Abstract
Glycosylation is a dynamic post-translational modification that changes during the development and progression of various malignancies. During the oncogenesis of breast carcinoma, the glycosyltransferase known as N-acetylglucosaminyltransferase Va (GnT-Va) transcript levels and activity are increased due to activated oncogenic signaling pathways. Elevated GnT-V levels leads to increased β(1,6)-branched N-linked glycan structures on glycoproteins that can be measured using a specific carbohydrate binding protein or lectin known as L-PHA. L-PHA does not bind to nondiseased breast epithelial cells, but during the progression to invasive carcinoma, cells show a progressive increase in L-PHA binding. We have developed a procedure for intact protein L-PHA-affinity enrichment, followed by nanospray ionization mass spectrometry (NSI-MS/MS), to identify potential biomarkers for breast carcinoma. We identified L-PHA reactive glycoproteins from matched normal (nondiseased) and malignant tissue isolated from patients with invasive ductal breast carcinoma. Comparison analysis of the data identified 34 proteins that were enriched by L-PHA fractionation in tumor relative to normal tissue for at least 2 cases of ductal invasive breast carcinoma. Of these 34 L-PHA tumor enriched proteins, 12 are common to all 4 matched cases analyzed. These results indicate that lectin enrichment strategies targeting a particular glycan change associated with malignancy can be an effective method of identifying potential biomarkers for breast carcinoma.
Keywords: breast, cancer, gnt-V, lectin, mass spectrometry, glycoprotein, glycan
Introduction
Breast carcinoma is the second leading cause of cancer deaths among women in the U.S. behind lung cancer.1 Early detection and diagnosis of breast cancer significantly improves 5-year survival rates.2 Currently, the only approved screening method for the detection for breast cancer is mammography; therefore, there is an urgent need for the identification of serum biomarkers for use in early breast cancer detection. In the past few years, several large-scale proteomic studies have begun to characterize the proteome of breast cancer.3–5 This high-throughput strategy leads to complex data sets, and while rich in information, it is often difficult to predict proteins that may be sensitive and specific biomarkers for the disease. The ability to add an additional, complementary means to enrich for potential marker proteins would increase the probability of identifying candidate biomarkers. Glycosylation is clearly the most complex set of post-translational modifications that proteins undergo during biosynthesis, and several specific types of glycan epitopes have been shown to be associated with various types of cancer.6,7
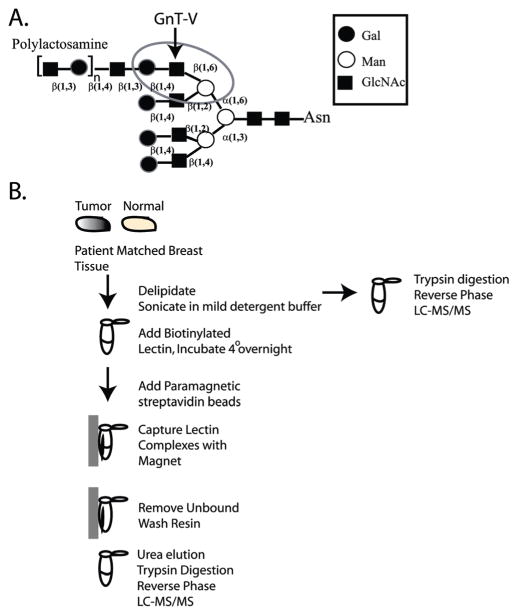
A particular type of N-linked glycosylation that is often increased in tumors is the N-linked β(1,6)-branched glycan8 that is bound by the lectin, L-PHA, when the glycan also expresses a distal β(1,4)-linked galactose9,10 (circled in Figure 1A). For example, staining of normal breast epithelia shows insignificant reactivity with L-PHA; yet, in breast carcinoma, staining by this lectin significantly increases.11,12 Expression of β(1,6)-branched glycan structures in both breast and colon carcinoma appears to be a qualitative change exhibited at the transition to malignancy, which is likely caused by up-regulation of the glycosyltransferase known as GnT-V (GnT-Va, Mgat5a) that synthesizes the N-linked β(1,6) branch.13 This glycosylation is often a step toward the formation of more complex poly N-acetyllactosamine glycan structures (Figure 1A) which serve as ligands for the class of animal lectin known as galectins that are often elevated in metastatic carcinoma.14 A recent study staining >700 primary breast tumors with L-PHA found that β(1,6)-branched oligosaccharides were an independent prognostic indicator for poor outcome in primary node-negative tumors.15 Experimental modulation of GnT-V activity, both in vivo and in vitro, results in changes in carcinoma invasiveness and metastasis, supporting the conclusion that increases in the post-translational modification of proteins by GnT-V is a mechanism by which tumor cell malignancy may increase.16–19
Figure 1.
(A) A tetra-antennary N-linked oligosaccharide showing the GnT-V β(1,6) GlcNAc addition that leads to the formation of polylactosamine structures. The L-PHA recognition site is circled. (B) Schematic flow diagram for the L-PHA enrichment protocol.
We have developed a simple glycoproteomic strategy to identify the glycoproteins from breast tissue that bind to L-PHA. Our experimental design utilizes intact proteins and allows the identification of glycoproteins whose glycans are bound by the lectin L-PHA (L-phytohemagglutinin), as well as those proteins/glycoproteins that may associate with the bound glycoproteins. In this report, we describe lectin-affinity glycan enrichment followed by NSI-MS/MS using a Finnigan LTQ linear ion trap mass spectrometer, and subsequent proteomic data analysis, to identify L-PHA-enriched glycoproteins that are elevated in breast tumor tissue compared with patient matched normal breast tissue. Our results demonstrate that this technique can be an effective method to identify proteins with tumor-specific glycosylation changes. Over 30 proteins were identified with elevated L-PHA reactivity in tumor tissue compared with normal tissue for at least 2 of 4 cases analyzed; 12 of these were elevated in all 4 cases of ductal breast carcinoma analyzed. These results demonstrate that L-PHA glycoproteomic fractionation can identify potential markers of malignancy common to a set of breast tumors with diverse clinical features.
Materials and Methods
Specimens
Tissue specimens, matched adjacent normal and tumor, from patients with histologically proven invasive ductal breast carcinoma were collected in accordance with approved institutional review board Human subject’s guidelines at Emory University Hospital, Atlanta, GA. Board certified clinical oncologists and pathologists carried out all clinical and histological analysis of the biopsies. All specimens for this study were immediately frozen at −70 °C until proteomic analysis. For the initial validation of our L-PHA affinity enrichment method, we analyzed four patients with matched normal and malignant breast tissue (Table 1).
Table 1.
Summary of Cases Analyzeda
| case | unique proteins total NL | unique proteins total TU | unique proteins L-PHA-NL | Unique Proteins L-PHA-TU | tumorb grade | HER2 status | ER status | PR status | LNc status | L-PHAd level |
|---|---|---|---|---|---|---|---|---|---|---|
| 10406 | ND | ND | ND | ND | II, moderate | Positive | 3+ Positive | Negative | Yes | 1 |
| 10119 | 362 | 363 | 88 | 80 | II, moderate | Negative | 3+ Positive | 3+ Positive | No | 4 |
| 11827 | 349 | 515 | 70 | 118 | III, poor | ND | ND | ND | Yes | 4 |
| 2417 | 347 | 491 | 53 | 214 | III, poor | 2+ Positive | Negative | Negative | Yes | 10 |
| 2207 | 362 | 476 | 145 | 193 | III, poor | 2+ Positive | 2+ Positive | 3+ Positive | No | 9 |
NL = normal, TU = tumor, ND = not determined.
Nottingham grade and differentiation.
Yes indicates that regional lymph nodes were histologically positive.
Scaled L-PHA level measured by densitometry of 10% of the L-PHA captured proteins by gel electrophoresis and sypro ruby staining; 1 = no change between normal and tumor and 10 = the highest tumor L-PHA staining.
