Significance
The plasticity of Polycomb repressive complex 2 (PRC2) in the context of tumorigenesis has remained a subject of contention. Here we demonstrate that the equilibrium between the oncogenic and tumor-suppressive activity of PRC2 in promoting breast cancer invasion is tightly regulated by hypoxia-inducible factor 1-α. PRC2 acts as a tumor-suppressor barrier to the hypoxia-driven invasion pathway, and the impaired PRC2 activity upon hypoxia promotes a chromatin switch at proinvasion matrix metalloproteinase gene loci. The study fundamentally changed our understanding of the role of PRC2 in breast cancer and also identified a previously unidentified function of enhancer of zeste 2 to complex with Forkhead box M1 to promote cancer invasion.
Keywords: PRC2, breast cancer, hypoxia, EZH2, FOXM1
Abstract
Despite the established oncogenic function of Polycomb repressive complex 2 (PRC2) in human cancers, its role as a tumor suppressor is also evident; however, the mechanism underlying the regulation of the paradoxical functions of PRC2 in tumorigenesis is poorly understood. Here we show that hypoxia-inducible factor 1, α-subunit (HIFI-α) is a crucial modulator of PRC2 and enhancer of zeste 2 (EZH2) function in breast cancer. Interrogating the genomic expression of breast cancer indicates high HIF1A activity correlated with high EZH2 expression but low PRC2 activity in triple-negative breast cancer compared with other cancer subtypes. In the absence of HIFIA activation, PRC2 represses the expression of matrix metalloproteinase genes (MMPs) and invasion, whereas a discrete Ezh2 complexed with Forkhead box M1 (FoxM1) acts to promote the expression of MMPs. HIF1-α induction upon hypoxia results in PRC2 inactivation by selective suppression of the expression of suppressor of zeste 12 protein homolog (SUZ12) and embryonic ectoderm development (EED), leading to a functional switch toward Ezh2/FoxM1-dependent induction of the expression of MMPs and invasion. Our study suggests a tumor-suppressive function of PRC2, which is restricted by HIF1-α, and an oncogenic function of Ezh2, which cooperates with FoxM1 to promote invasion in triple-negative breast cancer.
Breast cancer is a heterogeneous disease with diverse morphological features, molecular signaling, metastasis patterns, and clinical outcomes. Global gene-expression analyses attempting to deconstruct the biological heterogeneity in breast cancer have identified four distinct molecular intrinsic subtypes: luminal A, luminal B, HER2+ (human EGF receptor 2+), and basal-like (1–3). The defining factor underlying the biological heterogeneity of these breast cancer subtypes appears to be the subtype-specific transcriptional program (1, 4), emphasizing the distinct oncogenic driver events that may account for their differences in clinical behavior, with certain subtypes demonstrating better survival outcome than others (5). Among these breast cancer subtypes, triple-negative breast cancer (TNBC), which is defined by the lack of estrogen, progesterone, and Her2 receptors and is closely related to the basal-like subtype, is highly aggressive and has the worst clinical outcome (2, 3, 5).
The histone methyltransferase Ezh2 (enhancer of zeste 2) is the catalytic subunit of PRC2 (Polycomb repressive complex 2), which facilitates repression of its target genes via trimethylation of lysine residue 27 on histone 3 (H3K27me3) (6). In multiple malignancies including breast cancer, the up-regulation of Ezh2 is positively correlated with tumor grade, metastasis propensity, and poor survival rate (7). The oncogenic function of Ezh2 has been linked to both PRC2-dependent and PRC2-independent activities (8–13). Paradoxically, a context-dependent tumor-suppressive function of Ezh2 or PRC2 also has been reported in several malignancies (14–18). Moreover, high-grade breast, ovarian, and pancreatic cancers have been found to harbor low global H3K27me3, which is correlated with increased recurrence and poor survival (19–21). Particularly in breast cancer, EZH2 and H3K27me3 levels are found to be not correlated across different subtypes, with higher expression of EZH2 in basal-like/TNBC and HER2+ tumors, and high H3K27me3 level in luminal A, luminal B, and normal-like tumors (19, 22). Consequently, a high EZH2 expression is associated with poor disease outcome (19, 20, 23), and a high H3K27me3 level is associated with better outcome (19, 20, 22). Thus, the oncogenic function of Ezh2 in TNBC is not well coupled with the H3K27me3 level; instead, it might be more connected to its nonepigenetic silencing effect. Indeed, discrete functions of Ezh2, independent of PRC2, have been found to regulate NF-κB (8) and Notch pathways positively in TNBC (13). Furthermore, the inverse correlation between EZH2 and H3K27me3 levels seen in TNBC seems to indicate an impaired PRC2 activity in TNBC. Consistent with the clinical observation, a recent study has shown that deficient Ezh2/PRC2 activity is essential for TNBC tumorigenesis (17). Despite these findings in breast cancer, particularly in TNBC, the mechanism underlying the regulation of Ezh2 in relation to PRC2 activity or non-PRC2 activity is poorly understood.
In this study, we sought to address this gap in knowledge. By interrogating the transcriptional network and coordinated expression events in breast cancer, we identified a molecular mechanism by which PRC2 activity is restricted in TNBC. We discovered that HIF1-α (Hypoxia-inducible factor 1-α), which is highly activated in TNBC, is a crucial inhibitor of PRC2 activity. We also found that Ezh2 interacts with FoxM1 (Forkhead box M1), independent of PRC2, to promote invasion and the expression of MMP (matrix metalloproteinase) genes (hereafter, MMPs). Strikingly, PRC2 and the Ezh2/FoxM1 complex co-occupy the same MMP promoters, where they act antagonistically in regulating expression of MMPs. Upon hypoxia, HIF1-α induction underlies a functional switch from PRC2-mediated gene suppression to the Ezh2/FoxM1-mediated induction of MMP expression.
Results
Loss of PRC2-Mediated Gene Expression Is Accompanied by Up-Regulation of EZH2, HIF1A, and FOXM1 in TNBC.
Previous integrative genomic analyses have implicated a number of transcriptional networks in breast cancer, among which several transcription factors such as the HIF1-α– and FoxM1-regulatory pathways have been found to be particularly enriched in TNBC (4, 24). In addition, HIF1-α has been reported to bind to the promoters of EZH2 (25) and FOXM1 (26) to activate their expression, and all have been implicated in breast cancer invasion and metastasis (27–29). These findings suggest a possible functional convergence among these invasive drivers in TNBC progression.
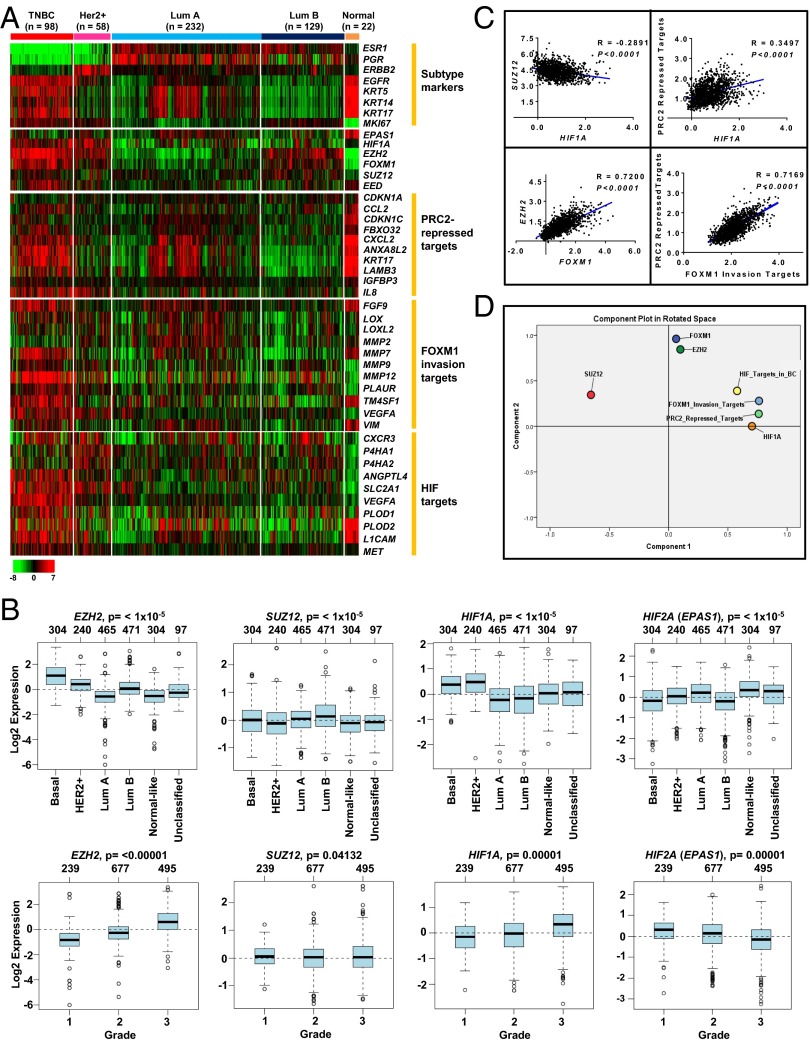
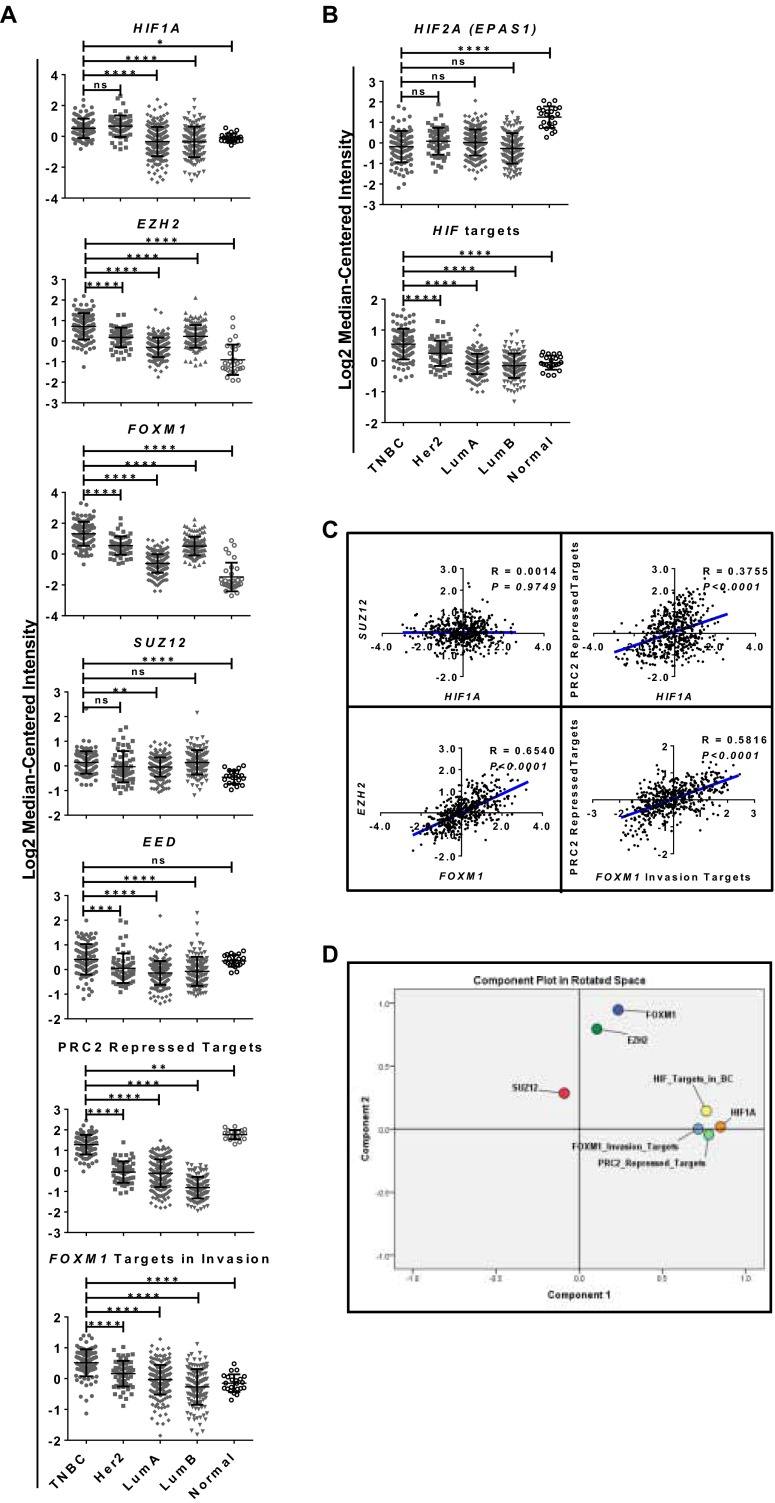
To uncover a potential interaction among the invasion-associated regulators HIF1-α, Ezh2/PRC2, and FoxM1 in breast cancer, we interrogated the gene-expression data of breast cancer in The Cancer Genome Atlas (TCGA) and examined their expression patterns in different subtypes of breast cancer together with their respective target gene sets, as reported previously (Fig. 1A) (30–32). Remarkably, gene-clustering analysis showed that HIF1A, FOXM1, and EZH2 expression were highly enriched in TNBC as compared with other subtypes (Fig. 1A and Fig. S1A). This result is consistent with the highly invasive nature of TNBC. Of significant interest, despite the higher expression of EZH2 in TNBC, the expression of another major PRC2 component, suppressor of zeste 12 protein homolog (SUZ12), did not show concomitant high expression in TNBC as compared with other subtypes of breast cancer (Fig. S1A). This observation was further validated independently using GOBO online analysis (co.bmc.lu.se/gobo) (33), in which, despite the higher expression of EZH2 in TNBC, SUZ12 expression was found to be higher in the luminal B breast cancer subtype but not in TNBC (Fig. 1B, Upper). Moreover, the concerted expression of EZH2 with progressive induction in breast tumors from grade 1 to grade 3 was not observed for SUZ12 (Fig. 1B, Lower). Consistently, PRC2-repressed target genes were de-repressed in TNBC compared with other subtypes, suggesting that PRC2 repressive activity is impaired in TNBC (Fig. 1A and Fig. S1A). In contrast, repressive PRC2 activity was robust in the luminal B subtype, which exhibited the lowest expression of PRC2-repressed targets. Of note, high expression of HIF target genes was found exclusively in TNBC and was correlated with the expression of HIF1A but not HIF2A (EPAS1) (Fig. 1A and Fig. S1A). Furthermore, HIF1A expression showed progressive induction in breast tumors from grade 1 to grade 3, and HIF2A (EPAS1) expression showed the opposite (Fig. 1B); this result is consistent with the antagonistic relationship between HIF1-α and HIF2-α reported previously in breast cancer (34). These observations indicate that the differential regulation of Ezh2 and PRC2 function in different subtypes of breast cancer might be associated with HIF1-α and that the up-regulation of Ezh2 in TNBC may implicate a PRC2-independent function, supporting previously reported non-PRC2 activity of Ezh2 in TNBC (8, 13).
