Abstract
With the availability of complete genome sequences of many cyanobacterial species, it is becoming feasible to study the broad prospective of the environmental adaptation and the overall changes at transcriptional and translational level in these organisms. In the evolutionary phase, niche-specific competitive forces have resulted in specific features of the cyanobacterial genomes. In this study, functional composition of the 84 different cyanobacterial genomes and their adaptations to different environments was examined by identifying the genomic composition for specific cellular processes, which reflect their genomic functional profile and ecological adaptation. It was identified that among cyanobacterial genomes, metabolic genes have major share over other categories and differentiation of genomic functional profile was observed for the species inhabiting different habitats. The cyanobacteria of freshwater and other habitats accumulate large number of poorly characterized genes. Strain specific functions were also reported in many cyanobacterial members, of which an important feature was the occurrence of phage-related sequences. From this study, it can be speculated that habitat is one of the major factors in giving the shape of functional composition of cyanobacterial genomes towards their ecological adaptations.
Keywords: Cyanobacteria, Functional profiling, COG categorization, Genome evolution, Ecological adaptation
1. Introduction
Cyanobacteria are oxygenic photosynthetic bacteria widely used as a model organism for the study of numerous biological processes including photosynthesis, nitrogen fixation, environmental stress tolerance and molecular evolution [1]. Cyanobacteria show broad diversity in terms of their morphology, habitat and functionalities [2], [3], which are reflected in their genome and/or other pertinent genomic features (e.g., genome size, coding region, GC content). Cyanobacterial species are subjected to a variety of niche-specific competitive forces in the environment that result in unique characteristics of each genome, in the course of evolution [2]. They are reported to follow two types of trends in terms of their genome size; certain species involve a mix of expansion and reduction, e.g. genus Acaryochloris [4], whereas closely related marine picocyanobacteria in general tends to follow genome reduction or genome streamlining and maintain only a minimal gene repertoire [5], [6], [7]. Earlier report suggests that, in billion years of evolution, significant changes occur in the genomes of cyanobacteria possibly due to adaptation towards their habitats and this might have resulted in redundancy of duplicated genes and increased mutation rates [2].
With respect to genome size and gene content, prokaryotic genomes are constantly shifting, owing to the factors like gene duplication, gene loss, horizontal gene transfer, and de novo origin of the genes (gene genesis) [8]. Overall, the genome size of the bacteria is maintained in equilibrium between the duplication or horizontal transfer, and mutations leading to elimination of function(s) followed by deletions (gene loss) [9]. It has been already reported that larger genomes preferentially accumulate genes involved in the regulation (e.g., genes involved in secondary metabolism along with those related to energy conversion are ecologically more adaptable to the environments, where the resources are varied and poor) and complete understanding of this will provide the detailed insight into the interaction between ecology and genome evolution [10]. Furthermore, microbial genomes are subjected to horizontal gene transfer related events and acquire foreign DNA from the surroundings more frequently in comparison to the higher organisms [11]. The events of horizontal gene transfer in combination with intra-genomic rearrangements and duplication are accountable for bacterial adaptations in different environmental niches and variance in closely related species. Phylogenomics studies have shown a complex evolutionary pattern for the microbes that undergo not only vertical descent or lateral gene transfer but also include a mix of recombination, gene duplication, gene invention, gene loss, gene degradation, convergence and selection processes [12], [13].
Keeping aforesaid facts in view and the availability of a large number of cyanobacterial sequenced genomes, here in this study we identified the functional composition of 84 cyanobacterial genomes and based upon the findings a correlation has been deduced with the adaptation of these organisms to various ecological niches. We attempted to compare the cyanobacterial species for the identification of the part/genes of their genome contributing to a particular cellular process as it may give reflection of both the cellular and ecological strategies associated with genome expansion and adaptation to the environmental conditions.
2. Material and methods
2.1. Genome sequences
The complete genome sequence of 84 cyanobacterial strains available at Integrated Microbial Genomes (IMG) database (https://img.jgi.doe.gov/cgi-bin/w/main.cgi) were retrieved for this study. Among these 84 genomes, 69 species occupy aquatic habitats (freshwater: 33 sps.; marine: 30 sps.; hot spring: 06 sps.) (genome size in a range of 1.44–8.73 Mb), 06 terrestrial species (genome size: 6.69–8.27 Mb), 05 members occupy multiple habitats (genome size: 4.66–9.06 Mb) while 04 species acquire other habitats (genome size: 4.68–5.13 Mb) (Table S1). The major characteristics of all the cyanobacteria (e.g., habitat, genome size, no. of genes etc.) included in this study have been given in Table S1.
