Significance
Cellular metabolism is tightly regulated by AMP-activated protein kinase (AMPK): the function of which is influenced by folliculin (FLCN), folliculin-interacting protein (FNIP)1, and FNIP2. FLCN is a known tumor-suppressor protein that is mutated in Birt–Hogg–Dubé syndrome, whereas FNIP1 and FNIP2 are binding partners of FLCN. Previous reports have suggested that the FLCN/FNIP1/FNIP2 complex acts a positive regulator of AMPK, whereas other reports suggest the opposite. Using a new mouse model of FNIP1 deficiency, our findings support the latter: we found that mutation of Fnip1 leads to B-cell deficiency and the development of a cardiomyopathy similar to mice and humans with gain-of-function mutations in AMPK.
Keywords: cellular metabolism, lymphocyte development, N-ethyl-N-nitrosourea, cardiomyopathy, autophagy
Abstract
Folliculin (FLCN) is a tumor-suppressor protein mutated in the Birt–Hogg–Dubé (BHD) syndrome, which associates with two paralogous proteins, folliculin-interacting protein (FNIP)1 and FNIP2, forming a complex that interacts with the AMP-activated protein kinase (AMPK). Although it is clear that this complex influences AMPK and other metabolic regulators, reports of its effects have been inconsistent. To address this issue, we created a recessive loss-of-function variant of Fnip1. Homozygous FNIP1 deficiency resulted in profound B-cell deficiency, partially restored by overexpression of the antiapoptotic protein BCL2, whereas heterozygous deficiency caused a loss of marginal zone B cells. FNIP1-deficient mice developed cardiomyopathy characterized by left ventricular hypertrophy and glycogen accumulation, with close parallels to mice and humans bearing gain-of-function mutations in the γ2 subunit of AMPK. Concordantly, γ2-specific AMPK activity was elevated in neonatal FNIP1-deficient myocardium, whereas AMPK-dependent unc-51–like autophagy activating kinase 1 (ULK1) phosphorylation and autophagy were increased in FNIP1-deficient B-cell progenitors. These data support a role for FNIP1 as a negative regulator of AMPK.
Birt–Hogg–Dubé (BHD) syndrome is a hereditary condition caused by germ-line or somatic mutations of the folliculin (FLCN) tumor-suppressor gene (1) and characterized by cutaneous hamartomas, pulmonary cysts, and an increased risk of renal cancer (2).
FLCN forms a complex with paralogous folliculin interacting proteins, folliculin-interacting protein (FNIP)1 and FNIP2, both of which bind the C terminus of FLCN with the potential to form homo- or heterotrimeric multimers (3–5). FLCN is a nonredundant component of the FLCN/FNIP1/FNIP2 complex, whereas FNIP1 and FNIP2 appear to be functionally redundant in several tissues. This distinction is illustrated by the fact that kidney-specific deletion of mouse Flcn or compound deletion of Fnip1 and Fnip2 is sufficient to cause renal tumors, and yet deletions of Fnip1 or Fnip2 alone are not (6). This redundancy is consistent with the equivalent expression of Fnip1 and Fnip2 mRNA in kidney tissue (6). In contrast, FNIP1 is nonredundant in other tissues, including bone marrow, myocardium, and skeletal muscle: all of which express relatively more Fnip1 than Fnip2 (6–9).
FNIP1 and FNIP2 also bind to the α, β, and γ subunits of the heterotrimeric AMP-activated protein kinase (AMPK) complex (3, 4). A critical regulator of cellular metabolism, AMPK senses and is activated by increased concentrations of AMP and ADP in the energy-depleted cell and subsequently phosphorylates an array of regulatory targets to restore cellular energy status (10). The multifaceted roles of AMPK include growth suppression by inhibiting synthesis of cellular macromolecules, in particular, through phosphorylation of the TSC2 tumor suppressor and inhibition of the mammalian target of rapamycin complex (mTORC)1 signaling pathway (11). AMPK also promotes autophagy via multiple pathways including mTORC1 and unc-51–like autophagy activating kinase 1 (ULK1) (11, 12), induces cell-cycle arrest by stabilizing p53 (13), and favors oxidative phosphorylation by up-regulating oxidative enzymes and promoting mitochondrial biogenesis (14).
Several reports have shown that the FLCN/FNIP1/FNIP2 complex influences both AMPK and mTOR, and yet the precise role of FNIP1 is uncertain. In one report (9), FNIP1-deficient skeletal muscle exhibited enhanced phosphorylation of the catalytic α subunit of AMPK (at residue Thr172—a requirement for its activation) (9) but reduced phosphorylation in another (6). Similarly, mTOR activity was reported to be increased in B-cell precursors in one Fnip1 mutant (8) but normal in a second model (7). Although phosphorylation of mTOR or the S6 ribosomal protein (a downstream mediator of mTOR signaling) was consistently increased in renal carcinomas of BHD syndrome patients or Flcn knockout mice (15–17), this observation may reflect a direct effect of transformation rather than the predisposing mutation.
To explore the role of the FLCN/FNIP1/FNIP2 complex in the regulation of metabolism and autophagy and better define its influence on AMPK, we investigated a loss-of-function allele of Fnip1 in mice, focusing on abnormalities in the development and function of B cells and of the myocardium.
Results
A Recessive B-Cell Deficiency Associated with a Splice Donor Variant of Fnip1.
As part of a broader mouse N-ethyl-N-nitrosourea (ENU) mutagenesis program (18), we designed a sensitized screen to identify genes required for lymphocyte development. Third-generation (G3) mutant mice were first treated with a sublethal dose of γ radiation (400 rad) and the following day, received an i.v. injection of 2 × 106 CD45.1+ bone marrow cells. Chimeric G3 mice were bled four weeks later, and the contribution of CD45.1+ donor cells to various lymphocyte compartments was determined by flow cytometry (Fig. 1A). One phenodeviant, named hamel, was characterized by complete repopulation of the CD19+ B-cell compartment by CD45.1+ donor-derived cells (Fig. 1A). This repopulation indicated a cell-intrinsic failure of hematopoietic precursors to repopulate the CD19+ compartment and was not apparent in CD4+, CD8+, NK1.1+, or CD11b+ compartments.
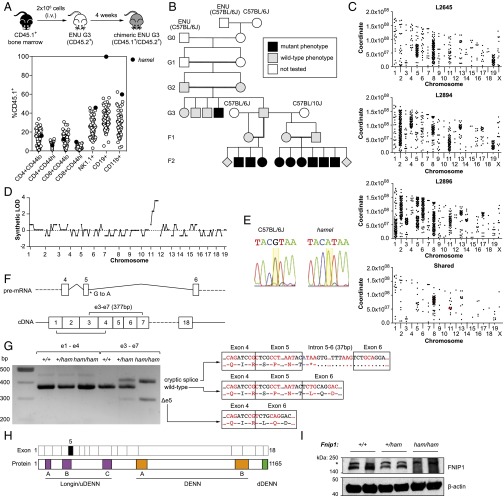
Fig. 1.
