Significance
Some peptide sequences are underrepresented or completely excluded from proteomes because they interfere with protein synthesis. Interference arises from interaction with the peptide exit tunnel of the ribosome. The low frequency of such interactions is an important, previously unrecognized feature of the ribosome.
Keywords: translation, bioinformatics, stalling peptides, underrepresented sequences, single-molecule methods
Abstract
Bioinformatic analysis of Escherichia coli proteomes revealed that all possible amino acid triplet sequences occur at their expected frequencies, with four exceptions. Two of the four underrepresented sequences (URSs) were shown to interfere with translation in vivo and in vitro. Enlarging the URS by a single amino acid resulted in increased translational inhibition. Single-molecule methods revealed stalling of translation at the entrance of the peptide exit tunnel of the ribosome, adjacent to ribosomal nucleotides A2062 and U2585. Interaction with these same ribosomal residues is involved in regulation of translation by longer, naturally occurring protein sequences. The E. coli exit tunnel has evidently evolved to minimize interaction with the exit tunnel and maximize the sequence diversity of the proteome, although allowing some interactions for regulatory purposes. Bioinformatic analysis of the human proteome revealed no underrepresented triplet sequences, possibly reflecting an absence of regulation by interaction with the exit tunnel.
The 20 naturally occurring amino acids can form 8,000 triplets, 160,000 quadruplets, and so forth. The bacterial proteome of about 5 million amino acids can contain, on a random basis, 600 copies of each triplet and 30 copies of each quadruplet. The resulting sequence diversity of the proteome underlies the great variety and specificity of protein structure and function (1). There is a seemingly limitless capacity for the evolution of catalytic activities, protein–ligand interactions, and protein–protein interactions (2). Bacteria are usually successful in adapting to their environment, no matter how harsh, and to the available nutrient source, no matter how unlikely. The question naturally arises of whether there are limitations. Here, we have asked whether all possible amino acid sequences are used in the bacterial proteome; whether some occur less frequently than others; and, if so, what are the possible reasons. We have exploited the comprehensive proteomic data now available by screening for sequences lacking in proteomic datasets. We have screened multiple bacterial proteomes to reveal triplet and quadruplet sequences that occur at frequencies either below [underrepresented sequences (URSs)] or above (overrepresented sequences) the frequencies expected on a random basis, and we have determined the cause of occurrence of URSs.
Identification of URSs in Escherichia coli
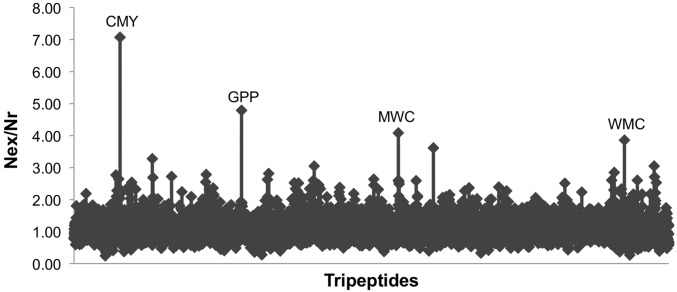
To identify potentially significant URSs, we analyzed all identified ORFs in the proteomes of 29 strains of Escherichia coli for which the entire genome had been sequenced, annotated, and deposited [using the Genome Information Broker DNA Data Bank of Japan (GIB-DDBJ) database, www.ddbj.nig.ac.jp/]. We calculated the ratio between the expected number of occurrences (Nex) and actual number (Nr) for all possible amino acid triplets (Fig. 1). Almost all triplets have a ratio of about 1 (mean = 1.08 ± 0.32 SD), indicating that most sequences occur nearly the expected number of times. Four triplets were identified as significant URSs (Nex/Nr > 4), one of which, CMY, occurs sevenfold less often than expected (18.5 SD above the mean). The other permutations of these three residues (YMC, MYC, etc.) occur at about the expected frequency, indicating that the underrepresentation of CMY is not a result of the reduced use of cysteine and/or methionine. By similar analysis, we could identify more than 5,000 quadruplet URSs that do not appear even once in the database. For example, extension of the CMY sequence to CMYW resulted in complete absence from the proteome. Analysis of the proteomes of 25 microorganism genera revealed different patterns of URSs (Fig. S1), whereas no tripeptide URSs exist in the human proteome (Fig. S2).
