Significance
T-cell receptors (TCRs) recognize pathogens during T-cell–mediated adaptive immune responses. Here we directly observed the organization of TCRs and their rearrangement on lymph node-resident T cells during an immune response. We found that TCRs were preclustered into nanometer-scale protein islands, which were organized into larger membrane territories, and that T-cell activation induced the formation of microclusters. Our data provide the first evidence, to our knowledge, of subdiffraction membrane organizations during immune responses in an animal model system.
Keywords: T-cell receptor, T-cell activation, protein cluster, superresolution, plasma membrane
Abstract
T cells become activated when T-cell receptors (TCRs) recognize agonist peptides bound to major histocompatibility complex molecules on antigen-presenting cells. T-cell activation critically relies on the spatiotemporal arrangements of TCRs on the plasma membrane. However, the molecular organizations of TCRs on lymph node-resident T cells have not yet been determined, owing to the diffraction limit of light. Here we visualized nanometer- and micrometer-scale TCR distributions in lymph nodes by light sheet direct stochastic optical reconstruction microscopy (dSTORM) and structured illumination microscopy (SIM). This dSTORM and SIM approach provides the first evidence, to our knowledge, of multiscale reorganization of TCRs during in vivo immune responses. We observed nanometer-scale plasma membrane domains, known as protein islands, on naïve T cells. These protein islands were enriched within micrometer-sized surface areas that we call territories. In vivo T-cell activation caused the TCR territories to contract, leading to the coalescence of protein islands and formation of stable TCR microclusters.
T cells play a central role in adaptive immunity against pathogens. T-cell receptors (TCRs) in the plasma membrane recognize pathogen-derived peptides bound to major histocompatibility complexes (MHCs) on the surface of antigen-presenting cells (APCs). The compartmentalization and distribution of TCRs reflect early signaling events (1, 2). TCR microcluster formation has been observed on artificial surfaces coated with antibodies against TCR or endogenous ligands, and on glass-supported lipid bilayers containing ligands. The microtubule-dependent inward movement of microclusters forms a mature immunologic synapse with its typical “bull’s eye” pattern (3–5).
New superresolution imaging techniques have extended the study of membrane architecture to the nanometer scale; these techniques include photoactivated localization microscopy [PALM (6) or FPALM (7)], (direct) stochastic optical reconstruction microscopy [STORM (8) or dSTORM (9)], and structured illumination microscopy [SIM (10, 11)]. In combination with electron microscopy (EM), these techniques have revealed 70- to 200-nm-wide TCR membrane domains on quiescent T cells (12–14), known as “protein islands” or “nanoclusters.” These protein islands coalesce into micrometer-scale clusters, or “microclusters,” after T-cell activation (13). Although many other membrane proteins form similar nanometer-sized compartments, including lipid-anchored proteins (15–17), integrins (18, 19), immune receptors (13, 14, 20, 21), growth factor receptors (22) and neurotransmitter receptors (23, 24), these have not been observed in tissue. In particular, nanometer-scale TCR organization and rearrangement after antigen recognition have not been observed in the physiological environment of T cells. As such, the physiological relevance of TCR clustering remains in question.
We investigated the subdiffraction organization of TCRs in mouse lymph nodes and its change in response to antigen exposure using light-sheet dSTORM and SIM. dSTORM provides higher spatial resolution than SIM, whereas SIM circumvents the spurious clustering artifacts in dSTORM. Comparison of lymph node sections from untreated and antigen-challenged mice revealed redistributions of TCR islands induced by antigenic peptides processed and presented by APCs in vivo. Our results demonstrate that T-cell activation is spatially controlled at nanometer and micrometer scales, ranging from protein islands to microclusters and newly described “territories.”
Results
Light Sheet dSTORM Visualized Nanometer-Scale TCR Islands on Naïve Lymph Node-Resident T Cells.
We developed prism-coupled light sheet dSTORM to overcome the high fluorescence background from bulk excitation in tissue (25). In brief, spatially aligned 642-nm and 405-nm lasers pass through a cylindrical lens and focus at the back aperture of a long-working distance illumination objective. The light sheet traverses through a glass prism and focuses at the interface between glass and tissue. Use of the glass prism increases the sample space to accommodate the use of a standard 35-mm culture dish (SI Appendix, Fig. S1 A–C). The ∼2-µm-thin light sheet is coplanar with the image plane. Optical sectioning is achieved by adjusting the position of the tissue sample in relation to the light sheet (SI Appendix, Fig. S1D).
