Significance
Significant progress has been made in our understanding of plant adaptive responses to maintain cellular pH under varied N supply forms. Rice is a plant adapted to grow in waterlogged or dryland environments, in contrast to other crops, such as wheat, soybean, and maize. The nitrate transporter OsNRT2.3b provides a molecular mechanism explaining plant adaptation to the ammonium-nitrate supply shift between the waterlogged and drained soil environments. The sensing of cytosolic pH by OsNRT2.3b can function to improve rice nitrogen use efficiency and pH balance, providing an explanation for plant adaptation to changes in the form of N supply.
Keywords: nitrate transporter, pH sensing, nitrogen use efficiency, yield, rice
Abstract
Cellular pH homeostasis is fundamental for life, and all cells adapt to maintain this balance. In plants, the chemical form of nitrogen supply, nitrate and ammonium, is one of the cellular pH dominators. We report that the rice nitrate transporter OsNRT2.3 is transcribed into two spliced isoforms with a natural variation in expression ratio. One splice form, OsNRT2.3b is located on the plasma membrane, is expressed mainly in the phloem, and has a regulatory motif on the cytosolic side that acts to switch nitrate transport activity on or off by a pH-sensing mechanism. High OsNRT2.3b expression in rice enhances the pH-buffering capacity of the plant, increasing N, Fe, and P uptake. In field trials, increased expression of OsNRT2.3b improved grain yield and nitrogen use efficiency (NUE) by 40%. These results indicate that pH sensing by the rice nitrate transporter OsNRT2.3b is important for plant adaption to varied N supply forms and can provide a target for improving NUE.
Intracellular pH is stringently regulated, because most metabolic enzymes can function only within a narrow range of pH. The cytosolic pH is maintained around neutrality, whereas in individual organelles, pH can range from 4.7 (lysosome) to 8.0 (mitochondria) (1). Mammalian cells balance cytosolic pH using Na+/H+ exchangers, Na+– cotransporters, Cl––, or anion exchangers (AEs) (1). When bacteria face an acid challenge, the proton-pumping respiratory chain complexes or proton-coupled ATPases, and secondary active transporters, such as anion-proton antiporters like the Cl–/H+ exchangers, are activated to maintain intracellular pH (2). In alkali conditions, bacterial Na+/H+ antiport and the generation and transport of CO2, , NH3, and are the main strategies for pH homeostasis (2).
In plants, cytosolic pH can vary from 7.3 to 8.0 (3). Plant roots acquire mineral nitrogen (N) as the source for growth as nitrate, ammonium, or both; the total amount and the ratio of the two N forms can determine cellular pH. In plants, the phloem is an important tissue for nutrient, mRNA, and signal transport, acting like a neural network connecting the shoot and root (4–8). Phloem pH homeostasis is important for maintaining the physiological balance of the whole plant, as well as the transport and signaling functions of the tissue.
Rice (Oryza sativa L.) is a major crop, feeding almost 50% of the world’s population. It has been traditionally cultivated under flooded anaerobic soil conditions, where ammonium is the main N source; however, specialized aerenchyma cells in rice roots can transfer oxygen from the shoots to the roots and release it to the rhizosphere, where bacterial conversion of ammonium to nitrate (nitrification) can occur (9). Nitrification in the waterlogged paddy rhizosphere can result in 25–40% of the total crop N being taken up in the form of nitrate, mainly through a high-affinity transport system (HATS) (10). The uptake of nitrate is mediated by cotransport with protons (H+) that can be extruded from the cell by plasma membrane H+-ATPases (11).
In this study, we analyzed the function of a nitrate transporter, OsNRT2.3, with natural variation of its expression in rice cultivars and the cytosolic pH regulatory motif in the protein. The high expression of one of the two splice forms of this protein, OsNRT2.3b, in rice resulted in better adaptation to changes of N supply forms in the environment and strong improvements in growth, yield, and nitrogen use efficiency (NUE). Our results have significant implications for the understanding of cytosolic pH balance in plant adaptation and its importance for crop improvement.
Results
Natural Variation in Two Splice Forms of Rice Nitrate Transporter OsNRT2.3.
We have shown that one rice gene encoding a component of nitrate high affinity transport system (HATS), OsNRT2.3, produces two different transcripts arising from alternative splicing that we term OsNRT2.3a and OsNRT2.3b (12, 13). Comparing the two mature OsNRT2.3 mRNAs shows that the predicted protein products differ by 30 amino acids (SI Appendix, Fig. S1 A and B). OsNRT2.3a encodes a plasma membrane protein of 516 amino acids that functions in long-distance nitrate transport in the xylem from root to shoot (12, 14), whereas OsNRT2.3b encodes a shorter 486-aa plasma membrane protein expressed moderately in the phloem of the shoot and faintly in the root (12) (SI Appendix, Fig. S1C).
We evaluated the expression of OsNRT2.3a and OsNRT2.3b in 10 rice cultivars (SI Appendix, Fig. S2 A and B) with differing N accumulation in their straw and found that under low N supply (0.63 mM NH4NO3; SI Appendix, Fig. S2C), the expression ratio of OsNRT2.3b and OsNRT2.3a in the straw has a strong corelationship with the N content. We identified two different groups of the cultivars showing this relationship, but this correlation was missing at 1.25 mM NH4NO3, the normal N supply level (SI Appendix, Fig. S2D).
Functional Characterization of OsNRT2.3a and OsNRT2.3b in Xenopus Oocytes.
