Abstract
Delivery of toxins, such as the ricin A chain, Pseudomonas exotoxin, and gelonin, using antibodies has had some success in inducing specific toxicity in cancer treatments. However, these antibody-toxin conjugates, called immunotoxins, can be bulky, difficult to express, and may induce an immune response upon in vivo administration. We previously reported delivery of a recombinant variant of gelonin (rGel) by the full-length prostate-specific membrane antigen (PSMA) binding aptamer, A9, to potentially circumvent some of these problems. Here, we report a streamlined approach to generating aptamer-rGel conjugates utilizing a chemically synthesized minimized form of the A9 aptamer. Unlike the full-length A9 aptamer, this minimized variant can be chemically synthesized with a 5′ terminal thiol. This facilitates the large scale synthesis and generation of aptamer toxin conjugates linked by a reducible disulfide linkage. Using this approach, we generated aptamer-toxin conjugates and evaluated their binding specificity and toxicity. On PSMA(+) LNCaP prostate cancer cells, the A9.min-rGel conjugate demonstrated an IC50 of ∼60 nM. Additionally, we performed a stability analysis of this conjugate in mouse serum where the conjugate displayed a t1/2 of ∼4 h, paving the way for future in vivo experiments.
Introduction
Delivery of plant and bacteria-based toxins via antibodies and other protein ligands have been explored as an alternative to small molecule and peptide chemotherapeutic cargoes. Many of these proteins have been engineered so they are unable to cross the cellular membrane without the assistance of a targeting ligand. This can greatly decrease off target effects and allow a higher tolerable dose. Variants of the Pseudomonas exotoxin, Shiga toxin, and ricin A chain have been explored as toxic cargoes [1]. Typically, these have been delivered chemically conjugated to an antibody by disulfide bonds. Later generations of immunotoxins have been produced as recombinant fusion proteins.
Aptamers that target cell surface receptors represent an alternative to other macromolecules (eg, antibodies) for the development of targeted toxins. A number of different aptamers that target cell surface receptors have been adapted for the delivery of a range of cargoes including small molecule drugs [2–4], nanoparticles [5,6], and even siRNA [7,8]. We previously utilized an aptamer, A9, which targets the prostate-specific membrane antigen (PSMA) to deliver the ribosomal toxin gelonin, a ricin A chain homolog, to PSMA-expressing prostate cancer cells [9]. On its own, gelonin lacks the ability to enter cells (IC50 ∼1–10 μM; [10]). However, when conjugated to A9, the conjugate demonstrated an IC50 of ∼30 nM on PSMA(+) LNCaP prostate cancer cells [9].
In our previous work, we generated aptamer-toxin conjugates using the full-length A9 aptamer. The aptamer was made via in vitro transcription, gel purified, and conjugated to recombinant gelonin (rGel) produced in Escherichia coli through the use of a heterobifunctional linker, SPDP [9]. The use of a full-length aptamer and the requirement for production by in vitro transcription limits the feasibility of generating large quantities (kg) of aptamer-toxin conjugates for potential use as a therapeutic. More immediately though, even the production of milligram quantities of aptamer toxin conjugates for small animal testing using in vitro transcribed RNA presents a significant challenge. There was need for the design of methods, which can be readily scaled to produce aptamer toxin conjugates.
Here, we report a streamlined method for the synthesis of aptamer rGel conjugate, which relies on the chemical synthesis of a minimized aptamer. More specifically, we synthesized a minimized variant of the A9 aptamer bearing a 5′ thiol. Following purification, the aptamer was efficiently converted to a thiol reactive pyridyl disulfide activated aptamer, which was then used to generate aptamer toxin (rGel) conjugates bearing a reducible disulfide linkage. Using this approach, we synthesized and evaluated an A9.min-rGel conjugate and compared its cell binding and cytotoxicity properties to that of a nontargeted control toxin conjugate. The IC50 of the aptamer-toxin conjugate was comparable to previously reported values [9]. Finally, in preparation for in vivo experiments, we evaluated the stability of the aptamer-toxin conjugate in whole serum. Our synthetic approach should prove applicable for the development of conjugates composed of other aptamers and other toxins. Additionally, the molecule produced here should prove useful for future studies in animals.
Method and Materials
Cells and cell culture
Cell lines were obtained from the ATCC and maintained according to their recommendations. LNCaP cells were grown and assayed in RPMI media with 10% fetal bovine serum (FBS). PC3 cells were grown in F12K media supplemented with 10% FBS. The PSMA expressing PC3-PSMA cell line was generated by stable transfection of cells with an EF1a-promoter-driven human PSMA expression cassette (pEF6-hPSMA). PSMA-expressing cells were enriched by FACS using an anti-PSMA antibody (Invitrogen, Carlsbad, CA). To maintain expression, cells were grown in their appropriate media supplemented with 5 μg/mL of blasticidin. All cell lines were routinely tested to insure they were free of mycoplasma contamination using MycoAlert mycoplasma detection kit (Lonza, Basel, Switzerland).
Aptamer synthesis
The A9.min (anti-PSMA) and C36 (nontargeting control) aptamers were synthesized in house by solid phase synthesis on an Expedite 8909 DNA synthesizer (Biolytic Lab Performance, Fremont, CA). All reagents were purchased from Glen Research (Sterling, VA) and Chemgenes (Wilmington, MA). A9.min (GGG ACC GAA AAA GAC CUG ACU UCU AUA CUA AGU CUA CGU UCC C) and C36 (GGC GUA GUG AUU AUG AAU CGU GUG CUA AUA CAC GCC) were synthesized using 2′-fluoro-modified pyrimidines and 2′OH purines on an inverted dT CPG column. All aptamers were synthesized bearing a 5′ thiol using a C6 S-S phosphoramidite with the final dimethoxytrityl group on to facilitate purification. Support cleavage and base deprotection was achieved by treatment with a 1:1 mixture of aqueous ammonium hydroxide (30%) and aqueous methylamine (40%) for 10 min at 65°C. Lyophilized samples were treated with a 250 μL mixture of TEA-3HF and TEA in NMP (2:1:0.67 v/v/v) for 2.5 h at 55°C to remove the 2′ TOM protecting groups. The deprotection reaction was quenched, and the RNA was precipitated by the addition of 550 μL trimethylmethoxysilane and washed with 100% ethanol before drying. The resulting pellet was resuspended in 200 μL 2 M TEAA (triethylammonium acetate, pH 7.5). All aptamers were purified by reversed-phase HLPC on a 10 × 50 mm Xbridge C18 column heated to 65°C (Waters, Milford, MA) using a linear gradient of acetonitrile in 0.1 M TEAA at pH 7.5.