For dSTORM imaging in the present study, a continuous 642-nm excitation light sheet was overlaid with a low-power 405-nm activation light sheet with various pulse durations. A sparse set of single molecule events was activated in each frame. Individual molecule localizations were determined by Gaussian-based centroid fitting using a custom MATLAB code. Drift correction was performed by subpixel cross-correlation (26–28), which achieved a level of precision comparable to that seen with the use of fiducial markers (SI Appendix, Figs. S2 and S3). The localization precision in lymph nodes was 35 ± 9 nm, with an average photon count of ∼400. The lateral resolution within the image plane was estimated as ∼50 nm (SI Appendix, Fig. S4).
The TCR distribution on naïve T cells was investigated in lymph nodes from 5c.c7 TCR transgenic mice without antigen exposure. CD4 T cells from 5c.c7 TCR transgenic mice recognize a peptide derived from moth cytochrome c (MCC; amino acids 88–103) presented by MHC class II molecules (specifically I-Ek) (29). Lymph nodes were fixed immediately after removal and cryosectioned for immunofluorescence staining. TCRs were detected with a monoclonal antibody (H57) against the β-chain directly conjugated with Alexa Fluor 647 dye (H57AF647). In a typical lymph node section, T cells surrounded B-cell follicles near the periphery of the tissue. In the T-cell zone, confocal imaging showed diffraction-limited distributions of TCRs at the plasma membrane (SI Appendix, Fig. S5).
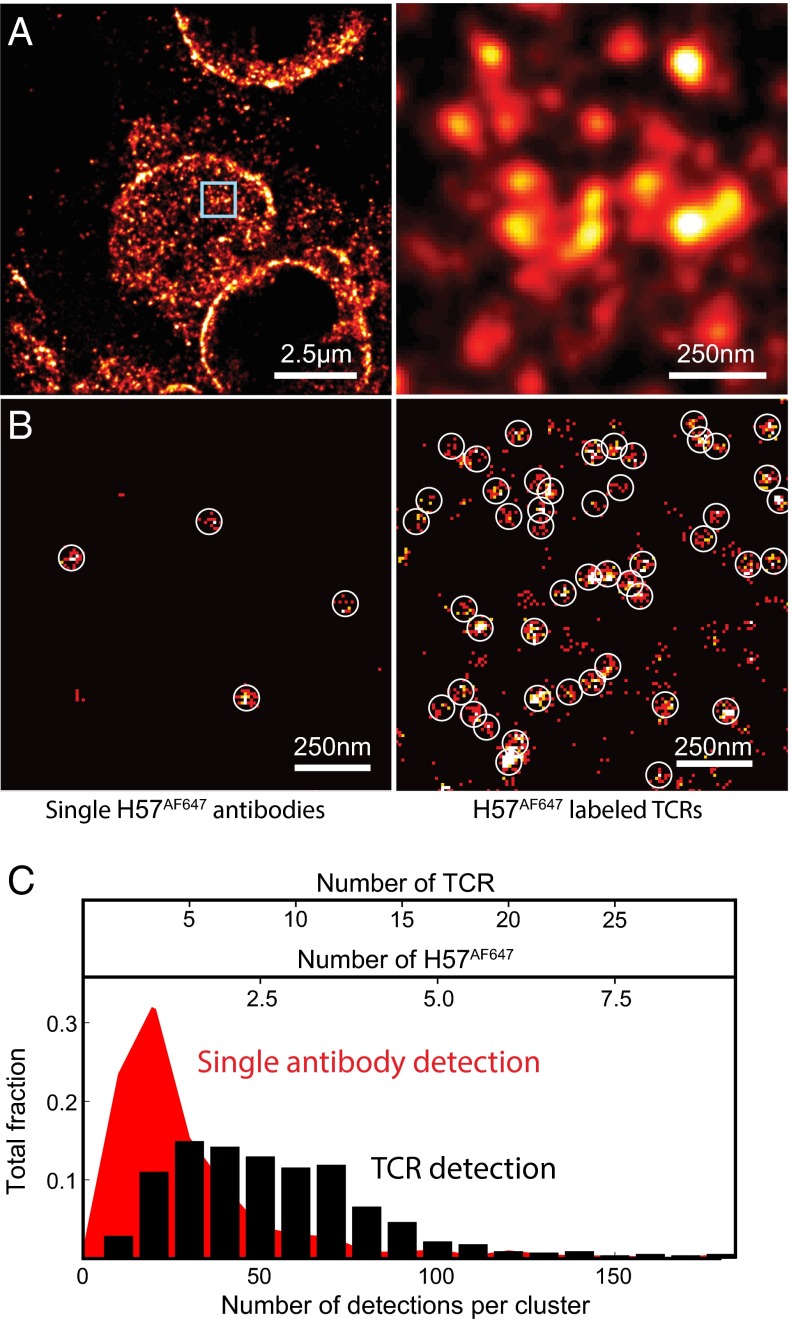
dSTORM imaging revealed nanometer-scale TCR islands on naïve lymph node-resident T cells (Fig. 1A). The simultaneous detection of the cell surfaces and cross-sections indicates specific staining at the plasma membrane of T cells. We tested whether TCR island and single H57AF647 antibody signals can be distinguished by comparing TIRF-based dSTORM images of isolated naïve T cells and single H57AF647 antibodies on glass surfaces. Circles with a diameter 2.5 times the localization accuracy, δ (87.5 nm), highlight detection events produced by single antibodies. In contrast to antibodies detected on the glass surface, TCRs in the plasma membrane of 5c.c7 T cells showed organized distributions (Fig. 1B). Based on the number of localization events, a small fraction of TCRs on the T-cell surfaces were detected as spatially isolated molecules. Here we defined TCR islands as assemblies of TCRs separated by less than 2.5 times the localization accuracy (overlapping circles in Fig. 1B). The majority of protein islands displayed larger dimensions, asymmetric shapes, and greater numbers of events (Fig. 1 B and C).
