Abstract
The brain-gut-microbiome axis refers to the interactions between the central nervous system, gastrointestinal system, and microorganisms that live in the gastrointestinal tract. Exploring these interactions provides a rationale for why gastrointestinal disorders commonly occur in children with Autism Spectrum Disorders (ASD). Signs of altered brain-gut interactions that are closely associated with functional GI disorders (FGIDs) commonly occur in children with ASD. Studies of microbiome in ASD suggest that changes in the gut microbiome may be associated with ASD and with GI disorders in children with ASD. Further studies into the brain-gut-microbiome axis could lead to new techniques for identifying GI disorders in children with ASD and novel therapies for treating ASD behaviors.
Keywords: brain-gut axis; central nervous system; developmental disorders; Autism Spectrum Disorders, ASD; GI disorders; functional GI disorders; gut microbiome; ASD behaviors; review
Introduction
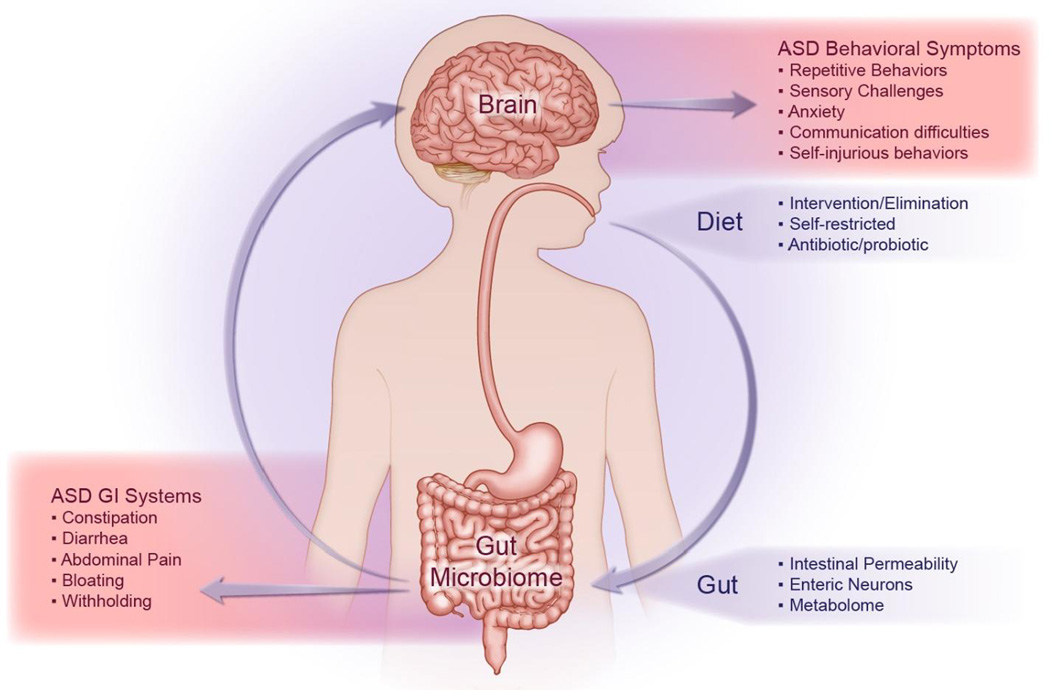
The brain-gut-microbiome axis refers to the highly integrated interactions and communications between the central nervous system (CNS), the gastrointestinal (GI) tract and the trillions of microorganisms that inhabit the gut (Figure 1). Emerging studies of these interactions indicate that alterations of this axis may play a regulatory role in health and wellbeing in various disorders, including Autism Spectrum Disorders (ASD). Based on the latest DSM-5 definition of ASD, an individual with a diagnosis of ASD displays persistent deficits in social communication and interactions as well as restricted and/or repetitive patterns of behavior, interests, or activities.1
Figure 1. The Brain-Gut-Microbiome Axis Plays a Critical Role in Autism Spectrum Disorder.
