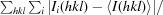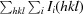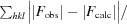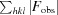Crystals of psCA3 undergo a space-group transition from P21212 to I222 following cryoannealing, resulting in higher resolution diffraction.
Keywords: β-carbonic anhydrase, Pseudomonas aeruginosa, cryoannealing, crystal packing
Abstract
Cryoannealing has been demonstrated to improve the diffraction quality and resolution of crystals of the β-carbonic anhydrase psCA3 concomitant with a change in space group. After initial flash-cooling in a liquid-nitrogen cryostream an X-ray diffraction data set from a psCA3 crystal was indexed in space group P21212 and was scaled to 2.6 Å resolution, but subsequent cryoannealing studies revealed induced protein rearrangements in the crystal contacts, which transformed the space group to I222, with a corresponding improvement of 0.7 Å in resolution. Although the change in diffraction resolution was significant, only minor changes in the psCA3 structure, which retained its catalytic ‘open’ conformation, were observed. These findings demonstrate that cryoannealing can be successfully utilized to induce higher diffraction-quality crystals while maintaining enzymatically relevant conformations and may be useful as an experimental tool for structural studies of other enzymes where the initial diffraction quality is poor.
1. Introduction
The packing and geometry of protein crystals are controlled by interrelated variables such as pH, ionic strength, temperature, salt concentration etc. which modulate the attractive and repulsive forces that are responsible for the interaction of protein molecules in the crystal lattice. Crystal contacts are often misinterpreted as physiological protein–protein interactions existing in multimeric (such as dimers and dimers of dimers) proteins; however, these contacts are sometimes solely the interactions observed between protein molecules in the unit cell or with molecules in the neighboring unit cell (Krissinel & Henrick, 2007 ▸). Additionally, the area of the protein surface patch involved in crystal contacts is normally smaller than those that may be involved in functional contacts. It has been suggested that most of these crystal contacts are formed at random as the crystal grows and the protein is removed from contact with solvent (Pellicane et al., 2008 ▸).
To prevent radiation damage, it is now common practice to cryocool protein crystals with liquid nitrogen (either in a pool of liquid nitrogen or in a stream of cold nitrogen) after the addition of a cryoprotectant and prior to data collection (Cosier & Glazer, 1986 ▸; Hope, 1988 ▸). Cryoannealing is the re-equilibration to room temperature (RT) of a flash-cooled crystal before flash-cooling it again (Yeh & Hol, 1998 ▸). As described in previous work (Hanson et al., 2003 ▸; Juers & Matthews, 2004 ▸; Harp et al., 1998 ▸; Kriminski et al., 2002 ▸), this approach has been shown to be beneficial as a procedure for improving diffraction and reducing crystal mosaicity. The initial cryocooling of a crystal can damage or modify the state of the protein-molecule interactions in the crystal lattice; however, submitting the crystal to one or more heating/cooling cycles can help it to recover from some of the deleterious effects of the initial cooling event (Harp et al., 1998 ▸).
Carbonic anhydrases (CAs; EC 4.2.1.1) are a family (classes α, β, γ, δ and ζ) of mostly zinc metalloenzymes that catalyze the reversible hydration/dehydration of carbon dioxide/bicarbonate (Frost & McKenna, 2013 ▸; Tripp et al., 2001 ▸). The β-class CAs (β-CAs) are found in prokaryotes, plants, algae and some animals, with the active-site zinc coordinated by two cysteines, a histidine and an exchangeable zinc-bound solvent (ZBS; Lotlikar et al., 2013 ▸; Joseph et al., 2010 ▸; Covarrubias, Larsson et al., 2005 ▸). In the ‘open’ active site, an arginine stabilizes an aspartate through a salt-bridge interaction (forming an Asp–Arg dyad), allowing the active-site zinc to be solvent-accessible (Cronk et al., 2001 ▸, 2006 ▸; Covarrubias, Bergfors et al., 2005 ▸; Covarrubias, Larsson et al., 2005 ▸).