Recessive B-cell deficiency associated with a splice donor variant in Fnip1. (A) Schematic of the competitive reconstitution screen and identification of the hamel phenotype. (B) Initial generations of the hamel pedigree, including mapping outcrosses. (C) Single-nucleotide variant distribution across the genomes of three affected mice and those shared by all three. Mutations affecting protein-coding sense are highlighted in red. (D) Simulated LOD score generated by chromosomal mapping. (E) Capillary-sequencing trace of a splice donor variant in Fnip1 (yellow highlight). (F) Schematic of the Fnip1 transcript (ENSMUST00000046835) and the location of the hamel mutation and positions of amplicons generated in G. (G) Fnip1 BM cDNA PCR amplification and sequencing, demonstrating the presence of two major alternate splice products in Fnip1 homozygous mutants. (H) Domain structure of the FNIP1 protein, with corresponding coding exons displayed above. Exon 5 (skipped in one alternate splice product) is highlighted in black fill. DENN, differentially expressed in neoplastic versus normal cells. (I) anti-FNIP1 Western blot on splenocyte lysates of the indicated genotypes. Asterisk indicates a nonspecific band.
The hamel pedigree was propagated by outcrossing male siblings of the proband to both C57BL/6J and C57BL/10J females and intercrossing the resulting progeny (Fig. 1B). By examining lymphocyte populations in the F2 generation before irradiation, it became clear that the hamel phenotype was a simple autosomal recessive B-cell deficiency. To identify the causative mutation, we performed whole-genome sequencing on three F2 mutants from the C57BL/6J outcross. Homozygous variants within each mouse were clustered in discrete blocks across the genome, with variants shared between all three largely confined to chromosomes 8 and 11 (Fig. 1C). Segregation analysis of 128 polymorphic variants in the C57BL/10J outcross showed that all six F2 mutants tested were homozygous for C57BL/6J-derived alleles on distal chr11 (but not chr8), corresponding to synthetic logarithm of odds (LOD) score of 4 (Fig. 1D). Of all of the shared homozygous variants on chr11, only one was predicted to change protein-coding sense: a critical splice donor variant in Fnip1 (GRCm38, chr11:54480685).
Capillary sequencing confirmed the presence of the Fnip1 splice donor variant (Fig. 1E), which occurred at the 3′ boundary of exon 5 (of a total of 18) (Fig. 1F). To measure the effects of the hamel variant on mRNA processing, PCR amplicons were generated from wild-type and mutant cDNA templates (Fig. 1F). Whereas the processing of exons 1–4 was equivalent between wild-type and mutant, amplification across exons 3–7 revealed at least two aberrant splice products (Fig. 1G). Sequencing revealed the smaller of the two products lacked exon 5 (leading to an in-frame deletion of 25 amino acids), whereas the larger had incorporated 37 bp of introns 5–6 before using a cryptic splice site (creating a premature termination codon) (Fig. 1G).
Fnip1 has been reported to play an essential role in B-cell development (7, 8). Mouse FNIP1 consists of 1,165 aa and shares 91% amino acid identity with human FNIP1 and 49% identity with mouse FNIP2 (Fig. 1H). Although the predicted molecular mass is 130 kDa, we were only able to detect a larger protein by Western blot (>140 kDa), which was absent from Fnip1 mutant bone marrow lysate (Fig. 1I). This finding is consistent with other reports (8, 19). The product of the Fnip1Δe5 splice variant (a 25-aa in-frame deletion) was not apparent by Western blotting using an antibody raised against an N-terminal peptide.
Early Block of B-Cell Development and a Reduction of Marginal Zone B Cells in Heterozygotes.
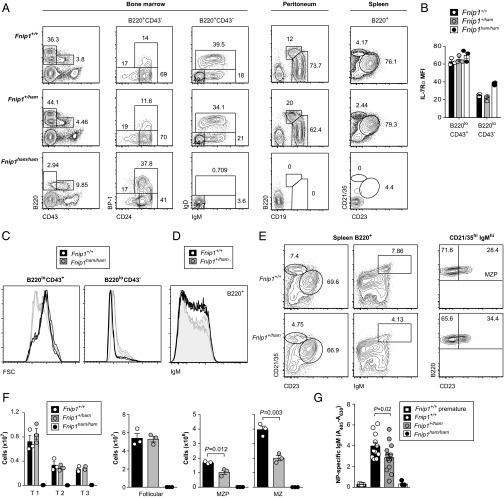
We next examined the major B-cell subsets in bone marrow, peritoneum, and spleen by flow cytometry. Although frequencies in wild-type and heterozygous littermates were largely indistinguishable, B cells were absent from the peritoneum and spleen of Fnip1 homozygous mutants (Fig. 2A). Analysis of bone marrow revealed an absence of IgM+ or IgD+ cells, although B220lo B-cell precursors were still present. Unlike cells deficient in the mTORC2 component Sin1 (20), these precursors expressed the receptor for the B-cell survival factor IL-7 (Fig. 2B). The precursors were also larger than wild-type counterparts at their terminal B220loCD43− stage (Fig. 2C) and smaller at the B220loCD43+ stage: observations that highlight the challenge of interpreting comparative assays that are not controlled for cell size.
Fig. 2.
An early block in B-cell development and a gene dose-sensitive MZ B-cell deficiency. (A) Frequencies of major B-cell subsets in bone marrow, peritoneum, and spleen. (B) Expression of the IL-7R α chain in early (B220loCD43+) and late (B220loCD43−) B-cell progenitors. (C) Forward scatter (FSC) profiles of the subsets in B indicate relative sizes of wild-type and Fnip1 mutant cells. (D) Reduced frequency of IgMhi cells in Fnip1+/ham heterozygotes (n = 3). (E) Reduced frequency of splenic MZ B cells (CD21/35hiCD23lo or CD21/35hiIgMhi) and MZ precursors (MZP) (CD21/35hiIgMhiCD23hi). (F) Absolute numbers of transitional (T1, CD93+CD23−; T2, CD93+CD23+IgMint; T3, CD93+CD23+IgMhi;), follicular (CD21/35intCD23hi), MZP, or MZ B cells in spleen. (G) Serum levels of NP-specific IgM antibodies before (preimmune) and after immunization with NP-Ficoll. Plots in A and E are representative of three mice per genotype. Symbols in B, F, and G represent individual mice, with bars representing the means (± SEM). P values calculated by unpaired two-tailed t test.
