Abstract
Persisters are dormant phenotypic variants of bacterial cells that are tolerant to killing by antibiotics1. Persisters are associated with chronic infections and antibiotic treatment failure1–3. In Escherichia coli, toxin/antitoxin (TA) modules have been linked to persister formation4–6. The mechanism of persister formation in Gram-positive bacteria is unknown. Staphylococcus aureus is a major human pathogen, responsible for a variety of chronic and relapsing infections such as osteomyelitis, endocarditis and infections of implanted devices. Deleting TA modules in S. aureus did not affect the level of persisters. Here we show that S. aureus persisters are produced due to a stochastic entrance into stationary phase accompanied by a drop in intracellular ATP. Cells expressing stationary state markers are present throughout the growth phase, increasing in frequency with cell density. Cell sorting revealed that expression of stationary markers is associated with a 100–1000 fold increase in the likelihood of survival to antibiotic challenge. The ATP level of the cell is predictive of bactericidal antibiotic efficacy and explains bacterial tolerance to antibiotics.
Antibiotic resistance is a major human health problem7. However, most pathogens that cause hard to treat chronic infections are not drug resistant1,3,8. There is mounting evidence that drug-tolerant persister cells contribute to this phenomenon2,9–12. Persister cells are phenotypic variants that survive lethal doses of antibiotics and are genetically identical to their drug susceptible kin. The mechanism of persister formation has been extensively studied in the closely related Gram-negative organisms Escherichia coli and Salmonella Typhimurium1,13,14. In E. coli, isolated persisters express toxin/antitoxin (TA) modules15, most of which code for mRNA endonucleases called interferases16. While deletion of individual interferases has no phenotype, a knockout of ten TAs produced a decrease in persisters in both a growing culture and in stationary phase4. A small fraction of persisters forms in E. coli when cells stochastically express the HipA toxin12. HipA is a protein kinase17 which phosphorylates glutamyl aminoacyl-tRNA synthetase, inhibiting protein synthesis18,19. Selection for increased drug tolerance in vitro led to the identification of a hipA7 mutant allele that produces up to 1000-fold more persisters than the wild type6. We recently identified hipA7 strains among patients with chronic urinary tract infections12. Similarly, hip mutants are common among isolates of P. aeruginosa from patients with cystic fibrosis11, and from patients with chronic Candida albicans infections20. In S. Typhimurium, TA modules are responsible for a sharp increase in persisters when the pathogen infects macrophages9. These findings provide a link between persisters and clinical manifestation of disease.
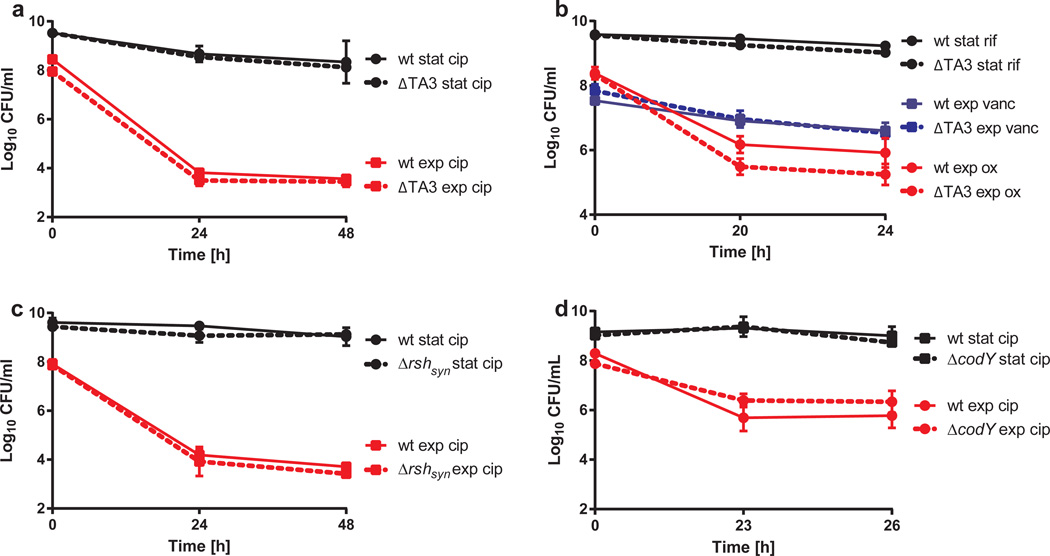
Little is known about the mechanism of persister formation in Gram-positive species. We first sought to examine the role of TAs in persister formation in S. aureus. There are three known type II TAs in S. aureus - mazEF, and relBE homologues axe1/txe1, and axe2/txe2. These are the only three TAs predicted in S. aureus 8325, the parental strain of HG001 and HG003. An additional phage associated toxin-antitoxin was identified in S. aureus Newman using the TAfinder tool21 but overexpression of the potential toxin did not inhibit growth (Supplemental Figure 1). Therefore we continued with analysis of the three active type II TAs. The toxins from all three modules are RNA endonucleases22. We constructed a triple knockout in the TAs (Δ3TA), and examined the strain’s ability to form persisters. Ciprofloxacin causes a characteristic biphasic killing of wild type S. aureus with a subpopulation of surviving persisters (Supplementary Figure 2, Figure 1a). Unexpectedly, knockout of all TAs had no effect on the level of persisters in exponentially growing or stationary phase cells (Figure 1a, Supplementary Figure 3). A similar result was obtained with oxacillin, vancomycin and rifampicin (Figure 1b). This is in stark contrast to E. coli, where a knockout of ten toxin endonucleases produces a decrease in persisters4. It remains possible that these TAs or as yet unannotated TAs play a role in persister formation under a specific environmental condition, but we see no evidence of a role for the TAs we examined, in persister formation under regular growth conditions.
