Abstract
APOBEC3A (A3A) inhibits the replication of a range of viruses and transposons and might also play a role in carcinogenesis. It is a single-domain deaminase enzyme that interacts with single-stranded DNA (ssDNA) and converts cytidines to uridines within specific trinucleotide contexts. Although there is abundant information that describes the potential biological activities of A3A, the interplay between binding ssDNA and sequence-specific deaminase activity remains controversial. Using a single-molecule atomic force microscopy spectroscopy approach developed by Shlyakhtenko et al. [(2015) Sci. Rep. 5, 15648], we determine the stability of A3A in complex with different ssDNA sequences. We found that the strength of the complex is sequence-dependent, with more stable complexes formed with deaminase-specific sequences. A correlation between the deaminase activity of A3A and the complex strength was identified. The ssDNA binding properties of A3A and those for A3G are also compared and discussed.
Graphical abstract
APOBEC3A (A3A) belongs to a family of single-stranded DNA (ssDNA) cytidine deaminases that inhibit the activity of a diverse number of retrotransposons1–3 and retroviruses4–8 and might also play a role in cancer.9 There are seven members of the APOBEC3 family in humans. A3A, A3C, and A3H have only one Zn-binding deaminase domain capable of converting cytidine into uridine in ssDNA.10 Unlike other single-domain family members, A3A has the unique capability to deaminate 5′-methyl cytidine into thymidine in ssDNA.2,11,12 Recently, both nuclear magnetic resonance (NMR) and crystal structures of A3A have been determined, and surfaces important for binding and deaminase activity have been identified.10,13
The relationship between A3A ssDNA binding and catalytic activities has been discussed.5,13 One study indicated that A3A binds and deaminates ssDNA substrates in a length-dependent manner, with the minimum being six to nine nucleotides.13 The same study indicated that the efficiency of binding of A3A to ssDNA increased with substrate length. However, higher concentrations of A3A were needed to achieve full deamination of longer targets. At the same time, several catalytically inactive A3A mutants still retained efficient ssDNA binding activity, suggesting that these two activities are separable. A considerable amount is known about the biophysical and biochemical properties of A3A, including binding specificity and deaminase activity;3,5,10,13 however, little is known about how different ssDNA sequences affect complex stability. This paper addresses this topic and particularly how deaminase-specific and nonspecific sequences affect the strength of A3A–ssDNA complexes.
To accomplish this goal, we applied a single-molecule atomic force spectroscopy (SMFS) technique, which has been widely used for the study of the protein–protein and protein–DNA interactions (see the recent review in ref 14 and references cited therein). We utilized SMFS to directly measure the strength of A3A–ssDNA complexes, based on the approach recently developed to study complexes of A3G with ssDNA.15 Our analysis of three different ssDNA targets revealed that A3A–ssDNA complexes are sequence-dependent, forming stronger complexes with deaminase-specific ssDNA targets than non-specific ones.
MATERIALS AND METHODS
DNA Preparation
Three 69-nucleotide ssDNA substrates with thiol attached to the 5′ end were used in this study: deaminase-specific sequence 1, 5′TACGTGTAGGAATTATATTAAAGAGAAAGTGATTTCATTTGAATGAATTTCATTTTGTTAGAATTGTTA3′; deaminase-specific sequence 2, 5′TACGTGTAGGAATTATATTAAAGAGAAAGTGAAACCCAAAGAATGAAAACCCAAATGTTAGAATTGTTA3′; nonspecific sequence 3, 5′TACGTGTAGGAATTATATTAAAGAGAAAGTGAAAAGAAGAGAATGAAAAAGAAGATGTTAGAATTGTA3′. Deaminase sites are underlined. All 5′-thiol-modified 69-nucleotide sequences were obtained from Integrated DNA Technologies, Inc. (Coralville, IA).
Protein Purification
Detailed procedures for A3A purification and ssDNA deamination activity quantification have been described previously.16,17
A3A Preparation
For force spectroscopy experiments, imidazole was removed from the protein’s storage buffer by dialyzing the protein with Slide-A-Lyzer dialysis cassettes (3.5K molecular weight cutoff, Thermo Fisher). The protein was dialyzed into 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 0.1% Triton X-100, and 10% glycerol buffer.
