Abstract
LIM domain-binding protein 1 (Ldb1) is a nuclear cofactor that interacts with LIM homeodomain proteins to form multiprotein complexes that are important for transcription regulation. Ldb1 has been shown to play essential roles in various processes during mouse embryogenesis. To determine the role of Ldb1 in mid- and hindbrain development, we have generated a conditional mutant with a specific deletion of the Ldb1 in the Engrailed-1-expressing region of the developing mid- and hindbrain. Our study showed that the deletion impaired the expression of signaling molecules, such as fibroblast growth factor 8 (FGF8) and Wnt1, in the isthmic organizer and the expression of Shh in the ventral midbrain. The midbrain and the cerebellum were severely reduced in size, and the midbrain dopaminergic (mDA) neurons were missing in the mutant. These defects are identical to the phenotype that has been observed previously in mice with a deletion of the LIM homeodomain gene Lmx1b. Our results thus provide genetic evidence supporting that Ldb1 and Lmx1b function cooperatively to regulate mid- and hindbrain development. In addition, we found that mouse embryonic stem cells lacking Ldb1 failed to generate several types of differentiated neurons, including the mDA neurons, serotonergic neurons, cholinergic neurons, and olfactory bulb neurons, indicating an essential cell-autonomous role for Ldb1 in the development of these neurons.
Introduction
The patterning of the mid- and hindbrain and generation of midbrain dopaminergic (mDA) neurons are complex processes regulated by a large number of signaling molecules and transcription factors [1–4]. Two closely related LIM homeodomain proteins, Lmx1a and Lmx1b, play important roles in these processes. The Lmx1b gene, expressed in the isthmic organizer, is essential for patterning of the mid- and hindbrain by controlling the expression of secreted signaling molecules, such as fibroblast growth factor 8 (FGF8) and Wnt1, in embryos of various vertebrate species, including chick, zebrafish, and mouse [5–7]. Lmx1b, together with Lmx1a, is also expressed in mDA neuron progenitors and required for the proliferation, specification, and differentiation of the mDA progenitors [8,9]. In addition, gain-of-function studies have shown that ectopic expression of Lmx1a or Lmx1b induces the generation of mDA neurons both in embryos and in differentiating embryonic stem (ES) cells [10–12].
The function of LIM homeodomain proteins is largely dependent on the formation of multiprotein complexes through interactions between these proteins and other nuclear factors [13,14]. A key component of these complexes is a transcription coregulator called “LIM domain-binding protein 1” [(Ldb1), also called “NLI” or “CLIM2”] [15–17]. A number of previous studies have revealed that Ldb1 is essential for the regulation of a variety of processes in mouse embryogenesis, including the head and heart formation, limb patterning, and forebrain and cerebellum development [18–21].
In this study, to determine the role of Ldb1 in mid- and hindbrain development, we generated a conditional mouse mutant to delete Ldb1 more specifically in the mid- and hindbrain regions during embryonic development by crossing the mutant Ldb1fl− [20] with mice expressing the Cre recombinase under the control of the regulatory element of the Engrailed-1 gene ([En1Cre], [22]). Our analysis of the Ldb1 mutant revealed a phenotype similar to that observed previously in Lmx1b null mutant [7]. Our results provide genetic evidence suggesting that Ldb1 cooperates with Lmx1b in regulation of mid- and hindbrain development. In addition, we provide evidence supporting that Ldb1 is also required for the generation of several types of terminally differentiated neurons, including the mDA neurons, from differentiation of the mouse ES cells.
Materials and Methods
Mouse lines and genotyping
For mouse care and experiment, we followed the guideline of the Korea University Animal Care and Use Committee, and our IRB number is KUIACUC-20111024-2.
To generate conditional mutants with a specific deletion of Ldb1 in the mid- and hindbrain regions, the Engrailed-1+/Cre (En1+/Cre) mouse line, which contains an insertion of the Cre recombinase gene into the En1 locus [22], was first crossed to Ldb1+− mice [18]. Offspring containing one Ldb1 null allele and one En1Cre allele (Ldb1+/− and En1+/Cre) were selected and mated to either heterozygous or homozygous Ldb1 floxed (Ldb1+/f or Ldb1f/f) mice [20] to produce Ldb1 conditional mutants (Ldb1f/− and En1+/cre) and controls (wild type, Ldb1+/f, and Ldb1+/f and En1+/Cre) for analysis. Mouse genotypes were determined by polymerase chain reaction (PCR) as described in previous studies [23] using the following primers: Ldb1 wild-type and floxed alleles: 5′-CAGCAAACGGAGGAAACGGAAGATGTCAG and 5′-CTTATGTGACCACAGCCATGCATGCATGTG; Ldb1 null allele: 5′-ACGAGTTCTTCTGAGGGGATC and 5′-TGCCACACAGAATCTGCTCTGAACGTCT; and En1Cre allele: 5′-CACCCTGTTACGTATAGCCG and 5′-GAGTCATCCTTAGCGCCG.
Whole-mount in situ hybridization
Embryos were fixed in 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) and processed for whole-mount in situ hybridization with digoxigenin-labeled RNA probes according to a well-established protocol [24]. The hybridization signal was detected with alkaline phosphatase-conjugated antidigoxigenin antibodies and BM purple substrate (Roche Diagnostics). Fgf8 probe (nucleotides 591–943, GenBank D38752) was previously described [25]. Shh and Wnt1 probes were a generous gift from Dr. Andrew McMahon.
