Significance
Singlet oxygen (1O2)- and EXECUTER1 (EX1)-dependent signaling triggers programmed cell death in seedlings and inhibits growth of mature plants of the fluorescent (flu) mutant of Arabidopsis. The EX1 protein has been located in chloroplasts to the grana margins close to where chlorophyll is synthesized and the disassembly of damaged photosystem II (PSII) and reassembly of active PSII take place. With the onset of 1O2-mediated signaling there is a rapid decline of EX1 that depends on the ATP-dependent zinc metalloprotease FtsH. Generation of 1O2 without the decline of EX1 is not sufficient to activate 1O2 signaling. As FtsH cleaves also the D1 reaction center protein of damaged PSII, EX1-dependent signaling seems not only spatially but also functionally linked to the repair of PSII.
Keywords: singlet oxygen, photosystem II, EXECUTER1-dependent signaling, stress, thylakoid FtsH protease
Abstract
Formation of singlet oxygen (1O2) has been implicated with damaging photosystem II (PSII) that needs to undergo continuous repair to maintain photosynthetic electron transport. In addition to its damaging effect, 1O2 has also been shown to act as a signal that triggers stress acclimation and an enhanced stress resistance. A signaling role of 1O2 was first documented in the fluorescent (flu) mutant of Arabidopsis. It strictly depends on the chloroplast protein EXECUTER1 (EX1) and happens under nonphotoinhibitory light conditions. Under severe light stress, signaling is initiated independently of EX1 by 1O2 that is thought to be generated at the acceptor side of active PSII within the core of grana stacks. The results of the present study suggest a second source of 1O2 formation in grana margins close to the site of chlorophyll synthesis where EX1 is localized and the disassembly of damaged and reassembly of active PSII take place. The initiation of 1O2 signaling in grana margins depends on EX1 and the ATP-dependent zinc metalloprotease FtsH. As FtsH cleaves also the D1 protein during the disassembly of damaged PSII, EX1- and 1O2-mediated signaling seems to be not only spatially but also functionally associated with the repair of PSII.
Plants exposed to environmental stress that interferes with the light-driven photosynthetic electron transport generate enhanced levels of highly reactive singlet oxygen (1O2) that has been implicated with damaging photosystem II (PSII), inhibiting photosynthesis and reducing the growth of plants (1). However, in addition to its potential toxicity, 1O2 may also activate signaling. This signaling role of 1O2 was first demonstrated in the conditional fluorescent (flu) mutant of Arabidopsis (2, 3). FLU is a nuclear-encoded chloroplast protein that functions as a negative feedback regulator of the Mg2+-branch of tetrapyrrole biosynthesis (4). Without this regulator, flu seedlings are unable to restrict the accumulation of the chlorophyll (Chl) precursor protochlorophyllide (Pchlide) in the dark. When these seedlings are transferred from the dark to the light they rapidly bleach and die due to the photosensitizing activity of excess amounts of free Pchlide that transfers its excitation energy onto ground-state triplet oxygen and transforms it into highly reactive 1O2 (2, 4). This property of the flu mutant has been exploited to study the physiological role of 1O2 by transferring light-grown flu plants to the dark and re-exposing them to light. By varying the length of the dark period the level of the photosensitizer Pchlide can be modulated noninvasively and conditions can be defined that minimize the cytotoxicity of 1O2 and reveal its signaling role. Under such mild stress conditions the release of 1O2 induces a rapid bleaching of flu seedlings, whereas in mature flu plants ready to bolt generation of 1O2 leads to growth inhibition and stress acclimation (5, 6). All these 1O2-mediated responses strictly depend on the activity of the chloroplast protein EXECUTER (EX) 1. In the flu/ex1 double mutant lacking EX1 these responses are abolished (3), even though flu/ex1 over-accumulates Pchlide in the dark and generates similar amounts of 1O2 during reillumination as the parental flu line (5). Hence, these responses are due to 1O2-mediated and EX1-dependent signaling rather than to 1O2 directly.
Singlet oxygen-mediated responses may also be activated under high light stress. Under these conditions 1O2 signaling occurs independently of EX1. Nonenzymatic oxidative breakdown products of β-carotene, a 1O2 scavenger associated with the reaction center (RC) of PSII, activate stress defense responses (7) or stress acclimation (8). Collectively, these results emphasize a key role of PSII as a major target of environmental stress that upon its damage leads to a significant reduction of plant productivity but at the same time is also involved in initiating 1O2-mediated signaling that allows plants to adapt to these stress conditions.
Formation of 1O2 that activates EX1-independent signaling has been located to the acceptor side of active PSII within the core of grana stacks (1, 7). It is still unclear whether this 1O2 may also activate EX1-dependent signaling. In the present work we have addressed this question by determining the localization of EX1 within the chloroplast. This approach was based on the premise that, due to its high reactivity and very short half-life, 1O2 interacts primarily with its nearest targets and thus in the case of EX1-dependent signaling should be generated close to where EX1 is localized. Our results suggest the existence of a previously unidentified source of 1O2 production in grana margins close to the site of chlorophyll synthesis where EX1 has been located and the disassembly of damaged PSII and reassembly of active PSII take place. Initiation of 1O2- and EX1-mediated signaling at this site depends on the FtsH protease that is also involved in the proteolytic breakdown of the PSII RC protein D1, thus linking repair of PSII with 1O2-mediated stress signaling.
Results
Localization of EX1 in Grana Margins.
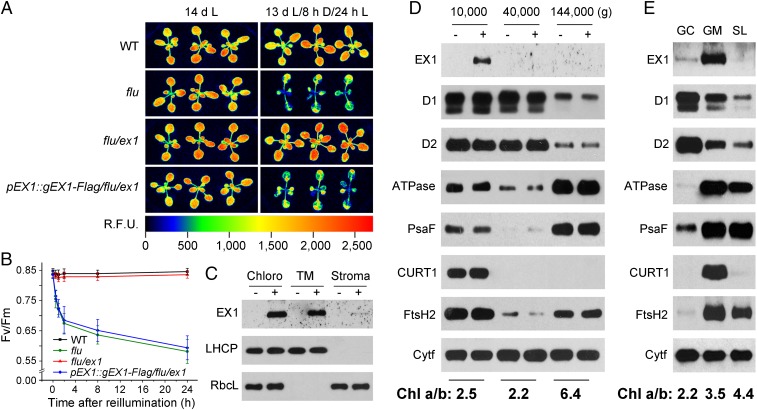
As a first step toward understanding how EX1 operates during 1O2 signaling its localization in chloroplasts was determined. Attempts to detect EX1 immunologically in plant homogenates or chloroplast lysates using antibodies against EX1 or EX1-specific peptides were unsuccessful. EX1 is a very low abundant protein as indicated by the initial failure of proteome studies to identify EX1 among the proteins of purified chloroplasts (9, 10). To enhance the sensitivity of EX1 detection, plants were transformed with DNA constructs that encode EX1 fused to different tags such as FLAG and GFP under control of the native EX1 (pEX1) or the CaMV 35S promoter, and tag-specific antibodies were then used to identify these EX1 proteins in plant homogenates. To ensure that tagged EX1 proteins retained their biological activity, the EXECUTER1 (gEX1) genes fused to tags were used to complement the flu/ex1 double mutant (Fig. 1 A and B). When grown under continuous light for 14 d (14 dL), seedlings of flu, flu/ex1, and the complemented pEX1::gEX1-Flag/flu/ex1 lines were indistinguishable and looked like wild-type (WT) control seedlings (Fig. 1A). When seedlings were transferred to the dark for 8 h (8 hD) and subsequently re-exposed to light for 24 h [13 dL/8 hD/24 hL (hours of light)], flu seedlings were severely affected by the release of 1O2 as shown by their reduced size and transient Chl fluorescence changes (Fig. 1A) and the decline of the maximum quantum efficiency of PSII (Fig. 1B). In contrast, flu/ex1 seedlings were phenotypically indistinguishable from wild type (WT) (Fig. 1 A and B), even though they accumulate similar excess amounts of Pchlide in the dark and generate the same amounts of 1O2 during reillumination as flu (5). Expression of the FLAG-tagged EX1 protein in flu/ex1 seedlings fully restored the 1O2-mediated responses of the parental flu line (Fig. 1 A and B). Similar results were obtained with flu/ex1 plants complemented with a gEX1-GFP gene (Fig. S1). These results demonstrate that the tags attached to EX1 do not interfere with its biological activity and that 1O2-mediated responses of flu seedlings are not caused directly by 1O2 but strictly depend on EX1 and are induced at 1O2 concentrations too low to visibly damage the plant (Fig. 1 A and B) (5).
Fig. 1.
The biological activity and localization of EX1-FLAG. (A) Complementation of flu/ex1 with a genomic DNA construct that encodes EX1 fused to the FLAG-tag (gEX1-Flag) under control of the native EX1 promoter (pEX1). WT, flu, flu/ex1, and the complemented flu/ex1 line (pEX1::gEX1-Flag/flu/ex1) were grown for 13 d under continuous light and either kept for another 32 h under continuous light (14 dL) or transferred to the dark for 8 h and re-exposed to light for 24 h (13 dL/8 hD/24 hL). Singlet oxygen-mediated growth inhibition and photoinhibition of PSII as revealed by transient Chl fluorescence changes in flu are suppressed in flu/ex1 and restored in the complemented flu/ex1 line. R.F.U.: Relative Fluorescence Unit. (B) Singlet oxygen-mediated changes of the maximum efficiency of PSII (Fv/Fm). Seedlings were grown for 5 d under continuous light, shifted to the dark for 8 h, and re-exposed to light for various lengths of time. PSII activity is inhibited in flu and the complemented flu/ex1 but not in flu/ex1 and WT, indicating that 1O2-mediated and EX1-dependent signaling occurs under nonphotoinhibitory light and 1O2 generated in seedlings with a flu background does not directly damage PSII. The results represent the mean and SD of Fv/Fm measurements of at least 30 individual seedlings. (C) Localization of EX1-FLAG in chloroplast membranes. Chloroplasts (Chloro) isolated from flu/ex1 complemented with pEX1::gEX1-Flag (+) and noncomplemented flu/ex1 control lines (−) were lysed and separated by centrifugation into membrane (TM) and stroma (Stroma) fractions. Proteins were solubilized, separated by SDS/PAGE, and analyzed on Western blots using antisera against FLAG, LHCP, and the large subunit of ribulose-1,5-bisphospate carboxylase (RbcL). (D) Localization of EX1-FLAG in grana stacks that retain their margin parts. Chloroplast membranes were treated with the detergent digitonin and separated by differential centrifugation into three subfractions that were pelleted at 10,000 × g, 40,000 × g and 144,000 × g. Proteins of these subfractions were analyzed by immunoblotting as shown in C, and the PSII RC proteins D1 and D2, the PSI-specific PsaF, ATPase, FtsH2, and the margin-specific CURT1 were used as markers to locate EX1-FLAG. Colocalization of EX1-FLAG and CURT1 indicates that EX1 is highly enriched in grana stacks that retain their margin parts. Cytochrome f (Cytf) was used as a loading control. (E) Localization of EX1-FLAG in grana margins. Detergent-treated chloroplast membranes were separated into grana core (GC), grana margins (GM), and stroma lamellae (SL). The same marker proteins as shown in D were used to locate EX1-FLAG. The various subfractions were also characterized by their Chl a/b ratios shown below D and E.