Fig. 1.
Expression analysis of PRC2 components in relation to HIF1A, FOXM1, and their targets in breast cancer subtypes. (A) Heat map showing the indicated gene expression in different subtypes of breast cancer using the TCGA dataset. The scale bar represents the absolute fold change. (B) Analysis of EZH2 and SUZ12 expression in indicated breast cancer subtypes and grades using GOBO online analysis. (C) Pearson’s correlation calculation showing multiple comparisons of transcript levels of HIF1A, EZH2, FOXM1, SUZ12, PRC2-repressed targets, and FOXM1 invasion targets in all tumors using the Curtis dataset (24) downloaded from ONCOMINE. Pearson’s correlation coefficients and P values are as indicated. (D) PCA analysis showing clustering of the differential transcript levels of HIF1A, EZH2, FOXM1, SUZ12, PRC2-repressed targets, FOXM1 invasion targets, and HIF targets in all tumors using the Curtis dataset (24).
Fig. S1.
(A) Scatter plots of transcript levels of HIF1A, EZH2, FOXM1, PRC2 members, PRC2-repressed targets, and FOXM1 targets in invasion from the heat map shown in Fig. 1A. Quantification of the transcript levels was determined as log median-centered intensity. A one-way ANOVA with Bonferroni posttest was used for statistical analysis. (B) Scatter plots of transcript levels of HIF2A (EPAS1) and HIF targets specific for breast cancer from the heat map shown in Fig. 1A. Quantification of the transcript levels was determined as log median-centered intensity. A one-way ANOVA with Bonferroni posttest was used for statistical analysis. (C) Pearson’s correlation calculation showing multiple comparisons of transcript levels of HIF1A, EZH2, FOXM1, SUZ12, PRC2-repressed targets, and FOXM1 invasion targets using the TCGA dataset downloaded from ONCOMINE. Pearson’s correlation coefficient and P value are as indicated. (D) PCA analysis showing clustering of the differential transcript levels of HIF1A, EZH2, FOXM1, SUZ12, PRC2-repressed targets, FOXM1 invasion targets, and HIF targets. All data represent mean ± SD; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
To establish further a possible coregulation of HIF1-α, Ezh2, and FoxM1 and their relationships with repressive PRC2 activity, we next sought to evaluate correlation analysis within these regulators. For this purpose, we calculated Pearson’s correlation (Fig. 1C and Fig. S1C) and principal component analysis (PCA) (Fig. 1D and Fig. S1D) in the TCGA dataset and in another larger, independent dataset from Curtis et al., which contains 2,000 breast tumor samples (24). As shown in Fig. 1 C and D and Fig. S1 C and D, these analyses revealed several interesting findings: (i) HIF1A showed either a negative or no correlation with SUZ12 but a positive correlation with PRC2-repressed targets, indicating a reverse relationship between HIF1A and repressive PRC2 activity; (ii) EZH2 and FOXM1 expression showed a strong positive correlation in both breast cancer datasets, suggesting a potential coordinated coregulation between these two regulators; (iii) FOXM1 invasive targets showed a strong positive correlation with PRC2-repressed targets, indicating that reduced repressive PRC2 activity is correlated with higher invasive activity of FoxM1.
Of interest, there are several well-documented reports on the inhibitory role of transcription factor HIF-1 during cancer progression (35–37). Taken together, our findings raise a hypothesis in which the impaired repressive PRC2 activity in TNBC, as indicated by the abundant expression of PRC2-repressed targets, may be attributed to high HIF1-α activity, whereas HIF1-α, Ezh2, and FoxM1 are positively coregulated to promote invasion.
HIF1-α Activation by Hypoxia Results in Inhibition of PRC2 Activity.
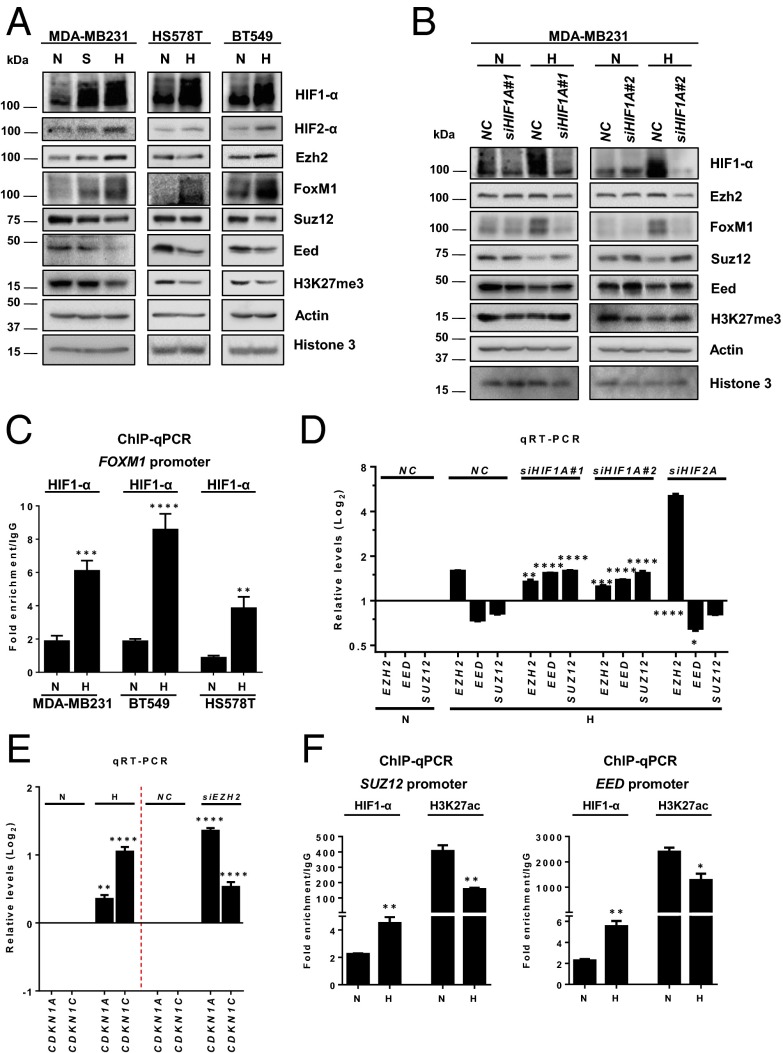
Having shown that an impairment of repressive PRC2 activity is correlated with HIF1A in TNBC, we sought to validate experimentally the functional impact of HIF1-α on the repressive PRC2 activity. To this end, MDA-MB231 cells were subjected to hypoxia or serum-starvation growth conditions; the latter condition is also known to activate HIF1-α (38). Cells that were serum starved or exposed to hypoxia for 48 h exhibited increased HIF1-α and HIF2-α proteins with concurrent up-regulation of FoxM1, which was particularly strong under the hypoxic condition (Fig. 2A, Left). In addition, we observed marked down-regulation of Eed (Embryonic ectoderm development), Suz12, and H3K27me3 in cells in which HIF1-α was induced by hypoxia and to a lesser extent in cells treated with serum starvation (Fig. 2A). The inhibitory effect of hypoxia on PRC2 activity was not restricted to MDA-MB231 cells but also was seen in other TNBC cell lines, e.g., HS578T and BT549 (Fig. 2A). In comparison, we did not see a consistent inactivation of PRC2 in luminal A and luminal B cell lines (Fig. S2), suggesting that hypoxia-induced inhibition of PRC2 is more relevant to TNBC. We further showed that this effect was HIF1-α dependent, because knockdown of HIF1A with two independent siRNA sequences restored the protein expression of Suz12, Eed, and H3K27me3 but abolished FoxM1 induction (Fig. 2B). Consistently with the HIF1-α–mediated induction of FoxM1, we also observed recruitment of HIF1-α on the FOXM1 promoter in multiple TNBC cell lines treated with hypoxia (Fig. 2C). Subsequent investigation into the transcript levels of PRC2 components revealed that hypoxia specifically induced EZH2 mRNA expression and, paradoxically, repressed SUZ12 and EED mRNA expression; this repression was reversed by HIF1A knockdown (Fig. 2D). In contrast, knockdown of HIF2A failed to rescue the hypoxia-induced repression of SUZ12 and EED and even further up-regulated EZH2 mRNA, further validating the role of HIF1-α, rather than HIF2-α, in repressing PRC2. As a functional readout of repressive PRC2 activity, we showed that hypoxia induced the expression of two known PRC2-repressed target genes, CDKN1A (encoding p21) and CDKN1C (encoding p57) (31, 39, 40), recapitulating the effect of EZH2 knockdown (Fig. 2E). Together, these findings suggest a previously unidentified mechanism whereby HIF1-α activation leads to PRC2 inactivation by selectively suppressing the expression of SUZ12 and EED but not EZH2.
Fig. 2.
Hypoxia-induced HIF1-α inhibits PRC2 activity. (A) Immunoblot analysis of the indicated proteins in the indicated cell lines cultured in normoxia (N), serum starvation (S), or hypoxia (H, 4% O2) for 48 h. (B) Immunoblot analysis of the indicated proteins in MDA-MB231 cells treated with control or two HIF1A siRNAs in normoxia or hypoxia. (C) ChIP-qPCR analysis of HIF1-α occupancy on the FOXM1 promoter in the indicated cell lines in normoxia or hypoxia for 48 h. Quantification of enrichment was represented as fold-enrichment over IgG control. A two-tailed unpaired Student’s t test was used for statistical analysis. (D) qRT-PCR analysis of the indicated genes in MDA-MB231 cells treated with control, HIF1A, or HIF2A siRNA in normoxia or hypoxia. Relative gene-expression levels normalized to respective levels in normoxia are shown. A one-way ANOVA with Bonferroni posttest was used for statistical analysis to compare the gene knockdown effect with the nonspecific control (NC) knockdown. (E) qRT-PCR analysis for CDKN1A and CDKN1C in MDA-MB231 cells in the indicated conditions. The gene expression was normalized, and statistical analysis was performed as above. (F) ChIP-qPCR analysis of HIF1-α or H3K27acetylation occupancy on SUZ12 or EED promoters in MDA-MB231 cells in the indicated conditions. Quantification of enrichment was represented as fold-enrichment relative to IgG. A two-tailed unpaired Student’s t test was used for statistical analysis. All data represent mean ± SEM; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Fig. S2.