Pseudomonas aeruginosa, a Gram-negative facultative aerobe, is an opportunistic human pathogen that causes life-threatening chronic infections and nosocomial infections in cystic fibrosis and endocarditis patients (Stover et al., 2000 ▸; Oliver et al., 2000 ▸; Reyes & Lerner, 1983 ▸). This pathogen expresses three β-class CAs (psCA1–psCA3) that have gained interest as possible drug targets in the development of antibacterial agents (Lotlikar et al., 2013 ▸).
In this paper, we describe the cryoannealing-induced space-group transition of psCA3 crystals from P21212 to I222 and the subsequent improved diffraction resolution and structure determination. The determination of the I222 crystal structure and its biological implications have been described elsewhere (Pinard et al., 2015 ▸).
2. Materials and methods
2.1. Expression and purification of psCA3
The cloning, expression and purification of psCA3 were carried out as described previously (Lotlikar et al., 2013 ▸). The recombinant gene was expressed in Escherichia coli BL21 (DE3) cells with a His tag and protein expression was induced by the addition of IPTG to a final concentration of 1 mM in the presence of 0.5 mM ZnSO4. The cells were then harvested, resuspended and lysed as described previously (Pinard et al., 2013 ▸). The psCA3 lysate was purified using a nickel column. Nonspecific proteins were washed off the column using 20 mM Tris–HCl pH 7.9, 60 mM imidazole, 150 mM NaCl buffer. The psCA3 protein was eluted with the same buffer but with 300 mM imidazole. To confirm the purity of the psCA3, the eluted fractions were run on an SDS–PAGE and stained with Coomassie Blue. Fractions containing psCA3 were pooled and dialyzed against 20 mM Tris–HCl pH 7.9, 150 mM imidazole, 100 mM NaCl, 10% glycerol for 2 h and then against 20 mM Tris–HCl pH 7.9, 50 mM imidazole, 50 mM NaCl, 5% glycerol for 1 h. Finally, psCA3 was dialyzed against 20 mM Tris–HCl pH 8.3 three times. The protein was then concentrated to ∼10 mg ml−1 using an Amicon 10 kDa filter (Millipore, Billerica, Massachusetts, USA).
2.2. Crystallization
Initial crystallization screening of psCA3 was performed in 96-well Intelli-Plate sitting-drop vapor-diffusion crystallization plates using a Gryphon robot (Art Robbins Instruments, Sunnyvale, California, USA). Crystal Screen 2, PEG/Ion and PEG/Ion 2 from Hampton Research, an in-house sodium citrate screen (screening conditions varied from 1.1 to 1.8 M sodium citrate, Tris–HCl pH 7.1–8.1) and an in-house ammonium sulfate screen (screening conditions varied from 1.8 to 2.8 M ammonium sulfate, 0.1 M malic acid, 0.1 M imidazole pH 7.0–8.5) prepared using a Rigaku Alchemist DT liquid-handling system were used.
The psCA3 crystals used were obtained from a precipitant consisting of 2.2 M ammonium sulfate, 0.1 M malic acid, 0.1 M imidazole pH 7.5. 0.5 µl drops were prepared by mixing protein solution at ∼10 mg ml−1 and precipitant solution in a 1:1 ratio and were equilibrated at 290 K against a 60 µl reservoir containing precipitant solution.
2.3. Data collection, indexing and processing
Diffraction data for the P21212 crystal form were collected at the F1 station at Cornell High Energy Synchrotron Source (CHESS F1; λ = 0.9177 Å) on an ADSC Q270 detector using a microfocused beam. Data were collected with a crystal-to-detector distance of 150 mm and a 0.5° oscillation angle with an exposure time of 20 s per image over 110 frames. The data for the I222 crystal form of psCA3 were collected as described previously (Pinard et al., 2015 ▸). Both crystal forms were indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▸).