Fig. S1.
Complementation of flu/ex1 with a genomic DNA construct that encodes EX1 fused to the GFP-tag (gEX1-GFP) under control of the native EX1 promoter (pEX1). WT, flu, flu/ex1, and the complemented flu/ex1 line (pEX1::gEX1-GFP) were grown for 21 d under continuous light (Left) or under an 8-h dark/16-h light regimen (Right).
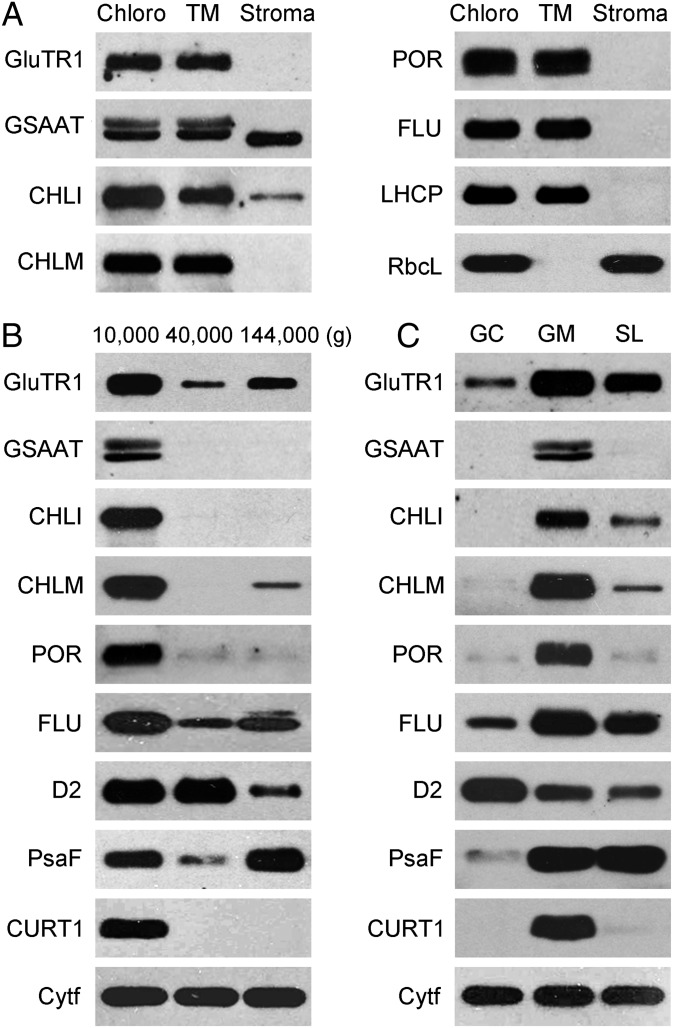
To localize EX1, chloroplasts isolated from light-grown complemented pEX1::gEX1-Flag/ex1 seedlings were lysed, and soluble stroma proteins were separated by centrifugation from insoluble thylakoid membrane proteins. Almost all of the FLAG-tagged EX1 proteins were recovered in the membrane fraction (Fig. 1C). This membrane fraction was then treated with the detergent digitonin to yield grana stacks and stroma lamellae and separated by differential centrifugation into three subfractions that were pelleted at 10,000 × g, 40,000 × g, and 144,000 × g (11, 12). Active PSII has been reported to be present mainly in the core of tightly appressed grana membranes, whereas photosystem I (PSI), ATPase, and damaged PSII reside predominantly in stroma lamellae (11, 13). Chl a/b ratio measurements and the distribution of marker proteins were used to identify the three fractions (Fig. 1D). The 10,000 × g pellet showed a Chl a/b ratio of 2.5, was highly enriched in the D1 and D2 RC proteins of PSII, was depleted in the PSI-specific PsaF protein and ATPase, and also contained the marker of grana margins, CURT1A (14). The 40,000 × g pellet was similarly enriched in D1 and D2 and depleted in PsaF and ATPase as the 10,000 × g pellet, had a slightly lower Chl a/b ratio of 2.2, but did not contain CURT1A. The 144,000 × g pellet had a Chl a/b ratio of 6.4. It was highly enriched in ATPase and the PSI-specific marker protein PsaF, but was virtually devoid of the margin-specific CURT1A protein. Based on the distribution of marker proteins and Chl a/b ratios, the 10,000 × g, 40,000 × g, and 144,000 × g fractions were identified as grana stacks that retained their margin parts, grana cores that lacked the margins, and stroma lamellae, respectively. Almost all of the FLAG-tagged EX1 present in chloroplast membranes was recovered in the 10,000 × g grana stack fraction, but was absent from the 40,000 × g grana core and the 144,000 × g stroma lamella fractions (Fig. 1D). Colocalization of EX1 with the margin-specific CURT1A protein during membrane fractionation suggests that EX1 is highly enriched in grana margins. To test this hypothesis, grana core and margins were separated and their polypeptide compositions were compared. The method for isolating grana margins (15) and separating them from grana cores is similar to the procedure used for the fractionation of thylakoid membranes (11, 12), except that the incubation of isolated membranes occurs at a higher digitonin concentration and for a longer time. The results of this fractionation procedure confirmed our previous suggestion: The purified margins were highly enriched in CURT1A and EX1, whereas only trace amounts of these proteins were found in the grana core and stroma lamellae fractions (Fig. 1E).
Identification of Proteins That Are Associated with EX1.
Grana margins play an important role during the repair of damaged PSII complexes (15–17). The repair cycle is primarily designed for the specific replacement of D1, which requires the disassembly and reassembly of PSII complexes and depends on rapid protein translocation between grana core, grana margin, and stroma lamella (15). Damaged D1 is proteolytically cleaved by the FtsH protease (18, 19). As most of the FtsH is found in grana margins (15, 17), this part of the grana stacks seems to be a main site of protein degradation (15). This conclusion is in line with the high enrichment of FtsH2 in the 10,000 × g grana stack fraction and isolated margins and its low concentration in the grana core fractions (Fig. 1 D and E).
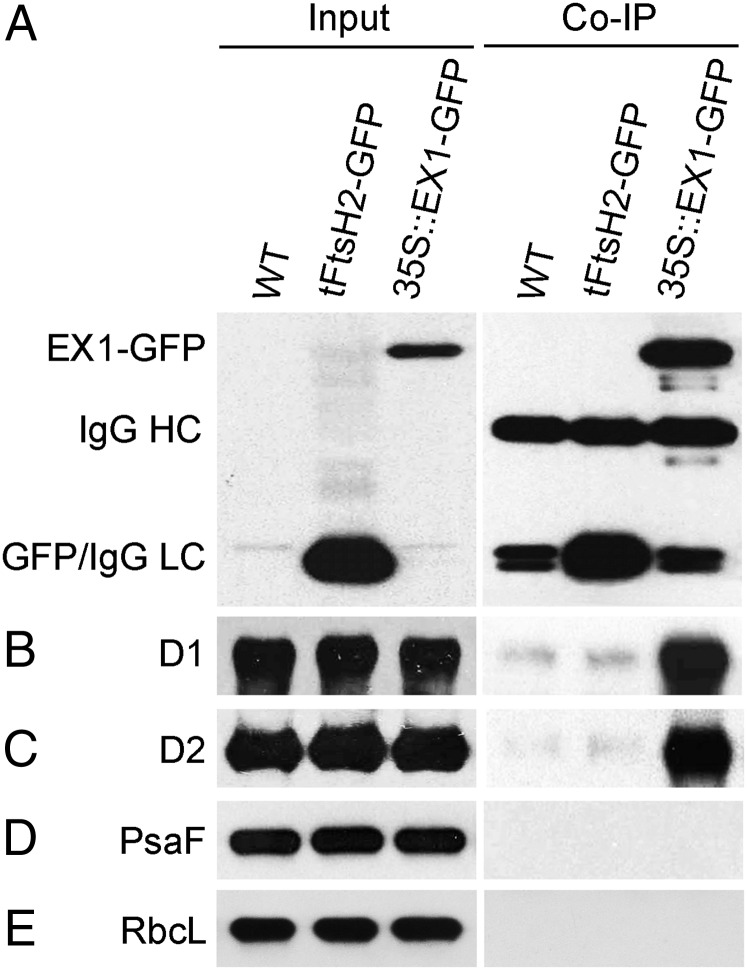
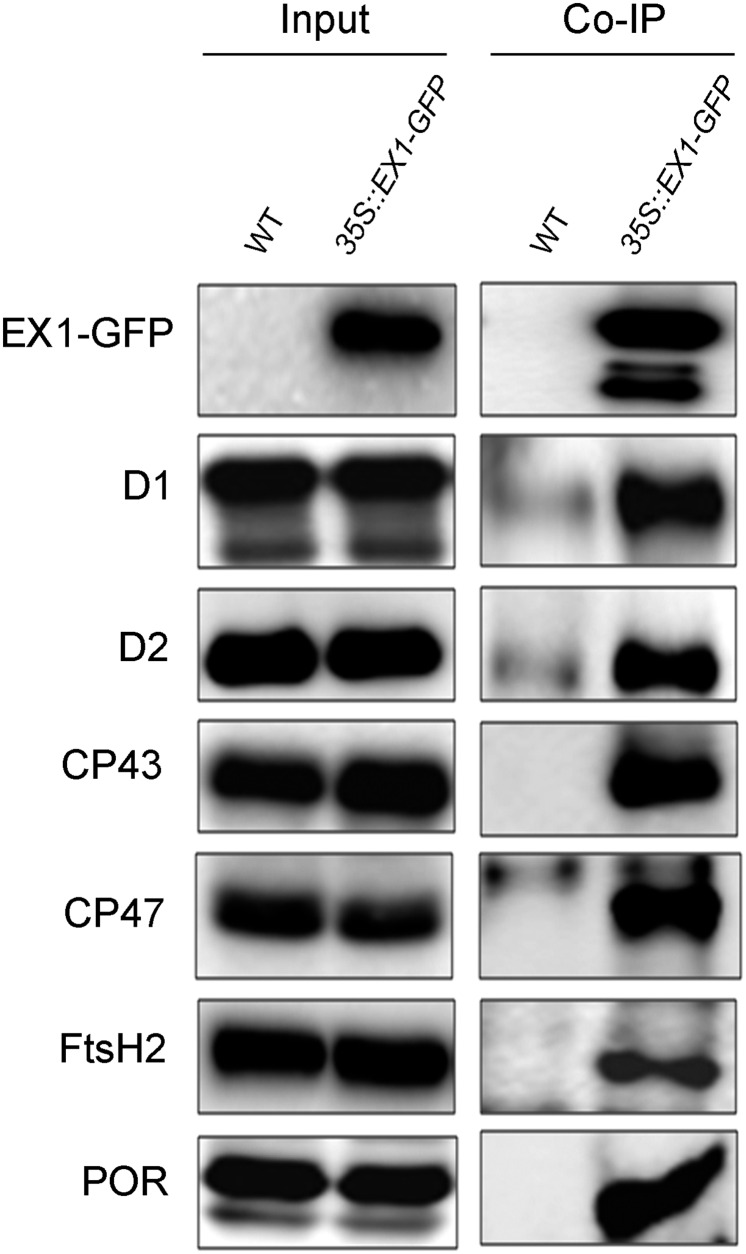
The concentration of PSII particles in grana margins has been shown to be significantly lower than in the grana core and to reach similar amounts as PSI (15) (Fig. 1E). To determine whether EX1 is associated with only one or both photosystems in the margin part of grana stacks, a coimmunoprecipitation assay was used with input samples from three different Arabidopsis lines: WT, ex1 complemented with EX1-GFP, and a line expressing a GFP attached to the signal sequence of the plastid-specific FtsH2 protease subunit that targets the GFP to chloroplasts (Fig. 2) (20). The D1 and D2 proteins of the PSII RC were coimmunoprecipitated together with the EX1-GFP but not with the plastid-targeted GFP. On the other hand, the PSI-specific marker protein PsaF was not found in the immunoprecipitate (Fig. 2). Protein analysis of the EX1-GFP immunoprecipitate by mass spectrometry and Western blots revealed that EX1 forms part of a larger physical unit in grana margins that, in addition to the RC of PSII and several PSII light-harvesting Chl a/b proteins, comprises also chloroplast-specific subunits of the FtsH protease, the NADPH-protochlorophyllide oxidoreductases (POR) B and C catalyzing the light-dependent reduction of Pchlide to Chlide (21, 22), the Tic110 protein translocation channel, the Ca2+-sensing protein and the GTP-binding Tu, and RAB GTPase homolog E1B elongation factors (Table S1 and Fig. S2).
