Abstract
Exposure to the agricultural fungicide vinclozolin during gestation promotes a higher incidence of various diseases in the subsequent unexposed F3 and F4 generations. This phenomenon is termed epigenetic transgenerational inheritance and has been shown to in part involve alterations in DNA methylation, but the role of other epigenetic mechanisms remains unknown. The current study investigated the alterations in small noncoding RNA (sncRNA) in the sperm from F3 generation control and vinclozolin lineage rats. Over 200 differentially expressed sncRNAs were identified and the tRNA-derived sncRNAs, namely 5′ halves of mature tRNAs (5′ halves), displayed the most dramatic changes. Gene targets of the altered miRNAs and tRNA 5′ halves revealed associations between the altered sncRNAs and differentially DNA methylated regions. Dysregulated sncRNAs appear to correlate with mRNA profiles associated with the previously observed vinclozolin-induced disease phenotypes. Data suggest potential connections between sperm-borne RNAs and the vinclozolin-induced epigenetic transgenerational inheritance phenomenon.
Keywords: transgenerational, noncoding RNA, sperm, vinclozolin, epigenetic inheritance
Introduction
Over a decade ago, the Skinner lab observed that gestating rats (F0 generation) transiently exposed during gonadal sex determination (i.e. embryonic day E8.5–E14.5) to the endocrine disrupter vinclozolin produced offspring (F1 generation) with higher incidences of various diseases, such as increased spermatogenic cell apoptosis and kidney abnormalities, through a mechanism that did not involve genetic inheritance nor relied on mutations of DNA sequences [ 1 , 2 ]. Interestingly, despite the exposure occurring solely in the parent generation (F0), the disease phenotype and frequency persisted in rats of the subsequent generations, including the great and great–great grand offspring (F3 and F4, respectively) [ 1 ]. Although the F1 generation embryo and germ cells of the grand offspring (F2 generation) had been exposed to vinclozolin in utero , the F3 generation rats and their progeny were never directly exposed, but maintained the transgenerational phenotype [ 1 , 3 , 4 ]. This phenomenon is termed “epigenetic transgenerational inheritance,” as the phenotype is transmitted across generations through altered epigenetic information in the germline, without continued direct environmental exposure (transgenerational inheritance) [ 1 , 3 ]. Since this initial discovery, a large number of environmental exposures and other endocrine disruptors [e.g. dioxin, bisphenol A (BPA), pesticides DDT and methoxychlor, hydrocarbons, and tributyltin] have also been shown to promote the epigenetic transgenerational inheritance of disease phenotypes [ 5–9 ]. Although germline DNA methylation has been investigated as a transgenerational epigenetic mechanism, the role of other epigenetic processes such as ncRNA have not been thoroughly investigated [ 4 ].
The vinclozolin-induced transgenerational inheritance of abnormal phenotypes is primarily paternally transmitted [ 1 ]. Therefore, the sperm of vinclozolin lineage rats transmit the altered epigenetic information between generations [ 2 ]. Differential DNA methylated regions (DMRs) have previously been identified in the sperm and male primordial germ cells (PGCs; isolated at E13 and E16) of F3 generation vinclozolin lineage rats [ 10 , 11 ]. A number of different epigenetic mechanisms have been suggested to have a role in epigenetic transgenerational inheritance [ 12 ]. In addition to transmitting the paternal genome to the zygote, sperm are known to contain a diverse population of RNAs, including small noncoding RNAs (sncRNAs), which are also delivered to the developing embryo following fertilization [ 13–17 ]. Previous studies have demonstrated that RNAs derived from the sperm of mice with certain phenotypes (e.g. paramutation induced white tail tip), when injected into a fertilized wildtype oocytes, were sufficient to induce the same phenotype in the next generation [ 18–21 ]. A similar approach was used to show the role of ncRNAs in the transgenerational inheritance of behavioral phenotypes [ 21 ]. These previous studies have suggested a possible role for sperm-borne ncRNAs in epigenetic transgenerational inheritance [ 4 ].
To investigate the relevance of sperm-borne ncRNAs in the context of vinclozolin-induced epigenetic transgenerational inheritance, sncRNA sequencing (sncRNA-Seq) using sperm of both F3 generation control lineage and vinclozolin lineage rats was performed. In this study, we identified >200 significantly differentially expressed sncRNAs in sperm, which belong to several different classes of sncRNAs. Notably, tRNA 5′ halves, a class of tRNA-derived sncRNAs (tsRNAs), were dramatically up-regulated. The gene targets of the altered miRNAs and tRNA 5′ halves were determined in silico and compared to previously observed mRNA transcriptomes associated with vinclozolin-induced disease phenotypes, as well as the genes proximal to DMRs found in F3 vinclozolin lineage male PGCs and sperm. Both the altered miRNAs and tRNA 5′ halves were predicted to target genes relevant to the vinclozolin transgenerational disease phenotypes, as well as a significant number of genes proximal to the DMRs. Observations suggest a correlation between sperm-borne ncRNA and the vinclozolin-induced epigenetic transgenerational inheritance phenomenon.