Labeling aptamers with AF488
Aptamers with a 5′ thiol modification were reduced in 0.1 M TEAA with 10 mM TCEP by heating for 3 min at 70°C followed by incubation at room temperature for 1 h. The reduced aptamers were desalted into PBS without Mg2+ or Ca2+ to remove excess TCEP using a Bio-Spin 6 micro spin column (Bio-Rad Hercules, CA) and reacted with a two to fivefold molar excess of maleimide-activated Alexa Fluor 488 (AF488; Life Technologies, Carlsbad, CA). The aptamer and dye were incubated at room temperature for 1 h or overnight at 4°C. Typical labeling reactions proceeded to ∼100% as determined by HPLC. Excess free dye was removed by desalting labeled aptamers into PBS with a Bio-Spin 6 column.
Flow cytometry assays
LNCaP cells were plated at 2 × 104 cells per well in tissue culture treated 96-well plates 48 h before assay. PC3 and PC3 PSMA cells were plated 18–24 h before assay at 3 × 104 in 96-well plates. Ninety microliters of fresh media containing 1 mg/mL deoxyribonucleic acid, sodium salt, salmon testes (ssDNA; Millipore, Billerica, MA) was added to the cells 1 h before the incubation with aptamers. AF488-labeled aptamers were diluted to a 10× stock in PBS without Mg2+ or Ca2+ and heated to 70°C for 3 min. The aptamers were allowed to renature at room temperature for 15 min before addition to the wells. Cells and aptamers were incubated together for 1 h at 37°C. Media were removed, and cells were washed twice with 50 μL PBS without Mg2+ or Ca2+ and lifted from the plate with 0.5% trypsin. Trypsin was inactivated with FACS buffer (Hank's balanced salt solution with 1% bovine serum albumin and 0.1% sodium azide). Cells were pelleted in a swing bucket centrifuge at 300 g for 5 min and resuspended in FACS buffer with bisbenzimide to differentiate live and dead cells. Assays were read on an iCyt Eclipse, EC800, flow cytometer and analyzed using Flow Jo.
For antibody control assays, 105 cells were stained with either an anti-PSMA antibody labeled with Alexa Fluor 488 or an isotype control (Sony Biotechnology, San Jose, CA). The cells were stained in 100 μL of FACS buffer for 30 min at room temperature. Cells were washed three times with 200 μL of FACS buffer and resuspended in FACS with bisbenzimide. Assays were read on an iCyt Eclipse, EC800, flow cytometer and analyzed using Flow Jo software.
Protein expression and purification
Recombinant rGel was expressed in E. coli as an N-terminal 6× His tag fusion from synthetic genes, as reported by Rosenblum et al., with amino acids 112–119 deleted and codons optimized for expression in E. coli (Genscript, Piscataway, NJ) in the pET28a backbone [11]. The protein also contained a Cys to Ala mutation, leaving only one Cys residue in the protein, which allows site specific conjugation [12]. The expression vector was transfected into Single Step KRX competent cells (Promega, Madison WI). A single colony was picked for a 5 mL starter culture grown overnight in LB with 50 μg/mL kanamycin. One milliliter of starter culture was added to each of four baffled flasks of 250 mL LB with 50 μg/mL kanamycin. Flasks were grown to an OD of 0.5, and expression was induced with 0.2% rhamnose overnight in a shaking incubator at 25°C.
Bacteria were pelleted in a fixed angle centrifuge at 4,000 g for 10 min and resuspended in sonication buffer (50 mM NaPO4 pH 8.0, 300 mM NaCl, 0.1% Triton X, 3 mM imidazole, and 1× mini-cOmplete protease [Sigma, St. Louis, MO]). Cells were lysed on ice by sonication using a microtip sonicator (Misonix, Farmingdale, NY) at an amplitude of 20 for ten 30 s intervals on ice with 30 s off between intervals to avoid overheating. After sonication, the bacteria were centrifuged at 4,000 g for 10 min to pellet the insoluble fraction. Six milliters of Ni-NTA (Qiagen, Venlo, Netherlands), for a bed volume of 3 mL, was equilibrated with three bed volumes of buffer A (1X Tris-buffered saline and Tween 20 (TBST), 300 mM NaCl, 10 mM imidazole). The resin was then transferred to a 50 mL conical with the soluble fraction and incubated with rotation for 1 h at 4°C. Resin and soluble fraction were applied to the column and washed with 10 bed volumes buffer A and 5 bed volumes buffer A+ (1X TBST, 300 mM NaCl, 20 mM imidazole). Protein was eluted with elution buffer (1X TBST, 300 mM NaCl, 250 mM imidazole). The purified protein was dialyzed into TBST overnight for storage.
The 6× His tag was removed via a thrombin cleavage site using thrombin CleanCleave Kit (Sigma). Briefly, protein was exchanged into cleavage buffer using Amicon Ultra 3k columns (Millipore) and incubated with thrombin resin for 1 h with rotation at room temperature and overnight at 4°C. Protein was recovered, dialyzed into PBS, and stored in 40% glycerol at −80°C.
Rabbit reticulocyte assay
We assessed the activity of recombinant gelonin using a rabbit reticulocyte lysate system (Promega) and Luciferase Control RNA (Promega). The reactions were set up according to the manual for a luciferase control reaction without mRNA. The recombinant protein was preincubated with lysate for 45 min before the addition of mRNA. After addition of luciferase mRNA, the reaction was incubated at room temperature for 90 min. Luciferase activity was monitored using a Synergy H4 hybrid plate reader (Biotek, Winooski, VT). Luciferase activating reagent was warmed to room temperature and injected into each well containing 2.5 μL of lysate reaction in a white 96-well plate and read 10 s after injection.
Pyridyl activation of aptamer
Aptamers with a 5′ thiol modification were reduced in 0.1 M TEAA with 10 mM TCEP. The reaction was heated to 70°C for 3 min and incubated at room temperature for greater than 1 h. Dithiodipyridine was dissolved in acetonitrile at 250 mM and added to the reaction for a final concentration of 25 mM. After 1 h at room temperature, the reaction was checked by analytical HPLC to confirm completion. Reactions routinely proceeded to ∼100%. Aptamers were desalted into 0.1 M NH4HCO3 using a Bio-Spin 6 column to remove excess dithiodipyridine and lyophilized for storage.