A3A-Cys Preparation
A3A cysteines C64 and C171 were identified as surface cysteines based on NMR data10 and were replaced with two alanines by site-directed mutagenesis in the pcDNA3.1-A3Ai-mycHis plasmid.18 An extra cysteine codon TGC was introduced into the 3′ end of six-His codons by overlap extension polymerase chain reaction. The encoded recombinant A3A-2A-mycHisCys (A3A-Cys), with cysteine at the C-terminus, was purified as described above.
Single-Molecule Force Spectroscopy (SMFS)
Tip Modification: Covalent Attachment at the N-Terminus of A3A
The details of AFM probe functionalization have been described in refs 19 and 20 and recently in ref 15. Briefly, the AFM probe (MSNL, Bruker, Santa Barbara, CA) was cleaned with methanol followed by UV irradiation for 1 h. The cleaned AFM probe was amino-functionalized with an APS solution.21 Then, the amino-functionalized AFM probe was incubated with 2 mM bifunctional linker SVA-PEG-SVA (MW = 3400; Laysan Bio Inc., Arab, AL) in a dimethyl sulfoxide (DMSO) solution for 3 h. The probe was washed with PBS buffer (pH 7.8), immersed in a 5 nM A3A solution, and left overnight. Before the experiment, 1 M Tris-HCl buffer (pH 7.4) was used to wash out the excess nonbound protein and to block nonreacted NHS groups on the surface of the probe. Finally, the probe was immersed in a probing buffer containing 50 mM HEPES (pH 7.4), 100 mM NaCl, 5 mM MgCl2, and 1 mM dithiothreitol (DTT).
Tip Modification: Covalent Attachment of A3A-cys at the C-Terminus
After the steps described above, the amino-functionalized AFM probe was incubated with 500 μM bifunctional SVA-PEG-MAL linker (MW = 3400; Laysan Bio Inc.) in a DMSO solution for 3 h. The probe was then washed with PBS buffer (pH 7.1) to remove DMSO and incubated overnight with 5 nM A3A-cys. The A3A-cys-immobilized probe was washed and left in the probing buffer. A3A-cys was treated with DTT to disrupt possible disulfide bonds. An ethyl acetate extraction method was used to remove DTT, as described in the protocol from Integrated DNA Technologies, Inc.
Surface Modification
This procedure was described previously.15,19,20,22 Briefly, the freshly cleaved mica surface was amino-functionalized with APS as described previously.15,21 Then, the amino-functionalized mica surface was coated with maleimide groups by treating the surface with 500 μM sulfo-GMBS [GMBS, N-y-maleimidobutyryl-oxysulfosucciminide ester (Thermo Fisher Scientific Inc., Waltham, MA)] in DMSO for 3 h. Then, the surface was washed with PBS buffer (pH 7.1) and incubated overnight with 30 μM 5′-thiol-modified 69-nucleotide oligonucleotides that were treated with DTT followed by an ethyl acetate extraction as described above. After the samples had been washed, the functionalized surface was immersed in probing buffer.
Single-Molecule Force Spectroscopy Experiments
They were conducted on the MFP3D instrument (Asylum Research, Santa Barbara, CA). The spring constant of the functionalized AFM probe (MSNL from Bruker) was measured using the thermal noise method and was in the range of 20–40 pN/nm. The force distance curves for interactions between ssDNA covalently attached to the surface through thiol groups at the 5′ end of ssDNA and A3A covalently attached via the N-terminus or C-terminus were recorded at room temperature in probing buffer. The parameters for the force spectroscopy experiments were set as follows: trigger force at contact, 100 pN; dwell time, 0.5 s; approach rate, 500 nm/s. The pulling velocity was varied in the range of 100–3000 nm/s.