2.4. Structure solution and refinement
The initial phases of the P21212 form were calculated using the previously solved I222 psCA3 structure described in Pinard et al. (2015 ▸), with water molecules deleted, as a template (PDB entry 4rxy) using the molecular-replacement algorithm in the PHENIX suite of programs (Adams et al., 2010 ▸). To avoid phase bias, both the zinc ion and the active-site Asp–Arg loop were also deleted from the phasing model.
The graphics program Coot (Emsley et al., 2010 ▸) was used to view electron-density maps and the psCA3 structure was manually adjusted. Refinement was continued until the R work and R free converged, and the geometry of the final psCA3 model was verified using MolProbity (Chen et al., 2010 ▸). The data-collection and refinement statistics for the P21212 form are summarized in Table 1 ▸.
Table 1. Crystallographic data-collection and refinement statistics for psCA3 in space groups P21212 and I222.
Values in parentheses are for the highest resolution bin.
| PDB code | 5jj8 | 4rxy † |
|---|---|---|
| Space group | P21212 | I222 |
| Crystal-to-detector distance (mm) | 150 | 150 |
| Wavelength (Å) | 0.917 | 0.917 |
| Temperature (K) | 100 | 100 |
| Unit-cell parameters (Å) | a = 88.4, b = 69.4, c = 77.7 | a = 71.2, b = 77.9, c = 87.7 |
| Resolution (Å) | 2.60 (2.64–2.60) | 1.90 (1.97–1.90) |
| Total No. of reflections | 44987 | 49728 |
| No. of individual reflections | 14242 | 16312 |
| Multiplicity | 3.2 (2.5) | 4.3 |
| Completeness (%) | 92.2 (80.0) | 98.2 (99.8) |
| 〈I/σ(I)〉 | 17.3 (4.5) | 14.3 (2.94) |
| R merge ‡ (%) | 15.3 (38.1) | 8.0 (52.6) |
| R cryst §/R free ¶ | 0.212/0.275 | 0.167/0.199 |
| R.m.s.d., bond lengths (Å) | 0.009 | 0.008 |
| R.m.s.d., bond angles (°) | 1.240 | 1.040 |
| Average B factors (Å2) | ||
| Main chain | 50.4 | 30.9 |
| Side chain | 53.2 | 36.3 |
| No. of protein atoms | 3392 | 1696 |
| No. of water molecules | 22 | 72 |
| Ramachandran statistics (%) | ||
| Favored | 93.5 | 98.1 |
| Allowed | 6.5 | 1.9 |
3. Results and discussion
During data collection at the F1 station at CHESS, various psCA3 crystals were screened for diffraction quality; these crystals were pre-soaked with 20% glycerol for various time intervals (1–30 s) and then flash-cooled to 100 K in a nitrogen cryostream. The quality of diffraction (based on the resolution of diffraction and the visual mosaic spread) varied, but the diffraction quality of several crystals was adequate for data collection. Hence, a data set was collected to 2.6 Å resolution and was indexed (χ2 errors of 0.68 and 13.04% for P21212 and I222, respectively) in space group P21212 (Table 1 ▸). After several more crystals were flash-cooled and initial diffraction images were collected, it was decided to try to cryo-reanneal the crystals. Upon several (more than three) heating/cooling cycles it was visually noted that the diffraction resolution and mosaic spread were substantially improved and a data set was collected. These data were subsequently indexed (χ2 errors of 19.68 and 0.25% for P21212 and I222, respectively) in space group I222 and scaled to 1.9 Å resolution (Table 2 ▸). It was concluded that during the cryoannealing process the crystal had undergone an induced space-group transition. The psCA3 I222 crystal structure was determined and the biological implications have been described elsewhere (Pinard et al., 2015 ▸). Here, we take a ‘with hindsight’ view of the conformational movements that occur within the psCA3 crystal lattice during the P21212 to I222 space-group transition.
