Significance
Despite intense interest over a period of decades, mechanisms of allelic exclusion have remained unsolved mysteries in the field of eukaryotic gene expression control. Parasitic African trypanosomes express variant surface glycoproteins (VSGs) in a monoallelic fashion and have long been a paradigm for studies in this area. We used an RNA interference screen for loss of exclusion and identified and characterized VSG exclusion 1 (VEX1). VEX1 sequestration restricts expression and prevents the simultaneous establishment of more than one active VSG gene. VEX1 also appears to reinforce sequestration-based exclusion through homology-dependent repression. Our results indicate a “winner-takes-all” mechanism that allows parasitic trypanosomes to express just one VSG gene at a time.
Keywords: epigenetic, monoallelic, silencing, telomere, Trypanosoma brucei
Abstract
Allelic exclusion underpins antigenic variation and immune evasion in African trypanosomes. These bloodstream parasites use RNA polymerase-I (pol-I) to transcribe just one telomeric variant surface glycoprotein (VSG) gene at a time, producing superabundant and switchable VSG coats. We identified trypanosome VSG exclusion-1 (VEX1) using a genetic screen for defects in telomere-exclusive expression. VEX1 was sequestered by the active VSG and silencing of other VSGs failed when VEX1 was either ectopically expressed or depleted, indicating positive and negative regulation, respectively. Positive regulation affected VSGs and nontelomeric pol-I–transcribed genes, whereas negative regulation primarily affected VSGs. Negative regulation by VEX1 also affected telomeric pol-I–transcribed reporter constructs, but only when they contained blocks of sequence sharing homology with a pol-I–transcribed locus. We conclude that restricted positive regulation due to VEX1 sequestration, combined with VEX1-dependent, possibly homology-dependent silencing, drives a “winner-takes-all” mechanism of allelic exclusion.
Cells often restrict expression to a single allele of a gene or gene family. This allelic exclusion underpins antigenic variation in pathogens, including trypanosomes that cause sleeping sickness (1) and Plasmodium parasites that cause malaria (2). Allelic exclusion is also essential for singular olfactory receptor expression and a sense of smell in metazoa (3). Although many factors have been identified that are required for the expression of one allele or for the silencing of other alleles in these systems, our understanding of the mechanisms by which expression and silencing are established and coordinated remains incomplete (1–3).
The African trypanosome Trypanosoma brucei is a flagellated parasitic protozoan transmitted among mammalian hosts by tsetse flies. In addition to causing trypanosomiasis in humans, a fatal and neglected tropical disease, these parasites also cause nagana in cattle. Antigenic variation is essential for persistent bloodstream infection in the face of host adaptive immune defenses and has long been a paradigm for studies on allelic exclusion (1); parasite immune evasion depends upon singular variant surface glycoprotein (VSG) gene expression and VSG switching. Although multiple subtelomeric VSGs are available for expression (4), only one is transcribed (5). Both active and silent VSGs are located at the ends of polycistronic transcription units known as expression sites (ESs) (6). Notably, VSG-ES promoters (6) recruit RNA polymerase-I (pol-I) that typically transcribes ribosomal RNA genes (7). Indeed, the active VSG-ES is associated with an extranucleolar focus of pol-I known as the expression-site body (ESB) (8–10). Although the active VSG-ES is specifically depleted of nucleosomes (11, 12), silent VSG-ESs are similarly located in the extranucleolar space in bloodstream-form cells, and neither active nor silent VSG-ESs show an appreciable association with the nuclear envelope (13). An HMG chromatin protein that is enriched and inversely correlated with nucleosome abundance at the ESB and in the nucleolus (14) appears to maintain open chromatin at these sites (15). In addition, a highly SUMOylated focus is specific to the site of the ESB (16).
Pol-I transcription at the active VSG-ES, combined with attenuation at other VSG-ESs (17), allows trypanosomes to produce a single superabundant VSG. Indeed, the active VSG generates the most abundant T. brucei mRNA and protein. The mRNA exceeds the next most abundant mRNA by >10-fold, and ∼10 million VSGs, constituting 10% of total cell protein (18), form a dense coat on each bloodstream-form cell (19). Antigenic variation itself occurs at low frequency and without immune selection (20) due to VSG rearrangement or coordinated transcription switching from one VSG-ES to another, the latter occurring in the absence of detectable change in the DNA sequence (1). Attempts to select for two simultaneously transcribed VSG-ESs indicated that double VSG expression is highly unstable (21).
Several reports link chromatin, chromatin-associated proteins, and telomere-binding proteins to VSG silencing (SI Appendix, Table S1). For example, a histone H3 variant, a bloodstream stage-specific modified DNA base known as J or hydroxymethyluracil (22, 23), the chromatin remodeling ISWI complex (24), the histone H3K76 trimethyltransferase DOT1B (25), and the telomere-associated protein RAP1 (26) all facilitate VSG-ES silencing. In addition, cohesin function facilitates maintenance of the active VSG-ES (27), and inositol phosphate signaling impacts VSG-ES regulation (28). Allelic exclusion, however, requires the establishment of differential expression states and coordination among members of a gene family, which are not understood (1–3). In the case of T. brucei, it remains unclear how pol-I action is concentrated at one telomeric VSG.