Closer examination of B220+ splenocytes in heterozygous Fnip1 mutants revealed a reduced frequency of IgM+ cells (Fig. 2D). This reduction corresponded to the marginal zone (MZ) B-cell population, defined as either CD21/35hiIgMhi or CD21/35hiCD23lo (Fig. 2E). Frequencies and absolute numbers were reduced in both the MZ and MZ precursor compartments, whereas transitional and follicular B-cell subsets were not affected (Fig. 2 E and F). This defect was also associated with a mild reduction in antigen-specific IgM following immunization with the T cell-independent antigen NP28-aminoethyl carboxymethyl–Ficoll (NP-Ficoll) (Fig. 2G).
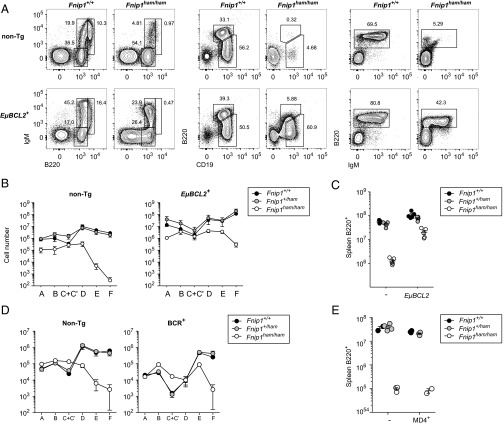
Given the importance of the antiapoptotic protein BCL2 in B-cell survival, we also measured the effects of a human EμBCL2 transgene on B-cell development in Fnip1 mutants. In contrast to previous reports (7), BCL2 overexpression only partially corrected B-cell numbers in the bone marrow, peritoneum, and spleen (Fig. 3 A–C). Consistent with earlier reports, expression of a prearranged BCR transgene did not correct peripheral B-cell numbers (Fig. 3 D and E) (7, 8).
Fig. 3.
Partial rescue of early B-cell development by an EμBCL2 transgene but not a BCR transgene. (A) Flow-cytometric plots of major B-cell populations in bone marrow (Left), peritoneum (Center), and spleen (Right) of wild-type and Fnip1 mutant mice with or without an EμBCL2 transgene. (B and D) Absolute numbers of cells in corresponding Hardy fractions (A, B220+CD43+BP-1−CD24−; B, B220+CD43+BP-1−CD24+; C+C′, B220+CD43+BP-1+CD24+; D, B220+CD43−IgM−IgD−; E, B220+CD43−IgM−IgD+; and F, B220+CD43−IgM+IgD+) in the presence or absence of an EμBCL2 or BCR transgene. (C and E) Absolute numbers of B220+ splenocytes. Plots in A are representative of five mice per genotype. Symbols in B and D represent the means (± SEM). Each symbol in C and E represents an individual mouse. Bars indicate means ± SEM.
Cardiac Hypertrophy, Ventricular Preexcitation, and Glycogen Accumulation in the Absence of FNIP1.
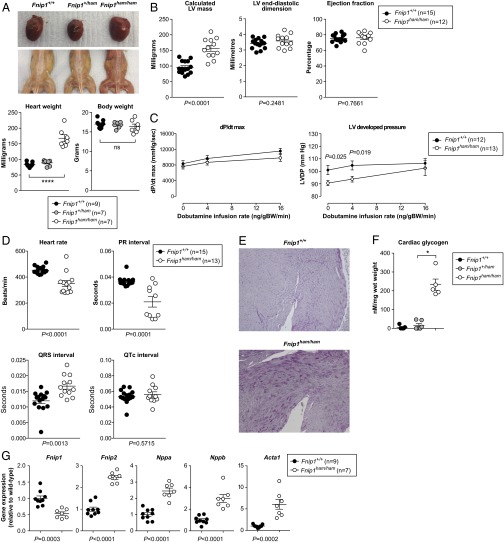
On dissection, the hearts of homozygous Fnip1 mutants were visibly enlarged (despite no difference in overall body weight) (Fig. 4A). Skeletal muscle from homozygous Fnip1 mutants was also darker in color than heterozygous or wild-type littermates (Fig. 4A), as has been reported previously (9).
Fig. 4.
Cardiac phenotype associated with the Fnip1 mutation. (A) Macroscopic appearance of heart and skeletal muscle of adult wild-type and Fnip1 mutant mice. Heart and body weight of 6-wk-old female mice are represented in the lower image. (B) Calculated LV mass, LV end-diastolic dimension (LVEDD), and LV ejection fraction. (C) Invasive hemodynamic measurements of LV dP/dt max, and LV developed pressure (LV systolic pressure – LV end-diastolic pressure) at baseline and in response to dobutamine stimulation in vivo. (D) Heart rate, PR, QRS, and corrected QT (QTc) intervals of 20-wk-old wild-type and Fnip1 mutant mice under isoflurane anesthesia. P values calculated by unpaired two-tailed t test. (E) Periodic acid–Schiff staining of LV myocardium (100× magnification). (F) Cardiac glycogen content. (G) Cardiac expression of Fnip1, Fnip2, and selected gene markers of cardiac stress. Symbols represent individual mice (A, B, D, F, and G) or the mean (C), with all error bars indicating means ± SEM. P values calculated by unpaired two-tailed t test (B–D and G) or one-way ANOVA with Bonferroni posttest (A and F). *Significance at α = 0.05.
Noninvasive determination of left ventricular (LV) mass and internal end-diastolic dimension by echocardiography revealed that cardiac enlargement was principally the result of hypertrophy, rather than chamber dilatation (Fig. 4B). LV systolic function was normal (Fig. 4B). Consistent with the echocardiographic findings, invasive measurement of LV pressure waveforms in vivo demonstrated preserved contractile reserve in response to inotropic stimulation [reflected in the maximal rate of rise in LV pressure (LV dP/dt max)], with some evidence for enhanced basal contractility (LV developed pressure = LV systolic pressure – LV end-diastolic pressure) (Fig. 4C). Electrocardiography recordings from homozygous Fnip1 mutant mice revealed marked shortening of the PR interval and prolongation of the QRS complex, consistent with overt ventricular preexcitation, together with slower heart rate but no change in corrected QT interval (Fig. 4D). Histological evaluation of mutant myocardium revealed marked accumulation of glycogen (Fig. 4E), confirmed on biochemical analysis (Fig. 4F). Pathological cardiac hypertrophy is associated with a return to a fetal gene expression profile and up-regulation of markers of cardiac stress (21). Consistent with this report, homozygous Fnip1 mutant mice manifested higher expression of Nppa, Nppb (encoding the natriuretic peptides A and B), and Acta1 (encoding skeletal muscle α-actin) (Fig. 4G). As expected, cardiac Fnip1 gene expression was reduced, whereas expression of its paralog, Fnip2, was increased (Fig. 4G).