Autism spectrum disorder (ASD) is commonly characterized by both behavioral symptoms (ex. repetitive and/or self-injurious behaviors) and gastrointestinal (GI) symptoms (ex. abdominal pain). While various factors can influence the gut microbiome, including diet and anti-microbial use, the interconnectivity of the enteric and central nervous systems can affect both behavioral and GI symptom manifestation in pediatric ASD.
Various studies reveal a close association between ASD and GI function. A recent meta-analysis indicates that gastrointestinal (GI) disorders occur at higher rates in children with ASD as compared to children with neurotypical development.2 Other studies indicate that the presence of GI disorders, particularly functional constipation with and without incontinence, can help identify children who are likely to meet criteria for ASD.3 Such findings suggest that ASD and GI disorders are so closely associated; that children with ASD should be screened for GI disorders and that children with particular GI disorders should be screened for ASD.
Evaluating and treating GI disorders in any population of children is challenging, but in children with ASD it is particularly difficult. Various behavioral and developmental deficits associated with ASD impair not only the ability of providers to treat GI disorders in children with ASD, but even identify when GI dysfunction is occurring in these children. Many ASD children lack the verbal skills necessary to communicate the symptoms and signs of abdominal discomfort. Often the only indication that a GI disorder is occurring in a child with ASD is a behavioral change or outburst.4 Of course, not every change or outburst of behaviors is due to GI dysfunction or abdominal discomfort. Medical providers often struggle with determining whether behavioral issues in children with ASD are due to an underlying GI disorder.
Emerging evidence suggests that a better understanding of the brain-gut-microbiome axis could not only help identify GI dysfunction in children with ASD, but potentially provide novel therapies for ASD. This article will review the reports of the types of GI disorders that commonly occur in children with ASD, of how these GI disorders may be related to altered brain-gut-microbiome interactions, and of evidence for a distinctive ASD microbiome. Finally, this article will explore how a better understanding of brain-gut-microbiome interactions could lead to novel treatment therapies for GI and/or behavioral disorders that are associated with ASD.
Functional Gastrointestinal Disorders in Autism Spectrum Disorder
Studies into the actual causes of GI disorders in children are limited. One study that prospectively evaluated for the potential causes of the GI symptoms in children with ASD indicates that the majority of GI complaints in children with ASD is not due to physiological or organic causes but is due to functional causes.5 Gorrindo et al recruited 40 children with ASD and 36 children with neurotypical development with GI complaints. Evaluation by pediatric GI specialists found functional GI disorders (FGIDs) as the most common cause for GI complaints in both groups. In the ASD group, 37 of the 40 children were found to have a FGID. Functional constipation was the most common GI disorder in the ASD study group. Consensus statements and practice recommendations by pediatric specialists further support that most GI symptoms in children with ASD are due to functional disorders similar to those seen in children with neurotypical development, and not due to a physiological condition unique to ASD.4, 6, 7
A diagnosis of a FGID is made when no organic or physiological conditions is found to be the cause of a GI symptom(s).8, 9 Published reports indicate that the most common GI symptoms reported by parents of children with ASD are abdominal pain and altered stool patterns.5, 6, 10, 11 FGIDs that can cause abdominal pain and altered stool patterns include a range of disorders, such as functional abdominal pain, functional constipation, irritable bowel syndrome, cyclic vomiting syndrome and functional dyspepsia.8, 9
FGIDs that are based upon the presence of abnormal stool patterns and abdominal pain are closely associated with signs of impaired brain-gut interactions, such as abnormal behaviors, sensory responses, and sleep patterns.11–14 Signs of impaired brain-gut function not only commonly occur in children with ASD, but are also associated with an increased incidence of GI symptoms in ASD population. Mazurek et al found increased anxiety behaviors and sensory responsivity in children with ASD who present with abdominal pain, constipation, or diarrhea when compared with ASD cases without GI symptoms.11 Peters et al found that children with ASD who have increased rigid compulsive behaviors are more likely to have issues with constipation and toileting.15 Maenner et al found in a study of 487 children with ASD that GI symptoms were associated with oppositional behaviors and abnormal sleep patterns.16 Interestingly, Maenner et al also found that oppositional behaviors and sleep disorders were so prevalent in children with ASD that their presence has limited utility in screening children with ASD for GI disorders.