Single-Molecule Force Spectroscopy Data Analysis
The selected specific individual rupture force events defined by the position of the rupture event corresponding to the tether lengths were analyzed using the Worm-Like Chain (WLC) approximation from Igor Pro 6.31 analysis software (Asylum Research), as described previously.15 Briefly, the force–distance (F–D) curves were fitted with the Worm-Like Chain (WLC) model using the following formula:
| (1) |
where F(x) is the force at distance x, kB is the Boltzmann constant, T is the absolute temperature, and Lp and Lc are the persistence length and the contour length, respectively. The persistence length was allowed to vary because of the existence of two flexible linkers: the PEG linker from the functionalized AFM probe and the ssDNA from the surface. Each experiment, consisting of hundreds of specific force–distance curves fitted with WLC, allowed us to determine rupture forces (F), persistence lengths (Lp), and contour lengths (Lc). The rupture forces and contour length distributions were assembled into histograms and fitted with Gaussian using Origin version 8.5 (Origin Lab, Northampton, MA).
RESULTS
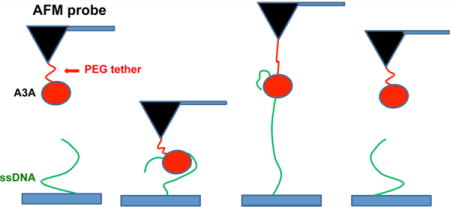
In the AFM force spectroscopy approach, as described in ref 15 and schematically shown in Figure 1, ssDNA covalently attached to the surface is probed by A3A immobilized on the AFM probe via a flexible tether (see Materials and Methods for specifics). The tether (PEG linker) provides A3A with orientational freedom for interacting with ssDNA. A3A initially is far from the ssDNA target (A) and then forms a complex with ssDNA when the probe approaches the surface (B). During the pulling stage (C), both PEG attached on the AFM probe and the ssDNA stretch, followed by rupture of the complex (D). Therefore, the rupture length defined by tether stretching is a characteristic feature of the specific rupture event. A typical overlay of multiple rupture force events is shown in Figure 2. The majority of the events are assembled around a single, characteristic rupture length, indicated with an arrow.
Figure 1.
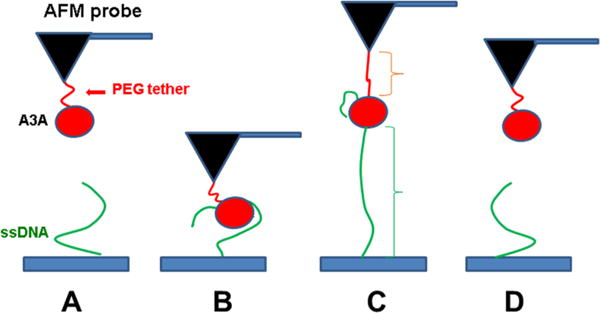
Schematic representation of the experimental setup. Protein is covalently attached to the AFM probe via a PEG tether (red), and ssDNA (green) is covalently attached at the 5′ end to the functionalized mica surface. (A) Initial position, where A3A and ssDNA are far from forming a complex. (B) As the probe approaches the surface, A3A captures ssDNA and forms a complex. (C) Retraction step causing stretching of the PEG tether and ssDNA. (D) Rupture of the complex.
Figure 2.
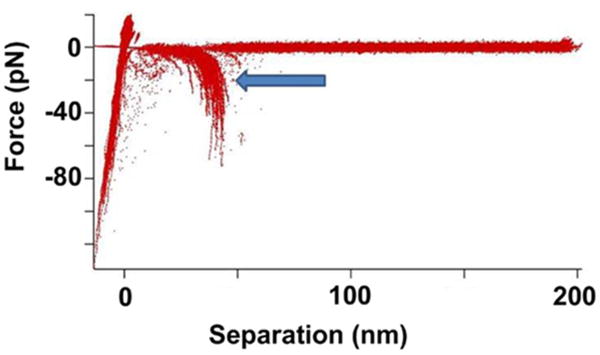
AFM spectroscopy experiments for probing interactions of A3A with ssDNA. An overlay of more than 100 force rupture events. The arrow points to the rupture event signature for the A3A complex with the 69-nucleotide ssDNA deaminase-specific sequence (arrow).