Sample Processing, L-PHA Enrichment, and MS
Frozen tissue samples were processed as follows: 100 mg of tissue was delipidated using a mixture of chloroform/methanol/water (4:8:3, v/v/v) as described previously.20,21 Delipidated and precipitated proteins were pelleted by centrifugation, and the pellet was given an additional wash with acetone and water (4:1) on ice for 15 min. Intact proteins were extracted from the delipidated tissues using a mild detergent solution as follows: 5 mg of delipidated protein powder was dissolved in 300 μL of 50 mM Tris-Cl, pH 7.5, 0.1% NP-40, 150 mM NaCl, 0.4 mM EDTA, and 1 protease inhibitor tablet, and the sample was sonicated three times for 10 s pulses at setting 5 (Vertis Virsonic microtip). The supernatant was taken after centrifugation at 10 000 rpm at 4 °C for 10 min. The protein concentration of the sample was determined by BCA assay, and 600 μg of total protein lysate was dialyzed overnight at 4 °C into 40 mM ammonium bicarbonate using a 4000 MWCO tube-O-dialyzer (GBiosciences). Minimal loss of protein occurred following dialysis due to the use of neutral nonbinding membrane, ≤ 5%. The sample was adjusted to 150 mM NaCl, 5 mM CaCl2, and 5 mM MgCl2 before the addition of the lectin. Biotinylated L-PHA (Vector Laboratories, Burlingame, CA) (10 μg) was added, and the sample was rotated at 4 °C overnight. Bound L-PHA reactive proteins were captured using 100 μL of paramagnetic streptavidin particles (Promega) at 4 °C for 2 h. After extensive washing in 1× PBS, captured proteins were eluted with 200 μL of 2 M urea/0.2 mM DTT/40 mM ammonium bicarbonate at 52 °C for 1 h. The eluted fraction was separated from the paramagnetic streptavidin particles using a magnetic stand. Ten percent fractions collected after elution were analyzed by gel electrophoresis and Sypro staining. Results indicate that 14% (84 μg) of the starting material (600 μg) elutes from the L-PHA column. Eluted proteins were carboxyamidomethylated by adding an equal volume of iodoacetamide (10 mg/mL in 40 mM ammonium bicarbonate) in the dark for 45 min and digested with 5 μg of sequencing grade trypsin (1:50, Promega) at 37 °C overnight. Tryptic peptides were acidified with 200 μL of 1% trifluoroacetic acid, and desalting was performed using C18 spin columns (Vydac Silica C18, The Nest Group, Inc.). Eluted peptides were dried in the speed-vac, resuspended in 78 μL of buffer A (0.1% formic acid) and 2 μL of buffer B (80% acetonitrile/0.1% formic acid), and filtered through a 0.2 μm filter (nanosep, PALL). Samples were loaded off-line onto a nanospray column/emitter (75 μm × 8.5 cm, New Objective) self-packed with C18 reverse-phase resin in a nitrogen pressure bomb for 10 min. Peptides were eluted via a 160-min linear gradient of increasing buffer B at a flow rate of approximately 250 nL/min directly into a linear ion trap (LTQ, Thermo Co., San Jose, CA) equipped with a nanoelectrospray ion source. The top eight ions from the full MS (300–2000 m/z) were selected for CID fragmentation at 34% with a dynamic exclusion of 2 fragmented mass ions using an exclusion time of 30 s.
Permethylation of Glycans
To facilitate the analysis of oligosaccharides by MS, N-linked glycans released by N-glycanase were permethylated as described previously.21 Briefly, following extraction from tissue samples, delipidated proteins were digested with trypsin and chymotrypsin. The chymotrypsin and trypsin was used for glycan analysis, not lectin glycoproteomic analysis. The resulting digests were enriched for glycopeptides, which were then treated with PNGaseF (Prozyme) to release N-linked glycans. Contaminants, buffer, salts, and residual peptides were removed from the released glycans by Sep-Pak C18 chromatography, and the resulting glycan preparation was permethylated prior to analysis by nanospray ionization mass spectrometry using a linear ion trap (LTQ; Thermo Finnagan). The total ion mapping (TIM) functionality of the Xcalibur software package (version 2.0) was used to obtain total glycan profiles for each sample. Through TIM analyses, automated MS and MS/MS spectra were obtained in small mass increments across a broad range of m/z values. For the analysis of tissue samples, TIM analysis was performed from m/z = 500–2000. This mass range collects MS profiles and MS/MS fragmentation spectra for glycans detected from their 1+ to 4+ charge states. Following data collection, resulting TIM profiles were filtered for the presence of characteristic glycan fragments in the associated MS/MS spectra. When the signal intensity of characteristic fragments is plotted as a function of elapsed scan time, a TIM chromatogram is generated that indicates the relative prevalence of specific glycan structures. For assessing the presence of β(1,6)-branched glycans extended with at least two N-acetyllactosamine repeats, TIM scans were filtered for the loss of a Hex-HexNAc-Hex-HexNAc fragment from the parent ion.
Proteomic Data Analysis
The raw peptide data was converted to mzXML using ReAdW, a software written at the Institute for Systems Biology in Seattle, WA (http://www.systemsbiology.org). MS/MS spectra were searched against the International Protein Index (IPI) human sequence database (IPI.HUMAN.v.3.26, 67 665 sequences) using MyriMatch.22 The MyriMatch search criteria included only tryptic peptides, all cysteines were presumed carboxyamidomethylated, and methionines were allowed to be oxidized. MyriMatch searches allowed a precursor error of up to 1.25 m/z and a fragment ion limit of within 0.5 m/z. All ambiguous identifications that matched to multiple peptide sequences were excluded. The identified proteins (2+ peptides required) from each individual tumor and normal sample were filtered and grouped using IDPicker software.23 IDPicker software incorporates searches against a separate reverse database, probability match obtained from MyriMatch, and DeltCN scores to achieve false-discovery rates typically <5%. Information about IDPicker tools can be found at http://www.mc.vanderbilt.edu/msrc/bioinformatics/. The raw data files were also analyzed using the TurboSequest algorithm24–26 (BioWorks 3.1, Thermo Finnigan) to achieve a false-discovery rate of less than 0.3% for proteins assigned by 2+ peptides using a separate inverted database. Our results indicate that the final MyriMatch/IDPicker proteins list and TurboSequest proteins list showed near complete agreement. All proteins reported in this manuscript were identified using both methods. We found no evidence of amino acid carbamylation following urea elution and DTT reduction. Variance for sample processing between normal and tumor samples was calculated by measuring the number of peptides identified for proteins that adhere to the L-PHA in a nonglycan-dependent manner, such as serum albumin. Our results indicate 12.6% ± 0.087 variance between normal and tumor for each case analyzed.
Biological Function Annotation
Proteins (defined by 2 or more peptides) showing differential binding to L-PHA for tumor compared to normal in at least 2 cases were converted to gene symbols and uploaded to DAVID 2007 (the Database for Annotation, Visualization and Integrated Discovery) for analysis.
Pull-down and Western Blot Experiments
One hundred micrograms of delipidated protein powder was solubilized in 1× TBS (Tris-buffered saline)/1% Triton X-100/protease inhibitor tablet for precipitation using antiperiostin (Abcam) (1 μg). Bound proteins were captured using protein G plus agarose or streptavidin paramagnetic particles before gel electrophoresis and transfer to PVDF membrane prior to probing using biotinylated L-PHA (1:5000). For L-PHA precipitations, 500 μg of delipidated protein powder was solubilized in 1× TBS/1% TritonX-100/protease inhibitors before adding 10 μg of biotinylated L-PHA and mixing overnight at 4 °C. Magnetic streptavidin beads (100 μL) were used to pull down the L-PHA bound complexes. After washing the beads, proteins were released by boiling in sample loading buffer and separated on 4–12% NuPage Bis-Tris gels and transferred to PVDF membrane before detection using either anti-periostin Ab (1:1000) (Abcam) or anti-haptoglobin Ab (1:200) (Santa Cruz Biotechnology). Blots were incubated with anti-rabbit HRP (1:5000) (Santa Cruz Biotechnology), anti-mouse HRP (1:5000) (Santa Cruz Biotechnology), or streptavidin-HRP (1:5000) (Vector Laboratories) before washing and detection using Western Lightening Plus (Perkin-Elmer).