Fig. 1.
Superresolution imaging of TCRs on naïve lymph node-resident T cells. (A) Light sheet dSTORM image of TCRs on naïve T cells in a lymph node section. (B) Cluster detection of single molecules of H57AF647 (Left) vs. TCR islands labeled with H57AF647 (Right) using TIRF-based dSTORM. Circles highlight detection events produced by single antibodies. (C) Characterization of the number of detected events per single antibody (red) and TCR protein island (black). The computed number of TCRs was based on the average number of events per single H57 antibody (∼20) and labeling efficiency of TCRs (∼30%) by immunostaining.
Antibodies are known to have low labeling efficiency (30). Here immunostaining of lymph node sections resulted in the binding of 2.5 H57AF647 antibodies per TCR island on average. We calculated the detection efficiency to determine the number of total (labeled and unlabeled) TCRs per island. In the plasma membrane of naïve T cells, we registered ∼40 H57AF647 per µm2. According to the previously reported surface concentrations of ∼140 TCRs per µm2 (2, 13), we estimated our detection efficiency to be ∼30%. Based on this efficiency, the calculated number of TCRs per island is consistent with literature values of 7–20 (Fig. 1C) (13). In addition, the island structures in tissue are similar to those found in previous in vitro studies, which reported 40- to 300-nm-wide TCR islands with distances of 130–260 nm between them (13, 16).
TCR islands were enriched within 0.5- to 2-µm-wide membrane regions, which we call “territories.” Within a territory, neighboring islands were distinct and separated by ∼200 nm (SI Appendix, Fig. S6). Territories have irregular and undefined shapes, and are surrounded by membrane regions with low or no detectable levels of TCRs.
Light Sheet dSTORM Revealed TCR Microclusters After in Vivo T-Cell Activation.
We next investigated changes to TCR organization after in vivo T-cell activation. Previous in vitro studies have shown concatenation of TCR islands into microclusters after antigen recognition on activating surfaces (13, 14). Here we studied additional physiological parameters, including the display of endogenous and antigenic peptides at physiological concentrations by host APCs, processing and transport of antigenic peptides, and mechanical and biochemical environments in the lymph node.
For T-cell activation, 5c.c7 TCR transgenic mice were injected i.p. with MCC peptides in alum adjuvant. Dendritic cells (DCs) either from the injection site or resident to the draining lymph nodes presented the MCC peptides and induced specific T-cell responses (SI Appendix, Fig. S7A). Mesenteric lymph nodes were immediately fixed and imaged at 2, 5, and 24 h after injection. The presence of MCC in the lymph node as early as 2 h postinjection (SI Appendix, Fig. S7B) indicates T-cell activation by DCs. Previous studies have shown that peptide injection causes rapid and synchronized activation of all T cells in TCR transgenic systems (31). Compared with T cells from untreated mice, T cells from injected mice showed increased heterogeneity in shape, size, and membrane roughness (Fig. 2A and SI Appendix, Fig. S7C).
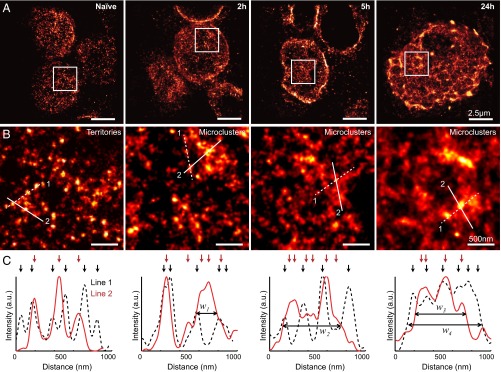
Fig. 2.
