Abstract
Aims: Protein S-glutathionylation is a widely distributed post-translational modification of thiol groups with glutathione that can function as a redox-sensitive switch to mediate redox regulation and signal transduction. The malaria parasite Plasmodium falciparum is exposed to intense oxidative stress and possesses the enzymatic system required to regulate protein S-glutathionylation, but despite its potential importance, protein S-glutathionylation has not yet been studied in malaria parasites. In this work we applied a method based on enzymatic deglutathionylation, affinity purification of biotin-maleimide-tagged proteins, and proteomic analyses to characterize the Plasmodium glutathionylome. Results: We identified 493 targets of protein S-glutathionylation in Plasmodium. Functional profiles revealed that the targets are components of central metabolic pathways, such as nitrogen compound metabolism and protein metabolism. Fifteen identified proteins with important functions in metabolic pathways (thioredoxin reductase, thioredoxin, thioredoxin peroxidase 1, glutathione reductase, glutathione S-transferase, plasmoredoxin, mitochondrial dihydrolipoamide dehydrogenase, glutamate dehydrogenase 1, glyoxalase I and II, ornithine δ-aminotransferase, lactate dehydrogenase, glyceraldehyde 3-phosphate dehydrogenase [GAPDH], pyruvate kinase [PK], and phosphoglycerate mutase) were further analyzed to study their ability to form mixed disulfides with glutathione. We demonstrate that P. falciparum GAPDH, PK, and ornithine δ-aminotransferase are reversibly inhibited by S-glutathionylation. Further, we provide evidence that not only P. falciparum glutaredoxin 1, but also thioredoxin 1 and plasmoredoxin are able to efficiently catalyze protein deglutathionylation. Innovation: We used an affinity-purification based proteomic approach to characterize the Plasmodium glutathionylome. Conclusion: Our results indicate a wide regulative use of S-glutathionylation in the malaria parasite and contribute to our understanding of redox-regulatory processes in this pathogen. Antioxid. Redox Signal. 15, 2855–2865.
Introduction
Redox-signaling pathways, protein structures, and enzymatic reactions are often controlled by thiol groups of cysteine residues acting as redox-sensitive switches both in proteins and in low-molecular-weight thiols such as the tripeptide glutathione. Glutathione (1–10 mM) (34) exists in a reduced (GSH) and an oxidized form (GSSG), with most of the glutathione being in the reduced form in unstressed cells. Glutathione is critical for maintaining a balanced intracellular redox state and for regulating oxidative signaling pathways (17). Protein S-glutathionylation represents the most abundant form of oxidative thiol modifications: The cysteine-sulfhydryl moiety of GSH forms a reversible mixed disulfide with a cysteine-sulfhydryl moiety of a protein (PSH), resulting in an S-glutathionylated protein (mixed protein glutathione disulfide, PSSG). Protein S-glutathionylation occurs during oxidative stress, but also under physiological conditions, and fulfills various important functions in redox regulation. Protein S-glutathionylation can initiate functional changes of enzymes, and regulates signaling transducers (11, 14). Numerous proteins such as carbonic anhydrase III, α-ketoglutarate dehydrogenase, as well as heat shock protein 70, c-Jun, and NF-κB have been reported to be reversibly activated or inhibited by S-glutathionylation of functionally or structurally critical cysteine residues [reviewed in (16)]. Further, during increased oxidative stress S-glutathionylation can protect thiol groups from overoxidation (49), and simultaneously functions as a storage form of glutathione inside the cell.
Besides thiol disulfide exchange between GSSG and a protein, protein S-glutathionylation can occur in the presence of GSH and oxidants. In this case, protein thiols are oxidized to sulfenic acids, which can react with GSH (1, 49). In addition, S-glutathionylation can be mediated by more reactive oxidized forms of glutathione, for example, S-nitrosoglutathione, or can be catalyzed enzymatically by glutaredoxin via a monothiol-mechanism (33), as well as by glutathione S-transferase (47). However, the predominant mechanism in vivo remains unclear. The reverse reaction, called deglutathionylation, can be efficiently catalyzed by glutaredoxins and thioredoxins via a dithiol exchange mechanism (4, 20, 23, 24).
Innovation
The rapidly growing and multiplying malaria parasite Plasmodium falciparum needs to adapt efficiently to various hostile environments in human and mosquito. Therefore, antioxidant defense and redox regulatory processes play a central role in this pathogen. Here we provide the first systematic study on cellular targets of protein S-glutathionylation in P. falciparum. Using multidimensional protein identification technology, we identified 321 proteins with a predicted function and 172 hypothetical proteins as targets of S-glutathionylation. Further, we studied the S-glutathionylation of selected proteins systematically by protein immunoblotting and functional analyses. We also provide evidence that not only Plasmodium glutaredoxin1 (Grx1), but also thioredoxin 1 (Trx1) as well as plasmoredoxin (Plrx) are able to efficiently catalyze protein deglutathionylation. Protein S-glutathionylation can be considered as redox regulation, if several aspects are met: (i) efficient reversibility, (ii) specificity toward certain cysteine residues, (iii) must occur in response to a physiological stimulus in intact cells, and (iv) protein activity as well as the respective cell function should be changed by S-glutathionylation (16, 43). Most of these criteria have been demonstrated for the examined proteins, including P. falciparum (Pf) GAPDH and PfOAT. However, the role of S-glutathionylation in response to different physiological and pathophysiological stimuli, like oxidative and pharmacological stress, the differential susceptibility of protein sulfhydryls to S-glutathionylation, as well as the mechanisms leading to protein (de)glutathionylation in P. falciparum in vivo remain to be elucidated in detail.
Malaria caused by P. falciparum is one of the deadliest diseases worldwide and affects nearly 250 million people annually, most of them being children in the world's most disadvantaged countries. During its developmental stages Plasmodium shows only minor transcriptional changes in response to external stimuli, suggesting that its proteins are mainly regulated by versatile post-transcriptional and post-translational modifications (21). Plasmodium possesses a functional glutathione system, including glutathione reductase (GR) (18), glutathione, a 2-Cys glutaredoxin (Grx1) (40), and a glutathione S-transferase (GST) (27). Thus, the parasite is enzymatically equipped to use protein S-glutathionylation as a redox-regulatory mechanism. However, despite its potential importance, protein S-glutathionylation has not yet been studied in Plasmodium.
Several recent articles describe the identification of S-glutathionylated proteins based on the enzymatic reduction of S-glutathionylated proteins by glutaredoxin, tagging with biotin-maleimide, affinity purification, and identification by proteomic analysis (2, 25, 32, 41). By using this specific and highly sensitive approach, we identified 321 proteins with a predicted function and 172 hypothetical proteins of the P. falciparum glutathionylome. Further, we studied the S-glutathionylation of selected proteins systematically by protein immunoblotting and functional analyses.