More than 1000 events were collected for each pulling rate, with an average yield of specific events of around 3–5%. The specificity of the interaction between A3A on the probe and ssDNA on the surface was verified by control experiments. In one control experiment, the functionalized mica surface without ssDNA attached was probed with A3A protein tethered to the AFM probe. The number of rupture events was less than 1%, with an estimated contour length of ~25 nm, as expected for a nonspecific interaction of PEG-A3A with the functionalized surface. In a second control experiment, the functionalized AFM tip without A3A protein (PEG only) was used to probe the interaction with ssDNA tethered to the mica surface. The yield of rupture events was only around 0.1%, which indicates that there are no specific interactions between ssDNA on the surface and the PEG linker attached to the probe.
The rupture events were analyzed in the framework of WLC approximation, as described in Materials and Methods, which allowed us to determine two major parameters of force spectroscopy data: rupture force (F) and contour length (Lc). Figure 3 shows distributions of rupture forces for interactions of A3A with two deaminase-specific sequences (A and B) and a nonspecific sequence (C). The data corresponding to the major part of the histogram were fitted to a Gaussian curve, and the maxima of the rupture forces were 75.8 ± 3.7 pN for deaminase-specific sequence 1, 70.5 ± 3.9 pN for deaminase-specific sequence 2, and 62.1 ± 5.9 pN for nonspecific sequence 3. These data clearly demonstrate the larger rupture force between A3A and a deaminase-specific sequence (75.8 ± 3.7 pN) compared to that of a nonspecific one (62.1 ± 5.9 pN). Statistically, these two values are substantially different, as the independent t test at 0.05 levels shows p ≪ 0.05. Note that when dynamic force spectroscopy is applied this difference becomes even more obvious. Indeed, Figure S1 of the Supporting Information shows the dependence of rupture forces F (piconewtons) on different loading rates ALR (piconewtons per second) for A3A in complex with sequence 1 (plot A) and sequence 3 (plot B). These data clearly demonstrate that the difference in rupture forces monotonously increases with loading rate. Altogether, these results indicate that the stability of the A3A–ssDNA complexes is sequence-dependent and the deaminase-specific sequence makes the most stable complex.
Figure 3.
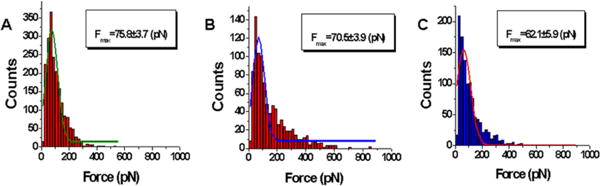
Quantitative analysis of the force spectroscopy data for rupture forces (F) obtained from probing events of specific and nonspecific sequences. Histograms for the rupture force distributions are approximations with a single Gaussian, and the parameters of the fit are shown in the boxes with the SEM indicated: (A) complex of A3A with specific sequence 1, (B) complex of A3A with specific sequence 2, and (C) complex of A3A with nonspecific sequence 3.
Figure 4 shows the histogram for the distributions of contour lengths (Lc). All histograms are fitted to Gaussian curves, and the maxima are 43.0 ± 1.1, 46.4 ± 0.5, and 45 ± 0.7 nm for sequences 1–3, respectively. These results indicate that rupture positions of A3A complexes are close.
Figure 4.
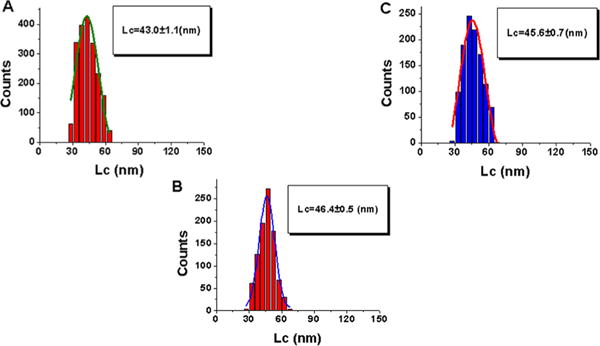
Quantitative analysis of contour lengths (Lc) obtained from force spectroscopy data for probing events of specific and nonspecific sequences. Histograms for the contour length distributions are approximations with a single Gaussian, and the parameters of the fit are shown in the boxes with the SEM indicated: (A) complex of A3A with specific sequence 1, (B) complex of A3A with specific sequence 2, and (C) complex of A3A with nonspecific sequence 3.
