Significance
We describe the first, to our knowledge, integrated dynamic DNA nanotechnology and 2D material electronics to overcome current limitations for the detection of DNA single-nucleotide polymorphism (SNP). Electrical detection of DNA has been advancing rapidly to achieve high specificity, sensitivity, and portability. However, the actual implementation of DNA detection is still in infancy because of low specificity, especially for analytically optimal and practically useful length of target DNA strands. Most of the research to date has focused on the enhancement of the sensitivity of DNA biosensors, whereas the specificity problem has remained unsolved. The low specificity is primarily attributed to the nonspecific binding during hybridization of the probe and the target DNA. Here, we have addressed these limitations by designing a functional prototype of electrical biosensors for SNP detection.
Keywords: bioelectronics, graphene FET DNA sensor, electrical biosensor, DNA strand displacement, SNP detection
Abstract
Single-nucleotide polymorphisms (SNPs) in a gene sequence are markers for a variety of human diseases. Detection of SNPs with high specificity and sensitivity is essential for effective practical implementation of personalized medicine. Current DNA sequencing, including SNP detection, primarily uses enzyme-based methods or fluorophore-labeled assays that are time-consuming, need laboratory-scale settings, and are expensive. Previously reported electrical charge-based SNP detectors have insufficient specificity and accuracy, limiting their effectiveness. Here, we demonstrate the use of a DNA strand displacement-based probe on a graphene field effect transistor (FET) for high-specificity, single-nucleotide mismatch detection. The single mismatch was detected by measuring strand displacement-induced resistance (and hence current) change and Dirac point shift in a graphene FET. SNP detection in large double-helix DNA strands (e.g., 47 nt) minimize false-positive results. Our electrical sensor-based SNP detection technology, without labeling and without apparent cross-hybridization artifacts, would allow fast, sensitive, and portable SNP detection with single-nucleotide resolution. The technology will have a wide range of applications in digital and implantable biosensors and high-throughput DNA genotyping, with transformative implications for personalized medicine.
DNA sequencing has opened new windows of opportunities for diagnosis of genetic disease (1), biological informatics (2), forensics (3), and environmental monitoring (4). Discrimination of a single mismatch in a long DNA strand is of significant importance and is essential to detect single nucleotide polymorphism (SNP). SNP is a single-nucleotide mutation in a gene sequence and varies among paired chromosomes, between individuals, and across biological species. SNP mutations can have dramatic influence on the health. They are markers for variety of diseases, including various forms of cancer, genetic disorders (5–7), and are of critical importance for successful practical implementation of the concept of personalized medicine (8). Thus, the development of biosensors detecting SNP mutations with high sensitivity and specificity is essential for effective personalized medicine approaches.
Current DNA sequencing, including SNP detection, is achieved primarily by enzyme-based methods, using DNA ligase (9), DNA polymerase (9), and nucleases (10). These methods generate highly accurate genotyping. However, the methods are expensive and time-consuming. One of the common enzyme-free methods to detect SNPs uses hybridization of the target DNA to a probe on a microarray and detects their binding events with fluorescence microscopy/spectroscopy. Hybridization-based methods for SNP detection have several disadvantages, including cross-hybridization between allele-specific probes (11). This limits the detection of a single mismatch in long probe–target hybridization as the longer probes have more frequent cross hybridization. For example, a single mismatch in the center of a 15-bp probe–target duplex can be detected because there is a critical difference in the hybridization affinity between a perfect-matched and a single-mismatched hybridization. However, when the probe length is 40 or 50 nt, a single mismatch produces a relatively small difference between a perfect-matched and a single-mismatched hybridization. Thus, SNP detection is difficult with the simple hybridization-based methods. The typical length of probes used in microarrays is over 20 nt; cross-hybridization significantly reduces its reliability and specificity. To reduce cross-hybridization, redundancies in the array design are used to confirm detections of the same SNP or probe sequences are modified to control hybridization affinity (12–14). If cross hybridization were reduced or eliminated, fewer probes would be needed to obtain the same level of reliable analysis with longer probes (60–80 nt) (15), and longer probes provide more sensitive detection (16, 17).
DNA strand displacement can be used to improve specificity even for a longer probe design. Strand displacement occurs when a DNA double helix exchanges one strand for another complementary strand (18). The newly introduced strand with higher affinity to one strand in the initial double helix displaces the other strand with lower affinity. Additionally, inosine or RNA can be used to control kinetics or Gibbs free energy of hybridization (19–22). Strand displacement has been a core technique in DNA nanomanipulation for over 20 years and has been used in several mechanical nanomachines (23), logic gates (24), and sensors (19, 25). Currently, strand displacement-based assays can discriminate SNPs efficiently, by controlling competition between the initially hybridized parts in the double-stranded or hairpin-structured probe and probe-to-target hybridization (12).
The broadly used SNP detection methods, even with the most advanced probe design, typically use the fluorescence-based readout (26). However, fluorescence-based sensors have life-time and background limitations. Moreover, they require fluorimeters or laser scanners to for the quantitative analysis of the optical signal and hence the application of optical probes depends on a sophisticated and expensive laboratory-setting. Electrical detection of DNA sequences using the field effect transistor (FET) presumably lowers the limit of detection to the femto-molar level (27). FETs are of particular interest because they can be used as highly sensitive bio-molecular sensors and could be efficiently integrated using electric chip designs, including silicon-based FET (28), 1D carbon nanotubes (29), and Si nanowires (30). Nanotubes and nanowires allow better sensitivity due to their high surface-area-to-volume ratio and the sensitive carrier mobility to the electric field (charge density). However, the complicated top-down fabrication processes and random array of the nanowires and nanotube are costly and unreliable. Graphene, a 2D material, is attractive because it is a single atom thick sheet that is easy to fabricate uniformly over a large area. Ambipolar field effect, high carrier mobility, low intrinsic electrical noise, mechanical strength, and flexibility collectively represent some of the advantages that make graphene a promising material for FET bio-sensing (31). First-generation graphene-based biosensors have been developed to successfully detect bacteria (32), glucose (33), protein (34), pH (35), and DNA (34, 36).