Whole-mount immunohistochemistry
Embryos were fixed in 4% PFA, 0.15% picric acid, and 0.1% Tween 20 in PBS for 5 h at 4°C. Fixed embryos were washed with PBS containing 0.1% Tween 20 (PBT) two times at 4°C, dehydrated in methanol, and bleached with 3% hydrogen peroxide in 80% methanol and 20% DMSO for 3 h at room temperature (RT). The embryos were washed at RT in 1% Tween 20/PBS for 3 h, 1% Triton X-100/PBT for 20 min, and 0.5% Triton X-100/PBT three times each for 15 min. Embryos were blocked in 5% skim milk, 0.5% Triton X-100, and 5% DMSO/PBT for 1 h at RT, followed by incubation with primary antibodies diluted in the same blocking solution for 2 days at RT. The following primary antibodies from Santa Cruz Biotech were used at a dilution of 1:100: goat anti-FGF8 and goat anti-sonic hedgehog (SHH). Afterward, embryos were washed in 0.1% Triton X-100/PBT and incubated with horseradish peroxidase-conjugated secondary antibodies (Dako) overnight at 4°C, washed in 1% Tween 20/0.5% Triton X-100/PBS, and developed for 10–30 min in DAB (Sigma).
Mouse ES cell culture
Ldb1−/− ES cells were generated as described previously [23]. ES cell culture medium is composed of Dulbecco's modified Eagle's medium mixed with 15% fetal bovine serum, 100 mM nonessential amino acids, 0.5% penicillin–streptomycin, 0.55 mM 2-mercaptoethanol, and 1,000 U/mL leukemia inhibitory factor (Chemicon). Both wild-type and Ldb1−/− ES cells were cultured in 0.1% gelatin-coated dishes at 37°C in an incubator containing 5% CO2. For differentiation of the ES cells to neurons, the adherent monolayer culture method was followed as previously described without any modifications [26].
Immunocytochemistry
Cells were fixed with 4% PFA in PBS (pH 7.4) for 20 min at RT, followed by washes in PBS. The cells were treated with 1% sodium dodecyl sulfate for 5 min for antigen retrieval. After more washes in PBS, the cells were incubated in a blocking solution containing 0.3% Triton X-100 and 3% bovine serum albumin (BSA) for 45 min and then in primary antibody diluted with the same blocking solution overnight at 4°C. After washes in PBS, the cells were incubated in secondary antibodies diluted in the blocking solution for 1 h at RT. After washes in PBS, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted with Fluoromount-G (Southern Biotech). The primary antibodies used were rabbit anti-MAP2 (Chemicon), rabbit anti-GFAP (Chemicon), mouse anti-TH (Chemicon), and rabbit anti-LDB1 (a gift from Dr. Paul Love, NIH, Bethesda, MD) [27]. Images were captured using a Zeiss LSM 510 confocal laser scanning microscope.
Histology and immunohistochemistry
Brains of E14.5 and E18.5 mouse embryos were dissected and fixed in 4% PFA/PBS overnight at 4°C. After washes in PBS, the tissue was either dehydrated in ethanol and embedded in paraffin or soaked with 30% sucrose/PBS and frozen in OCT compound (Sakura Finetek). For histological analysis, 5-μm-thick paraffin sections were cut and stained with hematoxylin and eosin (Sigma). For immunohistochemistry, frozen sections (14 μm) were cut and mounted on silane-coated slides (Muto Pure Chemicals Co., Ltd.). Sections were washed in PBS and incubated in a blocking solution containing 0.3% Triton X-100 and 3% BSA for 45 min. The sections were incubated with primary antibodies diluted with the same blocking solution overnight at 4°C. After washes in PBS, the sections were incubated in secondary antibodies diluted with the same blocking solution for 1 h at RT. After washes in PBS, the sections were stained with DAPI and mounted with Fluoromount-G. Images were taken by a Zeiss LSM 510 confocal laser scanning microscope. The primary antibodies used were rabbit antityrosine hydroxylase (Chemicon), rabbit anti-Pitx3 (Chemicon), rabbit anti-Nurr1 (Santa Cruz Biotech), rabbit anti-GABA (Sigma), rabbit anti-LDB1 (a gift from Dr. Paul Love, NIH, Bethesda, MD), and rabbit anti-caspase 3 (Merck Millipore).
Real-time reverse transcription PCR analysis
Total RNA was isolated from cells using a ToTALLY RNA™ Kit (Ambion) by following the manufacturer's guidance. One microgram of the RNA template was reverse transcribed by a Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics) according to the instruction from the manufacturer. Real-time PCR was performed using 2 μL aliquot of the reverse-transcribed product for each 20 μL sample reaction mixture containing 4 mM MgCl2, 10 pmole of upstream and downstream primers, and 2 μL of 10X Light Cycler Fast Start DNA Master SYBR Green 1 (Roche Diagnostics). Light Cycler software (version 3.5) was used to analyze the data. The list of primer sets used for reverse transcription PCR (RT-PCR) is shown in Table 1.
Table 1.