Results
Identification of S-glutathionylated proteins
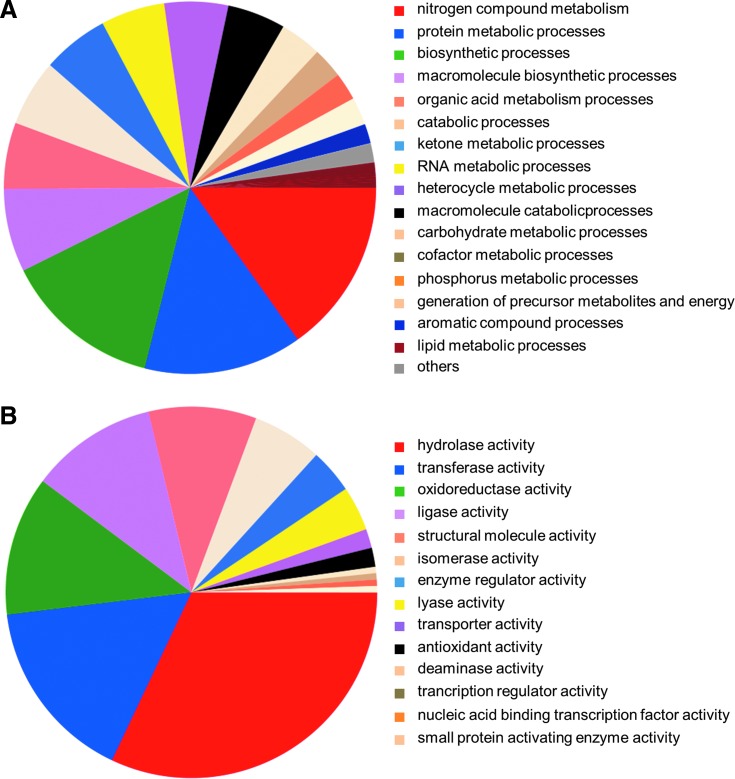
S-glutathionylated proteins in P. falciparum were detected by a specific and sensitive method based on enzymatic deglutathionylation of mixed protein-SSG disulfides by recombinant glutaredoxin in cell extracts, as first described by Lind et al. (32). Initially, all free thiols in the P. falciparum cell extracts were alkylated by N-ethylmaleimide (NEM), and then S-glutathionylated proteins were reduced with recombinant PfGrx1 using PfGR, GSH, and NADPH as back-up system. The emerging sulfhydryl groups of initially S-glutathionylated proteins were subsequently tagged with biotin-maleimide and purified by avidin affinity chromatography (Fig. 1). Isolated proteins were trypsin-digested and identified using an linear trap quadrupole (LTQ)-Orbitrap mass spectrometer. Three experimental repetitions were conducted to ensure high specificity. We reproducibly identified 321 targets for protein S-glutathionylation (listed in Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/ars). In addition, we identified 172 S-glutathionylated hypothetical proteins (listed separately in Supplementary Table S2). Only proteins that were identified in at least two experiments were included in the target lists. A functional profile of the identified proteins was generated according to their contribution to cellular metabolic pathways and their catalytic activity using the program PatternLab for Proteomics (9). Many S-glutathionylated proteins are involved in nitrogen compound and protein metabolism (Fig. 2A), and according to their catalytic activity most of the proteins have hydrolase, transferase, or oxidoreductase activity (Fig. 2B).
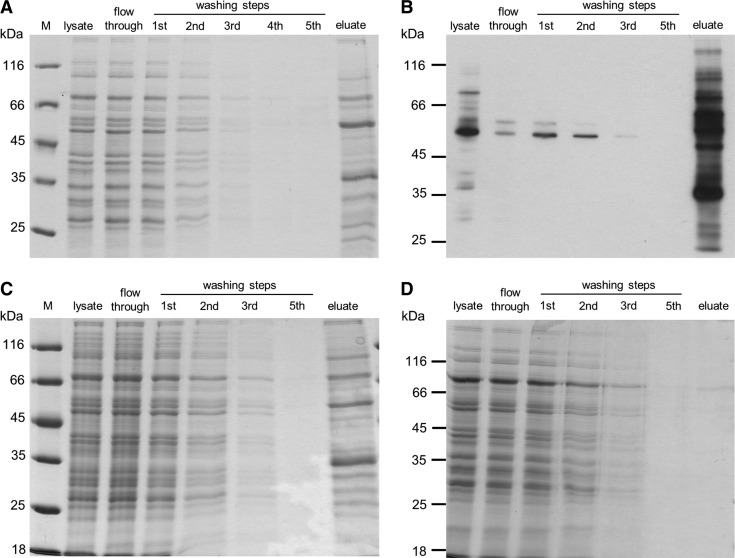
FIG. 1.
S-glutathionylated proteins in Plasmodium falciparum cell extract. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis profile and (B) western blot using an antibiotin antibody (Santa Cruz Biotechnology; 1:200) of S-glutathionylated proteins isolated via glutaredoxin-dependent deglutathionylation and affinity purification of biotin-maleimide-tagged proteins. (C) Control experiment using the mutant PfGrx1C32S for the deglutathionylation step. (D) Control experiment without both deglutathionylation and biotin-maleimide shows hardly any protein binding to the avidin resin. PfGrx1, glutaredoxin.
FIG. 2.
Functional classification of the S-glutathionylated proteins identified with wild-type glutaredoxin 1 (dithiol-plus monothiol-mechanism). Proteins are clustered according to cellular metabolic processes (A) and according to their catalytic activity (B). Only annotated proteins were used for analysis and classified using go annotations (downloaded from www.geneontology.org, accessed March 23, 2011) and the program PatternLab for Proteomics (24, 25). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Control experiments
To ensure specificity of the method, we performed two control experiments. In the first control experiment only the blocking of free thiols with NEM was carried out, aiming at identifying proteins that interact unspecifically with the avidin resin. In this approach we identified 13 proteins (Supplementary Table S1, column “w/o Grx1”; proteins bind unspecifically).
Besides reducing protein-SSG via a monothiol-mechanism, PfGrx1 can also reduce disulfides via a dithiol-mechanism and the emerging free sulfhydryl moieties can subsequently bind biotin-maleimide. To elucidate this aspect, we performed a parallel experiment using a mutant of PfGrx1 for the deglutathionylation step, where the resolving cysteine in the active site was mutated to serine (PfGrx1C32S). In contrast to the wild type, this mutant can only catalyze a deglutathionylation but not a disulfide reduction reaction and is therefore very specific for deglutathionylation (Supplementary Table S1, column “Grx1C32S”).
Verification of S-glutathionylation on selected proteins by protein immunoblotting
We confirmed S-glutathionylation for some of the captured proteins by incubating the recombinant proteins with different GSSG concentrations and performing antiglutathione (Virogen, Watertown, MA) Western blots under nonreducing conditions. Proteins were tested for antibody cross-reactivity, and reversibility of S-glutathionylation was examined by adding dithiothreitol (DTT). We confirmed that Pf thioredoxin peroxidase 1 (TPx1), Pf thioredoxin 1 (Trx1), Pf thioredoxin reductase (TrxR), PfGR, Pf plasmoredoxin (Plrx), PfGST, Pf mitochondrial dihydrolipoamide dehydrogenase (mLipDH), Pf glutamate dehydrogenase 1 GluDH1, Pf cytosolic glyoxalase I (cGloI), Pf cytosolic glyoxalase II (cGloII), Pf lactate dehydrogenase (LDH), PfGAPDH, and Pf phosphoglycerate mutase (PGM) can be reversibly modified by S-glutathionylation (Supplementary Fig. S1). Cross-reactivity of the antibody with examined proteins could either not be observed or was negligible when compared with the signal of the respective glutathionylated protein sample.