Fig. 1.
Identification of E. coli triplet URSs. The ratio of expected number to actual number (Nex/Nr) of all 8.33 × 107 triplets in the 29 redundant proteomes of E. coli used in this study identifies significant URSs as sequences whose expected use is at least fourfold greater than their actual use in the proteomes. Details on the method used for URS calculation are provided in Methods.
URSs Inhibit Protein Expression
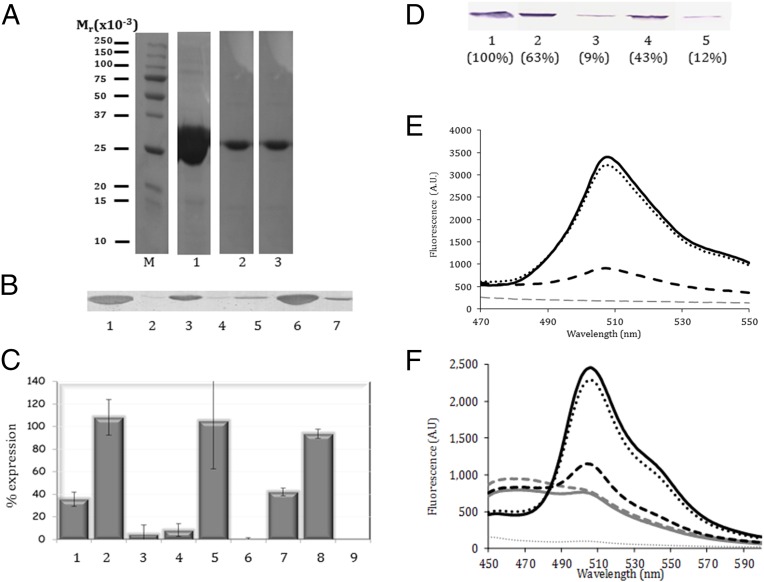
The occurrence of a URS could reflect an inhibitory effect upon protein production (3–6) or simply a lack of utility of the particular sequence in protein folding and function. To investigate the possibility of an inhibitory effect, we embedded the CMY sequence at three sites in the mntA (manganese ABC transporter protein A) gene (Table S1) and determined the effect on MntA expression in vivo. We chose this cyanobacterial gene because we had previously found that it was expressed at very high levels in E. coli and hoped that this level of translation would make identification of translation inhibition more significant. Depending on the site of insertion, the level of MntA protein was reduced 2.5- to 10-fold (Fig. 2 A–C). Insertion of the CMYW sequence at the same locations reduced expression to levels barely or completely undetectable, whereas insertion of the scrambled sequence WCMY at the same locations allowed almost full expression. The inhibitory effect of the presence of URSs was such that internal controls were not possible, because total protein expression was affected (Fig. S3A). All SDS/PAGE-based analysis was thus carefully performed on the basis of initiating expression with equal cell concentrations, and loading was performed on the basis of equal reaction volume. Similar inhibition of MntA expression with embedded URSs was also observed in an in vitro transcription/translation assay (Fig. 2D), showing that inhibition occurs at the level of transcription or translation. The same pattern of inhibition was obtained when the URSs were embedded in GFP (Fig. 2E and Fig. S3). Moreover, when the MntA protein containing an embedded URS was coexpressed with normal GFP, the fluorescence obtained from GFP was also diminished, even though no URS was present (Fig. 2F). This result indicates that the expression of the URS-embedded MntA protein significantly lowers the amount of available ribosomes for GFP expression, suggesting that ribosome recovery is also inhibited. The inhibitory effect of a URS was evidently strong enough to influence global gene expression. As a consequence, preliminary measurements show that total E. coli cell growth was diminished when URS-embedded MntA was expressed as well (Fig. S3D). The in vivo experiments are difficult to quantitate because expression was performed overnight and the bacteria may lose the plasmid, and thus regain viability. However, expression of GFP in HeLa cells and cell survival were unaffected by an E. coli URS embedded in GFP (Fig. S4 and Tables S2 and S3), showing that URSs are species-specific, consistent with our bioinformatic analysis.
Fig. 2.