A strong association between signs of impaired brain-gut function and GI dysfunction provides a rational for why GI disorders occur more frequently in ASD than neurotypical children. Further studies are needed to confirm how impaired brain-gut interactions could lead to GI disorders in children with ASD.
Evidence for a distinctive ASD Microbiome
Microbiome studies have generated great interest in evaluating and identifying altered brain-gut interactions in various patient populations, including children with ASD. In fact, the emerging interest in defining a role for gut bacteria in regulating CNS disease and vice versa has led to the term brain-gut-microbiome axis (Figure 1.) Differences in the types and composition of various bacterial organisms in children with ASD as compared to neurotypical siblings and/or healthy controls have been reported (Table 1).17–21 These studies have provided evidence that distinctive microbiomes may be associated with various ASD populations.
Table 1.
Recent studies related to the gut microbiome in autism spectrum disorder.
| Author, year |
Age, yrs |
Specimen type |
FGID* groups |
Antibiotic exclusion |
ASD1 +GI/−GI (n=) |
SIB2 +GI/−GI (n=) |
NT3 +GI/−GI (n=) |
ASD verification |
Trends in the gut microbiome of children with ASD |
|---|---|---|---|---|---|---|---|---|---|
| Finegold 2010 | 2–13 | Stool | No | 1 month | 33 | 7 | 8 | Study-based evaluation | ↑Bacteroidetes ↓Firmicutes ↑Desulfovibrio, Bacteroides vulgatus |
| Gondalia 2012 | 2–12 | Stool | Yes | 15 days | 28/23 | 4/49 | 0 | CARS | No difference |
| Williams 2011 | 3–5 | GI biopsy | No | None | 15 | 0 | 7 | ADI-R | ↓Bacteroidetes ↑ Firmicutes, Proteobacteria, ↑Sutterella |
| Kang 2013 | 3–16 | Stool | No | 1 month | 0/20 | 0 | 7/13 | ADOS | ↓Prevotella, Coprococcus, Veillonellaceae |
| Wang 2013 | 3–18 | Stool | Yes | None | 9/14 | 6/16 | 1/8 | CARS or DSM-IV | ↓Akkermansia muciniphila, Bifidobacterium, ↑Sutterella, Ruminococcus |
| deAngelis2013 | 4–10 | Stool | No | 1 month | 10 | 10 | 10 | ADOS | ↑ Sarcina, Clostridium, Barnesiella intestihominis |
| Son 2015 | 7–14 | Stool | Yes | 1 month | 25/34 | 13/31 | 0 | ADOS | No difference |
FGID- Functional GI disorder (denotes studies that stratified based on FGID diagnosis),
ASD- Autism spectrum disorder,
SIB- Unaffected sibling,
Unrelated neurotypical
Two studies have detected differences in bacterial composition between ASD populations and unrelated healthy controls.19, 21 Kang et al report that the microbiome in ASD subjects (n=20) was less diverse and possessed lower levels of Prevotella, Coprococcus, and unclassified Veillonellaceae as compared to the microbiome of neurotypical children (n=20).19 De Angelis et al also discovered gut microbial differences between children with ASD (n=10), Pervasive Developmental Disorder –Not Otherwise Specified (n=10), and neurotypical children (n=10).21 The microbiome from the children with an ASD diagnosis contained increased abundances of Sarcina, Clostridium, and Barnesiella intestihominis as compared to the microbiome of children with neurotypical development.21 While the exact species variations were not identical between these two studies, these findings indicate distinctive differences between the gut microbiome of children with ASD and those with neurotypical development. Because the GI function in the study participants was not evaluated, the studies could not determine whether the microbial differences may be more associated with GI dysfunction in the children with ASD than with diagnosis of ASD.