FNIP1 Is a Negative Regulator of AMPK.
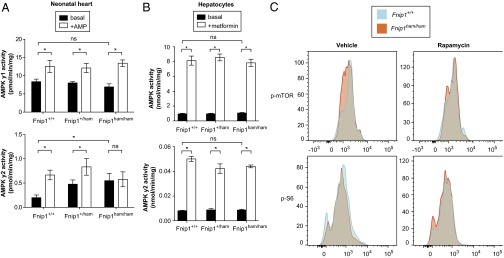
Given the recognized interaction between FNIP1 and AMPK subunits (3) and the cardiac phenotypic overlap of homozygous Fnip1 mutant mice with humans and mice expressing mutations in Prkag2 (encoding the γ2 regulatory subunit of AMPK) (22–25), we sought to assess the effect of the Fnip1 mutation on cardiac AMPK activity. The β subunit of AMPK contains a regulatory domain allowing AMPK to bind to, and be inhibited by, glycogen (26). To minimize any potential confounding effect of myocardial glycogen accumulation per se on AMPK activity—a factor recognized to contribute to bitemporal changes in AMPK activity in Prkag2 transgenic models (27)—we obtained cardiac tissue from neonatal Fnip1 mutant mice. We immunoprecipitated γ2- and γ1-containing AMPK complexes from these hearts and measured γ isoform-specific AMPK activity in the basal state and in the presence of the allosteric activating ligand AMP (Fig. 5A). We observed basal activation of γ2-containing AMPK complexes but reduced AMP responsivity in homozygous Fnip1 mutants. However, the activity and AMP responsiveness of γ1-containing AMPK complexes was comparable across genotypes.
Fig. 5.
Elevated AMPK activity in hearts, but not hepatocytes, of Fnip1 mutants. (A) AMPK γ1- and AMPK γ2-specific activity in neonatal heart tissue in the presence or absence of AMP. (B) Total AMPK and AMPK γ2-specific activity in hepatocytes in the presence or absence of metformin. (C) Bone marrow B-cell progenitors (B220+CD43+) were isolated and treated with rapamycin or vehicle control. Following permeabilization, cells were stained for phosphorylated mTOR (p-mTOR, Ser2448) or phosphorylated S6 ribosomal protein (p-S6, Ser235/236). (A and B) Error bars represent mean ± SEM (n = 4 per genotype). P values calculated by one-way ANOVA with Bonferroni posttest. *Significance at α = 0.05.
We also measured AMPK activity from primary isolated hepatocytes (Fig. 5B) but observed no differences in total or γ2-specific activity between wild-type and mutant (Fig. 5B), suggesting that Fnip1 and Fnip2 may be functionally redundant in the liver, as they are in the kidney (6).
We next measured phosphorylation of mTOR and S6 in bone marrow B-cell progenitors in the presence or absence of the mTORC1 inhibitor rapamycin. Unlike previous reports (8), but consistent with others (7), we did not observe a difference in phosphorylation of mTOR or S6 in B-cell progenitors (Fig. 5C).
Fnip1 Deficiency in B Cells Causes Increased Autophagy.
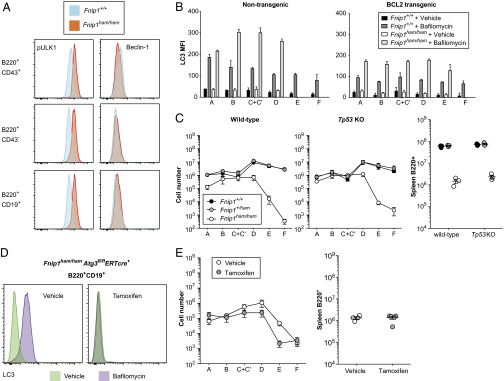
Given the diversity of pathways regulated by AMPK, we sought evidence of increased activation in a number of its downstream targets. Among these targets is the autophagy-inducing kinase ULK1, which is activated through phosphorylation at serine 555 by AMPK and inhibited by mTORC1. S555 p-ULK1 was consistently higher in Fnip1 mutant bone marrow cells than in wild type (Fig. 6A), suggesting that autophagy was enhanced. This hypothesis was supported by the observation that staining of membrane-bound LC3, an autophagosome marker, was also increased in Fnip1 mutant B cells (Fig. 6B). LC3 staining was only increased in the presence of bafilomycin (an inhibitor of autophagosome degradation) (Fig. 6B), implying that mutation of Fnip1 enhanced autophagosome formation, rather than inhibiting their degradation.
Fig. 6.
Autophagosome formation is increased in Fnip1 mutants but is not responsible for B-cell deficiency. (A) Bone marrow B cells were permeabilized and stained for S555 phospho-ULK1 (pULK1) and Beclin-1. (B) Bone marrow cells from wild-type or Fnip1 mutant mice (with or without the EμBCL2 transgene) were stained for LC3 in the presence or absence of bafilomycin (which inhibits degradation of LC3-II expressing autophagosomes). Hardy fractions were gated as in Fig. 3 (A, B220+CD43+BP-1−CD24−; B, B220+CD43+BP-1−CD24+; C+C′, B220+CD43+BP-1+CD24+; D, B220+CD43−IgM−IgD−; E, B220+CD43−IgM−IgD+; F, B220+CD43−IgM+IgD+). (C) Absolute numbers of bone marrow B-cell subsets and splenic B220+ cells in mice with or without a homozygous null allele of Tp53. (D) LC3-staining intensity in B220+CD19+ B cells from Fnip1ham/hamAtg3fl/flERT2cre+ mice treated with tamoxifen or vehicle control. Cells were incubated in the presence or absence of bafilomycin. (E) Absolute numbers of bone marrow B-cell subsets and splenic B220+ cells in Fnip1ham/hamAtg3fl/flERT2cre+ mice treated with vehicle (Atg3-sufficient) or tamoxifen (Atg3-deficient).
A key regulator of autophagosome formation, and a substrate of ULK1, is the protein Beclin-1. Beclin-1 is, in turn, inhibited by BCL2 (28), the same protein that when overexpressed was able to partially restore B-cell development in Fnip1 mutants. Although intracellular Beclin-1 was equivalent in wild-type and Fnip1 mutant B cells (Fig. 6A), overexpression of BCL2 substantially reduced LC3 staining in bafilomycin-treated wild-type and Fnip1 mutant cells (Fig. 6B). This reduction indicated that BCL2 overexpression could oppose the increased autophagic flux in Fnip1 mutant B cells while mediating increased survival.
