Significance
Many immune responses are mediated by direct cell–cell interactions and develop over multiple timescales. A mechanistic understanding of how diverse outcomes arise during these interactions entails identifying the relationships between various responses occurring at different stages by correlated measurements. Typical approaches that rely on population-wide correlations, however, reveal these connections broadly and mask the fine details that might be discernible only at the single-cell level. Here, we present a microfluidics-based cell–cell interaction assay that allows defined generation, real-time imaging, and longitudinal assay of lymphocyte interactions, thereby permitting direct correlative studies within each single cell. Our studies using this platform indicate a possible role for the strength of calcium signaling in selective regulation of cytotoxicity and interferon-gamma production of natural killer cells.
Keywords: microfluidics, cell pairing, single-cell analysis, cell–cell interactions, multiparameter assay
Abstract
Resolving how the early signaling events initiated by cell–cell interactions are transduced into diverse functional outcomes necessitates correlated measurements at various stages. Typical approaches that rely on bulk cocultures and population-wide correlations, however, only reveal these relationships broadly at the population level, not within each individual cell. Here, we present a microfluidics-based cell–cell interaction assay that enables longitudinal investigation of lymphocyte interactions at the single-cell level through microfluidic cell pairing, on-chip culture, and multiparameter assays, and allows recovery of desired cell pairs by micromanipulation for off-chip culture and analyses. Well-defined initiation of interactions enables probing cellular responses from the very onset, permitting single-cell correlation analyses between early signaling dynamics and later-stage functional outcomes within same cells. We demonstrate the utility of this microfluidic assay with natural killer cells interacting with tumor cells, and our findings suggest a possible role for the strength of early calcium signaling in selective coordination of subsequent cytotoxicity and IFN-gamma production. Collectively, our experiments demonstrate that this new approach is well-suited for resolving the relationships between complex immune responses within each individual cell.
Initiation and progression of immune responses require direct cell–cell interactions. Molecular interactions at the contact interface mediate cellular cross-talk and trigger a series of well-orchestrated downstream signaling events. The magnitude and dynamics of these signals promote and coordinate immune cell activation, and underlie a broad array of functional outcomes, such as target cell elimination, secretory activity, and proliferation (1, 2). Understanding how these early signaling events are transduced into functional responses demands correlated measurements at different stages as these responses unfold.
Typically, these relationships are probed by first activating cells in bulk cocultures, often mixing cell populations and initiating contacts via a brief centrifugal cosedimentation, and then piecing together measurements from independent assays performed at different time points. This approach may determine broad outcomes of intercellular interactions, but it suffers from three major limitations. First, individual assay measurements are averaged over many different combinations of interactions due to the uncontrolled and indefinite nature of cell–cell interactions in bulk cocultures. For example, varying times of initiation and duration of contacts (1, 3), number of interacting partners (4), and differences in antigen presenting cells (5) can all modulate immune cell responses. Unpredictable variation is therefore introduced into measured responses, masking the true intrinsic cellular heterogeneity and blurring the correlations. Second, conventional assays determine certain functional responses only indirectly using surrogate markers (for example, cytotoxicity by CD107a expression, and proliferation by fluorescent dye partitioning or Ki67 expression), as direct measurements require longitudinal tracking of individual cells and their interacting partners. Such indirect measurements usually limit the specificity of detection, conceivably resulting in erroneous interpretation of results to a certain degree. Finally and more importantly, relationships are resolved only at the population level and not within each single cell, as different cells are used in each individual assay. As such, results tacitly assume uniformity among the states of individual cells. However, it is increasingly evident that significant heterogeneity exists even among genetically identical cells, and that single-cell responses differ qualitatively from what is inferred from ensemble measures (6–8).
These limitations have motivated the development of several microscale tools to study immune cell interactions at the single-cell level. A common approach has been to isolate discrete numbers of cells in spatial confinements [such as microwells (9–12), microchambers (13), or droplets (14)] and monitor their interactions with multiple measurements. Although these approaches have made it feasible to resolve the relationships between different immune responses, particularly between cellular motility, cytotoxicity, and secretory activity (9, 10, 12), two primary limitations inherent in their operation principles have precluded the ability to study early signaling dynamics and their correlation to subsequent functional behaviors. First, generation of cell–cell interactions is governed by stochastic cell loading that limits conjugate formation efficiency and yields low-throughput-per-footprint area, which in turn severely limits the spatiotemporal resolution for dynamic imaging (i.e., number of cells that can be imaged with high temporal resolution). Second, due to the lack of control over the initiation and timing of interactions, contact formation can take time depending on chamber dimensions, migration behavior, and cell sizes, and cannot be verified unless imaged continuously. This variation in contacts not only makes imaging early dynamics impractical, but can also introduce variation into contact-dependent responses (taken at later stages) as a result of insufficient or varying contact durations. As an alternative, we and others have engineered approaches exploiting hydrodynamic (15) or electrical phenomena (16) to deterministically form interactions and control timings; however, these systems were geared toward assaying early events of intercellular interactions particularly (<20 min), and have not been optimized and extended to linked measurements at various timescales. Consequently, there exists a need for new approaches that can assay lymphocyte interactions longitudinally from the very onset and link diverse functional behaviors back to their early signaling signatures directly at the single-cell level.
To address this need, we report the use of a portable microfluidics platform to establish a cell–cell interaction assay that enables defined cell-pair generation, on-chip culture (∼24 h), and downstream assay without the need for any specialized or dedicated instruments, thereby allowing direct correlative studies within each single immune cell through longitudinal measurements beginning from the contact time. The device design further allows recovery of desired single-cell pairs using a micromanipulator for subsequent off-chip culture and analyses. We have demonstrated the utility of our assay by exploring the relationship between early signaling dynamics (here, calcium signaling) and prototypical effector functions directly within individual natural killer (NK) cells upon their interaction with tumor cells. Our findings indicate an inverse correlation between strength of early calcium signaling and IFN-gamma (IFN-γ) production of cytokine primed NK cells, and further suggest that the strength of early calcium signaling could be involved in selective coordination of subsequent cytotoxicity and IFN-γ production. Together, our results represent, to our knowledge, the first time the quantitative differences in calcium signals have been directly related to functional outputs at an individual cell level, and establish the microfluidic assay as a promising tool for longitudinal and quantitative investigation of lymphocyte interactions.
Results
Microfluidic Device Design and Optimization.
Monitoring early signaling events requires generation of defined cell pairs with controlled timings. We previously demonstrated a deformability-based cell-pairing approach that allowed sequential capture and pairing of fibroblasts in parallel (17). Here, as a first step to a microfluidic cell–cell interaction assay, we extended this cell-pairing technique to immune cells by developing microfluidic devices suitable for cell types with a wide size range (cell diameters ∼9–25 µm). The devices comprise weir-based hydrodynamic capture cups densely arrayed within a flow-through channel bonded to coverslips (SI Appendix, Fig. S1 A and B). The capture cup contains a two-cell trap connected to a single-cell trap via a narrow constriction (SI Appendix, Fig. S1C), and its structure is adjusted to maximize the ratio of the fluid flow into the cups versus around the cups. For this purpose, we placed support pillars around the capture cups (sides and back) to provide fluid flow through them for guiding cells into the traps for capture (SI Appendix, Figs. S1C and S2). The narrow openings at the sidewalls further provide additional flow path, and importantly help maintain similar flow resistances (therefore similar fluidic patterns) of the capture cups after the loading of the first cell population (that is, the flow resistance of the capture cups does not increase significantly due to the presence of trapped cells; SI Appendix, Fig. S2). This in turn facilitates sequential capture of the second cell population and their transfer from the single-cell traps to two-cell traps with similar fluidic operation. Whereas the single-cell trap was sized to capture only one cell, the two-cell trap was tailored to accommodate cell pairs with proper alignment within the traps. We placed capture cups within the array with proper spacing to minimize clogging and at the same time maximize cell capture. We designed mechanical filters made of pillars laid in an array format, and placed them before the trap array to prevent any cell clumps entering the array, thereby preventing clogging and device failure (Fig. S1B). The filter further helps cells enter into the trap array as single cells by facilitating the breakup of cell doublets and clumps. We incorporated a range of dimension variations within the mask design to ensure optimally matching devices for different types and sizes of cells. Through preliminary testing of various dimensions, we found that devices with a row spacing of ∼1.7–2× cell diameter and a column spacing of ∼1.1× cell diameter were most suitable among the available sizes.
Aside from successful cell pairing, establishing an assay for longitudinal immune cell interaction studies posed several additional technical challenges to be addressed. First, long-term observation of cell pairs requires isolation and culture of cells at spatially defined positions to track their interaction during the culture period. This is particularly challenging for immune cells as they are of suspension type and highly motile, and even more so for immune cell pairs as it further demands maintenance of cell–cell contacts. Second, multiparameter and longitudinal interrogation of interactions involve multiple measurements over time using various assays. Such extensive assaying requires the ability for repeated solution exchange for staining, washing, and media change cycles without disrupting or losing track of cell pairs, as would occur in bulk cocultures. Third, many microscale technologies that have been developed so far to deterministically form interactions necessitate specialized or dedicated equipment (for cell-scale control, perfusion, and on-stage incubation) which is often costly and laborious to operate, and further limits widespread adoption of these devices. In addition, the fact that the whole setup is committed to a single device for the duration of experiments limits the parallelization of experiments (i.e., number of devices that can be operated at the same time). A better approach would be to make the assay portable (without any disturbance to interactions), thereby making it compatible with standard laboratory equipment and parallelization.