Results
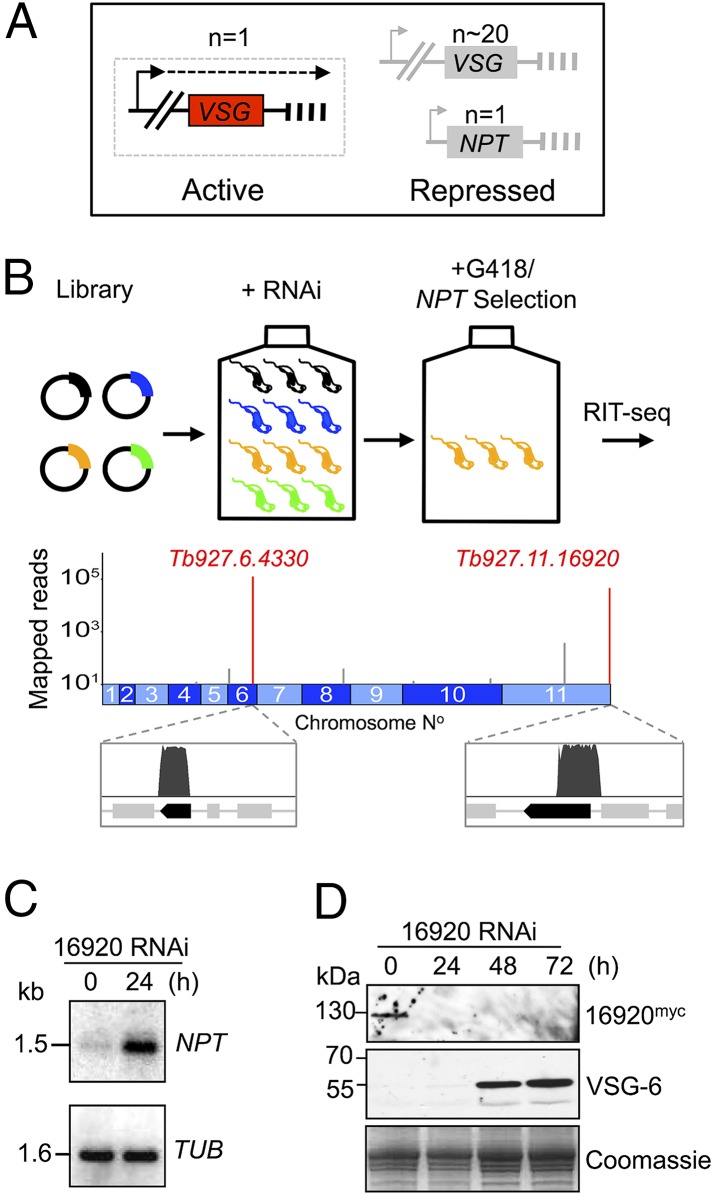
A Genetic Screen Reveals Tb927.11.16920 as a Candidate VSG Exclusion Regulator.
To identify T. brucei genes that control telomere-exclusive gene expression, we assembled an RNA interference (RNAi) library in bloodstream-form trypanosomes with a pol-I–transcribed telomeric reporter. The Neomycin PhosphoTransferase (NPT) reporter, on a telomere-mediated chromosome-fragmentation construct (29), incorporates an rDNA promoter and seeds a de novo telomere, comprising TTAGGG repeats, ∼2 kbp downstream (Fig. 1A). The rDNA promoter can be switched on and off through allelic exclusion when used to replace a native VSG-ES promoter (30) and is subject to repression when located close to a telomere (31). A reporter driven by an rDNA promoter was favored over a reporter driven by a VSG-ES promoter because defects in allelic exclusion were expected to result in a greater increase in NPT-reporter expression using the “stronger” rDNA promoter (32). Because VSG expression is essential in bloodstream-form T. brucei (33), we also reasoned that NPT activation, coupled to VSG silencing during a “telomere-switch,” would fail to yield viable cells, as would knockdowns previously linked to VSG silencing but associated with a severe growth defect following RNAi (SI Appendix, Table S1).
Fig. 1.
A genetic screen reveals a candidate VSG exclusion regulator. (A) The bloodstream-form strain for RNAi screening was based on a repressed rDNA-promoter/NPT-reporter cassette, with the promoter 2 kbp from the telomeric TTAGGG repeats. Arrowheads, pol-I promoters; dashed line, transcription; vertical bars, telomeres. (B) The schematic shows the RNAi screen for loss-of-exclusion (in orange cells), and the genome map indicates hits following RIT-seq (red spikes). Mapped reads are indicated relative to gene hits (dark bars). (C) Tb927.11.16920 knockdown was associated with NPT derepression as assessed by RNA blotting. TUB panel, loading control. (D) Knockdown of myc-epitope–tagged Tb927.11.16920 was associated with VSG-6 derepression as assessed by protein blotting. Coomassie-stained panel, loading control.
The population that emerged from the screen for defects in telomere-exclusive expression was subjected to RNAi target sequencing (RIT-seq), revealing two genes, Tb927.6.4330 and Tb927.11.16920, among ∼7,400 in the genome (Fig. 1B). To determine their impact on VSG exclusion, we assembled pairs of independent RNAi knockdown strains for each gene in cells with an active VSG-2 ES. Upon Tb927.11.16920 knockdown, we observed a moderate growth defect (SI Appendix, Fig. S1A) and derepression of the telomeric NPT reporter used in the screen (Fig. 1C), thereby validating this output from the screen. We confirmed efficient knockdown of myc-epitope–tagged Tb927.11.16920 (Fig. 1D) and saw derepression of silent telomeric VSGs using both protein blotting (Fig. 1D, VSG-6 panel) and microscopy (SI Appendix, Fig. S1B). These findings indicate that the telomeric NPT reporter is subject to the exclusion system operating in T. brucei. Analysis of Tb927.6.4430 supported previous reports of disrupted VSG silencing when telomere structure and/or function are compromised (26, 34); knockdown of this novel telomeric TTAGGG repeat-associated factor (SI Appendix, Fig. S2 A and B) was associated with reporter and VSG derepression (SI Appendix, Fig. S2 C and D), but these phenotypes were substantially weaker than those observed following Tb927.11.16920 knockdown (Fig. 1 C and D). Thus, Tb927.11.16920 emerged as the primary “hit” in our screen for VSG exclusion regulators, and we subsequently refer to this factor as VSG exclusion 1, or VEX1.
Like the majority of protein-coding genes identified in trypanosome genomes, Tb927.11.16920/VEX1 encodes a “hypothetical protein” with no prior functional assignment. Analysis of the predicted peptide sequence revealed a 101.23-kDa protein incorporating a putative SWIM-type zinc finger with a CxCx17CxH signature. Orthologous genes in other parasitic trypanosomatids also encode the zinc-finger motif (SI Appendix, Fig. S3), originally found in SWI2/SNF2 family ATPase, MuDR transposase, and MEK kinase (35), but these hypothetical proteins also lack any prior functional assignment.