Embedded URSs inhibit translation in vivo and in vitro. (A) SDS/PAGE analysis of MntA proteins expressed in BL-21(DE3)pLysS cells for 4 h and purified by chelation chromatography. Lane 1 shows WT-MntA, and lanes 2 and 3 show expression of MntA with the CMY URS embedded at sites 1 and 2 (Table S1). (B) Immunoblot analysis of a similar experiment, without protein isolation (by chelation chromatography), was performed using anti-His6 antibodies. The blot is representative of at least four independent experiments for each mutant. Lane 1 shows WT-MntA. Lanes 2–4 are the MntA with embedded CMY, WCMY, and CMYW at site 1. Lanes 5–7 are the MntA with embedded CMY, WCMY, and CMYW at site 2. (C) Quantification of MntA mutant protein expression compared with WT (100%). Three different URSs were embedded in the MntA protein: CMY (bars 1, 4, and 7), WCMY (bars 2, 5, and 8), and CMYW (bars 3, 6, and 9) at the three positions in the MntA protein (Table S1). Expression of the CMYW at sites 2 and 3 (bars 6 and 9) was below the level of detection. Results are the mean ± SD for two to four independent experiments. (D) Immunoblot of WT-MntA (lane 1) or MntA URS mutants (lanes 2–5) produced using an in vitro transcription/translation system. Lanes 2–4 are the CMY URSs embedded at the three sites (Table S1), whereas lane 5 is the CMYW URS at position 2. Parentheses indicate the amount of translated protein compared with MntA-WT as obtained by digital densitometry. (E) Fluorescence analysis of expression of GFP and GFP-URS proteins in vitro. Equal amounts of plasmids expressing WT-GFP (black solid line), mutated GFP with the CMY URS (black dashed line), mutated GFP with the MYC non-URS (black dotted line), or no plasmid (gray dotted line) were added to a transcription/translation mixture. After 30 min, a plasmid expressing the WT-GFP protein was added to the mixture, and the reaction was continued for 4.5 h. (F) Fluorescent analysis of the effect of coexpression of MntA-URS mutants on GFP expression in vitro. Equal amounts of plasmids expressing WT-MntA (black dotted line) or CMY URS containing mutants at site 1 (black dashed line), site 2 (gray dashed line), or site 3 (solid gray line) were added to a transcription/translation mixture. The negative control without any plasmid is shown by the gray dotted line. After 30 min, a plasmid expressing the WT-GFP protein was added to the mixture (black solid line for comparison without additional plasmids), and the reaction was continued for 4.5 h. If the presence of the URS in the target protein only inhibited its own expression, we would expect the same GFP fluorescence as in the absence of a URS-embedded protein. In E and F, following reaction termination, the expressed GFP was matured for 24 h and the level of GFP production was then assessed by fluorescence measurements of the reaction mixtures. The results are representative of four independent experiments. AU, arbitrary units.
URSs Induce Ribosomal Stalling
To determine whether inhibition of gene expression by URSs is due to an effect upon translation, we used single-molecule fluorescence methods for measuring translation rates. Continuous observation of fluorescence resonance energy transfer (FRET) between a Cy3B (donor dye)-labeled E. coli 30S and Black Hole Quencher 2 (a FRET quencher)-labeled 50S subunits (7, 8) in zero-mode waveguides (ZMWs) (9, 10) tracks the conformational dynamics of a single translating ribosome (8, 11–13). During each cycle of elongation, the ribosome undergoes two global conformational changes: a 30S body rotation relative to the 50S subunit that follows peptide bond formation and a reverse rotation upon elongation factor G–catalyzed translocation. The initial nonrotated state is characterized by a higher FRET value, which corresponds to a lower Cy3B donor signal intensity. The rotated state has a lower FRET value, and thus higher signal intensity. The number of high-low-high FRET cycles within an observation window (5 min in the experiments reported here) corresponds to the number of codons translated, whereas the lifetimes of the FRET states reflect the time required to translate each codon. In all experiments, a mixture of total, aminoacylated E. coli tRNAs was used to provide full coverage for all possible codon uses. We have used this approach previously to characterize stalling of translation induced by SecM and ErmCL peptides, as well as frame-shifting signals (14–16).