Protein conjugation
The toxin was exchanged into NaPO4+ Buffer (0.1 M NaPO4 pH 8.0 with 1 mM EDTA and 1 M NaCl) and concentrated to ∼300 μM using an Amicon Ultra 3K filter. Conjugation reactions were performed at a 5:1 protein to aptamer ratio, and the reaction was allowed to proceed at room temperature for 1 h followed by 4°C overnight. Conjugation reactions were monitored by acrylamide gel stained with SYBR gold.
Conjugate purification
Aptamer-rGel conjugates were purified using anion exchange spin columns (Pierce, Rockford, IL). Conjugates were first exchanged in PBS using Amicon Ultra 10K spin columns. Conjugates were then applied to the anion spin columns and washed with PBS containing 0.15 M NaCl. Conjugates were eluted in six 500 μL elutions of PBS with increasing amounts of NaCl. The first elution was 0.75 M NaCl, and the NaCl concentration was increased 0.05 M for each subsequent elution. The elutions were checked by acrylamide gel, and elutions containing conjugate with minimal or no free aptamer were buffered exchanged into PBS via an Amicon Ultra 30K spin column.
Viability assays
Cells were plated in 90 μL of media with 1 mg/mL ssDNA at 3 × 103 per well in a 96-well plate. Aptamer-conjugated toxin and free toxin were prepared in 10× stocks in PBS and added to wells. Cells were incubated at 37°C for 5 days with the conjugates. Cell viability was checked using the Alamar Blue cell viability reagent (Invitrogen) and plotted as a fraction of untreated control cell growth.
Serum stability assays
Whole blood was collected from C57BL/6 mice by terminal cardiac puncture. Whole blood was allowed to clot by incubating at room temperature for 20 min, and serum collection was performed by centrifugation in a fixed angle rotor at 2,000 g for 15 min. Aptamer directly labeled with AF488 or conjugated to rGel in 10 μL PBS was added to 90 μL of serum and incubated at 37°C. Conjugates were recovered by Clarity OTX columns according to the column protocol (Phenomenex, Torrance, CA).
Aptamers directly labeled with AF488 were recovered by methanol chloroform extraction [13]. Briefly, 50 μL of PBS, 50 μL of chloroform, and 100 μL of methanol were added to 10 μL of serum. After vortexing, 50 μL of dH2O and an additional 50 μL of chloroform were added and vortexed again. After centrifuging at 500 g for 20 min, the aqueous phase containing the aptamer was recovered. The recovered aptamer and conjugate were run on a 12% polyacrylamide denaturing gel, scanned using a Storm 840 imaging system (GE Healthcare, Marlborough, MA) and quantified using LICOR Lite software. First order rates were determined from the slope of the line produced from a linear fit of data.
Results
In vitro characterization of a minimized anti-PSMA binding aptamer
We chemically synthesized a 43 nucleotide minimized version of A9 (denoted here as A9.min) based on the minimal sequence reported by Archemix [14] and more recently reported by Rockey et al. [15]. The same minimized sequence was identified from an analysis of a doped selection performed by our own group using a library based on the full-length A9 ([16] and unpublished results). Because the bulk of the reported characterization of this aptamer relied on the inhibition of the enzymatic activity of PSMA [14,15], we initially preformed a cell-based analysis by flow cytometry to confirm the specificity and determine the apparent binding constant for this molecule on PSMA-positive cells.
In short, we synthesized the 2′-fluoro-modified (2′F) RNA aptamer, A9.min, and a nontargeting control sequence, C36 [6,17], in house, bearing a 5′ thiol modifier protected as a disulfide. Following reduction, the reduced aptamers were labeled with a maleimide functionalized derivative of AlexaFluor 488 producing the labeled molecules AF488-A9.min or AF488-C36. Conjugation was efficient and typically proceeded to 100% based on analysis by HPLC. The resulting conjugates were purified by desalting, and the absence of any unconjugated dye was confirmed by reverse phase HPLC.
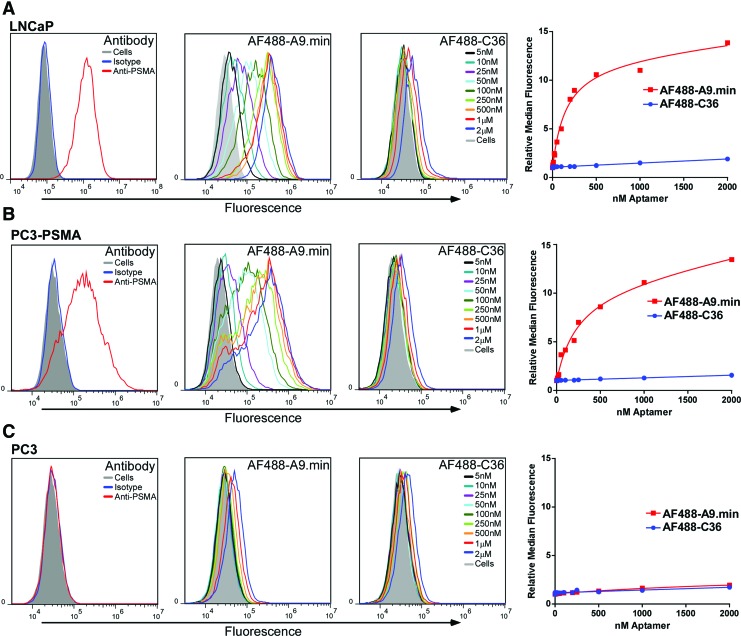
Cell staining experiments were performed using two PSMA-positive prostate cancer cell lines: LNCaP cells, which naturally express PSMA, and PC3-PSMA cells, a prostate line engineered by our lab to express this receptor. As a negative control, assays were also performed on PC3 cells, which are naturally PSMA negative. The PSMA expression status for each cell line was confirmed by flow cytometry using a commercially available anti-PSMA antibody or an isotype control (Fig. 1). For aptamer staining, cells were treated in the presence of a nonspecific inhibitor, ssDNA, to reduce variation in levels of nonspecific, background binding seen on the different cell lines under conditions which allow for endocytosis [16]. In short, increasing concentrations of the thermally equilibrated aptamer or control was added to adherent cells in media and allowed to incubate at 37°C for 1 h. After washing, the cells were lifted from the plate and analyzed by flow cytometry. The signal observed by cytometry is thus a combination of any remaining surface bound as well as internalized aptamers.