Figure 1. Toxin-antitoxin modules and stringent response do not control persister formation in S. aureus.
The contribution of a,b toxin-antitoxin modules, mazEF, axe1-txe1 and axe2-txe2, in strain Newman and c, the stringent response element rsh in strain HG001and d, the stringent response regulator, codY, in strain SH1000 to persister formation in S. aureus. Strains were grown for 4 hours to mid-exponential phase (exp) or overnight to stationary (stat) phase in MHB and challenged with either ciprofloxacin (cip), vancomycin (vanc), oxacillin (ox) or rifampicin (rif) (10× MIC). Aliquots were removed at indicated time points, washed and plated to enumerate survivors. All experiments were performed in biological triplicates. Standard deviations (SD) are indicated.
The stringent response has also been linked to persister formation in E. coli23. In S. aureus, Rsh, a homolog of RelA/SpoT, contains both a synthase and hydrolase domain for (p)ppGpp24. In response to starvation, the level of (p)ppGpp rises, leading to down-regulation of genes involved in proliferation, protein synthesis, and replication, and increased expression of genes involved in survival and stress responses. Most of these genes are regulated by the repressor CodY24. We tested persister levels in a codY mutant and in an rsh mutant (rshsyn) which has a mutated synthase domain and does not produce an increase in (p)ppGpp in response to amino acid starvation25. There was no effect of either a codY or an rshsyn mutation on the level of persisters in exponential and stationary phase of growth (Figure 1c and d). S. aureus possesses two minor ppGpp synthases that have been associated with the response to cell wall stress. We examined persister formation in a strain deleted in rshsyn, relP and relQ. Again, this strain produced persisters tolerant to ciprofloxacin or gentamicin at levels similar to those of the wild-type (Supplementary Figure 2). The mutant did not exhibit a growth defect and entered the stationary phase similarly to the wild-type suggesting that the stringent response is not a significant regulator of stationary phase under these conditions (Supplementary Figure 3).
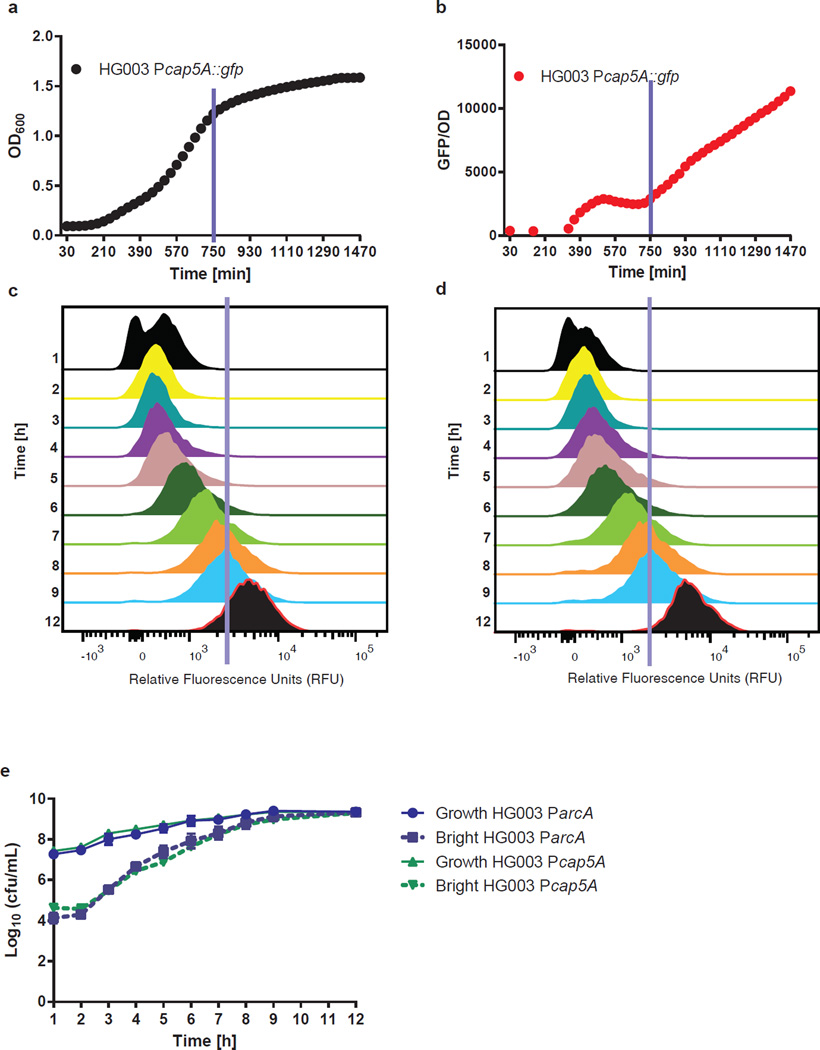
It is known that S. aureus exhibits complete tolerance to many antibiotics at stationary state which is another important distinction between this pathogen and E. coli2,26. It appears that S. aureus cells in a stationary state exhibit antibiotic tolerance similar to persisters. We reasoned that persisters in exponential phase may be cells that have entered the stationary phase early. To examine this we used two reporters of the stationary phase. The promoter of the capsular polysaccharide operon, Pcap5A, has been shown to be activated in the stationary phase27,28. Increase in relative fluorescence of a strain carrying Pcap5A-GFP over time in a growing culture confirmed the suitability of this promoter as a marker of the stationary phase (Figure 2a and b). The promoter of the arginine deiminase pathway, ParcA, was used as a second marker, since proteomic analysis showed that the ArcA protein accumulates specifically in the stationary phase, increasing in abundance 10.5-fold relative to exponential phase. Analysis of ParcA fused to gfp confirmed that this promoter is activated specifically in stationary phase (Supplementary Figure 4). Real-time qRT-PCR analysis showed that transcript levels of cap5A and arcA increase 3.88 and 25.38-fold in stationary phase, respectively. These promoters were inserted upstream of gfpuvr in plasmid pALC1434 to yield Pcap5A::gfp and ParcA::gfp.