AMPK promotes cell-cycle arrest by stabilizing p53 (13), and p53 is known to enhance autophagy and apoptosis in response to cellular stress (29). Similar stresses might be present at critical points in B-cell development, so we considered whether enhanced activation of p53 would be sufficient to explain B-cell deficiency in FNIP1 mutant mice. To test the role of p53, we created Fnip1 mutant mice with a parallel mutation in the gene encoding it (Tp53). B-cell numbers were no different, however, between p53-sufficient and p53-deficient Fnip1 mutants (Fig. 6C).
Autophagy plays a vital role during early B-cell development, highlighted by the failure of B-cell development in the absence of the autophagy regulator Atg5 (30). To determine whether enhanced, rather than reduced, autophagy was responsible for the observed block in B-cell development, we sought to inhibit autophagy completely using an Atg3 conditional allele. Atg3 plays an essential role in autophagosome formation, acting as an E2-like enzyme to catalyze conjugation of Atg8 and phosphatidylethanolamine. To create Atg3-deficient cells, we combined a floxed Atg3 allele with a tamoxifen-inducible ERT2cre transgene. Both the transgene and the floxed allele were bred onto the Fnip1 mutant background, and 10 d after administration of tamoxifen or vehicle, B-cell precursors were isolated for analysis. To confirm that Atg3 had been deleted successfully, cells from control (vehicle) or tamoxifen-treated mice were subjected to LC3 staining. Whereas LC3 staining was increased in the presence of bafilomycin in control cells, it was not increased in tamoxifen-treated cells (Fig. 6D). Despite this confirmation of successful Atg3 deletion, tamoxifen treatment did not correct B-cell numbers in Fnip1 mutant mice (Fig. 6E), suggesting that enhanced autophagy alone was not sufficient to block B-cell development.
Discussion
Our investigation of the hamel allele reinforces the nonredundant roles of FNIP1 in B-cell development (7, 8), skeletal muscle composition (9), and cardiac function (6). We reveal that BCL2 overexpression can partially, but not entirely, rescue B-cell development in the absence of FNIP1, and we provide evidence that FNIP1 is an endogenous negative regulator of AMPK. Our observations shed light on the inconsistencies in previous data and raise interesting questions about the metabolic demands of different cells and tissues. This difference in metabolic demand is exemplified by lineage-specific effects in B-cell development, where complete deficiency in FNIP1 blocks the development of rapidly proliferating progenitor cells, whereas partial deficiency in heterozygotes leads to decreased MZ B-cell numbers. MZ B cells are known to be sensitive to metabolic disturbances, with loss of the mTOR inhibitor TSC1 also associated with impaired MZ B-cell development (31).
A key controversy surrounding the FLCN/FNIP complex is its effect on mTOR activity (32). Several reports describe FLCN as a positive regulator of mTOR, and yet FLCN-deficient renal cancer cells show hyperactivity of mTOR (15–17). This hyperactivity may simply reflect the proliferative state of transformed cells or a compensatory change caused by the absence of the FLCN/FNIP complex. In the context of a loss-of-function Fnip1 mutation in B-cell precursors, we and others did not observe a difference in mTOR activation (7), whereas others saw an increase (8).
Despite their direct interaction, the role of FNIP1 in AMPK regulation was also unclear. Loss of FNIP1 in striated skeletal muscle was associated with increased AMPK activity in one report (9) but reduced activity in another (6). To investigate this phenomenon and minimize the effects of secondary changes, we directly examined AMPK activity in neonatal myocardium and observed an increase in basal activity of γ2-containing AMPK complexes. This biochemical finding and the cardiac phenotype of Fnip1 homozygous mutants are strikingly similar to that associated with gain-of-function mutations in Prkag2, encoding the γ2 subunit of AMPK. Humans bearing and mice engineered to carry such mutations exhibit a cardiomyopathy characterized by LV hypertrophy, ventricular preexcitation, and cardiomyocyte glycogen accumulation (23, 24, 33), as in our model. Intriguingly, Prkag2 mutations cluster within and around cystathionine β-synthase motifs, which occur as tandem pairs (Bateman domains) capable of binding adenine nucleotides to mediate energy sensing (34). Our findings suggest that in the neonatal heart, FNIP1-containing complexes play an inhibitory role in the selective regulation of γ2-containing AMPK complexes. The discrepancy with other reports in adults may be related to the progressive accumulation of glycogen, which is a known inhibitor of AMPK, rather than the primary effect of FNIP1 loss per se (26).
Conditional deletion of Flcn in myocardium also results in cardiomyopathy in adult mice (35). In this case, however, the phenotype is that of a lethal dilated cardiomyopathy characterized by ventricular dilatation, profound LV contractile dysfunction, pulmonary and hepatic congestion culminating in premature mortality by 3 mo, and reduced AMPK activity (measured by Thr172 phosphorylation). Deletion of the transcriptional coactivator Ppargc1a restored Thr172 phosphorylation in FLCN-deficient hearts, suggesting that increased Ppargc1a-dependent mitochondrial metabolism may lead to inactivation of AMPK and subsequent activation of mTORC1.
Our conclusion that the FLCN/FNIP complex has a primarily inhibitory function is also supported by earlier reports that FLCN repressed AMPK in Caenorhabditis elegans (36) and repressed AMPK and Ppargc1a induction in primary mouse embryonic fibroblasts (37). Loss of FLCN in these models resulted in constitutive activation of AMPK, which induced autophagy, inhibited apoptosis, and improved cellular energetics (36, 37). These models are however at odds with the suggestion that autophagy was reduced in BHD-associated tumor tissue (38).
The role of the FLCN/FNIP complex in negatively regulating AMPK was also apparent in Fnip1 mutant B cells, which showed increased phosphorylation of ULK1, a direct target of AMPK, and staining of LC3, a proxy for autophagy. We cannot yet explain how Fnip1 deficiency causes a block in B-cell development: overexpression of BCL2 can provide partial rescue, and although autophagy is increased, its inhibition—via temporal deletion of the core autophagy protein Atg3—fails to rescue the phenotype. Therefore, it remains to be seen whether another AMPK-regulated pathway is responsible or indeed if an AMPK-independent pathway is involved. These uncertainties and the important effects of FLCN and FNIP1 in cancer and heart disease highlight the need to focus future investigations on the role of AMPK and mTORC1 in early and late-stage development.
Materials and Methods
Mice.