Results
Transgenerationally Altered sncRNAs in F3 Generation Sperm
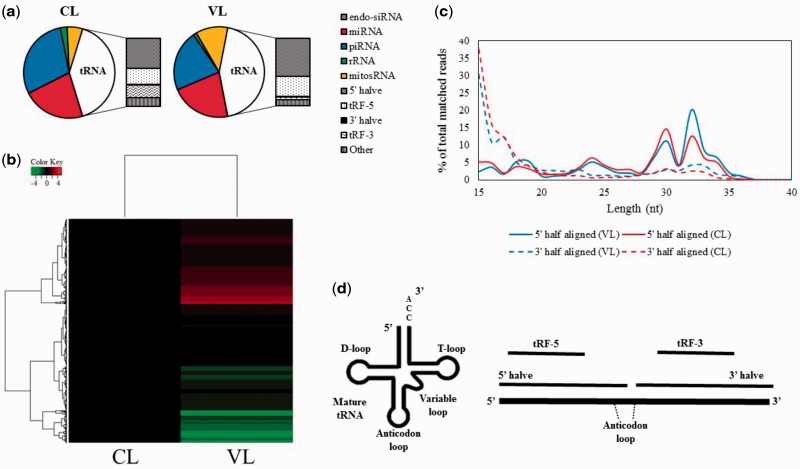
Both the control lineage and vinclozolin lineage F3 generation sperm possessed diverse populations of sncRNAs, consisting mainly of miRNAs, tsRNAs, mitochondrial genome-encoded small RNAs (mitosRNAs), and piRNAs ( Fig. 1 a). Using DESeq2, a differential expression analysis software, we compared control lineage and vinclozolin lineage sncRNA expression levels and identified 222 sncRNAs with significantly ( Padj ≤ 0.1) altered expression ( Fig. 1 b, Table 1 ). The magnitude of the changes (ratio) is presented in Table S1 . Although mostly unchanged, 21 of 251 miRNA observed in sperm (∼8%) displayed either up- or down-regulated expression. Similarly, both piRNAs and rRNA-derived sncRNAs appeared to be predominantly unaltered, with only 16% and 14% either up- or down-regulated, respectively ( Table 1 ). In male germ cells piRNAs can be classified as “pre-pachytene” piRNAs or “pachytene” piRNAs by their length, timing of expression, and 1st and 10th nucleotide preferences. Pre-pachytene piRNAs are typically 26–28 nt in length and prefer uracil and adenine at their 1st and 10th nucleotides, respectively. The pachytene piRNAs, however, expressed after the pre-pachytene piRNAs, are typically 30 nt long and only have a preference for uracil at their first nucleotide [ 22 ]. The majority of the dysregulated piRNAs displayed pachytene piRNA characteristics, indicating that they originate during the later stages of spermatogenesis ( Fig. S1 ). Interestingly, 19 out of 24 (79%) mitosRNAs were up-regulated in the vinclozolin lineage sperm. Unlike most other sncRNAs, mitosRNAs do not have a consensus length, and range from 12 to 137 nt depending on the individual species [ 23 ]. The length distribution of the mitosRNAs was compared to determine whether the processing of full-length mitochondrial RNAs in sperm was altered, but no significant differences were found (data not shown). Although sperm tails are generally removed from the preparation, the possibility that vinclozolin lineage sperm possess more mitochondria than the typical sperm may explain the uniform up-regulation of the mitosRNAs, but remains to be investigated.
Figure 1.