Fig. 2.
Identification of EX1-interacting proteins by coimmunoprecipitation. Input sample: Total proteins extracted from 5-d-old light-grown seedlings of WT and lines that express the plastid-targeted tFtsH2-GFP or EX1-GFP were separated by SDS/PAGE and probed on Western blots with antisera against GFP (A), D1 (B), D2 (C), PsaF (D), and RbcL (E). Co-IP sample: Immunoreactive proteins were precipitated from each of the three input samples by adding protein G beads with a conjugated monoclonal anti-GFP antibody. Bound proteins were solubilized and separated by SDS/PAGE and probed on Western blots as described above. The three unspecific bands cross-reacting with the GFP antibody in the WT immunoprecipitate sample represent the heavy (IgGHC) and two light (IgGLC) chains of IgG. In the tFtsH2-GFP immunoprecipitate, the IgGLC bands are superimposed by GFP. In the EX1-GFP immunoprecipitate sample, only EX1-GFP but not GFP is found. The PSII RC proteins D1 and D2 coimmunoprecipitate with EX1-GFP but not GFP or the PSI-specific PsaF. RbcL served as a nonbinding control.
Table S1.
Identification of proteins associated with EX1 by coimmunoprecipitation and mass spectrometry
| AGI code | Protein | No. of peptide matches | Sequence coverage, % | ||||||||
| 1 | 2 | 3 | 4 | BC | DC | 1 | 2 | 3 | 4 | ||
| AT4G33630 | EX1 | protein of unknown function (DUF3506) | 68 | 55 | 44 | 41 | 0 | 0 | 65.6 | 65.4 | 66.5 | 53 |
| ATCG00020 | PSBA | photosystem II reaction center protein A | 12 | 3 | 11 | 5 | 3 | 2 | 13.3 | 11.1 | 44.2 | 16 |
| ATCG00680 | PSBB | photosystem II reaction center protein B | 22 | 22 | 20 | 12 | 5 | 7 | 39.8 | 29.1 | 39.8 | 23 |
| ATCG00280 | PSBC | photosystem II reaction center protein C | 19 | 19 | 19 | 10 | 4 | 3 | 27.3 | 29.4 | 27.3 | 27 |
| ATCG00270 | PSBD | photosystem II reaction center protein D | 16 | 4 | 15 | 5 | 3 | 3 | 12.5 | 15.3 | 30.9 | 22 |
| AT1G71500 | PSB33 | photosystem b protein 33 | 10 | 6 | 8 | 0 | 0 | 0 | 48.1 | 34.5 | 40.8 | ND |
| AT4G27440 | PORB | protochlorophyllide oxidoreductase B | 9 | 8 | 2 | 8 | 3 | 4 | 32.4 | 39.7 | 15.2 | 24 |
| AT1G03630 | PORC | protochlorophyllide oxidoreductase C | 6 | 6 | 1 | 4 | 0 | 1 | 38.4 | 27.8 | 4 | 17 |
| AT5G42270 | VAR1 | FTSH5 | FtsH extracellular protease family | 12 | 13 | 1 | 12 | 3 | 7 | 26.4 | 31.3 | 8.7 | 21 |
| AT1G50250 | FTSH1 | FTSH protease 1 | 10 | 10 | 1 | 2 | 0 | 0 | 32.8 | 32.8 | 7 | 4 |
| AT2G30950 | VAR2 | FTSH2 | FtsH extracellular protease family | 9 | 9 | 1 | 14 | 6 | 2 | 30.8 | 30.8 | 9.8 | 24 |
| AT1G06430 | FTSH8 | FTSH protease 8 | 6 | 6 | 1 | 0 | 0 | 0 | 15.8 | 15.8 | 7.8 | ND |
| AT5G50920 | CLPC1 | CLPC homolog 1 | 9 | 11 | 6 | 21 | 12 | 8 | 31.5 | 17.7 | 18.7 | 32 |
| AT1G06950 | TIC110 | translocon at the inner envelope membrane of chloroplasts 110 | 19 | 13 | 5 | 22 | 4 | 4 | 33.9 | 23.1 | 15.9 | 29 |
| AT1G44575 | NPQ4, PSBS | chlorophyll A-B binding family protein | 3 | 3 | 3 | 6 | 2 | 3 | 23.8 | 18.9 | 23.8 | 25 |
| AT1G29930 | LHCB1.3 | chlorophyll A/B binding protein 1 | 16 | 15 | 10 | 6 | 4 | 6 | 52.8 | 52.8 | 50.2 | 18 |
| AT3G47470 | LHCA4 | light-harvesting chlorophyll-protein complex I subunit A4 | 6 | 6 | 6 | 5 | 1 | 1 | 4.8 | 39.8 | 39.8 | 32 |
| AT2G05100 | LHCB2.1 | photosystem II light harvesting complex gene 2.1 | 8 | 4 | 8 | 0 | 0 | 0 | 21.5 | 21.5 | 44.2 | ND |
| AT3G27690 | LHCB2.4 | photosystem II light harvesting complex gene 2.3 | 2 | 2 | 2 | 0 | 0 | 0 | 19.2 | 19.2 | 33.8 | ND |
| AT3G08940 | LHCB4.2 | light-harvesting complex photosystem II | 5 | 7 | 4 | 6 | 3 | 5 | 34.8 | 34.8 | 36.9 | 28 |
| AT1G54780 | TLP18.3 | thylakoid lumen 18.3 kDa protein | 2 | 2 | 2 | 3 | 0 | 1 | 37.9 | 28.8 | 37.9 | 14 |
| AT4G01050 | TROL | thylakoid rhodanese-like | 2 | 1 | 1 | 7 | 2 | 2 | 23.2 | 12.9 | 5.6 | 15 |
| AT5G42080 | DL1 | dynamin-like protein | 4 | 4 | 1 | 4 | 2 | 0 | 21.2 | 28.2 | 6.4 | 7.9 |
| AT5G55280 | FTSZ1-1 | homolog of bacterial cytokinesis Z-ring protein FTSZ 1–1 | 2 | 2 | 1 | 2 | 0 | 0 | 20.6 | 20.6 | 5.3 | 9.5 |
| AT3G19820 | DWF1 | cell elongation protein/DWARF1/DIMINUTO (DIM) | 4 | 4 | 1 | 5 | 1 | 0 | 14.6 | 16 | 5.7 | 9.6 |
| AT4G20360 | TRABE1B | RAB GTPase homolog E1B | 35 | 22 | 33 | 23 | 6 | 14 | 60.5 | 56.9 | 60.5 | 59 |
| AT5G60390 | GTP binding Elongation factor Tu family protein | 23 | 18 | 2 | 19 | 3 | 9 | 60.4 | 49 | 14.9 | 41 |
| AT5G13650 | SVR3 | SUPPRESSOR OF VARIEGATION 3 | 2 | 3 | 2 | 2 | 2 | 0 | 18.3 | 17.8 | 10.4 | 4.1 |
| AT5G23060 | CaS | calcium sensing receptor | 15 | 19 | 8 | 16 | 7 | 9 | 52.7 | 52.5 | 34.4 | 35 |
| AT5G66680 | DGL1 | dolichyl-diphosphooligosaccharide-protein glycosyltransferase 48-kDa subunit family protein | 2 | 3 | 1 | 3 | 0 | 0 | 15.8 | 17.6 | 6.2 | 9.6 |
Proteins were considered truly coprecipitated if found in three biological replicates using on-bead digestion or in-gel digestion of protein followed by mass spectrometry. Protein identity was considered if at least two peptide matches were found after MS/MS analysis. AGI, Arabidopsis Genome Initiative; BC, bead control without antibody; DC, dummy antibody control using myc antibody; ND, not determined.