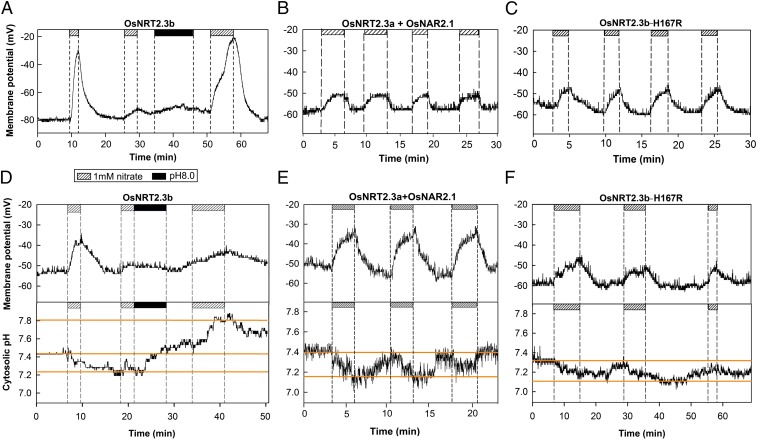
Previous studies have shown a major difference between OsNRT2.3a and OsNRT2.3b, with OsNRT2.3a requiring a partner protein, OsNAR2.1, for functional nitrate transport (13–15). In contrast, OsNRT2.3b does not require the OsNAR2.1 partner protein for nitrate membrane transport (13). To better understand the different properties of OsNRT2.3a and OsNRT2.3b in nitrate uptake, we expressed the two corresponding cDNAs in Xenopus oocytes. Oocyte expression is good for detecting the instant and dynamic responses of the transporters to nitrate supply changes by recording fluctuations in cell membrane potential and cytosolic pH. We found that oocytes coinjected with OsNRT2.3a and OsNAR2.1 RNA responded to repeated sequential nitrate treatments with an electrical depolarization of membrane potential, whereas OsNRT2.3b-injected oocytes did not show such repeated nitrate-elicited responses (Fig. 1 A, B, D, and E and SI Appendix, Fig. S3 A and B).
Fig. 1.
Functional analysis of OsNRT2.3b, OsNRT2.3a + OsNAR2.1, and H167R transporters expressed in Xenopus oocytes. (A–C) Oocyte plasma membrane potential changes in response to 1 mM nitrate treatment (shaded bar) and pH 8.0 saline wash (black bar) for a cell expressing OsNRT2.3b (A), OsNRT2.3a + OsNAR2.1 (B), and OsNRT2.3b-H167R mutant (C). (D–F) Double-barreled pH electrode recording of cytosolic pH from oocytes injected with OsNRT2.3b treated with 1 mM nitrate (shaded bar) and pH 8.0 saline (black bar) washing (D), with OsNRT2.3a + OsNAR2.1 (E), and with OsNRT2.3b-H167R (F) treated with 1 mM nitrate (shaded bar).
Double-barreled proton-selective microelectrode measurements revealed decreased cytosolic pH in oocytes during nitrate transport (shown from 6 to 9 min in the recording in Fig. 1D and in the first 7 min of the recording in SI Appendix, Fig. S3A), indicating that nitrate/proton cotransport resulted in cytosolic acidification (14, 16). The nitrate-elicited cytosolic acidification was reversed by washing the oocyte with pH 8 saline. After this alkaline wash, nitrate treatment could once again elicit a depolarization of the membrane potential of the OsNRT2.3b-expressing cells, with a critical threshold cytosolic pH for the response of ∼7.4 (shown from 33 to 41 min in Fig. 1D and from 39 to 46 min and from 58 to 62 min in SI Appendix, Fig. S3A). The relative initial acidic cytosolic pH (∼7.2) of the oocytes treated by a pH 7.0 bath solution also inhibited nitrate transport, but after a wash in pH 8.0 solution, the alkaline cytosolic pH restored nitrate transport (SI Appendix, Fig. S3C).
We repeated the experiments several times and pooled the data on simultaneous changes (delta) in membrane potential and cytosolic pH elicited by the nitrate treatments (SI Appendix, Fig. S3B). Statistical analysis indicated that the initial nitrate treatment at alkaline cytosolic pH (7.41 on average) resulted in large nitrate transport, whereas the relative acidic cytosolic pH (7.25 on average) at the beginning of the second nitrate treatment inhibited nitrate transport. This suggests that ∼0.16 pH units of cytosolic acidification prevented the nitrate transport activity of OsNRT2.3b (SI Appendix, Fig. S3B). There was no significant difference in the transporter activity of OsNRT2.3b at cytosolic pH 7.41 or 7.56 (SI Appendix, Fig. S3B).
We also observed that the membrane potential response to nitrate treatment at alkaline cytosolic pH, as in Fig. 1D, had a different slope than the responses shown in SI Appendix, Fig. S3 A and C at a more acidic cytosolic pH. Interestingly, the oocytes coinjected with OsNRT2.3a and OsNAR2.1 mRNA did not show any pH sensitivity in their nitrate-elicited electrical response (Fig. 1E).
Cytosolic pH Regulatory Motif Characterization in OsNRT2.3b Transport.
Some AE proteins participate in mammalian cell pH regulation, and regulatory motifs have been identified in their cytoplasmic domains (17–21). The differing sensitivity of OsNRT2.3a and OsNRT2.3b to nitrate transport-elicited changes in cytosolic pH prompted us to look for possible pH-sensing motifs. Using software for scanning protein fingerprints (www.ebi.ac.uk/Tools/pfa/fingerprintscan/), we identified two AE motifs (VYEAIHKI and LGLISGMTG) in the proteins of OsNRT2.3a and OsNRT2.3b. Analysis of some plant NRT2 protein sequences showed that these AE motifs were present in nitrate transporters from several different plant species (SI Appendix, Table S2).