Light sheet dSTORM imaging of TCRs in lymph nodes before and after in vivo T-cell activation. (A) Representative superresolution images of T cells from mice without peptide injection (naïve), and in mice at 2, 5, and 24 h after peptide injection. (B) High-magnification view of the TCR organizations in boxed regions in A. (C) Intensity profiles of lines in B. Arrows mark local intensity peaks that represent protein islands. Reduced distances between adjacent protein islands cause an overlapping of intensity peaks and a broadening of the structure (w1 = 240, w2 = 507, w3 = 533, and w4 = 753 nm).
In contrast to naïve T cells, a subset of TCRs in activated T cells was localized in 200- to 800-nm-wide structures (Fig. 2B). These structures are similar to the aforementioned microclusters (1, 32, 33). Unlike territories, protein islands could not be resolved within microclusters (Fig. 2B). Local fluorescence density peaks in superresolution images indicated reduced separation between protein islands (Fig. 2C). Microclusters could be formed through concatenation of territories and recruitment of additional protein islands. Naïve T cells contain only TCR territories, whereas activated T cells contain both territories and microclusters, suggesting that not all TCRs contribute to T-cell activation. Control injections of alum adjuvant without MCC peptide did not trigger TCR redistributions (SI Appendix, Fig. S8). Therefore, the observed TCR rearrangements are caused by the recognition of MCC peptide presented by MHC molecules on DCs. The locations of microclusters most likely coincide with the initial contact sites between T cells and DCs.
We confirmed the observed TCR organizations on freshly isolated T cells from mice before and after peptide injection using a TIRF-based dSTORM system with greater localization accuracy. Immunostaining was performed in T-cell suspension without membrane permeabilization, and T cells were spun onto glass slides for imaging. This procedure stains plasma membrane TCRs, but not the TCRs residing on extracellular microvesicles or intracellular membranes (e.g., endosomes, Golgi, endoplasmic reticulum). TIRF dSTORM resolved protein islands on naïve T cells and concatenating protein islands within microclusters on activated T cells (SI Appendix, Fig. S9). The interprotein–island separation distances appeared to be slightly increased compared with tissue. Tissue dissociation also yielded more homogeneous appearance and distributions of microclusters. The flattening of the plasma membrane could contribute to these differences. Costaining of T cells with a membrane dye (DiL) and H57AF647 verified that increased TCR localization was not due to membrane ruffling (SI Appendix, Fig. S10).
SIM Imaging of TCR Islands and Microclusters in Lymph Node-Resident T Cells.
We confirmed our light sheet dSTORM findings using SIM. On naïve lymph node-resident T cells, SIM detected segregated TCR islands with dimensions of ∼100–300 nm (Fig. 3 A–C). These islands are resolution-limited assemblies of TCRs, because SIM has a lower resolution than dSTORM (>100 nm in tissue). Each assembly entity could represent a single TCR island or multiple TCR islands. The interassembly separation distances were 200–500 nm. After in vivo activation, T cells showed clear changes in TCR distributions (Fig. 3 D–F). Decreased interassembly distances suggested closer packing of the protein islands. T cells with increased TCR clustering appeared more polarized, which could be mediated by T cell–APC contacts. These findings support the earlier light sheet dSTORM data from lymph nodes. In addition, SIM imaging of T cells freshly isolated from mice before and after peptide injection confirmed the TCR organizations observed in lymph node-resident T cells (SI Appendix, Fig. S9).