FET-based DNA biosensors currently use a single-stranded probe to detect hybridization. When the probe binds to the target strand, a double helix forms and the binding results in a measureable change in the electric charge over the active layer of the FET (27, 37). In currently available FET-DNA biosensors, the average length of the probe and the target is 10 to ∼25 nt (27, 29, 37, 38). In the case of a 25-nt probe, the signal difference between a fully complementary target and a single-mismatched one is less than 50%, and the target is significantly smaller than one reported with other biosensors using short probes (29).
Given the inherent disadvantages of the current probe’s design used in graphene FET biosensors, we reasoned that design and fabrication of biosensors using a strand displacement-based probe architecture on a graphene FET would provide improved specificity and resolution. In this paper, DNA strand displacement-based probe is successfully designed and used on a graphene FET biosensor for label-free detection of a single mismatch with higher specificity than that of a single-stranded probe DNA.
Architecture of double-stranded (DS) probes was conceived to facilitate the design compatibility of a graphene FET biosensor for the electrical sensing of DNA strand displacement. A DS probe containing targeted inosine substitutions and optimized toehold lengths was first tested with fluorescence/quencher technology to demonstrate an efficient single-mismatch discrimination. Then, the nonfluorescently labeled DS probe was attached on a graphene FET to reproduce the SNP discrimination based on the electrical sensing of DNA strand displacement. A liquid gate was used to obtain current–voltage (I-V) curve with DNA in buffer solution. I-V curve shifts and changes in resistance were monitored with fully complementary (perfect-match) and single-mismatched DNA sequences. With this combination of the electrical sensor and dynamic DNA nanotechnology, a single mismatch was detected in 47 nt of DNA with high resolution. To our knowledge, this is the first report of the successful electrical detection of strand displacement in long DNA strands by sensing the charge difference before and after strand displacement without any labeling or additional processes.
Description of SNP Detection Using Fluorescence-Based Observations of Strand Displacement
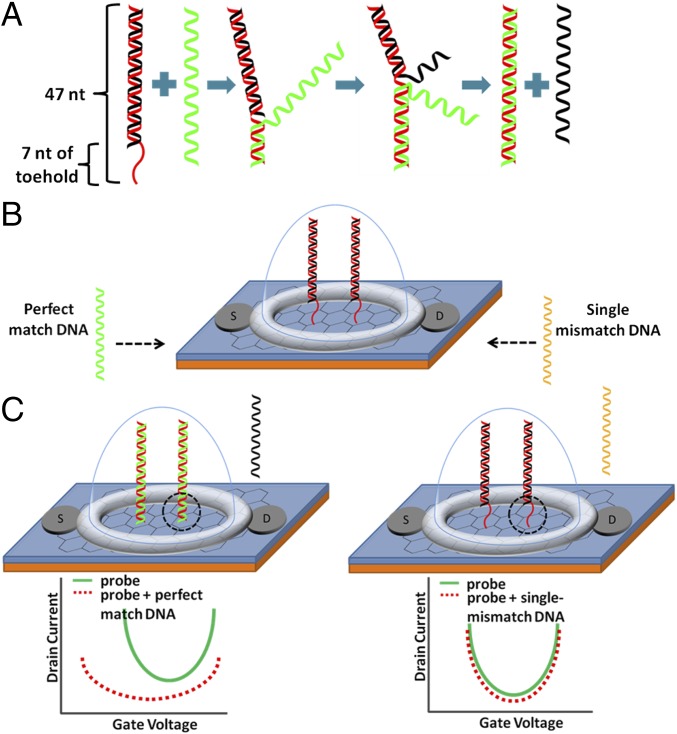
The schematics of strand displacement and single-mismatch detection are shown in Fig. 1. The DS probe is prepared by hybridization of two complementary strands. The red strand in Fig. 1 has a prolonged toehold section of 7 nt, and its total length is 47 nt. The black strand is 40 nt and contains four inosine (I) substitutions, where guanines were originally located, to weaken the affinity between the two strands. The 47-nt side is called the normal side (N); and the 40-nt side is called the weak side (W) because it contains I bases to weaken the double-helix affinity. The structure of the DS probe with specific sequences is shown in SI Appendix, Fig. S1. As shown in Fig. 1, when 47 nt of target strand (T), which is fully complementary with the normal strand (N), is introduced to DS probe, it displaces the weak strand (W) and hybridize with the normal strand (N). Inosine (I) bases allow the shortening of the toehold part. If W does not contain inosines, hybridization affinity between W and N becomes too strong to efficiently displace W with T with the 7 nt of toehold; without inosine’s substitutions, longer toehold is required to efficiently displace W strand by T. Thus, the strand displacement reaction can be summarized as the following (39):
When T has a single mismatch to N, the affinity between N and T is significantly decreased and the reaction rate is greatly reduced; DNA strands in the DS probe are not displaced, and it remains in its initial conformation (Fig. 1C) (19, 20).
Fig. 1.
Schematics of the SNP chip sensor. (A) DS probe action. The red strand is normal strand (N) containing 7 nt of toehold. Toehold is single-stranded at the initial state. When a perfect-match target strand (green strand) approaches a DS probe, the target strand displaces the weak strand (W) (black strand) by binding to the toehold. (B) Graphene FET sensor with DS probe. “S” and “D” represent source and drain, respectively, of FET. Gate voltage is applied directly on the liquid gate; the liquid gate is shown as a hemisphere (light blue) surrounding the DS probe. (C, Left) The green strand displaces the black strand, and the toehold portion becomes double-stranded (black dotted circle region). (C, Right) A target strand with single mismatch (yellow strand) does not allow the strand displacement properly (i.e., the yellow strand does not displace the black strand). Thus, the toehold region remains single-stranded (black dotted circle). (C) I-V relationship (I-V curve) attributable to charge difference during the strand displacement in the gene chip FET sensor. (C, Left) I-V curve for strand displacement with perfect match (C, Left). The I-V curve shifts leftward and downward. (C, Right) Single-mismatch target strand does not displace the black strand properly; thus, the I-V curve remains almost the same.