Table 2. Indexing-error statistics in space groups P21212 and I222.
| χ2 error (%) | ||
|---|---|---|
| Data set | Primitive orthorhombic lattice (P21212) | I-centered lattice (I222) |
| P21212 | 0.68 | 13.04 |
| I222 | 19.68 | 0.25 |
3.1. I222 and P21212 crystal-packing analysis
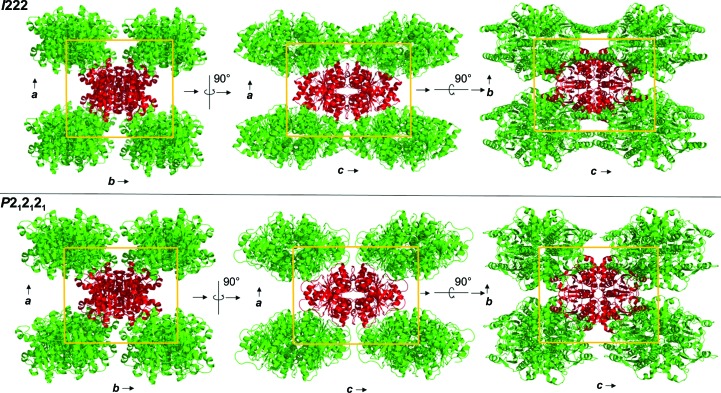
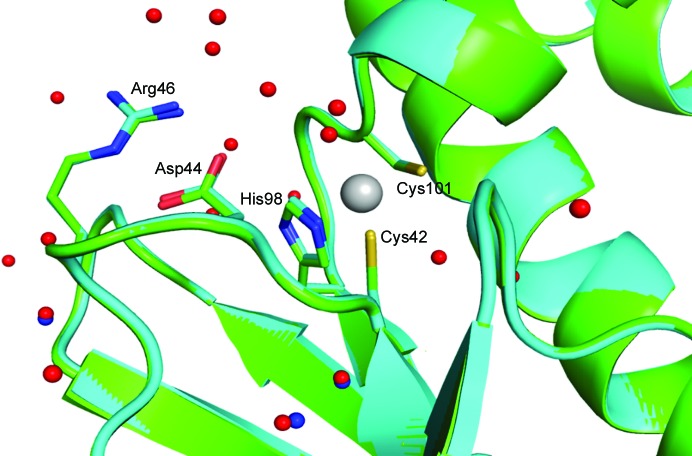
The structure of the I222 crystal form has been described in Pinard et al. (2015 ▸) and only the relevant details are included in this study. The structure contained one psCA3 molecule in its asymmetric unit, with eight molecules in the unit cell, with the biological dimer being present (Table 3 ▸, Fig. 1 ▸). The interactive web server application PDBePISA and visual inspection with Coot were used to analyze the crystal contacts between psCA3 molecules (Krissinel & Henrick, 2007 ▸; Emsley et al., 2010 ▸; Table 3 ▸). The majority of these interactions were polar in nature, with hydrogen bonds being the most dominant interactions. Interestingly, none of the crystal contact regions involved residues 44–48 (containing the Asp44–Arg46 salt-bridge dyad) which are involved in the pH-controlled opening/closing mechanism of the active site (Fig. 2 ▸).
Table 3. Crystal-packing information for psCA3 in the orthorhombic space groups I222 and P21212.
| Space group | I222 | P21212 |
|---|---|---|
| Resolution (Å) | 1.9 | 2.6 |
| Unit-cell parameters (Å) | a = 71.2, b = 77.9, c = 87.7 | a = 88.4, b = 69.4, c = 77.7 |
| V M (Å3 Da−1) | 2.51 | 2.49 |
| No. of molecules in asymmetric unit | 1 | 2 |
| No. of molecules in unit cell | 8 | 8 |
| Symmetry operators (A and B are chain identifiers) | (1) x, y, z | (1A,B) x, y, z |
| (2) x, −y, −z | (2A,B) 1/2 + x, 1/2 − y, −z | |
| (3) −x, y, −z | (3A,B) 1/2 − x, 1/2 + y, −z | |
| (4) −x, −y, z | (4A,B) −x, −y, z | |
| (5) 1/2 + x, 1/2 + y, 1/2 + z | ||
| (6) 1/2 + x, 1/2 − y, 1/2 − z | ||
| (7) 1/2 − x, 1/2 + y, 1/2 − z | ||
| (8) 1/2 − x, 1/2 − y, 1/2 + z | ||
| Unit-cell volume (Å3) | 486000 | 477000 |
Figure 1.