In our earlier studies with fibroblasts (17), we noticed that cell pairs remained within their traps after removing fluidic connections, which enabled us to transport them in a standard incubator for culture. However, we observed that cell pairs migrated out of their traps during on-chip culture duration, resulting in loss of contacts and cell identities. Here, we reasoned that by sizing the trap dimensions properly (especially trap openings), we could secure and maintain cell pairs within their traps, which would then allow us to address all of the aforementioned challenges. Accordingly, during the development stage of the microfluidic devices, we adjusted the trap dimensions such that the support pillar dimensions and openings were all less than the smallest cell radius (for example, for NK cells: support pillars ∼3–4 µm, side gap ∼3–5 µm), and the width of narrow constriction in between the single-cell and two-cell traps was on the order of cell radius (for NK cells: ∼5–6 µm). Proper optimization of these dimensions crucially enabled: (i) maintaining cell pairs in continuous contact, entrapped at spatially defined positions for up to ∼24 h on-chip culture and longitudinal tracking; (ii) disconnecting the devices from any fluidic hardware while maintaining cell pairs for portable operation, without the need for dedicated microscope incubators or perfusion systems; (iii) performing various assays via repeated solution exchange by simple pipetting without disturbing the cell pairs; and (iv) establishing a protocol to retrieve desired single-cell pairs using a micromanipulator.
Validation of Microfluidic Cell–Cell Interaction Assay.
We first validated the central working principles of the microfluidic cell–cell interaction assay––namely pairing cells with defined timings, culturing cell pairs on-chip up to 24 h while preserving identities and viabilities, conducting various on-chip assays on interacting cell pairs, and recovering desired single-cell pairs for off-chip culture or analyses––and established an assay workflow (Fig. 1). Cell conjugates are generated using a squeeze-based microfluidic cell-pairing approach (17) that achieves high-efficiency cell pairing in parallel using a multistep loading procedure. In this method, cell populations are loaded sequentially, first capturing them in the single-cell traps, followed by transfer into two-cell traps by squeezing them through a narrow constriction via flow-induced deformation (Fig. 1). Using optimized device geometries and loading parameters, we could achieve fill factors of >95% (percentage of traps occupied) and cell-pairing efficiencies between ∼40% and 80% using various cell types (SI Appendix, Figs. S1 D–G and S3). Cell loading was sample-efficient (∼104 cells, ∼5-µL volume), and cell pairing was fast and highly synchronous across the array by virtue of the loading procedure used. As such, this approach provided uniform parameters for all pairs such as only one interacting partner, guaranteed contacts, and uniformity of interactions timings. As we designed the trap structure to secure and keep the cells within their capture cups after pairing, we could disconnect the devices from any external fluidic hardware while maintaining cell–cell contact and cell-pair registration within the array (cell-pair loss <%1). Using this feature, we could transfer the device between a standard incubator for culture and microscope for imaging, and perform longitudinal imaging of cell pairs over ∼24-h culture period (Fig. 1 and SI Appendix, Fig. S4). Media exchange was achieved using gravitationally driven flow without any disturbance to the cell pairs, simply by pipetting solutions onto the input reservoir and removing solution from the outlet. During the culture period, cell pairs remained in contact within their traps even after 24 h (>95% at t = 6 h, >85% at t = 24 h including the loss in viability), and showed little loss in viability (viability loss= 9 ± 1.5%, n = 5; SI Appendix, Fig. S4). We further demonstrated that cells could proliferate on-chip after a day of culture, with both daughter cells remaining within the capture cups after division (SI Appendix, Fig. S5). Together, these results established two central working principles of the microfluidic cell–cell interaction assay: controlled initiation of interactions and on-chip culture of cell pairs up to 24 h.
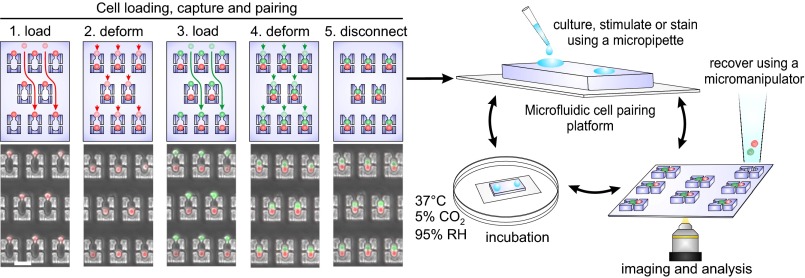
Fig. 1.
Microfluidics-based cell–cell interaction assay. Four-step cell-loading and pairing protocol (Left). (1) The first cell population (red) is captured in the single-cell traps. (2) Cells are transferred into two-cell traps through the constriction by flow-induced deformation. (3 and 4) The second cell population (green) is captured similarly and paired with already loaded cells using the same protocol. The transfer step (4) ensures that cell pairing is synchronous across the entire array. (5) Fluidic connections are removed to enable transferring the platform to different settings while maintaining cell pairing and registration. Transparent and nontransparent cells indicate the initial and final cell positions for each loading step; arrows display the fluidic paths between these positions. After disconnection, solutions can be introduced into the device using a pipette, devices can be transported between incubator and microscope, and desired pairs can be retrieved using micromanipulators (Right). (Scale bar, 50 µm.)
Next, we demonstrated the on-chip feasibility of carrying out various established assay protocols that are regularly used to characterize cell–cell interactions. This was enabled by the ability to introduce solutions by pipetting, together with the capacity to transfer devices in between different settings (incubator, refrigerator, or microscope stage) to carry out media exchange, staining, washing, and cell fixation steps on-chip. Our experiments centered on the interaction between an NK cell line, NK92MI, and a prototypical NK-sensitive target cell line K562. NK cells exhibit a wide range of responses (for example, Ca2+ signaling, cytotoxicity, and cytokine production) during their interaction with K562 cells. This provides the opportunity to validate microfluidic assay performance by assessing various responses with different timescales. We paired the NK92MI and K562 cells as described in Fig. 1, and although we obtained lower cell-pairing efficiencies (∼40%; SI Appendix, Fig. S1 F and G) primarily due to NK cell clusters (particularly cell doubles), we could consistently record from >200 cell pairs at the desired spatiotemporal imaging resolution in a single experiment. We first demonstrated that we can assay early signaling dynamics and molecular events in NK cell activation upon tumor cell encounter. To this end, we recorded the real-time activation dynamics immediately after pairing by measurement of cytosolic Ca2+ mobilization, which is a rapid and sensitive readout of immunoreceptor engagement (18, 19) (SI Appendix, Fig. S6A and Movie S1). We observed considerable cell-to-cell variation in Ca2+ dynamics and magnitudes, where responding cells generally assumed peak-plateau–type profiles, elevating within minutes of engagement and returning to baseline at the end of an hour. Similarly, we demonstrated the feasibility of assessing early molecular events such as the phosphorylation states of signaling molecules, whose number and dynamics govern downstream processes (20–23). As a representative example, we focused on the phosphorylation of extracellular signal-regulated kinase (ppERK), a key player of immunoreceptor signaling pathways mediating a variety of developmental and functional responses (20, 22), and measured its level 10 min after tumor cell contact (SI Appendix, Fig. S6 B–D). Similar to calcium responses, we again observed significant heterogeneity in responses with only ∼30% of the NK population showing an increase in ppERK levels within this 10-min window in comparison with control cells. These results established the capability of the microfluidic assay to efficiently assay early signaling events in lymphocyte interactions, and further highlighted the significant heterogeneity present in the operational states of cells and their early activation responses even with uniform interaction timings.
We then validated the ability to assay later-stage functional responses (for example, cytotoxic activity, cytokine production) and cellular traits (for example, surface molecule expression) of NK cells. We could assess NK cell cytotoxic activity directly on the tumor cells (instead of surrogate markers, such as CD107a), and could monitor real-time Ca2+ response and cytotoxic activity simultaneously using a cell-permeable fluorogenic granzyme B substrate that reports on the cytolytic activity following a lytic hit within an hour (SI Appendix, Fig. S7A and Movie S2). In these experiments, we observed that only a fraction of NK cells (∼20%) displayed cytotoxicity, which was similar to the percentages obtained in analogous bulk assays (SI Appendix, Fig. S7B). To assess NK cell cytokine production, we adapted intracellular cytokine staining on-chip, and assayed IFN-gamma (IFN-γ) production of NK92MI cells as a representative example (SI Appendix, Fig. S7C). For this purpose, NK92MI cells were stimulated for 20 h with IL-12 and IL-18 cytokines to induce secretion of IFN-γ (24, 25), as unstimulated cells did not exhibit any noticeable endogenous cytokine production (SI Appendix, Fig. S7D). Under these conditions, we observed robust IFN-γ production in ∼65% of the NK cells on-chip, yielding results similar to those obtained via analogous bulk assays (SI Appendix, Fig. S7E). Next, we showed the feasibility of coassaying expression of surface molecules on both partners together with Ca2+ dynamics and functional responses. As a demonstrative example, we measured the distribution of two important surface markers, lymphocyte function-associated antigen 1 on NK cells and intercellular adhesion molecule 1 on tumor cells (SI Appendix, Fig. S8A), the interaction of which mediates conjugate formation and induction of subsequent responses, particularly cytotoxicity (25–27). We observed wide distributions of both surface molecules conforming to log-normal profiles with coefficient of variations equal to 0.39 and 0.43, respectively, but their levels did not show significant correlation to Ca2+ mobilization or cytotoxic activity (SI Appendix, Fig. S8 B–D). Throughout all these measurements, NK cell:tumor cell pairs were maintained on-chip up to 24 h, with the majority of NK cells preserving viability (>99% at 6 h, 83 ± 3% after 24 h, n = 3; SI Appendix, Fig. S9). Together, these results established the third central working principle of our assay, namely that the microfluidic system can be used to characterize cell–cell interactions by performing measurements at various stages––from earliest timescales up to a day––encompassing a wide range of mechanistic (calcium, phosphorylation, cytokine production, and so on) and technical (ratiometric imaging, fixation, staining) parameters.