Immunoblots analysis of the indicated proteins in the indicated cell lines maintained in normoxia (N) or hypoxia (H) for 48 h.
The transcription factor HIF1-α has been shown previously to have both transcriptional activator and transcriptional repressor activities. We next investigated if HIF1-α represses the expression of SUZ12 and EED through direct binding to their respective promoters. As shown in Fig. 2F, HIF1-α binding was detected at the proximal promoters of SUZ12 and EED and was further enriched upon hypoxia treatment. In addition, we also observed reduced enrichment of the active H3K27 acetylation marker at both promoters upon hypoxia. Taken together, these findings demonstrate that activation of HIF1-α by hypoxia leads to transcriptional inactivation of PRC2 in TNBC.
Reciprocal Coregulation of EZH2 and FOXM1 Promotes Invasion Independently of PRC2.
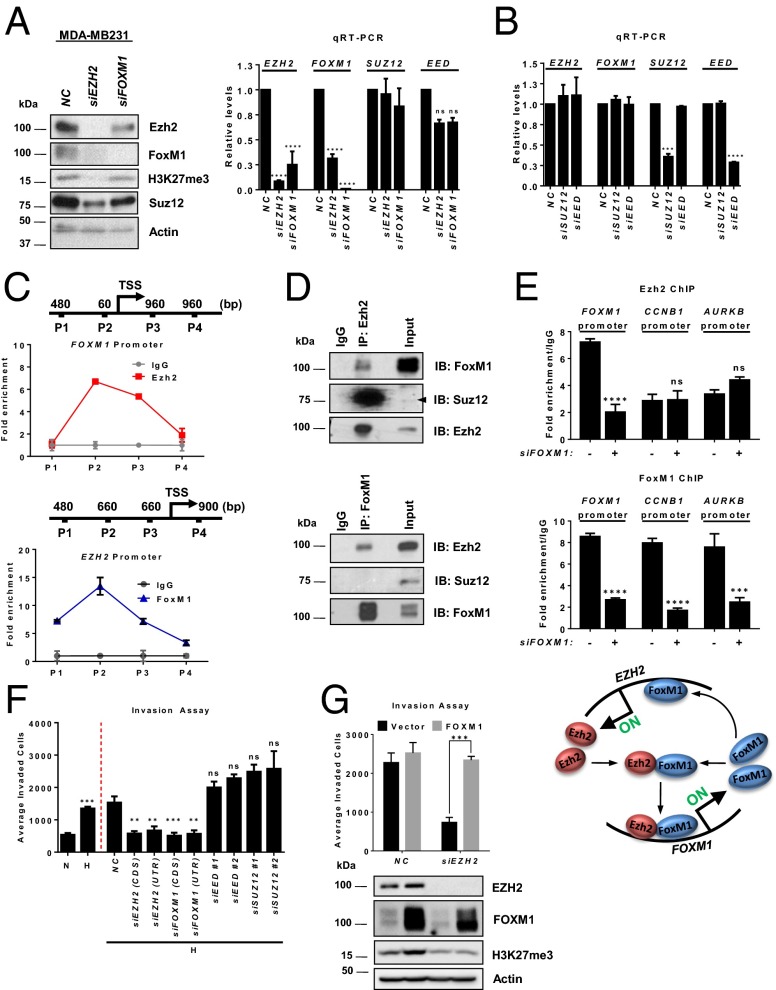
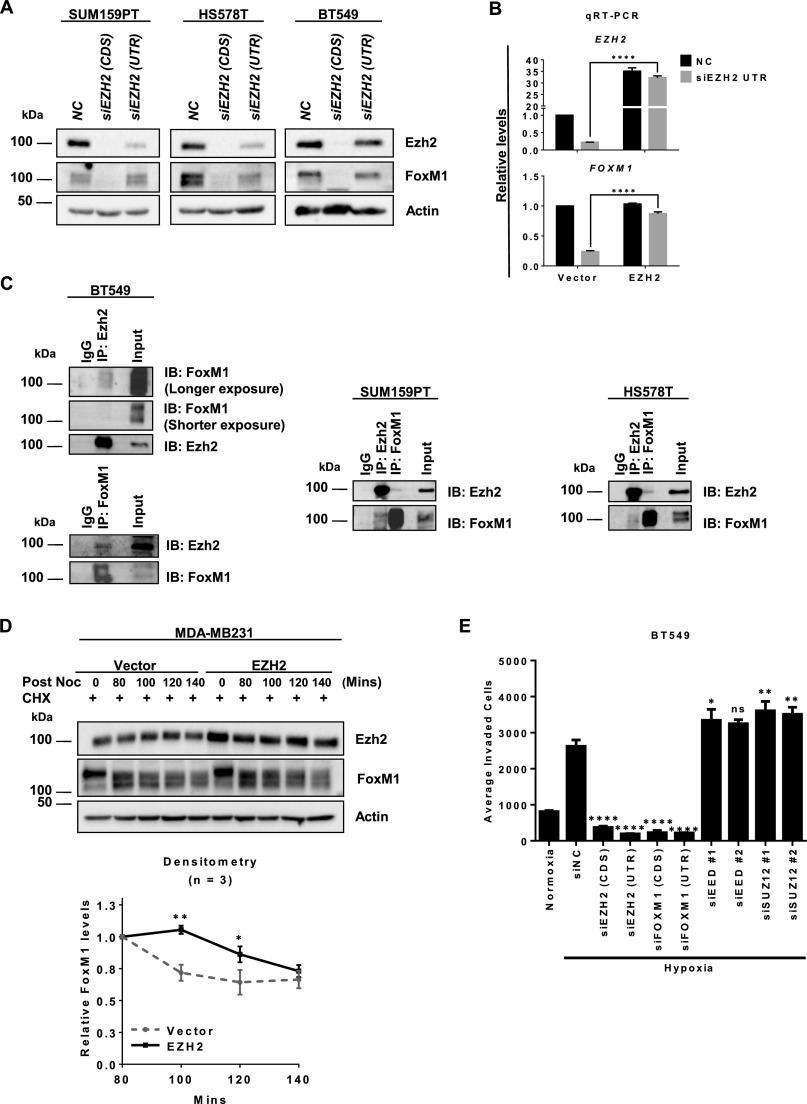
Both Ezh2 and FoxM1 have been implicated in breast cancer progression by regulating the cell cycle, invasion, and metastasis (23, 32, 41–44). Given the well-coordinated expression of EZH2 and FOXM1 in breast cancer as seen in Fig. 1 A and C and Fig. S1 A and C, we asked whether Ezh2 and FoxM1 coregulate each other. Remarkably, EZH2 knockdown efficiently ablated FOXM1 expression at both the transcript and protein levels, and vice versa, in multiple TNBC cell lines including MDA-MB231, SUM159PT, HS578T, and BT549 (Fig. 3A and Fig. S3A), but it did not affect the expression of SUZ12 or EED (Fig. 3A, Right). Moreover, in contrast to EZH2 knockdown, EED or SUZ12 knockdown had no effect on the expression of EZH2 and FOXM1 (Fig. 3B). The specificity of EZH2 siRNA was further validated by using another siRNA sequence targeting the 5′ UTR of EZH2, which allows ectopic EZH2 overexpression for a rescue experiment. As expected, both the down-regulation of EZH2 and FOXM1 mRNA expression upon knockdown of endogenous EZH2 were reversed by ectopic EZH2 overexpression (Fig. S3B). These results suggest a reciprocal Ezh2 and FoxM1 regulation in TNBC.
Fig. 3.
Reciprocal EZH2 and FOXM1 regulation independent of PRC2. (A) Immunoblot (Left) and qRT-PCR (Right) analyses of the indicated genes in MDA-MB231 cells treated with control, EZH2, or FOXM1 siRNA. Relative gene expression levels normalized to cells treated with control siRNA are shown. A one-way ANOVA with Bonferroni posttest was used to compare the knockdown effects with those of the control siRNA. (B) qRT-PCR analysis of the indicated genes in MDA-MB231 cells as in A treated with control, SUZ12, or EED siRNA. (C) ChIP-qPCR analysis of Ezh2 occupancy on FOXM1 promoter (Upper) and FoxM1 occupancy on EZH2 promoter (Lower) in MDA-MB231 cells. P1–P4 indicate primer pairs used to encompass the genomic regions analyzed for promoter occupancy. Quantification of enrichment is represented as fold-enrichment. (D) Co-IP assay showing a physical interaction between Ezh2 and FoxM1 in MDA-MB231 cells. (E) ChIP-qPCR analysis of Ezh2 (Top) or FoxM1 occupancy (Middle) on the indicated promoters in MDA-MB231 cells treated with control or FOXM1 siRNA. Quantification of enrichment is represented as fold-enrichment. A two-way ANOVA with Bonferroni posttest was used for statistical analysis comparing each pulldown with its own control siRNA. (Bottom) Illustration of Ezh2 and FoxM1 coregulation at the transcriptional level. (F) Matrigel invasion assay of MDA-MB231 cells treated as indicated. A one-way ANOVA with Bonferroni posttest was used for statistical analysis comparing knockdown effects with control siRNA. (G) Matrigel invasion assay (Upper) and immunoblot analysis (Lower) in MDA-MB231 cells treated as shown. A two-way ANOVA with Bonferroni posttest was used for statistical analysis. All data represent mean ± SEM; **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Fig. S3.
(A) Immunoblot analysis of the indicated proteins in the indicated cell lines treated with control or two EZH2 siRNAs. (B) qRT-PCR analysis of the indicated genes in MDA-MB231 cells as treated. Relative gene expression levels were normalized to control siRNA. A two-tailed unpaired Student’s t test was used for statistical analysis. (C) Co-IP assay showing the physical interaction between Ezh2 and FoxM1 in the indicated cell lines. (D) Immunoblot analysis of the indicated proteins in MDA-MB231 cells treated with nocodazole for 16 h followed by time-course intervals of nocodazole removal. (E) Matrigel invasion assay in BT549 cells treated as indicated. A two-way ANOVA with Bonferroni posttest was used for statistical analysis. All data represent mean ± SEM; *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001.
The above findings prompted us to investigate whether the Ezh2 and FoxM1 codependency might stem from transcriptional activation at the other’s promoters. ChIP assays using a series of PCR primers to probe for chromatin regions of respective promoters in MDA-MB231 cells revealed marked Ezh2 recruitment on the proximal FOXM1 promoter region [−60 bp from the transcription start site (TSS)] (Fig. 3C, Upper) and FoxM1 recruitment on the EZH2 promoter (−1,660 to −1,470 bp) (Fig. 3C, Lower).
Given the reciprocal regulation of Ezh2 and FoxM1, we next investigated whether Ezh2 acts independently of PRC2 to interact discretely with FoxM1. To do so, we performed Ezh2 and FoxM1 immunoprecipitation experiments in MDA-MB231 cells. As seen in Fig. 3D, Ezh2 pulldown coimmunoprecipitated with both FoxM1 and Suz12 (Fig. 3D, Upper), whereas FoxM1 pulldown coimmunoprecipitated only with Ezh2 but not with Suz12 (Fig. 3D, Lower). These results indicate that, in addition to PRC2, Ezh2 also forms an independent protein complex with FoxM1. This observation was further validated in other TNBC cell lines (Fig. S3C). We also found that Ezh2 was required to maintain the protein stability of FoxM1, because overexpression of EZH2 in MDA-MB231 cells was able to prevent premature FoxM1 degradation after the mitotic phase stimulated by the addition and removal of nocodazole (Fig. S3D). Consistent with the positive autoregulation of FOXM1 on its own promoter (45), we also observed enrichments of Ezh2 and FoxM1 on the same region in the FOXM1 promoter, and knockdown of FOXM1 reduced Ezh2 binding to the FOXM1 promoter (Fig. 3E). Interestingly, Ezh2 enrichment was not seen in the promoters of the well-known FoxM1 targets in cell-cycle regulation, CCNB1 and AURKB, indicating that the Ezh2–FoxM1 complex may not have an active role in cell proliferation. Taken together, these results suggest that Ezh2 and FoxM1 regulate each other at the transcriptional level and particularly that Ezh2 complexes with FoxM1 to maintain the stability of the latter protein, leading to the activation of FoxM1 expression as illustrated in Fig. 3E, Lower.