Fig. S2.
Identification of chloroplast proteins that coimmunoprecipitate with EX1-GFP. Coimmunoprecipitated proteins were separated by SDS/PAGE and detected by immunoblotting.
Does the Release of 1O2 in the flu Mutant Contribute to the Proteolytic Degradation of the D1 Protein?
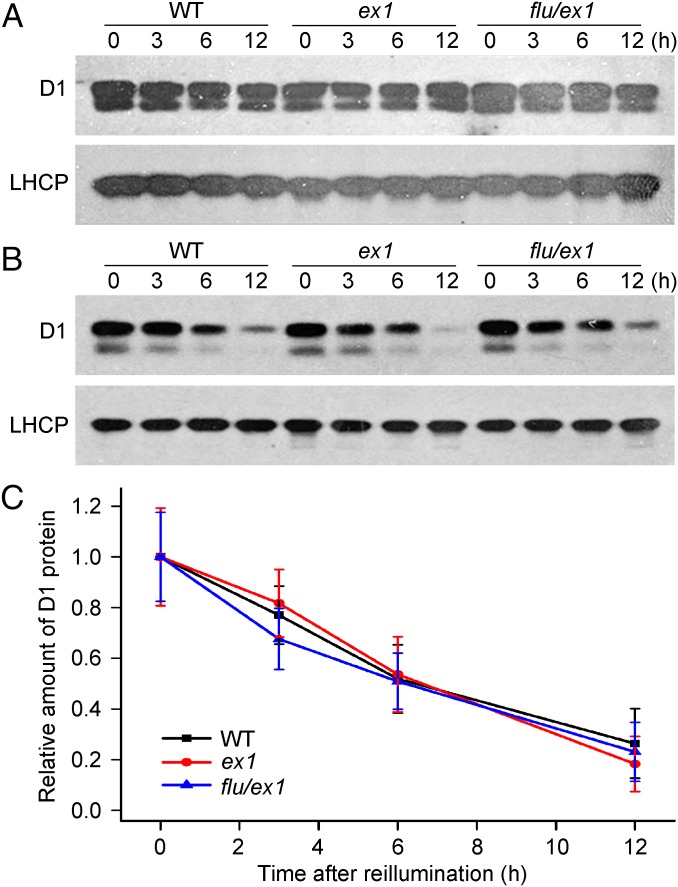
Colocalization of EX1, PSII, and the FtsH protease within the margin region suggests that 1O2-mediated and EX1-dependent signaling could be intimately associated with the proteolytic degradation of the D1 protein. Numerous signaling peptide fragments have been identified in plants and shown to control a wide range of physiological responses (23). Furthermore, one of the proteolytic D1 peptides in cyanobacteria has been suggested to be biologically active and control the synthesis of D1 (24). Thus, it seemed possible that proteolytic breakdown products of D1 in higher plants may act as a stable signaling component of the 1O2- and EX1-dependent signaling pathway. To see whether 1O2 generated in flu influences the degradation of D1, WT, ex1, and flu/ex1 seedlings grown under moderate light (90 μmol m−2⋅s−1 at 20 °C) were transferred to the dark for 8 h and re-exposed again to moderate light to generate 1O2. Thirty minutes before reillumination these seedlings were sprayed with lincomycin, an inhibitor of protein synthesis in chloroplasts used previously to block D1 synthesis (18). In mock-treated control seedlings the concentration of D1 remained constant during reillumination (Fig. 3A). However, in lincomycin-treated seedlings the D1 concentration rapidly declined (Fig. 3B). In the flu mutant lincomycin treatment did not interfere with the overaccumulation of Pchlide in the dark and the release of 1O2 during reillumination as shown by a comparison of lincomycin-treated and -untreated seedlings (Fig. S3). As flu/ex1 seedlings over-accumulate Pchlide in the dark and generate similar amounts of 1O2 during reillumination as flu (5), the possible effect of this 1O2 on the stability of D1 could be analyzed in these seedlings without interference by the rapid 1O2-mediated loss of chloroplast integrity as seen in the flu mutant (5). The rate of D1 degradation in lincomycin-treated seedlings of flu/ex1 was the same as in ex1 and WT, indicating that under moderate light the stability of D1 was not affected by 1O2 generated by the photosensitizer Pchlide in the flu mutant (Fig. 3C). Singlet oxygen-mediated and EX1-dependent signaling seems to be unrelated to the proteolytic breakdown of the D1 protein. Singlet oxygen generated in the flu mutant did not enhance the decline of D1 during reillumination of predarkened seedlings because its concentration was too low and/or the site of 1O2 production was too far away from the grana core where, supposedly, 1O2 oxidizes the D1 protein (1).
Fig. 3.
The turnover of the PSII RC protein D1 in the presence or absence of 1O2 generated in plants with a flu background. (A and B) Seedlings of WT, ex1, and flu/ex1 grown for 5 d under continuous light were transferred to the dark for 8 h and re-exposed to light. Thirty minutes before the end of the dark period seedlings were sprayed with a buffer containing no (A) or 5 mM lincomycin (B). During reillumination proteins were extracted from mock- and lincomycin-treated seedlings after various lengths of time, and concentrations of D1 were probed by Western blot analysis. The concentrations of the light-harvesting chlorophyll a/b protein (LHCP) in these samples were used as loading controls. (C) The amounts of D1 shown in B were quantified densitometrically and expressed relative to the loading control. The relative concentration of D1 at time “0” was set as “1.0.” The results represent the mean and SD of three independent experiments.
Fig. S3.
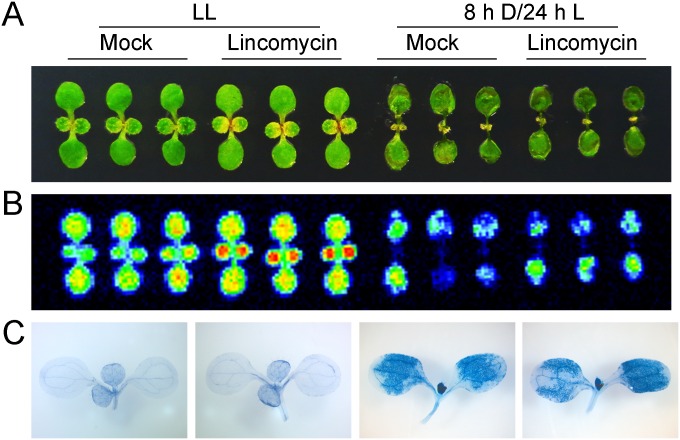
Singlet oxygen-mediated signaling in lincomycin-treated flu seedlings. (A) Light-grown flu seedlings were transferred to the dark for 8 h and re-exposed to light for 24 h (8 hD/24 hL). Thirty minutes before reillumination seedlings were sprayed with or without 5 mM lincomycin. Control flu seedlings kept under continuous light (LL) were sprayed with or without lincomycin in the light 24 h before images were taken. (B) Transient Chl fluorescence images taken from seedlings shown in A reveal a similar strong photoinhibition of PSII in mock- and lincomycin-treated flu seedlings subjected to the dark-to-light shift, but not in LL seedlings. (C) Singlet oxygen-mediated cell death as revealed by trypan blue staining occurs only in flu seedlings after a dark-to-light shift and is not affected by lincomycin.
FtsH2-Dependent Decline of EX1 Is Required for 1O2-Mediated and EX1-Dependent Signaling.
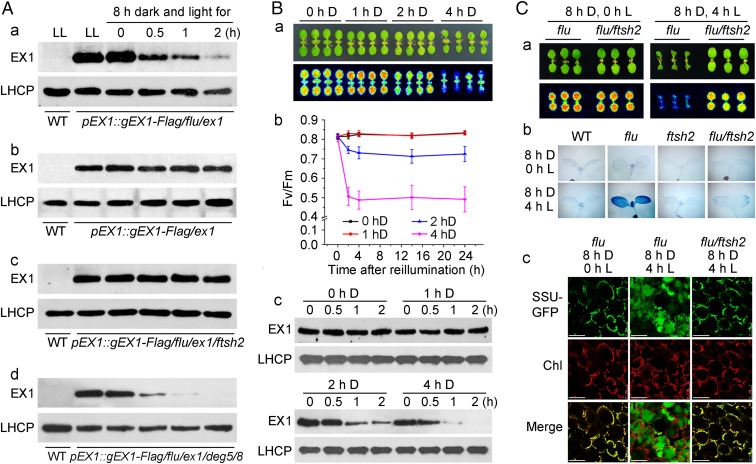
As these experiments failed to show a contribution of D1 degradation to 1O2-mediated and EX1-dependent signaling, it could be EX1 itself that at the beginning of 1O2 production activates signaling. This possibility was studied by analyzing the fate of EX1 after the release of 1O2 (Fig. 4A). In pEX1::gEX1-Flag/flu/ex1 seedlings the concentration of EX1-FLAG declined rapidly once 1O2 was generated (Fig. 4 A, a), whereas in complemented pEX1::gEX1-Flag/ex1 control seedlings that lack the flu mutation and do not generate 1O2 after a dark/light shift the concentration of EX1-FLAG remained constant throughout reillumination (Fig. 4 A, b). The decline of EX1 required protease activity. In the pEX1::gEX1-Flag/flu/ex1/ftsh2 transgenic line that lacks FtsH2, the EX1-FLAG concentration remained constant throughout reillumination (Fig. 4 A, c). As shown above, FtsH2 colocalizes with EX1 in grana margins, and it is also involved in proteolytic degradation of D1 (15, 17, 18). In addition to FtsH, under high light stress Deg proteases such as Deg5 and Deg8 also contribute to the proteolytic breakdown of D1 (25). However, inactivation of these proteases in the pEX1::gEX1-Flag/flu/ex1 background does not block the rapid decline of EX1 (Fig. 4 A, d).
Fig. 4.