The Fnip1hamel allele was generated on a pure C57BL/6J background by ENU mutagenesis as previously described (39) and maintained by heterozygote matings. Tg(BCL2)25Wehi (EμBCL2) (40), C57BL/6.SJL-PtprcaPepcb/BoyJ (CD45.1), and B6;129-Gt(ROSA)26Sortm1(cre/ERT)Nat/J (ERT2cre) strains were obtained from The Jackson Laboratory. Atg3tm1.1Ywh and Tp53tm1Tyj mice were obtained from X. Lu, Ludwig Institute for Cancer Research, Oxford, and MD4 BCR transgenic mice (BCR Tg) were provided by C. Goodnow, Garvan Institute of Medical Research, Sydney. C3H/HeN females were obtained from Taconic, C57BL/6J males used for mutagenesis were obtained from The Jackson Laboratory, and all other C57BL/6J mice were obtained from the Wellcome Trust Centre for Human Genetics breeding colony. For tamoxifen-induced deletion of Atg3, mice were given five consecutive daily i.p. injections of 75 mg/kg tamoxifen (T5648; Sigma) dissolved at 20 mg/mL in sunflower seed oil (88921-250ML-F; Sigma). All animal procedures were in accordance with guidelines of the UK Home Office or the institutional animal care and use committee of The Scripps Research Institute.
Sequencing.
Genomic DNA from three C57BL/6J outcrossed hamel mutants was subjected to whole-genome sequencing as described (41). Briefly, 100-bp paired-end libraries were prepared and sequenced on a single lane of an Illumina HiSEq. 2000. Reads were aligned to the mouse reference genome (GRCm38) using Stampy with BWA settings (42), and duplicate reads were identified and discarded using the MarkDuplicates tool (broadinstitute.github.io/picard). Variants were called by Platypus version 0.1.9 (43) and annotated using ANNOVAR with RefSeq and dbSNP 138 annotations (44). Total RNA was prepared from bone marrow using a Qiagen RNeasy mini kit, and cDNA was synthesized using the Invitrogen SuperScript III kit. Fnip1 splice products were amplified and sequenced using the following primers: exons 1–4 (341 bp), Fnip1_F1 (CCAGAAGCTCTTCAGCAAGC) and Fnip1_R1 (CTCCAAGCATATTGGCATCA); exons 3–7 (377 bp), Fnip1_F2 (GCTGAAGCCAGGAGGAGATA) and Fnip1_R2 (GGCTGCAGAACTGTGAAAGA).
Positional Cloning.
Male siblings of the index hamel mutant (C57BL/6J) were outcrossed to C57BL/10J females, and F1 daughters were backcrossed to their father. F2 mice were grouped into wild-type and B cell-deficient cohorts, and individual mice were typed at 128 microsatellite markers spaced across the genome (45). Expected and observed genotype frequencies used to estimate allele frequencies and LOD scores at each position.
Western Blotting.
Approximately 106 nucleated bone marrow cells were resuspended in 100 μL of PBS and incubated with 25 μL of 5× loading buffer (bromophenol blue 0.25%, DTT 0.5 M), glycerol 50% (vol/vol), SDS 10% (wt/vol), and Tris⋅Cl 0.25 M (pH 6.8) at 95°C for 5 min; 15 μL of lysate (∼20 μg of protein) was separated by electrophoresis on a precast 4–20% gel (Bio-Rad) and then transferred to a nitrocellulose membrane. After blocking with 5% milk in TBST solution (TBS, 0.1% Tween 20), the membrane was incubated with rabbit IgG polyclonal anti-FNIP1 antibody (1:1,000; ab61395; Abcam) raised against an N-terminal peptide (MAPTLFQKLFSKRTGLGAC). After incubation with HRP-conjugated anti-rabbit IgG (1:5,000), the membrane was visualized with ECL. HRP-conjugated anti–β-actin (1:50,000; A3854; Sigma) was used to reprobe the same membrane as a loading control.
Flow Cytometry.
Blood from the retroorbital plexus of isoflurane-anesthetized mice was collected in cluster tubes (Costar) containing 20 μL of 6% (wt/vol) EDTA in water; 50 μL of blood was subjected to two rounds of red blood cell lysis with ammonium chloride before staining. Lymphocyte suspensions from bone marrow (femurs and tibias from one hind leg), spleen, and thymus were counted (Z2 Coulter Counter; Beckman Coulter) and stained with a combination of the following mouse-reactive antibodies: FITC-conjugated IgM (goat polyclonal; 1020-02; SouthernBiotech); FITC-conjugated F4/80 (BM8); PE-conjugated IgD (11–26); PerCP-Cy5.5–conjugated TCRβ (H57-597); APC-conjugated CD8α (53-6.7), CD11b (M1/70), CD93 (AA4.1), IgM (II/41), Ly6G (RB6-8C5), NK1.1 (PK136), and TER119 (TER-119; eBioscience); FITC-conjugated CD4 (GK1.5), CD23 (B3B4), and HY-TCR (T3.70); PE-conjugated CD11b (M1/70), CD19 (1D3), CD21/35 (7G6), CD44 (IM7), γδ TCR (GL3), and Vα2 TCR (B20.1); PerCP-Cy5.5–conjugated B220 (RA3-6B2), CD8α (53–6.7), CD19 (1D3), and NK1.1 (PK136); CD44 (IM7; Horizon V500; BD); APC-conjugated CD3ε (145-2C11); and APC-Cy7–conjugated CD45.2 (104), PE-Cy7–conjugated CD19 (6D5), CD45.1 (A20; Pacific blue), and CD16/32 (93; purified; BioLegend). For intracellular antigens, cells were fixed and permeabilized with BD FACS/Perm solutions, and stained with rabbit monoclonal antibodies against the following: phosphorylated S6 ribosomal protein (Ser235/236, clone D57.2.2E, Alexa Fluor 647-conjugated), phosphorylated ULK1 (Ser555, clone D1H4), Beclin-1 (clone D40C5), phosphorylated mTOR (Ser2448, clone D9C2; Cell Signaling Technologies). Unconjugated antibodies were detected with Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (A-11008; Life Technologies). Where indicated, cells were pretreated with 100 nM rapamycin for 2 h. Samples were acquired on a FACSCalibur, FACSCanto, or LSRFortessa (BD), and data were analyzed with FlowJo software (Tree Star).
Immunization and ELISA.
Mice were injected with a single i.p. dose of 25 μg of NP-Ficoll (Biosearch Technologies) in 200 μL of PBS. Seven days later, blood was collected from the tail vein, and centrifuged to isolate serum. Round-bottom, 96-well ELISA plates were coated overnight at 4°C with 100 μL per well of 5 μg/mL NP25-BSA in carbonate buffer. While blocking plates with 5% (wt/vol) milk in PBS, 8 μL of sera was diluted in 1 mL of cold 1% milk. Serum samples were serially diluted in 1% milk. Plates were incubated with alkaline phosphatase-conjugated antibody to mouse IgM (Southern Biotechnology) and developed with p-nitrophenyl phosphate substrate. Absorbance was measured at 405 nm, with background absorbance at 630 nm subtracted.
LC3 Assay.