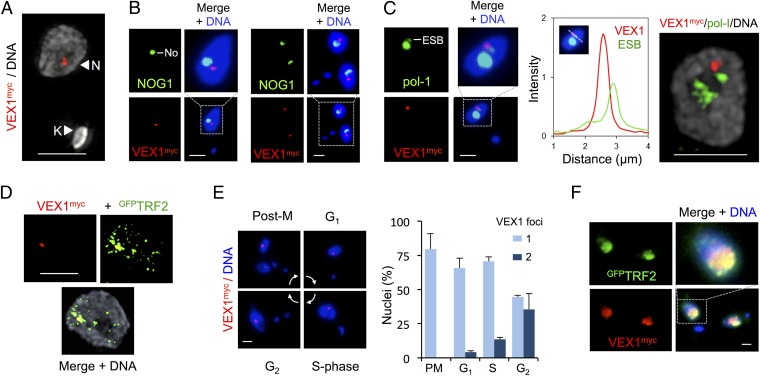
VEX1 Is Sequestered at the Active VSG-ES.
We next examined VEX1 subcellular localization. Epitope-tagged VEX1 was primarily concentrated in a single subnuclear focal compartment in bloodstream-form cells, as revealed by superresolution microscopy (Fig. 2A and Movie S1). The VEX1 compartment was extranucleolar (Fig. 2B) and closely associated, but not coincident, with the pol-I focus (Fig. 2C and Movie S2) that is the site of the single active VSG-ES (10). Because all competent VSG-ESs are telomeric, we also used superresolution microscopy to assess VEX1 localization relative to the telomeric TTAGGG repeat-binding factor TRF2 (34). This revealed punctate nuclear TRF2 staining and VEX1 staining that was coincident with a TRF2 punctum (Fig. 2D). VEX1 foci were observed at all cell-cycle phases, with segregated foci accumulating in G2 in particular, following DNA replication (Fig. 2E). VEX1 foci were no longer detected, however, following a 30-min exposure to the transcription inhibitor actinomycin D.
Fig. 2.
VEX1 associates with the active VSG-ES in bloodstream-form cells. (A) Three-dimensional structured-illumination immunofluorescence microscopy (3D SIM) projection of VEX1myc. N, nucleus; K, kinetoplast (mitochondrial genome). (B) Immunofluorescence microscopy of VEX1myc and a nucleolar (No) marker (NOG1). G1 (Left) and postmitotic (Right) cells are shown. (C) As in B but with a nucleolar plus ESB marker (pol-I). The linear intensity plot shows the distance between the center of the VEX1 focus and the center of the ESB; mean distance is 0.34 ± 0.09 μm (n = 7 G1 nuclei). (Right) A 3D-SIM projection. (D) A 3D-SIM projection of VEX1myc and GFPTRF2. (E) Immunofluorescence microscopy of VEX1myc during the cell cycle; phases are indicated and were inferred from DNA content. Numbers of VEX1 foci per nucleus were quantified at each cell cycle phase and are plotted on the Right. (F) Immunofluorescence microscopy of VEX1myc and GFPTRF2 in insect-stage cells. DNA was counterstained with DAPI. (Scale bars, 2 μm.)
VSG expression is inactivated during differentiation to the insect midgut stage and reactivated in the insect salivary gland (36). We observed a widespread punctate nuclear distribution of VEX1 in insect midgut-stage cells that substantially overlapped with telomeric TRF2 puncta (Fig. 2F). Thus, VEX1 is sequestered at the active VSG-ES in a transcription-dependent and life cycle stage-specific manner. VEX1 redistribution in insect-stage cells may allow VSG-ESs to compete for VEX1 sequestration as VSGs are reactivated in the insect salivary gland (37).
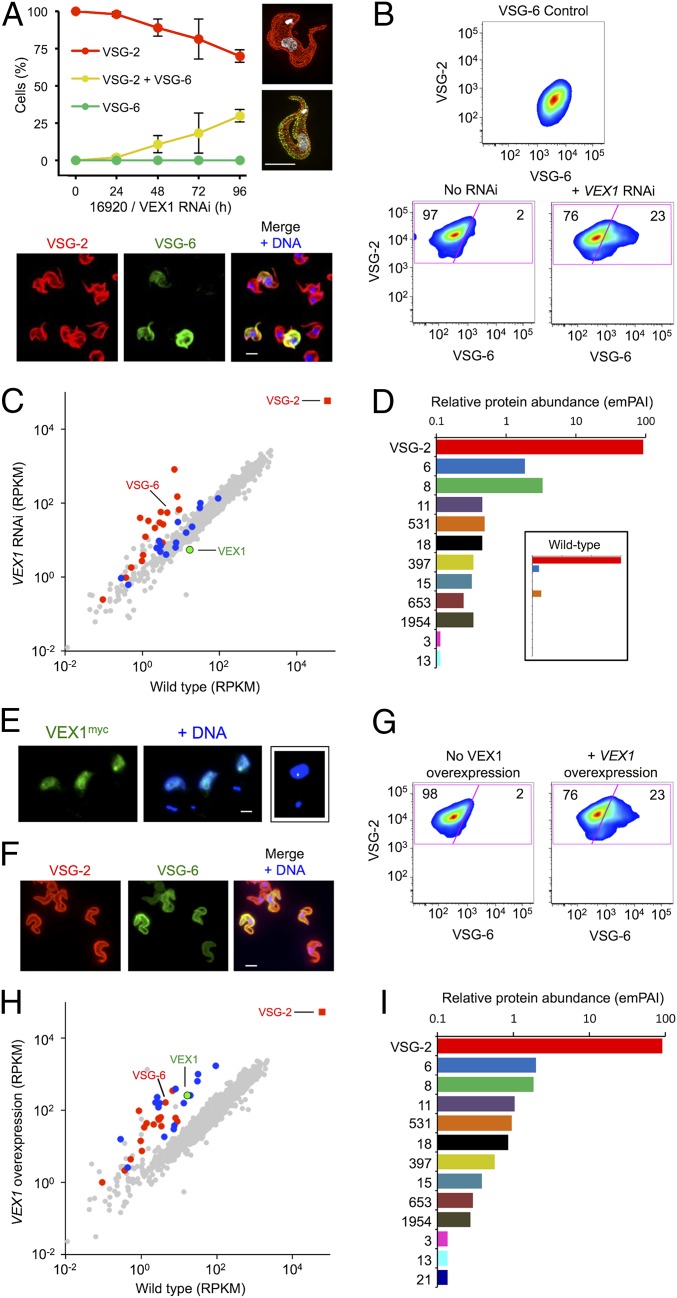
VEX1 Controls Telomeric VSG Exclusion.