Figure 2. Activation of stationary markers is heterogeneous.
a, Growth (OD600) and b, GFP expression of HG003 Pcap5A::gfp time. The blue lines represent entrance into stationary phase. Distribution of GFP signal in c, Pcap5A::gfp and d, ParcA::gfp at hourly intervals. The cut-off for the bright fraction is represented by a blue line. This cut-off represents the level of expression in a stationary phase culture. e, A subpopulation of stationary phase cells, defined as cells with stationary phase levels of expression of ParcA and Pcap5A, is always present and increases with population density. The blue line represents an estimation of the entrance into stationary phase. All experiments were performed in biological triplicates. SD are indicated. c, d are representative of one replicate.
Flow cytometry was then used to track cells expressing high levels of the stationary phase markers (termed bright) at hourly intervals from early exponential to stationary phase (Figure 2c and d). We found that a subpopulation of cells expresses stationary markers in early exponential phase, and their frequency increases with the rise in the density of the population (Figure 2e). This suggests that stationary phase does not initiate in a uniform manner but is a heterogeneous process.
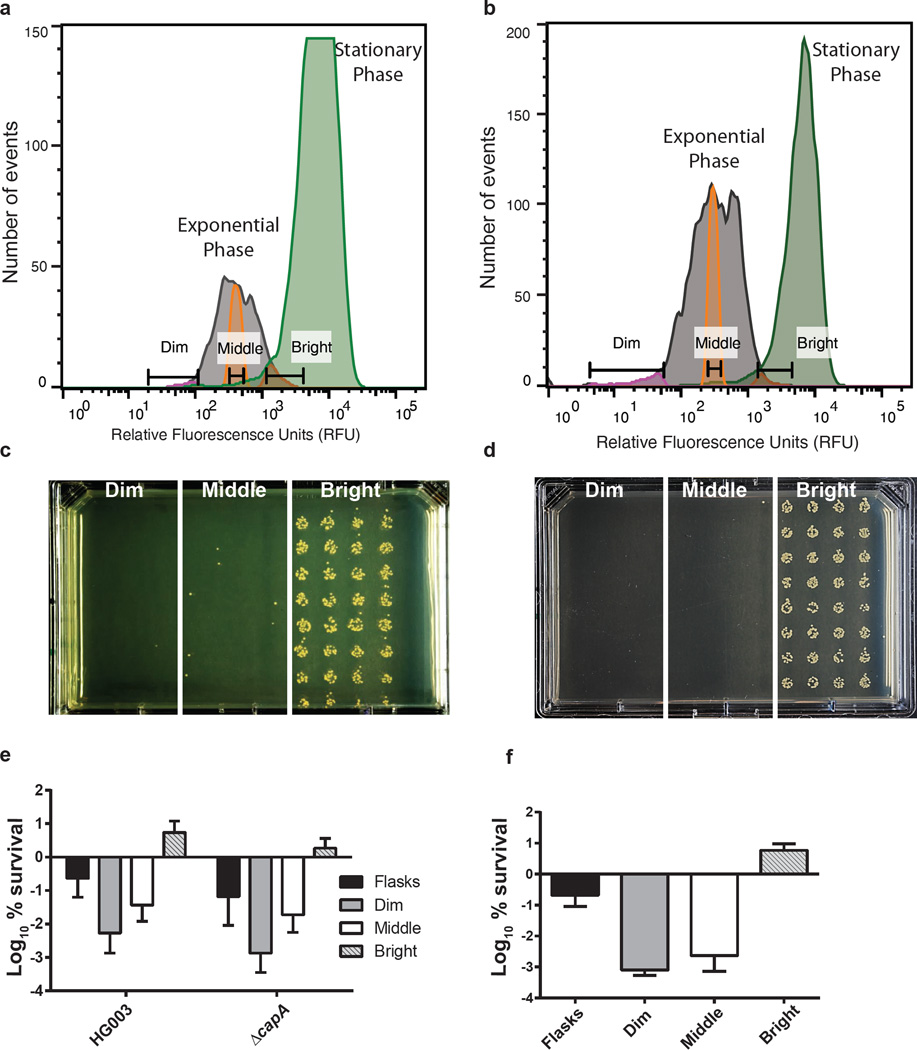
We next sought to determine if the subpopulation of stationary phase cells in a growing culture were in fact persisters. For this, we employed Fluorescence-Activated Cell Sorting (FACS). S. aureus HG003 Pcap5A::gfp or HG003 ParcA::gfp were grown to mid-exponential or stationary phase and analyzed by FACS (Figure 3a and b). In order to examine whether the bright cells were persisters, the exponential phase culture was exposed to a lethal dose of ciprofloxacin (10× MIC) for 24h. The culture was then re-analyzed by FACS, and cells were gated into bright, middle and dim populations based on expression of Pcap5A::gfp, or ParcA::gfp (Figure 3a and b). Cells were then sorted onto MH agar in 96 spots to enumerate survivors from each population (32 spots for each population: bright, middle, dim). The lethal dose of ciprofloxacin causes ~3 logs of killing in the total culture, so cells were sorted onto MH agar plates at 1, 10, 100, 1000, and 5000 per spot to achieve viable counts for each population (representative plate, Figure 3c and d). The bright population had 100–1000 fold more survivors than the middle and dim populations with both markers. We chose to compare only the middle and bright fractions for quantification as the dim fraction had < 100% sorting efficiency (Figure 3e and f).
Figure 3. Persister sorting using stationary markers Pcap5A and ParcA.