2.2. Functional characterization and Clusters of Orthologous Groups (COG) assignment
Functional characterization of cyanobacterial genomes was performed by using the Clusters of Orthologous Groups (COG) database [14]. For each cyanobacterial genome, all the genes were subjected to COG assignment using the Function Profile tool (IMG database) [15]. The Function Profile tool assists in the identification of the genes associated with a particular function in ‘query genome’ and thus, genes are expected to share at least the same functions associated with their COG matches. Once the genes were assigned to the COGs, they were clustered into 23 functional categories, which were further grouped into four major classes (Table 1).
Table 1.
Functional categories identified in the cyanobacterial genomes.
| S. no. | Function class | Identified COG categories |
|---|---|---|
| 1 | Metabolism | Amino acid transport and metabolism (E) Carbohydrate transport and metabolism (G) Nucleotide transport and metabolism (F) Energy production and conversion (C) Coenzyme transport and metabolism (H) Lipid transport and metabolism (I) Inorganic ion transport and metabolism (P) Secondary metabolite biosynthesis, transport and catabolism (Q) |
| 2 | Cellular processes and signalling | Cell wall/membrane/ envelope biogenesis (M) Cell motility (N) Cell cycle control, cell division, chromosome partitioning (D) Posttranslational modification, protein turnover, chaperones (O) Signal transduction mechanisms (T) Intracellular trafficking, secretion, and vesicular transport (U) Defense mechanisms (V) Cytoskeleton (Z) |
| 3 | Information storage & processing | RNA processing and modification (A) Chromatin structure and dynamics (B) Translation, ribosomal structure and biogenesis (J) Transcription (K) Replication, recombination and repair (L) |
| 4 | Poorly categorized | Function unknown (S) General function prediction only (R) |
2.3. COG categorization
Practical Extraction and Report Language (PERL) scripts were used for arranging all the COGs respective of their functional category for each cyanobacterial species. PERL scripts were also used for the analysis of the distribution pattern of each COG category across the members of the dataset.
2.4. Statistical analysis
All the statistical analyses involved in this study were performed by using the SPSS Version 16.0 software.
3. Results and discussion
3.1. General characteristics
The size of cyanobacterial genomes lies in a range of 1.44 Mb (CAt_ALOHA) to 9.06 Mb (Np_PCC_73102) (Table S1). All the genomes contain single circular chromosome as their major genetic material, whereas, an additional chromosome was present in three cyanobacterial species i.e., Cs_ATCC51142, Av_ATCC29413 and As_90. Members of the dataset occupied diverse habitats, i.e., aquatic (marine, freshwater or hot springs), terrestrial, and some represent multiple habitats (Table S1). It was also found that cyanobacteria exhibiting marine habitats tend to have lower genome size in comparison with those residing in other habitats (Table S1).