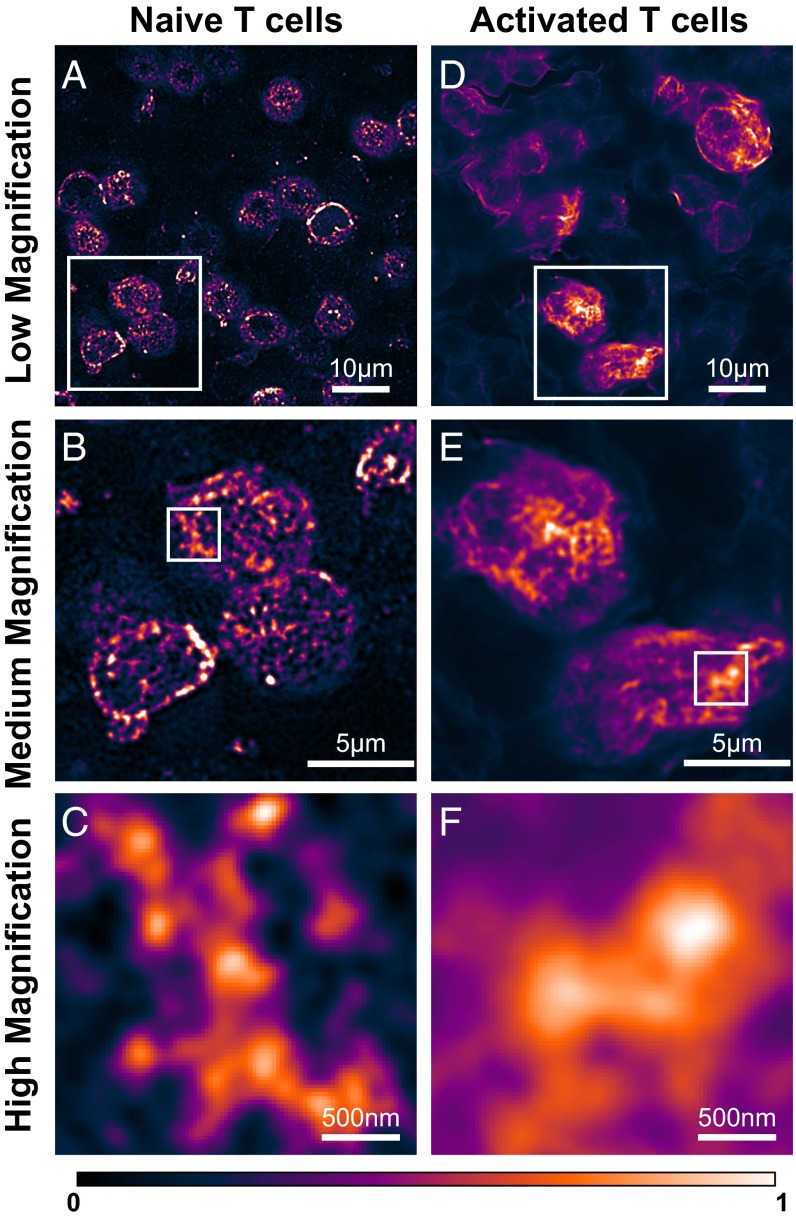
Fig. 3.
SIM imaging of TCRs in lymph nodes before and after in vivo T cell activation. (A) Low-magnification view of naïve T cells. (B) Medium-magnification view of the boxed region in A showing protein islands. (C) High-magnification view of the boxed region in B showing a TCR territory and protein islands within it. (D) Low-magnification view of in vivo activated T cells at 5 h after peptide injection. (E) Medium-magnification view of the boxed region in D showing areas with high TCR density on the plasma membrane. (F) High-magnification view of the boxed region in E showing a TCR microcluster.
Discussion
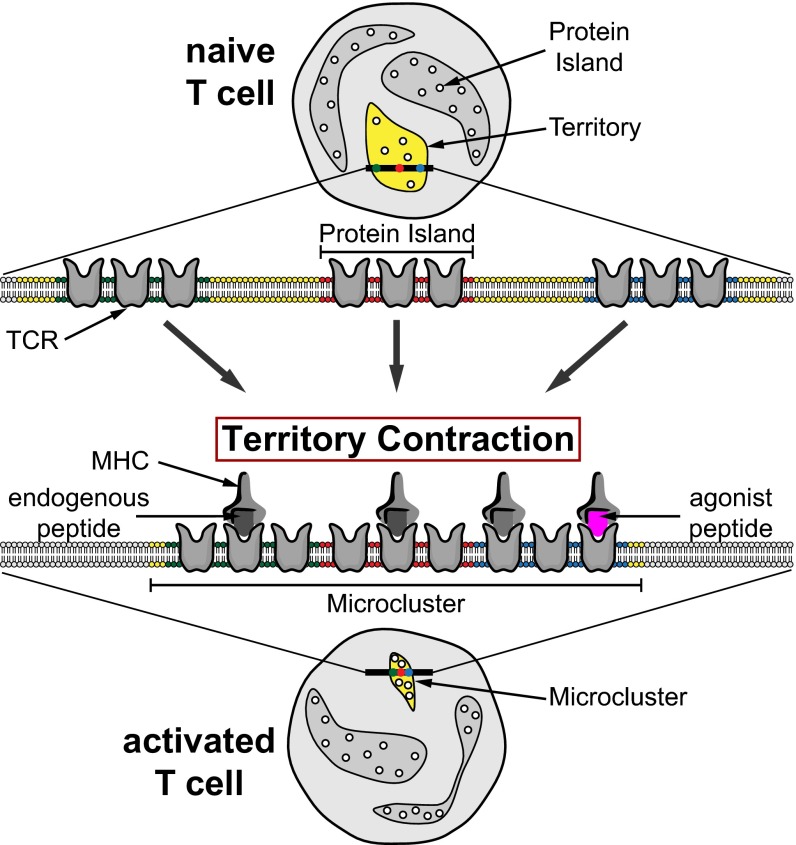
The complex organization and dynamics of the plasma membrane present an ongoing challenge. This study is the first, to our knowledge, to image plasma membrane receptors in the native tissue environment with subdiffraction resolution using dSTORM and SIM. For dSTORM, we achieved a high signal-to-noise ratio (SNR) by using ultrathin light sheet illumination. Our data prove the existence of nanometer- and micrometer-sized TCR membrane structures on lymph node-resident T cells. TCR islands on naïve T cells were enriched in surface areas that we call territories. On in vivo activation, T cells formed TCR microclusters with broad size distributions. The coexistence of microclusters and territories on activated T cells suggest that TCR redistribution is restricted to T cell–APC contact sites. Our observations suggest a hierarchical organization of TCRs on the plasma membrane of lymph node-resident T cells (Fig. 4). In naïve T cells, TCRs are preclustered into nanometer-scale protein islands, which are enriched in micrometer-scale territories. After antigen recognition, TCR territories contract to form microclusters. Territory contraction brings together distinct protein islands, which are required for signal transduction and T-cell activation.