Differentially expressed sncRNAs in vinclozolin lineage F3 sperm. ( a ) The relative amount of each sncRNA class found in the control lineage (CL; left) and vinclozolin lineage (VL; right) F3 sperm. Sequencing reads which aligned to tRNA genes were classified as 5′ halves, tRF-5, 3′ halves, or tRF-3 based on their length and tRNA half preference. Those tRNA-matching reads that did not fall into the aforementioned categories were classified as “Other.” ( b ) Heatmap of the sncRNA expression changes in VL F3 sperm relative to CL F3 sperm. The log 2 fold changes shown were obtained from DESeq2. Genes with Padj values of “NA” were not included in the heatmap. ( c ) tsRNA length distribution. Fragments were separated into the groups 5′ half or 3′ half depending on which side they matched to on their mature tRNA of origin. ( d ) tsRNA types. tsRNA that were approximately half the length of the mature tRNA (left schematic side derived from Ref. [ 25 ]) were classified as 5′ halves or 3′ halves, depending on the side of the anticodon loop to which they matched. Shorter fragments (>27 nt) were classified as tRF-5s or tRF-3s depending on their origins in the mature tRNA
Table 1.
differential expressed sncRNAs in vinclozolin lineage F3 sperm
| RNA type | Up-regulated genes (no. of genes) | Down-regulated genes (no. of genes) | Unchanged genes (no. of genes) |
|---|---|---|---|
| Mature miRNA | 13 | 8 | 230 |
| rRNA | 1 | 8 | 54 |
| piRNA | 16 | 68 | 513 |
| Mitochondrial RNA | 19 | 0 | 5 |
| 5′ halves | 16 | 0 | 14 |
| tRF-5 | 26 | 0 | 25 |
| 3′ halves | 0 | 2 | 2 |
| tRF-3 | 1 | 43 | 37 |
The number of sncRNA species, classified by RNA type, that were significantly ( Padj ≤ 0.1) up- or down-regulated in the VL F3 sperm sncRNA-Seq data, relative to the CL data. SncRNA species with a Padj ≤ 0.1 were termed “unchanged”; the species with “NA” Padj values were not included in this table
Numerous tsRNAs were identified in sperm ( Table 1 ), which is consistent with a previous report showing tsRNAs are abundant relative to other sncRNA classes in sperm [ 24 ]. To investigate this further, each sequencing read that aligned to a tRNA gene during annotation was matched to either the 5′ or 3′ half of the each known tRNA. The majority of reads that aligned to the 5′ half of the tRNA was 30–33 nt long, whereas the reads which aligned to the 3′ half had a much wider length distribution ( Fig. 1 c). The fragments that were 27 nt or longer were termed 5′ or 3′ halves depending on their origin within the mature tRNA, whereas those less than 26 nt in length were termed tRNA fragments from the 5′ end (tRF-5s) or tRNA fragments from 3′ end (tRF-3s) depending on their side preference (i.e. 5′ or 3′) [ 25 ] ( Fig. 1 d). Differential expression analysis via DESeq2 revealed that the 5′ halves and tRF-5s were exclusively up-regulated in F3 generation vinclozolin lineage sperm, whereas the 3′ halves and tRF-3s were predominantly down-regulated ( Table 1 ). Overall, levels of numerous sncRNAs were changed in vinclozolin lineage F3 sperm, with tsRNAs and mitosRNAs being the most altered ones.
miRNA and 5′ Halve tsRNA Gene Target Prediction
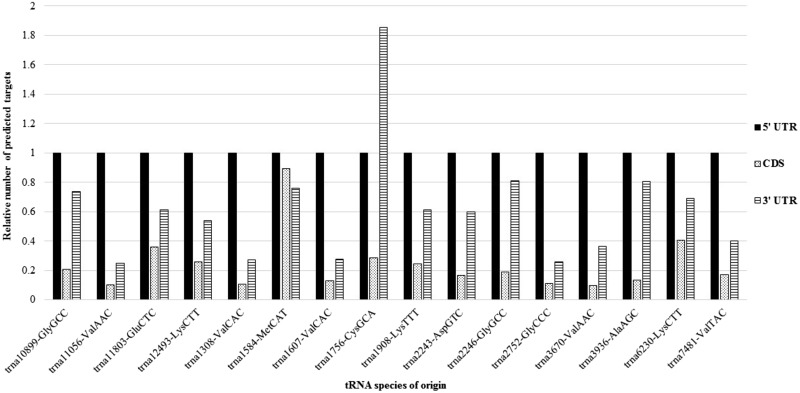