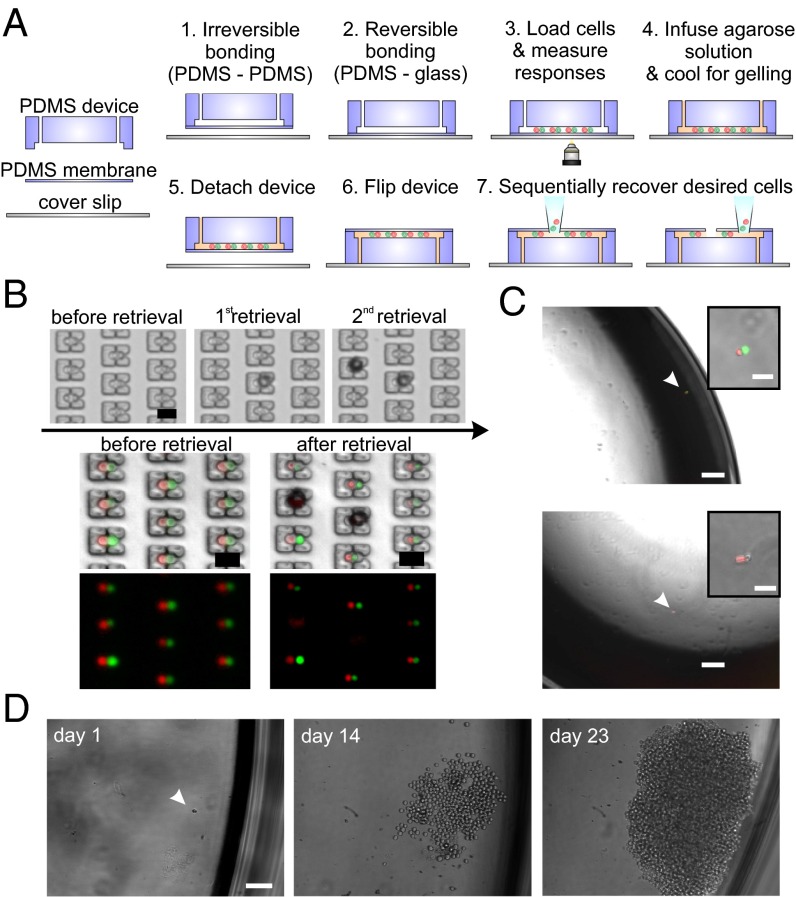
Finally, as it may be desirable to retrieve particular cell pairs for additional off-chip culture or analyses, we established a process flow for selective isolation of desired single-cell pairs using a micromanipulator. Micromanipulators are routinely used to retrieve target cells from standard culture plates or open-format microwell arrays (28). Cell recovery from closed-format microfluidic devices (such as those developed here), however, often requires more sophisticated cell manipulation strategies––generally at the expense of device footprint area––to route cells to the outlet [for example, via multiplexer architectures (29, 30), electrical manipulation (31), or optical sorting techniques (32, 33)]. Here, as our microfluidic devices share essential features with open-format devices, in particular portable device operation and registration of cells at spatially defined positions, we were able to adapt a semiautomated micromanipulator-based cell recovery approach previously established for microwell arrays (34). For this purpose, we introduced a design variation that involved bonding the polydimethyl siloxane (PDMS) devices to a thin (∼10–20-μm) PDMS sheet (membrane) instead of glass slides (Fig. 2A). This design alteration still allowed devices to be used for experiments on top of coverslips (providing rigid support for handling and ensuring flat surface for imaging) as the PDMS reversibly attaches to coverslips. For cell recovery purposes, the devices were detached from the coverslip and flipped to bring the thin membrane on top while still maintaining the cell pairs in their traps (cell-pair preservation >95%, n = 5; SI Appendix, Fig. S10). We could then use a glass microcapillary pipette with a tip broken back (creating a sharp opening to facilitate piercing the membrane) attached to a micromanipulator to remove desired cell pairs without disturbing the neighboring cells by adjusting the tip position on top and in close proximity of the desired pairs, piercing the membrane, applying suction, and transferring the cells to individual wells of standard multiwell plates (Fig. 2 B and C). Following retrieval, cells could then either be directly integrated into single-cell gene expression workflows (SI Appendix, Fig. S11A) or cultured off-chip for clonal expansion, reaching sufficient numbers to run in vitro assays, for example, for additional functional characterization and comparison of single-cell clones (SI Appendix, Fig. S11B). Collectively, these results established another important technical capability, namely the ability to recover single-cell pairs using standard micromanipulator approaches from our devices, and thus lay the groundwork to connect the comprehensive cell–cell interaction profiles acquired on-chip to additional off-chip genetic or transcriptional analyses, or even for using selected cells and their clones in in vivo studies.
Fig. 2.
Cell-pair recovery from microfluidic devices using a micromanipulator. (A) Procedure for device assembly and cell recovery. Devices are first bonded to thin PDMS membranes enclosing the channels, then to coverslips reversibly to facilitate handling during experimentation. Before cell recovery, devices can optionally be infused with ultralow-temperature agarose gel for further improving the cell entrapment and preventing any disturbance to neighboring cell pairs during micromanipulator operation. Devices are then detached from the coverslips, flipped, reattached to coverslips, and immersed in cell medium (not depicted in schematic). Desired cell pairs can then be removed sequentially using a micromanipulator. (B) Representative example of sequential cell pair retrieval. Single-cell pairs can be targeted and removed from their traps without disturbing the neighboring cells. (C) Representative images of NK92MI and K562 cell pairs recovered after live assay and transferred to standard multiwell plates. (Top) Image shows a cytolytic NK92MI cell (in red) where dead K562 cell is stained with Sytox Green nucleic acid stain. (Bottom) Image shows a noncytolytic NK92MI cell (in red) where the K562 is still alive (not stained). (Insets) Magnified images. (D) Representative image of an NK92MI cell retrieved after live assay and cultured off-chip for clonal expansion. [Scale bars: (B) 50 µm, (C) 200 µm, (Insets) 50 µm, (D) 100 µm.]
Single-Cell Correlation Analyses of Early Calcium Signaling and Functional Responses in NK Cells.
The ability to probe the cell–cell interactions with various measurements from initial contact onward affords the unique opportunity to perform correlative single-cell analyses of early signaling dynamics and functional responses in the same cells. Such experiments would be challenging, if not impossible, with any other assay format. Here, we exploited this feature to explore the direct relationship between early calcium signaling and prototypical effector responses (cytotoxicity and IFN-γ production) of NK cells. Calcium signaling in lymphocytes is essential for numerous functions (for example, motility, cytotoxic granule exocytosis, gene expression), and calcium patterns influence activation of different transcription factors and their target genes differentially (18, 19, 35). Therefore, in this study, we investigated whether and how the NK cell effector functions related to the quantifiable aspects of initial calcium signaling upon tumor cell encounter.
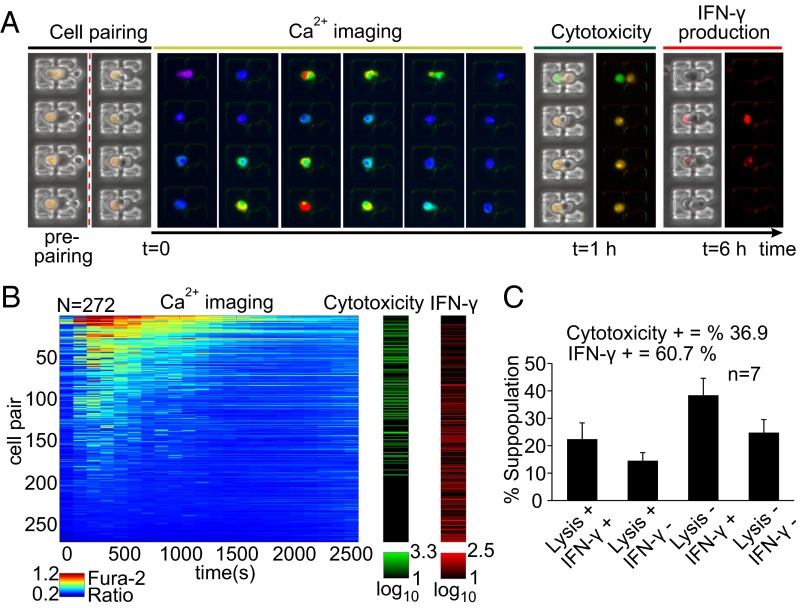
Before microfluidic assay, NK92MI cells were primed for 20 h with IL-12 and IL-18 to enhance their functions to display full effector activity, particularly secretion of cytokines (IFN-γ) and cytotoxic activity (24, 25, 36, 37). On-chip measurements included Ca2+ mobilization, recorded continuously for ∼45–60 min from initial engagement, measurement of cytotoxicity at the end of the initial hour, and assessment of IFN-γ production 6 h after pairing as previously described, yielding a multiparametric longitudinal dataset over >200 cell pairs per single experiment (Fig. 3 A and B). The NK cell population displayed various combinations of effector functions (Fig. 3C) with an average ∼35% of cells exhibiting cytotoxicity and ∼60% producing IFN-γ in total (n = 7), similar to the trends observed in analogous bulk assays (SI Appendix, Fig. S12). Although cells showed variation in size, the measured responses were not correlated to this variation, suggestive of other cell-intrinsic sources for the observed differences in responses (SI Appendix, Fig. S13).