Generation of 1O2 and the FtsH2-dependent decline of EX1 are both required for the activation of 1O2-mediated and EX1-dependent signaling. (A) Singlet oxygen and FtsH2-dependent decline of EX1-FLAG. In light-grown pEX1::gEX1-Flag/flu/ex1 seedlings transferred to the dark and re-exposed to light for up to 2 h, the concentration of EX1-FLAG rapidly declines (a) but not in pEX1::gEX1-Flag/ex1 seedlings that lack the flu mutation (b) or in FtsH2-deficient pEX1::gEX1-Flag/flu/ex1/ftsh2 seedlings (c). The decline of EX1-Flag requires FtsH2 but not the Deg proteases Deg5 and Deg8 (d). (B) The effect of increasing doses of 1O2 on the growth and transient Chl fluorescence changes (a), the maximum quantum efficiency (Fv/Fm) of PSII (b), and the 1O2-dependent decline of EX1-FLAG in complemented pEX1::gEX1-Flag/flu/ex1 seedlings grown under continuous light and shifted for 1, 2, and 4 h to the dark (“D”) and re-exposed to light for up to 2 h (c). The results shown in b represent the mean and SD of Fv/Fm measurements of at least 30 individual seedlings. Control plants were kept under continuous light (LL, 0 hD). LHCP was used as a loading control. (C) The FtsH2-dependent activation of 1O2-mediated and EX1-dependent signaling in flu seedlings. The 1O2-mediated suppression of PSII activity and growth inhibition of flu (a), 1O2-mediated cell death response (5) (b), and chloroplast leakage (5) (c) are abrogated in flu/ftsh2.
The rapid increase of 1O2 production in flu seedlings, following a dark-to-light shift, is paralleled by the rapid decline of EX1-FLAG and suggests that both processes are interconnected and may be involved in jointly activating the 1O2- and EX1-dependent signaling pathway. Two experiments strongly support this notion. First, by increasing the length of the dark treatment stepwise from 1 to 4 h one can enhance noninvasively the amount of Pchlide (5). The dose-dependent intensity of 1O2-mediated stress responses during reillumination (Fig. 4 B, a and b) is closely correlated with a dose-dependent decline of EX1 (Fig. 4 B, c). Second, light-grown flu and flu/ftsh2 mutants shifted to the dark start to accumulate similar excess amounts of the photosensitizer Pchlide (Fig. S4 A and B). During reillumination flu seedlings showed the expected 1O2-mediated stress response whereas flu/ftsh2 seedlings did not (Fig. 4 C, a–c). Thus, the decline of EX1 in the flu mutant and the constant high level of EX1 in flu/ftsh2, as shown in Fig. 4A, correlate with the onset of 1O2-mediated signaling in the former group of seedlings and the lack of signaling in the latter group. Contrary to light-grown seedlings exposed to a dark-to-light shift, in etiolated flu seedlings transferred to the light the 1O2-mediated cell death does not depend on EX1 (Fig. S4C) (26). Likewise, in similarly treated etiolated flu/ftsh2 seedlings the EX1-independent cell death response is also induced, indicating that the ftsh2 mutation does not suppress the overaccumulation of Pchlide and that in both mutant backgrounds Pchlide acts as a photosensitizer and generates similar amounts of 1O2 (Fig. S4C).
Fig. S4.
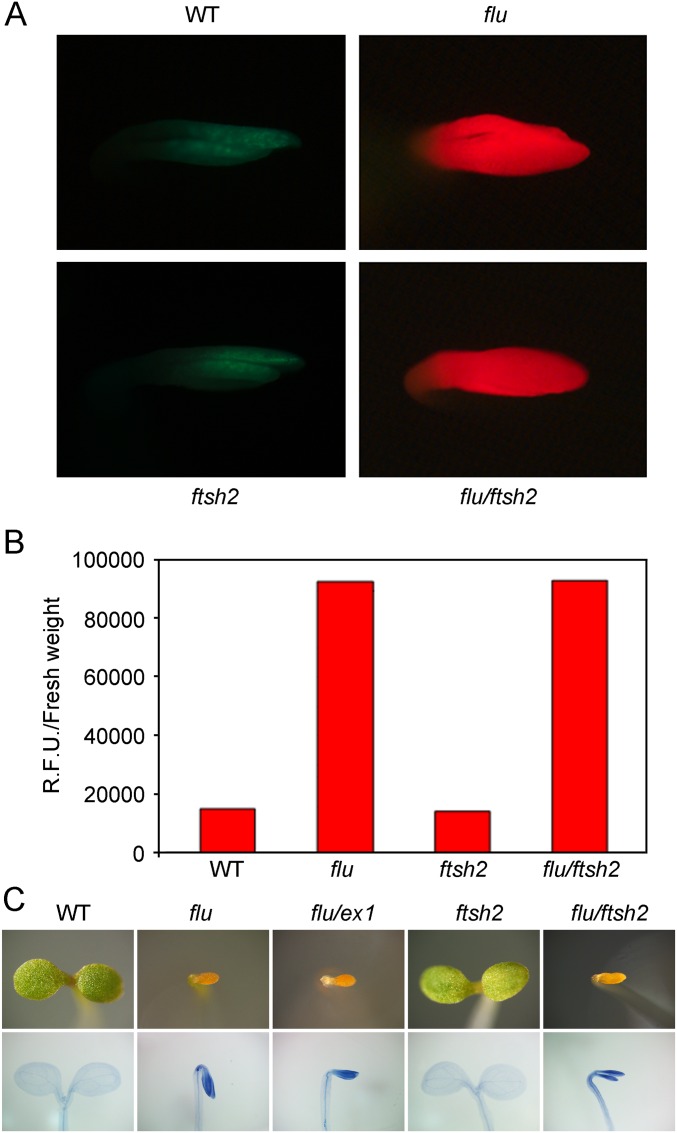
Overaccumulation of Pchlide and 1O2-mediated cell death in flu and flu/ftsh2 seedlings grown in the dark for 4 d. WT, flu/ex1, and ftsh2 seedlings were used as controls. (A) Four-day-old seedlings grown in the dark (etiolated seedlings) were exposed to blue light (400–450 nm), and the emitted fluorescence was recorded (2). The bright red fluorescence emitted by flu and flu/ftsh2 is caused by the excitation of free Pchlide. (B) Tetrapyrroles were extracted from etiolated seedlings and separated by HPLC, and the relative amounts of Pchlide were determined by their fluorescence. (C) Pictures of etiolated seedlings that were transferred to light for 48 h (Upper). Singlet oxygen-mediated cell death in these seedlings was revealed by trypan blue staining (Lower). In etiolated flu seedlings transferred to the light, 1O2-mediated cell death does not depend on EX1 and happens in both flu and flu/ex1 (26). Similarly treated flu/ftsh2 seedlings respond in the same way. In all three lines with a flu mutation, Pchlide acts as a photosensitizer and generates 1O2 that triggers the same death response. Thus, the photosensitizing activity of Pchlide and generation of 1O2 seem not to be affected by the ftsh2 mutation.
The Origin of 1O2 That Activates EX1-Dependent Signaling.
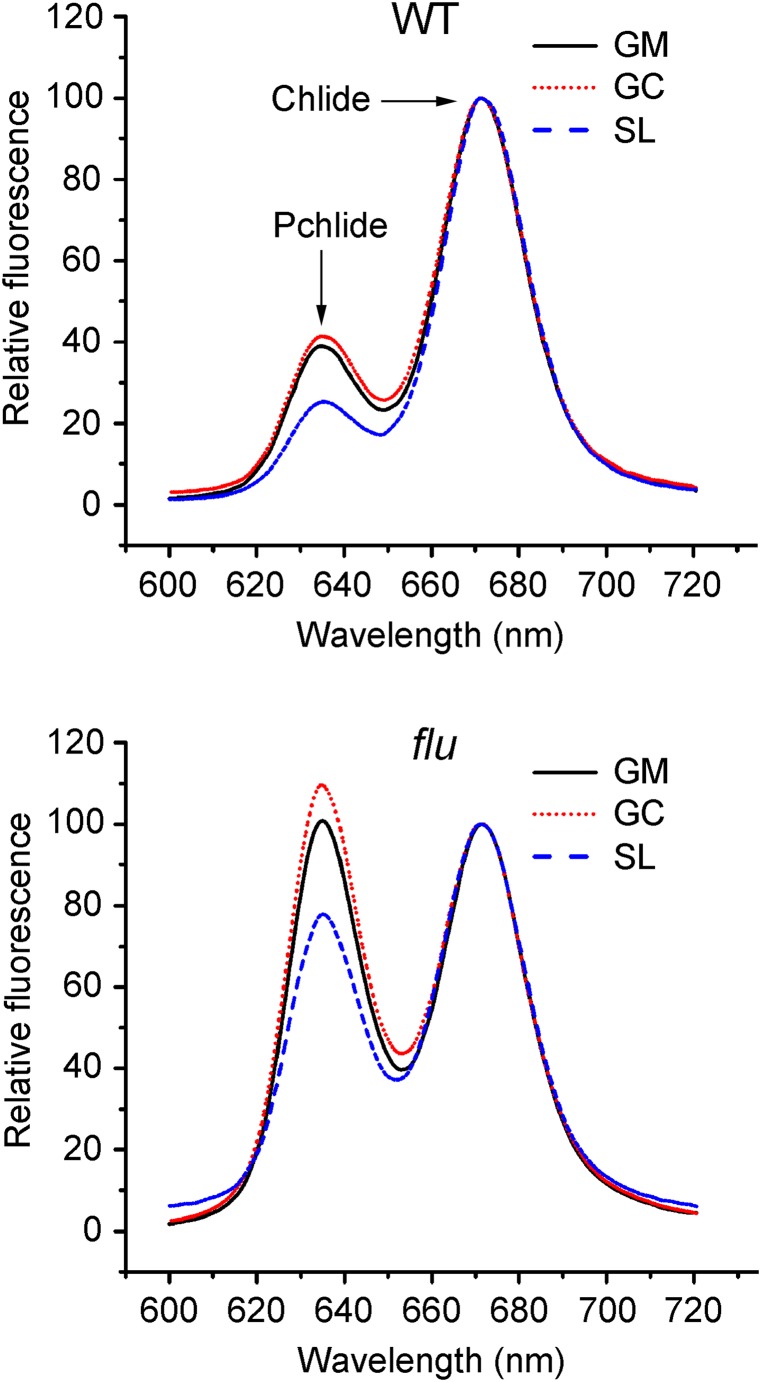
In light-grown plants placed under high light stress, formation of 1O2 has been primarily ascribed to the photosensitizing activity of excited 3Chl in the RC of active PSII that resides within the grana core (27, 28). Because of its very short half-life and diffusion range this 1O2 probably does not interact selectively with specific targets farther away in grana margins or stroma lamellae (29). Thus, it seems more likely that EX1-dependent signaling is activated by 1O2 generated in grana margins close to where EX1 is localized. A possible source of 1O2 production at this site may be the photosensitizing activity of tetrapyrroles destined for the assembly of PSII. So far, very little is known about the fate of Chl during the disassembly and reassembly of PSII. It has been proposed that Chl derived from damaged PSII is recycled and used for the reconstitution of active PSII (30). Also, newly synthesized Chl is likely to be used for the reassembly of active PSII; rapid turnover of Chl in green plants has previously been shown to be confined to the core of PSII (31). If true, enzymes of Chl biosynthesis should be localized close to the site of PSII repair in grana margins. To test this prediction, the localization of the following enzymes catalyzing the initial and final steps of Chl synthesis was determined: glutamyl-tRNA reductase (GluTR), glutamate-1-semialdehyde aminotransferase (GSAAT), Mg2+-chelatase subunit I (CHLI), Mg2+-protoporphyrin IX methyl transferase (CHLM), and PORs B and C. Although parts of GSAAT and CHLI were present also in the water-soluble stroma fraction of lysed chloroplasts, the other enzymes were almost exclusively associated with thylakoid membranes (Fig. 5A). After these membranes were treated with digitonin and fractionated by differential centrifugation, all enzymes tested were predominantly found in the 10,000 × g grana stack fraction that contains grana margins but not in the 40,000 × g grana core fraction that lacks the margins or in stroma lamellae. A similar localization was found for the negative feedback regulator of tetrapyrrole biosynthesis, FLU (Fig. 5B). FLU forms part of a membrane complex that in addition to FLU comprises the Mg2+-protoporphyrin IX monomethylester oxidative cyclase (CHL27) and PORs B and C and has been shown to interact in the dark also with GluTR (32). Hence, these results strongly support the notion that de novo synthesis of Chl in green plants occurs mainly in the margin region of grana stacks. This conclusion was confirmed by the analysis of the purified margin fraction (Fig. 5C). All enzymes tested and FLU were highly enriched in the margin region of grana stacks but were hardly detectable in the grana core fraction (Fig. 5C). As PORs B and C colocalize with EX1 and are very close to PSII and the FtsH protease (Table S1, Fig. S2, and Fig. 2), Ex1-dependent signaling seems to take its origin in grana margins adjacent to where Chl is released during the disassembly of damaged PSII, and newly synthesized Chl is integrated into the reconstituted PSII. Whereas EX1 is confined to this site, excess Pchlide that accumulates within flu seedlings following a light/dark shift is not restricted to the margin region but is distributed throughout the thylakoid membrane (Fig. 6). This finding re-emphasizes our previous conclusion that the concentration of 1O2 generated in light-adapted flu seedlings by Pchlide that accumulates during an 8-h dark period is too low to damage directly the plant and that the biological activity of this 1O2 strictly depends on the presence of EX1.