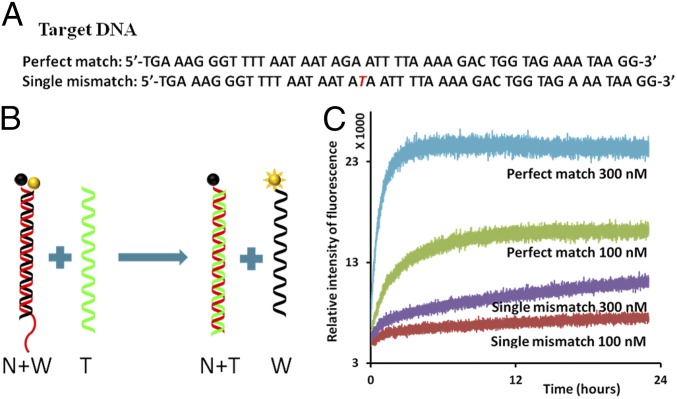
The strand displacement was monitored over time with the fluorescence labeling (Fig. 2). A Texas red fluorophore was labeled at the end of W and a fluorescence quencher was labeled at the end of N. The quencher absorbed the emission wave from the fluorescence when it was adjacent to fluorescence label, thus upon hybridization of N and W, the fluorescence was quenched. When the perfect-match T was added on the sample, strand displacement happened, the fluorophore and quencher were separated and the fluorescence signal became brighter. However, when the single-mismatch T strand was added, strand displacement happened at much slower rate and much lower fluorescence signals were measured compared with a perfect-match T experiment. The length of the toehold affected the reaction rate and hence different lengths of toeholds were tested (SI Appendix, Fig. S2). The test with 10-nt toehold showed vague discrimination of a single mismatch because affinity between N and T was too strong with 10 nt toehold. The two strands hybridized efficiently even with the single-mismatch.
Fig. 2.
Single-mismatch detection using fluorescently labeled nucleotides. (A) Sequences of the target DNA. (A, Top) Perfect match. (A, Bottom) Single mismatch. The mismatched nucleotide is marked in red. (B) Schematics of strand displacement: nucleotide with fluorophores (yellow ball) and nucleotide with quencher (black ball). Initially, the normal (N) (red) and weak (W) (black) strands are hybridized; fluorophore (yellow ball) and quencher (black ball) are adjacent so that fluorescence is quenched. When the green strand (perfect-match T) interacts with DS probe, strand displacement takes place, and the normal strand (N) and perfect-match target strand hybridize. The weak strand (W) remains single-stranded, and the fluorophore becomes active. (C) Real-time fluorescence measurement of the strand displacement. Interaction of the single-mismatch target strand with DS probe shows much less fluorescence activity than the interaction of the perfect match with DS probe. The concentration of DS probe was 10 nM, and the concentrations of T strands varied from 100 to 300 nM, shown under each plot.
To ascertain that the single-mismatch T strand did not hybridize partially from toehold part until the mismatched point, the discrimination was verified by DNA gel electrophoresis. For DNA gel electrophoresis, the structure of DS probe was modified to accumulate T onto the DS probe, not just for switching their positions. Activity of the DS probe is not affected when W and N are bound by the hinge part when the hinge part is introduced to conform the DS probe partial-triple strand after strand displacement (SI Appendix, Fig. S3) (39, 40). The details of the modification of DS probe are reported in the supplemental information. The gel image shows that the sample with the single-mismatch T stained weaker than the sample with perfect-match T (i.e., the single-mismatch T did not displace W effectively compared with the perfect-match T; SI Appendix, Fig. S3). Experimental protocols described above using fluorescence-based detection of DNA strand displacement represent the standard experimental approach for the DS probe design optimization and fine-tuning of toehold lengths and inosine base substitutions for SNP detection using graphene FET biosensors.
Detection of Strand Displacement on Graphene FET
A graphene FET with two electrodes and a liquid gate chamber was fabricated to examine electrical sensing of DNA using DNA strand displacement-based probes (Fig. 1). The toehold part of N, which is adjacent to the graphene surface (Fig. 1B, black dotted circle), became double-stranded after strand displacement and it changed the electrical signals as shown in the I-V curve and the corresponding electrical resistance. The graphene channel (4 × 6 mm) was transferred onto a silicon oxide-coated wafer using an established method (37, 41); 1-pyrenebutanoic acid succinimidyl ester (PASE) was used to link graphene and the amine group at the N side of DS probe (27, 42). The pyrene group of PASE and graphene were attached to each other by π-π stacking interactions to covalently link graphene/PASE to the amine group at the N side of DS prove.