3.2. COG assignment and functional genomics profiling of cyanobacteria
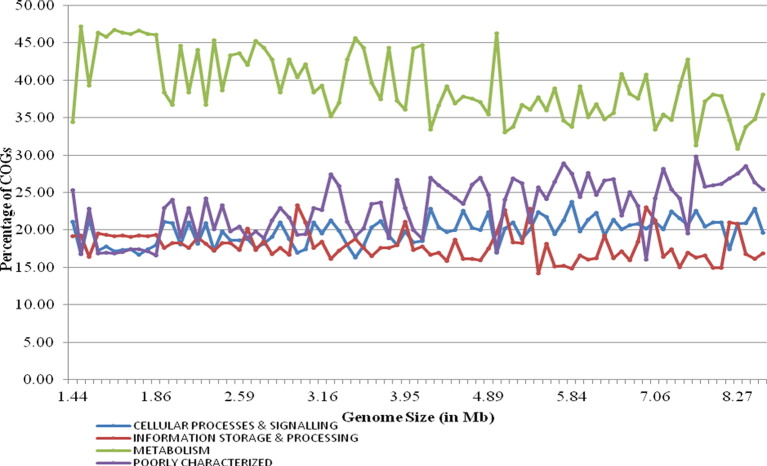
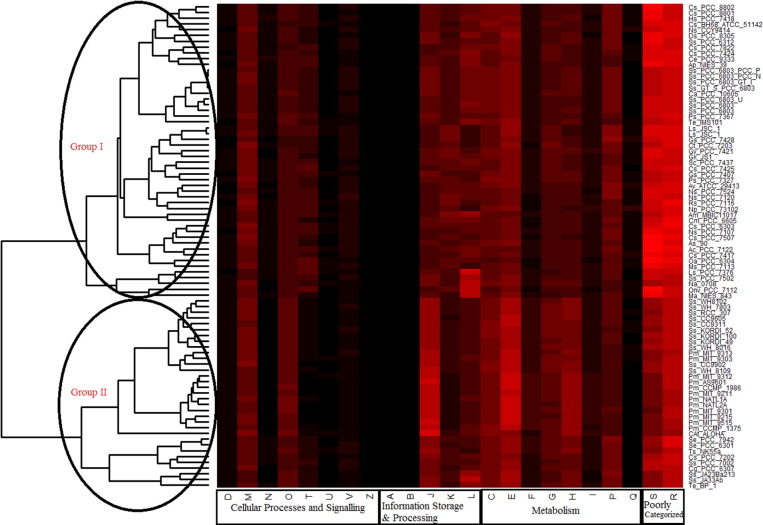
We were able to assign on an average of ~ 53% of the genes in any genome to a particular COG functional category by using the Function Profile tool. Among the cyanobacterial species, 2616 individual COGs from 23 different functional categories of four major classes, i.e., Metabolism, Cellular processes and signalling, Information storage & processing, and Poorly categorized were identified (Table 1). Only 400 COGs (15%) were common in all the 84 cyanobacteria, while remaining represent the functions of pan-genome for these organisms, in which ~ 06.68% functions are strain-specific, i.e., owned by only one member. Among all these 23 categories, Cell motility (N), Chromatin structure and dynamics (B), Cytoskeleton (Z), and RNA processing and modification (A) categories were not present in all the cyanobacteria (Table 1). While analyzing the distribution of each functional category, it was observed that across all cyanobacterial species, genes associated with metabolic functions gained maximum share (Fig. 1). The next most abundant functional category in most of the cyanobacteria (specifically those inhabiting freshwater and multiple habitat) was that of poorly categorized genes, i.e., Function unknown (S) and General function prediction only (R). Marine cyanobacteria preferred genes for Information Storage and Processing over Cellular Processes and Signalling, whereas, the later one is mostly preferred by the cyanobacteria from other habitats (Fig. 2). Further, hierarchical clustering on the basis of functional categories reflected two different groups among the cyanobacteria, the first group (group I) included the members from diverse habitats (freshwater, multiple, other), while the second group (group II) specifically included marine and hot-spring cyanobacteria with only three freshwater species (Fig. 2). Both the above mentioned groups showed different functional profiles. In general, habitat seems to influence the functional profile as members from similar habitats possess similar kind of functional profile (Fig. 2). Earlier, it has been reported that bacterial genomes contain specific functional gene inventories, which are in concurrence with their survival in the particular ecological niche [16], [17], [18]. In the bi-variate correlation analysis, the larger genomes showed a significant strong positive correlation with the functional categories R, T, K, S, Q, V and N (0.832**, 0.823**, 0.821**, 0.798**, 0.740**, 0.614**, 0.531**, respectively at 0.01 level (2-tailed)), whereas they exhibited a strong negative correlation with functional categories C, E, O, F, J, H (− 0.717**, − 0.783**, 0–0.786**, − 0.854**, − 0.887**, − 0.896**, respectively at 0.01 level (2-tailed)).
Fig. 1.
Percentage distribution of four major function classes in all cyanobacterial genomes.
Fig. 2.