Results
An initial set of 5 human ductal invasive breast carcinoma tissue samples was used to evaluate the use of the lectin L-PHA to bind and enrich for potential glycoprotein biomarkers to distinguish breast cancer from normal tissue. As shown in Table 1, these cases represent metastatic and nonmetastatic disease, cases positive and negative for amplified her2/neu, and cases that were both estrogen receptor (ER) and progesterone receptor (PR) positive and negative. Our approach is to analyze breast tissue, comparing normal and tumor tissue from the same patient, to identify potential glycoproteins that react with the lectin L-PHA. All cases showed increased levels of L-PHA binding indicative of β(1,6)-branched N-linked glycans except case 10406; therefore, case 10406 was not analyzed by NSI-MS. We conclude that 4 out of 5 cases or 80% of the tumor tissue analyzed had proteins with increased levels of β(1,6)-branched N-linked glycan structures relative to normal breast tissue. To isolate these L-PHA reactive glycoproteins, we developed a method using intact proteins for the lectin binding, which differs from the more common method of using glycopeptides (Figure 1B). In addition, we found that delipidation of the breast tissue prior to analysis significantly improved MS/MS results (Figure 1B).21
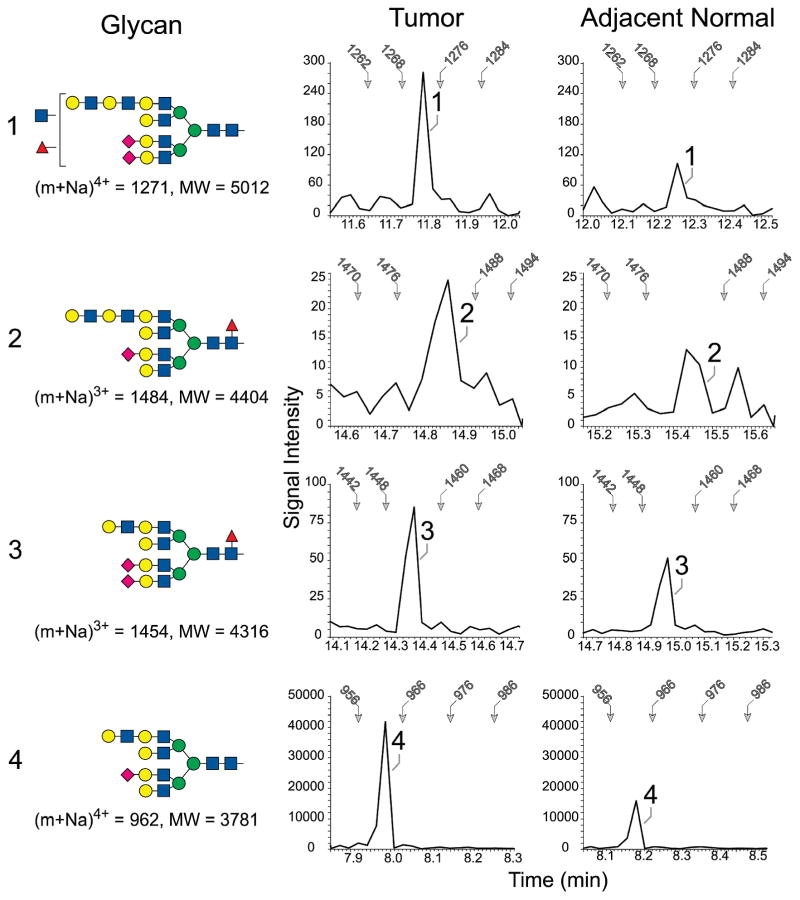
To verify that L-PHA binding accurately reports intrinsic differences in glycan expression between tumor and adjacent normal tissue, total N-linked glycans were profiled for tissues taken from case 2417 (Figure 2), which exhibited the greatest relative increase in tumor-associated L-PHA recognition (Table 1). The prevalence of glycans carrying a β(1,6) branch extended with N-acetyllactosamine was compared in tumor and adjacent normal preparations by quantifying the signal intensity of a specific fragment ion detected in TIM analysis. When the TIM profiles were filtered for the presence of a permethylated Hex-HexNAc-Hex-HexNAc fragment, the relative abundances of several β(1,6)-branched parent ions were measured and compared. The most prevalent of the detected glycans extended with N-acetylactosamine was increased more than 2.5-fold in tumor tissue relative to adjacent normal tissue, compared using equivalent protein amounts as starting material. The trimmed, high mannose glycan Man5GlcNAc2, that serves as an early precursor for complex terminal modifications including β(1,6)-branching, was found in equal prevalence between the two tissues (data not shown). Therefore, for case 2417, the increased abundance of L-PHA reactive proteins identified by MS/MS from tumor tissue compared with normal tissue correlates with increases in β(1,6)-branched N-glycan structures, confirming the specificity of the lectin enrichment analysis.
Figure 2.
Enrichment of tetra-antennary glycans extended with N-acetyllactosamine in tumor and adjacent normal tissue from case 2417. The four indicated N-linked glycans (1–4) were detected by NSI-MS/MS. In the profiles shown at the right of each glycan, the MS/MS spectra associated with the TIM scan for the indicated tissue were filtered to present the detected signal intensity of a signature tetrasaccharide fragment (Hex-HexNAc-Hex-HexNAc). The presence of this fragment indicates the detection of a glycan extended with at least two N-acetyllactosamine repeats at a scan time which predicts the m/z ratio for the parent ion. For reference, the scan time for specifc m/z values is indicated by arrows in each filtered profile. The shading and shapes for the glycan structures reflect standard nomenclature adopted by the Consortium for Functional Glycomics (CFG; GlcNAc, blue square; Gal, yellow circle, Man, green circle; Fuc, red triangle, NeuAc, pink diamond).
Proteins Showing Tumor-Specific Increased Reactivity with L-PHA
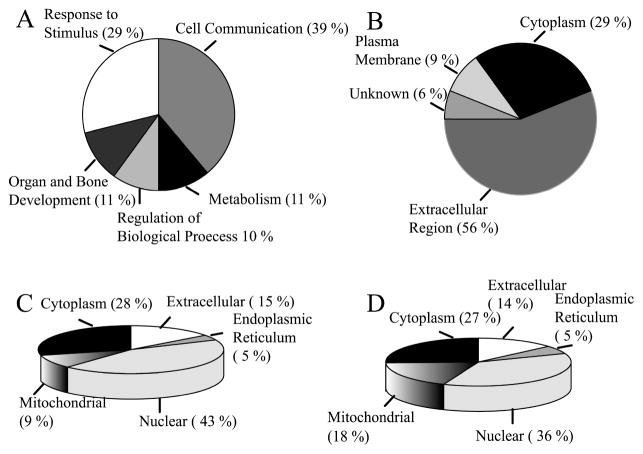
We detected a total of 258 L-PHA reactive proteins (2 or more peptides) from the 4 tumors showing elevated L-PHA reactivity that were analyzed by MS/MS. For each of the 258 unique proteins identified, we examined the number of peptides identified for that protein from NSI-MS/MS analysis of normal tissue and tumor tissue before and after L-PHA fractionation. Differences in protein abundance between normal and tumor were normalized by determining the ratio of peptides identified in patient matched normal and tumor tissue prior to L-PHA fractionation. Proteins were eliminated in each case if they did not show a minimum of 1.5-fold increase in peptides identified from tumor relative to normal tissue after L-PHA fractionation. From the list of proteins showing at least a 1.5-fold increase in tumor relative to normal tissue, proteins were considered “enriched” following L-PHA fractionation if they were identified from at least 2 cases. A total of 34 proteins had increased peptides and spectra present in L-PHA fractionations isolated from tumor compared with normal tissue for at least 2 separate cases of ductal invasive breast carcinoma (Table 2, number of spectra in parenthesis). The peptide sequences for these proteins are provided in Supplemental Table 1 in Supporting Information. As expected, the majority of these proteins are predicted to be glycoproteins by searching databases such as GenBank and IPI. Since we do not compete L-PHA bound proteins from the magnetic beads using a competitive sugar hapten, we have identified some proteins that are predicted to be nonglycosylated and are likely binding to L-PHA reactive glycosylated proteins. We utilized DAVID 2007 (Database for Annotation, Visualization and Integrated Discovery) to annotate the function of the proteins listed in Table 2 (Figure 3A). The top 2 functional classifications of glycoproteins showing differential binding to L-PHA in ductal invasive breast carcinoma relative to normal breast tissue are (i) binding proteins related to cell adhesion and cell-communication, and (ii) proteins that respond to external stimulus. These results indicate that largest group of proteins showing increases in β(1,6)-branched glycosylation function in mediating communication of the tumor cell with the extracellular matrix (COL6A1, COL6A2, COL6A3, COL14A1) and neighboring cells (HPX, BGN, DCN, VIM, POSTN, VTN, THBS1, OGN). The second highest group of proteins with elevated L-PHA reactive glycoproteins participates in the response of the tumor cells to environmental stress. This group includes the members of the lectin-induced complement pathway (C3 and C4B), activators of complement (HPR/HP), immunoglobulin/MHC complex (IGHA1, IGLV4-3, IGHM), enzyme inhibitors (SERPINA1, 14-3-3 zeta/delta, COL6A3, THBS1), enzyme activators of MMP (HPX), and enzymes that detoxify (PRDX1). This information contributes to our knowledge of the functions of glycoproteins acquiring β(1,6)-branched N-linked glycan structures in breast carcinoma and offers insights into how the acquisition of these structures may be associated with breast cancer progression. Several of the proteins enriched by L-PHA are predicted to participate in the development of the skeletal system and organ development (POSTN, ANXA2, DCN, COL6A3, PRDX1, and OGN). There are also several enzymes that participate in the glycolytic pathway that are enriched by L-PHA (PGK1, TPI1, ENO1, LDHA, and PKM2). Overall, these results suggest that proteins that are participating in cell communication, organ development, and metabolism derive β(1,6)-branched N-linked glycan structures in breast tumors. The abnormal elevated expression of these glycan structures on these proteins in breast epithelial cells may play roles in tumor progression and invasion.19
Table 2.