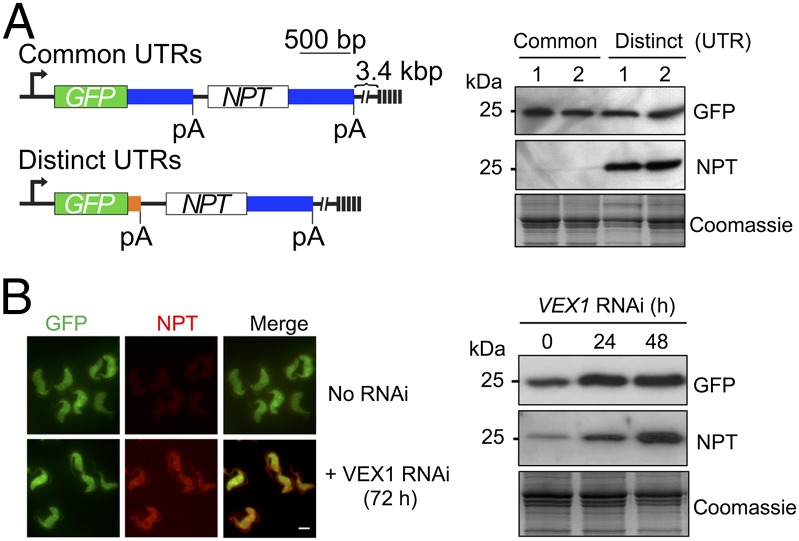
We next used microscopy to examine VSG exclusion over a time course following VEX1 knockdown in bloodstream-form cells. This analysis revealed an accumulation of cells simultaneously expressing VSG-2 and VSG-6 with no evidence for increased switching to VSG-6 (Fig. 3A). Flow cytometry confirmed multi-VSG expression and, again, no evidence for switching (Fig. 3B). To extend these findings, we carried out transcriptome analysis using pairs of wild-type subclones and pairs of independent knockdown strains, achieving >180× average genome coverage in each RNA-seq experiment. Assessment of VSG transcript abundance in wild-type cells revealed a >1,000-fold differential between the active VSG-2 transcript and the sum of all 18 “silent” pol-I promoter-associated VSGs; VSG-2 transcripts represented ∼7% of total mRNA. We also found that VEX1 produced a low-abundance transcript in wild-type cells within the lowest fifth percentile. Upon knockdown, VEX1 was depleted 3.1-fold on average (Fig. 3C and Dataset S1), whereas 19 genes (of ∼7,400) displayed >3-fold and significantly (P < 0.05) increased expression relative to wild type. These included many bloodstream ES-linked VSGs, metacyclic ES-linked VSGs, and genes immediately adjacent to VSGs (Fig. 3C and Dataset S1); metacyclic ES-linked VSGs are transcribed by pol-I in the insect salivary gland (36). Indeed, expression of all 18 silent pol-I promoter-associated VSGs increased >26-fold overall (Dataset S1).
Fig. 3.
VEX1 controls VSG allelic exclusion in bloodstream-form cells. (A) Immunofluorescence microscopy analysis of VSG expression. Cells were stained with α-VSG-2 and α-VSG-6 and counted daily during VEX1-RNAi. The 3D-SIM images show a wild-type control cell and a cell expressing both VSGs. The images below the plot show cells following VEX1-RNAi (72 h). (Scale bars, 5 μm.) (B) Flow-cytometry analysis of VSG expression following VEX1-RNAi (72 h). Numbers indicate percentage of cells in each quadrant. VSG-6 expressers serve as a control. n = 10,000 cells in each case. (C) RNA-seq analysis following VEX1-RNAi (72 h). Values are averages for a pair of independent strains (Dataset S1). Red circles, silent VSGs; red square, active VSG; blue circles, procyclins and procyclin-associated genes. RPKM, reads per kilobase of transcript per million mapped reads. (D) Quantitative mass spectrometry analysis of surface VSGs following VEX1-RNAi (72 h) (SI Appendix, Table S2). (Inset) Wild-type cells for comparison. emPAI, exponentially modified Protein Abundance Index. (E) Immunofluorescence microscopy of overexpressed and ectopic VEX1myc (72 h). (Scale bar, 2 μm.) (Right) Sequestered VEX1 for comparison. (F) Immunofluorescence microscopy analysis of VSG expression following VEX1 overexpression (72 h). (Scale bar, 5 μm.) (G) Flow cytometry following VEX1 overexpression (72 h). Other details are as in B. (H) RNA-seq analysis following VEX1 overexpression (72 h). Other details are as in C. (I) Quantitative mass spectrometry analysis of surface VSGs following VEX1 overexpression (72 h) (SI Appendix, Table S2).
We next released cell-surface VSGs and used quantitative proteomics to examine relative expression (Fig. 3D and SI Appendix, Materials and Methods and Table S2). VSG-2 on wild-type cells displayed a relative abundance of 99.6%; only two significant sequences mapped to other VSGs. On VEX1-knockdown cells, the VSG-2 relative abundance was 91% and 11 silent bloodstream and metacyclic ES-linked VSGs were also detected (Fig. 3D and SI Appendix, Table S2). Thus, VEX1 knockdown allows for silent VSGs to be transcribed, translated, and delivered to the cell surface.
Overexpressed VEX1 Derepresses VSG-ESs and Nontelomeric pol-I Loci.