In case of PfGST, we tested S-glutathionylation of its two cysteine residues by analyzing mutants lacking either one or both cysteine residues (PfGSTC86A, PfGSTC101A, PfGSTC86/101A). Glutathione was found to bind to both cysteine residues, since only the double mutant was not glutathionylated in western blots (Supplementary Fig. S1).
Adenylate kinase 1 (PfAK1) was identified in all S-glutathionylation experiments and found to bind unspecifically to the avidin material. As a specificity control, we tested whether PfAK1 can be S-glutathionylated in vitro. After incubation of PfAK1 with 10 mM GSSG, we performed nonreducing western blots, which confirmed that PfAK1 cannot be glutathionylated (Supplementary Fig. S1). These experiments verify the results from the identification approach and thus support the quality and sensitivity of the applied method. Protein deglutathionylation by dithiol oxidoreductases such as PfGrx1, PfTrx1, and PfPlrx was also studied exemplarily both by antiglutathione western blot and enzymatically (described below).
Regulation of PfOAT by GSH and GSSG
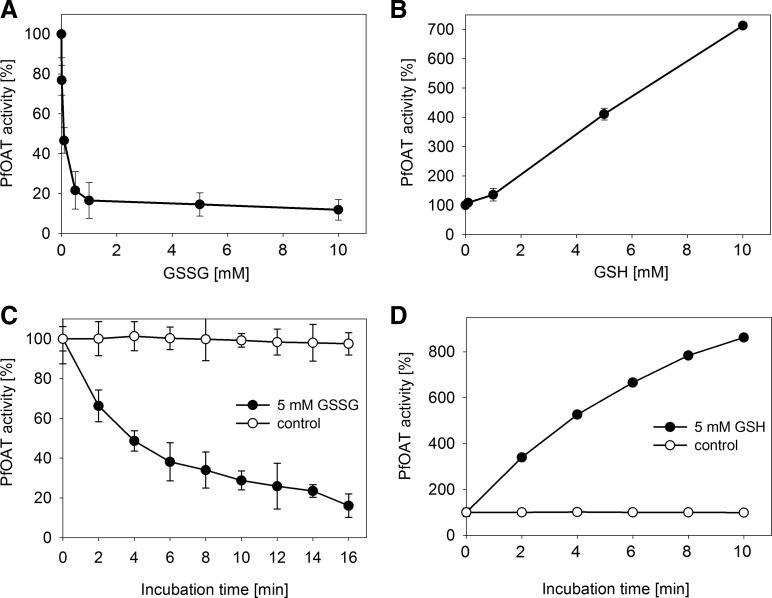
P. falciparum ornithine δ-aminotransferase (OAT) has been identified reproducibly as a target of S-glutathionylation (Supplementary Table S1). Incubation of PfOAT with GSSG results in a concentration- and time-dependent inhibition of PfOAT activity (Fig. 3A, C). The activity of PfOAT decreases rapidly, leaving <20% residual activity after 10 min of incubation with 1 mM GSSG. The effect is highly specific; already 10 μM GSSG inhibited PfOAT to 76% of its initial activity. Cysteine residues 154 and 163 can form a disulfide and are known to interfere with substrate binding of PfOAT (29). Thus, most likely S-glutathionylation of these cysteine residues inhibits PfOAT activity. The opposite effect can be observed after incubation of PfOAT with GSH, which leads to an increase in PfOAT activity (Fig. 3B, D). Incubation with 10 mM GSH enhances PfOAT activity to 700%, most likely due to the reduction of the Cys154-Cys163 disulfide. Reduction of this disulfide has previously been shown to mediate PfOAT activation by Trx (29).
FIG. 3.
Regulation of PfOAT activity by glutathione. (A) PfOAT was incubated with different concentrations of GSSG for 10 min at 37°C and (B) with different concentrations of GSH for 5 min at 37°C. (C) Time-dependent regulation of PfOAT with 5 mM GSSG and (D) 5 mM GSH. The samples were incubated at 37°C and aliquots were taken every 2 min to monitor the activity. Each value is a mean value from three independent determinations each including 6–8 measurements. Data are represented as mean±standard deviation. GSH, reduced glutathione; GSSG, oxidized glutathione disulfide; OAT, ornithine δ-aminotransferase.
Inactivation of PfGAPDH by GSSG
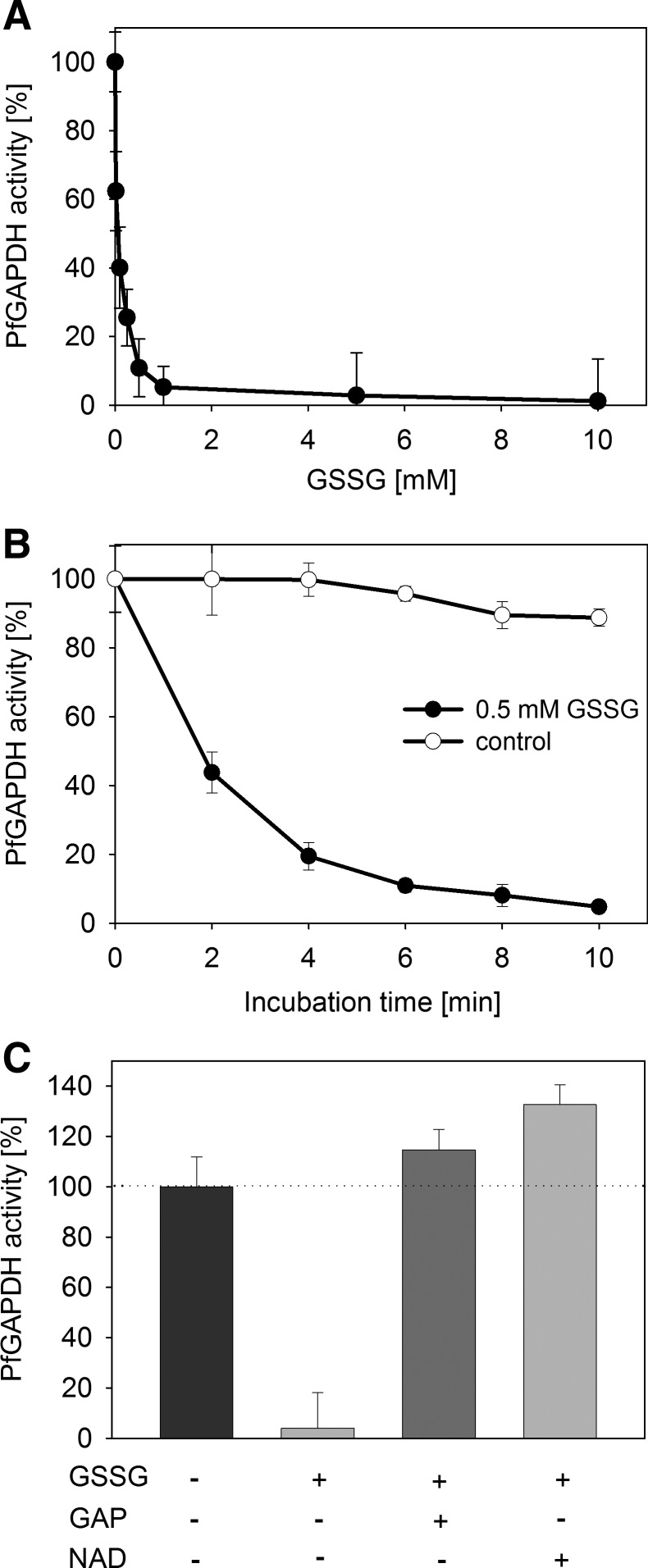
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is one of the best studied enzymes regulated by S-glutathionylation (28, 49), but Plasmodium GAPDH has not been reported to be redox-regulated before. Enzymatic assays demonstrate that PfGAPDH is nearly completely inhibited by GSSG, but not influenced by GSH. The degree of inhibition depends both on GSSG concentration and incubation time (Fig. 4A, B). Previous studies on GAPDH from other organisms reported that a highly conserved cysteine residue located in the active site is a target of thiol modifications that causes inhibition of the enzymatic activity (44, 49).