Next, we investigated the functional relevance of Ezh2/FoxM1 coregulation vs. repressive PRC2 in regulating TNBC invasion upon hypoxia. We showed that hypoxia increased cell invasion in MDA-MB231 cells and that this increase was ablated by EZH2 or FOXM1 knockdown (Fig. 3F). In contrast, knockdown of EED or SUZ12 enhanced the invasive capacity (Fig. 3F), suggesting a tumor-suppressive function of PRC2 in TNBC. The same result was obtained in BT549 cells (Fig. S3E). This observation was expected, because it is consistent with a recent report showing that PRC2 inactivation favors breast tumorigenicity (17). Importantly, the reduced cell invasion upon EZH2 knockdown was effectively rescued with ectopic overexpression of FOXM1, but the H3K27me3 level remained depleted (Fig. 3G), indicating that FoxM1 has an essential role in EZH2-mediated invasion that is independent of PRC2/H3K27me3. Taken together, our findings identify FoxM1 as a previously unidentified partner of Ezh2 in promoting TNBC invasion. In contrast, the repressive PRC2, which seems to exert a tumor-suppressive function in TNBC, might counteract the oncogenic property of the previously unidentified Ezh2/FoxM1 complex. However, these results raise the question of which downstream target genes are affected by this paradoxical role of Ezh2/PRC2 in response to hypoxia during TNBC invasion.
HIF1-α, Ezh2, and FoxM1 Promote the Expression of MMPs to Counteract PRC2-Mediated Repression.
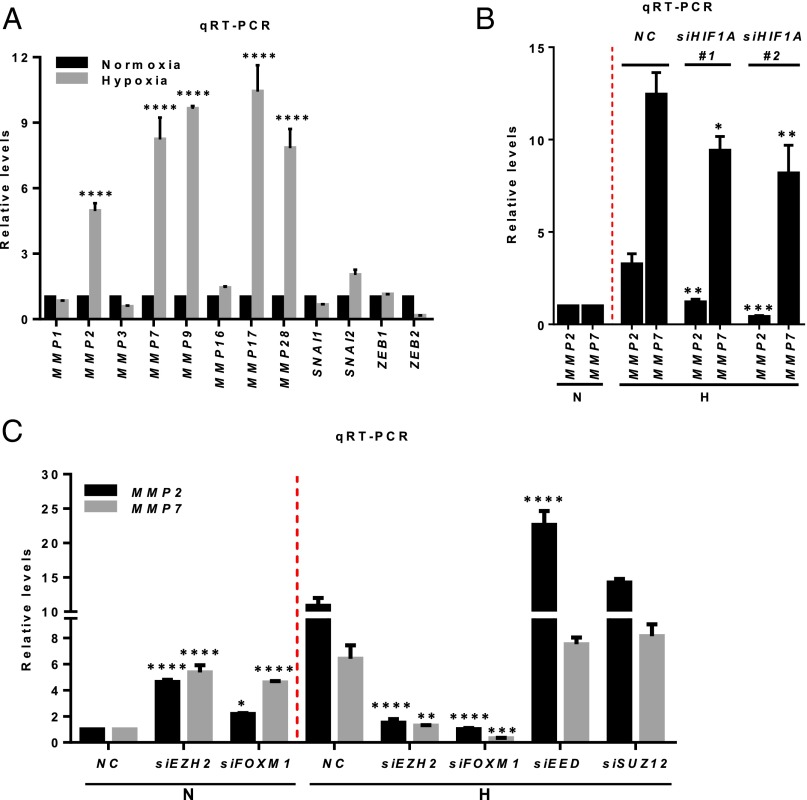
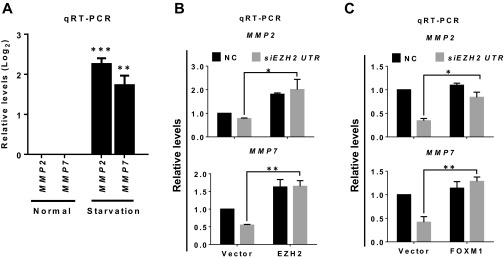
Hypoxia is known to regulate a cohort of genes, including members of the MMP family and genes that modulate epithelial-to-mesenchymal transition (EMT), and thus to promote cancer migration and invasion (28). We found that hypoxia induced marked up-regulation of MMP expression compared with normoxia, but this up-regulation was not seen for the EMT regulators SNAI1 (encoding SNAIL), SNAI2 (encoding SLUG), ZEB1 (Zinc finger E-box binding homeobox 1), and ZEB2 (Fig. 4A), suggesting that hypoxia has a selective effect on MMPs in the TNBC cellular context. Further analysis focusing on MMP2 and MMP7 validated that their hypoxia-induced expression was dependent on HIF1-α, because concomitant HIF1A knockdown reduced their expression (Fig. 4B). Strikingly, we found that the expression of MMPs was differentially regulated by Ezh2 in normoxia and hypoxia. EZH2 or FOXM1 knockdown induced the expression of MMP2 and MMP7 in normoxia but reduced their expression in hypoxia (Fig. 4C). In contrast, knocking down EED or SUZ12 increased the expression of MMP2 and MMP7 in hypoxia (Fig. 4C). These findings suggest that, although in normoxia Ezh2 functions in a PRC2-dependent manner to repress the expression of MMPs, in hypoxia it partners with FoxM1 to promote the expression of MMPs to counter PRC2-mediated repression. Furthermore, as did hypoxia, serum starvation induced the expression of MMPs (Fig. S4A). The induction of MMPs with serum starvation also was abolished by EZH2 siRNA treatment but was effectively rescued by ectopic overexpression of EZH2 (Fig. S4B) or FOXM1 (Fig. S4C). Collectively, these results demonstrate that in hypoxic or nutrition-deprived breast cancer cells, Ezh2 switches its effect on MMPs from repression to activation and that the activation requires FoxM1.
Fig. 4.
Hypoxia-induced HIF1-α regulates the expression of MMPs in an Ezh2/FoxM1-dependent manner. (A) qRT-PCR analysis of indicated genes in MDA-MB231 cells cultured in normoxia or hypoxia. (B) qRT-PCR analysis for MMP2 and MMP7 in MDA-MB231 cells treated with control or two HIF1A siRNAs as indicated. (C) qRT-PCR analysis of MMP2 and MMP7 in MDA-MB231 cells treated with siRNAs as indicated. All bars represent relative expression levels. The statistical analyses were performed using a one-way or two-way ANOVA with Bonferroni posttest. All data represent mean ± SEM; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Fig. S4.
(A) qRT-PCR analysis of the indicated genes in MDA-MB231 cells cultured in normoxia or with serum starvation for 48 h. Relative gene-expression levels were normalized to cells maintained in normoxia. A one-way ANOVA with Bonferroni posttest was used for statistical analysis. (B and C) qRT-PCR analysis of indicated genes in MDA-MB231 cells overexpressed with EZH2 (B) or FOXM1 (C) and treated with control or EZH2 siRNA cultured in serum starvation. Relative gene-expression levels were normalized to control siRNA of cells treated with empty vector. A two-tailed unpaired Student’s t test was used for statistical analysis. All data represent mean ± SEM; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Hypoxia-Induced HIF1-α Modulates PRC2 and Ezh2/FoxM1 Recruitment to the MMP Promoters.
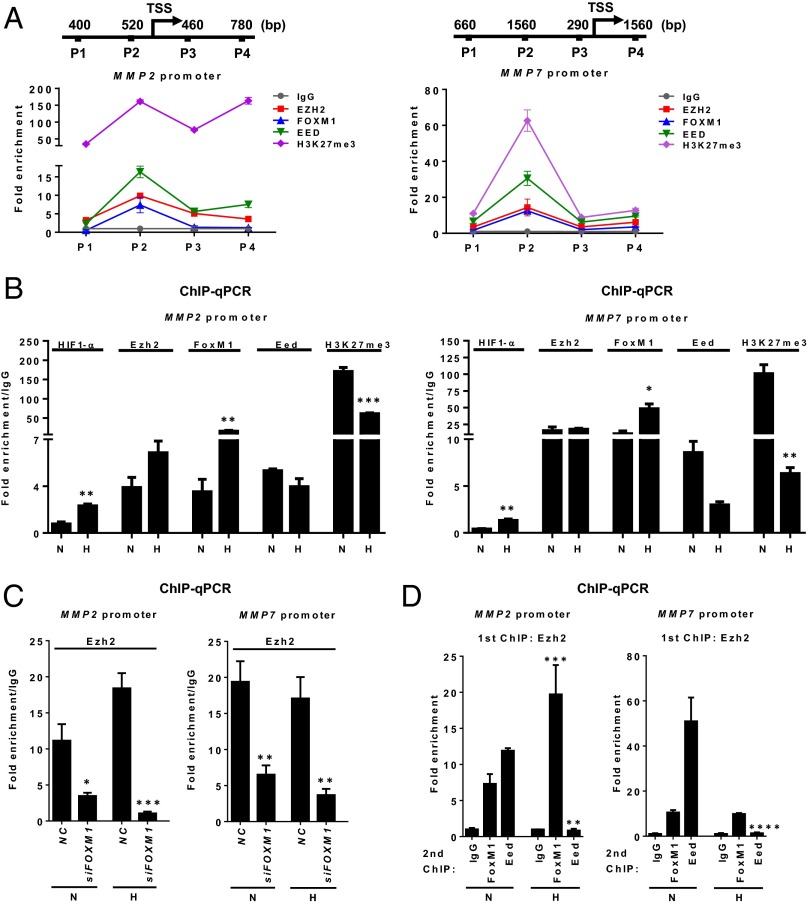
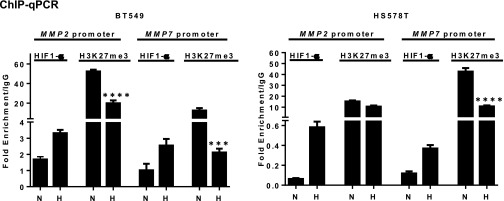
We next hypothesized that the regulation of MMP expression is under the direct antagonistic influence of both repressive PRC2 and a separate Ezh2/FoxM1 complex on MMP promoters and that this influence is modulated by hypoxia-induced HIF1-α. To test this hypothesis, we sought to investigate the occupancy on the MMP2 and MMP7 promoters by PRC2 and associated H3K27me3 and by Ezh2/FoxM1. As anticipated, in MDA-MB231 cells cultured in normoxia that expressed low levels of MMPs, ChIP analysis showed abundant Eed and H3K27me3 enrichment, as well as Ezh2 and FoxM1, in the MMP2 and MMP7 promoters (Fig. 5A). Upon hypoxia, the MMP2 and MMP7 promoters exhibited reduced Eed and H3K27me3 enrichment but increased recruitment of HIF1-α and FoxM1, although increased Ezh2 binding was seen only in the MMP2 promoter (Fig. 5B). The increased recruitment of HIF1-α coupled with decreased H3K27me3 enrichment on the promoters of MMP2/7 in response to hypoxia also was seen in other TNBC cell lines, e.g., BT549 and HS578T (Fig. S5). Collectively, these results demonstrate direct regulation of MMP promoters by both PRC2 and the Ezh2/FoxM1 complex, which is dynamically regulated by hypoxia-induced HIF1-α, resulting in depletion of PRC2/H3K27me3 and increased expression of MMPs through increased Ezh2/FoxM1 recruitment.
Fig. 5.
Hypoxia-induced HIF1-α modulates the PRC2 and Ezh2/FoxM1 complex co-occupancy on MMP promoters. (A) ChIP-qPCR analysis of Ezh2, FoxM1, Eed, or H3K27me3 occupancy on MMP2 (Right) or MMP7 (Left) promoter in MDA-MB231 cells. P1–P4 indicate primer pairs used to encompass the genomic regions analyzed for promoter occupancy. (B) ChIP-qPCR analysis of Ezh2, FoxM1, Eed, or H3K27me3 occupancy on MMP2 (Left) or MMP7 (Right) promoter in MDA-MB231 cells in normoxia or hypoxia. Quantification of enrichment is represented as fold-enrichment relative to IgG. (C) ChIP-qPCR analysis of Ezh2 occupancy on MMP2 (Left) or MMP7 (Right) promoter in MDA-MB231 cells treated with control or FOXM1 siRNA. (D) Sequential ChIP-qPCR analysis to assess in vivo colocalization of Ezh2, FoxM1, and Eed on MMP2 (Left) or MMP7 (Right) promoter in MDA-MB231 cells in normoxia or hypoxia. First ChIP and second ChIP antibodies are indicated in the chart title and x-axis labels, respectively. All statistical analyses were performed using one-way ANOVA with Bonferroni posttest. All data represent mean ± SEM; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Fig. S5.