L-PHA Reactive Proteins with Elevated Peptides and Spectra in Tumor Relative to Normal for at Least 2 Cases
| protein IDa | abb name | name | 10119-NL | 10119-TU | 11827-NL | 11827-TU | 2207-NL | 2207-TU | 2417-NL | 2417-TU | GO functionb | cell compartment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPI00000874.1 | PRDX1 | Peroxiredoxin-1 | 0 | 0 | 0 | 1(1) | 1(2) | 2(3) | 0 | 0 | GO:0008283 proliferation | Unknown |
| IPI00007960.4 | POSTN | Periostin | 0 | 4(9) | 0 | 6(14) | 0 | 11(26) | 1(1) | 10(51) | GO:0007155 adhesion | Ext |
| IPI00010790.1 | BGN | Biglycan | 4(7) | 1(3) | 0 | 0 | 3(7) | 6(12) | 1(3) | 7(14) | GO:0005488 binding | Ext |
| IPI00010896.3 | CLIC1 | Cholride intracellular channel protein | 0 | 0 | 0 | 0 | 0 | 1(2) | 0 | 1(2) | GO:0006821 CI transport | PM |
| IPI00012119.1 | DCN | Decorin | 3(7) | 1(5) | 0 | 0 | 4(14) | 6(18) | 2(6) | 8(18) | GO:0009887 organ morpho | Ext |
| IPI00021263.3 | KCIP-1 | 14-3-3 zeta/delta | 0 | 1(1) | 0 | 3(5) | 0 | 3(6) | 0 | 0 | GO:0005488 binding | Cyto |
| IPI00021841.1 | APOA1 | Apo-A1 precursor | 1(1) | 2(4) | 3(7) | 2(4) | 4(7) | 1(1) | 1(1) | 5(8) | GO:0005488 binding | Ext |
| IPI00022488.1 | HPX | Hemopexin | 0 | 0 | 0 | 0 | 3 | 5 | 0 | 6 | GO:0005488 binding | Ext |
| IPI00025465.1 | OGN | Osteoglycin | 3(7) | 5(8) | 0 | 1(2) | 0 | 8(25) | 0 | 7(12) | GO:0008283 proliferation | Unknown |
| IPI00072917.2 | COL6A3 | Collagen VI alpha 3 | 22(56) | 25(47) | 10(14) | 7(10) | 40(105) | 60(167) | 26(167) | 86(290) | GO:0007155 adhesion | Ext |
| IPI00164623.4 | C3 | 187 kDa protein | 4(8) | 1(2) | 0 | 0 | 4(6) | 11(18) | 0 | 25(44) | GO:0005488 binding | Ext |
| IPI00169383.3 | PGK1 | Phosphoglycerate kinase 1 | 0 | 0 | 0 | 1(2) | 0 | 2(3) | 0 | 3 | GO:0003824 catalytic | Cyto |
| IPI00176193.6 | COL14A1 | isoform 1 of collagen 14 | 1(2) | 9(15) | 0 | 0 | 14(43) | 22(72) | 6(13) | 28(98) | GO:0016337 cell–cell adhesion | Ext |
| IPI00216691.5 | PROF1 | Profilin Lactate | 0 | 0 | 0 | 1(2) | 0 | 1(4) | 0 | 2(3) | GO:0005488 binding | Cyto |
| IPI00217966.7 | LDHA | Dehydrogenase isoform 1 | 0 | 0 | 0 | 1(2) | 0 | 3(6) | 0 | 1(1) | GO:0003824 catalytic | Ext |
| IPI00218343.4 | TUBA1C | Tubulin alpha 6 | 0 | 1(1) | 0 | 2(5) | 3(3) | 7(18) | 1(1) | 7(12) | GO:0005488 binding | Cyto |
| IPI00291136.4 | COL6A1 | Collagen alpha 1 VI chain precursor | 6(13) | 5(10) | 1(2) | 2(3) | 7(23) | 13(36) | 2(3) | 14(43) | GO:0007155 adhesion | Ext |
| IPI00296099.6 | THBS1 | Thrombospondin-1 | 0 | 0 | 0 | 3(4) | 0 | 0 | 0 | 1(2) | GO:0006928 motility | Ext |
| IPI00298971.1 | VTN | Vitronectin | 0 | 1(1) | 0 | 1(1) | 0 | 0 | 0 | 2(3) | GO:0007155 adhesion | Ext |
| IPI00304840.4 | COL6A2 | Isoform 2C2 collagen alpha-1 VI | 7(19) | 8(15) | 5(7) | 3(7) | 10(22) | 12(31) | 5(13) | 14(62) | GO:0016337 cell–cell adhesion | Ext |
| IPI00304925.4 | HSPA1 | Heat Shock 70 kDa | 0 | 0 | 0 | 1(1) | 0 | 1(1) | 0 | 0 | GO:0006986 unfolded protein re | Ext |
| IPI00329801.12 | ANXA5 | Annexin A5 | 0 | 0 | 0 | 2(2) | 0 | 1(2) | 0 | 0 | GO:0005488 binding | cyto |
| IPI00382938.3 | IGLV4–3 | variable Ig | 0 | 1(1) | 0 | 1(1) | 3(5) | 5(12) | 0 | 8(32) | GO:0006955 immune res | Ext |
| IPI00418163.3 | C4B | C4B complement | 0 | 0 | 0 | 0 | 1(2) | 3(7) | 0 | 3(5) | GO:0005488 binding | Ext |
| IPI00419585.9 | PPIA | Peptidyl prolyl isomerase A | 0 | 0 | 0 | 2(5) | 0 | 0 | 0 | 1(2) | GO:0003824 catalytic | |
| IPI00430842.3 | IGHA1 | MHC class 1 protein | 0 | 0 | 0 | 0 | 2(5) | 4(10) | 2(2) | 7(14) | GO:0005488 binding | Ext |
| IPI00455315.4 | ANXA2 | Annexin A2 | 0 | 0 | 1(1) | 2(2) | 2(3) | 5(14) | 0 | 0 | GO:0004857 enzyme inhibitor | PM |
| IPI00465028.7 | TP1 | Triosphosphate iso | 0 | 1(1) | 0 | 1(2) | 4(3) | 3(4) | 0 | 3(3) | GO:004807 carb metabolism | Cyto |
| IPI00465248.5 | ENO1 | Alpha enolase | 0 | 0 | 0 | 3(5) | 4(17) | 3(5) | 0 | 5(10) | GO:0004634 glycolytic | Cyto |
| IPI00472610.2 | IGHM | IGHM protein | 0 | 0 | 0 | 0 | 8(37) | 10(54) | 6(13) | 11(63) | GO:0006955 immune res | PM |
| IPI00477597.1 IPI00478493.3 |
HPR/HP | Isoform-1 Haptoglobin-related protein precursor/Haptoglobin | 0 | 1(1) | 0 | 1(2) | 0 | 1(2) | 0 | 3(4) | GO:006952 defense | Ext |
| IPI00479186.5 | PKM2 | Pyruvate kinase M2 | 0 | 0 | 0 | 0 | 1(1) | 5(12) | 0 | 3(9) | GO:0005488 binding | Cyto |
| IPI00553177.1 | SERPINA1 | Alpha-1 antitrypsin inhibitor | 0 | 1(1) | 2(4) | 2(4) | 4(11) | 4(12) | 1(1) | 8(17) | GO:0005488 binding | Ext |
| IPI00827679.1 | VIM | vimentin like 50 kDa | 1(1) | 3(6) | 6(9) | 6(12) | 2(3) | 5(8) | 0 | 5(6) | GO:0006928 motility | Cyto |
International protein index database. Number of spectra in parenthesis beside the number of peptides.
Gene functions determined by the Gene Ontology Consortium.
Figure 3.