List of Primer Sets for Reverse Transcription Polymerase Chain Reaction
| Primer sequences | |||
|---|---|---|---|
| Gene | Upper primer sequence | Lower primer sequence | Length (bp) |
| GAPDH | 5′-GTGTTCCTACCCCCAATGTG-3′ | 5′-TGTGAGGGAGATGCTCAGTG-3′ | 400 |
| Nurr1 | 5′-CGGTTTCAGAAGTGCCTAGC-3′ | 5′–CTGGGTTGGACCTGTATGCT-3′ | 420 |
| Pitx3 | 5′-ACAAAGTGGAACCCCTATGAG-3′ | 5′-TTCTTGGCCAATCTGTAGGA-3′ | 255 |
| TH | 5′-TTGGCTGACCGCACATTTG-3′ | 5′-ACGAGAGGCATAGTTCCTGAGC-3′ | 336 |
| DAT | 5′-CACTCTGGGTATCGAC-3′ | 5′-ATGGCATAGGCCAGTTTCTC-3′ | 537 |
| Ldb1 | 5′-TGCTGACCATCACTTTCTGC-3′ | 5′-GGCTGAGGCTGTAGGTCTTG-3′ | 480 |
| Lhx2 | 5′-TCAACTGCTTCACATGCACA-3′ | 5′-TTTCCTGCCCTAAAAGGTTG-3′ | 590 |
| Lhx5 | 5′-GACCTCATCGGACAAGGAAA-3′ | 5′-ACCCCAACATCTCAGACTCG-3′ | 352 |
| Lhx6 | 5′-CTGTGCGGCAGACAAATCTA-3′ | 5′-CTCTCAATGTAGCCGTGCAA-3′ | 572 |
| Lhx8 | 5′-CAGTTCGCTCAGGACAACAA-3′ | 5′-CTTATTGGCAGCTGGGTCAT-3′ | 331 |
| Lmx1b | 5′-CTGCTGTGCAAGGGTGACTA-3′ | 5′-GGCTTGACAGAACCTCTTGG-3′ | 420 |
| OMP | 5′-GAAGCAGGATGGTGAGAAGC-3′ | 5′-CACAGAGGCCTTTAGGTTGG-3′ | 361 |
| Calretinin | 5′-CTCCTGAAGAAGGCCAACAG-3′ | 5′-GGTCTGGGAAGGAGTGTCAA-3′ | 488 |
| GAD2 | 5′-GGGTTTGAGGCACACATTGATAAG-3′ | 5′-GCGGAAGAAGTTGACCTTGTCC-3′ | 279 |
| ChAT | 5′-GCCAATCCATTCCCACTGAC-3′ | 5′-CATCCAAGACAAAGAACTGG-3′ | 198 |
| Sert | 5′-GTGACAGCCACCTTCCCTTA-3′ | 5′-CTAGCAAACGCCAGGAGAAC-3′ | 200 |
Results
Defects in development of the midbrain and cerebellum in Ldb1 conditional mutant
Homozygous Ldb1 null mutant embryos die at E9.5 [18]. To study the function of Ldb1 in the development of the mid- and hindbrain in later stages of embryos, we generated Ldb1 conditional mutant embryos that carried one floxed (f) allele [20] and one null (−) allele of the Ldb1 gene in addition to a knock-in allele containing a targeted insertion of the Cre recombinase gene into the En1 locus (En1Cre) [22] (Ldb1f/− and En1+/Cre). As revealed by X-gal staining of embryos from crossing between the En1+/Cre and the Rosa26R (Rosa26LoxP-stop-LoxP-LacZ) reporter line [28], the Cre recombinase was active in the mid- and hindbrain junction of the embryo as early as E8.5 (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd). As shown by in situ hybridization, the En1 expression domain at the mid- and hindbrain junction was reduced in size in Ldb1 conditional mutant embryos in comparison with wild-type embryos at E9.5 (Fig. 1A, A′). The conditional deletion of Ldb1 was also confirmed by the absence of immunostaining of Ldb1 in the midbrain of the mutant embryo (Supplementary Fig. S2). At E18.5, the Ldb1 conditional mutant embryo showed a clear truncation of the midbrain and missing of almost the entire cerebellum (Fig. 1B, B′, C, C′). Thus, the conditional Ldb1 deletion severely impaired the development of the midbrain and the cerebellum.
FIG. 1.