A study by Williams et al suggest that variations in gut microbiome between ASD and neurotypical children are not due GI dysfunction in children with ASD, but due to a distinctive microbiome composition associated with ASD.18 Evaluation of intestinal biopsies taken during colonoscopies as part of the clinical evaluation for various GI disorders in children with ASD (n=15) and neurotypical development (n=7) found microbial differences between the two groups of children. The microbiome of children with ASD demonstrated a decrease in relative abundances of Bacteroidetes and increased relative abundances of Firmicutes and Proteobacteria in ASD, including an increase in Sutterella sp.18 This suggests that differences in microbiome between ASD and neurotypical children are independent of GI dysfunction.
Other studies of microbiome indicates that the microbiome may vary between ASD children with and without FGIDs.20 As in Williams et al study, a study by Wang et al also found an increase in Sutterella sp. in stool samples from children with ASD (n=23) as compared to stool samples from typically developing siblings (n=22) and unrelated neurotypical children (n=9).20 When stool samples were stratified according to presence of FGIDs, the amount of Sutterella sp. did not differ between children with FGIDs and those without GI symptoms. However, children with ASD and FGID (n=9) were found to have an increase in R. torques as compared to children with ASD and no GI symptoms (n=14).20 These findings suggest that unique microbial patterns may exist for children with ASD, and further microbial differences may occur in children with ASD and FGIDs.
Further studies are needed to truly determine whether a distinctive ASD microbiome can be characterized. In 2010, Finegold, et al recruited 33 children with ASD, 7 unaffected siblings, and 8 healthy controls.17 Results suggested increased bacterial diversity in ASD with increased Bacteroidetes and decreased Firmicutes composition, findings that are contradictory to the biopsy-based study by Williams, et al.17, 18 In further contradiction, studies by Gondalia et al and Son et al revealed no significant differences in the gut microbiome of children with ASD as compared to their unaffected siblings.22, 23 It should be noted that neither of these studies included a group of unrelated children with neurotypical development, as was done in those studies that did detect microbial differences associated with ASD children (Table 1).
Larger well designed studies are needed to determine whether microbial composition may differ in children with ASD and may even stratify GI symptoms in this population. If future studies confirm a distinctive ASD microbiome, then knowledge of the specific microbiome differences could lead to the development of novel treatment therapies for behavioral issues and/or GI dysfunction in children with ASD.
Evidence for Therapeutic Microbiology Treatment Strategies
Studies of brain-gut-microbiome interactions provide hope for treating ASD behaviors and co-morbid conditions via manipulation of the gut microbiome. Both animal and clinical studies report changes in neurological function and behaviors by implementing therapies that alter the gut microbiome.20, 21 Use of probiotics in animal models of behavioral disorders provides evidence that changes in the gut microbiome can improve behaviors associated with anxiety and depression.24, 25 Messaoudi et al reported that a probiotic formulation of Lactobacillus helveticus and Bifidobacterium longum reduced anxiety behaviors in an established rat model designed to screen anti-anxiety agents.24 Bravo et al found that ingestion of Lactobacillus rhamnosus by mice led to reduction of anxiety and depression-related behaviors and that the changes in emotional behaviors were associated with alterations of central GABA receptor expression mediated by vagal nerve signaling.25 These studies provide evidence that manipulation of the gut microbiome may potentially be used to help treat abnormal behaviors.