LC3 staining was performed using the FlowCellect Autophagy LC3 Antibody-based Assay Kit (FCCH100171; Merck Millipore) according to the manufacturer’s instructions. Briefly, 1 × 106 cells were incubated with Reagent A (or 0.1 μM bafilomycin A1) at 37°C and 5% CO2 for 60 min and then washed with PBS and surface stained. Cells were washed and resuspended in 100 μL of Reagent B, centrifuged immediately, and resuspended in 100 μL of Assay buffer with 1:20 diluted FITC-conjugated anti-LC3 antibody. After 30 min at 4°C, samples were washed and resuspended in Assay buffer before acquisition on a flow cytometer.
Transthoracic Echocardiography.
Mice were placed on a heated platform with continual ECG monitoring and anesthetized using inhaled isoflurane (1.25–1.5%) delivered via nosecone; 2D echo images were acquired in the parasternal long-axis and orthogonal short-axis planes with a Vevo 2100 Imaging system (VisualSonics) and subsequently analyzed for ventricular dimensions and derived cardiac function blinded to genotype using Vevo software as described (46).
ECG Recordings.
Under light inhaled isoflurane anesthesia (1.25%) lead II ECG recordings were obtained using fine-needle electrodes positioned s.c. After a baseline 5-min equilibration period, ECG signals were filtered and amplified by a BioAmplifier (ADInstruments), ECG waveform data were then analyzed offline using LabChart software (version 7; ADInstruments). Both ECG recording and analysis were undertaken blinded to genotype.
Cardiac Histology.
Hearts were harvested, rinsed in PBS, and fixed in 10% formalin for 48 h. Tissues were subsequently embedded in paraffin, cut into 7-µm sections using a Leica RM2155 microtome and mounted onto polysine-coated glass slides (VWR). Sections were stained for glycogen using a periodic acid-Schiff staining kit (Sigma) according to the manufacturer’s instructions, mounted with water-free mounting medium (Neo-Mount; Merck Millipore) and glass coverslips (VWR) and subsequently imaged with a wide-field microscope (Nikon TE2000U).
RNA Extraction and Quantitative RT-PCR.
Hearts were rapidly harvested, snap-frozen in liquid nitrogen, and stored at −80 °C. Cardiac tissues were then powdered by mechanical disruption under liquid nitrogen. Approximately 30mg aliquots of cardiac tissue were homogenized with a blender and lysed in RLT buffer (Qiagen) containing β-mercaptoethanol. Total RNA was isolated using a column purification kit (Qiagen RNeasy) according to the manufacturer’s recommendations. After reverse transcription (Applied Biosystems), relative quantification of gene expression was evaluated in duplicate reactions by real time PCR (StepOnePlu; Applied Biosystems) using the comparative Ct method, inventoried TaqMan gene expression assays (Applied Biosystems) and β actin as an endogenous control.
Biochemical Determination of Cardiac Glycogen Content.
An aliquot of 80 mg of pulverized heart was homogenized in 300 μL lysis buffer containing 50 mM Tris, 0.25 M mannitol, 1 mM DTT, and 0.1 mM PMSF adjusted to pH 7.4 and supplemented with protease inhibitor and PhosSTOP tablets (Roche). After brief vortexing, lysates were sonicated twice for 30 s, then passed six times through a 21 G needle. To solubilize glycogen, 50 µL tissue homogenate was mixed with an equal volume of 2 M potassium hydroxide and incubated at 70 °C for 20 min, followed by centrifugation; 40 µL of this mixture was mixed with 180 µL of acetate buffer (0.3 M sodium acetate, pH 4.8) containing 0.5% amyloglucosidase and incubated at room temperature overnight. Blanks were set up for each sample without the enzyme. The next day, samples were spun and 20 µL of the supernatants were mixed with 200 µL of the assay mixture, containing 0.3 M triethanolamine (pH 7.5), 3 mM ATP, 3 mM MgSO4, and hexokinase/glucose-6-phosphate dehydrogenase (Roche). NADPH production was read (A340) after 30 min of incubation.
Invasive Left Ventricular Hemodynamics.
In vivo LV hemodynamics were performed in closed-chest, freely breathing mice under isoflurane general anesthesia (1.25–1.5%) as previously described (46). A 1.4F Millar Mikro-tip catheter (SPR-671; ADInstruments) was inserted into the left ventricle via the carotid artery and the jugular vein cannulated for inotropic drug administration. Hemodynamic indices were evaluated during a 15 min baseline recording and following infusion of dobutamine (4–16 ng g−1 body weight min−1). Pressure-tracing waveforms were subsequently analyzed off-line, blinded to genotype using LabChart Pro software (version 7; ADInstruments).
Neonatal Cardiac AMPK Activity Assay.
AMPK activity was measured by SAMS peptide (HMRSAMSGLHLVKRR) phosphorylation assay from AMPK complexes immunoprecipitated from rapidly harvested neonatal heart lysates using in-house γ2-specific (directed toward the C terminus) or γ1-specific AMPK antibodies in the presence or absence of 0.2 mM AMP, as described previously (47).
Primary Mouse Hepatocyte Isolation and AMPK Activity in Cell Lysates.
Primary hepatocytes were isolated and cultured overnight from adult mice as described in the presence or absence of metformin (2 mM for 2 h after overnight incubation) (48). To determine total AMPK activity, 50 μg of hepatocyte cell lysate was immunoprecipitated with an in-house pan-AMPK β antibody. 32P incorporation into SAMS peptide substrate was determined per minute per milligram of hepatocyte lysate. AMPK γ2 activity was measured using 250 μg of hepatocyte lysate immunoprecipitated with AMPK γ2-specific antibody. Each assay was performed in duplicate using four hepatocyte preparations from each genotype.