Expression of a, Pcap5A::gfp or b, ParcA::gfp in exponential following ciprofloxacin challenge (grey peak) and stationary phase (green peak) measured by FACS. Exponential phase cells were gated into 3 populations depending on expression of GFP - dim (pink peak), middle (orange peak) or bright (red peak - cells expressing stationary phase levels of reporter in exponential phase). (c,d) Cells were sorted based on dim, middle or bright GFP expression onto MHA plates at 1000 events/spot for both Pcap5A::gfp and ParcA::gfp. Representative plates are shown. Survivors from each population of HG003 or Δcap5A harboring e, Pcap5A::gfp and f, ParcA::gfp were counted following incubation overnight at 37°C. The asterisks indicate statistical significance between middle and bright populations, determined using Student’s t-test (P ** < 0.005 or P***<0.0005). All experiments were performed in biological triplicates. SD are indicated. a, b, c, d are representative of one replicate.
To determine if expression of capsular polysaccharide contributes to ciprofloxacin tolerance, we transformed plasmid Pcap5A::gfp into a cap5A mutant strain and repeated the cell sorting experiment. Disrupting the cap5A gene did not alter the expression profiles of Pcap5A::gfp (Supplementary Figure 5). Similarly, the bright cells in a cap5A mutant also exhibited a 100-fold enrichment for cells tolerant to ciprofloxacin in exponential phase compared to the middle fraction showing that entry into stationary phase rather than levels of the CapA protein affect persister formation (Figure 3e). We also examined persister formation in an arcA mutant and found it to be similar to the wild-type strain (Supplementary Figure 6). As a control for stationary phase reporters, we repeated the experiment using a promoter that is also expressed in exponential phase (Pspa::gfp). In this case, the bright population had no enrichment of persisters compared to the middle of the population (Supplementary Figure 5). This shows that expression of a stationary marker, rather than expression of GFP per se, determines whether a cell is a persister.
We wanted to further examine any potential role for the stringent response and tested expression of the persister markers in the rshsyn mutant background. Neither cap5A nor arcA promoter activity were significantly affected by mutation of rshsyn (Supplementary Figure 4). We reasoned that a decrease in the energy level of the cell in stationary phase could lead to antibiotic tolerance. Killing by bactericidal antibiotics results from corrupting active targets1. Aminoglycosides kill by causing mistranslation, which leads to the production of toxic peptides29; fluoroquinolones inhibit the re-ligation step of DNA gyrase and topoisomerase, causing double strand breaks30, and β-lactams lead to a futile cycle of peptidoglycan synthesis and autolysis31. A decrease in ATP would decrease the activity of ATP dependent antibiotic targets such as gyrase, topoisomerase, and RNA polymerase, leading to antibiotic tolerance, and ATP has previously been suggested to impact survival to antibiotics32–35.