FIG. 1.
Cell binding and PSMA specificity analysis of the anti-PSMA aptamer A9.min. (A) LNCaP, (B) PC3-PSMA, and (C) PC3 cells were incubated with an anti-PSMA antibody, and isotype controls or increasing concentrations of AF488-labeled A9.min or a nonfunctional aptamer control, AF488-labeled C36. LNCaP and PC3-PSMA cells both express PSMA. PC3 cells are a PSMA-negative cell line as confirmed by antibody staining. Cell staining experiments were performed in growth media containing 10% FBS supplemented with 1 mg/mL ssDNA as a non-specific blocking agent. Incubations were performed for 1 h after which the cells were washed to remove unbound aptamers, trypsinized from the plate and analyzed by flow cytometry. The identity and concentration of each plot in the histograms are as indicated. For each data set, we also generated a plot of the relative median fluorescence intensity versus concentration, which was used to determine the apparent binding constant (appKd) on each cell type. The appKd values are reported in Table 1. FBS, fetal bovine serum; PSMA, prostate-specific membrane antigen. Color images available online at www.liebertpub.com/nat
As shown in Fig. 1, AF488-A9.min stained both the PSMA expressing LNCaP cells and our engineered PC3-PSMA cell line, and displayed an apparent binding constant (appKd) of 190 and 320 nM respectively (Table 1). The control aptamer, AF488-C36, displayed essentially no staining under these conditions (appKd >>10uM; Table 1). Moreover, consistent with its specificity for PSMA, AF488-A9.min failed to stain the parental PC3 cells, which do not express PSMA, and displayed a level of binding similar to that observed for C36, our negative control (appKd >10uM; Table 1). For comparison the relative expression level of PSMA one each cell type is also shown (Fig. 1).
Table 1.
Summary of Apparent Binding Constants and IC50 Values for Fluorescently Labeled Aptamers, and Aptamer Toxin Conjugates
| Apparent Kd (μM)/IC50 | |||
|---|---|---|---|
| LnCap | PC3 PSMA | PC3 | |
| AF488-A9.min | 0.19 ± 0.010 | 0.32 ± 0.03 | >>10 |
| A9.min-rGel-AF488 | 0.12 ± 0.08/0.06* | 0.27 ± 0.0.2/0.09** | >>10/>1.0 |
| AF488-C36 | >>10 | >>10 | >>10 |
| C36-rGel-AF488 | >>10/>1.0 | >>10/>1.0 | >>10/>1.0 |
The apparent binding constants, shown in black, were determined from a best fit of the median fluorescent intensity versus aptamer concentration (Figs. 1 and 2C–E) on the cell line indicated. For constructs tested containing the nontargeting control aptamer, C36, or tested on non-PSMA expressing PC3 cells, reliable fits could not be obtained. The apparent Kd for these constructs are estimated and, being significantly greater than 10 μM (>>10). IC50 values, shown in red, were determined from the data shown in Fig. 3. Values were determined for the conjugate indicated but without the fluorescent label. For constructs tested containing the nontargeting control aptamer, C36, or tested on non-PSMA expressing PC3 cells, reliable fits could not be obtained and are estimated to be >1.0 μM (>1.0). *95% confidence interval 0.04–0.08. **95% confidence interval 0.06–0.12.
PSMA, prostate-specific membrane antigen.
Synthesis of A9.min-rGel conjugates
Having confirmed the binding specificity of A9.min on PSMA-positive cells, we next turned our attention to toxin conjugate synthesis. We produced recombinant gelonin (rGel) engineered to contain a single solvent accessible cysteine residue [11] as a His-tagged protein in E. coli followed by purification with a Ni-NTA column. The His tag was subsequently removed by thrombin cleavage.
We confirmed the function of the protein using a rabbit reticulocyte luciferase expression system to test the ability of these proteins to inhibit ribosomal activity (Supplementary Fig S1; Supplementary Data are available online www.liebertpub.com/nat), which yielded an IC50 value of ∼50 pM, consistent with literature values [11].
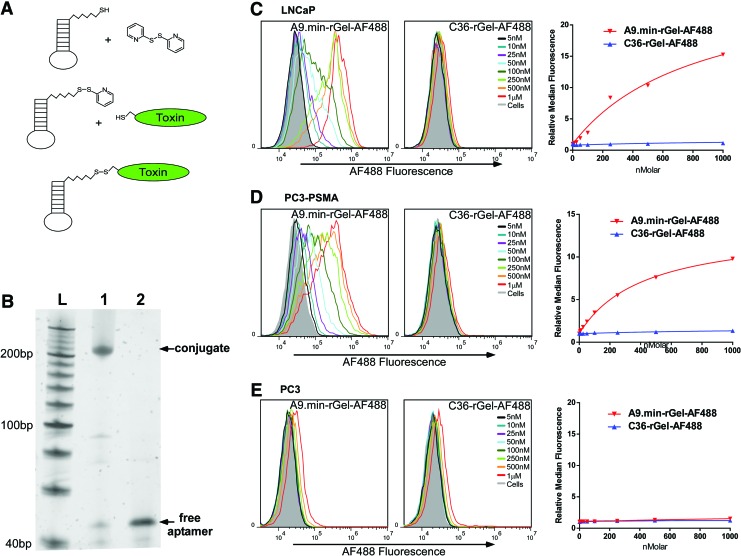
To chemically link rGel to A9.min or C36, the aptamers were first activated with a pyridyldisulfide group by reacting the reduced aptamer with 2′,2-dithiodypyridine (DTDP; Fig. 2A). The activation reaction generated the stable pyridyldisulfide-aptamer intermediate, which was monitored by analytical reverse phase HPLC (Supplementary Fig S2) and routinely proceeded to 100%. Excess DTDP was removed by simple desalting. Protein conjugation was subsequently achieved by incubating the activated aptamers with a three to fivefold molar excess of protein. Reactions typically proceeded to ∼90% overnight and were further purified by ion exchange chromatography to remove excess protein followed by size exclusion to remove any remaining aptamer (Fig. 2B; lane 1). Finally, we treated the purified aptamer conjugate with the reducing agent, TCEP, to confirm the presence of the disulfide linkage. Treatment resulted in complete release of the aptamer (Fig. 2B; lane 2).