The membrane topographical characteristic of OsNRT2.3a and OsNRT2.3b were predicted using transmembrane protein software (SI Appendix, Table S1). As displayed by TMPred (www.ch.embnet.org/software/TMPRED_form.html), the software predicted that VYEAIHKI is facing the cytosolic side in OsNRT2.3b, whereas it is at the external side in OsNRT2.3a (SI Appendix, Fig. S4 G and H); however, the LGLISGMTG motif is predicted at the external (LGL) and transmembrane (ISGMTG) regions in OsNRT2.3b (SI Appendix, Fig. S4 G and H). Therefore, we focused on detecting the role of VYEAIHKI in sensing the cytosolic pH in OsNRT2.3b.
Histidine residues are important for pH sensing (22). Given that the H residue of VYEAIHKI for the OsNRT2.3b protein might be located in the border between cytosol and transmembrane (SI Appendix, Fig. S4G), and VYEAI is predicted as transmembrane, we made a single site mutation (H167R) in this motif. Interestingly, OsNRT2.3b lost the cytosolic pH regulation by this mutation, even after repeated cycles of nitrate treatment (Fig. 1 C and F). The cytosolic pH decreased from 7.4 to ∼7.2 with nitrate treatments, but the electrical depolarization in membrane potential still occurred despite acidification by repeated nitrate treatments. It was no longer necessary to restore the membrane response to nitrate with an alkaline wash (Fig. 1 C and F).
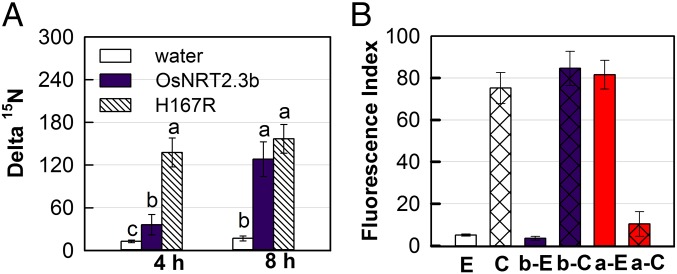
When oocytes were incubated in 15N-nitrate for only 4 h, the effect of H167R mutation on nitrate transport was clear, with the comparison of H167R and wild-type (WT) OsNRT2.3b showing that the mutation resulted in much greater nitrate accumulation (Fig. 2A). This also might result from changes in the Km of H167R protein for nitrate, given that the affinity was significantly increased when the membrane potential was in the −40 to −60 mV range, compared with OsNRT2.3b (SI Appendix, Fig. S5D). Thus, the H167R protein may be more efficient in nitrate transport in 4-h 15N- uptake experiments compared with OsNRT2.3b (Fig. 2A). After an 8-h 15N- incubation, the difference in nitrate accumulation between the two forms of the transporter had disappeared, suggesting that after the longer incubation, the accumulation of nitrate had reached a maximum in the oocytes (Fig. 2A).
Fig. 2.
Nitrate uptake assay in oocytes and the membrane orientation of the OsNRT2.3a/OsNRT2.3b pH-sensing motif in rice protoplasts determined by flow cytometry. (A) 15N- uptake by the oocytes injected with water, OsNRT2.3b, and H167R mutant mRNAs. Values are mean ± SE (n = 15). Cell viability was tested by electrophysiology after the incubation experiments. a, b, and c above bars indicate significant differences between mRNA and water injected cells (P < 0.05) estimated by one-way ANOVA. (B) Fluorescence index (FI) of OsNRT2.3b and OsNRT2.3a samples shown in SI Appendix, Fig. S4. FI is calculated as fluorescence protoplast number/total protoplast number. 6×-His tag–expressing protoplasts served as positive controls as shown: E for its external side fluorescence within only W5 solution and C for cytosolic side fluorescence with permeabilization buffer in W5 solution; b-E and b-C for external and cytosolic side fluorescence of OsNRT2.3b, respectively; and a-E and a-C for external and cytosolic side fluorescence of OsNRT2.3a, respectively.
The affinity of OsNRT2.3b, the H167R mutation, and OsNRT2.3a for nitrate showed similar Km values for at ∼0.45 mM when the membrane potential exceeded −100 mV (SI Appendix, Fig. S5), the normal level for plant cells (23–25). For OsNRT2.3a and H167R, nitrate Km increased as the potentials decreased from −100 mV to −40 mV (SI Appendix, Fig. S5D). The point mutation H167R influenced the voltage dependence of the turnover rate (SI Appendix, Fig. S5), with a change in charge density altering the voltage dependence of both Vmax and Km for nitrate (SI Appendix, Fig. S5 D and E). H167R behaves like the other spliced form of the transporter, OsNRT2.3a.
To confirm that the VYEAIHKI motif of OsNRT2.3a and OsNRT2.3b is indeed located on different sides of the plasma membrane, we performed flow cytometry with an anti-His(6)FITC–tagged antibody to determine the membrane topology in rice protoplasts. The 6-His signal of OsNRT2.3b H167 was found to be much lower at the external surface compared with cytosolic face signal obtained after intracellular fixation and permeabilization of the rice protoplasts (Fig. 2B and SI Appendix, Fig. S4 C and D). The opposite pattern was found in the 6-His signal of OsNRT2.3a H197, however; that is, the external surface of intact protoplast had more signal (Fig. 2B and SI Appendix, Fig. S4 E and F). These FITC data (Fig. 2B) confirm that the pH-sensing motif VYEAIHKI around residue H167 in OsNRT2.3b is on the cytosolic face (SI Appendix, Fig. S4G), but for OsNRT2.3a, the same motif is on the external face of the protoplast (SI Appendix, Fig. S4H).