Fig. 5.
Enzymes of Chl biosynthesis and EX1 colocalize in grana margins. For the localization of enzymes catalyzing the initial and final steps of Chl synthesis, the same chloroplast fractions were analyzed as described in Fig. 1 C–E: (A) Chloroplasts (Chloro), thylakoid membranes (TM), and stroma (Stroma). (B) The three subfractions of chloroplast membranes that were pelleted at 10,000 × g, 40,000 × g, and 144,000 × g. (C) Grana core (GC), grana margins (GM), and stroma lamellae (SL). The localization of the following enzymes was determined by SDS/PAGE and immunoblotting: glutamyl-tRNA reductase (GluTR), glutamate-1-semialdehyde aminotransferase (GSAAT), Mg2+-chelatase subunit I (CHLI), Mg2+-protoporphyrin IX methyl transferase (CHLM), and PORs B and C. FLU, D2, PsaF, and CURT1 were used as marker proteins for the identification of the various chloroplast fractions. Cyt f was used as a loading control.
Fig. 6.
The distribution of Pchlide and Chlide in chloroplast membranes of WT and the flu mutant. Plants were grown under continuous light until they were ready to bolt. At this stage they were transferred to the dark for 8 h. Chloroplast membranes were isolated under dim green safety light and treated with the detergent digitonin and separated by differential centrifugation into grana margins (GM), grana cores (GC), and stroma lamellae (SL) as shown in Fig. 1. Nonesterified porphyrins were extracted, and their fluorescence emission spectra were recorded using an excitation wavelength of 433 nm as described previously (2). The fluorescence emission maxima for Pchlide and Chlide were 635 and 678 nm, respectively.
Discussion
Our study implicates the margin region of grana stacks as the source of 1O2-dependent signaling that initially had been discovered in the flu mutant of Arabidopsis thaliana. The release of 1O2 in the flu mutant triggers drastic phenotypic changes that at first glance seemed to be the result of photo-oxidative damage caused by 1O2. However, this view had to be revised after the EX1 protein had been identified. Inactivation of this protein in a flu background abrogated the 1O2-mediated responses, but did not suppress the rapid release of 1O2 at the beginning of reillumination (3, 5). Localization of EX1 in grana margins close to where damaged PSII is repaired distinguishes 1O2- and EX1-mediated signaling from a second 1O2-dependent signaling pathway that does not depend on EX1. This latter signaling is initiated in plants under high light stress by 1O2 generated presumably by the photosensitizing activity of 3Chl in the RC of active PSII that resides in the grana core. Under high light stress that exceeds the light quenching capacity of the chloroplast and causes photoinhibition of photosynthesis, an enhanced generation of 1O2 leads to oxidative damage as indicated by nonenzymatic lipid peroxidation (33) and breakdown of β-carotene, a scavenger of 1O2 closely associated with the RC of PSII (7). Nonenzymatic oxidative breakdown products of β-carotene such as β-cyclocitral activate stress defense responses (7) and stress acclimation (8). In contrast, EX1-dependent signaling is triggered under nonphotoinhibitory light conditions. Unlike β-carotene, EX1 does not seem to act as a scavenger of 1O2 that reduces the impact of oxidative stress on photosynthetic membranes. EX1 is a very low abundant protein and is confined to only a small part of the thylakoid membrane system away from the grana core that is thought to generate most of the 1O2. Furthermore, 1O2 concentrations sufficient to induce EX1-dependent signaling in flu are too low to cause detectable damage in the absence of EX1.
With the onset of 1O2 formation there is also a rapid decline of EX1 in the flu mutant. Generation of 1O2 alone without this concomitant decline of EX1 was not sufficient to activate 1O2 signaling. The simultaneous onset of 1O2 formation and decline of EX1 suggests that, following the release of 1O2, EX1 becomes susceptible to proteolytic attack by FtsH. This is probably due to oxidation of EX1 by 1O2, but oxidation of another target in the close vicinity of EX1 that allows FtsH to attack EX1 cannot be excluded. As the detection of EX1 depends on the recognition of tags attached to EX1 by tag-specific antibodies, at present it is not known how FtsH affects EX1. EX1 could be cleaved to smaller fragments that are released from the membrane, or the membrane-bound EX1 may be processed proteolytically and either remains attached to the membrane or is translocated to the stroma. In any case, FtsH is expected to change the function of EX1. EX1 itself could act as a negative or positive regulator of 1O2 signaling. As a negative regulator, EX1 could block the 1O2-dependent signaling pathway. Activation of this pathway would then depend on the inactivation or removal of EX1 by proteolytic cleavage and degradation. For example, in plants there are several precedents of proteolytic degradation of negative regulators controlling phytohormone signaling (34), but there is little evidence supporting such a control mechanism during EX1-dependent signaling. On the contrary, in the flu/ex1 double mutant the release of 1O2 initiates signaling only after EX1 has been reintroduced into the complemented pEX1::gEX1-Flag/flu/ex1 line (Fig. 1 A and B). This result is not in agreement with EX1 being a suppressor of 1O2 signaling, but rather supports a role of EX1 as a positive regulator. Regardless of how FtsH affects EX1, the signaling capacity of EX1 or its proteolytic derivative(s) is expected to depend on the subsequent interaction with downstream signaling components that become active and trigger signaling.
The localization of EX1 suggests a new source of 1O2 that is closely associated with the repair of damaged PSII in grana margins and seems to depend on the photosensitizing activity of Chl that is needed for the reassembly of active PSII or some of its biosynthetic intermediates. As EX1-dependent signaling occurs also in young seedlings, it seems possible that it is linked not only to the repair of PSII but also to its biogenesis. The repair cycle operates to remove and replace damaged subunits of PSII, primarily the D1 subunit. During reassembly of active PSII the recycling and de novo synthesis of Chl must be tightly controlled and synchronized with the synthesis of the D1 polypeptide to minimize the disruptive effect of unbound photoreactive Chl and its intermediates. Various carrier proteins have been implicated with scavenging Chl released during PSII damage and recycling it during PSII repair. Also, enzymes involved in Chl biosynthesis have been hypothesized to form a multienzyme complex in vivo, which would allow an effective synthesis and channeling of Chl precursors (30, 35, 36). If this enzyme complex operates close to the assembly of PSII, Chl could be rapidly inserted into the apoprotein, thereby minimizing the risk of photooxidative damage. In agreement with this proposal, Chl biosynthetic enzymes and the negative feedback regulator of the tetrapyrrole biosynthesis pathway, FLU, are highly enriched in the grana margin region and colocalize with the site of PSII repair. The example of the flu mutant shows that deregulation of Chl synthesis in grana margins is sufficient to activate EX1-dependent, 1O2-mediated signaling. Likewise, a small disturbance of the equilibrium between the synthesis of Chl and the D1 protein during the assembly of PSII may enhance the level of unbound Chl or its intermediates and promote the production of 1O2. Contrary to what one would expect, in the flu mutant Pchlide not only accumulates in the immediate vicinity of EX1 but also is almost evenly distributed in margin and core regions of grana stacks and, to a slightly lesser extent, occurs also in unstacked stroma lamellae. This distribution of the photosensitizer supports our previous conclusion that the amounts of 1O2 generated in the flu mutant by the photosensitizing activity of tetrapyrroles are too low to directly affect the plant (5). To become biologically active, this 1O2 requires the presence of EX1 that seems to act as a sensor of 1O2 and amplifies and relays its potential signaling effect.