Although chromatin states may be effectively inherited, establishment of “allele-choice” is not understood. The association between VEX1 and the active VSG-ES, and maintenance of the active VSG-ES following VEX1 knockdown, pointed to a potential role for VEX1 in establishing the active VSG-ES. To explore this hypothesis, we assembled a pair of independent VEX1-overexpressing bloodstream-form strains. We observed a moderate growth defect associated with VEX1 overexpression (SI Appendix, Fig. S4 A and B), and, as predicted, these cells failed to effectively sequester VEX1. Although a VEX1 focus was often visible, we observed an additional dispersed signal through each nuclear compartment (Fig. 3E). Consistent with our hypothesis, when VEX1 was available to access a second telomeric VSG, this VSG was derepressed (SI Appendix, Fig. S4B, VSG-6 panel). Indeed, immunofluorescence microscopy (Fig. 3F) and flow cytometry (Fig. 3G) revealed cells simultaneously expressing both VSGs. In fact, the intensity of the cell-surface VSG-6 signal increased across the entire population of each clone, and clones overexpressing VEX1 lacking a myc tag yielded similarly increased VSG-6 expression (SI Appendix, Fig. S4C).
Transcriptome analysis revealed 65 genes that displayed more than threefold and significantly (P < 0.05) increased expression in VEX1 overexpresser strains relative to wild type. VEX1 mRNA was increased 16-fold on average, and other up-regulated genes included all 18 silent, pol-I promoter-associated VSGs, which increased >21-fold overall (Fig. 3H and Dataset S1). A striking difference compared with VEX1 knockdown was increased expression of 14 procyclin and procyclin-associated transcripts, also >21-fold overall (Fig. 3H, SI Appendix, Fig. S5A, and Dataset S1). These nontelomeric loci are also transcribed by pol-I, but normally produce abundant surface proteins in insect midgut-stage cells (7). Another difference was increased expression of active (Dataset S1) and silent VSG-ES–associated gene (ESAG) transcripts following VEX1 overexpression, in contrast to increased expression of the VSGs only following VEX1 knockdown (SI Appendix, Fig. S5B). Quantitative proteomic analysis of surface VSGs on VEX1-overexpressing cells revealed a VSG-2 abundance index reduced to 91%, with 12 additional bloodstream and metacyclic ES-linked VSGs also detected (Fig. 3I and SI Appendix, Table S2). We conclude that overexpressed VEX1 positively regulates both telomeric and nontelomeric pol-I–transcribed loci.
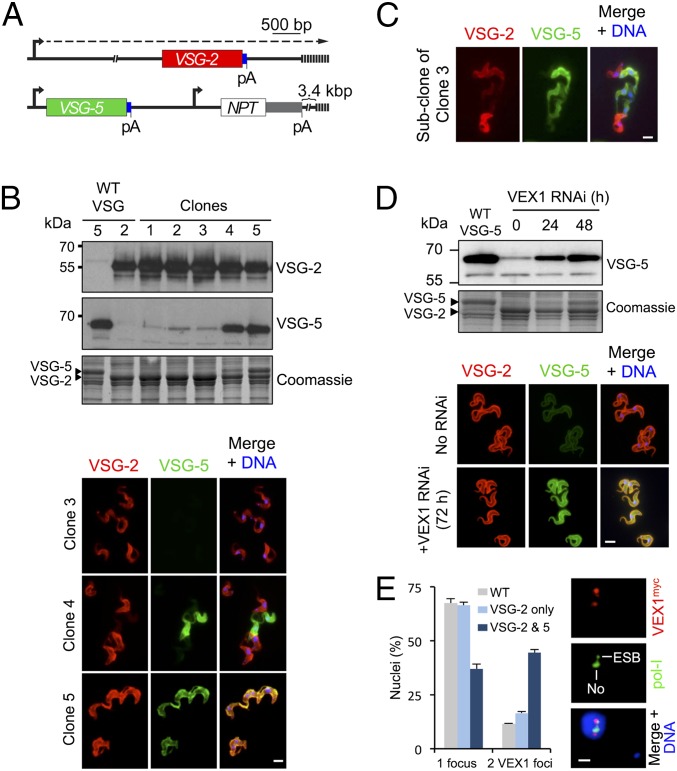
A VSG Reporter Is Subject to VEX1-Dependent Exclusion.
VEX1 was identified due to its ability to repress a reporter on a telomere-mediated chromosome-fragmentation construct (Fig. 1). We reasoned that manipulation of these constructs would allow us to identify the DNA sequences involved. A new construct was assembled that contained a VSG-5 gene with a downstream sequence [including the 3′-untranslated region (3′-UTR)] that is identical to that associated with the native active VSG-2 (Fig. 4A). This construct places a VSG-5–associated rDNA promoter ∼7 kbp from a de novo telomere (Fig. 4A). Consistent with competition between VEX1-mediated positive and negative regulation, we observed variable outcomes using this construct in bloodstream-form cells; all cloned populations continued to express VSG-2, but VSG-5 expression differed. Among 24 independent clones, 20 displayed repressed VSG-5 and -4 are shown (Fig. 4B); 3 displayed uniform repression (>99% of cells, clones 1–3) whereas some cells in one clone displayed VSG-5 expression that interfered with native VSG-2 expression (clone 4). The remaining clones displayed simultaneous expression of both VSGs (>99% of cells) that was stable over many generations and yielded approximately equal quantities of each VSG (Fig. 4B, clone 5, and SI Appendix, Fig. S6).
Fig. 4.
VEX1-dependent communication among recombinant and native VSGs. (A) The schematic shows the VSG-5 reporter in cells expressing native VSG-2. Blue boxes, common VSG 3′-UTRs; pA, poly-adenylation sites; other symbols as in Fig. 1A. (B) The protein blots show five bloodstream-form clones derived using this system. VSG-5– or VSG-2–expressing wild-type (WT) cells serve as controls. The Coomassie-stained panel serves as a loading control and also reveals the major VSGs. The immunofluorescence panels show VSG expression in clones 3–5. (C) Immunofluorescence analysis of VSG expression in a subclone of clone 3 following G418/NPT-selection (see A). (D) The protein blots and immunofluorescence microscopy show VSG-5 expression during VEX1 knockdown. (E) Immunofluorescence microscopy analysis of VEX1myc in clone 5 cells (expressing both VSG-2 and VSG-5). Nuclei with one or two VEX1 foci were quantified relative to wild type and cells equivalent to clone 3 above (expressing VSG-2 only). Immunofluorescence analysis of VEX1myc and pol-I reveals the location of the additional VEX1 focus relative to the ESB and nucleolus (No). The mean distance between the centers of the VEX1 foci is 1.1 ± 0.3 μm (n = 8 G1 nuclei). (Scale bars in B, C, and D, 5 μm; in E, 1 μm.) DNA was counterstained with DAPI.