High Expression of OsNRT2.3b Improved Rice Growth and NUE.
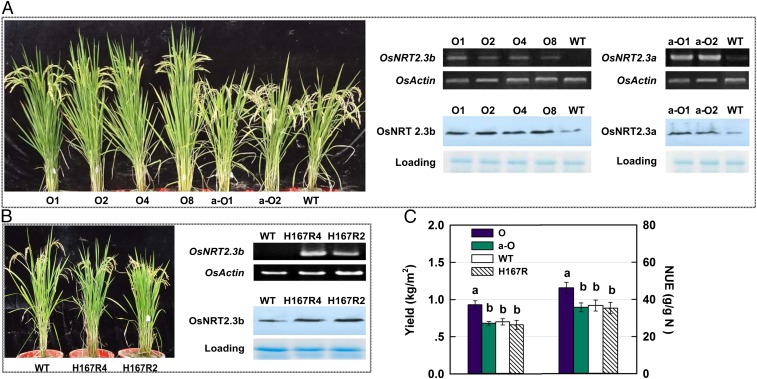
To investigate OsNRT2.3b function in rice NUE, we tested the effect of overexpressing ONRT2.3a, OsNRT2.3b, and the H167R mutated OsNRT2.3b in transgenic rice plants (Oryza sativa L ssp. Japonica, Nipponbare). We generated overexpression lines for OsNRT2.3b (O1, O2, O4, and O8), OsNRT2.3a (a-O1 and a-O2), and H167R (H167R2 and H167R4) (Fig. 3 and SI Appendix, Fig. S6 A–C and Tables S3 and S4). We backcrossed OsNRT2.3b overexpression lines with the WT to prove genetically that the phenotype of O8 line is linked to OsNRT2.3b overexpression in pot and field experiments (SI Appendix, Fig. S6D). We determined that the transgenic DNA insertion sites were in the noncoding regions of the genome (SI Appendix, Fig. S7 E and F). We observed that OsNRT2.3b-overexpressing plants showed improved growth, yield, and NUE relative to the WT, but not to the OsNRT2.3a- and H167R- overexpressing plants (Fig. 3 and SI Appendix, Fig. S6). We repeated these experiments in both pot (Fig. 3 A and B and SI Appendix, Fig. S6 A and B) and field experiments (SI Appendix, Figs. S6C, S8, and S9) under semitropical and tropical climate conditions (SI Appendix, Tables S5 and S6). Furthermore, irrespective of the promoter used to drive the overexpression [Ubiqutin (Ubi), O1 and O2 lines or 35S, O4 and O8 lines], we found the same phenotype for OsNRT2.3b-overexpressing plants under adequate and deficient N fertilizer supplies (SI Appendix, Figs. S8 and S9 and Table S6).
Fig. 3.
Growth, yield, and NUE of OsNRT2.3b, OsNRT2.3a, and H167R mutation overexpressing lines. (A) Phenotypes and transcriptional and translational expression of OsNRT2.3b- and OsNRT2.3a-overexpressing lines and Nipponbare WT. (B) Phenotypes and transcriptional and translational expression of WT and OsNRT2.3b-H167R mutant-overexpressing lines. (C) Average grain yield and NUE of OsNRT2.3b- (O), OsNRT2.3a- (a-O), and H167R- (H167R) overexpressing lines and WT in field plots. RT-PCR with the specific primers (SI Appendix, Table S10) and Western blot analyses with monoclonal antibodies were performed to identify protein expression levels. (NUE = grain yield/applied N fertilizer.) Values are mean ± SE (n = 3). a and b above bars indicate significant differences (P < 0.05) between the transgenic lines and WT estimated by one-way ANOVA.
Overexpression was checked at the mRNA and protein level and found to be increased for OsNRT2.3a, OsNRT2.3b, and H167R, with the same enhanced expression pattern in the transformed lines (Fig. 3 A and B). We observed strong expression of both OsNRT2.3b and H167R in root and leaf cells in the transgenic lines by RNA in situ blotting (SI Appendix, Fig. S7G). This overexpression pattern could improve plant nitrate uptake from the external environment and delivery to the shoot. We did not find any significant yield or NUE increase in the OsNRT2.3a- and H167R-overexpressing lines relative to the Nipponbare WT, however (Fig. 3 and SI Appendix, Figs. S6 and S8). Furthermore, we found the same improvement by OsNRT2.3b overexpression in a high-yielding and high-NUE cultivar background (SI Appendix, Fig. S9).
The grain yield of overexpressing lines showed that OsNRT2.3b improved rice growth to give 35–54% more in the O8 line under varied N supplies (Fig. 3C and SI Appendix, Fig. S8 and Table S6) in field plots. With only one-quarter of the normal local N fertilizer application (75 kg N per ha), the grain yield of O8 could reach that of WT under a more typical N supply (300 kg N per ha) (SI Appendix, Fig. S8 and Table S6). The most obvious improvement by OsNRT2.3b overexpression, but not by OsNRT2.3a or H167R overexpression, was in panicle size, including increased length, number of primary and secondary rachises (SI Appendix, Fig. S6B), number of seeds per panicle, and seed setting rates (SI Appendix, Table S6) under different N treatments.
Calculating the NUE using the yield produced divided by the N fertilizer supply, the value for O8 and other OsNRT2.3b-overexpressing plants was increased by 26–47% at 300 kg N per ha compared with WT, OsNRT2.3a, and H167R lines. The NUE was increased in overexpressor O8 up to 80% at 75 kg N per ha (Fig. 3C and SI Appendix, Fig. S8 and Table S6). In contrast, the OsNRT2.3b- and H167R-overexpressing lines did not show any N-dependent improvement in yield or NUE (Fig. 3C and SI Appendix, Fig. S8 and Table S6).