Packing diagram of psCA3 in space groups P21212 and I222, shown along the a–b, a–c and b–c axes. Unit-cell dimensions are denoted by a yellow box.
Figure 2.
Active-site overlay of psCA3 in space groups I222 (cyan) and P21212 (light green). Modeled water molecules for the I222 structure are shown in red and those for the P21212 structure are shown in blue. Note that the Asp44–Arg46 salt-bridge dyad is in the open active-site conformation in both structures. Active-site residues are labeled; the r.m.s.d. between Cα atoms for the two structures is 0.16 Å
The P21212 crystal form diffracted to a resolution of 2.6 Å, with unit-cell parameters a = 88.4, b = 69.4, c = 77.6 Å and an R merge of 15.3%. In this case psCA3 crystallized as a dimer in the asymmetric unit, with eight molecules in the unit cell (Table 3 ▸). The refined structure converged to an R cryst of 21.2% and an R free of 27.5%. The data-collection statistics and processing parameters are summarized in Table 1 ▸. We also note that an attempt was made to reprocess the reflection data for the P21212 form to show that these data preferentially indexed in the primitive orthorhombic rather than the I-centered orthorhombic Bravais lattice (I222 crystal form). This was confirmed via the larger χ2 error value associated with the I-centered lattice after auto-indexing (Table 2 ▸).
Crystal-packing analysis comparison between the two crystal forms revealed no significant differences (Table 4 ▸), with the only measurable difference being a modest 48 Å2 increase in the total interfacial contact area in comparison to the P21212 form (Table 4 ▸). The arrangements of the psCA3 molecules in each space group are shown in Fig. 1 ▸. Despite the protein rigid-body movements in the unit cell there was no significant movement of the 44–48 loop, which is the enzyme-functional on/off switch (Fig. 2 ▸). Hence, a functional open active-site configuration with the conserved Asp–Arg dyad was observed in both crystal forms. Overall, the P21212 psCA3 structure was very similar to that observed in the I222 crystal form (PDB entry 4rxy), with the r.m.s.d. for Cα atoms of chain A being 0.16 Å calculated over 204 (of 209) residues. The majority of the stabilization is most likely to be attributable to solvent-mediated inter-protein packing hydrogen bonds (implied by the number of ordered water molecules), which were not observed in the P21212 form. This protein/solvent rearrangement is presumably induced during the cryoannealing cycling.
Table 4. PDBePISA interface calculations for the I222 and P21212 crystal forms.
| P21212 | I222 | |||||
|---|---|---|---|---|---|---|
| Interface type | N at † | N res ‡ | Interface area (Å2) | N at † | N res ‡ | Interface area (Å2) |
| Dimer | 324 | 77 | 2892.6 | 322 | 77 | 2908.3 |
| Dimer–dimer | 127 | 33 | 1244.7 | 128 | 34 | 1245.1 |
| Packing contact I | 17 | 5 | 188.8 | 14 | 5 | 188.1 |
| Packing contact II | 12 | 5 | 139.4 | 11 | 5 | 140.1 |
| Packing contact III | 12 | 5 | 98.2 | 15 | 4 | 124.9 |
| Packing contact IV | 6 | 4 | 52.3 | 10 | 4 | 68.6 |
| Packing contact V | 3 | 1 | 37.1 | 3 | 1 | 26.2 |
| Total | 501 | 130 | 4653.1 | 503 | 130 | 4701.3 |
Number of interfacing atoms.
Number of interfacing residues.