FIG. 4.
Regulation of PfGAPDH activity by glutathione. (A) Concentration-dependent regulation of PfGAPDH when incubated with different GSSG concentrations for 5 min at 37°C. (B) Time-dependent regulation of PfGAPDH when incubated with 0.5 mM GSSG or with buffer (control) for 10 min at 37°C. Activity was monitored every 2 min. (C) Protection of PfGAPDH from GSSG-mediated inhibition by the substrates GAP and NAD+. PfGAPDH was incubated with 0.5 mM GAP or 1 mM NAD+ together with 10 mM GSSG for 5 min at 37°C. Each value is a mean value from at least three independent determinations each including five measurements. Data are represented as mean±standard deviation. GAP, glyceraldehyde 3-phosphate; PfGAPDH, P. falciparum glyceraldehyde 3-phosphate dehydrogenase.
The substrate glyceraldehyde 3-phosphate (GAP) forms a covalent intermediate with the active site cysteine of GAPDH, and protects it from oxidation (49). To test if the observed inhibition of PfGAPDH is mediated by the binding of GSSG to the catalytic cysteine residue (Cys153), we added either the substrate (GAP) or the cofactor (NAD+) to the incubation of PfGAPDH with GSSG. Both GAP and NAD+ efficiently protect PfGAPDH from inactivation by GSSG, thereby indicating that the activity decrease observed with only GSSG in the incubation is caused by S-glutathionylation of the catalytic cysteine (Fig. 4C).
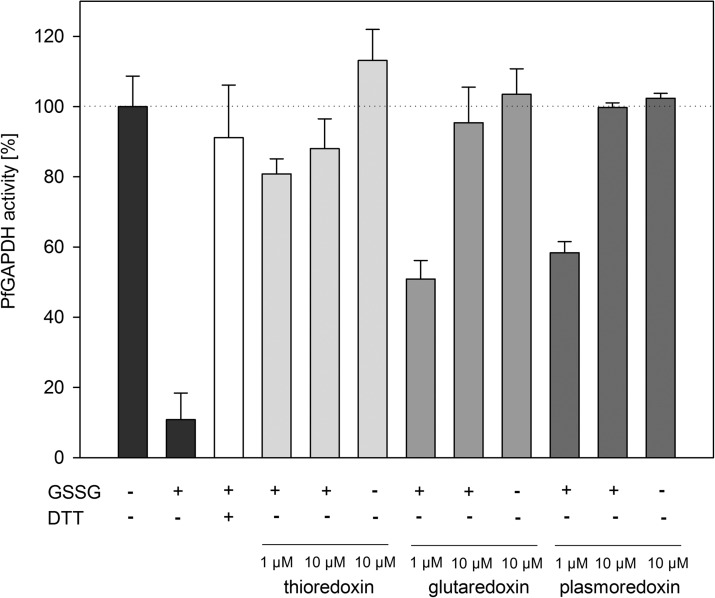
In general, deglutathionylation can be efficiently catalyzed by glutaredoxins (20, 23) and Trxs (24). We incubated glutathionylated PfGAPDH with 1–10 μM of prereduced PfGrx1, PfTrx1, or PfPlrx and monitored the activity before and after the incubation. All three redoxins are able to reactivate PfGAPDH and thus efficiently catalyze PfGAPDH deglutathionylation (Fig. 5).
FIG. 5.
Deglutathionylation of PfGAPDH by thioredoxin, glutaredoxin, and plasmoredoxin. PfGAPDH was incubated with 0.5 mM GSSG for 5 min at 37°C, excess GSSG was removed, and the S-glutathionylated PfGAPDH was incubated with DTT, or prereduced PfTrx, PfGrx, and PfPlrx for 5 min at 37°C. Each value is a mean value from at least two independent determinations each including six measurements. Data are represented as mean±standard deviation. Plrx, plasmoredoxin.
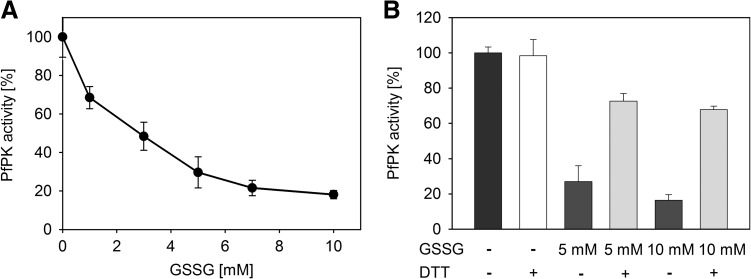
Influence of S-glutathionylation on the activity of PfPK
The activity of P. falciparum pyruvate kinase (PfPK) is inhibited by GSSG, but not influenced by GSH. The inhibitory effect of GSSG requires higher concentrations of GSSG compared with PfGAPDH and PfOAT, but shows a clearly concentration-dependent inhibition. Less than 20% residual activity of PfPK remains in the presence of 10 mM GSSG. This inhibition can be partially reversed to 70% of its initial activity by DTT (Fig. 6A, B). Further, PfPK can be enzymatically deglutathionylated by PfTrx1, PfGrx1, and PfPlrx as shown by protein immunoblotting (Fig. 7B).
FIG. 6.
Inhibition of PfPK by S-glutathionylation. (A) Concentration-dependent inhibition of PfPK by GSSG. (B) Reversibility of the GSSG-mediated inhibition of PfPK. Each value is a mean value from three independent determinations each including five measurements. Data are represented as mean±standard deviation.