Characterization of OsNRT2.3b Overexpression in Rice Showing a Function in pH Regulation.
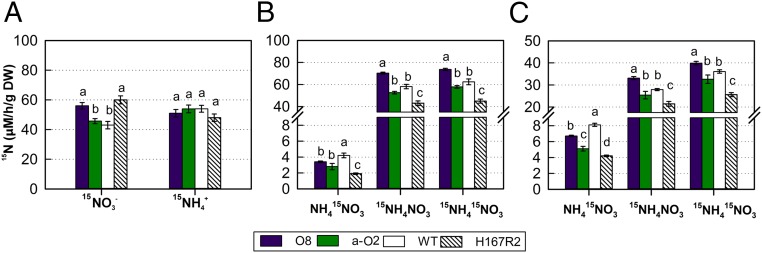
We measured the effect of OsNRT2.3b, OsNRT2.3a, and H167R overexpression on the 15N- and 15N- influx of hydroponically grown rice at pH 6 (Fig. 4A). The influx rate was increased significantly over that in WT in all transgenic lines except the OsNRT2.3a overexpressor (Fig. 4A and SI Appendix, Fig. S10A). In contrast, OsNRT2.3b, OsNRT2.3a, and H167R overexpression had no significant effect on short-term 15N- influx (Fig. 4A and SI Appendix, Fig. S10A). Compared with WT, the OsNRT2.3b, OsNRT2.3a, and H167R overexpressors showed less 15N-NH415NO3 in 5-min uptake experiments; in this short duration of N uptake, in NH4NO3 was the main form of N influx into the root (Fig. 4B and SI Appendix, Fig. S10B). Compared with WT, the O8 line and other OsNRT2.3b overexpressors showed more 15N-15NH4NO3 and total N uptake at pH 6 and pH 4; however, the OsNRT2.3a lines showed similar uptake and the H167R lines showed less uptake (Fig. 4 B and C and SI Appendix, Fig. S10B). An exception was line a-O1, which was lower than WT at pH 4 (SI Appendix, Fig. S10C). The same pattern was seen for 15N-15NH415NO3 uptake (Fig. 4 B and C and SI Appendix, Fig. S10 B and C). Expression in oocytes showed that OsNRT2.3b does not transport irrespective of the presence of nitrate (SI Appendix, Fig. S11 A and B).
Fig. 4.
Effects of OsNRT2.3b, OsNRT2.3a, and H167R overexpression on the root influx of and at pH 6 and 4 for 5 min. (A) The root 15N influx rate at 2.5 mM [supplied as Ca(NO3)2] or (supplied as NH4Cl) at pH 6.0. (B and C) The root 15N influx rate in NH415NO3, 15NH4NO3, and 15NH415NO3 supply at pH 6 (B) and pH 4 (C). The 15N influx was measured for 5 min. Values are mean ± SE (n = 5). a, b, and c above bars indicate significant differences (P < 0.05) between the transgenic lines and WT for the same treatment estimated by one-way ANOVA.
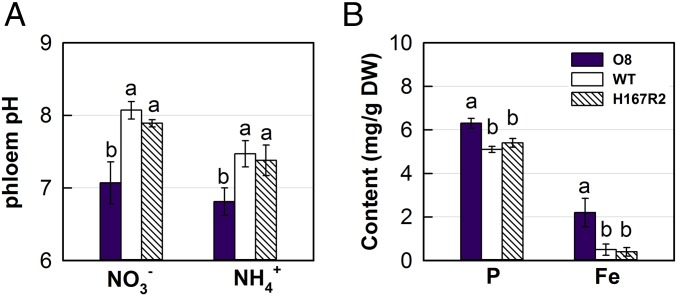
The phloem sap pH in overexpressor line O8 (collected as shown in SI Appendix, Fig. S12) was 7.1, whereas that of WT and H167R2 was near 8 under supply. In contrast O8 phloem sap pH was 6.8, but WT and H167R2 was ∼7.4, under supply (Fig. 5A). In nonsterile hydroponic culture with , some microbial nitrification produces low concentrations of nitrate (26); therefore, OsNRT2.3b overexpression may improve growth even under supply, owing to small amounts of nitrate in the solution. It is known that an acidic phloem sap benefits P and Fe translocation to the leaf (27). We found that the relatively low phloem sap pH increased total P and Fe in the leaves of the OsNRT2.3b lines compared with H167R2 and WT (Fig. 5B and SI Appendix, Fig. S13); however, phloem sap pH is a relative comparison, considering that the sample had passed through an insect.
Fig. 5.
Effects of OsNRT2.3b and H167R overexpression on phloem pH and P and Fe content. (A) Phloem pH under 2.5 mM [supplied as Ca(NO3)2] or (supplied as NH4Cl). (B) Total P and Fe content in leaves measured by ion chromatography analysis. Values are mean ± SE (n = 5). a, b, and c above bars indicate significant differences (P < 0.05) between the transgenic lines and WT estimated by one-way ANOVA.
The increases in N uptake and P and Fe content could benefit C metabolism in the OsNRT2.3b-overexpressing lines. Microarray and confirmatory quantitative RT-PCR showed that decreased expression of the genes involved in the photorespiratory pathway in line O8 relative to WT and H167R2 (SI Appendix, Fig. S14). Compared with WT and H167R-overexpressing lines, the OsNRT2.3b lines showed the same photosynthesis rate per unit of leaf area, higher total photosynthesis per leaf and intercellular CO2 concentration, and lower photorespiratory rate (SI Appendix, Fig. S15).