To determine whether VSG-5 repression was reversible, clone 3 cells were subcloned and selected for increased NPT-reporter expression (Fig. 4A). These populations expressed VSG-5 that interfered with VSG-2 expression (Fig. 4C), indicating that VSG-5 was subject to the exclusion system, being both repressible and able to repress. Also, VEX1 knockdown led to VSG-5 derepression and produced cells simultaneously expressing both VSGs (Fig. 4D). This VEX1-dependent VSG-5 reporter repression was confirmed using a second independent strain.
Simultaneous expression of two VSGs in clone 5 (Fig. 4B) indicated that VSG-5 could escape exclusion in the presence of VEX1. Indeed, simultaneous VSG expression, apparently at distinct telomeres, has been reported before (38). Because VEX1 is concentrated at the active VSG-ES (Fig. 2) and can positively regulate pol-I loci (Fig. 3 E–I), we wondered whether VSG-5 at a de novo telomere might compete for VEX1 and, in some cases, acquire sufficient material to establish a second “privileged” site. As suspected, the proportion of nuclei with two distinct VEX1 foci was increased in clone 5 cells (3.8-fold relative to “wild-type” control; Fig. 4E), and the additional VEX1 focus associated with VSG-5 expression was typically ESB-distal and perinucleolar (Fig. 4E); rDNA loci are also perinucleolar (39). Taken together with analysis of VEX1 overexpressers (Fig. 3H), these findings (Fig. 4 B and E) suggest that access to VEX1 increases pol-I transcription, that increased pol-I transcription can lead to the accumulation of VEX1, or both of the above. This is also consistent with the observation of a “preactive” VSG state when a VSG is in close proximity to the active VSG-ES (21).
VEX1 Mediates a Form of Homology-Dependent Silencing.
A serendipitous observation initially suggested to us that “homologous” DNA sequences might be important for exclusion. We had assembled a bicistronic and telomeric GFP:NPT reporter that fortuitously contained identical sequences downstream of both the GFP and NPT genes (Fig. 5A, Upper map). These sequences contain 3′-UTRs that are included in reporter constructs to guide mRNA polyadenylation and splicing; we did not expect them to be subject to repression or exclusion. However, we observed strong NPT repression when using this construct in bloodstream-form cells (Fig. 5A, Left, common UTR lanes). To determine whether this reflected homology-dependent interference, the T. brucei aldolase sequence downstream of the GFP gene was replaced with a T. brucei tubulin sequence (Fig. 5A, Lower map), which is unrelated to the aldolase sequence but also guides efficient mRNA processing. No NPT repression was observed when using this construct (Fig. 5A, Right, distinct UTR lanes). In cells containing the construct with common aldolase sequences, NPT repression was relieved following VEX1 knockdown (Fig. 5B); a result confirmed using a second independent strain. Thus, reporters with homologous sequences downstream displayed VEX1-dependent repression that no longer operated when homology was removed.
Fig. 5.
VEX1-dependent communication among homologous sequences. (A) The schematic shows reporters with common or distinct 3′-UTRs (blue, aldolase; orange, tubulin). The protein blots show reporter expression for pairs of bloodstream-form clones derived using each construct. Coomassie-stained panel, loading-control. Symbols as in Fig. 4A. (B) The immunofluorescence panels and protein blots show expression of reporters with common 3′-UTRs during VEX1 knockdown. (Scale bar, 5 μm.)
Discussion
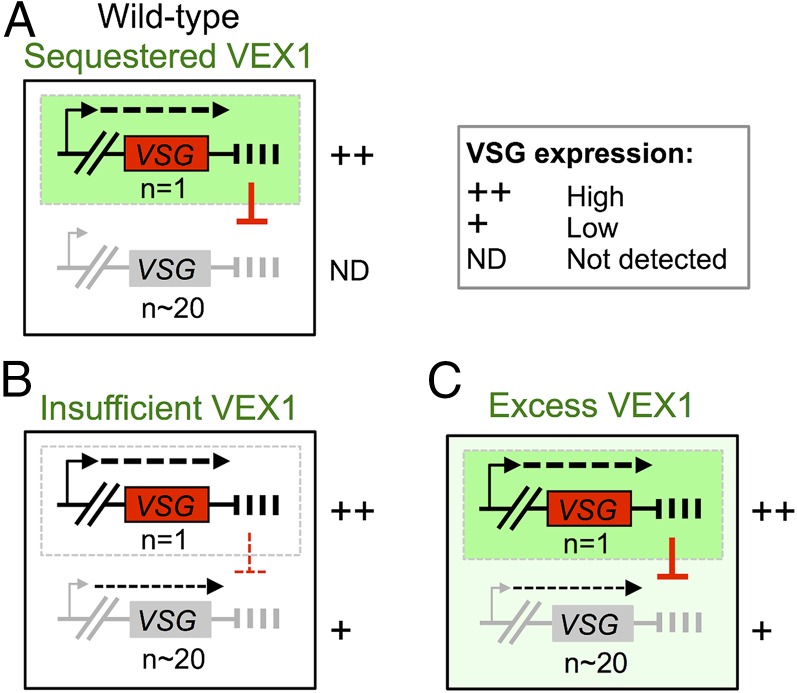
We identified trypanosome VEX1 using a genetic screen for defects in telomere-exclusive gene expression. VEX1 is sequestered at the active VSG-ES and coordinates VSG-positive and -negative regulation to sustain antigenic variation. Based on our findings, we propose a winner-takes-all model for the establishment of allelic exclusion by VEX1 (Fig. 6A). A similar model has been put forward for olfactory receptor gene choice (40) although no factor that displays similar properties to VEX1 has been identified in that system. Our results indicate that an established active VSG-ES is effectively inherited when VEX1 function is disrupted; this also appears to be the case when (telomeric) chromatin is disrupted by other means (22–24, 26, 34). One long-standing question, however, has been why are silent VSG-ESs only partially derepressed when (telomeric) chromatin is disrupted? This is the case even when substantial loss of viability is observed, following RAP1 knockdown, for example (26), and could be explained by failure to associate with sufficient transcription or RNA-processing factors. We suggest that, when VEX1 function is disrupted, directly or indirectly, negative regulation is relaxed but silent VSG-ESs lack VEX1-mediated positive regulation (Fig. 6B). Indeed, VSG-ES promoters appear to be substantially “weaker” than rDNA promoters (32) and may depend upon positive regulation by VEX1. In the case of excess VEX1, access to other VSG-ESs allows positive regulation, but these sites are still subject to negative regulation exerted by the active VSG-ES (Fig. 6C). Thus, we suggest that the “default” level for VSG expression is relatively low and that VEX1 drives the processes that increase expression at one locus and reduce expression elsewhere. We note that although a winner-takes-all mechanism may operate naturally, this can be perturbed either when VEX1 is artificially expressed in excess or, in some cases, when recombinant pol-I transcription units are introduced de novo.