Heatmap based on the percentage distribution of the genes in each functional category for all the cyanobacteria under consideration (color coding varies from black to red, where black represents the lowest value and red represents the highest one). (Note: Group I includes members from following habitats (in same order they appear in heatmap): Aquatic (Freshwater): Cs_PCC_8802, Cs_PCC_8801, Hs_PCC_7418; Aquatic (Marine): Cs_BH68_ATCC_51142, Ns_CCY9414; Aquatic (Freshwater): Ds_PCC_8305, Ss_PCC_6312, Cs_PCC_7822, Cs_PCC_7424; Terrestrial (sandy crust): Ce_PCC_9333; Aquatic (Freshwater): Ap_NIES_39, Ss_PCC_6803_PCC_P, Ss_PCC_6803_PCC_N, Ss_PCC_6803_GT_I, Ss_GT_S_PCC_6803, Ca_PCC_10605, Ss_PCC_6803_U, Ss_PCC_6803, Ss_PCC_6803; Other (Snail shell. Intertidal zone): Ps_PCC_7367; Aquatic (Marine): Te_IMS101; Aquatic (Freshwater): Ls_JSC_1, Ls_JSC_1, Gs_PCC_7428; Terrestrial (soil): Ct_PCC_7203; Multiple (Fresh water, Soil): Gv_PCC_7421; Other (epilithic biofilm): Gk_JS1; Aquatic (Freshwater): Sc_PCC_7437, Cs_PCC_7425; Other (Unknown): Gs_PCC_7407; Aquatic (Hot Spring): Ps_PCC_7327, Aquatic (Freshwater): Ns_PCC_7524; Multiple (Aquatic, Soil): Av_ATCC_29413, Ns_PCC_7120; Aquatic (Marine): Rs_PCC_7116; Multiple (Fresh water, Soil): Np_PCC_73102; Aquatic (Marine): Am_MBIC11017; Aquatic (Freshwater): Cm_PCC_6605, Cs_PCC_6303, Ns_PCC_7107, Cs_PCC_7507, As_90, Ac_PCC_7122; Terrestrial (soil): Cs_PCC_7417, Oa_PCC_6304, Ms_PCC_7113; Other (Unknown): Ls_PCC_7376; Aquatic (Freshwater): Ss_PCC_7502, Na_0708; Terrestrial (soil): Onv_PCC_7112; Multiple (Aquatic, Fresh water, Host): Ma_NIES_843 while Group II includes Aquatic (Marine): Ss_WH8102, Ss_WH_7803, Ss_RCC_307, Ss_CC9605, Ss_CC9311, Ss_KORDI_52, Ss_KORDI_100, Ss_KORDI_49, Ss_WH_8016, Pm_MIT_9313, Pm_MIT_9303, Ss_CC9902, Ss_WH_8109, Pm_MIT_9312, Pm_AS9601, Pm_CCMP_1986, Pm_MIT_9211, Pm_NATL1A, Pm_NATL2A, Pm_MIT_9301, Pm_MIT_9215, Pm_MIT_9515, Pm_CCMP_1375, CAt_ALOHA; Aquatic (Freshwater): Se_PCC_7942, Se_PCC_6301; Aquatic (Hot Spring): Ts_NK55a, Cs_PCC_7202; Aquatic (Marine): Ss_PCC_7002; Aquatic (Freshwater): Cg_PCC_6307; Aquatic (Hot Spring): Ss_JA23Ba213, Ss_JA33Ab, Te_BP_1).