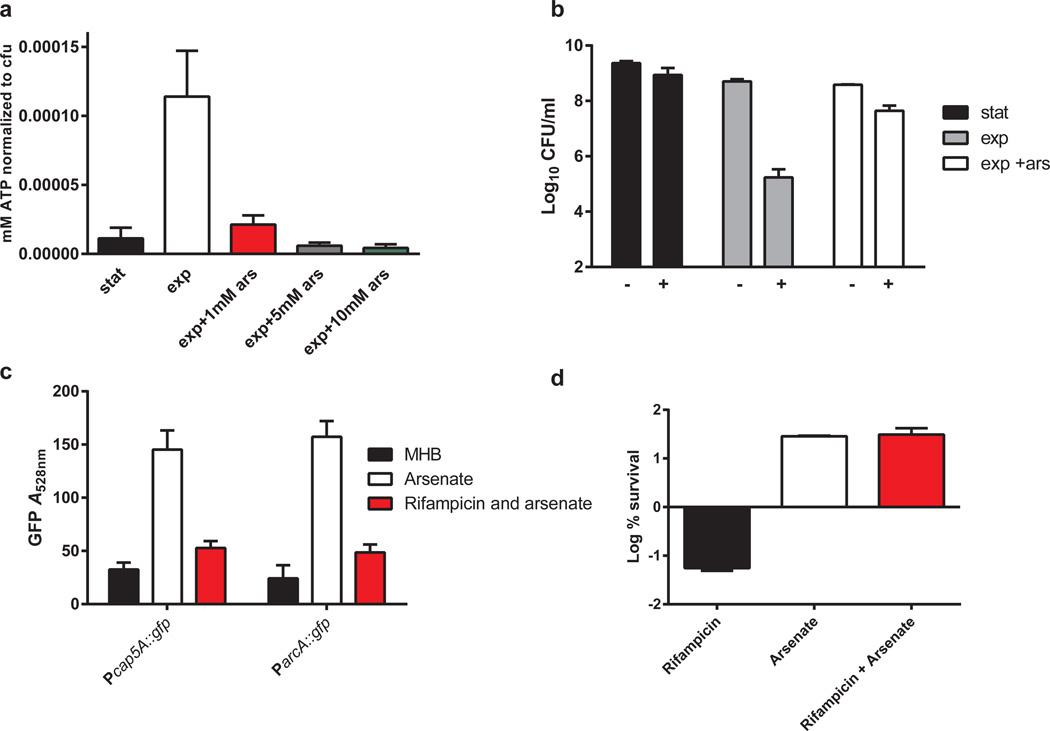
We examined ATP levels of an exponential and stationary phase population and indeed found that ATP levels decrease significantly in the stationary phase (Figure 4a). We then found that emulating stationary phase ATP levels in an exponential phase population by decreasing it with arsenate resulted in a 325-fold induction in persister formation (Figure 4b). ATP levels are lowered by arsenate as it forms a rapidly-hydrolysable ADP-As, producing a futile cycle36. Interestingly, we found that stationary phase-specific promoters were also activated in response to arsenate (Figure 4c). Hence, these promoters are activated in the stationary phase as ATP levels in the cells drop. The Pcap5A and ParcA promoters then enable single-cell detection of ATP, linking a decrease in the energy level to antibiotic tolerance in individual persisters. It was clear that cells with reduced ATP levels are antibiotic tolerant and express markers of this phenotypic state. What remained unclear was whether a transcriptional response was necessary for persister formation. To examine this, we again induced persister formation with arsenate, however, we also included a 15 minute pre-incubation with rifampicin at 1× MIC, which was sufficient to inhibit induction of stationary markers (Fig 4c) but did not cause cell death (Supplementary Figure 7). Inhibition of transcription did not impede persister induction (Figure 4d) (Supplementary Figure 7). This shows that although a specific transcriptional response that includes expression of Pcap5A and ParcA is induced in response to low ATP, this response is not required for antibiotic tolerance. Rather, tolerance of both stationary populations and persisters can be explained by a drop in ATP which will result in a decrease in the activity of drug targets. To further test whether ATP levels determine persister formation, we examined killing in a medium where ATP concentration is expected to increase. Supplementing TSB medium with glucose increased ATP significantly and resulted in a 100 fold reduction in persisters (Supplementary Figure 8).