Functional annotation and distribution of the L-PHA-enriched proteins identified from breast carcinoma. (A) Biological function of proteins listed in Table 2 as annotated by DAVID 2007. (B) Cellular compartment for L-PHA-enriched proteins assigned based on GO consortium. (C) Cellular compartment of proteins identified from normal breast tissue by total MS/MS analysis assigned by GO consortium. (D) Cellular compartment of proteins identified from tumor breast tissue assigned by GO consortium.
Interestingly, many of the proteins identified in Table 2 are connected with TGFβ signaling pathways. A recent manuscript identifies increased TGFβ protein levels in breast tumor tissue as a factor that correlates with shorter disease-free survival.27 Many of the proteins we identified after L-PHA enrichment in tumor tissue are either induced by TGFβ (POSTN, COL6A3, SERPINA1) or are known to bind TGFβ with nanomolar affinities (BGN, DCN). It will be interesting to determine how β(1,6)-branched N-linked glycosylation influences the binding of TGFβ to these proteins.
L-PHA Capture Facilitates the Identification of Biomarkers from the Extracellular Region
We have examined the predicted cellular distribution of proteins enriched by L-PHA listed in Table 2. The distribution is primarily extracellular (56%) and cytoplasmic (29%) with the remainder of proteins localized on the plasma membrane (9%) or unknown (6%) (Figure 3B). L-PHA-affinity enrichment significantly concentrates proteins identified from the extracellular region. For comparison, only 14–15% of the total proteins isolated from normal and tumor before L-PHA enrichment are extracellular (Figure 3C, D).
L-PHA Enrichment Can Identify Novel Markers for Breast Carcinoma
Approximately 74% of the 34 tumor L-PHA-enriched proteins have been previously reported in breast cancer studies (n = 25); while 26% (n = 9) have not been previously cited for breast cancer (Table 2). One of the novel, tumor-specific L-PHA reactive glycoproteins that we identified, known as osteoglycin (OGN), was present at similar levels of peptide abundance in normal and tumor breast tissue before L-PHA fractionation. However, OGN is consistently identified from tumor tissue after L-PHA fractionation. Osteoglycin has not been identified as a potential marker for breast cancer previously, likely due to the consistent levels of protein present in normal and tumor. Another interesting protein not previously associated with breast cancer is the 14-3-3 zeta protein. This protein is not predicted to be glycosylated and is probably enriched by L-PHA due to association with a protein that binds L-PHA. 14-3-3 zeta functions as an adapter protein that binds with other proteins controlling cell growth and proliferation. The cellular localization of 14-3-3 zeta protein has been reported to be largely cytoplasmic, but it has been reported to be present on the plasma membrane and in the Golgi (reviewed in ref 28). We find in 3 out of the 4 cases significant tumor-specific association with L-PHA for 14-3-3 zeta. Similar levels of 14-3-3 zeta present in normal and tumor tissue probably prevented previous identification of this protein as a marker for breast cancer. We conclude that selective enrichment using the lectin L-PHA has enabled the identification of novel markers for breast carcinoma and adds an additional level of biomarker selection.
L-PHA Enrichment Increases the Identification of Markers Common to Breast Carcinoma Cases with Diverse Clinical Features
The quest to identify markers for the early detection of many tumors has been hampered by tumor heterogeneity. Our approach, focusing on a specific post-translational modification that increases in parallel with malignant progression, has enabled the identification of 12 markers common to all 4 cases of breast carcinoma analyzed (Figure 4). A key element of our success is the targeting of β(1,6)-branched N-linked glycan structures that are normally expressed at a low level in nondiseased breast epithelial cells. This targeted approach has enabled the identification of proteins that change glycosylation only in breast carcinoma tumor tissue.
Figure 4.
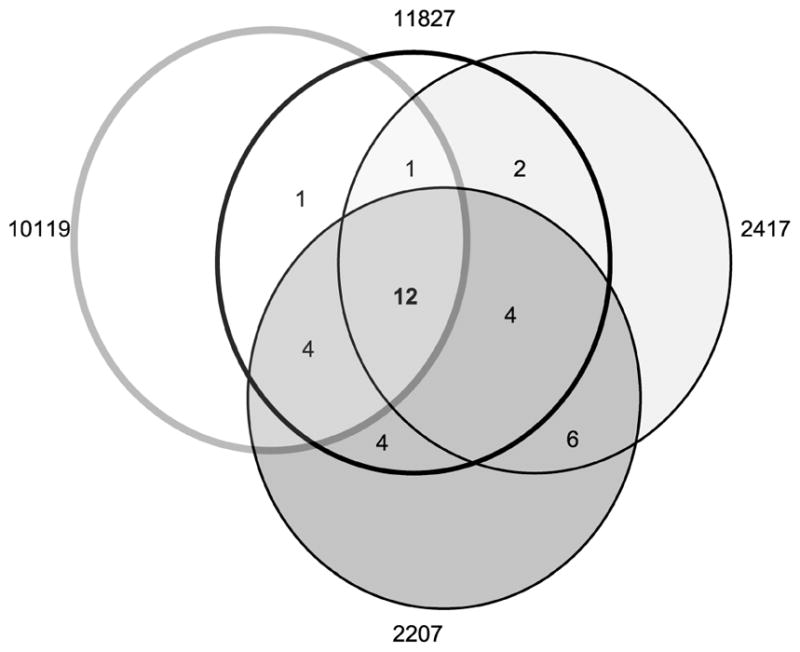
Venn diagram showing the number of L-PHA-enriched proteins identified in common for each case.
Validation of Glycoproteins with Differential L-PHA Reactivity in Normal and Tumor Tissue
We have selected 2 markers for further validation from Table 2. These glycoproteins were selected due to enrichment in tumor tissue for all cases analyzed in Table 2.
1. Periostin
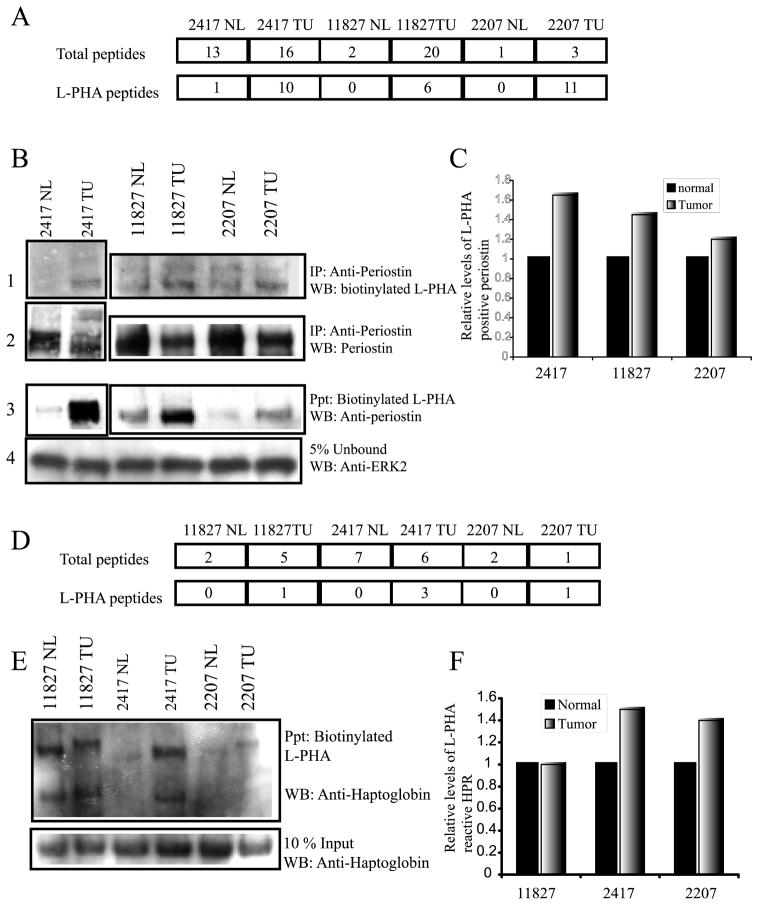
Periostin, POSTN, has been identified as a possible factor promoting breast cancer progression through induction of angiogenesis.29 Previous studies analyzing mRNA levels by pooled RNA sampling and immunohistochemistry arrays indicated that POSTN mRNA and protein levels were increased in malignant breast epithelial cells.29,30 We found in the total MS/MS analysis of matched normal and malignant breast tissue before L-PHA fractionation that there were roughly twice as many POSTN peptides identified from tumor tissue compared with normal tissue for cases 10119 and 2207. For case 11827, we detected a different isoform of periostin known as isoform 3, before L-PHA fractionation at 10 times the level found in normal tissue. However, for case 2417, we found equivalent levels of POSTN peptides identified in normal and tumor tissue before L-PHA enrichment using NSI-MS/MS (Figure 5A). These results are not quantitative and are only a qualitative assessment of abundance. However, they suggest that examining alterations in the abundance of POSTN protein alone would not be a selective marker for breast carcinoma. However, we identified POSTN peptides after L-PHA enrichment and MS/MS analysis in all 4 of the tumor tissues analyzed with only 1 peptide identified in normal tissue for case 2417 (Figure 5A). The high degree of tumor-specific association of POSTN with L-PHA suggests that the presence of β(1,6)-branched N-linked glycosylation on POSTN is a marker of breast cancer. To confirm these results, we have immunoprecipitated POSTN from normal and tumor breast tissue using an anti-periostin antibody and probed the blot using biotinylated L-PHA. Tissue amounts were limiting for case 10119 preventing further analysis of this case. For each case analyzed, POSTN reactivity with L-PHA is higher compared with the matched normal tissue control (Figure 5B, panel 1). To normalize for total periostin protein, we probed the membrane using the anti-periostin antibody (Figure 5B, panel 2). To confirm these results, we precipitated using biotinylated L-PHA followed by detection using the anti-periostin antibody (Figure 5B, panel 3). To ensure equivalent amounts of protein were present in the lectin precipitation reactions, 5% of the unbound material was analyzed by Western blot using an ERK2 antibody (Figure 5B, panel 4). To quantitate the relative increase in L-PHA reactive periostin, films for total periostin and L-PHA reactive periostin were scanned and analyzed by densitometry (Figure 5C). Immunoprecipitation and Western blot analysis used for validation were performed in the presence of 1% detergent; while the L-PHA pull-down for MS/MS analysis is performed under nondenaturing conditions. This technical difference may account for the detection of L-PHA reactive periostin in the Western blot data that was not observed in the MS/MS data. Qualitatively, the Western blot and the MS/MS data correlate, suggesting that POSTN acquires increased levels of β(1,6)-branched N-linked structures in the breast tumor tissue relative to normal breast tissue.