Fig. 6.
A winner-takes-all model for allelic exclusion by VEX1. (A) Wild-type bloodstream-form cells: VEX1 coordinates VSG-positive and -negative regulation. VEX1 (green) is recruited to a single VSG-ES through positive feedback involving pol-I transcription. VEX1 also exerts negative regulation through homology-dependent silencing (red symbol), which enhances differential expression and ensures that the winner takes all. (B) VEX1 knockdown: Insufficient VEX1 leads to relaxed homology-dependent silencing but also precludes further positive regulation. An established active VSG-ES is maintained in the absence of VEX1. (C) VEX1 overexpression: Ectopic VEX1 positively regulates all VSG-ESs, but negative regulation also persists and precludes further derepression.
We previously demonstrated that repression of pol-I–transcribed genes spreads only a short distance from the telomere in the absence of additional VSG-ES sequences (31). We now show that this repression is VEX1-dependent and also that homologous sequences can promote VEX1-dependent repression. Another recent report demonstrated that a VSG, transcribed at a chromosome-internal site by T7-phage polymerase, transiently silenced the native VSG (25). This same report demonstrated repression spreading along the VSG-ES in a DOT1B, histone methyltransferase-dependent manner (25). Spreading of a repressed domain may also explain why we see derepression of both bicistronic GFP and NPT reporters during VEX1 knockdown (Fig. 5B). In these experiments, sequences that are subject to repression also serve as repressive sequences when they are transcribed. VSG transcripts (SI Appendix, Fig. S7) and other VSG-associated sequences display a high degree of homology, and telomeric TTAGGG-repeat transcripts are also present in T. brucei (41), suggesting that transcripts could be involved. Further work will be required to delineate the mechanism, which, although not involving Argonaute1-based RNAi (42), could involve alternative RNA-based repression, as reported in other cell types (43). Thus, we tentatively suggest that VEX1-dependent VSG silencing is initiated by homologous transcripts and is then propagated along the chromatin fiber in a DOT1B-dependent manner.
In summary, we report the identification of VEX1, an allelic exclusion regulator that sustains antigenic variation in trypanosomes. We describe a winner-takes-all model whereby VEX1 sequestration establishes a single active VSG-ES that then mediates homology-dependent silencing at other VSG-ESs. Similar mechanisms involving positive and negative regulation, coordinated by sequestered regulators, could explain allelic exclusion in other cell types.
Materials and Methods
For details of T. brucei growth and manipulation, plasmids, nucleic acid analysis, Western blotting, microscopy, flow cytometry, and quantitative mass spectrometry, see SI Appendix, Materials and Methods.
T. brucei, Lister 427, MITat1.2 (VSG-2, aka VSG221), 1.5 (VSG-5, aka VSG118), and 1.6 (VSG-6, aka VSG121) cells were used for this study. RIT-seq was carried out on a MiSeq platform (Illumina) at The Beijing Genome Institute (BGI). RNA-seq was carried out on a HiSeq platform (Illumina) at the University of Dundee or at BGI. Three-dimensional structured illumination microscopy was carried out using a superresolution OMX Blaze system (GE Healthcare). Quantitative mass spectrometry was carried out using an Ultimate 3000 RSLCnano system (Thermo Scientific) coupled to a Linear Trap Quadropole OrbiTrap Velos Pro (Thermo Scientific).
Supplementary Material
Acknowledgments
We thank R. Clark of the Flow Cytometry and Cell Sorting Facility, which is supported by the Wellcome Trust (097418/Z/11/Z); M. Posch and B. Balagopal for assistance with superresolution microscopy; M. Febrer for assistance with Illumina sequencing; D. Lamont and K. Beattie of the Fingerprints Proteomics Facility for assistance with quantitative proteomics; A. Mehlert, L. Guther, and M. Ferguson for assistance and advice on VSG proteomics; J. Morris and P. Englund for the RNAi plasmid library; B. Dujon for the I-SceI gene; and M. Ferguson, J. Wright, and M. Field for comments on the manuscript. The work was funded by Project Grant 93010/B/10/Z (to D.H.); Investigator Award 100320/Z/12/Z (to D.H.); and a Strategic Award supporting Biological Chemistry & Drug Discovery (100476/Z/12/Z), all from The Wellcome Trust. Use of the superresolution microscope was funded by a Medical Research Council Next Generation Optical Microscopy Award (MR/K015869/1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RIT-seq and RNA-seq sequence data reported in this paper have been deposited in the European Nucleotide Archive, www.ebi.ac.uk/ena (accession nos. PRJEB8746 and PRJEB8747, respectively).
See Commentary on page 7017.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600344113/-/DCSupplemental.