FIG. 2.
Synthesis and binding characteristics of A9.min-rGel. (A) Synthesis scheme for the generation of aptamer-toxin conjugates using a 5′ thiol modified aptamer. (i) The reduced aptamer bearing a 5′ thiol is activated by reaction with dithiodipyridine (DTDP). (ii) The activated, thiol reactive aptamer is incubated with target protein bearing a single Cys residue. (iii) The resulting conjugate bears a reducible disulfide linkage. (B) Electrophoretic analysis of purified A9.min-rGel before (lane 1) and after treatment with the reducing agent TCEP (lane 2). (C) LNCaP, (D) PC3-PSMA, and (E) PC3 cells were incubated with increasing concentrations of an A9.min-rGel conjugate in which the protein was labeled with AF488 (A9.min-rGel-AF488) or a nonfunctional aptamer conjugate control (C36-rgel-AF488). LNCaP and PC3-PSMA cells both express PSMA. PC3 cells are a PSMA-negative cell line. Cell staining experiments were performed in growth media containing 10% FBS supplemented with 1 mg/mL ssDNA as a nonspecific blocking agent. Incubations were performed for 1 h after which the cells were washed to remove unbound aptamers, trypsinized from the plate, and analyzed by flow cytometry. The identity and concentration of each plot in the histograms are as indicated. For each data set, we also generated a plot of the relative median fluorescence intensity versus concentration which was used to determine the apparent binding constant (appKd) on each cell type. The appKd values are reported in Table 1. Color images available online at www.liebertpub.com/nat
Assessing cell binding and uptake of A9.min-rGel conjugates
To ensure that toxin conjugation did not impinge on aptamer function, we generated fluorescent variants of our aptamer-rGel conjugates and assayed cell binding using flow cytometry. In short, we used the amine reactive SDP ester of AlexaFluor488 to label the protein portion of the toxin conjugates. Importantly, the fluorescently labeled A9.min-rGel (A9.min-rGel-AF488) conjugate's cell staining activity closely mirrored that of the directly labeled AF488-A9.min, displaying apparent binding constants of 120 nM on LNCaP cells and 270 nM on PC3-PSMA cells (Table 1). Similarly, like the labeled control aptamer, AF488-C36, the control conjugate (C36-rGel-AF488) showed only minimal binding (appKd >10 μM) to all three cell types (Fig. 2C–E).
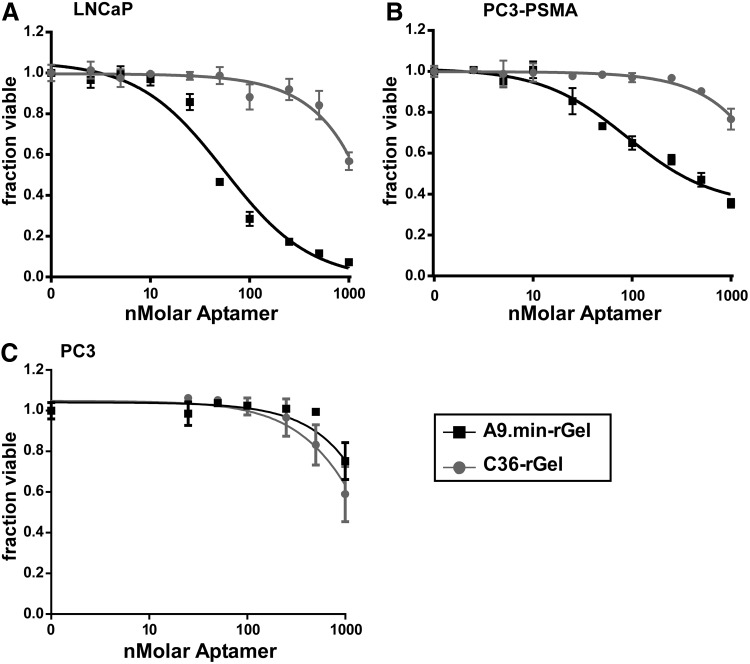
Measuring the cytotoxic activity of aptamer-rGel conjugates
Efficacy of gelonin as a cargo has been shown to be dependent upon both uptake and time [18]. To ensure the toxin had enough time to effectively inhibit cell growth, cell viability was measured after 5 days of treatment. We determined cell toxicity on our aptamer-rGel conjugates using the cell health indicator AlamarBlue. As shown in Fig. 3, the A9-rGel conjugate proved significantly more toxic to PSMA-expressing cells including both LNCaP and PC3-PSMA cells displaying IC50 values of 60 and 90 nM, respectively (Table 1). C36-rGel, on the other hand, proved considerably less toxic, displaying IC50 values of >1.0 μM (Table 1). Importantly, when we performed similar experiments using the non-PSMA expressing PC3 cells, both A9.min and C36 conjugates displayed similar IC50 values (>1.0 μM; Table 1), further confirming the requirement for PSMA expression.
FIG. 3.
Cell killing properties of A9.min-rGel. (A) LNCaP, (B) PC3-PSMA, and (C) PC3 cells were incubated with increasing concentrations of A9.min-rGel or a nonfunctional aptamer conjugate control C36-rGel. LNCaP and PC3-PSMA cells both express PSMA. PC3 cells are a PSMA-negative cell line. Assays were performed in growth media containing 10% FBS supplemented with 1 mg/mL ssDNA as a nonspecific blocking agent. Conjugates were added to cells at increasing concentrations and incubated for 5 days after which the fraction of viable cells remaining was determined. The IC50 values for each curve are reported in Table 1.