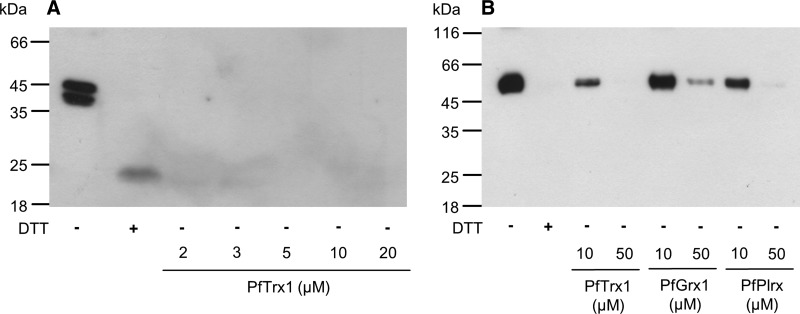
FIG. 7.
Enzymatic deglutathionylation of (A) PfTPx1 and (B) PfPK by P. falciparum redoxins. PfTPx1 and PfPK were incubated with 10 mM GSSG over night at 23°C, and excess GSSG was removed by gel filtration chromatography. Glutathionylated PfTPx1 (4.5 μM) and PfPK (9 μM) were incubated with different concentrations of reduced PfTrx1, PfGrx1, and/or PfPlrx for 10 min at 37°C.
S-glutathionylation of PfTPx1
We could not detect any effect of PfTPx1 S-glutathionylation on the Trx-dependent hydroperoxide reducing activity of the enzyme. However, this is most likely due to the presence of Trx in the assay, which can efficiently deglutathionylate PfTPx1 at low concentrations, as shown by anti-GSH immunoblotting of glutathionylated PfTPx1 with prereduced PfTrx1 (Fig. 7A). Under nonreducing conditions or conditions where only a limited amount of reducing agents is added, PfTPx1 runs as a dimer (Fig. 7A). Further, it has been hypothesized previously that the second (lower) band of PfTPx1 corresponds to the oxidized conformation of PfTPx1 (39). This putative redox-dependent conformational switch might represent a regulatory process that deserves to be studied in further detail (39).
S-glutathionylation has no major effect on the enzymatic activities of PfLDH, PfcGloI, PfcGloII, and PfTrxR
Although we could confirm that PfLDH can be reversibly S-glutathionylated (Supplementary Fig. S1), incubation of PfLDH with GSSG or GSH influenced neither the reduction of pyruvate to lactate nor the oxidation of lactate to pyruvate. Similar results were obtained for two Plasmodium glyoxalases, cGloI and cGloII. The enzymes can be reversibly S-glutathionylated (Supplementary Fig. S1), but the modification has no detectable effect on the activity of the enzymes.
Western blot analysis clearly confirms that PfTrxR can be S-glutathionylated (Supplementary Fig. S1), but in the Trx-reducing assay no activity change of S-glutathionylated PfTrxR compared with nonglutathionylated PfTrxR could be observed. To exclude that the missing effect is due to the deglutathionylating properties of Trx1, we measured the activity of S-glutathionylated PfTrxR by using the artificial substrate DTNB. However, no major change in activity (<10%) was detected in the DTNB assay, either. One possible explanation for these results is that while Trx is reduced by the C-terminal redox center alone, DTNB can also be reduced by the N-terminal redox center of PfTrxR (31). Since S-glutathionylation might predominantly affect the highly accessible C-terminal redox-center, no inhibitory effect is necessarily determined with the substrate DTNB.
Discussion
Protein S-glutathionylation is gaining more and more scientific attention as a redox switch. Here, we examined the glutathionylome of the malaria parasite P. falciparum by a highly specific and sensitive method based on enzymatic deglutathionylation of mixed protein-SSG disulfides by glutaredoxin in cell extracts. Several other methods have been developed for identifying substrates of protein S-glutathionylation. Radiolabeling of the cellular glutathione pool by 35S-cysteine followed by oxidative treatment to enhance S-thiolation is the most common method (19, 35). However, this method requires pretreatment of cells with protein synthesis inhibitors, potentially causing disturbances of cellular processes. Further, the approach does not distinguish between different types of S-thiolation and identifies proteins that are S-thiolated during treatment with oxidants, not reflecting S-glutathionylation under basal conditions [reviewed in (22)]. Another method, based on biotinylated glutathione, can be used in combination with affinity purification both in vivo and in vitro, but the biotin-tag can disturb the interaction between glutathione and the protein (22). Studies using antiglutathione antibodies in combination with immunoprecipitation are limited by their low sensitivity and thus a low number of detected proteins in cellular extracts (7). The identification of glutathionylated proteins by glutaredoxin reduction applied here allows the detection of proteins that are S-glutathionylated under physiological conditions, since the cells are not treated with protein synthesis inhibitors or oxidants (2, 32, 41). However, substantial knowledge on the specificity of deglutathionylation reactions by different dithiol oxidoreductases is missing. Further, it has to be considered that only proteins deglutathionylated by PfGrx1 are identified; proteins possibly deglutathionylated by, for example, Plasmodium Trx1, are not identified.
Using this strategy, we reproducibly identified 321 S-glutathionylated proteins with a predicted function in P. falciparum (Supplementary Table S1). An earlier study identified only 43 S-glutathionylated proteins in human ECV304 cells (32). Most likely the difference is due to the identification method used to detect isolated S-glutathionylated proteins in the elution fraction. In our case a highly sensitive LTQ-Orbitrap-based mass spectrometric analysis was used to detect target proteins directly. In contrast, Lind et al. separated the eluted proteins by two-dimensional gel electrophoresis and used MALDI-TOF to identify excised protein spots (32). This leads to a substantial decrease in sensitivity, because only highly abundant proteins in the range of a certain pI and molecular weight are recovered. In our approach, we identified proteins with a molecular weight ranging from below 10 kDa up to >200 kDa (Supplementary Table S1). Moreover, low abundance proteins are unlikely to be detected by excising protein spots from a gel, but are covered by our approach. High-resolution high-accuracy mass spectrometry also allowed the identification of more than 1000 proteins that undergo lysine acetylation (12), another post-translational modification.
To follow up our results on the glutathionylome of Plasmodium and prove the accuracy of the method, we validated the S-glutathionylation for 15 target proteins by western blot using recombinant proteins and an antiglutathione antibody. For the glutathionylated proteins, we could demonstrate that the modification is reversible, since glutathione could be removed by DTT (Supplementary Fig. S1). The antiglutathione antibody appears to be highly specific for glutathione, since we observe almost no cross-reactivity with the unglutathionylated proteins (discussed in Results section).