ChIP-qPCR analysis for HIF1-α or H3K27me3 occupancy on MMP2 and MMP7 promoters in the indicated cell lines maintained in normoxia or hypoxia for 48 h. Quantification of enrichment is represented as fold-enrichment over IgG control. A one-way ANOVA with Bonferroni posttest was used for statistical analysis. All data represent mean ± SEM; ***P ≤ 0.001, ****P ≤ 0.0001.
Furthermore, knockdown of FOXM1 reduced Ezh2 binding to the MMP promoters, indicating the dependency of Ezh2 on FoxM1 on the MMP promoters (Fig. 5C). To demonstrate the co-occupancy of Ezh2, FoxM1, and PRC2 on the same promoter, we performed pairwise sequential ChIP analyses. After the first Ezh2 immunoprecipitate, both FoxM1 and Eed bindings were also found to be enriched in Ezh2-bound MMP2 and MMP7 promoters under normoxia (Fig. 5D). Strikingly, the colocalization of these regulators on the MMP promoters was dynamically altered under hypoxia: FoxM1 was further enriched, but Eed enrichment was markedly reduced. These findings suggest that PRC2 and Ezh2/FoxM1 colocalize to the same MMP promoters, where they act antagonistically to regulate the expression of MMPs.
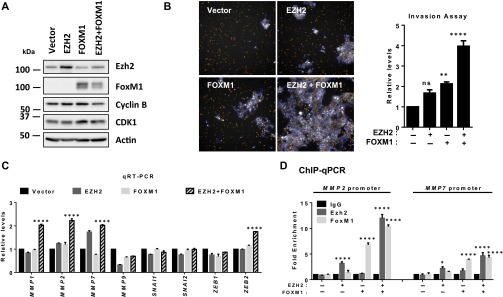
To demonstrate further the coordination of Ezh2 and FoxM1 in promoting MMP expression and invasion, we overexpressed EZH2, FOXM1, or both in an immortalized human mammary epithelial cell line, MCF10A (Fig. S6A), and evaluated their effects on the expression of MMPs and cellular outcomes. Overexpression of EZH2 or FOXM1 alone induced only a modest increase in invasion, whereas their coexpression resulted in marked increase in invasion (Fig. S6B). Consistent with the change in the invasive phenotype, we observed a marked increase in the mRNA expression of a subset of MMPs only in MCF10A cells overexpressing both EZH2 and FOXM1; single-gene overexpression seemed to be insufficient to induce the same effect (Fig. S6C). As a comparison, we did not see such a change in EMT-related genes, with the exception of ZEB2 (Fig. S6C). ChIP analysis further showed that overexpression of EZH2 or FOXM1 led to enriched recruitment to the MMP2 and MMP7 promoters and that coexpression of EZH2 and FOXM1 resulted in further enrichment of both promoters (Fig. S6D), highlighting the coordinated effect of Ezh2 and FoxM1 in promoting the expression of MMPs. Of note, although we detected marked enrichment of Ezh2 or FoxM1 in the MMP promoters in the cells overexpressing EZH2 or FOXM1 as single genes (Fig. S6D), we did not observe corresponding increases in the expression of MMPs (Fig. S6C). These findings further support the notion that a concerted effort between Ezh2 and FoxM1 is required to promote a robust expression of MMPs and invasion.
Fig. S6.
(A) Immunoblot analysis of the indicated proteins in MCF10A cells overexpressed with EZH2, FOXM1, or both. (B, Left) Images of the Matrigel invasion assay captured using a high content screening microscope. (Magnification: 10×.) (Right) Quantification of the average number of invading cells normalized to empty vector cells. A one-way ANOVA with Bonferroni posttest was used for statistical analysis. (C) qRT-PCR analysis of the indicated genes. Relative gene expression levels were normalized to the empty vector cells. A two-way ANOVA with Bonferroni posttest was used for statistical analysis. (D) ChIP-qPCR analysis for enrichment of Ezh2 or FoxM1 on the promoters of MMP2 and MMP7. Each ChIP pulldown was compared with empty vector cells. Quantification of enrichment was determined as fold-enrichment over IgG. A two-way ANOVA with Bonferroni posttest was used for statistical analysis. All data represent mean ± SEM; *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.001.
Pharmacological Inhibition of PRC2 Promotes the Expression of Ezh2/FoxM1-Mediated MMPs and Invasion.
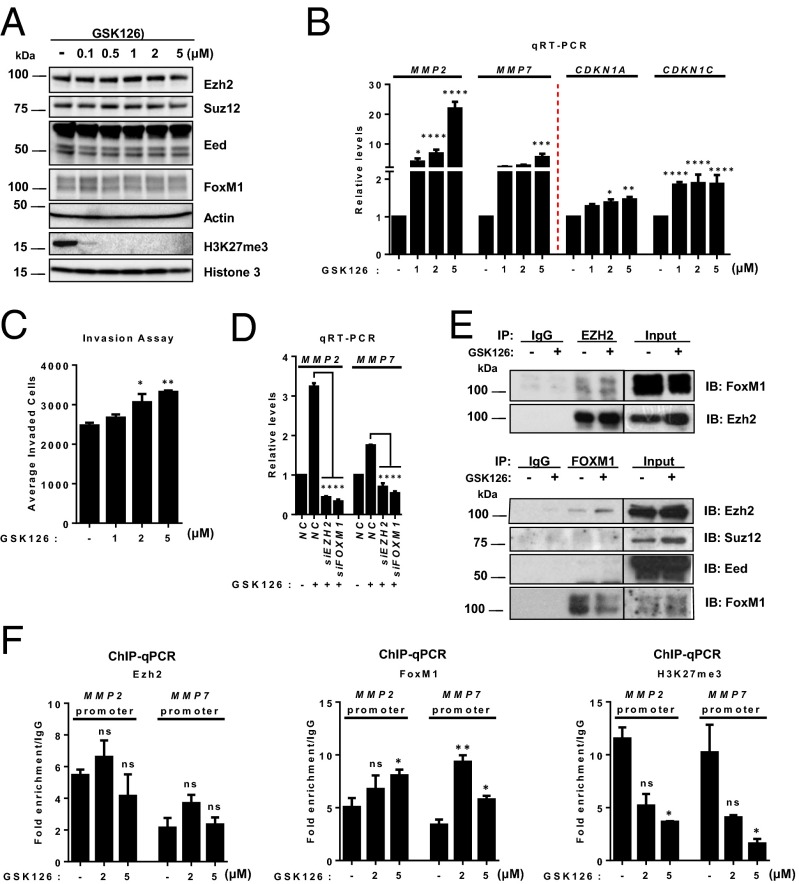
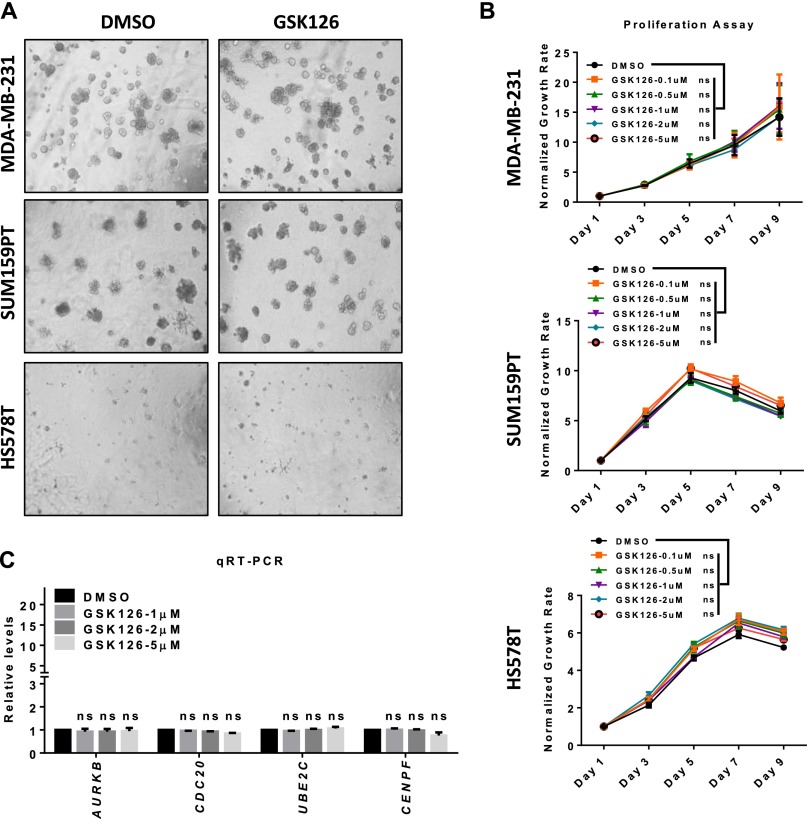
Having shown an inhibitory role of hypoxia-induced HIF1-α on PRC2 and H3K27me3, which led to increased in MMP expression and invasion, we sought to investigate whether direct inhibition of PRC2 activity by a small-molecule enzymatic inhibitor of Ezh2 would achieve a similar effect. For this purpose, we used GSK126, which is a selective inhibitor of Ezh2 with potent activity to deplete global H3K27me3 (46). Indeed, GSK126 treatment in MDA-MB231 cells resulted in nearly complete depletion of global H3K27me3 without affecting the expression of either FoxM1 or a major component of PRC2 (Fig. 6A). As anticipated, MMP2 and MMP7, as well as CDKN1A and CDKN1C, were markedly induced upon GSK126 treatment (Fig. 6B), which recapitulated the effect of EED or SUZ12 knockdown as shown in Fig. 4C. In line with the effect on MMPs, we detected increased invasion of MDA-MB231 cells in a GSK126 dose-dependent manner (Fig. 6C), whereas no significant changes were observed in 3D Matrigel growth and monolayer cell proliferation in a number of TNBC cell lines (MDA-MB231, SUM159PT, and HS578T) (Fig. S7 A and B). GSK126 also had no effect on cell-cycle–related genes (Fig. S7C). Importantly, we showed that GSK126-induced MMP2 and MMP7 expression was dependent on Ezh2 and FoxM1, because depletion of either EZH2 or FOXM1 effectively abolished the induction of MMPs by GSK126 (Fig. 6D).
Fig. 6.
Pharmacological inhibition of PRC2 activity promotes Ezh2/FoxM1-mediated MMP expression and invasion. (A) Immunoblot analysis of indicated proteins in MDA-MB231 cells treated with the indicated concentrations of GSK126 for 72 h. (B) qRT-PCR analysis of the indicated genes in MDA-MB231 cells upon GSK126 treatment for 72 h. (C) Matrigel invasion assay of MDA-MB231 cells treated as indicated. (D) qRT-PCR analysis for MMP2 and MMP7 in MDA-MB231 cells as indicated. (E) Co-IP assay showing the physical interaction between Ezh2 and FoxM1 in MDA-MB231 cells treated with DMSO or 5 μM GSK126. (F) ChIP-qPCR analysis of FoxM1 or H3K27me3 occupancy in MMP2 or MMP7 promoter in MDA-MB231 cells treated with DMSO or GSK126. Quantification of enrichment is represented as fold-enrichment. A two-way ANOVA with Bonferroni posttest was used for all the statistical analyses. All data represent mean ± SEM; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Fig. S7.
(A) 3D-Matrigel growth of the indicated cell lines treated with DMSO control or 5 μM GSK126 for 7 d. (Magnification: 10×.) (B) Proliferation assay of the indicated cell lines treated with DMSO control or GSK126 over the duration of 9 d. A two-way ANOVA with Bonferroni posttest was used for statistical analysis. (C) qRT-PCR analysis of the indicated genes in MDA-MB231 cells treated with GSK126. Relative gene-expression levels were normalized to DMSO control cells. A two-way ANOVA with Bonferroni posttest was used for statistical analysis. All data represent mean ± SEM.