This potential is further supported by reports that highlight a role for the brain-gut-microbiome axis in the pathogenesis of ASD phenotypes in animals and how manipulating the microbiome actually reverses many ASD behaviors.26–28 Using the maternal immune activation model of autism, Hsiao et al were able to document not only the associated GI dysfunction with this mouse model but the apparent correction of GI dysfunction following supplementation with a probiotic, in this case Bacteroides fragilis.27 Interestingly, the changes manifested even without the persistent colonization of B. fragilis. However, the most compelling results from this study were the impact on the behavioral symptoms, where improvement was seen in anxiety, sensorimotor gating, repetitive behaviors, and communication.27
Although it is not yet known whether findings in animal studies can be translated into children with ASD, results from clinical studies in non-ASD populations demonstrate that probiotics can alleviate abdominal pain, alter responses to stressful stimuli, and change brain activity.29–31 Two recent meta-analyses found that probiotics are more effective than placebo in the treatment of children and adolescents with abdominal pain-related FGID, especially with respect to patients with irritable bowel syndrome.29, 30 In addition to treating FGID, probiotics have also been found to help alleviate psychological distress in healthy volunteers.32 Most promising in illustrating how changes in the microbiome directly affects brain function, Tillisch et al showed, with functional magnetic resonance imaging, that four-week intake of a fermented milk product with a probiotic by healthy women affected activity of brain regions that control central processing of emotion and sensation.31
While studies support the feasibility of targeting brain-gut-microbiome interactions as a therapy for ASD, restraint and caution must be employed in developing and utilizing methods for altering the gut microbiome in children with ASD. Significant harm to children and families have occurred when people have attempted to develop new and novel therapies for ASD based upon unproven or incomplete findings.33 Efforts to avoid vaccines and chelate heavy metals that stemmed from misinformation and lack of medical evidence have caused deaths and have contributed to public health crises.34, 35 Currently, there are ways to alter the microbiome, such as the use of dietary interventions, probiotic supplementation, anti-microbials, and fecal microbiota transplant.36 Diet and probiotics are routinely used to treat various disorders, including ASD. Done appropriately under the direction of a medical provider or dietitian, these interventions pose little risk to children. However, use of anti-microbials (antibiotics and antifungal medications) and fecal microbiota transplantation are not without significant risk.37, 38 Adverse reactions, including development of drug resistant bacteria and introduction of life threatening enteric pathogens, in a child should give clinicians and families pause before implementing these treatments in children with ASD.37, 38 Much more information on the brain-gut-microbiome interactions is needed to safely develop specific therapies. Further studies are needed to establish specific ASD associated alterations of the brain-gut-microbiome axis and to evaluate what changes in microbiome function provide beneficial therapeutic outcomes for children with ASD.
Future Directions and Potentials of Brain-Gut-Microbiome Research
To be successful, future studies into the brain-gut-microbiome interactions that are associated with ASD will need to avoid the limitations of prior studies. Limitations of previously published studies include the lack of documented and uniform ASD diagnosis as well as documented rule out of ASD in the sibling and control groups. As GI dysfunction is common in children with ASD, future studies need to screen and characterize GI symptoms in both ASD and neurotypical groups in order to determine whether possible differences in microbiome are not due just solely to differences in gut function. Studies also need consistency in the exclusion criteria related to prior antibiotic, antifungal, and antiviral use, as these medications have a significant and lasting effect on the microbial community throughout the body. Future studies of the microbiome need to also consider the metabolome, i.e. the metabolites that are produced by the microbiome, such as changes in amino acid and antioxidants found and excreted in the blood. Studies analyzing amino acids and antioxidants in urine specimens indicate significant differences in gut bacterial metabolites between ASD children and healthy controls.39–43 To date, only one report has attempted to characterize the fecal metabolome between these two populations, and interestingly this study did find that the levels of free amino acids and volatile organic compounds were markedly affected in children with a diagnosis of Pervasive Developmental Disorder-NOS (n=10) and Autism (n=10) as compared to healthy controls (n=10).21 Differences in metabolites not only further support the efforts to identify a distinctive ASD microbiome; it also supports possible identification of ASD specific biomarkers.
Identifying biomarkers associated with ASD would provide better understanding of which individuals or specific populations may be at risk for disease development. Animal studies demonstrate that initial bacterial colonization of the gut can significantly impact brain development.26, 44 Evaluation of germ-free mice that are devoid of gut bacteria revealed increased baseline neuronal activity in the amygdala with potential implications on neurogenesis in the hippocampus, changes that are not completely reversed when the mice are introduced to the conventional environment and allowed to have bacteria populate their GI tract.44 Recent reports also highlight a role for the brain-gut-microbiome axis in CNS neurotransmission and in the pathogenesis of ASD phenotypes in animals.26, 28, 45 Finding a metabolomics profile associated with ASD could not only help with initial diagnosis of ASD but could also potentially be used to identify children at a young age who are at risk for developing ASD.
