Tao Li for help with data analyses. We thank Jing Yang, Juan-Hong Zhang, Chun-Yan Lin and Ji-Xiong Yang from Kunming Institute of Botany, Chinese Academy of Sciences, for their help with experiments. This work was supported by grants from the 1000 Talents Program (WQ20110491035).
Footnotes
Author Contributions X.Y. and R.T.C. conceived the experiments, X.Y., Y.H.T. and Y.Y.L. collected the samples, X.Y. and J.B.Y. conducted the experiments, X.Y., Y.H.T. and Y.S. analyzed the results, X.Y. and R.T.C. wrote the manuscript. All authors reviewed the manuscript.
References
- Manen J. F. et al. The history of extant Ilex species (Aquifoliaceae): evidence of hybridization within a Miocene radiation. Mol. Phylogenet. Evol. 57, 961–977 (2010). [DOI] [PubMed] [Google Scholar]
- Wu Z. Y., Raven P. H. & Hong D. Y. Flora of China. Vol. 11: Oxalidaceae through Aceraceae. (Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis., 2008). [Google Scholar]
- Stevens P. F. (2001 onwards). Angiosperm Phylogeny Website. Version 12. Available at: http://www.mobot.org/MOBOT/research/APweb/ (2012).
- Debat H. J. et al. Exploring the genes of yerba mate (Ilex paraguariensis A. St.-Hil.) by NGS and De Novo Transcriptome Assembly. PLoS ONE. 9, e109835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S. et al. Extract of Kuding tea prevents high-fat diet-induced metabolic disorders in C57BL/6 mice via liver X receptor (LXR) β antagonism. PLoS ONE. 7, e51007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. et al. Apoptosis inducing effects of Kuding tea polyphenols in human buccal squamous cell carcinoma cell line BcaCD885. Nutrients. 6, 3084–3100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu J. M., Tank D. C. & Donoghue M. J. A Southern hemisphere origin for campanulid angiosperms, with traces of the break-up of Gondwana. BMC Evol. Biol. 13, 80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuénoud P. et al. Molecular phylogeny and biogeography of the genus Ilex L. (Aquifoliaceae). Ann. Bot. 85, 111–122 (2000). [Google Scholar]
- Manen J. F., Boulter M. C. & Naciri-Graven Y. The complex history of the genus Ilex L. (Aquifoliaceae): evidence from the comparison of plastid and nuclear DNA sequences and from fossil data. Plant Syst. Evol. 235, 79–98 (2002). [Google Scholar]
- Ma P. F. et al. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst. Biol. 63, 933–950 (2014). [DOI] [PubMed] [Google Scholar]
- Yi T. S., Jin G. H. & Wen J. Chloroplast capture and intra-and inter-continental biogeographic diversification in the Asian–New World disjunct plant genus Osmorhiza (Apiaceae). Mol. Phylogenet. Evol. 85, 10–21 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang Q., Field T. S. & Antonelli A. Assessing the impact of phylogenetic incongruence on taxonomy, floral evolution, biogeographical history, and phylogenetic diversity. Am. J. Bot. 102, 566–580 (2015). [DOI] [PubMed] [Google Scholar]
- de Sousa F., Bertrand Y. J. K. & Pfeil B. E. Patterns of phylogenetic incongruence in Medicago found among six loci. Plant Syst. Evol. 1–21 (2016). [Google Scholar]
- Lam V. K. Y., Gomez, M. S. & Graham S. W. The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biol. Evol. 7, 2220–2236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley T. W. et al. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23, 2175–2190 (2006). [DOI] [PubMed] [Google Scholar]
- Yang J. B. et al. Comparative chloroplast genomes of Camellia species. PLoS ONE. 8, e73053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. B., Li D. Z. & Li H. T. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol. Ecol. Resour. 14, 1024–1031 (2014). [DOI] [PubMed] [Google Scholar]
- Xu C. et al. Accelerating plant DNA barcode reference library construction using herbarium specimens: improved experimental techniques. Mol. Ecol. Resour. 15, 1366–1374 (2015). [DOI] [PubMed] [Google Scholar]
- Zedane L. et al. Museomics illuminate the history of an extinct, paleoendemic plant lineage (Hesperelaea, Oleaceae) known from an 1875 collection from Guadalupe Island, Mexico. Biol. J. Linn. Soc. 117, 44–57 (2016). [Google Scholar]
- Peery R. M. Understanding angiosperm genome interactions and evolution: insights from sacred lotus (Nelumbo nucifera) and the carrot family (Apiaceae). University of Illinois at Urbana-Champaign (2015).
- Kim K. et al. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS ONE. 10, e0117159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle R. C. et al. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 66, 350–361 (2008). [DOI] [PubMed] [Google Scholar]
- Downie S. R. & Jansen R. K. A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 40, 336–351 (2015). [Google Scholar]
- Li L. et al. The large-leaved Kudingcha (Ilex latifolia Thunb and Ilex kudingcha CJ Tseng): a traditional Chinese tea with plentiful secondary metabolites and potential biological activities. J. Nat. Med. 67, 425–437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakoff S. H. et al. Capturing the biofuel wellhead and powerhouse: the chloroplast and mitochondrial genomes of the leguminous feedstock tree Pongamia pinnata. PLoS ONE. 7, e51687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. E. et al. The first complete chloroplast genome of the genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. 113, 1197–1210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. et al. Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics. 8, 1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangphatsornruang S. et al. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 17, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. et al. Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol. Ecol. Resour. 15, 337–348 (2015). [DOI] [PubMed] [Google Scholar]
- Rohwer J. G. Toward a Phylogenetic Classification of the Lauraceae: Evidence from matk Sequences. Syst. Bot. 25, 60–71 (2000). [Google Scholar]
- Zhang Y. J., Ma P. F. & Li D. Z. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS ONE. 6, e20596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J. & Doyle J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 19, 11–15 (1987). [Google Scholar]
- Patel R. K. & Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 7, e30619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S. K., Jansen R. K. & Boore J. L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20, 3252–3255 (2004). [DOI] [PubMed] [Google Scholar]
- Lohse M. et al. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. et al. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C. et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 (2000). [DOI] [PubMed] [Google Scholar]
- Frazer K. A. et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S. et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L. PAUP*: phylogenetic analysis using parsimony, version 4.0 b10. (2003).
- Guindon S. & Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- Darriba D. et al. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 9, 772–772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.