We next investigated the effect of GSK126 on the formation of the Ezh2/FoxM1 complex. Interestingly, coimmunoprecipitation (co-IP) analysis detected increased Ezh2 in the immunoprecipitate of FoxM1 in MDA-MB231 cells, and vice versa (Fig. 6E), suggesting an enhanced physical interaction between Ezh2 and FoxM1 upon GSK126 treatment. Furthermore, ChIP analysis showed that GSK126 treatment decreased the enrichment of H3K27me3 in MMP2 and MMP7 promoters but increased recruitment of FoxM1, although to a lesser extent, in Ezh2 (Fig. 6F). Taken together, these results provided direct evidence that PRC2/H3K27me3 activity has a suppressive role on the expression of MMPs and that the removal of this inhibition by directly inhibiting H3K27me3 is sufficient to induce the expression of MMPs by promoting Ezh2/FoxM1 recruitment to the MMP promoters.
Clinical Relevance of HIF1A, EZH2, and FOXM1 Expression vs. SUZ12 in Disease Outcomes.
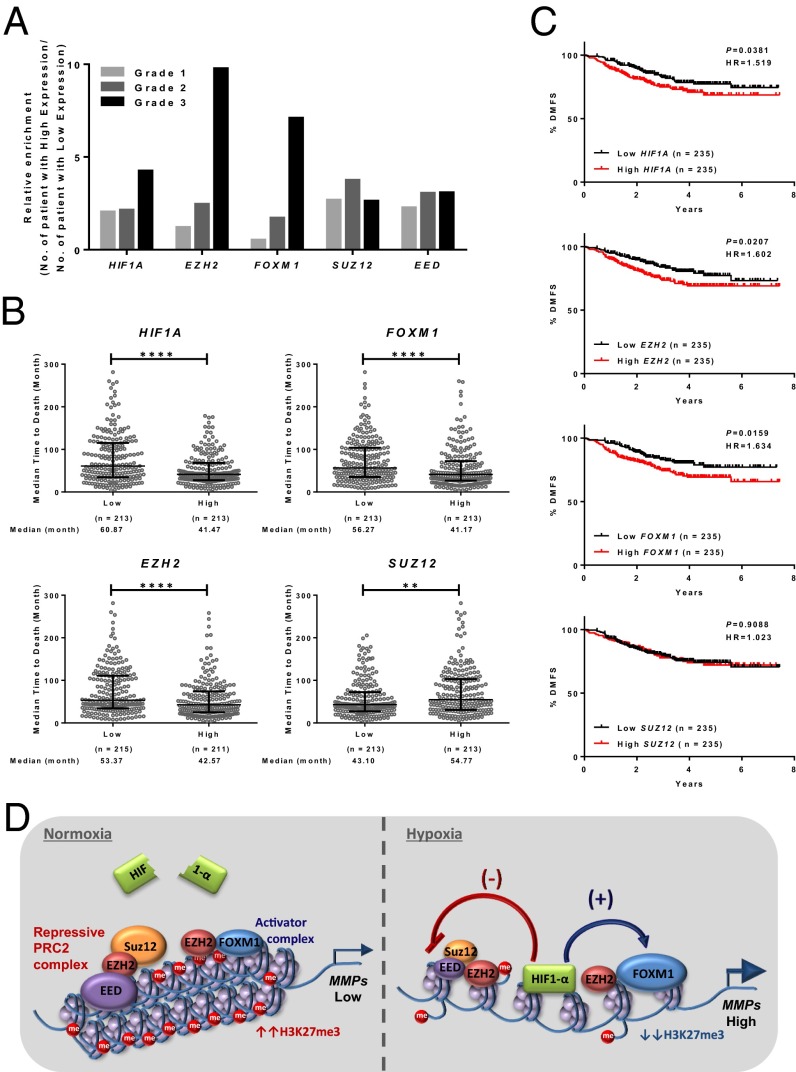
To demonstrate the clinical relevance of HIF1A, EZH2, and FOXM1 versus SUZ12 in breast cancer progression, we used the Curtis dataset (24), which consists of expression data of 1,556 tumor samples with clinical information. We found that HIF1A, EZH2, or FOXM1 expression displayed progressive up-regulation from grade 1 to grade 3 tumors, whereas SUZ12 did not show such a change (Fig. 7A). Furthermore, the time-to-death was significantly shorter for breast cancer patients whose tumors expressed higher levels of HIF1A, EZH2, or FOXM1 (Fig. 7B). In contrast, breast cancer patients with high expression of SUZ12 in their tumors had a significantly longer time-to-death; this finding is consistent with PRC2 having a tumor-suppressive role in breast cancer, as suggested in both our study and a recent report (17). Next, we investigated whether these oncogenic drivers correlated with patient distant metastasis-free survival (DMFS) in comparison with SUZ12. As shown in the Hatzis dataset (47), which contains clinical information about metastasis occurrence, Kaplan–Meier analysis indicated that breast cancer patients whose tumors had higher HIF1A expression had a significantly higher risk of developing distant metastasis than those with low expression (Fig. 7C). Consistently, EZH2 and FOXM1 showed patterns similar to those of HIF1A, but an opposite (albeit nonsignificant) trend was seen for SUZ12 (Fig. 7C). These data analyses further emphasize the clinical relevance of our in vitro findings.
Fig. 7.
Clinical outcome and mechanistic model. (A) Gene-expression data analysis for tumor grading in all primary breast tumors. The Curtis dataset (24) was used to stratify patients by high or low expression of HIF1-α, EZH2, FOXM1, SUZ12, or EED quantified as detailed in Materials and Methods. The y axis represents the number of patients whose tumors expressed high levels of the indicated gene relative to patients with low expression of that gene. (B) Analysis of gene-expression data for median time-to-death in all primary breast tumors in the Curtis dataset (24). Stratification of patients as having high or low expression based on the median cut-off was used to examine the time-to-death in patients with grade 3 tumor as detailed in Materials and Methods. Patients with high expression of HIF1A, EZH2, or FOXM1 have shorter time-to-death, and patients with high expression of SUZ12 have longer time-to-death. Statistical tests were performed using the Mann–Whitney u test; ***P ≤ 0.001, ****P ≤ 0.0001. (C) Analysis of gene-expression data for DMFS in all primary breast tumors. The Hatzis dataset (47) was used to stratify patients into groups having high or low expression (using a median cut-off) of HIF1A, EZH2, FOXM1, or SUZ12 and DMFS as detailed in Materials and Methods. Patients with high expression of HIF1A, EZH2, or FOXM1 have a higher risk of developing distant metastasis. P values were calculated by log-rank test, and the hazard ratio was determined by the log-rank method. (D) A model depicting the antagonistic relationship between repressive PRC2 and the activator Ezh2/FoxM1 complex in regulating MMP expression in both normoxia and hypoxia.
Taken together, our investigations uncovered several previously unidentified molecular insights into how a TNBC tumor acquires enhanced invasive capacity through functional integration of multiple transcriptional factors regulated by HIF1-α (summarized in Fig. 7D). We propose that the expression of invasive MMPs is counterbalanced by the presence of repressive PRC2 and the activating Ezh2/FoxM1 complex on their promoters, conferring a tight equilibrium of their expression. HIF1-α activation by hypoxia impairs the PRC2 activity but enhances the expression of FoxM1, resulting in a functional shift toward the transcriptional activation of MMPs mediated by both HIF1-α and the Ezh2/FoxM1 complex. Of note, the reciprocal regulation of Ezh2 and FoxM1 may further enhance the robustness of this model, leading to sustained expression of MMPs. This model is consistent with the clinical observation seen exclusively in TNBC patients, in whom high expression of HIF1A, EZH2, FOXM1, and invasion targets was accompanied by high expression of PRC2-repressed targets, indicating impaired PRC2 activity (Fig. 1A and Fig. S1A).
Discussion
The elucidation of molecular mechanisms that define the contribution of epigenetic programs to tumorigenesis is crucial for designing correct and effective therapeutic strategies. In this study, we used large-scale gene-expression data in breast cancer as a starting point to identify candidate oncogenic pathways that may orchestrate the PRC2 gene repressor program in different subtypes of breast cancers. Our analysis revealed that TNBC manifested defective PRC2 activity, as demonstrated by the abundant expression of PRC2-repressed targets in TNBC compared with other subtypes of breast cancer. We further demonstrated that reduced PRC2 activity in TNBC was functionally linked to HIF1-α activation, which subsequently promoted the Ezh2/FoxM1 complex toward increasing expression of MMPs and cellular invasion.
Using independent clinical datasets in breast cancer, we found that the transcript level of HIF1A was up-regulated selectively in TNBC patients, as is consistent with the up-regulation of invasion-associated genes. In addition to the established mechanism of HIF1-α activation by posttranslational modifications (48–50), several studies have reported transcriptional activation of HIF1A (51–53). We showed that up-regulation of HIF1A in TNBC correlated with high expression of several well-known PRC2-repressed targets, suggesting that HIF1-α has a selective inhibitory role for PRC2 activity. We further demonstrated that HIF1-α inhibited PRC2 activity through transcriptionally repressed PRC2 components, SUZ12 and EED, selectively. In contrast, HIF1-α was able to induce EZH2 mRNA expression, as is consistent with a previous report identifying hypoxic response element in EZH2 promoter (25). Of interest, a recent study has reported that PRC2/H3K27me3 inactivation induces HIF1A mRNA expression in multiple myeloma (54). Thus, it is possible that the high HIF1A mRNA expression in TNBC is related to the loss of PRC2 activity, thereby conferring a reciprocal negative feedback loop between HIF1-α and PRC2 in TNBC. This phenomenon warrants further investigation.
Another major finding in this study is the previously unidentified cross-talk between EZH2 and FOXM1. The two regulate each other and also form a complex to regulate the expression of MMPs and breast cancer cell invasion directly. We demonstrated that Ezh2 complexed with FoxM1 in the MMP promoters. Importantly, both reduced expression of MMPs and invasion could be rescued by ectopic FOXM1 expression, suggesting that Ezh2 depends on FoxM1 to enforce its oncogenic activity in this context. The codependency of EZH2 and FOXM1 in inducing the expression of MMPs and invasion was demonstrated further in the noncancerous mammary epithelial cell line MCF10A. We showed that marked induction of MMPs occurred only in cells that ectopically co-overexpressed EZH2 and FOXM1 and that this overexpression was associated with a strong increase in invasion.
The discovery of the functional cross-talk between EZH2 and FOXM1 in regulating MMPs and invasion provides important insights into the molecular functions of FOXM1 in breast cancer progression. Although FOXM1 has been connected to a wide spectrum of cellular processes in tumorigenesis, its precise contribution to malignancies is still the subject of speculation. As one of the key cell-cycle regulators, FOXM1 hyperactivation in the onset of tumorigenesis could reflect its role as an oncogenic driver. Alternatively, the hyperactivation of FOXM1 could simply be a passenger effect, given that cancer cells generally have a higher proliferative capacity than normal cells. This possibility is further illustrated by a study showing that an elevated level of FOXM1 in TNBC does not correlate with proliferation markers (29). These authors also reported that the poor prognostic value of TNBC does not correlate with its proliferative capacity but instead reflects its metastatic potential, suggesting that FOXM1 possesses an additional tumorigenic role beyond cell-cycle regulation. This observation is further supported by the effects of FOXM1 depletion, in which certain cancer cells are viable and proliferate but are severely reduced in tumorigenicity (55–57). Our works suggest that during hypoxia EZH2 may direct FOXM1 for a more invasive phenotype.
It is important to note that this effect of EZH2 acting through FOXM1 was independent of the catalytic activity of PRC2 on H3K27me3, as is consistent with growing reports that EZH2 has non-PRC2 roles usually associated with transcriptional activation (8–13). Paradoxically, Ezh2 also complexes with Suz12 and Eed to suppress MMP promoters directly, providing a predominant tumor-suppressor mechanism by inhibiting the expression of MMPs in normoxia. As such, the pharmacologic inhibition of Ezh2 histone methyltransferase activity or the knockdown of the PRC2 components SUZ12 and EED resulted in increased expression of MMPs and invasive capacity. Thus, to our knowledge, our study provides the first example showing the molecular interface between these two antagonistic Ezh2 complexes in regulating the transcription of MMPs associated with breast cancer invasion. In response to hypoxia, HIF1-α induction inhibited PRC2 activity to facilitate Ezh2/FoxM1-mediated activation of MMPs.
Our analysis of an antagonistic role of HIF1-α on repressive PRC2 activity eventually led to the recognition of the paradoxical nature of EZH2, which exhibited both tumor-suppressive and oncogenic properties. The paradoxical role of EZH2 in tumorigenesis has been observed in several human cancers (16). Although its oncogenic role in promoting cancer progression has been well documented, in certain contexts it also can function as a tumor suppressor (17, 20, 58–61). In addition to leukemia, a tumor-suppressive role for EZH2 has been found in solid tumors such as pancreatic (61) and renal cancer (60). Moreover, in a BRCA1-deficient mouse model of breast cancer, EZH2 depletion was found to facilitate tumor formation (20), indicating that EZH2 also has a tumor-suppressive role in breast cancer. This view is supported further by a recent study showing that impaired PRC2 activity promotes breast (17) and lung (18) tumorigenesis.