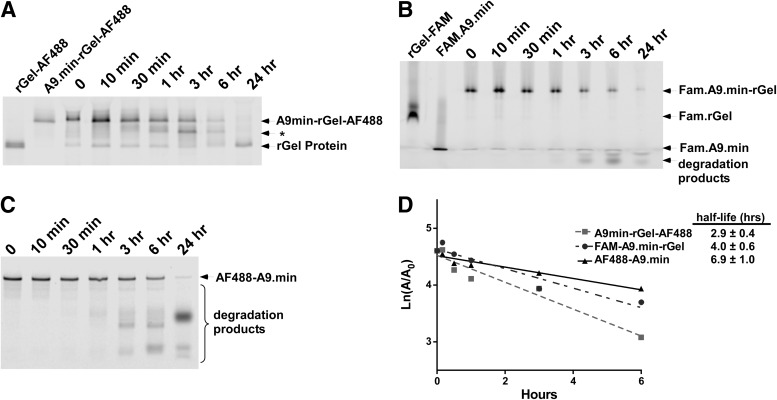
Serum stability of A9.min and A9.min-rGel conjugates
In preparation for downstream in vivo use, we assessed the stability of A9-rGel in whole mouse serum at 37°C. For these experiments, we initially utilized fluorescently labeled A9-rGel-AF488, which allowed us to target conjugate via the fluorescently labeled protein component of the conjugate following SDS page gel electrophoresis. In contrast to similar experiments performed in tissue culture media containing 10% heat inactivated FBS, where the aptamers display little, if any degradation over the course of our 5 day assays (data not shown), as shown in Fig. 4A, incubation in fresh mouse serum resulted in a progressive decrease in the amount of full-length conjugate with a stability half-life of ∼3 h. Interestingly, degradation resulted in the generation of a labeled species that displayed intermediate electrophoretic mobility between that of the full-length conjugate (A9.min-rGel-AF488) and fluorescently labeled protein alone (rGel-AF488) suggesting that the aptamer portion of the conjugate was being degraded. To better understand this, we performed two additional sets of analyses. First, we chemically synthesized an A9.min-rGel conjugate using a derivative of A9.min bearing a 5′ thiol but labeled at the 3′ end with fluorescein. Subsequent conjugation to rGel resulted in a fluorescently labeled conjugate, which allowed us to track the stability based on the aptamer (FAM.A9.min-rGel). As shown in Fig. 4B, D, FAM.A9.min-rGel displayed serum stability similar to that observed for protein labeled conjugate (A9.min-rGel-AF488) with a half-life of ∼4 h. For comparison we also assessed the serum stability of the aptamer only using AF488-A9.min, which displayed slightly increased stability Fig. 4C and half-life of ∼7 h (Fig. 4D).
FIG. 4.
Serum stability of A9.min and A9.min-rGel conjugates. (A) Analysis of A9.min-rGel conjugate in which the protein was labeled with AF488 (A9.min-rGel-AF488). (B) Analysis of an A9.min-rGel conjugate in which the aptamer was labeled at the 3′ end with FAM (FAM.A9.min-rGel). (C) Analysis of A9.min bearing a 5′ AF488 and a 3′ inverted dT residue (AF499-A9.min). Assays were performed in 100% fresh mouse serum for the time indicated. The identity of each band is as indicated. rGel-AF488 and rGel-FAM are fluorescently labeled rGel standards. FAM.A9.min is the 3′ FAM labeled A9.min aptamer and * indicates the position of the degradation product that displays intermediate electrophoretic mobility between that of the full-length conjugate and the protein alone. Sample workup before electrophoresis was performed as described in the Methods and Materials section. (D) Serum stability was determined from a plot of the area corresponding to the full-length conjugate or aptamer on the gel. The half-life for each sample is as indicated.
Discussion
The chemical nature of aptamers provides a means to incorporate almost any functional group within the molecule during synthesis. In this work, we present an improved method for the generation of aptamer-targeted toxin conjugates bearing a disulfide linkage. Indeed, in addition to the thiol modification strategy employed here, other functional groups including amine, aldehyde, aminooxy, azide, and alkyne are all commercially available and can facilitate conjugation. Here, we chose to exploit a 5′ thiol. In our approach, we incorporated this functional group as a disulfide and chose to leave the final dimethyoxytrityl protecting group (DMT) on the molecule. This facilitated reverse phase HPLC purification, as the full-length thiol modified RNA (bearing the final DMT group) displayed enhanced column retention and provided excellent separation from nonfull-length sequences (Supplementary Fig S2). Subsequent detritylation was not necessary, as the DMT was removed during reduction, and desalting further simplifying the approach. More importantly, the resulting reduced product reacted nearly quantatively with thiol reactive probes.
For toxin conjugation, we employed a strategy similar to that reported originally by Turner et al. for the conjugation of thiol modified oligonucleotides to peptides [19,20]. In this approach, we first activated our aptamers with a pyridyl thiol group by reaction with DTDP and then, following simple desalting, performed the toxin conjugation reaction, which also proceeded in high yield with only a modest excess of protein.
To demonstrate this approach, we utilized a minimized variant of the A9 anti-PSMA aptamer, A9.min, which could be readily synthesized chemically. Although this minimized molecule has previously been shown to inhibit PSMA enzymatic activity with an IC50 of ∼10 nM [14,15], to inhibit PSMA cell migration with an IC50 of ∼200 nM [21] and even to localize to tumors following systemic administration to mice bearing PSMA-positive tumors [21], assays characterizing the direct cell binding capabilities of this molecule and its specificity for PSMA in the literature were lacking detail. Therefore, we performed a set of analyses using a fluorescently labeled aptamer (AF488-A9.min) to directly determine the cell binding characteristics of this molecule. We assessed cell binding on two different PSMA-positive cell lines using flow cytometry with AF488-A9.min or a similarly labeled nontargeted control, AF488-C36. Cell staining was performed under conditions that allowed the aptamer to bind its target cell surface receptor and be endocytosed by the target cell. While we have previously found that binding assays performed on different cell types expressing the same receptor can yield similar values [6], and that such assays can be used as a proxy for assessing the Kd of an aptamer for its target on the cell surface [17] care must be taken in these interpretations, as differences in receptor density, expression level, and potential complications due to ligand depletion at low aptamer concentration may affect analysis. However, at a minimum, the approach provides a means to confirm target specificity and basally assess the binding characteristics of an aptamer for its target on live cells.
On live cells, AF488-labeled A9.min displayed clear PSMA expression-dependent binding on two PSMA-positive cell lines: LNCaP cells, which naturally express PSMA, and PC3-PSMA cells, a naturally PSMA-negative cell line, which we engineered to express this protein. Almost no binding was detected on the parental PSMA-negative PC3-cells (Fig. 1; Table 1). When we performed similar experiments using aptamer-toxin conjugates or controls, both the target specificity and the apparent binding constants remained similar, differing by only ∼2-fold (Fig. 2; Table 1). Perhaps more importantly, consistent with the observed cell binding characteristics, the A9.min-rGel conjugate displayed PSMA dependent toxicity on cells, with observed IC50s similar to those previously reported by us for conjugates generated using the full-length A9 [9] and values that roughly paralleled the apparent Kds (Table 1).