Figure 4. Reduction in ATP induces persister formation and expression of stationary phase markers.
a, Titering arsenate to produce stationary phase levels of ATP. Arsenate was added to an exponential phase population of S. aureus for 15 minutes before measuring ATP b, Decrease of ATP results in a 325-fold induction of persisters in exponential phase. On the x-axis, “–“ indicates cell count before addition of ciprofloxacin and “+” represents cell count after 24 hour incubation in 10 × MIC of ciprofloxacin (0.4 µg/ml) c, Pcap5A::gfp and ParcA::gfp are induced by depletion of ATP. d, inhibition of transcriptional response by addition of 0.1 µg/ml rifampicin 15 minutes prior to ATP depletion (30 minutes of 1 mM arsenate), with results represented as the Log % survival after 24 hours ciprofloxacin treatment. All experiments were performed in biological triplicates. SD are indicated.
Promoters of arcA and cap5A are induced when ATP drops in stationary phase or in the presence of arsenate. Cells expressing these markers are highly enriched for persisters. Low ATP can lead to tolerance of a stationary culture, and explains antibiotic tolerance of a persister sub-population. This work links the phenomena of population-wide tolerance and persister cell tolerance. A growing population contains cells that enter into stationary state early, and these become antibiotic tolerant persisters. Persisters form as cells lose ATP. The entrance into stationary state is stochastic, with the frequency of persisters increasing with cell density. Our measurements of ATP in single persister cells by FACS have been performed with two different reporters, ParcA-GFP and Pcap5A-GFP. Both are ATP sensors, but the detection requires transcription and translation of GFP. To establish direct causality, it would be interesting to perform single cell detection of ATP in persisters more directly, such as with a FRET-based sensor37, once it is adapted to S. aureus.
Interestingly, tolerance to clinically relevant daptomycin was also observed in stationary phase38. Also, a recent study shows that altered levels of inorganic phosphate and polyphosphate in daptomycin tolerant cells, which could also be related to depletion of ATP39. A recent study shows that population heterogeneity and capsular polysaccharide expressing sub-populations also occur in vivo in persistent carriers of S. aureus28. The role of ATP levels in recalcitrance of S. aureus infection should be examined and ATP levels of cells during infection may be an important determinant of the outcome of infection.
Understanding how persisters form will improve our ability to control chronic infections. We recently identified a compound capable of killing persisters, acyldepsipeptide (ADEP4). ADEP4 targets ClpP and converts it into a non-specific protease, which forces both growing and dormant cells to self-digest2. Importantly, ADEP4 dissociates the protease from its ATP-dependent chaperones and the dysregulated proteolysis does not require ATP. In combination with rifampicin (to decrease resistance development), ADEP4 eradicated a biofilm both in vitro and in a mouse model of a chronic S. aureus infection. This shows that persisters can be killed by a compound which does not require an ATP-dependent target. In this regard, it is interesting to note that stationary cells of S. aureus exhibit considerable tolerance to daptomycin, a membrane-acting antibiotic38,39. Why dormant cells would be tolerant to this compound is an interesting problem that remains to be solved.
This study suggests that a new mechanism of persister formation, loss of energy leading to drug tolerance, operates in S. aureus. It is possible that this is a general mechanism of tolerance which governs persister formation in other bacteria as well.
Methods
Bacterial strains and growth conditions
S. aureus were cultured in Mueller-Hinton broth (MHB) or Tryptic Soy Broth (TSB) with or without added glucose. TSB and TSB without glucose was buffered to pH 7.0 using 100mM MOPS. Bacteria were routinely grown at 37°C at 225 r.p.m. Media were supplemented with chloramphenicol 10 µg/ml to maintain plasmids where necessary. MSSA strains Newman, SH1000, and HG001 were used to analyze the role of TA modules and stringent response as mutations of interest had previously been constructed and characterized in these backgrounds25,40,41. The model strain HG003 was used for all subsequent experiments. For E. coli experiments, growth of the overexpression strain was compared to an empty vector control in a plate reader over 16 hours at 37°C in LB medium supplemented with 0.2% arabinose.
Strain construction
For construction of reporter plasmids, primers Pcap5A_1 (gcgcgaattctctatctgataataatcatc) Pcap5A_2 (ggcctctagactaatgtactttccattatt), Pspa_1 (gcaggaattctttccgaaattaaacctcagc) Pspa_2 (gcagtctagaattaataccccctgtatgta) and ParcA_1 (gcgcgaattcaaaatgtatattttgaccca) and ParcA_2 (ggcctctagatctatttcctccttttatct) flanked by restriction sites EcoRI and XbaI were used to amplify predicted promoter sequences of cap5A, spa and arcA, respectively. The promoter regions were cloned upstream of gfpuvr into the EcoRI and XbaI sites of plasmid pALC143442. A Newman strain was created containing deletions for all three known type II toxin-antitoxin systems (Newman ΔTA3). Using Newman ∆mazEF (ALC4072) as a starting strain, the axe1/txe1 and axe2/txe2 operons were deleted by sequential allelic exchange using the pMAD plasmids pALC6480 and pALC648143, respectively. Deletion of these genes was verified by PCR analysis and chromosomal DNA sequencing. For hypothetical toxin overexpression, the primers Ptox_1 gcgcgaattcatggaagaaactttaa and Ptox_2 gcgcggtaccttatgcaatttaaaaa were used to amplify the toxin and the fragment was digested with EcoRI and KpnI and cloned into the pBAD33 vector upstream of an arabinose inducible promoter, digested with the same restriction enzymes.