Acknowledgments
We thank Xin Lu and Chris Goodnow for mouse strains, Katharina Brandl for help with mapping, Anne Murray for editorial assistance, and Mercedes Gutierrez and Robert Green for animal care. We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics, funded by Wellcome Trust Grant 090532/Z/09/Z and MRC Hub Grant G0900747 91070, for the generation of sequencing data. This work was supported by the Bill & Melinda Gates Foundation and the National Institute of Allergy and Infectious Diseases, National Institutes of Health Grant HHSN272200700038C (to B.B.), Wellcome Trust Grant 100083/Z/12/Z (to O.M.S.), the Medical Research Council (R.J.C.), the UK National Institute for Health Research (A.Y.), and the BHF Centre of Research Excellence, Oxford (H.W. and H.A.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Vocke CD, et al. High frequency of somatic frameshift BHD gene mutations in Birt-Hogg-Dubé-associated renal tumors. J Natl Cancer Inst. 2005;97(12):931–935. doi: 10.1093/jnci/dji154. [DOI] [PubMed] [Google Scholar]
- 2.Birt AR, Hogg GR, Dubé WJ. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch Dermatol. 1977;113(12):1674–1677. [PubMed] [Google Scholar]
- 3.Baba M, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci USA. 2006;103(42):15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasumi H, et al. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene. 2008;415(1-2):60–67. doi: 10.1016/j.gene.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takagi Y, et al. Interaction of folliculin (Birt-Hogg-Dubé gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 2008;27(40):5339–5347. doi: 10.1038/onc.2008.261. [DOI] [PubMed] [Google Scholar]
- 6.Hasumi H, et al. Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc Natl Acad Sci USA. 2015;112(13):E1624–E1631. doi: 10.1073/pnas.1419502112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baba M, et al. The folliculin-FNIP1 pathway deleted in human Birt-Hogg-Dubé syndrome is required for murine B-cell development. Blood. 2012;120(6):1254–1261. doi: 10.1182/blood-2012-02-410407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park H, et al. Disruption of Fnip1 reveals a metabolic checkpoint controlling B lymphocyte development. Immunity. 2012;36(5):769–781. doi: 10.1016/j.immuni.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes NL, et al. Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy. Proc Natl Acad Sci USA. 2015;112(2):424–429. doi: 10.1073/pnas.1413021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 11.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331(6016):456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18(3):283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 14.Cantó C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baba M, et al. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst. 2008;100(2):140–154. doi: 10.1093/jnci/djm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, et al. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal neoplasia. PLoS One. 2008;3(10):e3581. doi: 10.1371/journal.pone.0003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasumi Y, et al. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci USA. 2009;106(44):18722–18727. doi: 10.1073/pnas.0908853106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnold CN, et al. ENU-induced phenovariance in mice: Inferences from 587 mutations. BMC Res Notes. 2012;5:577. doi: 10.1186/1756-0500-5-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betschinger J, et al. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153(2):335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazorchak AS, et al. Sin1-mTORC2 suppresses rag and il7r gene expression through Akt2 in B cells. Mol Cell. 2010;39(3):433–443. doi: 10.1016/j.molcel.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taegtmeyer H, Sen S, Vela D. Return to the fetal gene program: A suggested metabolic link to gene expression in the heart. Ann N Y Acad Sci. 2010;1188:191–198. doi: 10.1111/j.1749-6632.2009.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arad M, et al. Transgenic mice overexpressing mutant PRKAG2 define the cause of Wolff-Parkinson-White syndrome in glycogen storage cardiomyopathy. Circulation. 2003;107(22):2850–2856. doi: 10.1161/01.CIR.0000075270.13497.2B. [DOI] [PubMed] [Google Scholar]
- 23.Blair E, et al. Mutations in the gamma(2) subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: Evidence for the central role of energy compromise in disease pathogenesis. Hum Mol Genet. 2001;10(11):1215–1220. doi: 10.1093/hmg/10.11.1215. [DOI] [PubMed] [Google Scholar]
- 24.Gollob MH, et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344(24):1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 25.Patel VV, et al. Electrophysiologic characterization and postnatal development of ventricular pre-excitation in a mouse model of cardiac hypertrophy and Wolff-Parkinson-White syndrome. J Am Coll Cardiol. 2003;42(5):942–951. doi: 10.1016/s0735-1097(03)00850-7. [DOI] [PubMed] [Google Scholar]
- 26.McBride A, Ghilagaber S, Nikolaev A, Hardie DG. The glycogen-binding domain on the AMPK beta subunit allows the kinase to act as a glycogen sensor. Cell Metab. 2009;9(1):23–34. doi: 10.1016/j.cmet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folmes KD, et al. Distinct early signaling events resulting from the expression of the PRKAG2 R302Q mutant of AMPK contribute to increased myocardial glycogen. Circ Cardiovasc Genet. 2009;2(5):457–466. doi: 10.1161/CIRCGENETICS.108.834564. [DOI] [PubMed] [Google Scholar]
- 28.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 30.Miller BC, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4(3):309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 31.Benhamron S, Tirosh B. Direct activation of mTOR in B lymphocytes confers impairment in B-cell maturation and loss of marginal zone B cells. Eur J Immunol. 2011;41(8):2390–2396. doi: 10.1002/eji.201041336. [DOI] [PubMed] [Google Scholar]
- 32.Tsun ZY, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52(4):495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arad M, et al. Constitutively active AMP kinase mutations cause glycogen storage disease mimicking hypertrophic cardiomyopathy. J Clin Invest. 2002;109(3):357–362. doi: 10.1172/JCI14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott JW, et al. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113(2):274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasumi Y, et al. Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum Mol Genet. 2014;23(21):5706–5719. doi: 10.1093/hmg/ddu286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Possik E, et al. Folliculin regulates ampk-dependent autophagy and metabolic stress survival. PLoS Genet. 2014;10(4):e1004273. doi: 10.1371/journal.pgen.1004273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan M, et al. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J Clin Invest. 2014;124(6):2640–2650. doi: 10.1172/JCI71749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunlop EA, et al. FLCN, a novel autophagy component, interacts with GABARAP and is regulated by ULK1 phosphorylation. Autophagy. 2014;10(10):1749–1760. doi: 10.4161/auto.29640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgel P, Du X, Hoebe K, Beutler B. ENU mutagenesis in mice. Methods Mol Biol. 2008;415:1–16. doi: 10.1007/978-1-59745-570-1_1. [DOI] [PubMed] [Google Scholar]
- 40.Strasser A, Harris AW, Cory S. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1991;67(5):889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 41.Bull KR, et al. Unlocking the bottleneck in forward genetics using whole-genome sequencing and identity by descent to isolate causative mutations. PLoS Genet. 2013;9(1):e1003219. doi: 10.1371/journal.pgen.1003219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lunter G, Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21(6):936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rimmer A, et al. WGS500 Consortium Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46(8):912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siggs OM, Li X, Xia Y, Beutler B. ZBTB1 is a determinant of lymphoid development. J Exp Med. 2012;209(1):19–27. doi: 10.1084/jem.20112084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashrafian H, et al. Fumarate is cardioprotective via activation of the Nrf2 antioxidant pathway. Cell Metab. 2012;15(3):361–371. doi: 10.1016/j.cmet.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies JK, et al. Characterization of the role of gamma2 R531G mutation in AMP-activated protein kinase in cardiac hypertrophy and Wolff-Parkinson-White syndrome. Am J Physiol Heart Circ Physiol. 2006;290(5):H1942–H1951. doi: 10.1152/ajpheart.01020.2005. [DOI] [PubMed] [Google Scholar]
- 48.Woods A, et al. LKB1 is required for hepatic bile acid transport and canalicular membrane integrity in mice. Biochem J. 2011;434(1):49–60. doi: 10.1042/BJ20101721. [DOI] [PMC free article] [PubMed] [Google Scholar]