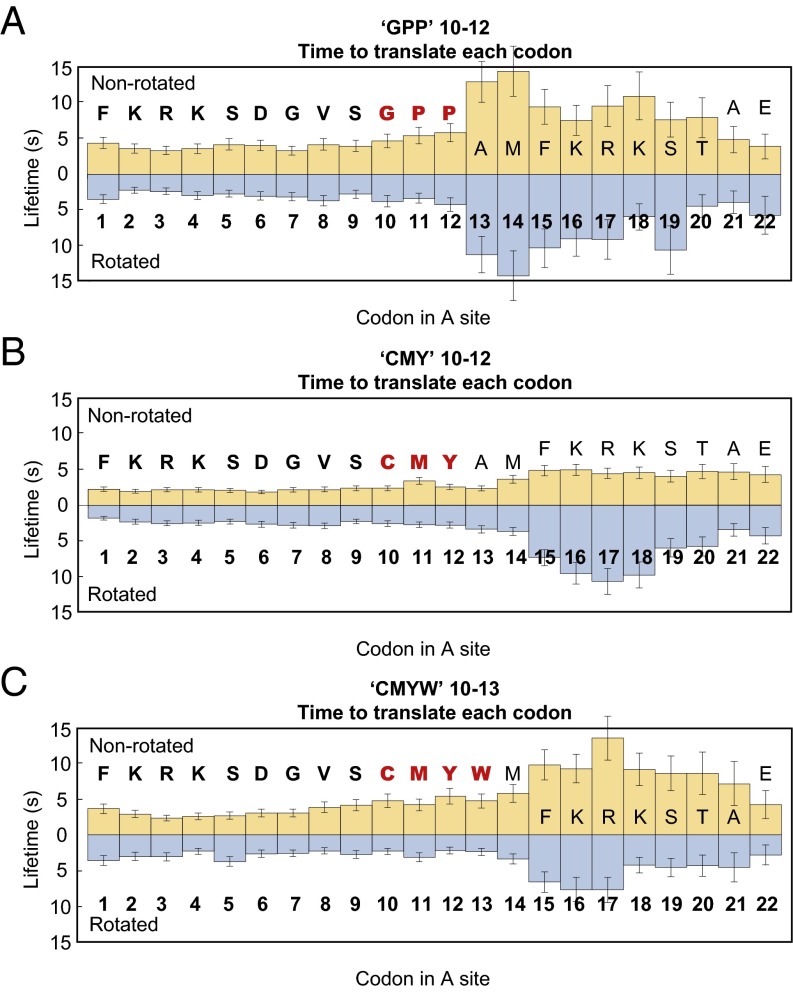
Effects of URS sequences on translation were assessed by insertion of the corresponding codons at codon 10 of bacteriophage T4 gp32 mRNA, whose translation rates and survival times were previously measured by observation of FRET in ZMWs (14). Translation in the presence of a codon for CMY, the most underrepresented triplet URS, began at rates of 4–6 s per codon (1.9–3.4 s in the nonrotated state and 1.8–3.7 s in the rotated state), comparable to translation rates observed for unaltered gp32 mRNA (14). Upon entry of codon 15 in the A site of the ribosome, with CMY residues incorporated in the nascent peptide two amino acids beyond the entrance to the peptide exit tunnel, there was a two- to threefold increase in rotated state lifetimes, and a gradual 1.3-fold increase in nonrotated state lifetimes (Fig. 3B). As translation continued, rotated state lifetimes increased, peaking at about 10 s over codon 17 and returning to normal by codon 21. Nonrotated state lifetimes remained constant at about 4 s per codon. Rotated and nonrotated state lifetimes were constant, and all were less than 3.7 s through 28 codons during translation of the WT gp32 mRNA (14).
Fig. 3.
CMY and GPP triplets and CMYW quadruplet cause a significant decrease in translation rates almost immediately upon entry into the exit tunnel. (A) Ribosomes translating the mRNA encoding the C10M11Y12 sequence proceed at a normal rate until codon 15 is positioned in the A site, and the CMY nascent peptide sequence is two amino acids beyond the entrance to the exit tunnel. At this stage, there is a two to threefold increase in rotated state lifetimes, and a gradual 1.3-fold increase in nonrotated state lifetimes. (B) Ribosomes translating the mRNA encoding the G10P11P12 sequence proceed at a rate of ∼6–8 s per codon until codon 13 is positioned in the A site, and the entire GPP sequence has entered the exit tunnel, at which point there is an abrupt ∼2.5-fold increase in nonrotated state lifetimes (∼13 s) and an approximately threefold increase in rotated state lifetimes (∼11 s). (C) As in the case of the CMY triplet, translation of the CMYW quadruplet causes a decrease in translation rate almost as soon as the peptide sequence has entered the exit tunnel. However, the CMYW peptide sequence also causes a rapid increase in nonrotated state lifetimes (0.6-fold) and acts to slow translation once it has been advanced into the exit tunnel by only one amino acid, a full codon before the effects of the CMY sequence are felt.