A similar set of data were obtained for the A3A-cys mutant that was attached to the AFM probe via cysteine located at the C-terminus. This mutant was obtained by replacing C64 and C171 with alanine residues, and an extra cysteine was added to the C-terminus10 (see details in Materials and Methods). A PEG linker, with the same length as in the case of N-terminal attachment, was used to provide orientational freedom to the protein during its interaction with the ssDNA target on the surface. The analysis of the data was performed in the same way that is described above. The force and contour length distributions are shown in Figures 5 and 6, respectively. As seen in Figure 5A, the histogram for the rupture force distribution for the interaction of A3A-cys with deaminase-specific sequence 2 fitted to a Gaussian curve has a maximum of 58.1 ± 6.1 pN (panel A). Nonspecific sequence 3 (panel B) produces a distribution with a maximum of 39.8 ± 1.9 pN. These data show that similar to the A3A data set, the interaction of A3A-cys is stronger with a specific sequence than with a nonspecific sequence, although the difference between forces is larger.
Figure 5.
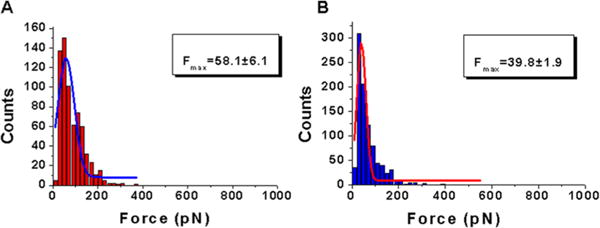
Quantitative analysis of the force spectroscopy data for rupture forces (F) obtained from probing A3A-cys complexes interacting with specific and nonspecific sequences. Histograms for the rupture force distributions are approximations with a single Gaussian, and the parameters of the fit are shown in the boxes with the SEM indicated: complexes of A3A-cys with (A) specific sequence 2 and (B) nonspecific sequence 3.
Figure 6.
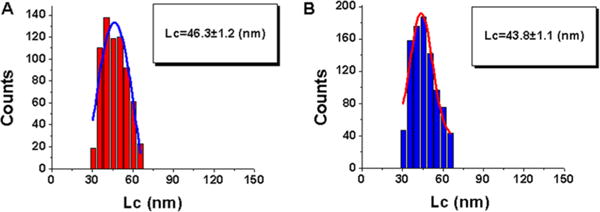
Quantitative analysis of contour lengths (Lc) obtained from force spectroscopy data for probing A3A-cys complexes interacting with specific and nonspecific sequences. Histograms for the contour length distributions are approximations with a single Gaussian, and the parameters of the fit are shown in the boxes with the SEM indicated: complex of A3A-cys with (A) specific sequence 2 and (B) nonspecific sequence 3.
The data for the contour length distribution for A3A-cys are shown in Figure 6. The Gaussian distributions have maxima at an Lc of 46.3 ± 1.2 nm for deaminase-specific sequence 2 and at an Lc of 43.8 ± 1.1 nm for nonspecific sequence 3, which are similar in value.
DISCUSSION
Effect of DNA Sequence on the Stability of Complexes with A3A and A3A-cys
The results mentioned above indicate that the stability of A3A complexes is sequence-specific. The data in the first column of Table 1 represent the values of the rupture forces for complexes between A3A and deaminase-specific and nonspecific sequences. The highest force was observed for A3A interacting with deaminase-specific sequence 1, while the lowest force was observed for interactions with nonspecific sequence 3. The difference in strengths of the A3A complexes formed with sequences 1 and 2 is consistent with data from prior studies,10,13 in which A3A appeared to bind slightly tighter to TTCA (sequence 1) than to CCCA (sequence 2). Taken together with these prior reports, our studies indicate that hydrophobic interactions may contribute to the specific binding of A3A to preferred ssDNA substrates. The same sequence-specific trend was obtained for the A3A-cys mutant, which showed higher rupture forces for complexes with deaminase-specific sequence 2 in comparison to nonspecific sequence 3.