Discussion
The recently released database of rice genomic variations (28) identifies single nucleotide polymorphisms and insertions/deletions in 1,479 rice accessions, including both landraces and improved varieties from 73 countries. The cross-population likelihood method (XP-CLR) was used to identify genetic selection signals. The XP-CLR data for these rice accessions showed that only OsNRT2.3 of the rice nitrate transporter genes was under selection pressure during evolution and was much stronger in Indica II compared with Indica I rice cultivars (29). Our expression data confirm the XP-CLR results, showing that there were two selection patterns in rice; that is, one group of rice cultivars had selection pressure on the expression ratio of OsNRT2.3b to OsNRT2.3a linking with N accumulation, whereas the other group lost this selection. The division of rice cultivars in the responses of OsNRT2.3b and OsNRT2.3a expression to N supply may result from their different growth and cultivation conditions (29).
Comparison of the predicted protein sequences of HvNRT2.1 and the rice and Arabidopsis NRT2s using the FingerPRINTScan software (www.ebi.ac.uk/Tools/pfa/fingerprintscan/) revealed AE family signature motifs in both OsNRT2.3a/b and AtNRT2.7 (30). Because AE proteins participate in pH regulation (17, 31), these motifs may be important for pH sensing in the plant nitrate transporters. In view of the critical importance of histidine residues within pH-sensing motifs (22), we chose the histidine within the VYEAIHKI as a target for single site mutagenesis, and it was changed to an arginine residue. The functional analysis in oocytes showed that the single amino acid site mutation, H167R of OsNRT2.3b, had lost cytosolic pH regulation even after repeated cycles of nitrate treatment (Fig. 1D). This result suggests the VYEAIHKI motif as a pH-regulation site in the sequence of some plant nitrate transporters.
Along with the external and cytosolic pH, the proton gradient for nitrate transport also depends on the membrane potential. Nitrate transport is driven by the electrochemical gradient for protons, and our data in oocytes suggest that it is this parameter, as well as cytosolic pH, that can directly regulate OsNRT2.3b nitrate transport activity (Fig. 1D and SI Appendix, Fig. S3 A–C).
The form of N supply for plants is well known to influence plant pH balance (32). The pH-sensing switch of OsNRT2.3b is a key factor, providing an explanation for the phenotype of OsNRT2.3b transgenic plants, because OsNRT2.3a and the H167R mutation lines had lost this super phenotype and behaved like WT (Figs. 3–5 and SI Appendix, Figs. S6, S8, and S9 and Table S6). A synergism between ammonium and nitrate nutrition in plants has been widely observed. The simultaneous availability of nitrate and ammonium enhances the uptake of ammonium and total N influx, whereas this mix depresses short-term nitrate influx compared with either ammonium or nitrate alone in rice (33). This synergism between N supply forms was enhanced by overexpression of OsNRT2.3b, with increased ammonium and total N influx compared with WT (Fig. 4 A–C and SI Appendix, Fig. S10 A–C). The influx and assimilation of nitrate and ammonium requires cytosolic pH homeostasis, with regulation partly contributed by the OsNRT2.3b sensing motif. The proton-cotransport mechanism for the entry of nitrate into cells provides cytosolic acidification, whereas ammonium transport can cause an alkalinization (34), which may enhance proton-coupled nitrate transport. The assimilation of ammonium produces at least one H+ per , whereas assimilation produces almost one OH− per (32). Either H+ or OH− produced in excess of that required to maintain cytoplasmic pH is exported from the cell in an energy-requiring step (e.g., plasma membrane H+ pumping ATPase) (11, 16). The advantages of a mixed and supply for plant pH balance have long been recognized (32, 35). The short-term synergism between ammonium and nitrate to maintain cytosolic pH can explain the measured increase in 15N- uptake when the plant was supplied with a mixed N source (Fig. 4 B and C).
In WT plants, OsNRT2.3b expression was low (12, 13) (SI Appendix, Fig. S7G). The transgenic plants with OsNRT2.3b overexpression driven by strong promoters had more general tissue expression (SI Appendix, Fig. S7G). The synergism between ammonium and nitrate was enhanced by overexpression of the pH-sensing transporter OsNRT2.3b more generally in cells, but this did not occur in OsNRT2.3a or H167R overexpressors when the pH sensor was lost. The lower N uptake in the H167R lines may be related to (i) the changes in Km and Vmax for nitrate introduced by the H167R mutation from OsNRT2.3b, making the protein behave more like OsNRT2.3a (SI Appendix, Fig. S5); (ii) alterations of expression pattern by different promoters; (iii) formation of a homodimer by the transporter, impairing native OsNRT2.3b function; and (iv) inactivation of the pH sensor along with flipping the location of the protein on the membrane by the H167R substitution. More work is needed to uncover the full phenotype of H167R lines.
Because nitrate assimilation depends on photorespiration (36, 37), the relationship between the assimilation of nitrate and ammonium and photorespiration (36–38) is closely coupled to the shuttling of malate between the cytoplasm and chloroplast to balance pH (39). Compared with the WT and H167R overexpressors, the OsNRT2.3b overexpressors had a higher intercellular CO2 level and a lower photorespiratory rate in the leaves (SI Appendix, Fig. S15 C and D), which might result in increased biomass and grain yields (40, 41) (Fig. 3C and SI Appendix, Fig. S6 A–C). Why OsNRT2.3b overexpression would increase leaf intercellular CO2 remains unclear, however. One possible explanation may be the linkage of nitrate assimilation and photorespiration, given that nitrate in comparison with ammonium nutrition is reported to increase photorespiration under high light (36). OsNRT2.3b overexpression increased the uptake of ammonium more than that of nitrate (Fig. 4 B and C), which might result in a lower photorespiratory rate. Another possible explanation is the strong ectopic expression of OsNRT2.3b in leaf mesophyll cells (SI Appendix, Fig. S7G), which may enhance the cytosolic pH balance in leaf mesophyll cells and influence the intercellular dissolved CO2 level (40).