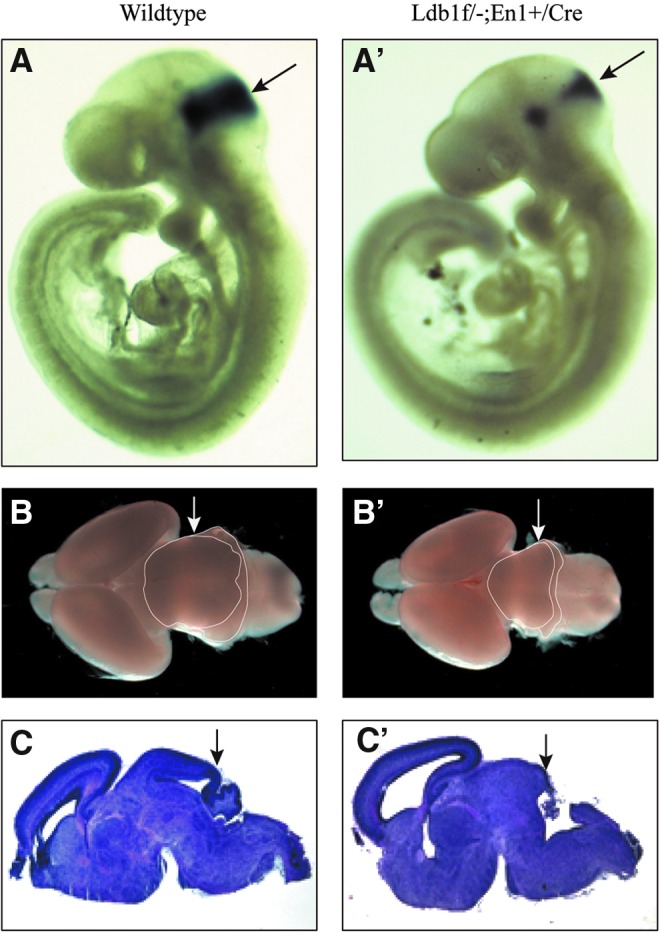
Defects in the development of the mid- and hindbrain structures in Ldb1 conditional mutant. (A, A′) Whole-mount in situ hybridization showing a reduction in the expression of En1 in the mid- and hindbrain junction in an E9.5 Ldb1 mutant embryo (A′) compared with a control (A). (B, B′) Dorsal view of brain dissected from E18.5 control (B) and Ldb1 mutant (B′) embryos showing a truncation of the midbrain and the cerebellum in the mutant. (C, C′) Hematoxylin and eosin-stained sagittal sections of the brain showing a severe reduction in size of the dorsal midbrain and missing of the cerebellum in the Ldb1 mutant (C′) compared with the control (C) at E18.5.
Impaired Fgf8, Wnt1, and Shh gene expression in the isthmus organizer and ventral midbrain in Ldb1 conditional mutant
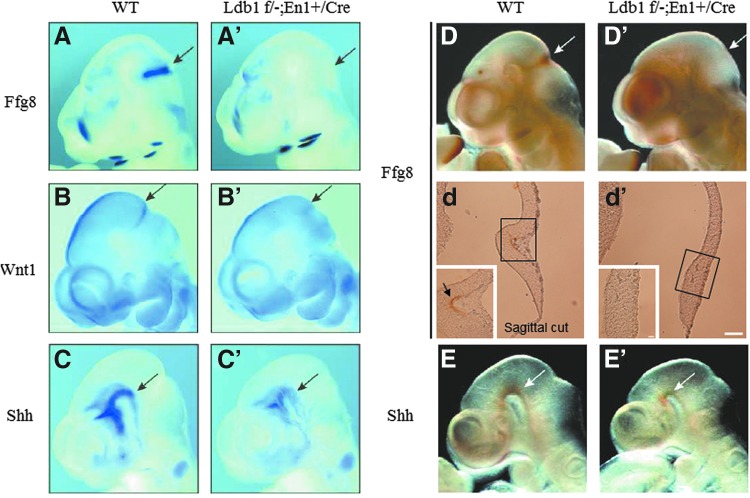
Signaling molecules, such as Fgf8, Wnt1, and Shh, from the isthmus organizer or the ventral midbrain play important roles in the development of the mid- and hindbrain (see [3,4] for a detailed review). By performing whole-mount in situ hybridization analysis, we examined the expression of these molecules in Ldb1 conditional mutant embryos. At E10.5, while Fgf8 RNA was detected in the forebrain and the pharyngeal regions, it was absent at the mid- and hindbrain junction in Ldb1 mutant embryos compared with wild-type controls (Fig. 2A, A′). Similarly, the expression of Wnt1 was unaffected in the forebrain and dorsal midbrain, but the expression in a sharp semicircular domain at the mid- and hindbrain junction was missing in the mutant embryos compared with the controls (Fig. 2B, B′). Shh was expressed in the ventral diencephalon and ventral midbrain. The Shh mRNA expression in the ventral midbrain was reduced in Ldb1 mutant embryos compared with the controls (Fig. 2C, C′). We also performed whole-mount immunostaining of embryos with antibodies directed against Fgf8 and Shh. The staining revealed that Fgf8 and Shh proteins were either missing or reduced in the midbrain in Ldb1 mutant embryos compared with the controls (Fig. 2D, D′, E, E′). The absence of Fgf8 expression in Ldb1 mutant embryos was further confirmed on sagittal sections (Fig. 2d, d′). These results indicate that the conditional deletion of Ldb1 impairs the expression of multiple signaling molecules required for mid- and hindbrain development.
FIG. 2.
Reduced expression of Fgf8, Shh, and Wnt1 in Ldb1 mutant embryos. Whole-mount in situ hybridization (A–C, A′–C′), whole-mount immunostaining (D, E, D′, E′), and sagittally sectioned brain (d, d′) of E10.5 embryos show missing of Fgf8 and Wnt1 expression in the isthmus in Ldb1 mutant embryos (A′, B′, D′) compared with wild-type controls (A, B, D). The expression of Shh in the ventral region of the midbrain was reduced in Ldb1 mutant embryos (C′, E′) compared with controls (C, E). Fgf8, fibroblast growth factor 8.