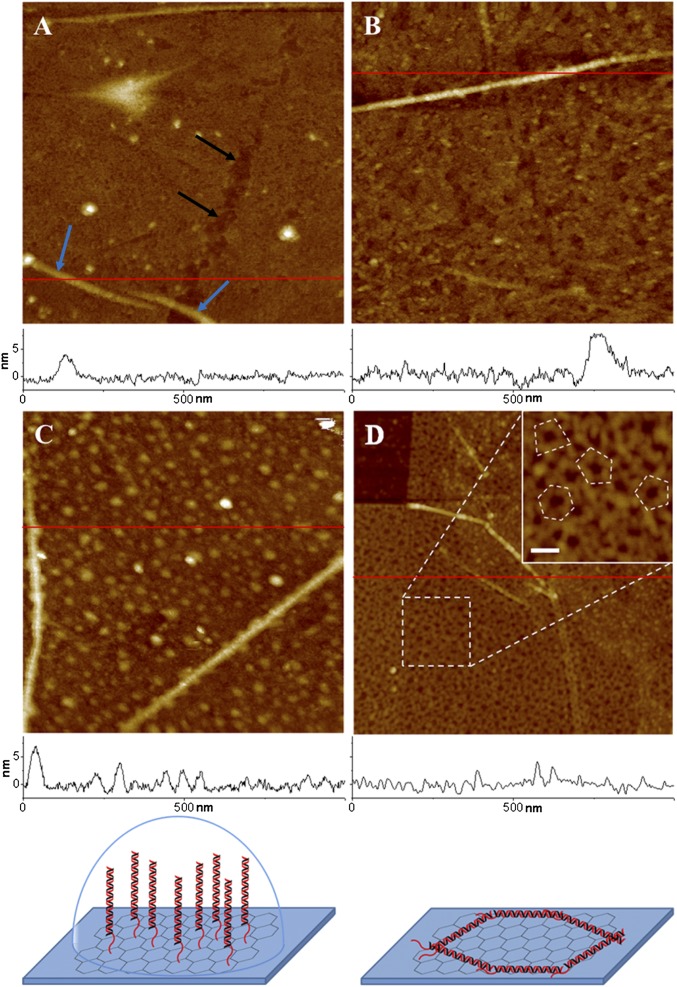
The process of graphene functionalization was monitored at each step using an atomic force microscope (AFM). As shown in AFM images (Fig. 3A), topography of a bare graphene surface is mostly flat with some defects and wrinkles. Graphene wrinkles ∼4 to ∼7 nm in height are observed; these heights and shapes are in good agreement with the previously published results (32, 43, 44). PASE functionalization of graphene surface does not change the graphene surface roughness or morphology (Fig. 3B). However, after immobilizing DNA on the graphene FET device through PASE and amine reaction, surface morphology was dramatically changed with the appearance of distinct globular structures (Fig. 3C). The average height of these globular structures is 3.6 ± 1.4 nm and varies between 2–6 nm (SI Appendix, Fig. S4). The appearance of these structures seem consistent with the conformation of the standing DNA strands in fluid; a typical height of dsDNA lying flat, as detected by AFM imaging in air is ∼2 nm (45), which is significantly shorter than the height of globular structures in fluid observed in our experiments. DNA strands on the graphene FET device were further imaged in air after drying the graphene surface (Fig. 3D). After drying, the appearance of dotted globular structures of dsDNA observed in fluid condition have changed to distinctive rod shapes of ∼2 nm in height as shown in details of the inset image (Fig. 3D). These observations were validated by the analyses of AFM images of graphene and PASE-functionalized surfaces with and without DNA, which were also imaged in air (SI Appendix, Fig. S5). A DS probe consisted of a 40-bp double-strand section and a 7-nt single-strand overhang; in total the estimated length is ∼15 nm. Consistently, the rod shapes of the DNA strands in the air AFM images show about 15 to ∼20 nm of length (SI Appendix, Fig. S5C). Therefore, AFM images of devices in fluid and air conditions indicated that the PASE–amine functionalization strategy was working appropriately.
Fig. 3.
AFM images of graphene transistor surface with and without DNA strands. (A) Graphene surface in fluid is mostly flat with some defects (black arrows) and graphene wrinkles (blue arrows). (B) PASE-coated graphene surface in fluid showing a flat surface with a similar wrinkle height of 7 nm, as seen in A. (C) After binding of dsDNA on the PASE-coated graphene surface in fluid, graphene’s smooth surface is covered with DNA strands of ∼2–6 nm in height. The height of the graphene wrinkles remains the same. (D) DNA strands are visualized better in an air AFM image with distinctive appearance of DNA structures. (D, Inset) Image showing more details of DNA structures at higher magnification. The randomly lying DNA probes in the Inset are outlined with a dotted line. Surface height profiles at the red line are plotted at the bottom of each image. Cartoons at the bottom represent models of formation of DNA structure in liquid and air. The right cartoon renders the random polygonal structure of DNA in air. All images have a scan area of 1 × 1 μm and a z range of 20 nm, except for the Inset. The z range and the scale bar of the Inset are 10 and 50 nm, respectively.
The conformation and position of the DS probe on the graphene surface is a critically important factor for the electrical detection of strand displacement. Only 7 nt of single-stranded toehold of the 47-bp DNA becomes double-stranded after the strand displacement compared with the initial DS prove (Fig. 1). If the DS probe is lying down or absorbed on the surface, the signal difference would be too small to be detectable by recognizing the charge difference. The AFM image in Fig. 3C shows that DS probe is observed as islands in liquid, whereas it was grain-boundary-shaped in air (Fig. 3D). When the surface was fully covered by PASE and ethanolamine, the DS probe was not exposed and not absorbed into graphene except amine–amide bonding; otherwise, nucleotides can be absorbed to graphene by π-π stacking interaction. Additionally, molecular dynamics simulations have shown that DNA established an upright conformation to the silica surface, fluctuating only around 10° from vertical at stable state (46). We thus concluded that the positioning of DS probes is perpendicular to the graphene surface in liquid and the functionalization strategy was successful.
The source and drain electrodes were applied by silver paste, and silicone rubber was used before the DNA probe functionalization to insulate the electrodes and create a solution reservoir (41). Sample in buffer solution was placed in the reservoir, and a gate voltage was applied directly to the top of the buffer solution (27, 31, 34, 37). When the surface charge was changed by strand displacement, the charge built up and the I-V curve shifted to left side and the resistance was increased (27, 37, 41). Functionalization of the DS probe changed electrical signals of FET (SI Appendix, Fig. S6).The I-V curve was measured after PASE was fixed on the graphene channel with 1× PBS buffer solution as liquid gate. After DS probe was bonded on the PASE, the measurement was repeated again. The I-V curve shifted to left side after the bonding of DS probe. The resulting unique U-shaped I-V curve is attributable to the ambipolar characteristic of graphene. Additionally, the electrical resistance increased and shifted the I-V curve lower.