In conclusion, the present study shows how overexpression of the cytosolic pH-sensing motif from one side of the membrane to the other can alter pH homeostasis to benefit the rice plant to improve yield and NUE. The improved pH homeostasis may enable plants to adapt to growth environment changes and the ammonium-nitrate shift between waterlogged and drained soils.
Materials and Methods
Functional Analysis in Oocytes.
Oocyte preparation, mRNA synthesis and injection, 15N-nitrate uptake, and electrophysiology are described in SI Appendix.
Transgenic Plants.
Two lines of T7 OsNRT2.3a-overexpressing plants (a-O1 and a-O2), four lines of T7 OsNRT2.3b- overexpressing plants (O1, O2, O4, and O8), and two lines of T7 H167R overexpressing plants (H167R2 and H167R4) were used in the experiments (SI Appendix, Tables S3 and S4). The transformation process and all of the other experiments are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Ping Wu (Zhejiang University) for support in the field experiments in Changxing; Yu Liu for part of the 15N analysis; Xiaorong Fan for some of the oocyte experiments, 15N analyses in plants, and phloem sap collection at the John Innes Centre; Xiudong Xia and Shengyuan Li (Nanjing Agricultural University) for the field experiments; I. Bedford (John Innes Centre) for the leaf hopper experiments; and Ali Pendle (John Innes Centre) for technological support in the RNA in situ hybridization. Research in China was funded by the 973 Program (Grant 2011CB100300), the National Natural Science Foundation, the Transgenic Project (Grant 2016ZX08001003-008), the 111 Project (Grant 12009), the Innovative Research Team Development Plan of the Ministry of Education of China, and the Priority Academic Program Development of the Jiangsu Higher Education Institutions project. A.J.M. is supported by Grants BB/J004553/1 and BB/L010305/1 from the Biotechnology and Biological Sciences Research Council and the John Innes Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525184113/-/DCSupplemental.
References
- 1.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11(1):50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 2.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9(5):330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinière A, Desbrosses G, Sentenac H, Paris N. Development and properties of genetically encoded pH sensors in plants. Front Plant Sci. 2013;4:523. doi: 10.3389/fpls.2013.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molnar A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328(5980):872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 5.Dunoyer P, et al. Small RNA duplexes function as mobile silencing signals between plant cells. Science. 2010;328(5980):912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- 6.Wu S, Gallagher KL. Transcription factors on the move. Curr Opin Plant Biol. 2012;15(6):645–651. doi: 10.1016/j.pbi.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Wang A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu Rev Phytopathol. 2015;53:45–66. doi: 10.1146/annurev-phyto-080614-120001. [DOI] [PubMed] [Google Scholar]
- 8.Notaguchi M, Higashiyama T, Suzuki T. Identification of mRNAs that move over long distances using an RNA-Seq analysis of Arabidopsis/Nicotiana benthamiana heterografts. Plant Cell Physiol. 2015;56(2):311–321. doi: 10.1093/pcp/pcu210. [DOI] [PubMed] [Google Scholar]
- 9.Li YL, Fan XR, Shen QR. The relationship between rhizosphere nitrification and nitrogen-use efficiency in rice plants. Plant Cell Environ. 2008;31(1):73–85. doi: 10.1111/j.1365-3040.2007.01737.x. [DOI] [PubMed] [Google Scholar]
- 10.Kirk GJD, Kronzucker HJ. The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: A modelling study. Ann Bot (Lond) 2005;96(4):639–646. doi: 10.1093/aob/mci216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, et al. Adaptation of plasma membrane H(+)-ATPase of rice roots to low pH as related to ammonium nutrition. Plant Cell Environ. 2009;32(10):1428–1440. doi: 10.1111/j.1365-3040.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- 12.Tang Z, et al. Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol. 2012;160(4):2052–2063. doi: 10.1104/pp.112.204461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng H, et al. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot. 2011;62(7):2319–2332. doi: 10.1093/jxb/erq403. [DOI] [PubMed] [Google Scholar]
- 14.Yan M, et al. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 2011;34(8):1360–1372. doi: 10.1111/j.1365-3040.2011.02335.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, et al. Identification and functional assay of the interaction motifs in the partner protein OsNAR2.1 of the two-component system for high-affinity nitrate transport. New Phytol. 2014;204(1):74–80. doi: 10.1111/nph.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu G, Fan X, Miller AJ. Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol. 2012;63:153–182. doi: 10.1146/annurev-arplant-042811-105532. [DOI] [PubMed] [Google Scholar]
- 17.Kurschat CE, et al. Alkaline-shifted pHo sensitivity of AE2c1-mediated anion exchange reveals novel regulatory determinants in the AE2 N-terminal cytoplasmic domain. J Biol Chem. 2006;281(4):1885–1896. doi: 10.1074/jbc.M509734200. [DOI] [PubMed] [Google Scholar]