Defects in generation of mDA neurons in Ldb1 conditional mutant
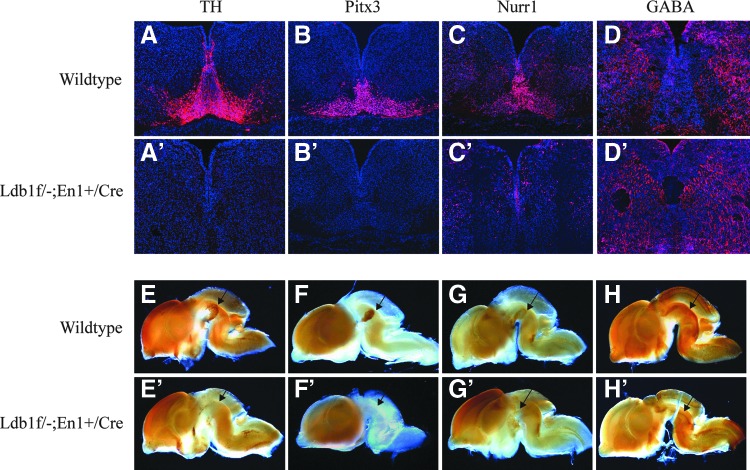
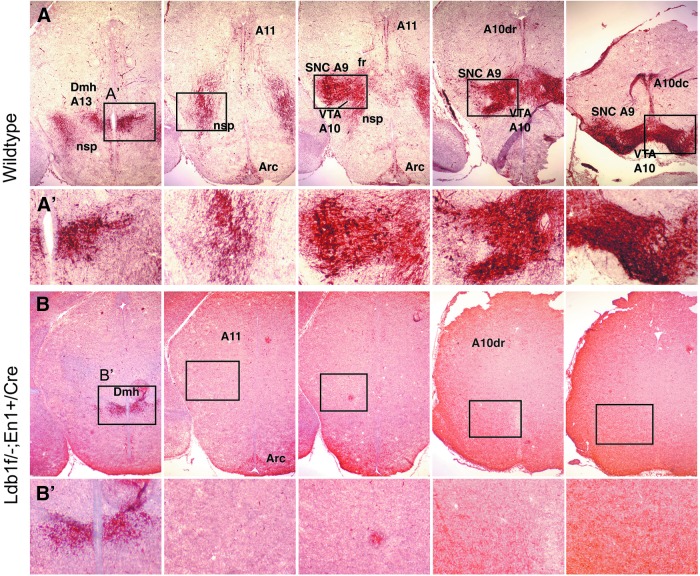
The isthmus organizer and signaling molecules, such as Fgf8, Wnt1, and Shh, are important for the patterning of the mid- and hindbrain and the specification of mDA neurons. To investigate whether the defect in mid- and hindbrain patterning and the impaired expression of the signaling molecules affect the generation of mDA neurons in Ldb1 mutant embryos, we examined the expression of a number of the factors that are involved in mDA neuron fate determination, differentiation, or dopamine biosynthesis. At E14.5, neurons positive for tyrosine hydroxylase (TH), Pitx3, or Nurr1 were detected abundantly in the ventral region of the midbrain in wild-type control embryos (Fig. 3A, B, C), but these cells were either missing or greatly reduced in number in Ldb1 mutant embryos (Fig. 3A′, B′, C′). However, there was not a significant difference in the number of GABA-positive neurons between control and Ldb1 mutant embryos (Fig. 3D, D′). In line with the results from staining of sections, sagittal view of the brains stained by whole-mount immunohistochemistry also showed that neurons positive for TH, Pitx3, and Nurr1 were absent in the ventral midbrain in Ldb1 mutant embryos (Fig. 3E′, F′, G′) compared with the controls (Fig. 3E, F, G). No difference in the number of GABA+ cells was detected between the control and Ldb1 mutant embryos (Fig 3H, H′). The absence of mDA neurons in Ldb1 mutant embryos was further confirmed by anti-TH staining of a series of coronal sections through the ventral midbrain of E18.5 embryos (Fig. 4A, B). We also examined the expression of cleaved caspase 3 to determine whether there was an increase of cell apoptosis in association with the defect in the generation of mDA neurons. At E14.5, we did not detect a clear increase in the number of cells labeled positive by cleaved caspase 3 (Supplementary Fig. S3).
FIG. 3.
Defects in the expression of mature mDA neuron markers in Ldb1 mutant embryos at E14.5. (A–D, A′–D′) Coronal brain sections were stained with anti-TH, -Pitx3, -Nurr1, and -GABA antibodies. Staining of TH, Pitx3, and Nurr1 was missing or greatly reduced in Ldb1 mutant embryos (A′, B′, C′) compared with the controls (A, B, C). No change in GABA staining was detected in Ldb1 mutant (D′) compared with the control (D). (E–H, E′–H′) Similar results were shown by whole-mount immunostaining of brains from wild-type (E–H) and Ldb1 mutant (E′–H′) embryos. mDA, midbrain dopaminergic.
FIG. 4.
Missing of mDA neurons in Ldb1 mutant embryos. Anti-TH staining of a series of sections through the midbrain of E18.5 embryos shows missing of major groups of mDA neurons in Ldb1 mutant (B) compared with the control (A). Sections from the mutant were overdeveloped to ensure no staining was detected. Higher magnified images of the boxed area in (A) and (B) are shown in (A′) and (B′). Arc, arcuate nucleus; Dmh, dorsomedial hypothalamus; nsp, nigra striatal projection; SNC, substantial nigra compacta; VTA, ventral tegmental area. A9, A10, A10dr (dorsorostral), A10dc (dorsocaudal), and A11 indicate the various TH+-cell groups assigned by the alphanumeric system of Dahlstrom and Fuxe.