The Debye length should be considered when detecting electrical charge in ionic solution (47). It can be written for aqueous solution at room temperature as
where λ is the Debye length expressed in nanometer, IB is Bjerrum length which is about 0.7 nm, zi is valencies of the various types of ions, and ρi is number densities or number of molecules per volume (48). Note that the Debye length is an estimate of the distance where Coulomb interactions are ignored. The size of the region near a point charge where opposite-charge counterions can be found is also ignored. The Debye length represents the net length of the electrostatic effect in ionic solution. Charges are electrically screened outside the sphere whose radius is the Debye length. In the 1× PBS solution, which is generally used as a DNA buffer solution, its Debye length is <1 nm. More diluted PBS such as 0.1× or 0.01× PBS allows detection of the longer part of the hybridization. However, the DS probe requires a high ionic concentration to stably operate the strand displacement. If the ionic concentration of buffer solution is too low, its double-helix structure can be unstable and could fail in strand displacement.
For measuring the effect of strand displacement, 12.5 mM MgCl2 and 30 mM Tris buffer were used to increase Debye length. This MgCl2 concentration is known to be equivalent to about 1× PBS for DNA helix stabilization (49). MgCl2 is 2:1 electrolyte (e.g., Mg2+:2Cl−), and its Debye length of 12.5 mM MgCl2 is calculated by the above equation to be ∼1.6 nm. The electrical effect of DNA is reported to be rapidly decreased, and only a few sequences that are close to the graphene surface determine the electrostatic potential on the sensor (50). Thus, the first few sequences would have effective charges on the surface. The tests were also conducted with 1× PBS and compared with MgCl2 buffer solution. The MgCl2 buffer solution generated clearer discrimination (SI Appendix, Fig. S7).
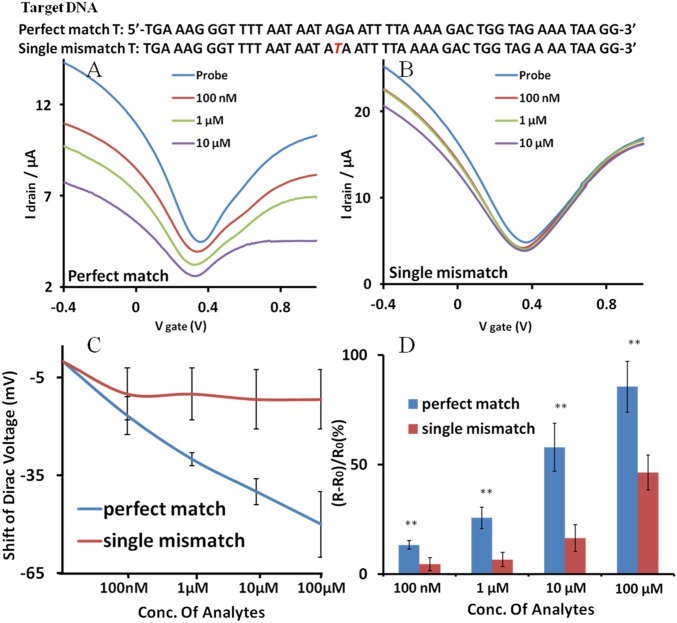
To examine the specificity of the graphene FET sensor, perfect-match and single-mismatch samples were tested. Target strands in different concentrations (100 nM to 100 μM) were incubated on the sensor for 8 h (Fig. 4). When the perfect-match T was treated on the graphene sensor, the U-shaped I-V curve shifted down and to the left, which indicates increasing resistance and imposition of the n-doping effect (27, 37). The corresponding resistance change was observed as discussed below. With 100 nM of T strands, which is equivalent to about 3.011 × 1012 of T molecules in 50 µL of buffer solution, DS probe showed clear discrimination of single mismatch (Fig. 4). As the concentration of the perfect-match T was increased, I-V curve kept shifting left and down and the shape of the curve became flatter. As shown in the Fig. 4C, the Dirac point of the I-V curve was shifted approximately −50 mV with 100 μM of perfect-match T and approximately −11.6 mV with single-mismatch T (4.3-fold difference). The more detailed graph of Dirac point shifts is shown in SI Appendix, Fig. S8. Different values of source voltages were also tested. The I-V curve also showed the same trends of Dirac point shifts (SI Appendix, Fig. S9). Significantly, the single-mismatch T made much smaller shifts and the I-V curve was saturated. It is reasonable to believe that a single-mismatch T could not result in the proper strand displacement, whereas a perfect-match T could induce a proper strand displacement. Another set of data with different dimension of the graphene channel is presented in the SI Appendix and also showed a convincing distinction of single mismatches (SI Appendix, Fig. S10). Our results show a clear discrimination of a single mismatch in the 47-bp DNA probe, suggesting that it could be possible to detect single mismatch in a longer DNA strand.
Fig. 4.
I-V relationship of the graphene FET sensor for the strand displacement reactions. (A) The perfect-match T shifted the I-V curve according to the indicated concentrations. (B) The single-mismatch T shifts the I-V curve significantly less. The DNA sequences of T used in the experiments are shown over the I-V curve. (C) Dirac voltage shift of the FET sensor. The Dirac voltage is expressed as a function of the concentration of the added target DNAs. (D) Distinguishable resistance change of the channel layer caused by strand displacement at different concentrations of the T DNAs. Statistical values (means ± SD) were based on three sets of independent data points (n = 3). **P < 0.01. Conc, concentration.
The resistance change of the channel was measured and compared between the perfect-match and the single-mismatch targets at different concentrations (Fig. 4). When DNA is accumulated on the graphene surface, its resistance increases (38). Immobilization of DS probe and addition of target strands increased the resistance of the channel. The resistance increased 40% to ∼60% when the probe was anchored on the graphene surface. As shown in the Fig. 4C, perfect-match T increased the resistance significantly more than single-mismatch T. The largest difference was observed at a target concentration of 100 nM, with the resistance changes of ∼26.0% and ∼6.8% (3.8-fold difference) for the perfect-match and the single-mismatch T, respectively. The minimum difference was observed at the target concentration of 100 µM with the resistance changes of ∼84.9% and ∼46.0% (1.8-fold difference), for the perfect match and the single mismatch, respectively.