Persister assays
Strains were grown to mid-exponential or stationary phase (~16h) in MHB in 14 ml round bottom snap-cap culture tubes. Cells were plated for CFU counts and challenged with antibiotics ciprofloxacin, rifampicin, vancomycin, gentamicin or oxacillin (4.0 µg/ml, 0.4 µg/ml, 10 µg/ml and 1.5 µg/ml respectively) At indicated times, an aliquot of cells was removed, washed with 1% NaCl, and plated to enumerate survivors. All experiments were performed in biological triplicates. Averages and standard deviations are representative of three biological replicates. Rifampicin resistant mutants arise spontaneously at the frequency of ~2.3 × 10−8. Rifampicin killing in exponential phase selected for the proliferation of rifampicin resistant mutants, which had repopulated the exponential phase cultures by 24h (Supplemental Figure 9). For this reason, levels of persisters tolerant to rifampicin were examined in stationary phase only.
Arsenate and rifampicin persister assays
Strains were grown to mid-exponential phase in MHB media. Where indicated, rifampicin 0.01 µg/ml was added for 15 minutes and/or arsenate 1 mM for 30 minutes prior to ciprofloxacin challenge for 24h (10× MIC).
Flow cytometry and FACS analysis using gfp reporters
Fluorescent protein level was analyzed with a BD Aria II flow cytometer (BD Biosciences) with a 70-micron nozzle. Cell population was detected using forward and side scatter parameters (FSC and SSC), and fluorescence was analyzed with emitting laser of 488 nm and bandpass filter of 525/15 nm, using a FACS ARIA II (Becton Dickinson, CA). Strains harboring plasmids Pcap5A::gfp, ParcA::gfp or Pspa::gfp were grown to mid-exponential and stationary phase in MHB containing 10 µg/ml chloramphenicol. For growth curve construction, the population was gated so that over 90% of the stationary phase population were designated ‘bright’. These gates were applied to all timepoints. At each timepoint, cfu was measured and the number of stationary phase cells was calculated by multiplying the percentage of cells in the bright fraction by the total cell number. An overnight culture was sub-cultured 1:100 into fresh MHB and grown for 3 hours. 300 µl of this was added to 3 ml of MHB to begin the growth curve. This sub-culturing step removed any carry-over of stationary phase cells from the stationary phase culture. For FACS analysis of persisters, strains were exposed to ciprofloxacin for 24h. Before the challenge, an aliquot of the culture was diluted and plated for cfu. Challenged cells were washed and plated to enumerate survivors. Cells pre- and post- antibiotic challenge were analyzed by FACS. A gate was drawn based on stationary phase expression of Pcap5A::gfp or ParcA::gfp. Exponential phase cells expressing stationary phase levels of Pcap5A::gfp or ParcA::gfp were termed ‘bright’. Two gates were drawn within the exponential phase Pcap5A::gfp expression peak and termed ‘middle’ and ‘dim’ respectively. To calculate the percent survival of each population following antibiotic challenge, first, we calculated the sorting efficiency from each population prior to antibiotic challenge. Events (cells) from each population were sorted in a 96-well format with 32 spots for each population; dim, middle and bright. 1 event per spot (for 32 spots) and colonies were counted following incubation. For the middle and bright fractions we achieved 100% sorting efficiency (32 colonies), however the sorting efficiency for the dim fraction was lower, ~90% or 29 colonies. This indicated that not all events in the dim fraction were cells. For this reason we chose to focus on the differences between the bright population and the middle or bulk of the population. Following antibiotic challenge, cells (events) from each population were sorted onto MH agar plates in a 96-well format at 1, 10, 100, 1000, 5000 per spot (32 spots / population) to enumerate survivors. A similar method was applied for all reporters. Ciprofloxacin treatment did not affect expression of any reporters used in this study. A control experiment was performed where samples were sonicated for 5 minutes in a sonicating water bath prior to cell sorting. Sonication had no impact on the sorting results, confirming that cell aggregation was not influencing FACS experiments. Cells were analyzed and sorted using FACS-Diva software. Figures were generated using FlowJo software. Experiments were performed in triplicate. Error bars represent the standard deviations of the means, and statistical significance was determined by the Student’s t test.
Proteomic analysis
Biological duplicates were grown in MHB and harvested in the mid-exponential and stationary phase of growth. Samples were labelled and fractionated and mass spectrometry was performed as previously described2.
Real-Time qRT-PCR
RNA was isolated from exponential phase population after 4 hours of growth and stationary phase after 16 hours of growth using a QIAGEN® RNA purification kit. Samples were treated with Turbo DNase and RNA integrity was confirmed on a bioanalyzer. Reverse transcriptase was used to convert to cDNA as per manufacturer’s instructions. Serial 10-fold dilutions of genomic DNA were used to construct standard curves for each set of primers. qRT-PCR was performed using SYBR® green enzyme. Fold change was calculated based on the cycle number required to achieve a predesignated quantity of signal normalized to a 16S rRNA control.