3.3. Variations among different functional categories and their possible roles in ecological adaptation
Marine species showed completely different functional genome profile as compared to the rest of the cyanobacteria. In marine cyanobacteria, Amino acid transport and metabolism (E) was the most abundant functional category followed by Translation, ribosomal structure and biogenesis (J), while the functional category, Function unknown (S) and General function prediction only (R) were the most abundant in the cyanobacteria from other habitats (freshwater and terrestrial) (Fig. 2). It was observed that the category E was not so abundant in the freshwater or terrestrial cyanobacteria. As, it has been already reported that the bacteria in the nutrient-limited environment prefer production of only those proteins, which are indispensable for their survival, owing to the fact that biosynthesis of proteins are expensive [19], [20]. To ensure this, bacteria follows signal-dependent regulation which make certain that particular genes and proteins are expressed or activated only at the time of their need and it is essential for the bacteria occupying diverse range of habitats each with its own set of nutrients and adversaries [21]. In this study, cyanobacteria present in habitats of diverse and scarce nutritional sources (like soil, sandy crust, freshwater etc.) showed an abundance of genes involved in signal transduction mechanism (T). Cyanobacteria from freshwater and terrestrial habitat possess significant fractions of genes involved in transcription (K) in comparison to rest of the members and the probable reason for this is the fact that adaptations to specific niche require deep transcriptional reshaping [22]. Also, the expression of many other genes especially in specific conditions depends on the transcription processes [23]. Regulatory genes, like those involved in, transcription and signal transduction mechanisms are dominant in the genomes of organisms in which successful control of the metabolic functions can lead to good growth conditions under stressful environments [24], [25], [26]. Among all the 84 cyanobacteria selected, 4.98–7.86% genes were involved in the Energy Production and Conversion (C). Interestingly, the marine cyanobacterial species possesses slightly higher number of such genes (C). Energy generation is a fundamental phenomenon for microbes to drive physiologically significant mechanisms for the survival in sub-normal or extreme environments [27], [28]. RNA processing and modifications (A) category genes were present in 13 species (Ce_PCC_9333, Ps_PCC_7327, Cs_PCC_7424, Oa_PCC_6304, Ns_PCC_7524, Cs_PCC_7822, Gs_PCC_7428, Cs_PCC_7417, Av_ATCC_29413, Cs_PCC_7507, Ms_PCC_7113, Ns_PCC_7120 and Np_PCC_73102), while Cytoskeleton (Z) category was present only in Ls_PCC_7376.
Genes of functional category Inorganic ion transport and metabolism (P), Cell motility (N), Defense mechanisms (V), Replication, recombination and repair (L) were more abundant in the group I as compared to the group II (Fig. 2). These genes often correspond to the features that may provide a competitive advantage to the organisms for adaptation against the environmental conditions [29]. Multiple copies of the genes of the category ‘Inorganic ion transport and metabolism’ assists in enhanced uptake of the trace metals crucial for the survival, as the availability of indispensable trace metals (e.g., copper and iron) in various ecological niches may be scarce, and may be difficult to utilize by the organisms [30]. Thus, improving the functional capabilities within the organisms might be advantageous. Genes involved in Defense mechanisms (V) enhance the adaptability of the microbe to diverse ecological niches. Stressful habitats, like, presence of damaging contaminations, nutrition deficiency or a high or low temperature may lead to DNA damage and thus, the presence of extra copies of genes involved in replication/repair enhances the robustness of the repair systems within the organisms [29]. Furthermore, presence of additional copies of genes involved in cell motility may also be beneficial, making movement more feasible for organisms in these diverse environment [29]. In the group I, a significant proportion of genes of unknown functions were present and identified, and only general prediction has been made about their functions. About 12% of the genes (an average) were from each of these categories, i.e. unknown functions and general predictions, in the members of the group I (Fig. 2).
3.4. Strain-specific functional categories
Strain-specific functions were identified in 53 members of cyanobacteria among which only 14 members belong to marine habitat (Table S2). Many strain-specific COGs with poorly characterized functions were identified and shown in Table S2. Unique genes are reported to be distinctive for particular environments and are also subjected to wide horizontal gene transfer [3], [31], [32]. Based upon the findings, it can be speculated that these genes possibly play a significant role in the adaptation of these cyanobacterial species to the different ecological areas. Occurrence of some phage-related sequences, i.e., uncharacterized homolog of phage Mu protein gp47 (Cs_PCC_6303), phage terminase large subunit (Ns_PCC_7524), phage P2 baseplate assembly protein gpV (Se_PCC_7942), Mu-like prophage FluMu protein gp28 (Ss_PCC_6312), Mu-like prophage protein gpG (Ss_PCC_6312), phage-related protein, predicted endonuclease (Ss_WH_8016) and phage anti-repressor protein (Ms_PCC_7113) were found as additional characteristics of species-specific COGs (Table S2). Prophage is recognized as the main contributor of microbial diversification that helps in the survival of the bacteria under different ecological conditions, genomic rearrangements, and mediating transfer of virulence factors [33], [34], [35]. No unique gene was identified for COG categories, RNA processing and modification (A) and Chromatin structure and dynamics (B). This finding is in concurrence with earlier publications reporting conservation of genes involved in information processing and signalling, in large evolutionary distances [36], [37], [38], [39]. The probable reason lies in the fact that they encode for the basic functionalities of the cell (i.e., transcription, translation, repair etc.) and any changes in the gene sequence will lead to the disruption of normal cellular machinery [23].