Figure 5.
Analysis of periostin and haptoglobin-related protein or haptoglobin by Western blot. (A) Number of peptides identified for periostin (isoform 1 and isoform 3) before L-PHA fractionation (total) and after lectin fractionation (L-PHA). (B) Precipitation of periostin using an anti-periostin antibody followed by detection using biotinylated L-PHA and streptavidin HRP (panel 1) (lower band is periostin). Total levels of periostin precipitated are confirmed by detection of the blot using anti-periostin antibody (panel 2). Reverse precipitation with L-PHA first followed by detection using an anti-periostin antibody (panel 3). Protein inputs for the L-PHA precipitations were normalized by the detection of ERK2 in 5% of the unbound fraction (panel 4). (C) Densitometry quantification of the relative increase in L-PHA reactive periostin normalized for total periostin. (D) Number of peptides identified for haptoglobin-related protein (HPR) precursor and haptoglobin (HP) by MS/MS before (total) and after L-PHA fractionation (L-PHA). (E) L-PHA precipitation followed by detection using an anti-haptoglobin antibody shows increased reactivity for the beta chains of tumor HPR/HP for cases 2417 and 2207 and, in all cases, a migratory shift to a higher molecular weight. Protein levels present in the L-PHA precipitations were determined by analysis of total haptoglobin in a 10% input blot. (F) Levels of L-PHA reactive haptoglobin were determined following densitometry analysis of L-PHA reactive HPR/HP and total HPR/HP determined from the 10% input levels.
2. Haptoglobin-Related Protein Precursor or Haptoglobin
Haptoglobin-related precursor protein (HPR) has 90% sequence identity to the conventional form of haptoglobin (HP) found in serum. The adult liver expresses low levels of HPR mRNA,31 and very little HPR protein has been detected in serum.32 This suggests that, if HPR was found in the serum of breast cancer patients, it would likely originate from the breast tumor. HPR shares antigenic epitopes with the pregnancy-associated plasma protein-A that is secreted from uterine epithelial cells or placenta during pregnancy and has been reported as an independent prognostic factor useful for detecting the recurrence of breast cancer.33,34 We have identified peptides common to haptoglobin (HP), haptoglobin precursor protein, isoform 1 of HPR, and isoform 2 of HPR from breast carcinoma and normal tissues. The peptides we identified after L-PHA precipitation for each tumor are common to both HP and HPR. Therefore, we cannot distinguish at this time whether the β(1,6)-branched N-linked glycan structure is present on HP or HPR. To validate the identification of L-PHA reactive HP/HPR as a breast tumor-specific marker, we performed L-PHA precipitation followed by Western blotting using an anti-haptoglobin antibody that recognizes HP as well as HPR (Figure 5E). To normalize the levels of L-PHA-positive haptoglobin versus total haptoglobin, 10% of the protein input for the L-PHA precipitation was analyzed for haptoglobin (Figure 5E, lower panel). Our results revealed an increased association of the beta chain of haptoglobin with L-PHA for the tumor tissue compared with normal breast tissue in cases 2417 and 2207. For case 11827, we observed a similar intensity in beta haptoglobin between normal and tumor on the Western blot. In all 3 cases, the tumor haptoglobin displayed a shift to a larger molecular weight compared with normal breast tissue from the same patient. Therefore, we conclude that in agreement with our MS/MS data, breast tumor haptoglobin has elevated β(1,6)-branched N-linked glycan structures compared with normal tissue.
Discussion
Elevation of β(1,6)-branched N-linked glycans in breast cancer has been previously cited as a poor prognostic indicator.15 In this study, we have used L-PHA, a lectin that specifically recognizes these glycan structures, to pull out potential biomarkers for breast carcinoma. Using this type of targeted glycoproteomic approach enabled us to identify markers common to breast cancer tissues with different stages, hormone receptor status, lymph node status, and her2/neu status. Our ability to analyze the relative abundance of biomarkers in normal and tumor tissue from the same patient, before and after L-PHA fractionation, eliminates possible bias that may be introduced from differences in individual gene expression profiles. Our targeted glycoproteomic approach has enabled the identification of several potential markers for breast carcinoma. Therefore, future studies focused on defining the normal and tumor glycome of various tissues can be useful for the development of new lectin targeting strategies to identify glycoproteins with tumor-specific glycan alterations.
TGFβ Connection
We have identified several proteins either induced by TGFβ or known to associate with TGFβ, suggesting a link between β(1,6)-branched N-linked glycosylation in breast tumors and the TGFβ pathway. Changes in downstream signaling controlled by TGFβ have been documented for breast cancer.35 However, unlike other types of malignancy that have evaded the normal growth-inhibitory functions of TGFβ through inactivating mutations in TGFβ receptors, the mechanisms of breast cancer resistance to TGFβ-mediated growth inhibition remain poorly understood (reviewed in ref 36). We have found increased β(1,6)-branched N-linked glycosylation on several extracellular proteins known to interact with TGFβ. Differential glycosylation of these ligands may initiate alterations in TGFβ signaling. The extracellular proteoglycans decorin and biglycan have been shown to bind TGFβ reducing its bioavailability for TGFβRI and TGFβRII during skeletal muscle differentiation.37 Therefore, it may be possible that the aberrant glycosylation of small leucine-rich proteoglycans such as decorin (DCN) and biglycan (BGN) increases the amount of TGFβ sequestered in the extracellular matrix complexes making less TGFβ available for canonical TGFβ receptor activation. Increased expression of TGFβ in fibroblasts has been shown to induce the expression of several glycoproteins implicated in the pathogenesis of breast cancer such as collagen VI α3, tenascin, and PAI-1.38 In this study, we have identified collagen VI α3 (COL6A3) as one of the proteins enriched by L-PHA in tumor tissue. Collagen VI up-regulation and secretion is associated with increased cell survival via resistance to apoptosis through down-regulation of Bax and prevention of β1 integrin-mediated apoptosis.39 Logically, increased levels of TGFβ in the tumor should initiate the cytostatic effect often associated with TGFβ signaling. However, elevated TGFβ levels in the tumor can have an opposite effect by inducing collagen VI expression that can oppose cytostatic effects by blocking apoptosis.39 Interestingly, collagen VI has been shown to associate in a ternary complex with DCN and BGN by co-immunoprecipitation experiments.40 COL6A3, DCN, and BGN were enriched by L-PHA in at least 2 cases (Table 2), suggesting a possible link between the formation of this complex and the presence of β(1,6)-branched N-linked glycans. Experiments investigating the impact of β(1,6)-branched glycosylation on the formation of this complex and the resulting effect on cell survival in breast epithelial cells will be necessary to evaluate this hypothesis as a possible mechanism for breast tumor cells evasion of TGFβ-induced cytostatic response.