It has been well documented that in photoinhibited PSII RC damaged D1 proteins are proteolytically cleaved and replaced by newly synthesized D1 polypeptides (37, 38) and that photoinhibited PSII enhances 1O2 production (39, 40) presumably by the photosensitizing activity of the P680 Chl of PSII RC, but the role of this 1O2 during the rapid turnover of the D1 protein is still under debate (29, 39, 40). Singlet oxygen generated by PSII is believed to interact primarily with its nearest target, the D1 protein of PSII RC that binds the P680 Chl (29, 40). However, so far oxidation of the D1 protein by 1O2 has not been shown in vivo, and thus it remains unclear whether this step is required to initiate D1 degradation (1). As an alternative function of 1O2 during the repair of photodamaged PSII, it has been implicated with inhibiting the de novo synthesis of the D1 protein due to the inactivation of the elongation step at stroma lamellae (41). This suggestion, however, is difficult to reconcile with the very short half-life and diffusion range of 1O2. It seems unlikely that 1O2 produced by PSII inside the grana stacks can specifically target components of the repair cycle of PSII that are localized far away in stroma lamellae (29). However, if indeed the repair cycle is prone to attack by 1O2, as suggested by several studies (42, 43), there may be another source of 1O2 associated with PSII that is closer to the site of D1 de novo synthesis. The results of our study may help to resolve this controversy. The close proximity of elongation factors, PORs B and C, FtsH, and PSII in grana margins as shown by the coimmunoassay suggests that 1O2 generated at this site could easily interact with elongation factors and suppress D1 synthesis. At increasing light intensities the relative proportion of grana margins expands at the expense of grana cores because of an enhanced breakdown of D1 and a greater need for the repair of damaged PSII (15, 16). Under these more severe stress conditions higher amounts of Chl may be released during the disassembly of damaged PSII, and the balance between the synthesis of D1 and Chl in grana regions may be perturbed and strongly shifted toward a higher accumulation of unbound Chl or its precursors, giving rise to an enhanced production of 1O2 that oxidizes elongation factors and inhibits synthesis of D1.
Materials and Methods
Plant Materials and Growth Conditions.
Seedlings of WT and flu, ex1, flu/ex1, and complemented ex1 and flu/ex1 mutant lines, all in a Columbia-0 (Col-0) background, were grown for 5 d under continuous light (90 μmol photons m−2⋅s−1) and room temperature (20 °C) on 0.6% agar plates containing 1/2 MS medium and 1× Gamborg vitamins, except for hygromycin-resistant transformed T1 and T2 seedlings that were selected on agar plates containing 0.5% sucrose. Mature plants were grown on soil for 3 wk under similar light and temperature conditions. In plants with a flu background, transferring light-grown plants to the dark for 8 h and then re-exposing them to light generated 1O2.
Complementation of ex1 Mutants.
The genomic EX1 (At4G33630) region of Arabidopsis thaliana (Col-0) was amplified by PCR using the sense primer 5′-GGGAATATTTAACTCTGCTTAC-3′ and the antisense primer 5′-GGCTTGAAGCCTCAGGCG-3′. The PCR product was cloned into the entry vector pCR8/GW/TOPO (Invitrogen) and subsequently transferred to the destination vectors pGWB504 and pGWB510 (44) using the Gateway LR reaction (Invitrogen). The gene constructs pGWB504(pEX1::gEX1-GFP) and pGWB510(pEX1::gEX1-Flag) were translocated into Agrobacterium tumefaciens strain GV3101::pMP90 by electroporation and transferred into flu/ex1 and ex1 plants by the floral dip method (45).
Isolation and Fractionation of Chloroplasts.
Chloroplasts were isolated and separated into membrane and stroma fractions as described by Kauss et al. (32). Isolation and fractionation of thylakoid membranes into 10,000 × g, 40,000 × g, and 144,000 × g fractions were performed as described by Lu et al. (11) except that SIGMAFAST Protease Inhibitor (catalog no. S8830) was used instead of the Pefabloc SC protease inhibitor (Roche). The 40,000 × g pellet designated previously as “grana margin-enriched fraction” did not contain the grana margin-specific marker protein CURT1A. Almost all of CURT1A was confined to the 10,000 × g pellet. CURT1A was identified immunologically with a CURT1A antiserum that recognized the 11,000-kDa polypeptide in WT but not in the quadruple curt1abcd mutant (14). Throughout the fractionation procedure, starting with chloroplast isolation, SIGMAFAST Protease Inhibitor was added to all buffers (1 tablet per 300 mL buffer). Fractionation of thylakoid membranes into grana core, grana margin, and stroma lamella was performed according to Puthiyaveetil et al. (15). Thylakoid membranes were suspended and diluted with a suspension buffer containing 0.1 M sorbitol, 50 mM Hepes (pH 7.5), 2 mM MgCl2, and 1× SIGMAFAST Protease Inhibitor to a Chl concentration of 0.6 mg/mL. After 40–60 min of dark treatment at room temperature (RT), samples were gently mixed with an equal volume of 2% (wt/vol) digitonin that was dissolved in the same suspension buffer and then transferred back to the dark for 10 min at RT. To remove insolubilized material, samples were centrifuged at 1,000 × g for 1 min at 4 °C. The supernatants were then centrifuged with a fixed angle rotor at 40,000 × g for 30 min at 4 °C to pellet the grana core. The supernatant fractions of this separation were centrifuged again with the same rotor at 140,000 × g for 90 min at 4 °C, resulting in a loose pellet (grana margin) and a tight pellet (stroma lamella) (15). The grana core, grana margin, and stroma lamella pellets were resuspended in the suspension buffer described above, using a fine brush. The yield ratios of grana core:grana margin:stroma lamella were ∼60: 20: 20. The Chl a/b ratios of grana core, grana margin, and stroma lamella were ∼2.2, 3.5, and more than 4.0, respectively. Before the isolation and fractionation of chloroplasts, mature Arabidopsis plants were transferred for 8 h to the dark, and the handling of chloroplast membrane samples was done under dim light.
Western Blot Analysis of Tagged EX1.
To detect EX1-Flag or EX1-GFP in the chloroplast or its subfractions, Chl had to be removed. Three hundred microliters of suspended thylakoid membranes (equivalent to 300 μg Chl) were mixed with 200 μL 0.1 M Na2CO3 and 0.1 M DTT, and absolute acetone was added to a final volume of 5 mL. The suspension was then kept for 5 min on ice before centrifuging it at 3,000 × g for 3 min. The pellet was dried and then dissolved in 300 μL 1× Laemmli SDS sample buffer. To detect EX1-FLAG or EX1-GFP in total leaf extracts, a minimum of 50, 7-d-old Arabidopsis seedlings grown on soil under continuous light were harvested and ground under liquid nitrogen. A 400-μL suspension buffer (Hepes 20 mM, pH 7.4; EDTA 2 mM; EGTA 2 mM; NaF 25 mM; Na3VO4 1 mM; Glycerol 10%; NaCl 100 mM; Triton X-100 0.5%; 1× SIGMAFAST Protease Inhibitor) was added. The suspension was kept for 10 min at room temperature and was then centrifuged at 10,000 × g for 30 min. The supernatant was transferred to a new tube, and 1.6 mL acetone was added. This sample was placed on ice for 5 min and then centrifuged at 3,000 × g for 3 min. The Chl content of the supernatant was measured, and the pellet was dissolved in 1× Laemmli SDS sample buffer (1.0 μg Chl/μL sample buffer).
Coimmunoprecipitation.
Leaves of 2- to 3-wk-old plants were homogenized under liquid nitrogen. The resulting powder was suspended in a buffer containing 20 mM Hepes⋅KOH (pH 7.4), 2 mM EDTA, 2 mM EGTA, 25 mM NaF, 1 mM Na3VO4, 10% glycerol, 100 mM NaCl, and 0.5% Triton X-100 and put on ice for 10 min before centrifuging it at maximum speed in an Eppendorf table centrifuge for 45 min at 4 °C. The supernatant was filtered through a 0.22-μm Millipore Express polyethersulfone membrane. A small aliquot was taken as an INPUT sample, whereas the remaining part was used for coimmunoprecipitation (Co-IP). Protein G beads (Novex, Dynabeads Protein G, RET: 10003D) were first conjugated with monoclonal anti-GFP antibody (Sigma, G1546) and were then added to the Co-IP sample. The samples were first agitated at room temperature for 1.5–2 h, and then the beads were washed four times for 6 min with a buffer containing 50 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 10% glycerol, and 0.05% Triton X-100. Finally, the washed beads were suspended in 100 μL of 1× Laemmli SDS sample buffer and incubated for 20 min at 70 °C. SDS/PAGE then separated solubilized proteins.
In-Gel Digestion, Mass Spectrometry, and Protein Identification.
SDS/PAGE-separated immunoprecipitated proteins, and gel slices were subjected to trypsinolysis using a modified procedure of Dogra et al. (46). Gel slices were destained with 100 mM NH4HCO3/50% acetonitrile, reduced in 10 mM DTT for 60 min at 56 °C, and alkylated in 65 mM iodoacetamide for 45 min at room temperature. The gels were dehydrated in 100% acetonitrile in vacuo and preincubated in 50 µL of trypsin solution (0.02 µg/µL Trypsin Gold-Promega) in digestion buffer (25 mM NH4HCO3/10% acetonitrile) for 10 min at room temperature. The gel slices were completely covered with digestion buffer and incubated overnight at 37 °C. The peptides were extracted in extraction buffer (0.1% trifluoroacetic acid/50% acetonitrile) by vortexing and brief sonication. The extracted peptides were dried and reconstituted in 0.1% formic acid for liquid chromatography–mass spectrometry analysis.
Peptides were trapped and separated using Waters nanoAcquity UPLC (Waters Corporation) coupled with a SCIEX Triple TOF 5600. The peptides were trapped by a 2G-V/MTTrap symmetry C18 column (5-µm particles, 180-µm i.d. × 20-mm length) at a flow rate of 3 µL/min and separated on a BEH130 C18 analytical column (1.7-µm particles, 100-µm i.d. × 250-mm length) at 250 nL/min (Waters Corporation). The liquid gradient was run from 8% to 25% of solvent B (acetonitrile in 0.1% formic acid) over 65 min. The eluate was delivered to the mass spectrometer with a nanospray, and the high sensitivity scanning over a mass range of m/z 350–1,800 was carried out. Tandem MS (MS/MS) measurements of the top 20 most intense precursor ions were done. The MS data were searched against the Arabidopsis Information Resource database (www.arabidopsis.org) using ProteinPilot 4.5 software with paragon algorithm 4.5.0.0, 1654 (SCIEX). Iodoacetamide-induced alkylation of cysteine residues was considered as fixed modification, and all other biological modifications were also recorded wherever found. Proteins were considered identified when a confidence level of 95% (expectation value below 0.05) and at least two peptide matches were recorded. These criteria resulted in a false-positive rate below 1% at peptide level.