3.5. Acquisition of plasmids
Varying numbers (1–9) of plasmids were found in 36 cyanobacterial species (Table S1), and most of them belong to freshwater, terrestrial or other habitats. Remarkably, it is worth to note that plasmids were present in all the terrestrial species. Plasmids are reported to play a vital role in adaptation [40], and their acquisition is suggested as a factor of adaptability towards habitats [35]. They also have pivotal role in bacterial evolution and diversification and reported to play significant role in horizontal gene transfer in bacteria [41], [42]. In fact, the key factors responsible for maintaining the plasmids in bacterial populations are horizontal transfer, compensatory adaptation and a positive selection for the genes encoded by them [41]. The presence of plasmids in these cyanobacterial species suggested that they are gained and maintained by these species in the course of evolution, as a step towards adaptation to their respective ecological surroundings.
In a nutshell, we observed that cyanobacteria with different habitats (freshwater, terrestrial or rocks) tends to have larger genome size as compared to the marine species and preferentially accumulate genes for regulation, motility and secondary metabolism in contrast to the genes responsible for informational consequences that are abundant in marine members. Broad metabolic diversity was also observed in large sized cyanobacteria. Consequences of having large genomes in bacteria were realized with the fact that bigger genomes are more successful in the environment of scarce but diverse nutritional resources [43], [44], [45]. The characteristics of gene gain within the genomes may help in understanding the interaction between the ecological conditions and genomic evolution. Although, it's clear that micro-evolutionary processes (functional divergence) in amalgamation with macro-evolutionary processes (horizontal gene transfer or genome shrinkage) are accountable for survival and adaptation of bacterial population to varied ecological niches [23], [42], [46]. Based upon the functional genomics characteristics, it can be suggested that cyanobacterial species develop specific mechanisms for the adaptation to a particular ecological niche. Earlier, genome shrinkage is thought to be a key element underlying the ecological success of marine Prochlorococcus sps in oligotrophic open ocean environments [47].
4. Conclusion
In conclusion, we can suggest that the interaction between cyanobacteria and particular habitats is responsible for genome expansion. These cyanobacteria have gained a number of genes (most of them are uncharacterized), though they will definitely have some important role in the survival of the organism and adaptability towards the specific environment. Also, metabolic diversity was observed for larger genomes of cyanobacteria present in diverse habitats. These cyanobacteria have also gained a large number of plasmids and phage related sequences in some of their species. Identification of the number of genes with unidentified functions and especially those which are exclusive in particular cyanobacteria provides an opportunity for the microbiologists to investigate the unique traits that are still not annotated and might facilitate with certain novel information. Overall, this study provides a better understanding of the functional profile of cyanobacteria inhabiting diverse ecological niches and also aims to identify the shifts in their functional profile towards ecological adaptation.
Conflict of interest
Authors declared no conflict of interest.
Acknowledgements
Authors are grateful to the Indian Council of Agricultural Research, India for financial support in the form of projects “National Agricultural Bioinformatics Grid” (NABG), NAIP and “Centre for Agricultural Bioinformatics (CABin)”.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2016.06.005.
Appendix A. Supplementary data
Details of all the cyanobacterial genomes included in this study.
Table S2. Information about strain- specific functional categories identified in cyanobacteria under consideration.