ATP assays
ATP levels of stationary and mid-exponential cultures with the addition of various concentrations of arsenate were measured using a Promega BacTiter Glo kit according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We would like to thank Dr. Christiane Wolz for the gift of the HG001, HG001 rshsyn and triple mutant rshsyn, relP, relQ strains. We thank Dr. Richard Lee and Dr. Michael LaFleur for critical discussions. This work was supported by NIH grant R01AI110578 to KL and by a Charles A. King fellowship to BC.
Footnotes
Author Contributions
BPC and SER designed the study, performed experiments, analyzed results and wrote the paper. ABG ASN and EAZ performed experiments. NPG created the triple TA mutant strain. GC and JNA designed the study and analyzed results. ALC designed the study. KL designed the study, analyzed results and wrote the paper.
References
- 1.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 2.Conlon BP, et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlon BP. Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease. BioEssays : news and reviews in molecular, cellular and developmental biology. 2014;36:991–996. doi: 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- 4.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hede K. Antibiotic resistance: An infectious arms race. Nature. 2014;509:S2–S3. doi: 10.1038/509S2a. [DOI] [PubMed] [Google Scholar]
- 8.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 9.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaFleur MD, Qi Q, Lewis K. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrobial agents and chemotherapy. 2010;54:39–44. doi: 10.1128/AAC.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulcahy LR, Burns JL, Lory S, Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher MA. Molecular basis of heritable multidrug tolerance. Nature. 2015 doi: 10.1038/nature14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balaban NQ, Gerdes K, Lewis K, McKinney JD. A problem of persistence: still more questions than answers? Nat Rev Microbiol. 2013;11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 14.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 15.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 16.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nature reviews. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 17.Correia FF, et al. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J Bacteriol. 2006;188:8360–8367. doi: 10.1128/JB.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular mechanism of bacterial persistence by HipA. Molecular cell. 2013;52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 19.Kaspy I, et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nature communications. 2013;4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- 20.LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Y, et al. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Research. 2011;39:D606–D611. doi: 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donegan NP, Thompson ET, Fu Z, Cheung AL. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J Bacteriol. 2010;192:1416–1422. doi: 10.1128/JB.00233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 24.Geiger T, et al. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun. 2010;78:1873–1883. doi: 10.1128/IAI.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiger T, et al. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 2010;78:1873–1883. doi: 10.1128/IAI.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan Y, Lazinski D, Rowe S, Camilli A, Lewis K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. MBio. 2015;6 doi: 10.1128/mBio.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beenken KE, et al. Global gene expression in Staphylococcus aureus biofilms. Journal of bacteriology. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George SE, et al. Phenotypic Heterogeneity and Temporal Expression of the Capsular Polysaccharide in Staphylococcus aureus. Mol Microbiol. 2015 doi: 10.1111/mmi.13174. [DOI] [PubMed] [Google Scholar]
- 29.Davis BD, Chen LL, Tai PC. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:6164–6168. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik M, Zhao X, Drlica K. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol Microbiol. 2006;61:810–825. doi: 10.1111/j.1365-2958.2006.05275.x. [DOI] [PubMed] [Google Scholar]
- 31.Cho H, Uehara T, Bernhardt TG. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwan BW, Valenta JA, Benedik MJ, Wood TK. Arrested protein synthesis increases persister-like cell formation. Antimicrobial Agents and Chemotherapy. 2013;57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinbrecher T, et al. Peptide-lipid interactions of the stress-response peptide TisB that induces bacterial persistence. Biophysical journal. 2012;103:1460–1469. doi: 10.1016/j.bpj.2012.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorr T, Vulić M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. Plos Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, et al. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 2013;15:1734–1744. [Google Scholar]
- 36.Moore SA, Moennich DM, Gresser MJ. Synthesis and hydrolysis of ADP-arsenate by beef heart submitochondrial particles. J Biol Chem. 1983;258:6266–6271. [PubMed] [Google Scholar]
- 37.Imamura H, et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechner S, Lewis K, Bertram R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. Journal of molecular microbiology and biotechnology. 2012;22:235–244. doi: 10.1159/000342449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mechler L, et al. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2015;59:5366–5376. doi: 10.1128/AAC.00643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donegan NP, Thompson ET, Fu Z, Cheung AL. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. Journal of Bacteriology. 2010;192:1416–1422. doi: 10.1128/JB.00233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geiger T, Kastle B, Gratani FL, Goerke C, Wolz C. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. Journal of Bacteriology. 2014;196:894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung AL, Nast CC, Bayer AS. Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infection and immunity. 1998;66:5988–5993. doi: 10.1128/iai.66.12.5988-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donegan NP, Cheung AL. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J Bacteriol. 2009;191:2795–2805. doi: 10.1128/JB.01713-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.