An Enrichment of L-PHA Reactive Glycoproteins That Have Functions in the Skeletal System
Many of the proteins that were enriched by L-PHA in tumor tissue have proposed functions in the skeletal system, and a gene encoding one of these proteins, periostin (POSTN), has been identified as a gene expressed in the myoepithelial cells of breast tumors.41 This gene has also been reported to be expressed in normal bone chondrocytes and preosteoblasts; both are mesenchymal cell types.42 We find in our analysis of breast tissue after L-PHA fractionation that we detect POSTN almost exclusively in the tumor. This is the first identification of a difference in the glycosylation of POSTN for breast cancer tissue relative to normal breast tissue. Prior to L-PHA fractionation we detect POSTN in both normal and tumor tissue, likely due to stromal expression. We do not separate epithelial and stromal cells before L-PHA fractionation; therefore, our data strongly support the notion that POSTN with complex β(1,6)-branched N-linked glycan structures is expressed mainly in the breast tumor epithelial cells since there is very little L-PHA reactive POSTN isolated from normal breast tissue. The activation of POSTN expression in osteoblasts has been linked to twist, a bHLH transcription factor that controls the expression of embryonic mesenchymal genes during development.43 Twist expression has been reported to be increased in lobular infiltrating breast cancer.44,45 Therefore, these observations may explain how POSTN is expressed in breast epithelial cells. Also, TGFβ has been shown to increase the expression of POSTN in cardiac development providing another possible explanation for how POSTN may be expressed in breast cancer epithelial cells.46
GnT-V, the gene that adds the β(1,6) GlcNAc branch leading to the formation of β(1,6)-branched N-linked glycans, has recently been implicated in the maintenance of bone density as GnT-V (−/−) mice show a loss of bone mineral density.18 It is possible that POSTN may be a preferred substrate for GnT-V in osteoblasts and chondrocytes during development. Therefore, the abnormal expression of both GnT-V and POSTN in breast carcinoma epithelial cells would lead to significant changes in breast epithelial cell adhesion and migration, promoting tumor progression.
Another protein, osteoglycin (OGN), which was enriched by L-PHA in breast tumor tissue relative to normal tissue, was originally identified in bone as an osteoinductive factor.47 OGN along with DCN and BGN are members of a group of small lecuine-rich repeat proteoglycans (SLRPs) that are important during skeletal development. BGN and DCN are also involved in the development and maintenance of bone. More pronounced loss of bone mass is present in the double knockout of BGN and DCN than for each individual gene knockout, suggesting that both of these SLRPs play a role in the maintenance of bone.48 Interestingly, DCN is also found to be expressed in the myoepithelial cells of the breast.30 Exactly how the presence of β(1,6)-branched N-linked glycosylation on these proteins within the breast tissue may influence malignancy is unknown. One can postulate that increased β(1,6)-branched N-linked glycans may influence the formation of collagen fibrils making it easier for tumor cells to migrate and invade through the basement membrane. Many proteins that we have identified as highly L-PHA reactive in tumor tissue relative to normal tissue have reported functions in skeletal development. This suggests that L-PHA reactive N-linked structures may be promoting mesenchymal functions within breast epithelial cells. Future experiments will test if GnT-V expression levels can affect the epithelial to mesenchymal transition (EMT).
We are continuing to identify L-PHA reactive proteins from breast carcinoma tissue samples with future experiments also focused on determining if these L-PHA reactive markers can be identified from patient serum.
Supplementary Material
Acknowledgments
This work is supported by a grant from the Georgia Cancer Coalition (to M.P.) and from NIH/NIGMS (to M.T.) and a grant from NCI R01CA064462 and the NCRR Center for Biomedical Glycomics P41RR018502. L.W. is a Georgia Cancer Coalition Distinguished Cancer Scientist and K.L.A. is a postdoctoral fellow supported by a grant from the American Cancer Society and the Canary Fund (PF-05-233-01-SIED).
Abbreviations
- GnT-V
N-acetylglycosyltransferase
- V L-PHA
Phaseolus Vulgaris Leucoagglutinin
- NSI
nanospray ionization MS/MS tandem mass spectrometry
- DTT
dithiothreitol
- PBS
phosphate buffered saline
- TBS
Tris buffered saline
- EDTA
ethylenediamine tetraacetic acid
- TGFβ
transforming growth factor beta
Footnotes
Supporting Information Available: Supplemental Table 1 showing the peptide sequences of the identified proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55(1):10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Ries L, Eisner M, Kosary CL. SEER Cancer Statistics Review. National Cancer Institute; Bethesda, MD: 2005. [Google Scholar]
- 3.Pucci-Minafra I, Cancemi P, Fontana S, Minafra L, Feo S, Becchi M, Freyria AM, Minafra S. Expanding the protein catalogue in the proteome reference map of human breast cancer cells. Proteomics. 2006;6(8):2609–2625. doi: 10.1002/pmic.200500627. [DOI] [PubMed] [Google Scholar]
- 4.Celis JE, Gromov P, Cabezon T, Moreira JM, Ambartsumian N, Sandelin K, Rank F, Gromova I. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol Cell Proteomics. 2004;3(4):327–344. doi: 10.1074/mcp.M400009-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Hudelist G, Singer CF, Pischinger KI, Kaserer K, Manavi M, Kubista E, Czerwenka KF. Proteomic analysis in human breast cancer: identification of a characteristic protein expression profile of malignant breast epithelium. Proteomics. 2006;6(6):1989–2002. doi: 10.1002/pmic.200500129. [DOI] [PubMed] [Google Scholar]
- 6.Kim YJ, Varki A. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj J. 1997;14(5):569–576. doi: 10.1023/a:1018580324971. [DOI] [PubMed] [Google Scholar]
- 7.Hakomori S. Tumor-associated carbohydrate antigens defining tumor malignancy: basis for development of anti-cancer vaccines. Adv Exp Med Biol. 2001;491:369–402. doi: 10.1007/978-1-4615-1267-7_24. [DOI] [PubMed] [Google Scholar]
- 8.Pierce M, Arango J. Rous sarcoma virus-transformed baby hamster kidney cells express higher levels of asparagine-linked tri- and tetraantennary glycopeptides containing [GlcNAc-beta(1,6)Man-alpha (1,6)Man] and poly-N-acetyllactosamine sequences than baby hamster kidney cells. J Biol Chem. 1986;261(23):10772–10777. [PubMed] [Google Scholar]
- 9.Cummings RD, Kornfeld S. Fractionation of asparagine-linked oligosaccharides by serial lectin-Agarose affinity chromatography. A rapid, sensitive, and specific technique. J Biol Chem. 1982;257(19):11235–11240. [PubMed] [Google Scholar]
- 10.Cummings RD, Kornfeld S. Characterization of the structural determinants required for the high affinity interaction of asparagine-linked oligosaccharides with immobilized Phaseolus vulgaris leukoagglutinating and erythroagglutinating lectins. J Biol Chem. 1982;257(19):11230–11234. [PubMed] [Google Scholar]
- 11.Fernandes B, Sagman U, Auger M, Demetrio M, Dennis JW. Beta 1–6 branched oligosaccharides as a marker of tumor progression in human breast and colon neoplasia. Cancer Res. 1991;51(2):718–723. [PubMed] [Google Scholar]
- 12.Dennis JW, Laferte S. Oncodevelopmental expression of–GlcNAc beta 1–6Man alpha 1--6Man beta 1–branched asparagine-linked oligosaccharides in murine tissues and human breast carcinomas. Cancer Res. 1989;49(4):945–950. [PubMed] [Google Scholar]
- 13.Buckhaults P, Chen L, Fregien N, Pierce M. Transcriptional regulation of N-acetylglucosaminyltransferase V by the src oncogene. J Biol Chem. 1997;272(31):19575–19581. doi: 10.1074/jbc.272.31.19575. [DOI] [PubMed] [Google Scholar]