In control experiments, single-strand probes were tested using only the N side of DS probe, to confirm that the discrimination of single mismatch in 47 bp resulted from using DS probe. The single-mismatch target strands were tested with the concentration ranging from 10 pM to 1 nM. SI Appendix, Fig. S11 shows that when a single-mismatch target strand was hybridized with the N side of the probe, this probe’s signal transferred as much as that of the DS probe with the perfect-match target. The concentration of the T strand required to saturate the I-V curve transfer was much lower because it does not have the W side of DS probe. When it was a DS probe, the T strand required more energy (higher concentration) to displace the W strand. The single-mismatch T strand shared identical 27 and 19 nt with the perfect-match target and the melting temperature of hybridization of single-mismatch T and N side is 55–65 °C, with a concentration range of 10 pM to 10 μM (51). The stable hybridization of single-mismatch target and the N strand made the discrimination of the single mismatch impossible when the probe was single-stranded. These results indicate the superior capability of DS probe on graphene FET to discriminate a single mismatch in long DNA sequences.
Originally, microarrays used photolithography to fabricate microsized spots inspired by transistors array in electronics. The proposed graphene FET biosensor can also be integrated in the form of microarray. These array sensors would not need fluorescence labeling or optical components and would reduce the required number of spots for individual sequences by detecting strand displacement of longer DNA strands. It seems likely that specificity of the graphene FET sensor described in this work is sufficiently high. This would alleviate the need for complex algorithms to analyze vague data for the detection of SNP with current technology. As such, the further development and implementation of this technology would allow more affordable and accurate diagnosis of myriad of diseases, including cancer and degenerative, genetic, and other various disorders.
Conclusions
Label-free detection of a single mismatch in a 47-bp probe with high resolution was achieved by using DNA strand displacement on a graphene FET chip. This detection was possible by electrical sensing of the inherent charge of DNA with the graphene transistor. Using a double-stranded probe capable of detecting a single mismatch in a 47-bp double helix, the performance of the sensors appears to exceed significantly the performance of a single-stranded probe. The structure of double-stranded part in the probe was changed by strand displacement, and this difference was readable in the electrostatic gating effect. The SNP-discrimination results are readily detectable in the I-V curve and the electrical resistance. Electrical detection of a single mismatch was also correlated with the real-time fluorescence measurement. The work provides a significantly improved platform for SNP detection by combining high-fidelity probe design and a detection scheme that has not been reported previously to our knowledge. Our results demonstrate the practical utility of the biosensor technology based on a combination of dynamic DNA nanotechnology and 2D nanoelectronics. This technology opens opportunities for the development of more reliable and efficient diagnostic tools, including design and development of miniaturized, point-of-care, and implantable biosensors, for early detection of potentially life-threatening human diseases.
Methods
Materials.
PASE, ethanolamine, MgCl2, and conducting silver paste were obtained from Sigma Aldrich. Graphene was from ACS Material. Silicone rubber was from Dow Corning, PBS and Tris solutions were from Thermo Fisher Scientific, and poly(methyl methacrylate) (PMMA) was from MicroChem. Ammonium persulfate was from MP Biomedicals, and DNAs were from IDT. DNA gels from Lonza were obtained, respectively. All of the DNA sequences for this experiment appear in SI Appendix, Table S1. Ultrapure water was from a Millipore purification system.
Fluorescence Test.
The normal strand (N) side, which was tagged with fluorescence quencher, and the weak strand (W) side, which was tagged with fluorescence label, are mixed in a ratio of 1:1 in 1× PBS solution and annealed from 20 to 90 °C and cooled to 4 °C over 3 h. Texas red has an excitation maximum wavelength of 596 nm and an emission maximum wavelength of 613 nm. Perfect-match and single-mismatch T strands are all suspended in 1× PBS. The hybridized DS probe was diluted in 1× PBS and tested using a Tecan Infinite 200 M plate-reading spectrometer at 27 °C, with an accuracy we estimate to be approximately ±1.5 °C. Excitation and emission of Texas red are observed at 590 and 620 nm, respectively. Each experiment began with a 50-µL sample volume with a device concentration of 10 nM (10 nM DS probe and 100 and 300 nM T) in black 96-well plates. Clear microplate sealing films were applied over the sample wells to avoid evaporation. The same test was conducted in 1× PBS buffer solution.
Fabrication of Graphene FET.
The top side of graphene on a copper film was spin-coated with PMMA to protect the top side of graphene, whereas the bottom side of graphene was etched away. PMMA acted as the supporting layer of the graphene after etching the copper. The back side of graphene was removed by oxygen plasma etching. The sample was cut into 4 × 6 mm pieces with scissors. Copper was etched by floating on 0.1 M ammonium persulfate for about 5 h and rinsed in deionized (DI) water overnight. Graphene supported by PMMA was then transferred on a SiO2-coated silicon wafer. PMMA layer was removed by acetone at 60 °C for 1 h. The sample was annealed at 300 °C for 2 h under a hydrogen/argon atmosphere (52). To fabricate transistor, conducting silver paste was used as source and drain electrodes at the two ends of the graphene. Silicone rubber was used to insulate source and drain electrodes from liquid and construct solution reservoir.
Immobilization of DS Probe.
PASE (5 mM) in dimethylformamide (DMF) was treated on the graphene for 1 h and rinsed with pure DMF and DI water; 50 µM of DS probe was added on PASE-modified graphene for 2 h. The graphene FET with DS probe functionalization was rinsed with 1× PBS; 100 mM ethanolamine solution was treated to saturate the possibly unreacted amino group on PASE and rinsed with 1× PBS solution. The volume of all treated chemicals and samples was 50 µL.
Visualization of DNA and Graphene Surface.