In summary, our findings uncovered a previously unidentified mechanism of PRC2 regulation in breast cancer and demonstrated an intricate equilibrium of PRC2 and Ezh2/FoxM1 in regulating MMP expression. Our data suggest that the reduced PRC2 activity upon hypoxia might promote cancer invasion through Ezh2/FoxM1-mediated induction of MMPs, and this suggestion was supported further by clinical data analysis showing that high expression of HIF1A, EZH2, or FOXM1 was associated with high tumor grade, high metastasis propensity, and worse survival outcome, whereas high expression of SUZ12 conferred a protective role with a more favorable survival outcome. Thus, catalytic inhibitors of EZH2, which are under clinical development, might not be appropriate for treating TNBC. Instead, a rational design based on the molecular pictures we have provided could pave the way for new therapeutic options for TNBC.
Materials and Methods
Cell Culture and Drug Treatment.
All cell lines used in this study were obtained from the American Type Culture Collection and were cultured in an incubator set at 37 °C in 5% CO2 and 95% atmospheric air. MDA-MB231, HS578T, and BT549 cells were maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (Invitrogen). SUM159PT cells were maintained in Ham’s F-12 medium (Invitrogen) supplemented with 5% (vol/vol) FBS, 5 μg/mL insulin (Invitrogen), and 1 μg/mL hydrocortisone (Invitrogen). MCF10A cells were maintained in mammary epithelial growth medium (MEGM) (Invitrogen) supplemented with 5% (vol/vol) horse serum (Invitrogen), 20 ng/mL EGF (Invitrogen), 0.5 mg/mL hydrocortisone, 100 ng/mL cholera toxin (Invitrogen), and 10 μg/mL insulin. All media were supplemented with 5,000U/mL penicillin/streptomycin mixture (Invitrogen). For hypoxia, cells were grown in a hypoxia incubator at 37 °C in 4% O2 and 5% CO2. For drug treatment, cells were treated with GSK126 (Pharmaron, Inc.) for 72 h.
Gene-Expression Dataset Analysis.
To determine the expression levels of HIF1A, EZH2, and FOXM1 and the expression of their target genes in breast cancer patients, normalized mRNA expression data derived from the Agilent expression platform, “BRCA.exp.547.med.txt.”, was obtained from the TCGA breast cancer online portal (https://tcga-data.nci.nih.gov/docs/publications/brca_2012/). PAM50 subtype classifications were available for 547 of the primary breast invasive carcinoma tumor samples, and these samples were further classified into four intrinsic breast cancer subtypes: TNBC (98), HER2-enriched (58), luminal A (232), and luminal B (129). To establish the clinical relevance further, two independent microarray datasets of breast cancer cohorts, Curtis (24) and Hatzis (47), were downloaded from Oncomine (https://www.oncomine.org//). Hierarchical clustering analysis was generated using Cluster 3.0, and a gene heat map was further visualized using TreeView (Eisen).
Transfection of siRNA and Plasmid Vectors.
siRNA and plasmid transfections were conducted using Lipofectamine RNAiMAX (Invitrogen) and Lipofectamine 2000 (Invitrogen), respectively, according to the manufacturer’s instructions. Target-specific siRNA and nontargeting control siRNA were purchased from Integrated DNA Technologies Singapore with the following sequences: HIF1A: 5′-UCAAGUUGCUGGUCAUCAG-3′; EZH2: 5′-GACUCUGAAUGCAGUUGCU-3′; EZH2 5′ UTR: 5′-CGGUGGGACUCAGAAGGCA-3′; FOXM1: 5′-GGACCACUUUCCCUACUUU-3′; FOXM1 5′ UTR: 5′-CAAGUCAGCUUUCCUGCA-3′; EED #1: HSC.RNAI.N003797.12.1_; EED #2: HSC.RNAI.NOO3797.12.2_; SUZ12 no. 1: HSC.RNAI.NO15355.12.1_; and SUZ12 no. 2: HSC.RNAI.N015355.12.2_.
To generate stable overexpression cell lines, target genes from their respective transient expression plasmids were subcloned into PMN retroviral expression vectors (a gift from Linda Penn's laboratory, Ontario Cancer Institute, Ontario, Canada). EZH2 plasmid had been described previously (8) and FOXM1 plasmid was a gift from Eric Lam, Imperial College London, London. Cells were infected with retrovirus packaged with PMN-EZH2 or PMN-FOXM1 or with the PMN-empty vector for 48 h, followed by cell sorting for selection of GFP overexpressed cells.
Histone Extraction and Immunoblots.
Whole-cell extract was prepared as described previously (31). For histone extraction, cells were lysed in Triton extraction buffer (1× PBS, 0.5% Triton X-100, and 2 mM PMSF). The histone pellet was further collected for acid extraction in 0.2 N HCl overnight at 4 °C. For protein lysate, briefly, cells were lysed in RIPA buffer and further sonicated using an XL2000 Microson Ultrasonic Processor (Misonix). Equal amounts of histone extract (2 μg) or protein extract (30 μg) were separated on SDS-polyacrylamide gels and transferred to PVDF membranes. These membranes were further blocked with 5% (wt/vol) milk, and the immunoblots were probed with the following antibodies: anti-EED (07-368; 1:5,000), anti-EED (09-774; 1:5,000), and anti-H3K27me3 (07-449; 1:1,000) were purchased from Upstate Biotechnology, Millipore Corporation. Anti-Cyclin B (sc-245; 1:1,000) and anti-FOXM1 (sc-500; 1:500) were purchased from Santa Cruz. Anti-CDC2 (CST-9112; 1:1,000), anti-EZH2 (CST-3147; 1:1,000), and anti–HIF1-α (CST-3716; 1:1,000) were purchased from Cell Signaling. Anti–HIF1-α (Ab-2185; 1:1,000) and anti-SUZ12 (Ab-12073; 1:1,000) were purchased from Abcam. Anti–HIF2-α (NB100-122; 1:500) was purchased from Novus Biologicals, and anti-Actin (1:200,000) was purchased from Sigma-Aldrich.
Co-IP.
Co-IP was performed as described previously (8). Briefly, cells were lysed with 1 mL of immunoprecipitation lysis buffer [20 mM Tris⋅HCl (pH 7.4), 2 mM EDTA, 25 mM NaF, and 1% Triton X-100] supplemented with protease inhibitors (Roche). The lysates were immunoprecipitated with antibody pulldown and Protein A/G agarose beads (Roche) overnight at 4 °C. The agarose beads were further washed three times with washing buffer [50 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS]. The immune complexes were subsequently eluted with sample buffer containing 1% SDS and DTT for 5 min at 95 °C. The sample was subsequently separated by SDS-polyacrylamide gels and transferred to PVDF membranes for immunoblotting. Antibodies used for pulldown were nonspecific IgG (sc-2025 or sc-2027) and anti-FOXM1 (sc-500 or sc-502) purchased from Santa Cruz. Anti-EZH2 (39901 or 39875) was purchased from Active Motif. The subsequent immunoblots were probed with anti-EED (09-774; 1:5,000) purchased from Upstate Biotechnology, Millipore Corporation, anti-EZH2 (CST-3147; 1:1,000) purchased from Cell Signaling, anti-FOXM1 (sc-500 or sc-502; 1:500) purchased from Santa Cruz, and anti-SUZ12 (Ab-12073; 1:1,000) purchased from Abcam.
Quantitative RT-PCR, ChIP, and Sequential ChIP.
Total RNA was isolated using TRIzol (Invitrogen) and extracted with the RNAeasy Mini Kit (Qiagen). A total of 1,000 μg RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), and the product subsequently was subjected to quantitative real-time PCR (qRT-PCR) with the KAPA SyBr Fast qPCR Kit (KAPA Biosystems). All reactions were performed in triplicate using an Applied Biosystems 7500 Fast Real-Time PCR system in a 96-well plate format. Primer sequences can be found in Table S1. For quantification of mRNA levels, ACTB mRNA level were used as an internal control.
Table S1.
qRT-PCR primer sequences
| Gene | Forward primer | Reverse primer |
| ACTB | GCACAGAGCCTCGCCTT | GTTGTCGACGACGAGCG |
| AURKB | AGGAGAACTCCTACCCCTGG | AGATGGGGTGACAGGCTCTT |
| CCNB1 | TGGAAAAGTTGGCTCCAAAG | GATTTTGCAGAGCAAGGCAT |
| CDC2 | CCCTCCTGGTCAGTACATGGA | GTTGAGTAACGAGCTGACCCC |
| CDC20 | ATTCGCATCTGGAATGTGTG | TGTAATGGGGAGACCAGAGG |
| CDKN1A | CATGGGTTCTGACGGACAT | AGTCAGTTCCTTGTGGAGCC |
| CDKN1C | AAGAAATCGGAGATCAGAGGC | AGCTGCACTCGGGGATTT |
| CENPF | AAGCCCAGCTCATTTTGTTC | GCGCCAGAACTGTACTCTCC |
| EZH2 | AGTGTGACCCTGACCTCTGT | AGATGGTGCCAGCAATAGAT |
| FOXM1 | AGGTGTTTAAGCAGCAGAAACG | GCTAGCAGCACCTTGGGGGCAA |
| MMP1 | CCTCGCTGGGAGCAAACA | TTGGCAAATCTGGCGTGTAAT |
| MMP2 | TGAGCTATGGACCTTGGGAGAA | CCATCGGCGTTCCCATAC |
| MMP3 | GATGCAGCCATTTCTGATAAGGA | CCATGGAATTTCTCTTCTCATCAAA |
| MMP7 | GGGACATTCCTCTGATCCTAATGC | GAATTACTTCTCTTTCCATATAGTTTCTGAATGC |
| MMP9 | GGACGATGCCTGCAACGT | CAAATACAGCTGGTTCCCAATCT |
| MMP16 | ATTGACTGGATGAAGAAGCCCC | TAAGTGATGTGCTTGTGCTGCC |
| MMP17 | AAGGCAAGTACTTCTGGCGG | ACGATCTTGTGGTCGCTGGT |
| MMP28 | CATCAGAGCGTTTCAGTGGG | AGTCACTGATC TCTCAGCC |
| SNAI1 | CCCCAATCGGAAGCCTAACT | GCTGGAAGGTAAACTCTGGATTAGA |
| SNAI2 | CTTGCCCTCACTGCAACAGA | TCTGCAGATGAGCCCTCAGA |
| SUZ12 | CGGAATCTCATAGCACCAAT | CTCATAGCTTGTCAGCTCAT |
| UBE2C | AGCAGGAGCTGATGACCCT | GGCGTGAGGAACTTCACTGT |
| ZEB1 | TGAGCAAAACCATGATCCTAATGT | CAGGTGCCTCAGGAAAAATGA |
| ZEB2 | GCCGAGTCCATGCGAACT | CCATGATCGGCTGCTTCAT |
ChIP was performed as described previously (31). Briefly, precleared chromatin was immunoprecipitated with the following antibodies: anti-EZH2 (39901; Active Motif), anti-FOXM1 (sc-500; Santa Cruz), anti-EED (09-733; Millipore), anti-H3K27me3 (9744; Cell Signaling), anti–HIF1-α (Ab-2185), and a nonspecific IgG (sc-2027; Santa Cruz). For sequential ChIP, after the first antibody pulldown, the chromatin was further eluted with 10 mM DTT by gentle shaking at 37 °C for 30 min, followed by a second antibody pulldown. The immunoprecipitated DNA and input DNA were quantified by qRT-PCR analysis; primer sequences can be found in Table S2. Quantification of promoter-binding enrichment was defined as the percentage of the whole-cell lysate relative to the input DNA. The fold-enrichment was calculated by normalizing the specific antibody-enriched against the nonspecific IgG-enriched chromatins. All ChIP qRT-PCR promoter primer sequences were synthesized and purchased from AITbiotech.
Table S2.