In addition to early detection of ASD, biomarkers associated with brain-gut-microbiome interactions could help provide a method to diagnose co-morbid GI conditions in children with ASD. These subjects are often verbally impaired and are unable to express their symptoms, especially abdominal pain. GI dysfunction and discomfort often presents with a sudden change in behaviors or unprovoked behavioral outbursts.4 Finding a biomarker associated with abdominal pain would greatly enhance the ability of medical providers to identify GI dysfunction as a potential cause of a sudden change in behaviors or recurring behavioral outbursts, including self-injurious behaviors, in children with ASD.
Conclusion
Future studies of the brain-gut-microbiome axis could greatly enhance our understanding of ASD, such as explaining why GI symptoms are so common in children with ASD. Also, this line of study could help identify potential targets for therapy for GI disorders and for behavioral issues associated with ASD. Finally, the identification of biomarkers associated with GI dysfunction and with an increased risk for developing ASD could significantly impact the current strategies for evaluation of individuals with a suspected ASD diagnosis.
Footnotes
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Conflict of Interest
Ruth Ann Luna, Tor C. Savidge, and Kent C. Williams declare that they have no conflict of interest
References
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders : DSM-5. 5th. Washington, D.C.: American Psychiatric Association; 2013. American Psychiatric Association. DSM-5 Task Force. [Google Scholar]
- 2.McElhanon BO, McCracken C, Karpen S, Sharp WG. Gastrointestinal Symptoms in Autism Spectrum Disorder: A Meta-analysis. Pediatrics. 2014;133(5):872–883. doi: 10.1542/peds.2013-3995. [DOI] [PubMed] [Google Scholar]
- 3.Peeters B, Noens I, Philips EM, Kuppens S, Benninga MA. Autism Spectrum Disorders in Children with Functional Defecation Disorders. J Pediatr. 2013 doi: 10.1016/j.jpeds.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Buie T, Fuchs GJ, 3rd, Furuta GT, Kooros K, Levy J, Lewis JD, et al. Recommendations for evaluation and treatment of common gastrointestinal problems in children with ASDs. Pediatrics. 2010;125(Suppl 1):S19–S29. doi: 10.1542/peds.2009-1878D. [DOI] [PubMed] [Google Scholar]
- 5.Gorrindo P, Williams KC, Lee EB, Walker LS, McGrew SG, Levitt P. Gastrointestinal dysfunction in autism: parental report, clinical evaluation, and associated factors. Autism Res. 2012;5(2):101–108. doi: 10.1002/aur.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buie T, Campbell DB, Fuchs GJ, 3rd, Furuta GT, Levy J, Vandewater J, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 2010;125(Suppl 1):S1–S18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- 7.Furuta GT, Williams K, Kooros K, Kaul A, Panzer R, Coury DL, et al. Management of constipation in children and adolescents with autism spectrum disorders. Pediatrics. 2012;130(Suppl 2):S98–S105. doi: 10.1542/peds.2012-0900H. [DOI] [PubMed] [Google Scholar]
- 8.Rasquin A, Di Lorenzo C, Forbes D, Guiraldes E, Hyams JS, Staiano A, et al. Childhood functional gastrointestinal disorders: child/adolescent. Gastroenterology. 2006;130(5):1527–1537. doi: 10.1053/j.gastro.2005.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyman PE, Milla PJ, Benninga MA, Davidson GP, Fleisher DF, Taminiau J. Childhood functional gastrointestinal disorders: neonate/toddler. Gastroenterology. 2006;130(5):1519–1526. doi: 10.1053/j.gastro.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 10.Ibrahim SH, Voigt RG, Katusic SK, Weaver AL, Barbaresi WJ. Incidence of gastrointestinal symptoms in children with autism: a population-based study. Pediatrics. 2009;124(2):680–686. doi: 10.1542/peds.2008-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazurek MO, Vasa RA, Kalb LG, Kanne SM, Rosenberg D, Keefer A, et al. Anxiety, sensory over-responsivity, and gastrointestinal problems in children with autism spectrum disorders. J Abnorm Child Psychol. 2013;41(1):165–176. doi: 10.1007/s10802-012-9668-x. [DOI] [PubMed] [Google Scholar]