Fig. 4.
Two-level organization model for TCR distribution in the plasma membrane. The illustration demonstrates the rearrangement of TCRs from naïve T cells (Upper) to activated T cells (Lower). The rearrangement is facilitated by the contraction of TCR territories (yellow) enriched with protein islands (small circles). The contraction can be initiated by the presence of agonist peptides (purple) and facilitated by endogenous peptides (gray).
Potential limitations of the present study include artifacts due to postexcision fixation, low labeling efficiencies, and reduced resolution. These limitations require the consideration of alternative procedures and interpretations. Postexcision fixation of lymph nodes by immersion could affect T-cell morphology and plasma membrane organizations. For optimal results, it should be replaced in future studies by perfusion fixation (34). The low labeling efficiency of immunofluorescence staining (∼30%) hinders quantitative analysis of the dimensions of TCR islands and microclusters. Some localization clusters in naïve T cells could represent monomers or islands within which only one TCR was detected. TCR transgenic mice with T cells expressing TCRs fused to photoactivatable fluorescent proteins could be used to increase the detection efficiency. In addition, the resolution achieved here does not allow us to distinguish between TCRs at the plasma membrane and in intracellular or extracellular vesicles close to the cell surface (35, 36). Although our data from freshly isolated lymph node T cells with and without peptide injections showed similar changes to the data from tissue sections, contributions from vesicles cannot be excluded. TCRs are continuously recycled by endocytosis and exocytosis (36–41). TCR down-regulation by endocytosis is a well-documented mechanism to reduce T-cell signaling (37–41). More recently, secretion of exosomes in the immunologic synapse has been reported for T cells activated on glass-supported lipid bilayers (36). Taken together, these processes can contribute to the TCR enrichment that we observed in lymph node-resident T cells.
Traditional central supramolecular activation clusters (cSMACs), a hallmark of mature synapses, were not apparent on lymph node-resident T cells. This observation is consistent with multifocal TCR structures observed by EM (42), transient TCR accumulations in vivo (35), and subregions identified within the cSMAC (33, 43, 44). The stable microclusters described here may resemble these previously described structures and could share some of their functions, such as signal maintenance, signal inhibition, endocytosis, and exocytosis. The sustained presence of microclusters in our study suggests that the plasma membrane retains its TCR reorganization beyond initial T cell–APC contacts (45). Based on in vitro studies, TCR clustering is most likely maintained by cytoskeletal structures and lipid phase separation (1, 2).
TCR territories provide a mechanism for microcluster formation. TCR-enriched territories are likely responsible for the fast and amplified assembly of the signaling cascade, making them surface “hot spots” for APC recognition. Previous studies have shown that T cells can recognize a single agonist peptide and become fully activated by ∼10 peptides (46, 47). Our present data and previous data suggest that recognition of a single agonist peptide within a protein island can lead to contraction of the corresponding territory into a microcluster. It is likely that TCR interactions with endogenous peptide-MHC complexes on the APC contribute to this mechanism, to ensure sufficient cell contact and TCR recruitment (48). A territory would serve as the smallest activation unit to simultaneously ensure antigen specificity and sensitivity.
Conclusions
In conclusion, we have investigated the organization and rearrangement of TCRs on lymph node-resident T cells before and after in vivo activation. We achieved subdiffraction resolution and resolved TCR protein islands and territories on naïve T cells. In mice injected with agonist peptides, we observed the formation of microclusters. We have summarized our findings in a model that considers TCR organization on nanometer and micrometer scales. Our study provides multiple lines of evidence for the spatial organizations of TCRs in vivo. Our imaging approach is an important step toward studying many biological systems in which the cellular architecture controls their physiological functions at nanometer scale.