Table 1.
Summary of the Force Spectroscopy Data Analysis
| Protein | ssDNA sequence | force (pN)a | contour length Lc (nm)b |
|---|---|---|---|
| A3A | deaminas-specific sequence 1 | 75.8 ± 3.7 | 43.0 ± 1.1 |
| A3A | deaminas-specific sequence 2 | 70.5 ± 3.9 | 46.4 ± 0.5 |
| A3A | nonspecific sequence 3 | 62.1 ± 5.9 | 45.6 ± 0.7 |
| A3A-cys | deaminase-specific sequence 2 | 58.1 ± 6.1 | 46.3 ± 1.1 |
| A3A-cys | nonspecific sequence 3 | 39.8 ± 1.9 | 43.8 ± 1.1 |
Maximal rupture force values obtained from a Gaussian fit.
Maximal contour length values obtained from a Gaussian fit.
Correlations between ssDNA Deaminase Activity and A3A–ssDNA Complex Stability
The data in Figure S2 of the Supporting Information show the deamination activity of A3A compared to that of the A3A-cys mutant. Although both A3A and A3A-cys reach a plateau, A3A fully deaminates the ssDNA substrate at a protein concentration approximately 3-fold lower than that of A3A-cys (i.e., wild-type A3A is 3-fold more active). As seen in Table 1, the maximal rupture forces for protein–DNA (sequence 2) complexes are larger for A3A (70.5 ± 3.9 pN) than for the A3A-cys mutant (58.1 ± 6.1 pN). A3A-cys was made by substituting cysteine at position 64 with alanine. According to prior studies,13 residues 57–70 in loop 3 show conformational changes when bound to ssDNA but are not thought to be involved directly in binding ssDNA. Thus, the cysteine to alanine substitution in position 64 may change the structure of loop 3 and indirectly change the potential of interaction of A3A with ssDNA, leading to deceleration of the deaminase reaction for A3A-cys.
A comparison of the A3A complex strength with deaminase-specific sequences 1 and 2 indicates a correlation between complex stability and deaminase activity. Indeed, our observation of a small difference in the strength of A3A complexes with sequences 1 and 2 is consistent with data presented in refs 10 and 13 that showed that the deaminase activity of A3A bound to TTCA (sequence 1) was slightly more efficient than that of A3A bound to CCCA (sequence 2).
It is important to compare the force spectroscopy and deamination activity data for A3A and A3G proteins. According to prior studies,11 A3A is at least 10-fold more efficient as an enzyme than A3G. We have shown recently15 that the rupture force for A3G is around 54.3 ± 2.9 pN, which is considerably lower than the value of 75.8 ± 3.7 pN obtained for A3A. These data suggest that the higher deaminase activity of A3A may be attributable to stronger interactions with ssDNA substrates.
The probing experiments with A3A and A3A-cys show only one maximum in the contour length distribution, contrary to the case for A3G, which had two distinct maxima on the contour length distribution.15 This suggests that unlike the two-Zn-coordinating domain enzyme, A3G, the one-domain A3A has a single binding mode with ssDNA. Moreover, the similar Lc values for A3A and A3A-cys indicate that this mode is independent of the type of attachment of A3A to the probe because both the C- and N-terminal attachments have identical Lc positions.
Supplementary Material
Acknowledgments
This work was supported by National Institute of General Medical Sciences Grants GM091743 to R.S.H. and Y.L.L. and GM118006 to Y.L.L. R.S.H. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- A3A and APOBEC3A
apolipoprotein B editing catalytic subunit-like 3A
- ssDNA
single-stranded DNA
- APS
1-(3-aminopropyl)silatrane
- SMFS
single-molecule force spectroscopy
- SEM
standard error of the mean
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00214.
Dynamic force spectroscopy for the interaction of A3A with deaminase-specific sequence 1 and nonspecific sequence 3 (Figure S1) and deaminase activity assay for A3A and A3A-cys (Figure S2) (PDF)
Notes
The authors declare the following competing financial interest: R.S.H. is a co-founder of ApoGen Biotechnologies.