We studied the influence of the observed S-glutathionylation on the enzymatic activity of three proteins: PfGAPDH, PfOAT, and PfPK. We chose GAPDH as an intensely studied enzyme in mammals and plants, which is inactivated by S-glutathionylation of its highly conserved active site cysteine (28, 33, 49). Plasmodium GAPDH was also detected to be susceptible to glutathionylation of the active site cysteine, and is inactivated by low glutathione concentrations, which are likely to reflect physiological concentrations (Fig. 4). PfGAPDH can be efficiently deglutathionylated and thus activated by PfTrx1, PfGrx1, and PfPlrx (Fig. 5). PfTrx1, PfGrx1, and PfPlrx have been shown to be involved in redox regulation of several proteins before (45). Besides its function in glycolysis, the highly redox-sensitive GAPDH is involved in several cellular processes such as transcription (50), apoptosis (26), and calcium homeostasis (37). The importance of the enzyme is underlined by the finding that GAPDH is essential for the survival of human pathogens such as Trypanosoma brucei (8).
Malaria parasites require glycolysis, since the second most common human enzyme disorder, PK-deficiency, protects against Plasmodium infection in human erythrocytes (3). P. falciparum PK is reversibly inactivated by S-glutathionylation (Fig. 6), indicating a role of cysteine residues in the catalytic mechanism. This has been suggested for human PK from red blood cells before, which is highly susceptible to inactivation by increased oxidative stress, but its activity can be recovered by glutathione (36). Despite its enormous potential as an antimalarial drug target, Plasmodium PK is barely studied (10), but has been identified as a putative target of PfTrx1, PfGrx1, and PfPlrx (45). This is also the case for PfOAT, which is redox-regulated by PfTrx1 via two critical cysteine residues that interfere with substrate binding (29). This motivated us to test whether the activity of PfOAT is affected by S-glutathionylation. Interestingly, PfOAT is differentially regulated by GSH and GSSG presumably by different mechanisms (Fig. 3): GSSG inhibits PfOAT by S-glutathionylation of functionally critical cysteine residues (presumably Cys136 and Cys154), whereas GSH is likely to reduce the disulfide between these cysteines, leading to an activation of PfOAT comparable to that induced by PfTrx1.
Further, we could show very recently that both enzymatic activities of the bifunctional enzyme glucose 6-phosphate dehydrogenase 6-phosphogluconolactonase from P. falciparum are regulated by S-glutathionylation (30).
Not all enzymes examined here changed their activity as a consequence of S-glutathionylation: PfLDH as well as the glyoxalases PfcGloI and PfcGloII can be S-glutathionylated, but their enzymatic activity is not regulated by the modification (Supplementary Table S1 and Fig. S1). Interestingly, human cytosolic glyoxalase I is strongly inhibited by S-glutathionylation on cysteine 139 (corresponding to Cys311 in PfcGloI) (6). However, S-glutathionylation of PfLDH, PfcGloI, and PfcGloII might influence their oligomerization behavior or protein–protein interactions, which remains to be studied in detail.
The mechanism of protein (de)glutathionylation in vivo is still under investigation and not completely understood. The thiol disulfide oxidoreductase glutaredoxin is the best studied enzyme in terms of protein (de)glutathionylation and is supposed to catalyze the main part of protein deglutathionylation in mammalian cells (13), but also Trx has recently been reported to efficiently deglutathionylate protein targets in Saccharomyces cerevisiae (24). Here, we examined whether PfGrx1, PfTrx1, and/or PfPlrx can efficiently catalyze the deglutathionylation of PfGAPDH, PfTPx1, and PfPK using protein immunoblotting and enzymatic assays. Our results demonstrate a deglutathionylating activity not only for PfTrx and PfGrx, but also for PfPlrx, which is a new function attributed to this unique plasmodial protein.
Materials and Methods
Cultivation of P. falciparum and preparation of parasite cell extract
Intraerythrocytic stages of P. falciparum (3D7) were maintained in culture and prepared using standard procedures. Details are described in the Supplementary Materials and Methods.
Preparation of cell extracts, blocking of residual protein thiols, deglutathionylation, biotin-NEM tagging of deglutathionylated thiols, and purification of biotinylated proteins
Biotin-NEM-tagged proteins were obtained as previously described (2, 25, 32, 41), with slight modifications as described in Supplementary Materials and Methods.
Sample preparation for mass spectrometry
Protein fractions were prepared for MS-analysis using standard procedures as described in Supplementary Materials and Methods.
Multidimensional protein identification technology
The trypsin-digested protein samples were analyzed using multidimensional protein identification technology with slight variations depending on the protein amount of the sample as described in the Supplementary Materials and Methods.
Analysis of tandem mass spectra
MS/MS spectra were analyzed using the following software analysis protocol. Poor quality spectra were removed from the dataset using an automated spectral quality assessment algorithm (5). MS/MS spectra remaining after filtering were searched with the ProLuCID algorithm (48) against the NCBI-RefSeq P. falciparum database (8/24/2005) concatenated to a decoy database in which the sequence for each entry in the original database was reversed (38). All searches were parallelized and performed on a Beowulf computer cluster consisting of 100 1.2 GHz Athlon CPUs (42). Differential modifications of maleimide (+125.0477), biotin-maleimide (+451.1889), and iodoacetamide (+57.02146) were considered on cysteines. Peptides within 3 amu mass tolerance of the precursor mass and have one or two tryptic ends were considered during the database searches. ProLuCID results were assembled and filtered using the DTASelect (version 2.0) program (15, 46). DTASelect 2.0 uses a linear discriminant analysis to dynamically set XCorr and DeltaCN thresholds for the entire dataset to achieve a user-specified false-positive rate (5% in this analysis). The false-positive rates are estimated by the program from the number and quality of spectral matches to the decoy database.
Construction of P. falciparum expression plasmids and heterologous overexpression
Recombinant proteins from P. falciparum (PfOAT, PfmLipDH, PfcGloI and II, PfGrx1 und PfGrx1C32S, PfPlrx, PfTPx1, PfTrx1, PfTrxR, PfGR, PfGST, PfPK, PfGAPDH, PfLDH, PfPGM, and PfAK1) were produced and purified as described in the Supplementary Materials and Methods.
Protein immunoblotting analysis
Reduced proteins (0.4 mg/ml) were incubated with 0.1–10 mM GSSG in 50 mM Tris, 1 mM EDTA, pH 7.4 for 5 min at 37°C. Deglutathionylation experiments were performed by incubating S-glutathionylated proteins (0.4 mg/ml) with 2–50 μM reduced PfTrx1, PfGrx1, and/or PfPlrx for 10 min at 37°C. To detect recombinant S-glutathionylated proteins, western blots were performed under nonreducing conditions, using a monoclonal antiglutathione antibody (Virogen; diluted 1:500 in 5% nonfat milk with Tris-buffered saline Tween-20). Details are described in Supplementary Materials and Methods.
Enzyme activity assays and enzyme treatment
The enzymatic activity for PfGADPH, PfPK, PfLDH PfOAT, PfTPx1, PfcGloI, PfcGloII, and PfTrxR was measured with standard spectrophotometrically methods as described in the Supplementary Materials and Methods.