ChIP-qPCR primer sequences
| Gene | Primer ID | Forward primer | Reverse primer |
| AURKB | GGGGTCCAAGGCACTGCTAC | GGGGCGGGAGATTTGAAAAG | |
| CCNB1 | CGCGATCGCCCTGGAAACGCA | CCCAGCAGAAACCAACAGCCGT | |
| EZH2 | P1 | TTGGTGGCCTAGACAGATGC | ACAAGCAATCATGTGCCCAG |
| P2 | CCAACAGTTCATAGGTGACC | AAGCTACCTCAACCCAGAGG | |
| P3 | AGACCAGCCTGACCAAGACC | GAGTTTCGCTCTGGTTGTCC | |
| P4 | GCGGGAGGAATCGAGAAGCA | ATCCTCATTCAACCCGTCGG | |
| EED | TGGGCGCGATTTGCGACAGT | ACATATTCCTCCGCCTCCCT | |
| FOXM1 | P1 | GCCATTGTATCTTCAGGGCC | CGCGAAACCTGTCTTTTGCC |
| P2 | CCGGAGCTTTCAGTTTGTTC | CGGAATGCCGAGACAAGG | |
| P3 | AGCTGGGCATGGTGGTGCAT | GCTTCGCTCTTGTCACCCAG | |
| P4 | CTTATCTCCAACCCAGTCCC | ACATGAGCCTAAAGGCCCAG | |
| MMP2 | P1 | TGCTGGAGTTCCCCATCA | GCATGTTAAAGGAAGCACCCA |
| P2 | AACCAGTCTTGCCCAATTTC | CCTTCTGCAAATGTGTAAGCC | |
| P3 | AAAACGGACAAAGAGTTGGC | TGTTTACCGAAGCCCCTCC | |
| P4 | ATGTTGTCTTGTGAGCGTGC | AACAAGGGAGCAGATATCCC | |
| MMP7 | P1 | ATACCTATGAGAGCAGTCAT | CATCGTTATTGGCAGGAAGC |
| P2 | GCATATTTGGAGTGTTTCCC | CTCAGTGCTTTTTCTATCCC | |
| P3 | CAAAATGACATGAATCCACC | ATATCAGTAGTTGTGTGGGC | |
| P4 | AAACATTCCCTGATGCCCTT | TAGAGCGAGCAGTCAATGTG | |
| SUZ12 | P1 | AAAGCAAGACCTGGCCAGGCG | TAGTAGAGACAGGGTTTCACC |
| P2 | TCAGT TCAAAGAGGC GGCCGG | TCACCCTACCCTGGCCTCGCT |
Phenotypic Assays: Proliferation Assay, 3D Matrigel Growth, Migration Assay, and Invasion Assay.
To measure the rate of cellular proliferation, cells were assayed with CellTiter-Glo (CTG) (Promega) according to the manufacturer’s protocol. Cells were lysed with CTG solution, and the chemiluminescent signal was detected with a microplate reader (Tecan). The growth rate was calculated by normalizing the CTG values obtained with those taken on day 1 and plotted against time.
For 3D Matrigel growth, an eight-well chamber slide (BD Biosciences) was overlaid with 45 μL of growth factor-reduced Matrigel (BD Biosciences) and incubated for 20 min at 37 °C. For each breast cancer cell line, 5 × 103 cells were seeded in each chamber with 400 μL complete culture medium supplemented with 4% (wt/vol) Matrigel. The culture medium was replaced at 2-d intervals, and phase-contrast images were captured for 7 d at 3-d intervals.
For the invasion assay, a 24-well Falcon FluoroBlok Transwell insert (BD Biosciences) with a pore size of 8 μm was overlaid with 250 μg/mL of Matrigel and incubated for 6 h at 37 °C. For MDA-MB231 and BT549 cells, 2.5 × 104 cells were seeded in each insert with DMEM supplemented with 0.5% FBS. For MCF10A cells, 5 × 104 cells were seeded in each insert with MEGM supplemented with 0.5% horse serum. The chemoattractant used was the respective complete culture medium for each cell line and was added into the outer chamber. Invaded cells were fixed after 48-h incubation time using 3.7% (vol/vol) formaldehyde (Sigma-Aldrich) and were stained with 25 μg/mL propidium iodide. Ten fields per insert were scanned, and average invaded cells were counted in triplicate using a Cellomics ArrayScan reader (Thermo Fisher Scientific).
Statistical Analysis: Pearson’s Correlation, PCA Analysis, Tumor Grade, Time-to-Death, and DMFS.
The data were presented as the mean values ± SEM. Comparisons between groups were evaluated by Student’s t test, one-way ANOVA, or two-way ANOVA as indicated in the respective figures. Values of at least P ≤ 0.05 (or as indicated in each figure legend) were considered to be statistically significant. For correlation analysis of differential gene expression, Pearson’s correlation coefficient was performed using GraphPad Prism 6. The statistical significance of the correlation was expressed as a P value. PCA was performed using IBM SPSS statistics 20 to analyze further the clustering of differential gene expression.
For tumor grading, the Curtis gene-expression data (24) with corresponding tumor-grade information were used. Patients were stratified into groups with high or low expression of HIF1A, EZH2, FOXM1, SUZ12, or EED by calculating the median level of gene expression in the respective tumor grade. Quantification of enrichment for each gene expression was defined as the number of patients whose tumors expressed high expression for a particular gene relative to the number of patients whose tumors expressed low expression for the corresponding gene.
For survival analysis, the time-to-death was examined specifically in deceased patients with grade 3 tumors. The median follow-up time for death was calculated, and a Mann–Whitney test was performed to determine whether the observed differences between patients with high or low expression of HIF1A, EZH2, FOXM1, or SUZ12 were statistically significant. For DMFS, Hatzis gene-expression data (47) with corresponding metastatic status were used. The survival curves were performed by the Kaplan–Meier method, and the P value was calculated by log-rank test with a hazard ratio between high or low expression of HIF1A, EZH2, FOXM1, or SUZ12 determined by the log-rank method.
Acknowledgments
This work was supported by the Agency for Science, Technology, and Research of Singapore. S.M. was supported by the Cancer Science Institute of Singapore PhD Graduate Programme in Cancer Biology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602079113/-/DCSupplemental.
References
- 1.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandhu R, Parker JS, Jones WD, Livasy CA, Coleman WB. Microarray-based gene expression profiling for molecular classification of breast cancer and identification of new targets for therapy. Lab Med. 2010;41(6):364–372. [Google Scholar]
- 6.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang T, et al. Prognostic value of high EZH2 expression in patients with different types of cancer: A systematic review with meta-analysis. Oncotarget. 2016;7(4):4584–4597. doi: 10.18632/oncotarget.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee ST, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Kim E, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan J, et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013;121(22):4512–4520. doi: 10.1182/blood-2012-08-450494. [DOI] [PubMed] [Google Scholar]
- 11.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung HY, et al. PAF and EZH2 induce Wnt/β-catenin signaling hyperactivation. Mol Cell. 2013;52(2):193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez ME, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci USA. 2014;111(8):3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 15.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42(8):665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 16.Hock H. A complex Polycomb issue: The two faces of EZH2 in cancer. Genes Dev. 2012;26(8):751–755. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wassef M, et al. Impaired PRC2 activity promotes transcriptional instability and favors breast tumorigenesis. Genes Dev. 2015;29(24):2547–2562. doi: 10.1101/gad.269522.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serresi M, et al. Polycomb repressive complex 2 is a barrier to KRAS-driven inflammation and epithelial-mesenchymal transition in non-small-cell lung cancer. Cancer Cell. 2016;29(1):17–31. doi: 10.1016/j.ccell.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Holm K, et al. Global H3K27 trimethylation and EZH2 abundance in breast tumor subtypes. Mol Oncol. 2012;6(5):494–506. doi: 10.1016/j.molonc.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae WK, et al. The methyltransferase EZH2 is not required for mammary cancer development, although high EZH2 and low H3K27me3 correlate with poor prognosis of ER-positive breast cancers. Mol Carcinog. 2015;54(10):1172–1180. doi: 10.1002/mc.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Y, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog. 2008;47(9):701–706. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Healey MA, et al. Association of H3K9me3 and H3K27me3 repressive histone marks with breast cancer subtypes in the Nurses’ Health Study. Breast Cancer Res Treat. 2014;147(3):639–651. doi: 10.1007/s10549-014-3089-1. [DOI] [PubMed] [Google Scholar]
- 23.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100(20):11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curtis C, et al. METABRIC Group The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CJ, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19(1):86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia LM, et al. Transcriptional up-regulation of FoxM1 in response to hypoxia is mediated by HIF-1. J Cell Biochem. 2009;106(2):247–256. doi: 10.1002/jcb.21996. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, et al. Clinical and prognostic relevance of EZH2 in breast cancer: A meta-analysis. Biomed Pharmacother. 2015;75:218–225. doi: 10.1016/j.biopha.2015.07.038. [DOI] [PubMed] [Google Scholar]
- 28.Gilkes DM, Semenza GL. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 2013;9(11):1623–1636. doi: 10.2217/fon.13.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yau C, Wang Y, Zhang Y, Foekens JA, Benz CC. Young age, increased tumor proliferation and FOXM1 expression predict early metastatic relapse only for endocrine-dependent breast cancers. Breast Cancer Res Treat. 2011;126(3):803–810. doi: 10.1007/s10549-011-1345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, et al. HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc Natl Acad Sci USA. 2015;112(45):E6215–E6223. doi: 10.1073/pnas.1520032112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wierstra I. The transcription factor FOXM1 (Forkhead box M1): Proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res. 2013;118:97–398. doi: 10.1016/B978-0-12-407173-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 33.Ringnér M, Fredlund E, Häkkinen J, Borg Å, Staaf J. GOBO: Gene expression-based outcome for breast cancer online. PLoS One. 2011;6(3):e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stiehl DP, et al. Non-canonical HIF-2α function drives autonomous breast cancer cell growth via an AREG-EGFR/ErbB4 autocrine loop. Oncogene. 2012;31(18):2283–2297. doi: 10.1038/onc.2011.417. [DOI] [PubMed] [Google Scholar]
- 35.To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1alpha from HIF-2alpha in NBS1 repression. EMBO J. 2006;25(20):4784–4794. doi: 10.1038/sj.emboj.7601369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koshiji M, et al. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17(6):793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y, et al. Role and mechanism of hypoxia-inducible factor-1 in cell growth and apoptosis of breast cancer cell line MDA-MB-231. Oncol Lett. 2010;1(4):657–662. doi: 10.3892/ol_00000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan T, et al. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res. 2011;9(4):418–429. doi: 10.1158/1541-7786.MCR-10-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X, et al. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS One. 2009;4(4):e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren G, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72(12):3091–3104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 42.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27(58):7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad A, et al. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2010;122(2):337–346. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- 45.Halasi M, Gartel AL. A novel mode of FoxM1 regulation: Positive auto-regulatory loop. Cell Cycle. 2009;8(12):1966–1967. doi: 10.4161/cc.8.12.8708. [DOI] [PubMed] [Google Scholar]
- 46.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 47.Hatzis C, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang BH, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272(31):19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 49.Mahon PC, Hirota K, Semenza GL. FIH-1: A novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15(20):2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isaacs JS, et al. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277(33):29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- 51.Rius J, et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453(7196):807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belaiba RS, et al. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol Biol Cell. 2007;18(12):4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pagé EL, Robitaille GA, Pouysségur J, Richard DE. Induction of hypoxia-inducible factor-1alpha by transcriptional and translational mechanisms. J Biol Chem. 2002;277(50):48403–48409. doi: 10.1074/jbc.M209114200. [DOI] [PubMed] [Google Scholar]
- 54.Kikuchi J, et al. Phosphorylation-mediated EZH2 inactivation promotes drug resistance in multiple myeloma. J Clin Invest. 2015;125(12):4375–4390. doi: 10.1172/JCI80325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halasi M, Gartel AL. Suppression of FOXM1 sensitizes human cancer cells to cell death induced by DNA-damage. PLoS One. 2012;7(2):e31761. doi: 10.1371/journal.pone.0031761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, et al. FoxM1 in tumorigenicity of the neuroblastoma cells and renewal of the neural progenitors. Cancer Res. 2011;71(12):4292–4302. doi: 10.1158/0008-5472.CAN-10-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhat UG, Jagadeeswaran R, Halasi M, Gartel AL. Nucleophosmin interacts with FOXM1 and modulates the level and localization of FOXM1 in human cancer cells. J Biol Chem. 2011;286(48):41425–41433. doi: 10.1074/jbc.M111.270843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18(2):298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanharanta S, et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19(1):50–56. doi: 10.1038/nm.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mallen-St Clair J, et al. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev. 2012;26(5):439–444. doi: 10.1101/gad.181800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]