References
- 1.Wiegand HL, Cullen BR. Inhibition of alpharetrovirus replication by a range of human APOBEC3 proteins. Journal of virology. 2007;81:13694–13699. doi: 10.1128/JVI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulliard Y, Narvaiza I, Bertero A, Peddi S, Rohrig UF, Ortiz M, Zoete V, Castro-Diaz N, Turelli P, Telenti A, Michielin O, Weitzman MD, Trono D. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. Journal of virology. 2011;85:1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger G, Durand S, Fargier G, Nguyen XN, Cordeil S, Bouaziz S, Muriaux D, Darlix JL, Cimarelli A. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 2011;7:e1002221. doi: 10.1371/journal.ppat.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Love RP, Xu H, Chelico L. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J Biol Chem. 2012;287:30812–30822. doi: 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pham P, Calabrese P, Park SJ, Goodman MF. Analysis of a Single-stranded DNA-scanning Process in Which Activation-induced Deoxycytidine Deaminase (AID) Deaminates C to U Haphazardly and Inefficiently to Ensure Mutational Diversity. J Biol Chem. 2011;286:24931–24942. doi: 10.1074/jbc.M111.241208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aydin H, Taylor MW, Lee JE. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure. 2014;22:668–684. doi: 10.1016/j.str.2014.02.011. and references cited therein. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Schmitt K, Guo K, Santiago ML, Stephens EB. The Role of the Single Deaminase Domain APOBEC3A in Virus Restriction, Retrotransposition, DNA Damage and Cancer. J Gen Virol. 2016;97:1. doi: 10.1099/jgv.0.000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. and references cited therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. and references cited therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair. 2010;9:579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wijesinghe P, Bhagwat AS. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 2012;40:9206–9217. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra M, Hercik K, Byeon IJ, Ahn J, Hill S, Hinchee-Rodriguez K, Singer D, Byeon CH, Charlton LM, Nam G, Heidecker G, Gronenborn AM, Levin JG. Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res. 2014;42:1095–1110. doi: 10.1093/nar/gkt945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Zhang T, Pan Y, Ciacchi LC, Xu B, Wei G. AFM-based force spectroscopy for bioimaging and biosensing. RSC Adv. 2016;6:12893. [Google Scholar]
- 15.Shlyakhtenko LS, Dutta S, Banga J, Li M, Harris RS, Lyubchenko YL. APOBEC3G Interacts with ssDNA by Two Modes: AFM Studies. Sci Rep. 2015;5:15648. doi: 10.1038/srep15648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, Parniak MA, Johnson JR, Krogan NJ, Somasundaran M, Ali A, Schiffer CA, Harris RS. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem Biol. 2012;7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shlyakhtenko LS, Lushnikov AJ, Li M, Harris RS, Lyubchenko YL. Interaction of APOBEC3A with DNA assessed by atomic force microscopy. PLoS One. 2014;9:e99354. doi: 10.1371/journal.pone.0099354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathore A, Carpenter MA, Demir O, Ikeda T, Li M, Shaban NM, Law EK, Anokhin D, Brown WL, Amaro RE, Harris RS. The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J Mol Biol. 2013;425:4442–4454. doi: 10.1016/j.jmb.2013.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Lyubchenko YL. Early stages for Parkinson’s development: alpha-synuclein misfolding and aggregation. J Neuroimmune Pharmacol. 2009;4:10–16. doi: 10.1007/s11481-008-9115-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim BH, Palermo NY, Lovas S, Zaikova T, Keana JF, Lyubchenko YL. Single-molecule atomic force microscopy force spectroscopy study of Abeta-40 interactions. Biochemistry. 2011;50:5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shlyakhtenko LS, Lushnikov AY, Li M, Lackey L, Harris RS, Lyubchenko YL. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. J Biol Chem. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong Z, Mikheikin A, Krasnoslobodtsev A, Lv Z, Lyubchenko YL. Novel polymer linkers for single molecule AFM force spectroscopy. Methods. 2013;60:161–168. doi: 10.1016/j.ymeth.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