Materials and Methods
Mice, Antibodies, Peptides, and Fluorophores.
5c.c7/Rag2−/− (Taconic) and B10.A mice (Taconic) were crossed to obtain homozygous 5c.c7 TCR transgenic mice. The animal protocol was approved by the Salk Institutional Animal Care and Use Committee (protocol #13-00006). Armenian hamster monoclonal antibody H57 anti-murine TCR β-chain (Bio-X-Cell) was used to label TCRs. Intraperitoneal injection of MCC peptides was used for in vivo activation of T cells. Mesenteric lymph nodes were removed from mice, cryosectioned, and labeled with H57 conjugated with Alexa Fluor 647 (SI Appendix, Materials and Methods).
Superresolution Fluorescence Imaging.
Light Sheet dSTORM.
Single-molecule superresolution imaging was performed using a prism-coupled light sheet system as described previously (25). Single molecule events within 2.5 times the localization precision (35 nm) from consecutive frames were grouped into a single localization event. Drift correction was performed using subpixel cross-correlation (SI Appendix, Materials and Methods).
TIRF dSTORM.
The same excitation and activation lasers as used for light sheet dSTORM were used for TIRF illumination on an inverted Nikon Eclipse microscope. Drift correction was performed using near-infrared gold nanoshells as fiducials (SI Appendix, Materials and Methods).
SIM.
The Zeiss ELYRA PS.1 system was used for SIM imaging. Tissue samples were embedded in Vectashield for index matching (SI Appendix, Materials and Methods).
Analysis.
Superresolution images were reconstructed by an in-house MATLAB code that performs Gaussian fit to single molecule images in both lateral and horizontal directions. Drift correction using subpixel cross-correlation was performed using a MATLAB code adapted from Guizar-Sicairos et al. (49). Fourier ring correlation analysis was performed using a MATLAB code adapted from Nieuwenhuizen et al. (50).
Supplementary Material
Acknowledgments
We thank James Fitzpatrick, Matt Jones, and Sarah Dunn for assisting with tissue imaging at the Salk Biophotonics Core; Kimberly McIntyre for assisting with tissue sectioning at the Salk Histology Core; and Greg Lemke for providing antibodies and advice for the immunofluorescent staining of dendritic cells in tissue. We also thank the Nomis Foundation, the Waitt Foundation, and the James B. Pendleton Charitable Trust for their support. This project was supported by National Institutes of Health Grants DP2GM105455 (to B.F.L.) and DP2EB020400 (to H.C.), the Waitt Advanced Biophotonics Core Facility of the Salk Institute (with funding from National Institutes of Health Cancer Center Support Grant P30 014195, a National Institute of Neurologic Disorders and Stroke Neuroscience Core Grant, and the Waitt Foundation), and National Cancer Institute Grant CA014195. Y.S.H. was supported by California Institute for Regenerative Medicine Grant TG2-01158.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512331113/-/DCSupplemental.
References
- 1.Bunnell SC, et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. 2002;158(7):1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campi G, Varma R, Dustin ML. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J Exp Med. 2005;202(8):1031–1036. doi: 10.1084/jem.20051182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395(6697):82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 4.Grakoui A, et al. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285(5425):221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto-Tane A, et al. Dynein-driven transport of T cell receptor microclusters regulates immune synapse formation and T cell activation. Immunity. 2011;34(6):919–931. doi: 10.1016/j.immuni.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 7.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91(11):4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47(33):6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 10.Gustafsson MGL. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102(37):13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198(Pt 2):82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 12.Schamel WW, et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202(4):493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lillemeier BF, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11(1):90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman E, et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 2011;35(5):705–720. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102(43):15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci USA. 2006;103(50):18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Hancock JF. Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim Biophys Acta. 2015;1853(4):841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Cambi A, et al. Organization of the integrin LFA-1 in nanoclusters regulates its activity. Mol Biol Cell. 2006;17(10):4270–4281. doi: 10.1091/mbc.E05-12-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Zanten TS, et al. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci USA. 2009;106(44):18557–18562. doi: 10.1073/pnas.0905217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattila PK, et al. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013;38(3):461–474. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Pageon SV, et al. Superresolution microscopy reveals nanometer-scale reorganization of inhibitory natural killer cell receptors upon activation of NKG2D. Sci Signal. 2013;6(285):ra62. doi: 10.1126/scisignal.2003947. [DOI] [PubMed] [Google Scholar]