Fig. 3.
Longitudinal multiparametric characterization of cell–cell interactions from initial contact. (A) Measurements include intracellular Ca2+ mobilization for ∼45–60 min from t = 0 (initiation of interactions, red dotted line), assessment of cytotoxicity at t = 1 h, and detection of IFN-γ production at t = 6 h. (B) Multiparametric dataset collected from cytokine primed (IL-12, IL-18) NK92MI cells paired with K562 cells (N = 272). Calcium dynamics are displayed in heatmap (to the left, in “jet” colormap) where each row corresponds to a single NK92MI cell. Cytotoxicity (granzyme substrate fluorescence activity) and IFN-γ production are displayed as green and red colormaps, respectively. Range for each colormap displayed below corresponding colormap. Results are representative of seven independent experiments. (C) Subpopulation percentages of different functional groups. Results are from seven independent experiments.
To reveal any relationships between Ca2+ signaling and effector functions, we first examined the association of Ca2+ signaling with cytotoxicity. All cytolytic NK cells exhibited Ca2+ fluxes (SI Appendix, Fig. S14A) in agreement with the role of Ca2+ signaling in delivery of lytic granules (38). We also observed Ca2+ fluxes in some noncytolytic NK cells (SI Appendix, Fig. S14A), which was likely due to the regulatory action of calcium in many signal transduction pathways in addition to cytotoxicity (19). The Ca2+ waveforms and integrated Ca2+ levels in cytolytic and noncytolytic Ca2+ fluxing cells were comparable; thus, these experiments suggested that the type of Ca2+ responses was not predictive of cytotoxic behavior (SI Appendix, Fig. S14 B and C).
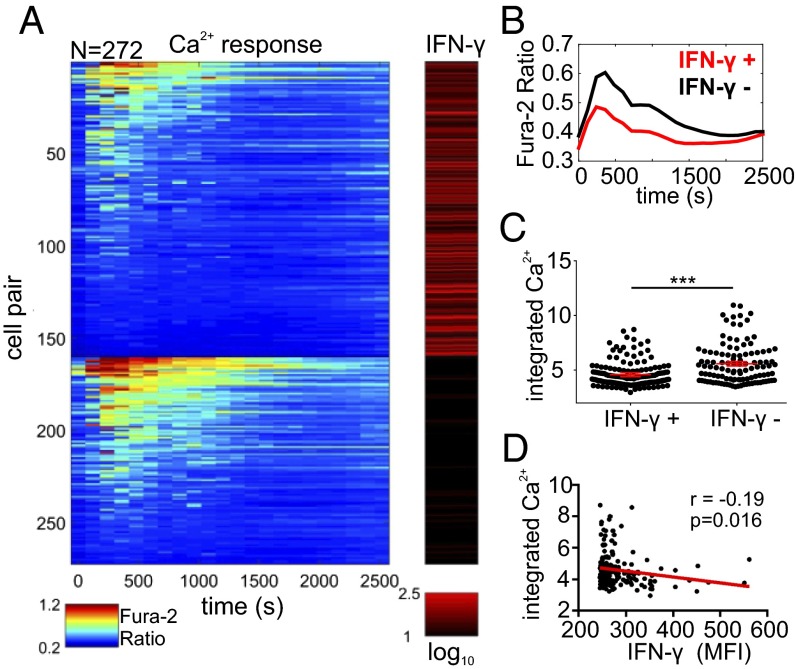
Next, we examined the relationship between the Ca2+ signaling and IFN-γ production, and divided the cells into two subpopulations based on IFN-γ activity (IFN-γ producers and nonproducers). Despite the vast heterogeneity in Ca2+ responses in both groups, we observed that the IFN-γ–producing cells displayed lower Ca2+ responses on average than nonproducer cells as determined by the average waveforms and integrated Ca2+ levels (Fig. 4 A–C). Additionally, examination of the Ca2+ responses within the IFN-γ–producing cells also showed a reproducible (n = 7) correlation between higher IFN-γ levels and lower integrated Ca2+ levels that could be approximated by a weak linear relationship (Fig. 4D; r = −0.19, P < 0.02). To further elucidate this Ca2+ dependence more explicitly, we performed additional unsupervised clustering of Ca2+ responses, and our analysis organized NK cells into two clusters (SI Appendix, Fig. S15). Whereas the first cluster comprised entirely cells characterized with relatively higher calcium signals (referred to as “strong Ca2+ responders” throughout the text), the second cluster included both weak calcium responders and nonresponders (that is, no significant Ca2+ activity; SI Appendix, Fig. S15A). To better delineate differences between these weakly responding versus nonresponding cells, we further divided this second cluster into two subgroups that we classified as “weak Ca2+ responders” and “Ca2+ nonresponders” (SI Appendix, Fig. S15A). The strong calcium responders differed significantly in IFN-γ production compared with weak Ca2+ responders and Ca2+ nonresponders, where they showed both a lower percentage of IFN-γ producing cells (SI Appendix, Fig. S15B) and lower relative IFN-γ levels (SI Appendix, Fig. S15 C and D). Indeed, strong Ca2+ responders constituted only 22 ± 9.2% of the total IFN-γ producers (SI Appendix, Fig. S15E). On the other hand, weak Ca2+ responders and Ca2+ nonresponders yielded similar percentages of IFN-γ–producing cells and similar relative IFN-γ levels, suggestive of a certain Ca2+ signaling threshold before any differences in IFN-γ production is observed (SI Appendix, Fig. S15 B–D). Together, these data suggested an inverse link between strength of early Ca2+ signaling and IFN-γ production in cytokine primed NK cells upon target cell encounter, and support a model in which NK cells with lower Ca2+ signals are more likely to produce IFN-γ.
Fig. 4.
Relationship between Ca2+ response and IFN-γ production of cytokine primed (IL-12, IL-18) NK92MI cells interacting with K562 cells. (A) Clustering of Ca2+ responses of NK92MI cells based on IFN-γ production. Within each group, cells are ordered based on the integrated Ca2+ levels. Calcium dynamics and IFN-γ production are displayed in jet and red colormaps, respectively. Range for each colormap displayed below corresponding colormap (N = 272). (B) Comparison of average Ca2+ traces of the IFN-γ+ and IFN-γ− groups. (C) Comparison of single-cell integrated Ca2+ levels of the IFN-γ+ and IFN-γ− groups. Each dot represents single-cell data. Solid red lines represent mean ± SEM of corresponding data points. ***P < 0.001, Mann–Whitney test. (D) Linear correlation analysis between integrated Ca2+ and relative IFN-γ levels (MFI). Red line indicates the linear fit with coefficient r = −0.19, linear trend is significant, P < 0.05. (A–D) Results are representative of seven independent experiments.
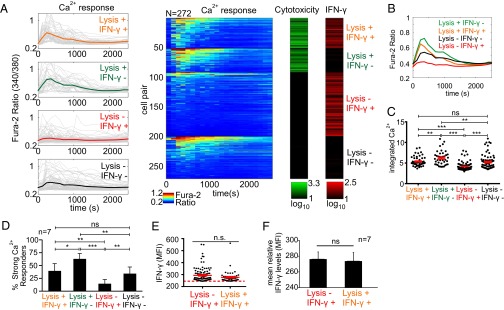
Given this inverse relationship between IFN-γ production and strength of Ca2+ signaling (Fig. 4) together with the requirement of Ca2+ signaling for cytotoxicity (SI Appendix, Fig. S14), we next explored how cells within different effector response groups compared with respect to their initial calcium signaling. To this end, we clustered the NK cell responses into four groups based on exhibited effector functions (“Lysis+ IFN-γ+,” “Lysis+ IFN-γ−,” “Lysis− IFN-γ+,” “Lysis− IFN-γ−”) and contrasted Ca2+ signaling among these groups (Fig. 5A). All groups exhibited appreciable heterogeneity in their Ca2+ signaling, and we observed the presence of similar Ca2+ flux profiles within each group (that is, each group included cells with very high and very low Ca2+ signaling profiles). However, despite this vast heterogeneity, quantification of integrated Ca2+ levels and subpopulation averages revealed discernible patterns. Importantly, whereas Lysis+ IFN-γ− subgroup displayed the highest average Ca2+ waveform and integrated Ca2+ levels, Lysis− IFN-γ+ subgroup displayed the lowest, suggesting a possible dichotomy between these two functions with regard to strength of initial Ca2+ signaling (Fig. 5 B and C and SI Appendix, Figs. S16 and S17). The average Ca2+ waveforms and integrated Ca2+ levels for Lysis+ IFN-γ+ and Lysis− IFN-γ− subgroups remained in between these two limits with moderate levels, and were in general indistinguishable from one another (Fig. 5 B and C and SI Appendix, Figs. S16 and S17). However, the similar average waveforms between these two subgroups were mostly due to the prevalence of nonresponder cells (that is, no significant Ca2+ activity) within the Lysis− IFN-γ− subgroup (∼30–45% nonresponders, n = 7), and responder cells in this subgroup generally displayed higher Ca2+ levels and an average waveform than the Lysis+ IFN-γ+ subgroup (SI Appendix, Fig. S18). We could also quantify these differences in Ca2+ responses among different effector groups by examining the prevalence of strong Ca2+ responders within each group. Lysis+ IFN-γ− and Lysis− IFN-γ+ groups showed the highest and lowest strong responder percentages, respectively, and Lysis+ IFN-γ+ and Lysis− IFN-γ− subgroups remained in between (Fig. 5D and SI Appendix, Fig. S17). Overall, these results suggested that NK cells with moderate Ca2+ responses are more likely to execute both effector functions. Despite the differences in their initial calcium signaling, we found that Lysis+ IFN-γ+ group produced similar levels of IFN-γ at the subpopulation level in comparison with Lysis− IFN-γ+ group (Fig. 5 E and F). The Lysis− IFN-γ+ group produced slightly more IFN-γ on average, although the difference was not significant (Fig. 5 E and F). This slight difference was likely the result of higher percentage of strong Ca2+ responders (which are associated with none or low IFN-γ production) in Lysis+ IFN-γ+ group. However, as both groups comprised mainly weak Ca2+ responders and/or Ca2+ nonresponders, their IFN-γ production was comparable. Taken together, these findings suggest that the strength of early Ca2+ signaling upon target cell engagement is a factor that may selectively coordinate subsequent cytotoxicity and IFN-γ production in NK cells, and warrants further investigation to determine (i) whether this calcium correlation causes (and not only correlates with) this functional dichotomy, and if so, (ii) which calcium-dependent pathways can be exploited for targeted manipulation. Collectively, these studies represent for the first time, to our knowledge, that calcium signals are directly related to functional outputs at the single-cell level, and show the ability of our microfluidic assay to identify the relationships between various responses that occur at different stages by single-cell measurements and correlation analyses.