Topographic images of DNA on graphene surface were acquired using a Multimode AFM equipped with a Nanoscope V controller (Bruker). Silicon cantilevers with a spring constant of 42 N/m (PPP-NCHR; Nanosensor) were used for imaging in air using tapping mode. Silicon nitride cantilevers with spring constants of 0.08 N/m (OMCL-TR400; Olympus) were used for imaging in fluid using peak force-tapping mode. The Nanoscope software was used for analyzing imaging data.
Strand Displacement on the Chip.
The strand displacement reaction was conducted by dropping perfect-match and single-mismatch T strands with concentrations that are indicated in the legends in Fig. 4 of data and incubated overnight in the reservoir on the graphene FET chip. Then, the chip was rinsed gently with 1× PBS. All of the volume of treated samples was 50 µL.
Electrical Measurements.
I-V curves and resistance were measured in a semiconductor parameter analyzer equipped with a probe station. Silver wire was used as an electrode, which applied gate voltage (Vg) to the 12.5 mM MgCl2/30 mM Tris buffer solution. Tests were also conducted with 1× PBS buffer solution. Vg was swept from −0.5 to 1 V, and drain–source voltage (Vds) was picked between 0.05 and 0.3 V. Drain–source current (Ids) was measured at an assigned Vds. Resistances were measured between 0 ∼ 100 mV of Vds.
Supplementary Material
Acknowledgments
The work is supported by National Institute on Drug Abuse Grants R01DA025296 and R01DA024871 and departmental development funds from the Department of Mechanical and Aerospace Engineering, University of California, San Diego.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603753113/-/DCSupplemental.
References
- 1.Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci USA. 1991;88(1):189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapiro J. Genome informatics: The role of DNA in cellular computations. Biol Theory. 2006;1(3):288–301. [Google Scholar]
- 3.Jin L, Chakraborty R. Population structure, stepwise mutations, heterozygote deficiency and their implications in DNA forensics. Heredity (Edinb) 1995;74(Pt 3):274–285. doi: 10.1038/hdy.1995.41. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, et al. DNA electrochemical biosensors for environmental monitoring. A review. Anal Chim Acta. 1997;347(1–2):1–8. [Google Scholar]
- 5.Singh M, Singh P, Juneja PK, Singh S, Kaur T. SNP-SNP interactions within APOE gene influence plasma lipids in postmenopausal osteoporosis. Rheumatol Int. 2011;31(3):421–423. doi: 10.1007/s00296-010-1449-7. [DOI] [PubMed] [Google Scholar]
- 6.Kujovich JL. Factor V Leiden thrombophilia. Genet Med. 2011;13(1):1–16. doi: 10.1097/GIM.0b013e3181faa0f2. [DOI] [PubMed] [Google Scholar]
- 7.Wolf AB, Caselli RJ, Reiman EM, Valla J. APOE and neuroenergetics: An emerging paradigm in Alzheimer’s disease. Neurobiol Aging. 2013;34(4):1007–1017. doi: 10.1016/j.neurobiolaging.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain KK. Personalized medicine. Curr Opin Mol Ther. 2002;4(6):548–558. [PubMed] [Google Scholar]
- 9.Newton CR, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17(7):2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olivier M. The Invader assay for SNP genotyping. Mutat Res. 2005;573(1-2):103–110. doi: 10.1016/j.mrfmmm.2004.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rapley R, Harbron S. Molecular Analysis and Genome Discovery. Wiley Online Library; Hoboken, NJ: 2004. [Google Scholar]
- 12.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14(3):303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 13.Xiao Y, et al. An electrochemical sensor for single nucleotide polymorphism detection in serum based on a triple-stem DNA probe. J Am Chem Soc. 2009;131(42):15311–15316. doi: 10.1021/ja905068s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Y, et al. Fluorescence detection of single-nucleotide polymorphisms with a single, self-complementary, triple-stem DNA probe. Angew Chem Int Ed Engl. 2009;48(24):4354–4358. doi: 10.1002/anie.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou C-C, Chen C-H, Lee T-T, Peck K. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 2004;32(12):e99. doi: 10.1093/nar/gnh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Relógio A, Schwager C, Richter A, Ansorge W, Valcárcel J. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 2002;30(11):e51. doi: 10.1093/nar/30.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes TR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19(4):342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 18.Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat Chem. 2011;3(2):103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 19.Khodakov DA, Khodakova AS, Linacre A, Ellis AV. Toehold-mediated nonenzymatic DNA strand displacement as a platform for DNA genotyping. J Am Chem Soc. 2013;135(15):5612–5619. doi: 10.1021/ja310991r. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto K, et al. A peptide nucleic acid (PNA) heteroduplex probe containing an inosine-cytosine base pair discriminates a single-nucleotide difference in RNA. Chemistry. 2013;19(16):5034–5040. doi: 10.1002/chem.201204183. [DOI] [PubMed] [Google Scholar]
- 21.Mo AH, et al. An on-demand four-way junction DNAzyme nanoswitch driven by inosine-based partial strand displacement. Nanoscale. 2014;6(3):1462–1466. doi: 10.1039/c3nr05365b. [DOI] [PubMed] [Google Scholar]
- 22.Hwang MT, et al. DNA nano-carrier for repeatable capture and release of biomolecules. Nanoscale. 2015;7(41):17397–17403. doi: 10.1039/c5nr05124j. [DOI] [PubMed] [Google Scholar]