Defects in generation of dopaminergic and other differentiated neuronal cell types from the Ldb1−/− ES cells
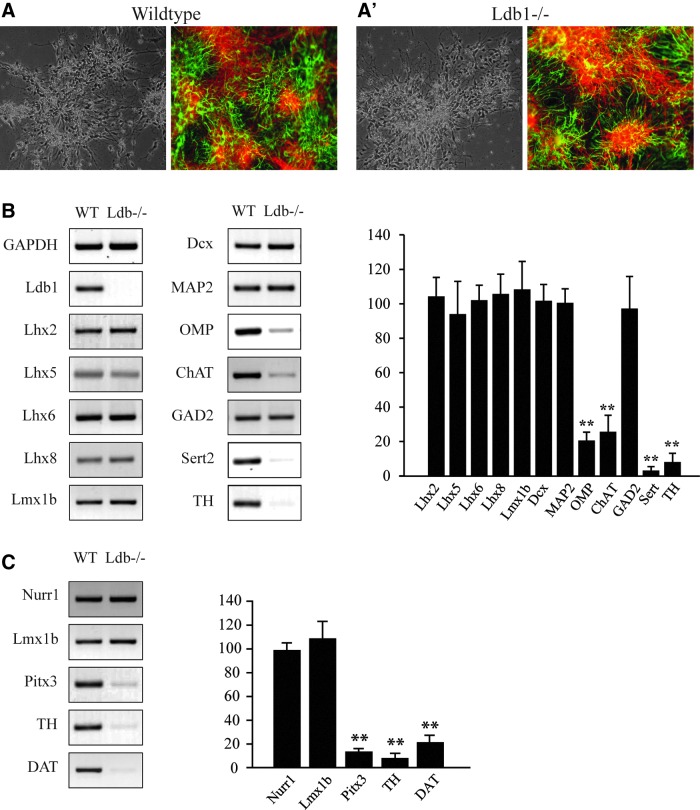
Our previous study revealed that Ldb1−/− ES cells maintain neuronal differentiation potential [23]. As shown in Figure 5A and A′, at 2 weeks after differentiation, cells derived from both the Ldb1−/− and wild-type ES cells grown as adherent monolayer culture expressed the general neuronal cell marker MAP2 or the glial cell marker GFAP. To determine whether the deletion of Ldb1 affects the potential of the ES cells to generate dopaminergic neurons, we examined the expression of multiple markers of the dopaminergic neurons in cells derived from wild-type or Ldb1−/− ES cells after differentiation by quantitative RT-PCR. Our analysis revealed that the expression of all these markers, including Nurr1, Pitx3, DAT (encoding a dopamine transporter), and TH, was significantly reduced in cells derived from the Ldb1−/− ES cells compared with those derived from the control ES cells (Fig. 5B, C). In addition, the generation of other differentiated neuronal cell types, such as the cholinergic neurons, serotonergic neurons, and olfactory bulb neurons from the Ldb1−/− ES cells, was also impaired, as shown by the reduction in expression of their respective markers, choline acetyltransferase (ChAT), serotonin transporter (Sert), and olfactory marker protein (OMP) (Fig. 5B). In contrast, the expression of members of the LIM homeodomain gene family, including Lhx1, Lhx2, Lhx5, Lhx6, Lhx8, and Lmx1b, the general neuronal cell marker MAP2, and the neural stem cell marker Dcx was not significantly changed in cells derived from the Ldb1−/− ES cells compared with those derived from the control ES cells (Fig. 5B).
FIG. 5.
Difference in gene expression between neurons derived from wild-type (WT) and Ldb1−/− ES cells. (A, A′) Both wild-type and Ldb1−/− ES cells were successfully differentiated into neurons (MAP2+) and glial cells (GFAP+) by the adherent monolayer culture method. Cells were immunostained with anti-MAP2 (green) and anti-GFAP (red) antibodies. (B) Left, quantitative RT-PCR products of molecular markers expressed in neurons derived from wild-type and Ldb1−/− ES cells. Right, graph shows the relative expression levels of the various markers compared with wild type, normalized with GAPDH, and presented as mean ± SEM. (C) Left, quantitative RT-PCR products of mDA neuron-specific markers expressed in neurons derived from wild-type and Ldb1−/− ES cells. Right, graph shows the relative expression levels of the markers compared with wild type, normalized with GAPDH, and presented as mean ± SEM. The mark “**” indicates a difference with statistical significance examined by Student's t-test (P < 0.01) between neurons derived from wild-type and Ldb1−/− ES cells. RT-PCR, reverse transcription polymerase chain reaction.
Discussion
Our study revealed that a conditional deletion of Ldb1 in the En1-expressing region of the developing mid- and hindbrain severely impaired the function of the isthmic organizer as demonstrated by the absence in the expression of Fgf8 and Wnt1 (Supplementary Fig. S4). Consistent with the crucial roles of FGF8 and Wnt1 in patterning of the mid- and hindbrain in mouse embryonic development [29–33], the midbrain was drastically reduced in size, and the cerebellum was almost entirely missing in Ldb1 conditional mutant. In a previous study, identical phenotype was observed in Lmx1b mutant embryos [7]. The identical phenotypes observed in both Lmx1b and Ldb1 mutants support the idea that Lmx1b and Ldb1 work cooperatively to maintain or regulate the function of the isthmic organizer for mid- and hindbrain patterning.