Supplementary Material
PDB reference: psCA3, 5jj8
Acknowledgments
The authors would like to thank the UF Center for Structural Biology for support of the X-ray facility. We would also like to thank the MacCHESS staff for their help during X-ray diffraction data collection at the Cornell High Energy Synchrotron (CHESS) Facility, Ithaca. CHESS is supported by the NSF and NIH/NIGMS via NSF award DMR-1332208, and the MacCHESS resource is supported by NIGMS award GM-103485. This manuscript has been authored by UT-Battelle LLC under DOE Contract No. DE-AC05-00OR22725 with the US Department of Energy. The publisher, by accepting the article, acknowledges that the US Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for US Government purposes.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst. 19, 105–107.
- Covarrubias, A. S., Bergfors, T., Jones, T. A. & Högbom, M. (2006). J. Biol. Chem. 281, 4993–4999. [DOI] [PubMed]
- Covarrubias, A. S., Larsson, A. M., Högbom, M., Lindberg, J., Bergfors, T., Björkelid, C., Mowbray, S. L., Unge, T. & Jones, T. A. (2005). J. Biol. Chem. 280, 18782–18789. [DOI] [PubMed]
- Cronk, J. D., Endrizzi, J. A., Cronk, M. R., O’Neill, J. W. & Zhang, K. Y. J. (2001). Protein Sci. 10, 911–922. [DOI] [PMC free article] [PubMed]
- Cronk, J. D., Rowlett, R. S., Zhang, K. Y. J., Tu, C., Endrizzi, J. A., Lee, J., Gareiss, P. C. & Preiss, J. R. (2006). Biochemistry, 45, 4351–4361. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Frost, S. & McKenna, R. (2013). Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications. Dordrecht: Springer.
- Hanson, B. L., Schall, C. A. & Bunick, G. J. (2003). J. Struct. Biol. 142, 77–87. [DOI] [PubMed]
- Harp, J. M., Timm, D. E. & Bunick, G. J. (1998). Acta Cryst. D54, 622–628. [DOI] [PubMed]
- Hope, H. (1988). Acta Cryst. B44, 22–26. [DOI] [PubMed]
- Joseph, P., Turtaut, F., Ouahrani-Bettache, S., Montero, J. L., Nishimori, I., Minakuchi, T., Vullo, D., Scozzafava, A., Köhler, S., Winum, J. Y. & Supuran, C. T. (2010). J. Med. Chem. 53, 2277–2285. [DOI] [PubMed]
- Juers, D. H. & Matthews, B. W. (2004). Acta Cryst. D60, 412–421. [DOI] [PubMed]
- Kriminski, S., Caylor, C. L., Nonato, M. C., Finkelstein, K. D. & Thorne, R. E. (2002). Acta Cryst. D58, 459–471. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lotlikar, S. R., Hnatusko, S., Dickenson, N. E., Choudhari, S. P., Picking, W. L. & Patrauchan, M. A. (2013). Microbiology, 159, 1748–1759. [DOI] [PMC free article] [PubMed]
- Oliver, A., Cantón, R., Campo, P., Baquero, F. & Blázquez, J. (2000). Science, 288, 1251–1254. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pellicane, G., Smith, G. & Sarkisov, L. (2008). Phys. Rev. Lett. 101, 248102. [DOI] [PubMed]
- Pinard, M. A., Lotlikar, S. R., Boone, C. D., Vullo, D., Supuran, C. T., Patrauchan, M. A. & McKenna, R. (2015). Bioorg. Med. Chem. 23, 4831–4838. [DOI] [PubMed]
- Pinard, M., Lotlikar, S., Patrauchan, M. & McKenna, R. (2013). Acta Cryst. F69, 891–894. [DOI] [PMC free article] [PubMed]
- Reyes, M. P. & Lerner, A. M. (1983). Rev. Infect. Dis. 5, 314–321. [DOI] [PubMed]
- Stover, C. K. et al. (2000). Nature (London), 406, 959–964.
- Tripp, B. C., Smith, K. & Ferry, J. G. (2001). J. Biol. Chem. 276, 48615–48618. [DOI] [PubMed]
- Yeh, J. I. & Hol, W. G. J. (1998). Acta Cryst. D54, 479–480. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: psCA3, 5jj8