The two- to threefold increase in rotated state lifetimes, and thus the decrease in translation rates, resulted in significant translational arrest, but with a two-codon lag. The number of translating ribosomes remained constant through codon 16, with, at most, 2% of ribosomes ceasing translation upon each amino acid incorporation event; from codons 17–21, there was an increase in “attrition,” with 10–14% of ribosomes ceasing translation upon each incorporation event. The “attrition rate” then dropped to 4% per codon, and by codon 25, it returned to values observed in the absence of CMY. Due to attrition, only 4% of ribosomes were still translating in the presence of CMY by codon 30 (Fig. S5A). By contrast, in the absence of CMY, over 30% of ribosomes were still active beyond codon 30 (14).
Translation in the presence of a codon for GPP, the second most underrepresented triplet URS, began at a rate of ∼6–8 s per codon (3.3–4.7 s in the nonrotated state and 2.2–3.8 s in the rotated state) (Fig. 3A). Immediately upon entry of the full GPP sequence in the exit tunnel, with codon 13 in the A-site, there was a threefold increase in rotated state lifetimes (to about 11 s) and a 2.5-fold increase in nonrotated state lifetimes (to about 13 s). There was a further increase in lifetimes at codon 14 (to about 14 s for both rotated and nonrotated states) and concomitant decrease in translation rate (28 s per codon). After the GPP sequence had advanced by two amino acids into the exit tunnel (codon 15 and beyond), rotated and nonrotated state lifetimes decreased (to about 10 s for both states between codons 15–17 and to 3–6 s by codon 21). The decrease in the translation rate resulted in immediate translational arrest: 12% of ribosomes were unable to translate past codon 13, and by codon 29, translation ceased altogether (Fig. S5B).