References
- 1.Horn D. Antigenic variation in African trypanosomes. Mol Biochem Parasitol. 2014;195(2):123–129. doi: 10.1016/j.molbiopara.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guizetti J, Scherf A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol. 2013;15(5):718–726. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monahan K, Lomvardas S. Monoallelic expression of olfactory receptors. Annu Rev Cell Dev Biol. 2015;31:721–740. doi: 10.1146/annurev-cellbio-100814-125308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertz-Fowler C, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008;3(10):e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Lange T, Borst P. Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature. 1982;299(5882):451–453. doi: 10.1038/299451a0. [DOI] [PubMed] [Google Scholar]
- 6.Zomerdijk JC, et al. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 1990;9(9):2791–2801. doi: 10.1002/j.1460-2075.1990.tb07467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Günzl A, et al. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell. 2003;2(3):542–551. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109(1):5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- 9.Chaves I, et al. Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei. Proc Natl Acad Sci USA. 1998;95(21):12328–12333. doi: 10.1073/pnas.95.21.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414(6865):759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 11.Figueiredo LM, Cross GA. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot Cell. 2010;9(1):148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanne TM, Rudenko G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot Cell. 2010;9(1):136–147. doi: 10.1128/EC.00281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navarro M, Peñate X, Landeira D. Nuclear architecture underlying gene expression in Trypanosoma brucei. Trends Microbiol. 2007;15(6):263–270. doi: 10.1016/j.tim.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Narayanan MS, Rudenko G. TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res. 2013;41(5):2981–2992. doi: 10.1093/nar/gks1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aresta-Branco F, Pimenta S, Figueiredo LM. A transcription-independent epigenetic mechanism is associated with antigenic switching in Trypanosoma brucei. Nucleic Acids Res. 2016;44(7):3131–3146. doi: 10.1093/nar/gkv1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López-Farfán D, Bart JM, Rojas-Barros DI, Navarro M. SUMOylation by the E3 ligase TbSIZ1/PIAS1 positively regulates VSG expression in Trypanosoma brucei. PLoS Pathog. 2014;10(12):e1004545. doi: 10.1371/journal.ppat.1004545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassem A, Pays E, Vanhamme L. Transcription is initiated on silent variant surface glycoprotein expression sites despite monoallelic expression in Trypanosoma brucei. Proc Natl Acad Sci USA. 2014;111(24):8943–8948. doi: 10.1073/pnas.1404873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross GA. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71(3):393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- 19.Vickerman K. On the surface coat and flagellar adhesion in trypanosomes. J Cell Sci. 1969;5(1):163–193. doi: 10.1242/jcs.5.1.163. [DOI] [PubMed] [Google Scholar]
- 20.Doyle JJ, Hirumi H, Hirumi K, Lupton EN, Cross GA. Antigenic variation in clones of animal-infective Trypanosoma brucei derived and maintained in vitro. Parasitology. 1980;80(2):359–369. doi: 10.1017/s0031182000000810. [DOI] [PubMed] [Google Scholar]
- 21.Chaves I, Rudenko G, Dirks-Mulder A, Cross M, Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18(17):4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds D, et al. Histone H3 variant regulates RNA polymerase II transcription termination and dual strand transcription of siRNA loci in Trypanosoma brucei. PLoS Genet. 2016;12(1):e1005758. doi: 10.1371/journal.pgen.1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz D, Zaringhalam M, Papavasiliou FN, Kim HS. Base J and H3.V regulate transcriptional termination in Trypanosoma brucei. PLoS Genet. 2016;12(1):e1005762. doi: 10.1371/journal.pgen.1005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanne T, et al. Identification of the ISWI chromatin remodeling complex of the early branching eukaryote Trypanosoma brucei. J Biol Chem. 2015;290(45):26954–26967. doi: 10.1074/jbc.M115.679019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batram C, Jones NG, Janzen CJ, Markert SM, Engstler M. Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. eLife. 2014;3:e02324. doi: 10.7554/eLife.02324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137(1):99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landeira D, Bart JM, Van Tyne D, Navarro M. Cohesin regulates VSG monoallelic expression in trypanosomes. J Cell Biol. 2009;186(2):243–254. doi: 10.1083/jcb.200902119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cestari I, Stuart K. Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc Natl Acad Sci USA. 2015;112(21):E2803–E2812. doi: 10.1073/pnas.1501206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horn D, Spence C, Ingram AK. Telomere maintenance and length regulation in Trypanosoma brucei. EMBO J. 2000;19(10):2332–2339. doi: 10.1093/emboj/19.10.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudenko G, Blundell PA, Dirks-Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83(4):547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 31.Glover L, Horn D. Repression of polymerase I-mediated gene expression at Trypanosoma brucei telomeres. EMBO Rep. 2006;7(1):93–99. doi: 10.1038/sj.embor.7400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horn D, Cross GA. Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. EMBO J. 1997;16(24):7422–7431. doi: 10.1093/emboj/16.24.7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheader K, et al. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc Natl Acad Sci USA. 2005;102(24):8716–8721. doi: 10.1073/pnas.0501886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jehi SE, et al. Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity. Nucleic Acids Res. 2014;42(20):12899–12911. doi: 10.1093/nar/gku942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarova KS, Aravind L, Koonin EV. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem Sci. 2002;27(8):384–386. doi: 10.1016/s0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- 36.Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338(6112):1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tetley L, Turner CM, Barry JD, Crowe JS, Vickerman K. Onset of expression of the variant surface glycoproteins of Trypanosoma brucei in the tsetse fly studied using immunoelectron microscopy. J Cell Sci. 1987;87(Pt 2):363–372. doi: 10.1242/jcs.87.2.363. [DOI] [PubMed] [Google Scholar]
- 38.Baltz T, et al. Stable expression of two variable surface glycoproteins by cloned Trypanosoma equiperdum. Nature. 1986;319(6054):602–604. doi: 10.1038/319602a0. [DOI] [PubMed] [Google Scholar]
- 39.Landeira D, Navarro M. Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J Cell Biol. 2007;176(2):133–139. doi: 10.1083/jcb.200607174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanchate NK, et al. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350(6265):1251–1255. doi: 10.1126/science.aad2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudenko G, Van der Ploeg LH. Transcription of telomere repeats in protozoa. EMBO J. 1989;8(9):2633–2638. doi: 10.1002/j.1460-2075.1989.tb08403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janzen CJ, et al. Expression site silencing and life-cycle progression appear normal in Argonaute1-deficient Trypanosoma brucei. Mol Biochem Parasitol. 2006;149(1):102–107. doi: 10.1016/j.molbiopara.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16(2):71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.