References
- 1.Koksharova O.A., Wolk C.P. Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 2002;58(2):123–137. doi: 10.1007/s00253-001-0864-9. [DOI] [PubMed] [Google Scholar]
- 2.Larsson J., Nylander J.A., Bergman B. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol. Biol. 2011;11:187. doi: 10.1186/1471-2148-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck C., Knoop H., Axmann I.M., Steuer R. The diversity of cyanobacterial metabolism: genome analysis of multiple phototrophic microorganisms. BMC Genomics. 2012;13:56. doi: 10.1186/1471-2164-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swingley W.D., Chen M., Cheung P.C., Conrad A.L., Dejesa L.C., Hao J., Honchak B.M., Karbach L.E., Kurdoglu A., Lahiri S., Mastrian S.D., Miyashita H., Page L., Ramakrishna P., Satoh S., Sattley W.M., Shimada Y., Taylor H.L., Tomo T., Tsuchiya T., Wang Z.T., Raymond J., Mimuro M., Blankenship R.E., Touchman J.W. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc. Natl. Acad. Sci. U. S. A. 2008;105(6):2005–2010. doi: 10.1073/pnas.0709772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dufresne A., Salanoubat M., Partensky F., Artiguenave F., Axmann I.M., Barbe V., Duprat S., Galperin M.Y., Koonin E.V., Le Gall F., Makarova K.S., Ostrowski M., Oztas S., Robert C., Rogozin I.B., Scanlan D.J., Tandeau de Marsac N., Weissenbach J., Wincker P., Wolf Y.I., Hess W.R. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocap G., Larimer F.W., Lamerdin J., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., Johnson Z.I., Land M., Lindell D., Post A.F., Regala W., Shah M., Shaw S.L., Steglich C., Sullivan M.B., Ting C.S., Tolonen A., Webb E.A., Zinser E.R., Chisholm S.W. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424(6952):1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 7.Prabha R., Singh D.P., Gupta S.K., Rai A. Whole genome phylogeny of Prochlorococcus marinus group of cyanobacteria: genome alignment and overlapping gene approach. Interdiscip. Sci. 2014;6(2):149–157. doi: 10.1007/s12539-013-0024-9. [DOI] [PubMed] [Google Scholar]
- 8.Snel B., Bork P., Huynen M.A. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12(1):17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- 9.Wernegreen J.J., Ochman H., Jones I.B., Moran N.A. Decoupling of genome size and sequence divergence in a symbiotic bacterium. J. Bacteriol. 2000;182(13):3867–3869. doi: 10.1128/jb.182.13.3867-3869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konstantinidis K.T., Tiedje J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. U. S. A. 2004;101(9):3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capella-Gutierrez S., Kauff F., Gabaldón T. A phylogenomics approach for selecting robust sets of phylogenetic markers. Nucleic Acids Res. 2014;42(7) doi: 10.1093/nar/gku071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen J.A. Horizontal gene transfer among microbial genomes: new insights from complete genome analysis. Curr. Opin. Genet. Dev. 2000;10:606–611. doi: 10.1016/s0959-437x(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 13.Meier-Kolthoff J.P., Hahnke R.L., Petersen J., Scheuner C., Michael V., Fiebig A., Rohde C., Rohde M., Fartmann B., Goodwin L.A., Chertkov O., Reddy T., Pati A., Ivanova N.N., Markowitz V., Kyrpides N.C., Woyke T., Göker M., Klenk H.P. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Rao B.S., Smirnov S., Sverdlov A.V., Vasudevan S., Wolf Y.I., Yin J.J., Natale D.A. The COG database: an updated version includes eukaryotes. BMC Bioinf. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markowitz V.M., Chen I.M., Palaniappan K., Chu K., Szeto E., Grechkin Y., Ratner A., Jacob B., Huang J., Williams P., Huntemann M., Anderson I., Mavromatis K., Ivanova N.N., Kyrpides N.C. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daubin V., Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lerat E., Ochman H. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res. 2005;33(10):3125–3132. doi: 10.1093/nar/gki631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francino M.P. The ecology of bacterial genes and the survival of the new. Int. J. Evol. Biol. 2012;2012 doi: 10.1155/2012/394026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibbing M.E., Fuqua C., Parsek M.R., Peterson S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010;8(1):15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoehler T.M., Jørgensen B.B. Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 2013;11(2):83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 21.Seshasayee A.S.N. University of Cambridge; 2009. A Computational Study of Bacterial Gene Regulation and Adaptation on a Genomic Scale. (Ph.D. Thesis) [Google Scholar]