In addition to the defect in patterning of the mid- and hindbrain regions, the mDA neurons were largely missing in the Ldb1 conditional mutant, as shown by the absence of the staining of TH, Pitx3, and Nurr1. Lmx1b and the closely related Lmx1a are required for the specification, proliferation, and differentiation of the mDA progenitors in the ventral midbrain [8,9,34]. The function of Lmx1a and Lmx1b in the mDA progenitors may also be dependent on Ldb1. However, the missing of mDA neurons analyzed in this study could also be caused by the disruption of the signaling of Fgf8, Wnt1, and Shh and the early patterning defect of the midbrain. A clear demonstration of the cell-autonomous role for Ldb1 in development of the mDA progenitors in vivo will require the generation and analysis of mutant with a more specific deletion of the Ldb1 in the mDA progenitors using other mouse line, such as the ShhCre line, to drive the expression of the Cre recombinase.
To address the issue whether Ldb1 plays a cell-autonomous role in the specification and differentiation of mDA neurons, we instead took an alternative approach by assessing the potential of the Ldb1−/− mouse ES cells for the generation of mDA neurons. Our result showed that while the Ldb1−/− ES cells still retained the ability to generate cells expressing the general neuronal or glial cell markers, they failed to produce cells expressing specific markers for mDA neurons. A previous gain-of-function study has revealed that overexpression of Lmx1a induces robust generation of mDA from the mouse ES cells [10]. Based on our result, this induction of ES cells for generation of mDA neurons by the Lmx1a may be dependent on the presence of Ldb1. Despite the impaired expression of specific markers of mDA neurons, RT-PCR analysis showed that Lmx1b and other LIM homeodomain genes, including Lhx2, Lhx5, Lhx6, and Lhx8, were expressed in neurons derived from the Ldb1−/− ES cells. This indicates that while these genes are expressed, the proper function of their products is critically dependent on Ldb1. Previous studies have shown that Ldb1 plays a central role in the formation of protein complexes that are essential for the function of the various LIM homeodomain proteins [13,20,21,35]. Lmx1b has been shown to interact with Ldb1 [36]. Therefore, although the expression of Lmx1b was not significantly affected, its function in development of mDA neurons was impaired by the deletion of the Ldb1.
Interestingly, in addition to mDA neurons, the cells derived from the Ldb1−/− ES cells were also impaired in expression of specific markers for other terminally differentiated neuronal cell types, including the cholinergic (ChAT+), serotonergic (Sert+), and olfactory bulb (OMP+) neurons. It has been shown that various LIM homeodomain factors, such as Lhx8, Lmx1b, and Lhx2, are required, respectively, for the generation of these neurons [37–40]. Our results present here suggest that the generation of these neurons may also require Ldb1. Recently, analysis of the Ldb1/Nkx2.1-Cre conditional mutant showed that Ldb1 is indeed required in vivo for the development of the cholinergic neurons in the telencephalon [21]. The role of Ldb1 in development of the serotonergic and olfactory bulb sensory neurons in vivo remains to be determined.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Wolfgang Wurst for providing the En1Cre mice, Dr. Andrew McMahon for providing the probes of Shh and Wnt1 for in situ hybridization, and Dr. Paul Love for providing the Ldb1 antibody. This work was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (grant number HI14C3347), the National Research Foundation of Korea (NRF-2012M3A9C7050135), and the Intramural Research Program of the NICHD/NIH.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ang SL. (2006). Transcriptional control of midbrain dopaminergic neuron development. Development 133:3499–3506 [DOI] [PubMed] [Google Scholar]
- 2.Prakash N. and Wurst W. (2006). Development of dopaminergic neurons in the mammalian brain. Cell Mol Life Sci 63:187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenas E, Denham M. and Villaescusa JC. (2015). How to make a midbrain dopaminergic neuron. Development 142:1918–1936 [DOI] [PubMed] [Google Scholar]
- 4.Blaess S. and Ang SL. (2015). Genetic control of midbrain dopaminergic neuron development. Wiley Interdiscip Rev Dev Biol 4:113–134 [DOI] [PubMed] [Google Scholar]
- 5.Adams KA, Maida JM, Golden JA. and Riddle RD. (2000). The transcription factor Lmx1b maintains Wnt1 expression within the isthmic organizer. Development 127:1857–1867 [DOI] [PubMed] [Google Scholar]
- 6.O'Hara FP, Beck E, Barr LK, Wong LL, Kessler DS. and Riddle RD. (2005). Zebrafish Lmx1b.1 and Lmx1b.2 are required for maintenance of the isthmic organizer. Development 132:3163–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo C, Qiu HY, Huang Y, Chen H, Yang RQ, Chen SD, Johnson RL, Chen ZF. and Ding YQ. (2007). Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development 134:317–325 [DOI] [PubMed] [Google Scholar]
- 8.Deng Q, Andersson E, Hedlund E, Alekseenko Z, Coppola E, Panman L, Millonig JH, Brunet JF, Ericson J. and Perlmann T. (2011). Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development 138:3399–3408 [DOI] [PubMed] [Google Scholar]
- 9.Yan CH, Levesque M, Claxton S, Johnson RL. and Ang SL. (2011). Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci 31:12413–12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T. and Ericson J. (2006). Identification of intrinsic determinants of midbrain dopamine neurons. Cell 124:393–405 [DOI] [PubMed] [Google Scholar]