- 18.Kopito RR. Molecular biology of the anion exchanger gene family. Int Rev Cytol. 1990;123:177–199. doi: 10.1016/s0074-7696(08)60674-9. [DOI] [PubMed] [Google Scholar]
- 19.Alper SL. The band 3-related anion exchanger (AE) gene family. Annu Rev Physiol. 1991;53:549–564. doi: 10.1146/annurev.ph.53.030191.003001. [DOI] [PubMed] [Google Scholar]
- 20.Tanner MJA. The structure and function of band 3 (AE1): Recent developments (review) Mol Membr Biol. 1997;14(4):155–165. doi: 10.3109/09687689709048178. [DOI] [PubMed] [Google Scholar]
- 21.Bruce LJ, Unwin RJ, Wrong O, Tanner MJA. The association between familial distal renal tubular acidosis and mutations in the red cell anion exchanger (band 3, AE1) gene. Biochem Cell Biol. 1998;76(5):723–728. doi: 10.1139/bcb-76-5-723. [DOI] [PubMed] [Google Scholar]
- 22.Stewart AK, et al. Transmembrane domain histidines contribute to regulation of AE2-mediated anion exchange by pH. Am J Physiol Cell Physiol. 2007;292(2):C909–C918. doi: 10.1152/ajpcell.00265.2006. [DOI] [PubMed] [Google Scholar]
- 23.Fan XR, et al. A comparison of nitrate transport in four different rice (Oryza sativa L.) cultivars. Sci China Ser C. 2005;48:897–911. [PubMed] [Google Scholar]
- 24.Miller AJ. Real time measurement of cytoplasmic ions with ion-selective microelectrodes. Methods Mol Biol. 2013;953:243–254. doi: 10.1007/978-1-62703-152-3_16. [DOI] [PubMed] [Google Scholar]
- 25.Miller AJ, Smith S. Measuring intracellular ion concentrations with multi-barrelled microelectrodes. In: Shabala S, Cuin TA, editors. Plant Salt Tolerance: Methods and Protocols. Humana; Totowa, NJ: 2012. pp. 67–77. [DOI] [PubMed] [Google Scholar]
- 26.Padgett PE, Leonard RT. Contamination of ammonium-based nutrient solutions by nitrifying organisms and the conversion of ammonium to nitrate. Plant Physiol. 1993;101(1):141–146. doi: 10.1104/pp.101.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomine S, Lanquar V. Iron transport and signaling in plants. In: Geisler M, Venema K, editors. Transporters and Pumps in Plant Signaling, Signaling and Communication in Plants. Springer; Berlin: 2011. pp. 99–131. [Google Scholar]
- 28.Zhao H, et al. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 2015;43(Database issue) D1:D1018–D1022. doi: 10.1093/nar/gku894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie W, et al. Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc Natl Acad Sci USA. 2015;112(39):E5411–E5419. doi: 10.1073/pnas.1515919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chopin F, et al. The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell. 2007;19(5):1590–1602. doi: 10.1105/tpc.107.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart AK, Kerr N, Chernova MN, Alper SL, Vaughan-Jones RD. Acute pH-dependent regulation of AE2-mediated anion exchange involves discrete local surfaces of the NH2-terminal cytoplasmic domain. J Biol Chem. 2004;279(50):52664–52676. doi: 10.1074/jbc.M408108200. [DOI] [PubMed] [Google Scholar]
- 32.Raven JA, Smith FA. Nitrogen assimilation and transport in vascular land plants in relation to intracellular pH regulation. New Phytol. 1976;76(3):415–431. [Google Scholar]
- 33.Kronzucker HJ, Siddiqi MY, Glass ADM, Kirk GJD. Nitrate-ammonium synergism in rice: A subcellular flux analysis. Plant Physiol. 1999;119(3):1041–1046. doi: 10.1104/pp.119.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosegarten H, Grolig F, Wieneke J, Wilson G, Hoffmann B. Differential ammonia-elicited changes of cytosolic pH in root hair cells of rice and maize as monitored by 2′,7′-bis-(2-carboxyethyl)-5 (and -6)-carboxyfluorescein-fluorescence ratio. Plant Physiol. 1997;113(2):451–461. doi: 10.1104/pp.113.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raven JA. Acquisition of nitrogen by the shoots of land plants: Its occurrence and implications for acid-base regulation. New Phytol. 1988;109(1):1–20. [Google Scholar]
- 36.Bloom AJ. Photorespiration and nitrate assimilation: A major intersection between plant carbon and nitrogen. Photosynth Res. 2015;123(2):117–128. doi: 10.1007/s11120-014-0056-y. [DOI] [PubMed] [Google Scholar]
- 37.Rachmilevitch S, Cousins AB, Bloom AJ. Nitrate assimilation in plant shoots depends on photorespiration. Proc Natl Acad Sci USA. 2004;101(31):11506–11510. doi: 10.1073/pnas.0404388101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stitt M, et al. Steps towards an integrated view of nitrogen metabolism. J Exp Bot. 2002;53(370):959–970. doi: 10.1093/jexbot/53.370.959. [DOI] [PubMed] [Google Scholar]
- 39.Backhausen JE, Kitzmann C, Scheibe R. Competition between electron acceptors in photosynthesis: Regulation of the malate valve during CO2 fixation and nitrite reduction. Photosynth Res. 1994;42(1):75–86. doi: 10.1007/BF00019060. [DOI] [PubMed] [Google Scholar]
- 40.Moroney JV, Jungnick N, Dimario RJ, Longstreth DJ. Photorespiration and carbon concentrating mechanisms: Two adaptations to high O2, low CO2 conditions. Photosynth Res. 2013;117(1-3):121–131. doi: 10.1007/s11120-013-9865-7. [DOI] [PubMed] [Google Scholar]
- 41.Kebeish R, et al. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol. 2007;25(5):593–599. doi: 10.1038/nbt1299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.