- 14.Lagana A, Goetz JG, Cheung P, Raz A, Dennis JW, Nabi IR. Galectin binding to Mgat5-modified N-glycans regulates fibronectin matrix remodeling in tumor cells. Mol Cell Biol. 2006;26(8):3181–3193. doi: 10.1128/MCB.26.8.3181-3193.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handerson T, Camp R, Harigopal M, Rimm D, Pawelek J. Beta1,6-branched oligosaccharides are increased in lymph node metastases and predict poor outcome in breast carcinoma. Clin Cancer Res. 2005;11(8):2969–2973. doi: 10.1158/1078-0432.CCR-04-2211. [DOI] [PubMed] [Google Scholar]
- 16.Guo HB, Lee I, Kamar M, Akiyama SK, Pierce M. Aberrant N-glycosylation of beta1 integrin causes reduced alpha5beta1 integrin clustering and stimulates cell migration. Cancer Res. 2002;62(23):6837–6845. [PubMed] [Google Scholar]
- 17.Guo HB, Lee I, Kamar M, Pierce M. N-acetylglucosaminyl-transferase V expression levels regulate cadherin-associated homotypic cell-cell adhesion and intracellular signaling pathways. J Biol Chem. 2003;278(52):52412–52424. doi: 10.1074/jbc.M308837200. [DOI] [PubMed] [Google Scholar]
- 18.Cheung P, Pawling J, Partridge EA, Sukhu B, Grynpas M, Dennis JW. Metabolic homeostasis and tissue renewal are dependent on beta1,6GlcNAc-branched N-glycans. Glycobiology. 2007;17(8):828–837. doi: 10.1093/glycob/cwm048. [DOI] [PubMed] [Google Scholar]
- 19.Guo HB, Randolph M, Pierce M. Inhibition of a specific N-glycosylation activity results in attenuation of breast carcinoma cell invasiveness-related phenotypes: inhibition of epidermal growth factor-induced dephosphorylation of focal adhesion kinase. J Biol Chem. 2007;282(30):22150–22162. doi: 10.1074/jbc.M611518200. [DOI] [PubMed] [Google Scholar]
- 20.Seppo A, Moreland M, Schweingruber H, Tiemeyer M. Zwitterionic and acidic glycosphingolipids of the Drosophila melanogaster embryo. Eur J Biochem. 2000;267(12):3549–3558. doi: 10.1046/j.1432-1327.2000.01383.x. [DOI] [PubMed] [Google Scholar]
- 21.Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282(12):9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 22.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6(9):3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 25.Yates JR, III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67(8):1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 26.Yates JR, III, McCormack AL, Link AJ, Schieltz D, Eng J, Hays L. Future prospects for the analysis of complex biological systems using micro-column liquid chromatography-electrospray tandem mass spectrometry. Analyst. 1996;121(7):65R–76R. doi: 10.1039/an996210065r. [DOI] [PubMed] [Google Scholar]
- 27.Desruisseau S, Palmari J, Giusti C, Romain S, Martin PM, Berthois Y. Determination of TGFbeta1 protein level in human primary breast cancers and its relationship with survival. Br J Cancer. 2006;94(2):239–246. doi: 10.1038/sj.bjc.6602920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 29.Shao R, Bao S, Bai X, Blanchette C, Anderson RM, Dang T, Gishizky ML, Marks JR, Wang XF. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 2004;24(9):3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigoriadis A, Mackay A, Reis-Filho JS, Steele D, Iseli C, Stevenson BJ, Jongeneel CV, Valgeirsson H, Fenwick K, Iravani M, Leao M, Simpson AJ, Strausberg RL, Jat PS, Ashworth A, Neville AM, O’Hare MJ. Establishment of the epithelial-specific transcriptome of normal and malignant human breast cells based on MPSS and array expression data. Breast Cancer Res. 2006;8(5):R56. doi: 10.1186/bcr1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bensi G, Raugei G, Klefenz H, Cortese R. Structure and expression of the human haptoglobin locus. EMBO J. 1985;4(1):119–126. doi: 10.1002/j.1460-2075.1985.tb02325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fawcett HA, al-Hawi Z, Brzeski H. Identification of the products of the haptoglobin-related gene. Biochim Biophys Acta. 1990;1048(2–3):187–193. doi: 10.1016/0167-4781(90)90055-7. [DOI] [PubMed] [Google Scholar]
- 33.Kuhajda FP, Katumuluwa AI, Pasternack GR. Expression of haptoglobin-related protein and its potential role as a tumor antigen. Proc Natl Acad Sci USA. 1989;86(4):1188–1192. doi: 10.1073/pnas.86.4.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhajda FP, Piantadosi S, Pasternack GR. Haptoglobin-related protein (Hpr) epitopes in breast cancer as a predictor of recurrence of the disease. N Engl J Med. 1989;321(10):636–641. doi: 10.1056/NEJM198909073211003. [DOI] [PubMed] [Google Scholar]
- 35.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10(3):203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 36.Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580(12):2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 37.Droguett R, Cabello-Verrugio C, Riquelme C, Brandan E. Extracellular proteoglycans modify TGF-beta bio-availability attenuating its signaling during skeletal muscle differentiation. Matrix Biol. 2006;25(6):332–341. doi: 10.1016/j.matbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Berking C, Takemoto R, Schaider H, Showe L, Satyamoorthy K, Robbins P, Herlyn M. Transforming growth factor-beta1 increases survival of human melanoma through stroma remodeling. Cancer Res. 2001;61(22):8306–8316. [PubMed] [Google Scholar]
- 39.Ruhl M, Sahin E, Johannsen M, Somasundaram R, Manski D, Riecken EO, Schuppan D. Soluble collagen VI drives serum-starved fibroblasts through S phase and prevents apoptosis via down-regulation of Bax. J Biol Chem. 1999;274(48):34361–34368. doi: 10.1074/jbc.274.48.34361. [DOI] [PubMed] [Google Scholar]
- 40.Reinboth B, Thomas J, Hanssen E, Gibson MA. Beta ig-h3 interacts directly with biglycan and decorin, promotes collagen VI aggregation, and participates in ternary complexing with these macromolecules. J Biol Chem. 2006;281(12):7816–7824. doi: 10.1074/jbc.M511316200. [DOI] [PubMed] [Google Scholar]
- 41.Grigoriadis A, Mackay A, Reis-Filho JS, Steele D, Iseli C, Stevenson BJ, Jongeneel CV, Valgeirsson H, Fenwick K, Iravani M, Leao M, Simpson AJ, Strausberg RL, Jat PS, Ashworth A, Neville AM, O’Hare MJ. Establishment of the epithelial-specific transcriptome of normal and malignant human breast cells based on MPSS and array expression data. Breast Cancer Res. 2006;8(5):R56. doi: 10.1186/bcr1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blumer MJ, Schwarzer C, Perez MT, Konakci KZ, Fritsch H. Identification and location of bone-forming cells within cartilage canals on their course into the secondary ossification centre. J Anat. 2006;208(6):695–707. doi: 10.1111/j.1469-7580.2006.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oshima A, Tanabe H, Yan T, Lowe GN, Glackin CA, Kudo A. A novel mechanism for the regulation of osteoblast differentiation: transcription of periostin, a member of the fasciclin I family, is regulated by the bHLH transcription factor, twist. J Cell Biochem. 2002;86(4):792–804. doi: 10.1002/jcb.10272. [DOI] [PubMed] [Google Scholar]
- 44.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Auersperg N, Wong AS, Choi KC, Kang SK, Leung PC. Ovarian surface epithelium: biology, endocrinology, and pathology. Endocr Rev. 2001;22(2):255–288. doi: 10.1210/edrv.22.2.0422. [DOI] [PubMed] [Google Scholar]
- 46.Norris RA, Kern CB, Wessels A, Moralez EI, Markwald RR, Mjaatvedt CH. Identification and detection of the periostin gene in cardiac development. Anat Rec, Part A. 2004;281(2):1227–1233. doi: 10.1002/ar.a.20135. [DOI] [PubMed] [Google Scholar]
- 47.Bentz H, Nathan RM, Rosen DM, Armstrong RM, Thompson AY, Segarini PR, Mathews MC, Dasch JR, Piez KA, Seyedin SM. Purification and characterization of a unique osteoinductive factor from bovine bone. J Biol Chem. 1989;264(34):20805–20810. [PubMed] [Google Scholar]
- 48.Corsi A, Xu T, Chen XD, Boyde A, Liang J, Mankani M, Sommer B, Iozzo RV, Eichstetter I, Robey PG, Bianco P, Young MF. Phenotypic effects of biglycan deficiency are linked to collagen fibril abnormalities, are synergized by decorin deficiency, and mimic Ehlers-Danlos-like changes in bone and other connective tissues. J Bone Miner Res. 2002;17(7):1180–1189. doi: 10.1359/jbmr.2002.17.7.1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.