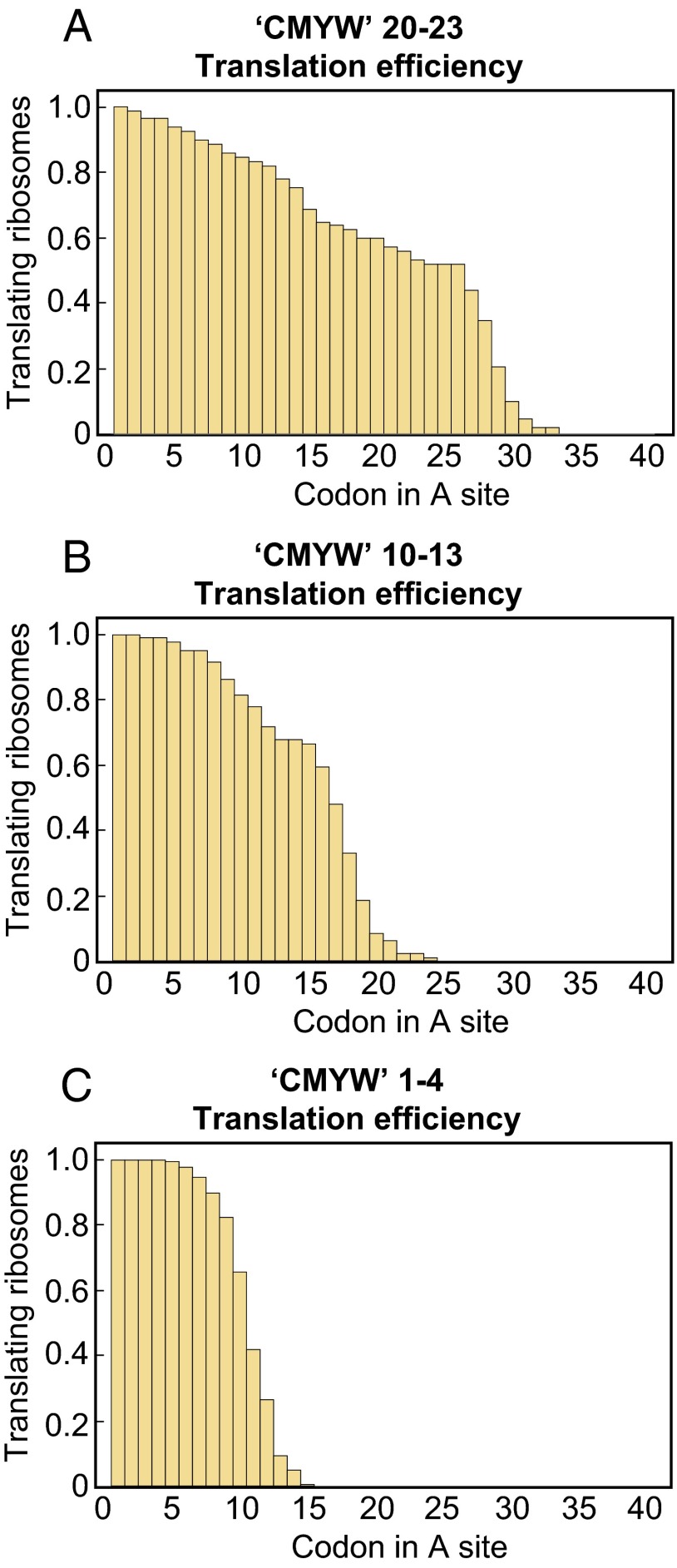
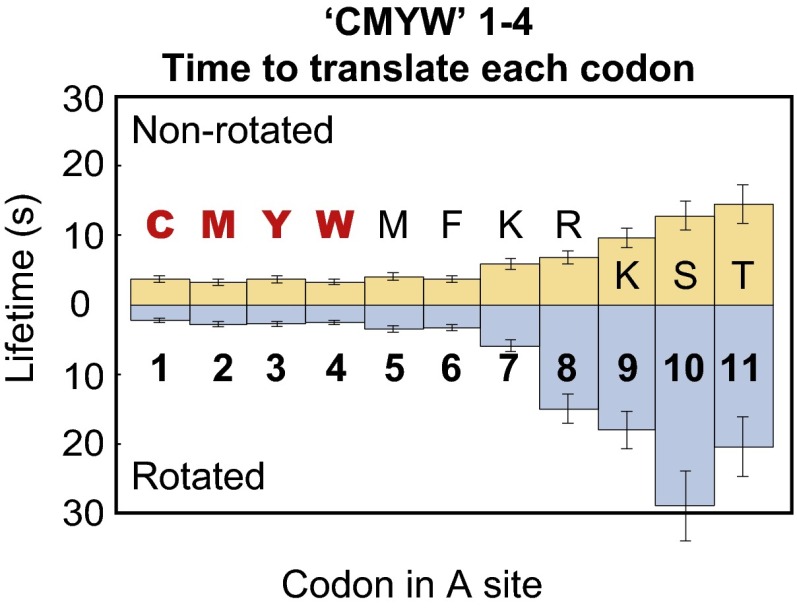
Translation in the presence of a codon for CMYW, a quadruplet completely absent from the E. coli proteome, exhibited more pronounced stalling than either CMY or GPP. The translation rate decreased at codon 15 (with the CMYW sequence advanced by one amino acid into the peptide exit tunnel; Fig. 3C), as in the case of the CMY triplet, but was soon followed by complete translational arrest; no ribosome translating the CMYW sequence progressed past codon 24 (Fig. 4A). When the CMYW sequence was shifted by 10 codons downstream, the decrease in translation rate and the region of translational arrest were shifted by 10 codons as well (Fig. 4B and Fig. S6). When the CMYW sequence was placed immediately after the initiator methionine, the translation rate decreased at codon 6 (with the CMYW sequence advanced by two amino acids into the exit tunnel; Fig. 5), with 16–23% of ribosomes stalling at each codon thereafter, and all ribosomes stalled by codon 16 (Fig. 4C). The profound inhibition by CMYW was due to the sequence and not the amino acid composition, because the reverse sequence, WYMC, had no effect on translation (Fig. S7).
Fig. 4.
Shifting the CMYW sequence results in a shift in the final stall position. (A) One hundred percent of the ribosomes translating mRNA encoding the C11M12Y13W14 sequence have ceased translating by codon 25. (B) One hundred percent of the ribosomes translating the mRNA encoding the C21M22Y23W24 sequence have ceased translating by codon 34. (C) One hundred percent of the ribosomes translating the mRNA encoding the C1M2Y3W4 sequence have ceased translating by codon 16.