Fig. 5.
Comparison of Ca2+ responses of cytokine primed (IL-12, IL-18) NK92MI cells within different effector response groups. (A) Clustering of response profiles into four functional groups. Lysis+ indicates cytolytic activity, IFN-γ+ indicates cytokine production. Time plots displaying Ca2+ profiles are shown to the left. Each gray trace represents the response of a single NK92MI cell, and color-coded bold traces show group averages. Corresponding Ca2+ heat maps are shown to the right (jet colormap), each row corresponding to an individual cell. Cytotoxicity and IFN-γ production displayed as green and red colormaps, respectively. Range for each colormap displayed below corresponding colormap. (B) Comparison of average Ca2+ traces of the functional groups. (C) Comparison of integrated Ca2+ levels among the functional groups. Each dot represents single-cell data. Solid red lines represent mean ± SEM of corresponding data points. **P < 0.01, ***P < 0.001; n.s., not significant, one-way ANOVA with Tukey’s posttest. (D) Comparison of the strong Ca2+ responder percentages within each functional group. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant, repeated-measures one-way ANOVA with Tukey’s posttest. (E) Comparison of single-cell relative IFN-γ levels (MFI) between Lysis− IFN-γ+ and Lysis+ IFN-γ+ groups. Red dashed line represents the threshold for positive cytokine production events. Solid red lines represent mean ± SEM of corresponding data points. Only IFN-γ positive events are shown. Each dot represents single-cell data. n.s., not significant, Mann–Whitney test. (F) Comparison of mean relative IFN-γ levels between Lysis− IFN-γ+ and Lysis+ IFN-γ+ groups. n.s., not significant, Student’s t test. (A–C, E) Results are representative of seven independent experiments. (D and F) Results are from seven independent experiments.
Discussion
Here, we presented a microfluidics-based cell–cell interaction assay for longitudinal interrogation of contact-dependent immune cell interactions at the single-cell level. Several important advantages distinguish our microfluidic assay from other approaches. The microfluidic platform achieves highly efficient generation of hundreds of cell conjugates using a deterministic cell-pairing approach, creating homogeneously defined one-to-one interactions with enduring contacts, simultaneous timings, and identical contact durations. Such controlled pairing grants access to earliest signaling events in lymphocyte activation cascade from the moment of contact. It further prevents any variation that would have been otherwise introduced into functional responses measured at later stages due to the differences in interaction parameters. The specially tailored trap structure ensures spatial isolation of cell pairs (while in continuous contact) and portable use of the devices, permitting longitudinal tracking and assaying of interaction outcomes with relative ease using standard laboratory equipment and procedures. These features offer the ability to probe into complete activation window––from initiation of interactions up to 1 d––for direct correlation analyses over hundreds of single immune cells in a single experiment. The ability to retrieve desired cell pairs further provides an opportunity to integrate on-chip measurement profiles with off-chip follow-up analyses. This feature could be particularly useful for paired genetic and/or transcriptional analyses of interacting cells with desired outcomes, for example to link particular lymphocyte activation profiles to genetic/transcriptional state of antigen presenting cells, or to coidentify tumor-reactive immune cell receptor specificities (e.g., T-cell receptors) together with their corresponding target antigens (39). We also envision that with additional design variations such as larger traps or compartmentalization of cell pairs, the presented approach should also enable studying cellular interactions between more than two cells (for example, T cells, dendritic cells, and regulatory T cells) or for longer timescales (>24 h), for example to track the lineages of immune cell pairs upon cell division following their interactions.
In this study, we demonstrated the utility of our assay by performing the first direct single-cell correlation study between early activation dynamics and later-stage functional responses over same individual NK cells interacting with tumor cells. While these relationships have been studied extensively before, most of our knowledge relating immune cell activation to function hitherto has been based on population-wide correlations and inferences from snapshot analyses (e.g., FACS or immunoblots), and has not been previously analyzed and resolved at the single-cell level. On this front, our experiments yielded important observations into the heterogeneities and correlations of these responses at the single-cell level. One critical observation was that there exists large heterogeneity in NK cell activation and function. Other studies have already made that point, but one could have argued that cell-to-cell variability was the result of nonuniform timings, insufficient durations of interactions, or different number of partners. Here, we demonstrated that this phenotypic variability is intrinsic even with uniform interactions and timings. This observation cautions against the correlative analyses between early and late responses that are generally performed using ensemble comparisons and neglecting the contributions of nonuniform interaction parameters. Another important observation was the weak inverse relationship between the strength of initial Ca2+ signaling and IFN-γ production in cytokine primed NK cells. In connection with this observation, we further noticed that NK cells displaying both cytotoxicity and IFN-γ production generally exhibited moderate Ca2+ responses. Elucidating the mechanisms that segregate the signals from cytokine production from NK-cell killing is an active area of research and has implications for controlling these responses separately in clinical settings. Previous studies have successfully identified some of the different receptor–ligand engagements (25, 40) and signaling molecules [for example, Vav1 (41), Carma-1 (42), and Fyn-ADAP (43)] as important regulators of this functional dichotomy. Our findings in this study further support a model in which calcium signals can play a possible quantitative role in this functional separation (for example, by setting different thresholds for cytotoxic and cytokine programs). We further hypothesize that the relationships between calcium signals and effector functions observed in NK cells could be extrapolatable to T cells and may help explain the dichotomy observed between cytotoxic activity and cytokine production reported for CD8 T cells in several studies (9, 44–46). Similarly, exploring many other early regulators of immune cell functions [for example, proximal signaling molecules (47), kinase activities (48), or transcriptional factors (49) using fusion proteins or reporter mouse (50)] should also allow better resolving the contributions of the dynamics and magnitudes of a wide variety of molecules to functional diversification at the single-cell level. Establishment of this new tool now opens the door to pursue these and many other previously intractable hypotheses on immune cell communication, activation, and function at the single-cell level.
Materials and Methods
Microfluidic Device Fabrication.
Master wafers for the microfluidic devices were fabricated using a two-layer SU8 process. The first layer was created to pattern support pillars (2–3 µm for small devices using SU8-2002, 3–5 µm for medium-sized devices using SU8-2005, and 5–6 µm for large devices using SU8-2005). The wafers were then UV-exposed through the first chrome mask with support pillar patterns, developed and baked following manufacturer’s protocols. The capture cup patterns were then formed on top of the support pillars by spinning a second layer of SU8 with subsequent UV exposure through capture cup mask (8–11 µm for small devices using SU8-2007, 14–17 µm for medium-sized devices using SU8-2010, and 16–20 µm for large devices using SU8-2010). After developing and baking, the wafers were hard-baked at 150 °C for 30 min, and were treated with Trichloromethylsilane (Sigma-Aldrich) for 2 h in a saturated vacuum chamber to prevent adhesion of PDMS (Sylgard 184, Dow Corning) during the molding process.
Microfluidic devices were made using soft lithography by pouring thoroughly mixed and degassed PDMS over the master wafers or plastic masters (51) to a thickness of 3–4 mm, followed by curing at 80 °C overnight (>12 h) to ensure complete curing of the PDMS devices to prevent any effect of unpolymerized PDMS on cells during on-chip culture. Following curing, PDMS devices were peeled off, individual devices were cut to proper sizes, and holes for tubing connections were punctured. The devices were then plasma-bonded to cover glasses of 0.17-mm thickness that were compatible with high numerical aperture fluorescence imaging, and stored in 80 °C oven until experiments (usually for an additional overnight). For researchers who would like to use the microfluidic assay in their applications, the predesigned devices and molds are available upon request.
Microfluidic Setup and Cell-Pairing Procedure.