Supplementary Material
List of Abbreviations
- AK
adenylate kinase
- cGloI
cytosolic glyoxalase I
- cGloII
cytosolic glyoxalase II
- GAP
glyceraldehyde 3-phosphate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GluDH1
glutamate dehydrogenase
- GR
glutathione reductase
- Grx
glutaredoxin
- GSH
reduced glutathione
- GSSG
oxidized glutathione disulfide
- GST
glutathione S-transferase
- LDH
lactate dehydrogenase
- LTQ
linear trap quadrupole
- mLipDH
mitochondrial dihydrolipoamide dehydrogenase
- OAT
ornithine δ-aminotransferase
- PGM
phosphoglycerate mutase
- PK
pyruvate kinase
- Plrx
plasmoredoxin
- protein-SSG
mixed protein-glutathione disulfide
- TPx
thioredoxin peroxidase
- Trx
thioredoxin
- TrxR
thioredoxin reductase 1
Acknowledgments
The authors gratefully acknowledge Claudia Köhler for supporting the work on PK, Elisabeth Fischer and Marina Fischer for their great technical assistance, and Judith Helena Prieto for her help with the protein functional classification. This study was supported by the Deutsche Forschungsgemeinschaft (Grant BE 1540/15–1 to K.B.).
Author Disclosure Statement
The authors have nothing to declare.
References
- 1.Ahmetagic S, N. Salkić N. Cickusic E. Zerem E. Mott-Divkovic S. Tihic N. Smriko-Nuhanovic A. Hepatitis C virus genotypes in chronic hepatitis C patients and in first time blood donors in northeastern Bosnia and Herzegovina. Bosn J Basic Med Sci. 2009;9:278–282. doi: 10.17305/bjbms.2009.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Applegate MA. Humphries KM. Szweda LI. Reversible inhibition of alpha-ketoglutarate dehydrogenase by hydrogen peroxide: glutathionylation and protection of lipoic acid. Biochemistry. 2008;47:473–478. doi: 10.1021/bi7017464. [DOI] [PubMed] [Google Scholar]
- 3.Ayi K. Min-Oo G. Serghides L. Crockett M. Kirby-Allen M. Quirt I. Gros P. Kain KC. Pyruvate kinase deficiency and malaria. N Engl J Med. 2008;358:1805–1810. doi: 10.1056/NEJMoa072464. [DOI] [PubMed] [Google Scholar]
- 4.Bedhomme M. Zaffagnini M. Marchand CH. Gao XH. Moslonka-Lefebvre M. Michelet L. Decottignies P. Lemaire SD. Regulation by glutathionylation of isocitrate lyase from Chlamydomonas reinhardtii. J Biol Chem. 2009;284:36282–36291. doi: 10.1074/jbc.M109.064428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bern M. Goldberg D. McDonald WH. Yates JR., 3rd Automatic quality assessment of peptide tandem mass spectra. Bioinformatics. 2004;20(Suppl.1):i49–i54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 6.Birkenmeier G. Stegemann C. Hoffmann R. Gunther R. Huse K. Birkemeyer C. Posttranslational modification of human glyoxalase 1 indicates redox-dependent regulation. PLoS One. 2010;5:e10399. doi: 10.1371/journal.pone.0010399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan JP. Miller JI. Fuller W. Wait R. Begum S. Dunn MJ. Eaton P. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol Cell Proteomics. 2006;5:215–225. doi: 10.1074/mcp.M500212-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Caceres AJ. Michels PA. Hannaert V. Genetic validation of aldolase and glyceraldehyde-3-phosphate dehydrogenase as drug targets in Trypanosoma brucei. Mol Biochem Parasitol. 2010;169:50–54. doi: 10.1016/j.molbiopara.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho PC. Yates JR., III Barbosa VC. Analyzing shotgun proteomic data with Pattern-Lab for proteomics. Curr Protoc Bioinformatics. 2010;30 doi: 10.1002/0471250953.bi1313s30. 13.13.1–13.13.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan M. Sim TS. Functional analysis, overexpression, and kinetic characterization of pyruvate kinase from Plasmodium falciparum. Biochem Biophys Res Commun. 2005;326:188–196. doi: 10.1016/j.bbrc.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Chen CA. Wang TY. Varadharaj S. Reyes LA. Hemann C. Talukder MA. Chen YR. Druhan LJ. Zweier JL. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature. 2010;468:1115–1118. doi: 10.1038/nature09599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhary C. Kumar C. Gnad F. Nielsen ML. Rehman M. Walther TC. Olsen JV. Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 13.Chrestensen CA. Starke DW. Mieyal JJ. Acute cadmium exposure inactivates thioltransferase (glutaredoxin), inhibits intracellular reduction of protein-glutathionyl-mixed disulfides, and initiates apoptosis. J Biol Chem. 2000;275:26556–26565. doi: 10.1074/jbc.M004097200. [DOI] [PubMed] [Google Scholar]
- 14.Clavreul N. Adachi T. Pimental DR. Ido Y. Schoneich C. Cohen RA. S-glutathiolation by peroxynitrite of p21ras at cysteine-118 mediates its direct activation and downstream signaling in endothelial cells. FASEB J. 2006;20:518–520. doi: 10.1096/fj.05-4875fje. [DOI] [PubMed] [Google Scholar]
- 15.Cociorva D. Tabb DL. Yates JR. Validation of tandem mass spectrometry database search results using DTA Select. Curr Protoc Bioinformatics. 2007;16 doi: 10.1002/0471250953.bi1304s16. 13.4.1. [DOI] [PubMed] [Google Scholar]
- 16.Dalle-Donne I. Rossi R. Giustarini D. Colombo R. Milzani A. S-glutathionylation in protein redox regulation. Free Radic Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson DA. Forman HJ. Glutathione in defense and signaling: lessons from a small thiol. Ann NY Acad Sci. 2002;973:488–504. doi: 10.1111/j.1749-6632.2002.tb04690.x. [DOI] [PubMed] [Google Scholar]
- 18.Farber PM. Arscott LD. Williams CH., Jr. Becker K. Schirmer RH. Recombinant Plasmodium falciparum glutathione reductase is inhibited by the antimalarial dye methylene blue. FEBS Lett. 1998;422:311–314. doi: 10.1016/s0014-5793(98)00031-3. [DOI] [PubMed] [Google Scholar]
- 19.Fratelli M. Demol H. Puype M. Casagrande S. Eberini I. Salmona M. Bonetto V. Mengozzi M. Duffieux F. Miclet E. Bachi A. Vandekerckhove J. Gianazza E. Ghezzi P. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc Natl Acad Sci USA. 2002;99:3505–3510. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallogly MM. Starke DW. Mieyal JJ. Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glutaredoxins and potential mechanisms of regulation. Antioxid Redox Signal. 2009;11:1059–1081. doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganesan K. Ponmee N. Jiang L. Fowble JW. White J. Kamchonwongpaisan S. Yuthavong Y. Wilairat P. Rathod PK. A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates. PLoS Pathog. 2008;4:e1000214. doi: 10.1371/journal.ppat.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao XH. Bedhomme M. Veyel D. Zaffagnini M. Lemaire SD. Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Mol Plant. 2009;2:218–235. doi: 10.1093/mp/ssn072. [DOI] [PubMed] [Google Scholar]