- 11.Lin W, Metzakopian E, Mavromatakis YE, Gao N, Balaskas N, Sasaki H, Briscoe J, Whitsett JA, Goulding M, Kaestner KH. and Ang SL. (2009). Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol 333:386–396 [DOI] [PubMed] [Google Scholar]
- 12.Nakatani T, Kumai M, Mizuhara E, Minaki Y. and Ono Y. (2010). Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev Biol 339:101–113 [DOI] [PubMed] [Google Scholar]
- 13.Hobert O. and Westphal H. (2000). Functions of LIM-homeobox genes. Trends Genet 16:75–83 [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Malik N. and Westphal H. (2006). Functions of LIM-homeodomain proteins in the development of the nervous system. In: Transcription Factors in the Nervous System. Krieg H, ed. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, pp 75–94 [Google Scholar]
- 15.Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB. and Westphal H. (1996). Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384:270–272 [DOI] [PubMed] [Google Scholar]
- 16.Jurata LW, Kenny DA. and Gill GN. (1996). Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci U S A 93:11693–11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bach I, Carriere C, Ostendorff HP, Andersen B. and Rosenfeld MG. (1997). A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev 11:1370–1380 [DOI] [PubMed] [Google Scholar]
- 18.Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D. and Westphal H. (2003). Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development 130:495–505 [DOI] [PubMed] [Google Scholar]
- 19.Tzchori I, Day TF, Carolan PJ, Zhao Y, Wassif CA, Li L, Lewandoski M, Gorivodsky M, Love PE, et al. (2009). LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136:1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR. and Westphal H. (2007). LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci U S A 104:13182–13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Flandin P, Vogt D, Blood A, Hermesz E, Westphal H. and Rubenstein JL. (2014). Ldb1 is essential for development of Nkx2.1 lineage derived GABAergic and cholinergic neurons in the telencephalon. Dev Biol 385:94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA. and Joyner AL. (2000). Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev 14:1377–1389 [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang M, Gorivodsky M, Kim M, Westphal H. and Geum D. (2008). The neuronal differentiation potential of Ldb1-null mutant embryonic stem cells is dependent on extrinsic influences. Stem Cells 26:1490–1495 [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson DG, Bhatt S, Chavrier P, Bravo R. and Charnay P. (1989). Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature 337:461–464 [DOI] [PubMed] [Google Scholar]
- 25.Gorivodsky M. and Lonai P. (2003). Novel roles of Fgfr2 in AER differentiation and positioning of the dorsoventral limb interface. Development 130:5471–5479 [DOI] [PubMed] [Google Scholar]
- 26.Ying QL, Stavridis M, Griffiths D, Li M. and Smith A. (2003). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21:183–186 [DOI] [PubMed] [Google Scholar]
- 27.Li L, Jothi R, Cui K, Lee JY, Cohen T, Gorivodsky M, Tzchori I, Zhao Y, Hayes SM, et al. (2011). Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol 12:129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21:70–71 [DOI] [PubMed] [Google Scholar]
- 29.Chi CL, Martinez S, Wurst W. and Martin GR. (2003). The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development 130:2633–2644 [DOI] [PubMed] [Google Scholar]
- 30.Meyers EN, Lewandoski M. and Martin GR. (1998). An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet 18:136–141 [DOI] [PubMed] [Google Scholar]
- 31.McMahon AP. and Bradley A. (1990). The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62:1073–1085 [DOI] [PubMed] [Google Scholar]
- 32.Thomas KR. and Capecchi MR. (1990). Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature 346:847–850 [DOI] [PubMed] [Google Scholar]
- 33.McMahon AP, Joyner AL, Bradley A. and McMahon JA. (1992). The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69:581–595 [DOI] [PubMed] [Google Scholar]
- 34.Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL. and Burbach JP. (2000). A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci 3:337–341 [DOI] [PubMed] [Google Scholar]
- 35.Thaler JP, Lee SK, Jurata LW, Gill GN. and Pfaff SL. (2002). LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 110:237–249 [DOI] [PubMed] [Google Scholar]
- 36.Marini M, Bongers EM, Cusano R, Di Duca M, Seri M, Knoers NV. and Ravazzolo R. (2003). Confirmation of CLIM2/LMX1B interaction by yeast two-hybrid screening and analysis of its involvement in nail-patella syndrome. Int J Mol Med 12:79–82 [PubMed] [Google Scholar]
- 37.Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL. and Westphal H. (2003). The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci U S A 100:9005–9010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL. and Chen ZF. (2003). Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci 6:933–938 [DOI] [PubMed] [Google Scholar]
- 39.Kolterud A, Alenius M, Carlsson L. and Bohm S. (2004). The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development 131:5319–5326 [DOI] [PubMed] [Google Scholar]
- 40.Hirota J. and Mombaerts P. (2004). The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci U S A 101:8751–8755 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.