- 23.Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406(6796):605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 24.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314(5805):1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, et al. A DNA-Origami chip platform for label-free SNP genotyping using toehold-mediated strand displacement. Small. 2010;6(17):1854–1858. doi: 10.1002/smll.201000908. [DOI] [PubMed] [Google Scholar]
- 26.He S, Song B, Li D, Zhu C, Qi W, Wen Y, Wang L, Song S, Fang H, Fan C. A graphene nanoprobe for rapid, sensitive and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010;20(3):453–459. [Google Scholar]
- 27.Cai B, et al. Ultrasensitive label-free detection of PNA-DNA hybridization by reduced graphene oxide field-effect transistor biosensor. ACS Nano. 2014;8(3):2632–2638. doi: 10.1021/nn4063424. [DOI] [PubMed] [Google Scholar]
- 28.Souteyrand E, et al. Direct detection of the hybridization of synthetic homo-oligomer DNA sequences by field effect. J Phys Chem B. 1997;101(15):2980–2985. [Google Scholar]
- 29.Ko JW, et al. Multi-order dynamic range DNA sensor using a gold decorated SWCNT random network. ACS Nano. 2011;5(6):4365–4372. doi: 10.1021/nn102938h. [DOI] [PubMed] [Google Scholar]
- 30.Gao A, et al. Silicon-nanowire-based CMOS-compatible field-effect transistor nanosensors for ultrasensitive electrical detection of nucleic acids. Nano Lett. 2011;11(9):3974–3978. doi: 10.1021/nl202303y. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, et al. Nanoelectronic biosensors based on CVD grown graphene. Nanoscale. 2010;2(8):1485–1488. doi: 10.1039/c0nr00142b. [DOI] [PubMed] [Google Scholar]
- 32.Mohanty N, Berry V. Graphene-based single-bacterium resolution biodevice and DNA transistor: Interfacing graphene derivatives with nanoscale and microscale biocomponents. Nano Lett. 2008;8(12):4469–4476. doi: 10.1021/nl802412n. [DOI] [PubMed] [Google Scholar]
- 33.Kwak YH, et al. Flexible glucose sensor using CVD-grown graphene-based field effect transistor. Biosens Bioelectron. 2012;37(1):82–87. doi: 10.1016/j.bios.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 34.Green NS, Norton ML. Interactions of DNA with graphene and sensing applications of graphene field-effect transistor devices: A review. Anal Chim Acta. 2015;853:127–142. doi: 10.1016/j.aca.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 35.Ohno Y, Maehashi K, Yamashiro Y, Matsumoto K. Electrolyte-gated graphene field-effect transistors for detecting pH and protein adsorption. Nano Lett. 2009;9(9):3318–3322. doi: 10.1021/nl901596m. [DOI] [PubMed] [Google Scholar]
- 36.Dontschuk N, et al. A graphene field-effect transistor as a molecule-specific probe of DNA nucleobases. Nat Commun. 2015;6:6563. doi: 10.1038/ncomms7563. [DOI] [PubMed] [Google Scholar]
- 37.Dong X, Shi Y, Huang W, Chen P, Li L-J. Electrical detection of DNA hybridization with single-base specificity using transistors based on CVD-grown graphene sheets. Adv Mater. 2010;22(14):1649–1653. doi: 10.1002/adma.200903645. [DOI] [PubMed] [Google Scholar]
- 38.Yin Z, et al. Real-time DNA detection using Pt nanoparticle-decorated reduced graphene oxide field-effect transistors. Nanoscale. 2012;4(1):293–297. doi: 10.1039/c1nr11149c. [DOI] [PubMed] [Google Scholar]
- 39.Landon PB, et al. DNA zipper-based tweezers. Langmuir. 2012;28(1):534–540. doi: 10.1021/la201267e. [DOI] [PubMed] [Google Scholar]
- 40.Landon PB, et al. Energetically biased DNA motor containing a thermodynamically stable partial strand displacement state. Langmuir. 2014;30(46):14073–14078. doi: 10.1021/la503711g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen T-Y, et al. Label-free detection of DNA hybridization using transistors based on CVD grown graphene. Biosens Bioelectron. 2013;41:103–109. doi: 10.1016/j.bios.2012.07.059. [DOI] [PubMed] [Google Scholar]
- 42.Zheng C, et al. Fabrication of ultrasensitive field-effect transistor DNA biosensors by a directional transfer technique based on CVD-grown graphene. ACS Appl Mater Interfaces. 2015;7(31):16953–16959. doi: 10.1021/acsami.5b03941. [DOI] [PubMed] [Google Scholar]
- 43.Calado VE, Schneider GF, Theulings AMMG, Dekker C, Vandersypen LMK. Formation and control of wrinkles in graphene by the wedging transfer method. Appl Phys Lett. 2012;101(10):103116. [Google Scholar]
- 44.Zhu W, et al. Structure and electronic transport in graphene wrinkles. Nano Lett. 2012;12(7):3431–3436. doi: 10.1021/nl300563h. [DOI] [PubMed] [Google Scholar]
- 45.Lu C-H, Yang H-H, Zhu C-L, Chen X, Chen G-N. A graphene platform for sensing biomolecules. Angew Chem Int Ed Engl. 2009;48(26):4785–4787. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- 46.Wong K-Y, Pettitt BM. Orientation of DNA on a surface from simulation. Biopolymers. 2004;73(5):570–578. doi: 10.1002/bip.20004. [DOI] [PubMed] [Google Scholar]
- 47.Stern E, et al. Importance of the Debye screening length on nanowire field effect transistor sensors. Nano Lett. 2007;7(11):3405–3409. doi: 10.1021/nl071792z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Israelachvili JN. Intermolecular and Surface Forces. Revised 3rd Ed Academic; Amsterdam, The Netherlands: 2011. [Google Scholar]
- 49.Owczarzy R, Moreira BG, You Y, Behlke MA, Walder JA. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47(19):5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- 50.Zhang G-J, et al. DNA sensing by silicon nanowire: Charge layer distance dependence. Nano Lett. 2008;8(4):1066–1070. doi: 10.1021/nl072991l. [DOI] [PubMed] [Google Scholar]
- 51.Owczarzy R, You Y, Groth CL, Tataurov AV. Stability and mismatch discrimination of locked nucleic acid-DNA duplexes. Biochemistry. 2011;50(43):9352–9367. doi: 10.1021/bi200904e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Regan W, et al. A direct transfer of layer-area graphene. Appl Phys Lett. 2010;96(11):113102. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.