- 12.Waters AM, Schilpzand E, Bell C, Walker LS, Baber K. Functional gastrointestinal symptoms in children with anxiety disorders. J Abnorm Child Psychol. 2013;41(1):151–163. doi: 10.1007/s10802-012-9657-0. [DOI] [PubMed] [Google Scholar]
- 13.Walker LS, Greene JW. Children with recurrent abdominal pain and their parents: more somatic complaints, anxiety, and depression than other patient families? J Pediatr Psychol. 1989;14(2):231–243. doi: 10.1093/jpepsy/14.2.231. [DOI] [PubMed] [Google Scholar]
- 14.Jarrett M, Heitkemper M, Cain KC, Burr RL, Hertig V. Sleep disturbance influences gastrointestinal symptoms in women with irritable bowel syndrome. Dig Dis Sci. 2000;45(5):952–959. doi: 10.1023/a:1005581226265. [DOI] [PubMed] [Google Scholar]
- 15.Peters B, Williams KC, Gorrindo P, Rosenberg D, Lee EB, Levitt P, et al. Rigid-compulsive behaviors are associated with mixed bowel symptoms in autism spectrum disorder. J Autism Dev Disord. 2014;44(6):1425–1432. doi: 10.1007/s10803-013-2009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maenner MJ, Arneson CL, Levy SE, Kirby RS, Nicholas JS, Durkin MS. Brief report: Association between behavioral features and gastrointestinal problems among children with autism spectrum disorder. J Autism Dev Disord. 2012;42(7):1520–1525. doi: 10.1007/s10803-011-1379-6. [DOI] [PubMed] [Google Scholar]
- 17.Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16(4):444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Williams BL, Hornig M, Buie T, Bauman ML, Cho Paik M, Wick I, et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6(9):e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8(7):e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism. 2013;4(1):42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8(10):e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Son JS, Zheng LJ, Rowehl LM, Tian X, Zhang Y, Zhu W, et al. Comparison of Fecal Microbiota in Children with Autism Spectrum Disorders and Neurotypical Siblings in the Simons Simplex Collection. PLoS One. 2015;10(10):e0137725. doi: 10.1371/journal.pone.0137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gondalia SV, Palombo EA, Knowles SR, Cox SB, Meyer D, Austin DW. Molecular Characterisation of Gastrointestinal Microbiota of Children With Autism (With and Without Gastrointestinal Dysfunction) and Their Neurotypical Siblings. Autism Research. 2012 doi: 10.1002/aur.1253. [DOI] [PubMed] [Google Scholar]
- 24.Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr. 2011;105(5):755–764. doi: 10.1017/S0007114510004319. [DOI] [PubMed] [Google Scholar]
- 25.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108(38):16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Theije CG, Wopereis H, Ramadan M, van Eijndthoven T, Lambert J, Knol J, et al. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun. 2014;37:197–206. doi: 10.1016/j.bbi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 29.Korterink JJ, Ockeloen L, Benninga MA, Tabbers MM, Hilbink M, Deckers-Kocken JM. Probiotics for childhood functional gastrointestinal disorders: a systematic review and meta-analysis. Acta Paediatr. 2014;103(4):365–372. doi: 10.1111/apa.12513. [DOI] [PubMed] [Google Scholar]