- 22.Piveteau P., Depret G., Pivato B., Garmyn D., Hartmann A. Changes in gene expression during adaptation of Listeria monocytogenes to the soil environment. PLoS One. 2011;6(9):e24881. doi: 10.1371/journal.pone.0024881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caffrey B.E., Williams T.A., Jiang X., Toft C., Hokamp K., Fares M.A. Proteome-wide analysis of functional divergence in bacteria: exploring a host of ecological adaptations. PLoS One. 2012;7(4):e35659. doi: 10.1371/journal.pone.0035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López-Maury L., Marguerat S., Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008;9(8):583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 25.de Nadal E., Ammerer G., Posas F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011;12(12):833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- 26.Picard F., Loubière P., Girbal L., Cocaign-Bousquet M. The significance of translation regulation in the stress response. BMC Genomics. 2013;14:588. doi: 10.1186/1471-2164-14-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyson G.W., Chapman J., Hugenholtz P., Allen E.E., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428(6978):37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 28.Chen L., Hu M., Huang L., Hua Z., Kuang J., Li S., Shu W. Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J. 2014 doi: 10.1038/ismej.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bratlie M.S., Johansen J., Sherman B.T., Huang da W., Lempicki R.A., Drabløs F. Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics. 2010;11:588. doi: 10.1186/1471-2164-11-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morel F.M., Price N.M. The biogeochemical cycles of trace metals in the oceans. Science. 2003;300(5621):944–947. doi: 10.1126/science.1083545. [DOI] [PubMed] [Google Scholar]
- 31.Zhaxybayeva O., Gogarten J.P., Charlebois R.L., Doolittle W.F., Papke R.T. Phylogenetic analyses of cyanobacterial genomes: Quantification of horizontal gene transfer events. Genome Res. 2006;16(9):1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi T., Falkowski P.G. Genome evolution in cyanobacteria: the stable core and the variable shell. Proc. Natl. Acad. Sci. U. S. A. 2008;105(7):2510–2515. doi: 10.1073/pnas.0711165105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner P.L., Waldor M.K. Bacteriophage control of bacterial virulence. Infect. Immun. 2002;70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brüssow H., Canchaya C., Hardt W.D. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung J., Park W. Comparative genomic and transcriptomic analyses reveal habitat differentiation and different transcriptional responses during pectin metabolism in Alishewanella species. Appl. Environ. Microbiol. 2013;79(20):6351–6361. doi: 10.1128/AEM.02350-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mushegian A.R., Koonin E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. U. S. A. 1996;93(19):10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova K.S., Aravind L., Galperin M.Y., Grishin N.V., Tatusov R.L., Wolf Y.I., Koonin E.V. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9(7):608–628. [PubMed] [Google Scholar]
- 38.Lake J.A., Jain R., Rivera M.C. Mix and match in the tree of life. Science. 1999;283:2027–2028. doi: 10.1126/science.283.5410.2027. [DOI] [PubMed] [Google Scholar]
- 39.Azuma Y., Ota M. An evaluation of minimal cellular functions to sustain a bacterial cell. BMC Syst. Biol. 2009;3:111. doi: 10.1186/1752-0509-3-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heuer H., Smalla K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 2012;36(6):1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- 41.San Millan A., Peña-Miller R., Toll-Riera M., Halbert Z.V., McLean A.R., Cooper B.S., MacLean R.C. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 2014;5:5208. doi: 10.1038/ncomms6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiedenbeck J., Cohan F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011;35(5):957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- 43.Bentkowski P., Van Oosterhout C., Mock T. A model of genome size evolution for prokaryotes in stable and fluctuating environments. Genome Biol. Evol. 2015;7(8):2344–2351. doi: 10.1093/gbe/evv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zelezniak A., Andrejev S., Ponomarova O., Mende D.R., Bork P., Patil K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. U. S. A. 2015;112(20):6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown M.V., Ostrowski M., Grzymski J.J., Lauro F.M. A trait based perspective on the biogeography of common and abundant marine bacterioplankton clades. Mar. Genomics. 2014;15:17–28. doi: 10.1016/j.margen.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Dutta C., Pan A. Horizontal gene transfer and bacterial diversity. J. Biosci. 2002;27(1 Suppl 1):27–33. doi: 10.1007/BF02703681. [DOI] [PubMed] [Google Scholar]
- 47.Dufresne A., Garczarek L., Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of all the cyanobacterial genomes included in this study.
Table S2. Information about strain- specific functional categories identified in cyanobacteria under consideration.