Devices were initially infused with 70–80% ethanol for bubble-free filling. Ethanol was then washed out using PBS, and PDMS surfaces were then blocked with 7.5% BSA at 37 °C for >30 min. The fluidic setup included two syringe pumps providing flow at different flow rates, one at 0.1–3 μL min−1 for infusion of solutions and passive trapping of cells, and other at high flow rates (50–500 μL min−1) to squeeze cells into traps by increased fluidic pressure during cell loading. Tygon Microbore tubing (Cole Parmer) connected to a four-way valve (UpChurch Scientific) was plugged into the outlet of the device with remaining connections attached to two 1-mL glass syringes (Hamilton) on the syringe pumps and a waste reservoir. Only one port (outlet) of the device was connected to the tubing, and the other port (inlet) was left unconnected to use the devices in open-reservoir format. This configuration enabled cells and reagents to be directly pipetted onto the inlet reservoir and withdrawn into the device, thereby reducing cell and reagent loss due to dead spaces (syringe and tubing volumes) and also eliminating cell settling and clumping in stationary syringes. After assembly of the fluidic connections, devices were rinsed with cell media to remove the BSA solution and prime the channels before introducing the cells.
Cell loading and pairing was accomplished using a multistep loading procedure. Initially, the first cell population was captured in the single-cell traps by pipetting ∼5 µL of cell suspension on the inlet reservoir and withdrawing into the channel. After the cell traps were saturated, the inlet reservoir was washed out by cell media and any additional cells passed by the array without disturbing already trapped cells. Then, the flow was briefly increased to transfer cells from single-cell traps into two-cell traps through the narrow constriction by flow-induced deformation. This step was fast (∼1–2 s) and highly parallel across the entire array, and flow was immediately reduced back after the transfer step. Next, the second cell population was then loaded similarly, first capturing cells in the single-cell traps followed by transfer into the two-cell traps, immediately in front of the already trapped cells. The squeezing-based cell loading ensured that cell pairing occurred at once across the whole array, synchronizing the initiation of interactions (that is, cells make contact as soon as the second cell population is squeezed into two-cell traps). The two-cell trap design further ensured the prealignment of the pairs within cups to promote productive contacts, and as it was sized to hold the two cells in close proximity, this further helped cells to preserve their contact over the entire assay duration based on the natural and dynamic course of the interaction (i.e., cells can stop their communication depending on information exchange; for example, NK cells can separate from target cells following lysis). In addition, it is also possible to induce a very slight gravitational flow by adjusting the liquid volumes on the inlet and outlet reservoirs to help maintain contacts. (Although in our experiments we observed lasting contacts without having the need to induce gravitational flow, this feature could be useful with other cell types as an additional option.) Proper choice of trap dimensions (height of support pillar layer) and backside support pillars helped maintain the cells within the traps during cell loading procedures, particularly preventing the already trapped first cell population to escape from traps when loading the second cell population (even during increased flow rates) or maintaining cell pairs within their traps during tubing removal. (Although removal of tubing creates a brief flow from inlet toward outlet due to negative pressure generated, the backside support pillar blocks any fluidic path in that direction and maintains cell pairs within cups, preventing any cell loss during this stage.) However, we still observed some cell loss while loading K562 cells, during the transfer step from single-cell traps into two-cell traps, where K562 cells could squeeze out of the traps through the side openings (∼5-µm gap) after entering into the two-cell traps if the high flow rates are not reduced back immediately. This was not observed with NK92MI cells, and was mostly due to the higher deformability of K562 cells. Instead of addressing this issue using a new design with narrower side openings (<5 µm), we minimized this loss (∼5–10%) by empirically adjusting the time for brief increase in fluid flow to prevent cell escape.
Fluidic Modeling.
Flow patterns were simulated with the Comsol Multiphysics software using the laminar flow model. The dimensions of the capture cups were identical to the actual devices. Only a single capture cup structure is simulated taking advantage of the symmetry conditions, and minor losses due to curvatures were ignored to reduce the computation time and memory use. The initial boundary was assigned a flow velocity of 100 µm/s (corresponding to the average flow rate in the channel before the trap zone), and the exit boundary was set as the outlet. The side boundaries were set to symmetry boundary condition while the trap surfaces and remaining walls were assigned no-slip conditions. Flow resistance ratios were determined using the inverse ratios of the volumetric flow rates calculated by integrating the flow rates over parametrized surfaces defined at corresponding entrance locations of each illustrated fluidic path (SI Appendix, Fig. S1E).
Cell Culture, Stimulations, and Preparations.
The NK92MI and K562 cell lines were obtained from the American Type Culture Collection. NK92MI cells were maintained in alpha minimum essential medium without ribonucleosides and deoxyribonuclosides, supplemented with 2 mM l-glutamine, 0.2 mM inositol, 0.02 mM folic acid, 0.1 mM 2-mercaptoethanol, 100 Units/mL penicilin G sodium, 100 µg/mL streptomycin sulfate, 12.5% heat-inactivated FBS, 12.5% horse serum. K562, Jurkat, and B16 cells were maintained in RPMI medium 1640 supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 0.1 mM 2-mercaptoethanol, 100 Units/mL penicilin G sodium, and 100 µg/mL streptomycin sulfate. Coculture experiments were carried out in NK cell media. Where indicated, NK92MI cells were primed with IL-12 (10 ng/mL) and IL-18 (100 ng/mL) (all from R&D Systems) for 20 h before experiments to promote IFN-γ production and enhance cytotoxicity.
Before cell loading microfluidic assays, cells were suspended either in PBS or cell medium containing 0.1–1% BSA and 50 U/mL DNase solution to reduce aggregation. Cells were pipetted regularly to minimize cell adhesion and maintain a homogeneous single-cell suspension.
On-Chip Cell Culture.
For culturing cells in the microfluidic devices, tubing connected to outlet of the device was removed gently by pulling with tweezers while still maintaining cell pairing within the traps. Additional 20–30 µL cell media was pipetted onto the inlet reservoir, and devices were placed inside a humidifying chamber prepared using a regular 10-cm cell culture Petri dish along with wet tissues. Devices could then be transferred to different settings, and were maintained inside a humidified incubator at 37 °C and 5% CO2 during culture up to 24 h while refreshing medium every 3–5 h. Sufficient gas exchange was facilitated by the permeability of PDMS where final device heights were set at 3–4 mm to additionally reduce the diffusion path. These conditions helped ensure sufficient on-chip culture conditions for cell pairs where >80–95% of cells remained viable throughout the 24-h period.
On-Chip Cytotoxicity Assays.
Real-time microfluidic cytotoxicity assays were performed by adapting the PanToxiLux assay (OncoImmunin) following manufacturer’s protocols. This assay uses a cell-permeable enzyme substrate that fluoresces in the presence of Granzyme B and/or upstream caspases, and hence informs if a target cell has received a lytic hit and is amenable for real-time detection of cytolytic activity. Upon pairing, cells were incubated in the presence of the substrate for 1 h, and cytotoxicity was determined by target-cell fluorescence via imaging.
Analogous bulk cytotoxicity experiments were conducted as a control for microfluidic assays. K562 cells were stained with 5 µM Cell Tracker Orange CMTMR (Invitrogen) for 1 h at 37 °C in humidified 5% CO2 incubator before experiments. The labeled cells were washed twice in PBS, and resuspended in culture medium. Bulk cytotoxicity was determined by coculturing 104 K562 cells with NK92MI cells in target effector ratios of 1:1, 1:5, and 1:10 in 200 µL cell medium with 0.5 µM SYTOX green (nucleic acid stain to identify dead cells). Each condition was performed in triplicates. Cocultures were incubated for 4 h at 37 °C in humidified 5% CO2 incubator, and percentage of target cell lysis was determined by counting target cells stained with SYTOX green using fluorescence microscopy, and quantified over >500 cells.
On-Chip Phosphorylation Measurements.
For ppERK measurements, cell pairs were generated, tubing connections were removed, and devices were then placed inside a humidifying chamber and transferred to a humidified incubator at 37 °C and 5% CO2 for 10 min. Following the 10-min incubation period, cells were fixed by 4% paraformaldehyde for 15 min at room temperature, and permeabilized with 0.5% Triton X-100 for additional 10 min. Cells were then washed with PBS, blocked with blocking solution (PBS with 1% BSA, 10% donkey serum) for 30 min, and stained with anti-ppERK antibodies (D12.14.4E, Cell Signaling) at 4 °C overnight by transferring the devices into a refrigerator. After staining, cells were washed with PBS with 1% BSA, and analyzed with fluorescence microscopy.
On-Chip Intracellular IFN-γ Staining.
Cytokine primed (IL-12, IL-18) NK92MI cells were washed twice in PBS, and resuspended in cytokine-free media. Cells were then loaded on-chip either alone or paired with K562 cells, and then cultured on-chip for 4–6 h in the presence of 1 μL/mL Golgi plug (Invitrogen) by placing devices inside a humidifying chamber and transferring them into a humidified incubator at 37 °C and 5% CO2. Following the incubation period, cells were fixed and permeabilized as described before, followed by blocking with PBS containing 10% normal donkey serum, 0.5% Triton X-100, and 1% BSA for 30 min. After blocking, cells were incubated with antihuman IFN-γ antibodies (NL285R, R&D Systems) in blocking buffer at 4 °C overnight in dark, following manufacturer’s protocols. Cells were then washed with PBS with 1% BSA, and analyzed by fluorescence microscopy.
Analogous bulk intracellular cytokine staining for IFN-γ was also performed as a control for microfluidic assays. Cells were prepared similarly, incubated with 1 μL/mL Golgi plug (Invitrogen) in standard multiwell plates for 6 h at 37 °C in humidified 5% CO2 incubator in triplicates. After the incubation period, cells were processed and analyzed as described before.
On-Chip Antibody Staining.