The ability of ribosomal toxins such as rGel, and many other cytotoxic molecules, is dependent on the ability to access the cytosol of the cell. For ribosomal plant toxins such as gelonin, ricin, or saporin, and bacterial toxins such as PE-38, this is an innate property of the toxin itself (reviewed in [22]) and not the targeting molecule. In the case of gelonin, cytosolic access, not cell uptake, has previously been demonstrated to be the limiting step in cell killing [18]. As aptamers are unlikely to gain access to the cytosol, ligand release within the endosomal pathway may be an important role in freeing the toxin.
For the conjugates described here, we chose to utilize a reducible disulfide linkage, as this has previously been shown to be a means to abet cargo release following endocytosis and in the mechanism by which ribosomal toxins such as ricin employ to separate the toxin A-chain from the cell surface targeting B-chain [23]. Although we note that debate remains regarding the oxidation state of the endosome and the actual mechanism of release [24,25]. While stable in solution, one potential limitation of disulfide-linked conjugates in vivo is the disulfide exchange with serum proteins resulting in cargo release. Disulfide exchange in serum rates have reported half-lives of ∼15 h [26,27]. In the case of antibody drug conjugates, which utilize this linkage, the long systemic circulation half-lives of these molecules, ∼10–20 days [27,28], has led investigators to utilize hindered disulfides that bear a methyl group adjacent to one or both sides of the disulfide linkage to further increase serum stability [26,27]. However, aptamers display clearance rates in minutes [29] which can be extended by conjugation to high molecular weight (20–40 kDa) PEG, which have reported half-lives ranging from 3 to 12 h [29–31]. rGel has a molecular weight of 28 kDa, thus we would expect our aptamer-rGel conjugates to behave similarly. Future work will need to examine this.
In our serum stability studies, we observed a half-life for our A9-rGel conjugate of ∼3 h. Breakdown of the AF488-labeled rGel conjugate appeared to be a consequence of aptamer degradation and not only reduction of the linkage, as determined by the presence of an intermediate species when analyzed by SDS-PAGE. To better visualize the conjugate while tracking the aptamer, we utilized a conjugate produced with a 3′FAM group on the aptamer. These conjugates displayed a half-life of ∼4 h, slightly longer than the protein-labeled conjugate, but still significantly shorter than the rate expected for disulfide exchange. The discrepancy between the two conjugates may be in part due to the different labeling methods, and the variation in the 3′ terminus of the aptamers, notably an inverted dT to an FAM group. However, the aptamer alone, labeled with an AF488 maleimide, displayed a serum half-life of ∼7 h. Taken together, these data suggest that the aptamer-toxin conjugates are limited by the stability of the aptamer and not the disulfide linkage. Future improvement will thus be needed to address this fact in addition to potentially stabilizing the disulfide.
In summary, we report a more streamlined approach to develop disulfide-linked targeted toxin conjugates using a minimized, chemically synthesized aptamer for targeting. To demonstrate this, we conjugated a minimized anti-PSMA aptamer, A9.min, to the ribosomal toxin gelonin and demonstrated that the conjugate maintains its specificity for PSMA-positive cells using fluorescent conjugates by flow cytometry. More importantly, we demonstrate that these conjugates display target specific toxicity. We then performed an analysis to assess the serum stability of this molecule in preparation for in vivo efficacy studies, which suggests that the limitation of the current conjugate lies in the stability of the aptamer. Future work should focus on utilizing more stabilized variants of this aptamer [14] and exploring alternative linker strategies, which may provide for optimal release.
Finally, we note that although we have chosen PSMA as the target, our approach can be readily applied to any number of different aptamers and cargoes. There are, in fact, currently a number of cell surface binding aptamers reported in the literature that could be employed for this purpose (reviewed in [32]). However, each aptamer may need to be adapted for this purpose. For example, as the length of the aptamer has a direct effect on the yield during chemical synthesis, shorter aptamers, <50 nucleotides in length, are preferred. However, in unpublished work we have synthesized, purified, and validated 2′F modified aptamers bearing a 5′ thiol modification for subsequent conjugation of up to 75 nucleotides in length using the methods described here. An additional consideration is serum stability. As noted above, the PSMA aptamer used here displayed a half-life of ∼7 h in fresh mouse serum, which would be expected to be shorter in the more nuclease-rich environment of human serum. Thus, future efforts focused on developing more stabilized molecules would seem prudent. Indeed, a more stable variant of this molecule, backfilled using a combination of 2′OMe, 2′H, and a phosphorothioate linkage, is reported to have a 20 h degradation half-life in human serum [14]. However, this 20 h half-life is still significantly shorter than those reported for other aptamers composed of 100% modified RNA that have been reported display <1% degradation over 3 days [33].
Similarly, while the current work has focused on the use of recombinant gelonin, rGel, as the cytoxic cargo, other toxins such as the ricin A-chain, which already bears a free cysteine residue, or saporin and PE-38, which can be engineered with one, could all be used in a similar manner to generate aptamer-toxin conjugates. In this respect, our conjugate synthesis method and the experiments performed here may serve as a model for others to perform thorough in vitro analyses to assess the cell-binding capabilities of both aptamers themselves and of the resulting aptamer-toxin conjugates.
Supplementary Material
Author Disclosure Statement.
No competing financial interests exist.