Extraction and Measurement of Tetrapyrroles.
Tetrapyrroles were extracted from plant samples and analyzed as described previously (2).
Measurements of the Maximum Quantum Efficiency of PSII and Transient Chl Fluorescence Changes.
Images of transient Chl fluorescence changes were taken, and the ratio of variable to maximum chlorophyll fluorescence (Fv/Fm) was determined with a FluorCam 800MF system (Photon Systems Instruments) following protocols provided by Photon Systems Instruments.
Staining of Dead Cells and 1O2-Mediated Chloroplast Leakage in the flu Mutant.
Trypan blue staining was performed as described by op den Camp et al. (2) and chloroplast leakage was monitored according to Kim et al. (5).
Acknowledgments
We thank Drs. D. Leister, T. Nakagawa, W. Sakamoto, and S. Greiner for providing curt1abcd quadruple mutant seeds and CURT1A antibody, pGWB504 and pGWB510 destination vectors, seeds of the tFtsH2-GFP–expressing line, and antiserum against cytochrome f, respectively; G. Wang and Dr. Y.-M. She for help with mass spectrometry; Drs. B. Grimm and B. Hedtke for providing antisera against glutamyl-tRNA reductase, glutamate-1-semialdehyde aminotransferase, Mg2+-chelatase subunit I, and Mg2+-protoporphyrin IX methyl transferase; Drs. T. Kleine and G. Wanner for efforts to detect EX1 by scanning electron microscopy; Drs. F. Landgraf, Y. Kato, K. v. Wijk, and G. Friso for help and contributions during the initial phase of this work; and S. Puthiyaveetil for information concerning the isolation of grana margins. This research was supported by the Boyce Thompson Institute; National Institutes of Health Grant R01-GM085036; the Swiss Federal Institute of Technology and the Swiss National Science Foundation (K.A.); and the Chinese Academy of Sciences and National Natural Science Foundation of China (NSFC) Grant 31570264 (to C.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603562113/-/DCSupplemental.
References
- 1.Fischer BB, Hideg É, Krieger-Liszkay A. Production, detection, and signaling of singlet oxygen in photosynthetic organisms. Antioxid Redox Signal. 2013;18(16):2145–2162. doi: 10.1089/ars.2012.5124. [DOI] [PubMed] [Google Scholar]
- 2.op den Camp RG, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell. 2003;15(10):2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner D, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science. 2004;306(5699):1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 4.Meskauskiene R, et al. FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98(22):12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim C, et al. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell. 2012;24(7):3026–3039. doi: 10.1105/tpc.112.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S, Apel K, Kim C. Singlet oxygen-mediated and EXECUTER-dependent signalling and acclimation of Arabidopsis thaliana exposed to light stress. Philos Trans R Soc Lond B Biol Sci. 2014;369(1640):20130227. doi: 10.1098/rstb.2013.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramel F, et al. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109(14):5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramel F, et al. Light-induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell. 2013;25(4):1445–1462. doi: 10.1105/tpc.113.109827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peltier JB, et al. Proteomics of the chloroplast: Systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell. 2000;12(3):319–341. doi: 10.1105/tpc.12.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleffmann T, et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14(5):354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y, Hall DA, Last RL. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell. 2011;23(5):1861–1875. doi: 10.1105/tpc.111.085456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuello J, Quiles M. Fractionation of thylakoid membranes into grana and stroma thylakoids. Methods Mol Biol. 2004;274:1–9. doi: 10.1385/1-59259-799-8:001. [DOI] [PubMed] [Google Scholar]
- 13.Albertsson P. A quantitative model of the domain structure of the photosynthetic membrane. Trends Plant Sci. 2001;6(8):349–358. doi: 10.1016/s1360-1385(01)02021-0. [DOI] [PubMed] [Google Scholar]
- 14.Armbruster U, et al. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell. 2013;25(7):2661–2678. doi: 10.1105/tpc.113.113118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puthiyaveetil S, et al. Compartmentalization of the protein repair machinery in photosynthetic membranes. Proc Natl Acad Sci USA. 2014;111(44):15839–15844. doi: 10.1073/pnas.1413739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khatoon M, et al. Quality control of photosystem II: Thylakoid unstacking is necessary to avoid further damage to the D1 protein and to facilitate D1 degradation under light stress in spinach thylakoids. J Biol Chem. 2009;284(37):25343–25352. doi: 10.1074/jbc.M109.007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshioka-Nishimura M, et al. Quality control of photosystem II: Direct imaging of the changes in the thylakoid structure and distribution of FtsH proteases in spinach chloroplasts under light stress. Plant Cell Physiol. 2014;55(7):1255–1265. doi: 10.1093/pcp/pcu079. [DOI] [PubMed] [Google Scholar]
- 18.Bailey S, et al. A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol Chem. 2002;277(3):2006–2011. doi: 10.1074/jbc.M105878200. [DOI] [PubMed] [Google Scholar]
- 19.Kato Y, Miura E, Ido K, Ifuku K, Sakamoto W. The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 2009;151(4):1790–1801. doi: 10.1104/pp.109.146589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato Y, Miura E, Matsushima R, Sakamoto W. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol. 2007;144(2):952–960. doi: 10.1104/pp.107.099002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Q, Frick G, Armstrong G, Apel K. POR C of Arabidopsis thaliana: A third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol Biol. 2001;47(6):805–813. doi: 10.1023/a:1013699721301. [DOI] [PubMed] [Google Scholar]
- 22.Frick G, Su Q, Apel K, Armstrong GA. An Arabidopsis porB porC double mutant lacking light-dependent NADPH:protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J. 2003;35(2):141–153. doi: 10.1046/j.1365-313x.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- 23.Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. Plant peptides in signalling: Looking for new partners. Trends Plant Sci. 2009;14(5):255–263. doi: 10.1016/j.tplants.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Stelljes C, Koenig F. Specific binding of D1 protein degradation products to the psbAI promoter in Synechococcus sp. strain PCC 7942. J Bacteriol. 2007;189(5):1722–1726. doi: 10.1128/JB.01428-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato Y, Sun X, Zhang L, Sakamoto W. Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 2012;159(4):1428–1439. doi: 10.1104/pp.112.199042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Przybyla D, et al. Enzymatic, but not non-enzymatic, 1O2-mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1-dependent stress response program in the flu mutant of Arabidopsis thaliana. Plant J. 2008;54(2):236–248. doi: 10.1111/j.1365-313X.2008.03409.x. [DOI] [PubMed] [Google Scholar]
- 27.Vass I, Cser K. Janus-faced charge recombinations in photosystem II photoinhibition. Trends Plant Sci. 2009;14(4):200–205. doi: 10.1016/j.tplants.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Telfer A, Bishop SM, Phillips D, Barber J. Isolated photosynthetic reaction center of photosystem II as a sensitizer for the formation of singlet oxygen. Detection and quantum yield determination using a chemical trapping technique. J Biol Chem. 1994;269(18):13244–13253. [PubMed] [Google Scholar]
- 29.Ohad I, Berg A, Berkowicz SM, Kaplan A, Keren N. Photoinactivation of photosystem II: Is there more than one way to skin a cat? Physiol Plant. 2011;142(1):79–86. doi: 10.1111/j.1399-3054.2011.01466.x. [DOI] [PubMed] [Google Scholar]
- 30.Komenda J, Sobotka R, Nixon PJ. Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol. 2012;15(3):245–251. doi: 10.1016/j.pbi.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Feierabend J, Dehne S. Fate of the porphyrin cofactors during the light-dependent turnover of catalase and of the photosystem II reaction-center protein D1 in mature rye leaves. Planta. 1996;198(3):413–422. [Google Scholar]
- 32.Kauss D, Bischof S, Steiner S, Apel K, Meskauskiene R. FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this pathway. FEBS Lett. 2012;586(3):211–216. doi: 10.1016/j.febslet.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 33.Triantaphylidès C, et al. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008;148(2):960–968. doi: 10.1104/pp.108.125690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santner A, Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010;61(6):1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez-Prieto MA, et al. The small CAB-like proteins of the cyanobacterium Synechocystis sp. PCC 6803: Their involvement in chlorophyll biogenesis for Photosystem II. Biochim Biophys Acta. 2011;1807(9):1143–1151. doi: 10.1016/j.bbabio.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Wang P, Grimm B. Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth Res. 2015;126(2-3):189–202. doi: 10.1007/s11120-015-0154-5. [DOI] [PubMed] [Google Scholar]
- 37.Aro EM, Virgin I, Andersson B. Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143(2):113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- 38.Adir N, Zer H, Shochat S, Ohad I. Photoinhibition: A historical perspective. Photosynth Res. 2003;76(1-3):343–370. doi: 10.1023/A:1024969518145. [DOI] [PubMed] [Google Scholar]
- 39.Hideg E, Kálai T, Hideg K, Vass I. Photoinhibition of photosynthesis in vivo results in singlet oxygen production detection via nitroxide-induced fluorescence quenching in broad bean leaves. Biochemistry. 1998;37(33):11405–11411. doi: 10.1021/bi972890+. [DOI] [PubMed] [Google Scholar]
- 40.Vass I. Molecular mechanisms of photodamage in the Photosystem II complex. Biochim Biophys Acta. 2012;1817(1):209–217. doi: 10.1016/j.bbabio.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Nishiyama Y, Allakhverdiev SI, Murata N. A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim Biophys Acta. 2006;1757(7):742–749. doi: 10.1016/j.bbabio.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 42.Nishiyama Y, et al. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001;20(20):5587–5594. doi: 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishiyama Y, Allakhverdiev SI, Yamamoto H, Hayashi H, Murata N. Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry. 2004;43(35):11321–11330. doi: 10.1021/bi036178q. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa T, et al. Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71(8):2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 45.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 46.Dogra V, Ahuja PS, Sreenivasulu Y. Change in protein content during seed germination of a high altitude plant Podophyllum hexandrum Royle. J Proteomics. 2013;78:26–38. doi: 10.1016/j.jprot.2012.10.025. [DOI] [PubMed] [Google Scholar]