Cell pairs were generated, and incubated on-chip for 30 min at 37 °C and 5% CO2 in a humidified incubator, followed by fixation and antibody staining following manufacturer’s protocols at suggested dilutions (in PBS with 1% BSA) for recommended durations (∼30 min). Cells were then washed with PBS with 1% BSA before imaging, and analyzed with fluorescence microscopy. The following antibodies were used in our assays: anti–human-CD11a (eBioscience) and anti–human-CD54 (eBioscience).
On-Chip Calcium Imaging.
Cells were stained with 1–3 µM Fura-2/AM (Life Technologies) ratiometric calcium indicator dye in serum-free media at 37 °C for 1 h. Cells were then washed with PBS with 1% BSA, and resuspended in phenol red-free cell media for on-chip experiments. Imaging experiments were performed using 10× magnification on an automated inverted microscope (Nikon Eclipse Ti, Nikon) fitted with a cooled CCD camera (CoolSNAP HQ2, Photometrics). NK cells were loaded into the traps first, and exposure times were determined while cells were at rest, and chosen close to saturation (but not saturated) for 380 nm, and well below the saturation for 340-nm channel. Imaging was started before cell pairing to record a baseline measurement, and images were acquired for 1 h, taken every 90 or 120 s.
Cell Recovery Using a Micromanipulator.
For cell recovery experiments, PDMS devices were plasma-bonded to thin (10–20-µm) PDMS membranes (made on 4-inch silicon wafers by spin coating at 1,000–2,000 rpm for 10 min) to form enclosed channels. Devices were then placed on coverslips (forming reversible bonds) for cell pairing and experiments. Following experiments and recording the address positions of desired cells, devices were detached from the coverslips using tweezers, flipped and reattached to coverslips to bring the thin membrane on top for easy micromanipulator access. Optionally, before this flipping step, the devices could be infused with ultralow-temperature agarose solution [0.5–1% (wt/vol) in PBS; A2576, Sigma-Aldrich; gelling at 4 °C for 15 min] to further stabilize cell entrapment and reduce disturbance to neighboring cells during micromanipulator operation. The devices were then placed in wells of a six-well plate, their positions in wells were fixed using agarose gel [2% (wt/vol)], and cell media was added to wells until devices were immersed completely. The six-well plates were then transferred to the micromanipulator station.
Cells were retrieved from their traps using an semiautomated micromanipulator (CellCelector, AVISO GmbH) by piercing the membrane and applying suction following an adaptation of the operation procedures described before (34). Glass capillaries were pulled and broken back manually to induce an angled opening at the tip to facilitate easy membrane piercing. For each device, final z-axis position to move to pierce the membrane was calibrated and optimized by preliminary trials. To retrieve cells, a glass capillary tip was positioned on top and in close proximity of the trap entrance where the desired cell pairs are located, and cells were retrieved by initiating a program routine with preinputted micromanipulator parameters (micromanipulator speed, aspirated volume, and time delay after aspiration). Images were recorded before and after the micromanipulator operation to confirm target-cell removal from the devices. After removal, cells were transferred to wells of a 96-well plate and the efficiency of cell recovery was determined by manually counting the number of cells in 96-well plates. Cell recovery efficiency was critically dependent on the preparation of the glass capillary tips where we observed higher yields with capillaries with sharp edges and ∼50–70-µm diameter. Whereas successful recovery of both partners was usually achieved with up to ∼35% efficiency (∼50% for successful recovery of the NK cell partner, n > 100), these efficiencies could further be improved by a more systematic characterization and optimization of parameters affecting the removal and transfer of cells (for example, capillary diameter, uniform beveling angle of capillary tips, aspirated volume, time delay after aspiration, and thickness of PDMS membrane).
For clonal expansion experiments, single NK cells were cultured first in 1:1 mixture of fresh media and conditioned media supplemented with IL-2 (100 U/mL), IL-12 (10 ng/mL), and IL-18 (100 ng/mL) for more effective initial proliferation for several cell division cycles. Afterward, NK cell clones were transferred into cell culture inserts (0.4-µm pore size, Millicell, Millipore) and cocultured with other NK cells (as feeder cells) in a two-compartment culture system. This configuration allowed exchange of cell-secreted factors from feeder cells while preventing passage and mixing of cells. Once NK cell clones reached adequate numbers to survive without feeder cells, they were transferred to culture plates and kept in culture until they reach sufficient numbers for bulk assays (∼1 million cells, ∼45–60-d expansion period).
For single-cell qPCR experiments, cells were first transferred to 96-well plates filled with PBS buffer. After verifying cell presence in individual wells, cells are transferred to PCR tubes, centrifuged, and resuspended in 10 µL lysis buffer (Qiagen). Samples were then immediately frozen on dry ice and stored at −80 °C before reverse transcription and cDNA synthesis. Whole transcriptome amplification and cDNA synthesis were performed using the protocol described before (52). Predesigned primers were obtained from Sigma (Kicqstart SYBR green primers) and qPCR was performed using Bio-Rad iQ SYBR Green Supermix.
Longitudinal Multiparametric Measurements over Cell Pairs.
Fura-2 stained NK92MI cells were paired with K562 cells, and cells were imaged simultaneously for calcium signaling and cytotoxicity for 1 h as described above. At the end of the 1-h period, cell medium with 1 μL/mL Golgi Plug was pipetted onto the reservoir and withdrawn into the device. The tubing at the outlet was disconnected, and the device was placed inside a humidifying chamber and transferred into a humidified incubator at 37 °C and 5% CO2 for 6-h culture. Following the incubation period, cells are fixed, permeabilized, and blocked on-chip as previously described. After the blocking period, cells were incubated in staining solution with antihuman IFN-γ at 4 °C overnight in dark. Cells were then washed with PBS with 1% BSA, and analyzed using fluorescence microscopy for IFN-γ production.
Analogous experiments exploring the effector responses (cytotoxicity and IFN-γ production) of NK92MI cells were also performed in bulk assays. NK92MI cells were stained with CellTracker Blue and cultured overnight with IL-12 (10 ng/mL) and IL-18 (100 ng/mL). NK92MI cells were washed, counted, and cocultured in NK media with K562 cells at the indicated ratios (250,000 total cells per well) in a 96-well round-bottom plate. Cells were cultured for 1 h, then GolgiStop (BD Biosciences) was added to the media for an additional 5 h. Cells were then washed and stained with anti-CD107a FITC (BD Pharmingen). Cells were washed, fixed, permeabilized, and stained with anti-IFNg PE (R&D Systems) using Fixation Buffer/Permeabilization Wash Buffer according to the manufacturer’s protocol (Biolegend). Samples were analyzed by flow cytometry using a BD Fortessa.
Data Analysis and Statistics.
Images were analyzed in ImageJ (rsb.info.nih.gov/ij/), and regions of interest (ROIs) were defined manually and chosen to cover the cell bodies. Mean fluorescent intensities (MFIs) for selected ROIs were exported as MS Excel file for additional analysis. A custom-written script was used to analyze the signals in MATLAB (MathWorks) after background substraction. Time-lapse Ca2+ signals were determined by the ratio of fluorescence emission intensities at wavelengths 340 and 380 nm. Different measurements (calcium, cytotoxicity, and IFN-γ production) were matched for each cell to construct their measurement profiles for subsequent quantitation of parameters and statistical analyses.
Unsupervised clustering was performed using k-means algorithm using the sqEuclidean distance as the distance measure. Number of clusters and quality of clustering were determined by optimizing over the average silhouette scores. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software) using the specific tests indicated in figure legends. All data were reported as mean ± SD unless otherwise stated in corresponding figure legends.
Supplementary Material
Acknowledgments
We thank Kurt Broderick and Dennis Ward for their help with the fabrication process, Brittany A. Goods and Sarvesh Varma for their help with qPCR experiments, and Dr. J. Christopher Love for providing the automated micromanipulator setup and sharing established protocols. Master wafers for microfluidic devices were fabricated in the Microsystems Technology Laboratory at Massachusetts Institute of Technology (MIT). This research was supported by the Singapore–MIT Alliance, the Frank Quick Faculty Research Innovation Fellowship, and American Association for Cancer Research–Pancreatic Cancer Action Network.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515364113/-/DCSupplemental.
References
- 1.Davis DM. Mechanisms and functions for the duration of intercellular contacts made by lymphocytes. Nat Rev Immunol. 2009;9(8):543–555. doi: 10.1038/nri2602. [DOI] [PubMed] [Google Scholar]
- 2.Corse E, Gottschalk RA, Allison JP. Strength of TCR-peptide/MHC interactions and in vivo T cell responses. J Immunol. 2011;186(9):5039–5045. doi: 10.4049/jimmunol.1003650. [DOI] [PubMed] [Google Scholar]
- 3.Iezzi G, Karjalainen K, Lanzavecchia A. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity. 1998;8(1):89–95. doi: 10.1016/s1074-7613(00)80461-6. [DOI] [PubMed] [Google Scholar]
- 4.Underhill DM, Bassetti M, Rudensky A, Aderem A. Dynamic interactions of macrophages with T cells during antigen presentation. J Exp Med. 1999;190(12):1909–1914. doi: 10.1084/jem.190.12.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothoeft T, Gonschorek A, Bartz H, Anhenn O, Schauer U. Antigen dose, type of antigen-presenting cell and time of differentiation contribute to the T helper 1/T helper 2 polarization of naive T cells. Immunology. 2003;110(4):430–439. doi: 10.1111/j.1365-2567.2003.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, et al. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity. 2013;39(5):846–857. doi: 10.1016/j.immuni.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertek F, Xu C. Digital response in T cells: To be or not to be. Cell Res. 2014;24(3):265–266. doi: 10.1038/cr.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zikherman J, Au-Yeung B. The role of T cell receptor signaling thresholds in guiding T cell fate decisions. Curr Opin Immunol. 2015;33(0):43–48. doi: 10.1016/j.coi.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varadarajan N, et al. A high-throughput single-cell analysis of human CD8⁺ T cell functions reveals discordance for cytokine secretion and cytolysis. J Clin Invest. 2011;121(11):4322–4331. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamanaka YJ, et al. Single-cell analysis of the dynamics and functional outcomes of interactions between human natural killer cells and target cells. Integr Biol (Camb) 2012;4(10):1175–1184. doi: 10.1039/c2ib20167d. [DOI] [PubMed] [Google Scholar]
- 11.Christakou AE, et al. Live cell imaging in a micro-array of acoustic traps facilitates quantification of natural killer cell heterogeneity. Integr Biol (Camb) 2013;5(4):712–719. doi: 10.1039/c3ib20253d. [DOI] [PubMed] [Google Scholar]
- 12.Vanherberghen B, et al. 2013. Classification of human natural killer cells based on migration behavior and cytotoxic response. Blood 121(8):1326-1334.