- 23.Gao XH. Zaffagnini M. Bedhomme M. Michelet L. Cassier-Chauvat C. Decottignies P. Lemaire SD. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii: kinetics and specificity in deglutathionylation reactions. FEBS Lett. 2010;584:2242–2248. doi: 10.1016/j.febslet.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Greetham D. Vickerstaff J. Shenton D. Perrone GG. Dawes IW. Grant CM. Thioredoxins function as deglutathionylase enzymes in the yeast Saccharomyces cerevisiae. BMC Biochem. 2010;11:3. doi: 10.1186/1471-2091-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamnell-Pamment Y. Lind C. Palmberg C. Bergman T. Cotgreave IA. Determination of site-specificity of S-glutathionylated cellular proteins. Biochem Biophys Res Commun. 2005;332:362–369. doi: 10.1016/j.bbrc.2005.04.130. [DOI] [PubMed] [Google Scholar]
- 26.Hara MR. Agrawal N. Kim SF. Cascio MB. Fujimuro M. Ozeki Y. Takahashi M. Cheah JH. Tankou SK. Hester LD. Ferris CD. Hayward SD. Snyder SH. Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 27.Harwaldt P. Rahlfs S. Becker K. Glutathione S-transferase of the malarial parasite Plasmodium falciparum: characterization of a potential drug target. Biol Chem. 2002;383:821–830. doi: 10.1515/BC.2002.086. [DOI] [PubMed] [Google Scholar]
- 28.Holtgrefe S. Gohlke J. Starmann J. Druce S. Klocke S. Altmann B. Wojtera J. Lindermayr C. Scheibe R. Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol Plant. 2008;133:211–228. doi: 10.1111/j.1399-3054.2008.01066.x. [DOI] [PubMed] [Google Scholar]
- 29.Jortzik E. Fritz-Wolf K. Sturm N. Hipp M. Rahlfs S. Becker K. Redox regulation of Plasmodium falciparum ornithine delta-aminotransferase. J Mol Biol. 2010;402:445–459. doi: 10.1016/j.jmb.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 30.Jortzik E. Mailu BM. Preuss J. Fischer M. Bode L. Rahlfs S. Becker K. Glucose 6-phosphate dehydrogenase 6-phosphogluconolactonase: a unique bifunctional enzyme from Plasmodium falciparum. Biochem J. 2011;436:641–650. doi: 10.1042/BJ20110170. [DOI] [PubMed] [Google Scholar]
- 31.Kanzok SM. Rahlfs S. Becker K. Schirmer RH. Thioredoxin, thioredoxin reductase, and thioredoxin peroxidase of malaria parasite Plasmodium falciparum. Methods Enzymol. 2002;347:370–381. doi: 10.1016/s0076-6879(02)47037-1. [DOI] [PubMed] [Google Scholar]
- 32.Lind C. Gerdes R. Hamnell Y. Schuppe-Koistinen I. von Lowenhielm HB. Holmgren A. Cotgreave IA. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys. 2002;406:229–240. doi: 10.1016/s0003-9861(02)00468-x. [DOI] [PubMed] [Google Scholar]
- 33.Lind C. Gerdes R. Schuppe-Koistinen I. Cotgreave IA. Studies on the mechanism of oxidative modification of human glyceraldehyde-3-phosphate dehydrogenase by glutathione: catalysis by glutaredoxin. Biochem Biophys Res Commun. 1998;247:481–486. doi: 10.1006/bbrc.1998.8695. [DOI] [PubMed] [Google Scholar]
- 34.Meister A. Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 35.Michelet L. Zaffagnini M. Vanacker H. Le Marechal P. Marchand C. Schroda M. Lemaire SD. Decottignies P. In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J Biol Chem. 2008;283:21571–21578. doi: 10.1074/jbc.M802331200. [DOI] [PubMed] [Google Scholar]
- 36.Ogasawara Y. Funakoshi M. Ishii K. Pyruvate kinase is protected by glutathione-dependent redox balance in human red blood cells exposed to reactive oxygen species. Biol Pharm Bull. 2008;31:1875–1881. doi: 10.1248/bpb.31.1875. [DOI] [PubMed] [Google Scholar]
- 37.Patterson RL. van Rossum DB. Kaplin AI. Barrow RK. Snyder SH. Inositol 1,4,5-trisphosphate receptor/GAPDH complex augments Ca2+ release via locally derived NADH. Proc Natl Acad Sci USA. 2005;102:1357–1359. doi: 10.1073/pnas.0409657102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng J. Elias JE. Thoreen CC. Licklider LJ. Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 39.Rahlfs S. Becker K. Thioredoxin peroxidases of the malarial parasite Plasmodium falciparum. Eur J Biochem. 2001;268:1404–1409. doi: 10.1046/j.1432-1327.2001.02005.x. [DOI] [PubMed] [Google Scholar]
- 40.Rahlfs S. Fischer M. Becker K. Plasmodium falciparum possesses a classical glutaredoxin and a second, glutaredoxin-like protein with a PICOT homology domain. J Biol Chem. 2001;276:37133–37140. doi: 10.1074/jbc.M105524200. [DOI] [PubMed] [Google Scholar]
- 41.Reynaert NL. Ckless K. Guala AS. Wouters EF. van der Vliet A. Janssen-Heininger YM. In situ detection of S-glutathionylated proteins following glutaredoxin-1 catalyzed cysteine derivatization. Biochim Biophys Acta. 2006;1760:380–387. doi: 10.1016/j.bbagen.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Sadygov RG. Eng J. Durr E. Saraf A. McDonald H. MacCoss MJ. Yates JR., 3rd Code developments to improve the efficiency of automated MS/MS spectra interpretation. J Proteome Res. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 43.Shelton MD. Chock PB. Mieyal JJ. Glutaredoxin: role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 44.Shenton D. Grant CM. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sturm N. Jortzik E. Mailu BM. Koncarevic S. Deponte M. Forchhammer K. Rahlfs S. Becker K. Identification of proteins targeted by the thioredoxin super family in Plasmodium falciparum. Plos Pathogens. 2009;5:e1000383. doi: 10.1371/journal.ppat.1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabb DL. McDonald WH. Yates JR., 3rd DTA select and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Townsend DM. Manevich Y. He L. Hutchens S. Pazoles CJ. Tew KD. Novel role for glutathione S-transferase pi. Regulator of protein S-glutathionylation following oxidative and nitrosative stress. J Biol Chem. 2009;284:436–445. doi: 10.1074/jbc.M805586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu T. Venable JD. Park SK. Cociorva D. Lu B. Liao L. Wohlschlegel J. Hewel J. Yates JR. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5:S174. [Google Scholar]
- 49.Zaffagnini M. Michelet L. Marchand C. Sparla F. Decottignies P. Le Marechal P. Miginiac-Maslow M. Noctor G. Trost P. Lemaire SD. The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J. 2007;274:212–226. doi: 10.1111/j.1742-4658.2006.05577.x. [DOI] [PubMed] [Google Scholar]
- 50.Zheng L. Roeder RG. Luo Y. S phase activation of the histone H2B promoter by OCA-S, a co-activator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.