- 30.Tiequn B, Guanqun C, Shuo Z. Therapeutic effects of Lactobacillus in treating irritable bowel syndrome: a meta-analysis. Intern Med. 2015;54(3):243–249. doi: 10.2169/internalmedicine.54.2710. [DOI] [PubMed] [Google Scholar]
- 31.Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144(7):1394–1401. 1401, e1391–e1394. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anthes E. Study Finds No Link between Autism and Gut Microbes. Simons Foundation Autism Research Initiative. 2012 [Google Scholar]
- 33.James S, Stevenson SW, Silove N, Williams K. Chelation for autism spectrum disorder (ASD) Cochrane Database Syst Rev. 2015;5:CD010766. doi: 10.1002/14651858.CD010766.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pegorie M, Shankar K, Welfare WS, Wilson RW, Khiroya C, Munslow G, et al. Measles outbreak in Greater Manchester, England, October 2012 to September 2013: epidemiology and control. Euro Surveill. 2014;19(49) doi: 10.2807/1560-7917.es2014.19.49.20982. [DOI] [PubMed] [Google Scholar]
- 35.Brown MJ, Willis T, Omalu B, Leiker R. Deaths resulting from hypocalcemia after administration of edetate disodium: 2003–2005. Pediatrics. 2006;118(2):e534–e536. doi: 10.1542/peds.2006-0858. [DOI] [PubMed] [Google Scholar]
- 36.Frye RE, Slattery J, MacFabe DF, Allen-Vercoe E, Parker W, Rodakis J, et al. Approaches to studying and manipulating the enteric microbiome to improve autism symptoms. Microbial ecology in health and disease. 2015;26:26878. doi: 10.3402/mehd.v26.26878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aroniadis OC, Brandt LJ. Fecal microbiota transplantation: past, present and future. Curr Opin Gastroenterol. 2013;29(1):79–84. doi: 10.1097/MOG.0b013e32835a4b3e. [DOI] [PubMed] [Google Scholar]
- 38.Sandler RH, Finegold SM, Bolte ER, Buchanan CP, Maxwell AP, Vaisanen ML, et al. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J Child Neurol. 2000;15(7):429–435. doi: 10.1177/088307380001500701. [DOI] [PubMed] [Google Scholar]
- 39.Ming X, Stein TP, Barnes V, Rhodes N, Guo L. Metabolic perturbance in autism spectrum disorders: a metabolomics study. J Proteome Res. 11(12):5856–5862. doi: 10.1021/pr300910n. 2012s; [DOI] [PubMed] [Google Scholar]
- 40.Yap IK, Angley M, Veselkov KA, Holmes E, Lindon JC, Nicholson JK. Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J Proteome Res. 2010;9(6):2996–3004. doi: 10.1021/pr901188e. [DOI] [PubMed] [Google Scholar]
- 41.Mavel S, Nadal-Desbarats L, Blasco H, Bonnet-Brilhault F, Barthelemy C, Montigny F, et al. 1H-13C NMR-based urine metabolic profiling in autism spectrum disorders. Talanta. 2013;114:95–102. doi: 10.1016/j.talanta.2013.03.064. [DOI] [PubMed] [Google Scholar]
- 42.Cozzolino R, De Magistris L, Saggese P, Stocchero M, Martignetti A, Di Stasio M, et al. Use of solid-phase microextraction coupled to gas chromatography-mass spectrometry for determination of urinary volatile organic compounds in autistic children compared with healthy controls. Anal Bioanal Chem. 2014;406(19):4649–4662. doi: 10.1007/s00216-014-7855-z. [DOI] [PubMed] [Google Scholar]
- 43.Emond P, Mavel S, Aidoud N, Nadal-Desbarats L, Montigny F, Bonnet-Brilhault F, et al. GC-MS-based urine metabolic profiling of autism spectrum disorders. Anal Bioanal Chem. 2013;405(15):5291–5300. doi: 10.1007/s00216-013-6934-x. [DOI] [PubMed] [Google Scholar]
- 44.Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ, et al. Microbes & neurodevelopment - Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun. 2015 doi: 10.1016/j.bbi.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Alemi F, Kwon E, Poole DP, Lieu T, Lyo V, Cattaruzza F, et al. The TGR5 receptor mediates bile acid-induced itch and analgesia. J Clin Invest. 2013;123(4):1513–1530. doi: 10.1172/JCI64551. [DOI] [PMC free article] [PubMed] [Google Scholar]