- 13.Elitas M, Brower K, Lu Y, Chen JJ, Fan R. A microchip platform for interrogating tumor-macrophage paracrine signaling at the single-cell level. Lab Chip. 2014;14(18):3582–3588. doi: 10.1039/c4lc00676c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konry T, Golberg A, Yarmush M. Live single cell functional phenotyping in droplet nano-liter reactors. Sci Rep. 2013;3:3179. doi: 10.1038/srep03179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dura B, et al. Profiling lymphocyte interactions at the single-cell level by microfluidic cell pairing. Nat Commun. 2015;6:5940. doi: 10.1038/ncomms6940. [DOI] [PubMed] [Google Scholar]
- 16.Abonnenc M, et al. Lysis-on-chip of single target cells following forced interaction with CTLs or NK cells on a dielectrophoresis-based array. J Immunol. 2013;191(7):3545–3552. doi: 10.4049/jimmunol.1300890. [DOI] [PubMed] [Google Scholar]
- 17.Dura B, Liu Y, Voldman J. Deformability-based microfluidic cell pairing and fusion. Lab Chip. 2014;14(15):2783–2790. doi: 10.1039/c4lc00303a. [DOI] [PubMed] [Google Scholar]
- 18.Lewis RS. Calcium signaling mechanisms in T lymphocytes. Annu Rev Immunol. 2001;19(1):497–521. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- 19.Feske S. Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. 2007;7(9):690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- 20.Cantrell D. T cell antigen receptor signal transduction pathways. Annu Rev Immunol. 1996;14(1):259–274. doi: 10.1146/annurev.immunol.14.1.259. [DOI] [PubMed] [Google Scholar]
- 21.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21(2):140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27(1):591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love PE, Hayes SM. ITAM-mediated signaling by the T-cell antigen receptor. Cold Spring Harb Perspect Biol. 2010;2(6):a002485. doi: 10.1101/cshperspect.a002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fehniger TA, et al. Differential cytokine and chemokine gene expression by human NK cells following activation with IL-18 or IL-15 in combination with IL-12: implications for the innate immune response. J Immunol. 1999;162(8):4511–4520. [PubMed] [Google Scholar]
- 25.Fauriat C, Long EO, Ljunggren H-G, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115(11):2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barber DF, Faure M, Long EO. LFA-1 contributes an early signal for NK cell cytotoxicity. J Immunol. 2004;173(6):3653–3659. doi: 10.4049/jimmunol.173.6.3653. [DOI] [PubMed] [Google Scholar]
- 27.Bryceson YT, March ME, Barber DF, Ljunggren H-G, Long EO. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J Exp Med. 2005;202(7):1001–1012. doi: 10.1084/jem.20051143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogunniyi AO, Story CM, Papa E, Guillen E, Love JC. Screening individual hybridomas by microengraving to discover monoclonal antibodies. Nat Protoc. 2009;4(5):767–782. doi: 10.1038/nprot.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HS, Devarenne TP, Han A. A high-throughput microfluidic single-cell screening platform capable of selective cell extraction. Lab Chip. 2015;15(11):2467–2475. doi: 10.1039/c4lc01316f. [DOI] [PubMed] [Google Scholar]
- 30.Kellogg RA, Gómez-Sjöberg R, Leyrat AA, Tay S. High-throughput microfluidic single-cell analysis pipeline for studies of signaling dynamics. Nat Protoc. 2014;9(7):1713–1726. doi: 10.1038/nprot.2014.120. [DOI] [PubMed] [Google Scholar]
- 31.Kirschbaum M, Jaeger MS, Duschl C. Correlating short-term Ca(2+) responses with long-term protein expression after activation of single T cells. Lab Chip. 2009;9(24):3517–3525. doi: 10.1039/b911865a. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, et al. Enhanced cell sorting and manipulation with combined optical tweezer and microfluidic chip technologies. Lab Chip. 2011;11(21):3656–3662. doi: 10.1039/c1lc20653b. [DOI] [PubMed] [Google Scholar]
- 33.Landry ZC, Giovanonni SJ, Quake SR, Blainey PC. Optofluidic cell selection from complex microbial communities for single-genome analysis. Methods Enzymol. 2013;531:61–90. doi: 10.1016/B978-0-12-407863-5.00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi JH, et al. Development and optimization of a process for automated recovery of single cells identified by microengraving. Biotechnol Prog. 2010;26(3):888–895. doi: 10.1002/btpr.374. [DOI] [PubMed] [Google Scholar]
- 35.Christo SN, Diener KR, Hayball JD. The functional contribution of calcium ion flux heterogeneity in T cells. Immunol Cell Biol. 2015;93(8):694–704. doi: 10.1038/icb.2015.34. [DOI] [PubMed] [Google Scholar]
- 36.Chehimi J, et al. Enhancing effect of natural killer cell stimulatory factor (NKSF/interleukin-12) on cell-mediated cytotoxicity against tumor-derived and virus-infected cells. Eur J Immunol. 1993;23(8):1826–1830. doi: 10.1002/eji.1830230814. [DOI] [PubMed] [Google Scholar]
- 37.Novick D, Kim S, Kaplanski G, Dinarello CA. Interleukin-18, more than a Th1 cytokine. Semin Immunol. 2013;25(6):439–448. doi: 10.1016/j.smim.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Pores-Fernando AT, Zweifach A. Calcium influx and signaling in cytotoxic T-lymphocyte lytic granule exocytosis. Immunol Rev. 2009;231(1):160–173. doi: 10.1111/j.1600-065X.2009.00809.x. [DOI] [PubMed] [Google Scholar]
- 39.Siewert K, et al. Unbiased identification of target antigens of CD8+ T cells with combinatorial libraries coding for short peptides. Nat Med. 2012;18(5):824–828. doi: 10.1038/nm.2720. [DOI] [PubMed] [Google Scholar]
- 40.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colucci F, et al. Functional dichotomy in natural killer cell signaling: Vav1-dependent and -independent mechanisms. J Exp Med. 2001;193(12):1413–1424. doi: 10.1084/jem.193.12.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross O, et al. Multiple ITAM-coupled NK-cell receptors engage the Bcl10/Malt1 complex via Carma1 for NF-kappaB and MAPK activation to selectively control cytokine production. Blood. 2008;112(6):2421–2428. doi: 10.1182/blood-2007-11-123513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajasekaran K, et al. Signaling by Fyn-ADAP via the Carma1-Bcl-10-MAP3K7 signalosome exclusively regulates inflammatory cytokine production in NK cells. Nat Immunol. 2013;14(11):1127–1136. doi: 10.1038/ni.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sad S, Kägi D, Mosmann TR. Perforin and Fas killing by CD8+ T cells limits their cytokine synthesis and proliferation. J Exp Med. 1996;184(4):1543–1547. doi: 10.1084/jem.184.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snyder JE, et al. Measuring the frequency of mouse and human cytotoxic T cells by the Lysispot assay: Independent regulation of cytokine secretion and short-term killing. Nat Med. 2003;9(2):231–235. doi: 10.1038/nm821. [DOI] [PubMed] [Google Scholar]
- 46.Einarsdottir T, Lockhart E, Flynn JL. Cytotoxicity and secretion of gamma interferon are carried out by distinct CD8 T cells during Mycobacterium tuberculosis infection. Infect Immun. 2009;77(10):4621–4630. doi: 10.1128/IAI.00415-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singleton KL, et al. Spatiotemporal patterning during T cell activation is highly diverse. Sci Signal. 2009;2(65):ra15. doi: 10.1126/scisignal.2000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157(7):1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sung M-H, et al. Switching of the relative dominance between feedback mechanisms in lipopolysaccharide-induced NF-κB signaling. Sci Signal. 2014;7(308):ra6. doi: 10.1126/scisignal.2004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croxford AL, Buch T. Cytokine reporter mice in immunological research: perspectives and lessons learned. Immunology. 2011;132(1):1–8. [Google Scholar]
- 51.Desai SP, Freeman DM, Voldman J. Plastic masters-rigid templates for soft lithography. Lab Chip. 2009;9(11):1631–1637. doi: 10.1039/b822081f. [DOI] [PubMed] [Google Scholar]
- 52.Trombetta JJ, et al. Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr Protoc Mol Biol. 2014;107:4.22.1–4.22.17. doi: 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.