- 22.Ariotti N, et al. Epidermal growth factor receptor activation remodels the plasma membrane lipid environment to induce nanocluster formation. Mol Cell Biol. 2010;30(15):3795–3804. doi: 10.1128/MCB.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78(4):615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair D, et al. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J Neurosci. 2013;33(32):13204–13224. doi: 10.1523/JNEUROSCI.2381-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu YS, et al. Light-sheet Bayesian microscopy enables deep-cell super-resolution imaging of heterochromatin in live human embryonic cells. Opt Nanoscopy. 2013;2:7. doi: 10.1186/2192-2853-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319(5864):810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisler C, et al. Drift estimation for single marker switching based imaging schemes. Opt Express. 2012;20(7):7274–7289. doi: 10.1364/OE.20.007274. [DOI] [PubMed] [Google Scholar]
- 28.Mlodzianoski MJ, et al. Sample drift correction in 3D fluorescence photoactivation localization microscopy. Opt Express. 2011;19(16):15009–15019. doi: 10.1364/OE.19.015009. [DOI] [PubMed] [Google Scholar]
- 29.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med. 1992;176(4):1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández-Suárez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9(12):929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 31.Zell T, et al. Single-cell analysis of signal transduction in CD4 T cells stimulated by antigen in vivo. Proc Natl Acad Sci USA. 2001;98(19):10805–10810. doi: 10.1073/pnas.191567898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25(1):117–127. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokosuka T, et al. Spatiotemporal regulation of T cell costimulation by TCR-CD28 microclusters and protein kinase C theta translocation. Immunity. 2008;29(4):589–601. doi: 10.1016/j.immuni.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bajénoff M, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25(6):989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman RS, Beemiller P, Sorensen CM, Jacobelli J, Krummel MF. Real-time analysis of T cell receptors in naive cells in vitro and in vivo reveals flexibility in synapse and signaling dynamics. J Exp Med. 2010;207(12):2733–2749. doi: 10.1084/jem.20091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choudhuri K, et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507(7490):118–123. doi: 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das V, et al. Activation-induced polarized recycling targets T cell antigen receptors to the immunological synapse; involvement of SNARE complexes. Immunity. 2004;20(5):577–588. doi: 10.1016/s1074-7613(04)00106-2. [DOI] [PubMed] [Google Scholar]
- 38.Patino-Lopez G, et al. Rab35 and its GAP EPI64C in T cells regulate receptor recycling and immunological synapse formation. J Biol Chem. 2008;283(26):18323–18330. doi: 10.1074/jbc.M800056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Oro U, et al. Regulation of constitutive TCR internalization by the zeta-chain. J Immunol. 2002;169(11):6269–6278. doi: 10.4049/jimmunol.169.11.6269. [DOI] [PubMed] [Google Scholar]
- 40.Minami Y, Samelson LE, Klausner RD. Internalization and cycling of the T cell antigen receptor: Role of protein kinase C. J Biol Chem. 1987;262(27):13342–13347. [PubMed] [Google Scholar]
- 41.Liu H, Rhodes M, Wiest DL, Vignali DA. On the dynamics of TCR:CD3 complex cell surface expression and downmodulation. Immunity. 2000;13(5):665–675. doi: 10.1016/s1074-7613(00)00066-2. [DOI] [PubMed] [Google Scholar]
- 42.Brossard C, et al. Multifocal structure of the T cell–dendritic cell synapse. Eur J Immunol. 2005;35(6):1741–1753. doi: 10.1002/eji.200425857. [DOI] [PubMed] [Google Scholar]
- 43.Saito T, Yokosuka T, Hashimoto-Tane A. Dynamic regulation of T cell activation and co-stimulation through TCR-microclusters. FEBS Lett. 2010;584(24):4865–4871. doi: 10.1016/j.febslet.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 44.Tseng SY, Waite JC, Liu M, Vardhana S, Dustin ML. T cell-dendritic cell immunological synapses contain TCR-dependent CD28-CD80 clusters that recruit protein kinase C theta. J Immunol. 2008;181(7):4852–4863. doi: 10.4049/jimmunol.181.7.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fahmy TM, Bieler JG, Edidin M, Schneck JP. Increased TCR avidity after T cell activation: A mechanism for sensing low-density antigen. Immunity. 2001;14(2):135–143. [PubMed] [Google Scholar]
- 46.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419(6909):845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 47.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat Immunol. 2004;5(5):524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 48.Wülfing C, et al. Costimulation and endogenous MHC ligands contribute to T cell recognition. Nat Immunol. 2002;3(1):42–47. doi: 10.1038/ni741. [DOI] [PubMed] [Google Scholar]
- 49.Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008;33(2):156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- 50.Nieuwenhuizen RP, et al. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10(6):557–562. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.