Fig. 5.
Translation rate of the CMYW 1–4 sequence begins to decrease at codon 7. Ribosomes translating the C1M2Y3W4 sequence proceed at a normal rate until codon 7 enters the A site, by which point the CMYW sequence has been pushed by two amino acids into the exit tunnel. This sequence is the shortest bacterial stall sequence found to date and, due to its length, can be interacting solely with the lower region of the exit tunnel.
Discussion
Short-sequence use has been analyzed previously (17–20), and different reasons for a lack of some sequences have been suggested. Our bioinformatics results identify triplet and quadruplet sequences that slow translation and lead to stalling almost immediately upon entry into the exit tunnel. These sequences evidently interact with the wall of the tunnel. Although other nascent peptides have been shown to induce arrest (21), these sequences have not been previously identified, most likely because they could not be identified in screens of existing sequences (21–25). The hypothesis that the URSs are omitted from the bacterial proteome to avoid translation inhibition could only be proven experimentally, which we show here in vivo, in vitro, and in single-molecule measurements. We suggest that there may exist other URSs, in bacteria or elsewhere, that may have other effects, such as inducing promiscuous interactions between critical cellular components. These additional effects will have to be explored by other experiments.
The three URSs studied here likely interact with the wall of the exit tunnel in different ways. Translation slows immediately upon entry of the GPP sequence into the exit tunnel, whereas the CMY triplet must be advanced by a full two amino acids into the tunnel before any effect is observed. Stalling caused by GPP may include a component of polyproline-induced stalling, with slow peptidyl transfer causing a decrease in nonrotated state lifetimes after two prolines have been incorporated. In the case of CMY, the effect is mostly seen in increased rotated state lifetimes, suggesting an increased energy barrier to translocation. The addition of Trp in CMYW, results in an almost twofold increase in nonrotated state lifetimes, and although the effects on rotated state lifetimes are similar to the values measured for CMY, they begin earlier, when the CMYW sequence has moved by translation of only a single codon into the exit tunnel.
The point at which CMY and CMYW exert their effects suggests interaction with the region of the exit tunnel near ribosomal residues A2062 and U2585 (26), a region that has been implicated by mutagenesis and by structural and single-molecule studies in interaction with SecM, a sequence shown to perform a regulatory role by stalling translation (14, 15). The last two amino acids of SecM, Gly-Pro, suggest a similarity in mechanism also with GPP. There are, however, fundamental differences between URS- and SecM-induced stalling. The SecM stalling sequence is 17 amino acids long and must interact with both the entrance and the L4-L22 constriction point of the tunnel for stalling, whereas the URSs studied here interact only with the entrance. Other regulatory sequences, such as ErmCL or TnaC, differ by the requirement for a tunnel-bound cofactor to induce stalling (27).
It is noteworthy that no URSs were detected by bioinformatic analysis of the human proteome. Perhaps the human ribosome lacks features of the exit tunnel entrance involved in URS-induced stalling or translation in higher organisms involves additional factors that can compensate for sequence-induced stalling. The occurrence of these features in bacteria may represent a compromise between avoiding URSs and enabling regulation by natural stall sequences.
Methods
URSs were identified using an in-house script that analyzed multiple proteome datasets for the number of unique triplet (or larger) sequences in comparison to the calculated expected number of the same sequence. The expected number of a certain sequence was obtained based on the products of the representative frequencies of each amino acid in the same databases. URSs were embedded by PCR-directed mutagenesis into two cloned target genes at different positions (Figs. S8 and S9). These vectors were then transformed into the BL-21(DE3)pLysS E. coli expression strain. Levels of protein expression were assessed in vivo and in vitro by SDS/PAGE, immunoblotting, or fluorescence. SDS/PAGE and immunoblot bands were digitally quantified using GelAnalyzer software (www.gelanalyzer.com/). Coexpression in the presence of GFP was assessed by fluorescence using standard methods at 508 nm. All labeled ribosomes, factors, and tRNAs were prepared and purified as described (12). Unless noted otherwise, all experiments were performed under buffer conditions described in SI Methods. Data collection from ZMW chips was conducted using instrumentations and techniques described previously (9, 28). Statistical analysis on those traces was also conducted as described before (9, 12), using custom software written in MATLAB (MathWorks).