References
- 1.Pastan I, Hassan R, FitzGerald DJ. and Kreitman RJ. (2007). Immunotoxin treatment of cancer. Annu Rev Med 58:221–237 [DOI] [PubMed] [Google Scholar]
- 2.Ray P, Cheek MA, Sharaf ML, Li N, Ellington AD, Sullenger BA, Shaw BR. and White RR. (2012). Aptamer-mediated delivery of chemotherapy to pancreatic cancer cells. Nucleic Acid Ther 22:295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao N, Pei SN, Qi J, Zeng Z, Iyer SP, Lin P, Tung CH. and Zu Y. (2015). Oligonucleotide aptamer-drug conjugates for targeted therapy of acute myeloid leukemia. Biomaterials 67:42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang YF, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y. and Tan W. (2009). Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. Chembiochem 10:862–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA. and Langer R. (2004). Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res 64:7668–7672 [DOI] [PubMed] [Google Scholar]
- 6.Wilner SE, Wengerter B, Maier K, de Lourdes Borba Magalhaes M, Del Amo DS, Pai S, Opazo F, Rizzoli SO, Yan A. and Levy M. (2012). An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids 1:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO. and Giangrande PH. (2009). Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 27:839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Tiemann K, Chomchan P, Alluin J, Swiderski P, Burnett J, Zhang X, Forman S, Chen R. and Rossi J. (2013). Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res 41:4266–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu TC, Marks JW, 3rd, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD. and Levy M. (2006). Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res 66:5989–5992 [DOI] [PubMed] [Google Scholar]
- 10.Rosenblum MG, Marks JW. and Cheung LH. (1999). Comparative cytotoxicity and pharmacokinetics of antimelanoma immunotoxins containing either natural or recombinant gelonin. Cancer Chemother Pharmacol 44:343–348 [DOI] [PubMed] [Google Scholar]
- 11.Rosenblum MG, Kohr WA, Beattie KL, Beattie WG, Marks W, Toman PD. and Cheung L. (1995). Amino acid sequence analysis, gene construction, cloning, and expression of gelonin, a toxin derived from Gelonium multiflorum. J Interferon Cytokine Res 15:547–555 [DOI] [PubMed] [Google Scholar]
- 12.Better M, Bernhard SL, Fishwild DM, Nolan PA, Bauer RJ, Kung AH. and Carroll SF. (1994). Gelonin analogs with engineered cysteine residues form antibody immunoconjugates with unique properties. J Biol Chem 269:9644–9650 [PubMed] [Google Scholar]
- 13.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, et al. (2010). Rational design of cationic lipids for siRNA delivery. Nat Biotechnol 28:172–176 [DOI] [PubMed] [Google Scholar]
- 14.Diener JL, Hatala P, Killough JR, Wagner-Whyte J, Wilson C. and Zhu S. (2006). Stabilized aptamers to psma and their use as prostate cancer therapeutics. (Google Patents)
- 15.Rockey WM, Hernandez FJ, Huang SY, Cao S, Howell CA, Thomas GS, Liu XY, Lapteva N, Spencer DM, et al. (2011). Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther 21:299–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magalhaes ML, Byrom M, Yan A, Kelly L, Li N, Furtado R, Palliser D, Ellington AD. and Levy M. (2012). A general RNA motif for cellular transfection. Mol Ther 20:616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wengerter BC, Katakowski JA, Rosenberg JM, Park CG, Almo SC, Palliser D. and Levy M. (2014). Aptamer-targeted antigen delivery. Mol Ther 22:1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirie CM, Hackel BJ, Rosenblum MG. and Wittrup KD. (2011). Convergent potency of internalized gelonin immunotoxins across varied cell lines, antigens, and targeting moieties. J Biol Chem 286:4165–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner JJ, Williams D, Owen D. and Gait MJ. (2006). Disulfide conjugation of peptides to oligonucleotides and their analogs. Curr Protoc Nucleic Acid Chem Chapter 4:Unit 4 28. [DOI] [PubMed] [Google Scholar]
- 20.Turner JJ, Arzumanov AA. and Gait MJ. (2005). Synthesis, cellular uptake and HIV-1 Tat-dependent trans-activation inhibition activity of oligonucleotide analogues disulphide-conjugated to cell-penetrating peptides. Nucleic Acids Res 33:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dassie JP, Hernandez LI, Thomas GS, Long ME, Rockey WM, Howell CA, Chen Y, Hernandez FJ, Liu XY, et al. (2014). Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther 22:1910–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antignani A. and Fitzgerald D. (2013). Immunotoxins: the role of the toxin. Toxins (Basel) 5:1486–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lord JM, Roberts LM. and Robertus JD. (1994). Ricin: structure, mode of action, and some current applications. FASEB J 8:201–208 [PubMed] [Google Scholar]
- 24.Austin CD, Wen X, Gazzard L, Nelson C, Scheller RH. and Scales SJ. (2005). Oxidizing potential of endosomes and lysosomes limits intracellular cleavage of disulfide-based antibody-drug conjugates. Proc Natl Acad Sci U S A 102:17987–17992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Chen H, Vlahov IR, Cheng JX. and Low PS. (2006) Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proc Natl Acad Sci U S A 103:13872–13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C, Wang S, Brulisauer L, Leroux JC. and Gauthier MA. (2013) Broad control of disulfide stability through microenvironmental effects and analysis in complex redox environments. Biomacromolecules 14:2383–2388 [DOI] [PubMed] [Google Scholar]
- 27.Kellogg BA, Garrett L, Kovtun Y, Lai KC, Leece B, Miller M, Payne G, Steeves R, Whiteman KR, et al. (2011). Disulfide-linked antibody-maytansinoid conjugates: optimization of in vivo activity by varying the steric hindrance at carbon atoms adjacent to the disulfide linkage. Bioconjug Chem 22:717–727 [DOI] [PubMed] [Google Scholar]
- 28.Morell A, Terry WD. and Waldmann TA. (1970). Metabolic properties of IgG subclasses in man. J Clin Invest 49:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C. and McCauley TG. (2004). Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res 21:2234–2246 [DOI] [PubMed] [Google Scholar]
- 30.Diener J, Daniel Lagasse H, Duerschmied D, Merhi Y, Tanguay JF, Hutabarat R, Gilbert J, Wagner D. and Schaub R. (2009). Inhibition of von Willebrand factor-mediated platelet activation and thrombosis by the anti-von Willebrand factor A1-domain aptamer ARC1779. J Thromb Haemost 7:1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vater A, Sell S, Kaczmarek P, Maasch C, Buchner K, Pruszynska-Oszmalek E, Kolodziejski P, Purschke WG, Nowak KW, Strowski MZ. and Klussmann S. (2013). A mixed mirror-image DNA/RNA aptamer inhibits glucagon and acutely improves glucose tolerance in models of type 1 and type 2 diabetes. J Biol Chem 288:21136–21147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J. and Rossi JJ. (2014). Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol Ther Nucleic Acids 3:e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siller-Matula JM, Merhi Y, Tanguay J-F, Duerschmied D, Wagner DD, McGinness KE, Pendergrast PS, Chung J-K, Tian X. and Schaub RG. (2012). ARC15105 is a potent antagonist of von Willebrand factor mediated platelet activation and adhesion. Arterioscler Thromb Vasc Biol 32:902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.