Supplementary Material
Acknowledgments
N.A. thanks Ada Yonath, Roger Kornberg, Ilana Agmon, Yael Mandel-Gutfreund, Oded Beja, and Gadi Schuster for helpful discussions and support. This work was supported by the Technion VPR Research Fund (N.A.) and by NIH Grants GM51266 and GM09968701 (to J.D.P. and E.V.P.) and a Stanford Interdisciplinary Graduate Fellowship (to J.C.).
Footnotes
Conflict of interest statement: N.A., S.P.N., and T.S. filed a patent application on the use of underrepresented sequences in novel antimicrobials.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606518113/-/DCSupplemental.
References
- 1.Worth CL, Gong S, Blundell TL. Structural and functional constraints in the evolution of protein families. Nat Rev Mol Cell Biol. 2009;10(10):709–720. doi: 10.1038/nrm2762. [DOI] [PubMed] [Google Scholar]
- 2.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11(8):572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 3.Bashan A, Yonath A. Correlating ribosome function with high-resolution structures. Trends Microbiol. 2008;16(7):326–335. doi: 10.1016/j.tim.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Tsai A, O’Leary SE, Petrov A, Puglisi JD. Unraveling the dynamics of ribosome translocation. Curr Opin Struct Biol. 2012;22(6):804–814. doi: 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mankin AS. Nascent peptide in the “birth canal” of the ribosome. Trends Biochem Sci. 2006;31(1):11–13. doi: 10.1016/j.tibs.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Wekselman I, et al. Ribosome’s mode of function: Myths, facts and recent results. J Pept Sci. 2009;15(3):122–130. doi: 10.1002/psc.1077. [DOI] [PubMed] [Google Scholar]
- 7.Dorywalska M, et al. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33(1):182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci USA. 2008;105(40):15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464(7291):1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai A, et al. The impact of aminoglycosides on the dynamics of translation elongation. Cell Reports. 2013;3(2):497–508. doi: 10.1016/j.celrep.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17(7):793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Petrov A, Tsai A, O’Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20(6):718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Tsai A, Petrov A, Puglisi JD. Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. J Am Chem Soc. 2012;134(13):5734–5737. doi: 10.1021/ja2119964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai A, Kornberg G, Johansson M, Chen J, Puglisi JD. The dynamics of SecM-induced translational stalling. Cell Reports. 2014;7(5):1521–1533. doi: 10.1016/j.celrep.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson M, Chen J, Tsai A, Kornberg G, Puglisi JD. Sequence-dependent elongation dynamics on macrolide-bound ribosomes. Cell Reports. 2014;7(5):1534–1546. doi: 10.1016/j.celrep.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, et al. Dynamic pathways of -1 translational frameshifting. Nature. 2014;512(7514):328–332. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otaki JM, Ienaka S, Gotoh T, Yamamoto H. Availability of short amino acid sequences in proteins. Protein Sci. 2005;14(3):617–625. doi: 10.1110/ps.041092605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otaki JM, Gotoh T, Yamamoto H. Potential implications of availability of short amino acid sequences in proteins: An old and new approach to protein decoding and design. Biotechnol Annu Rev. 2008;14:109–141. doi: 10.1016/S1387-2656(08)00004-5. [DOI] [PubMed] [Google Scholar]
- 19.Tuller T, Chor B, Nelson N. Forbidden penta-peptides. Protein Sci. 2007;16(10):2251–2259. doi: 10.1110/ps.073067607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acquisti C, Poste G, Curtiss D, Kumar S. Nullomers: Really a matter of natural selection? PLoS One. 2007;2(10):e1022. doi: 10.1371/journal.pone.0001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woolstenhulme CJ, et al. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA. 2013;110(10):E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem. 2009;284(50):34809–34818. doi: 10.1074/jbc.M109.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peil L, et al. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci USA. 2013;110(38):15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339(6115):82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 25.Sothiselvam S, et al. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc Natl Acad Sci USA. 2014;111(27):9804–9809. doi: 10.1073/pnas.1403586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito K, Chiba S. Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 27.Ito K, Chiba S, Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun. 2010;393(1):1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc Natl Acad Sci USA. 2014;111(2):664–669. doi: 10.1073/pnas.1315735111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.