Due to their potential relevance to the epigenetic transgenerational inheritance phenomenon [ 26 ], we mainly focus further investigations on the 16 altered 5′ halves, as well as the 21 altered miRNAs. The miRNAs are known to regulate gene expression by acting as post-transcriptional regulators [ 27 ]. Although less established, various types of tsRNAs, including 5′ halves, have also shown post-transcriptional regulatory abilities [ 28–33 ]. To assess whether the targets of these sncRNAs were relevant to any of the effects of ancestral vinclozolin exposure that had been reported previously [ 1 , 2 ], the putative sncRNA targets were analyzed in silico . The transgenerationally altered sperm miRNAs were matched to the 3′ untranslated region (UTR) sequences available for rat, utilizing both RNAhybrid and miRanda [ 34–37 ]. Without an established tRNA halve-mediated silencing mechanism available for 5′ halves, the search was expanded to 5′ UTRs and coding sequences (CDS) in addition to 3′ UTRs. Only RNAhybrid was used, as it accounts for the length of the target sequence, which ensured that there would not be a bias toward the longer (and therefore more prone to random matches) 3′ UTR and CDS over the 5′ UTR [ 35 ]. The total number of targets for each transgenerationally altered sperm miRNA and 5′ halves is provided in Table S2 . The down-regulated miRNAs had significantly more predicted targets than the up-regulated miRNA, suggesting that they may have more functional significance. The 5′ UTR sequences were targeted by the 5′ halves much more frequently on average than the 3′ UTRs and CDS, with the CDS typically having the fewest putative targets ( Table S2 ; Fig. 2 ). Around two-thirds of the predicted gene targets identified in the 5′ halve–5′ UTR RNAhybrid analysis were targeted by more than one 5′ halve, whereas roughly 50% and 25% of the 5′ halve–3′ UTR and 5′ halve–CDS gene target predictions, respectively, were targeted more than once ( Table S3 ). This suggests that if sperm 5′ halves do possess silencing capabilities, they likely exercise this ability through the 5′ UTRs of their target mRNAs. As little is known about the potential function of 5′ halves, the gene target predictions based on the 3′ UTR and CDS were still considered in our subsequent analyses.
Figure 2.
Relative numbers of predicted 5′ halve gene targets in 5′ UTR, CDS, and 3′ UTR sequences. The number of predicted gene targets for each differentially expressed 5′ halve species within the 5′ UTR, CDS, and 3′ UTR was normalized to the number of targets within the 5′ UTR, for each 5′ halve species. Relative to CDS and 3′ UTR, 5′ UTR sequences had the highest incidence of RNAhybrid predicted 5′ halve binding sites. A P -value ≤0.01 was used as a cutoff
Gene Category and Pathway Analysis
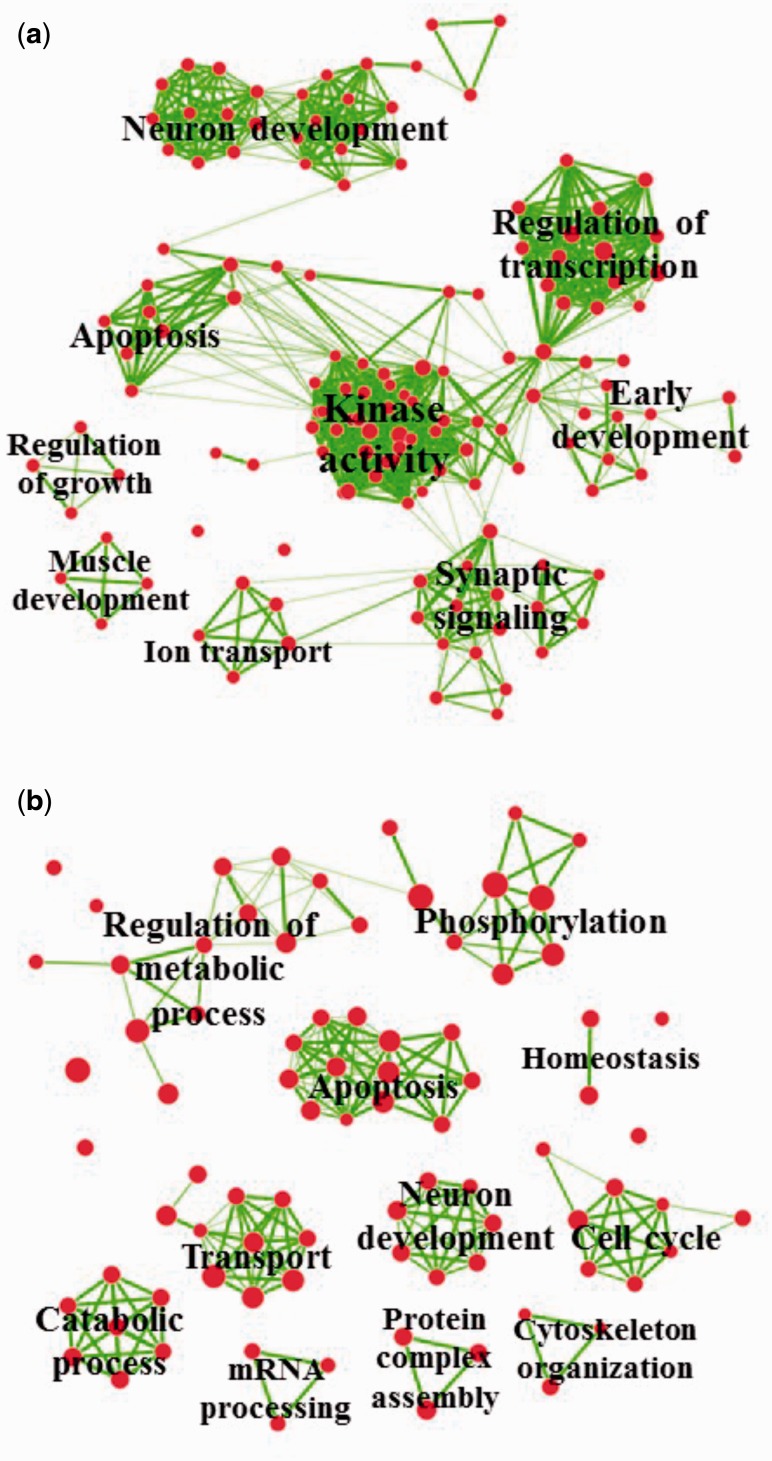
To investigate the putative targets of the transgenerationally altered sperm miRNAs and 5′ halves, a gene ontology (GO) term enrichment analysis was performed on the genes that were targeted by more than one sncRNA. The miRNA target genes that had at least two miRNAs were split into redundant up- and down-regulated miRNA target lists. Searching the Database for Annotation, Visualization and Integrated Discovery (DAVID) for enriched biological process (BP), molecular function (MF), cellular component (CC), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways revealed several significant (Benjamini ≤ 0.05) results for the redundant down-regulated miRNA targets, but none for the up-regulated miRNA ( Table S4 ). An enrichment map of the significant BP genes ( Fig. 3 a) shows that the redundantly targeted genes are relevant to apoptosis, early development, kinase activity, regulation of transcription, and the nervous system (i.e. its development and synaptic signaling). Significant CC genes also revealed that many of the genes localize in various nervous system cells (e.g. “synapse part”) and the only significant KEGG pathway identified was “axon guidance” ( Table S4 ).
Figure 3.
GO term analyses of dysregulated sncRNAs in the vinclozolin lineage F3 sperm. ( a ) Enrichment map of the BP terms associated with redundantly predicted gene targets of down-regulated miRNAs. The genes which were predicted targets of at least two down-regulated miRNAs were analyzed via DAVID for significantly enriched BP terms. The results of the DAVID analysis were used to generate an enrichment map, grouping related BP terms into clusters. The text overlaying each cluster describes the unifying “theme” of that cluster (chosen by the authors). The individual BP terms are described in more detail in Table S4 . ( b ) Enrichment map of the BP terms associated with redundantly predicted gene targets of 5′ halves. The genes which were predicted targets of at least three dysregulated 5′ halves were analyzed via DAVID for significantly enriched BP terms. A Benjamini value ≤0.05 was used to define significance. The results of the DAVID analysis were used to generate an enrichment map, grouping related BP terms into clusters. The text overlaying each cluster describes the unifying “theme” of that cluster (chosen by the authors). The individual BP terms are described in more detail in Table S5
A similar analysis was performed on the predicted gene targets of the transgenerationally altered 5′ halve tsRNAs using genes that were targeted by at least three 5′ halves. The results of the 5′ UTR–, CDS–, and 3′ UTR–5′ halve analyses were analyzed separately. For every GO term category analyzed, the 5′ UTR-derived redundant gene targets had the largest number of significantly enriched terms ( Table S5 ). Apoptosis, neuronal development, regulation of metabolic processes, phosphorylation, cell cycle, and transport were the predominant gene categories of the significantly enriched BP terms ( Fig. 3 b). In terms of significance, endocytosis, neurotrophin signaling pathway, and oocyte meiosis were the top enriched KEGG pathways ( Table S5 ). A portion of the enriched terms for the redundant targets of the down-regulated miRNAs were also related to apoptosis and nervous system development, suggesting that the alterations of miRNAs and 5′ halves may play similar roles in vinclozolin-induced epigenetic transgenerational inheritance.
Correlation with Vinclozolin-Induced Transgenerational Sperm DNA Methylation Alterations
A correlation between the transgenerationally altered sperm miRNAs and 5′ halves observed was made with the F3 generation vinclozolin sperm differential DMRs previously described [ 10 , 38 ]. The majority of transgenerational DMR is not associated with genes and is intergenic [ 38 ]. Of the 21 miRNAs and 16 5′ halves that were found to be differentially expressed, 16 and 14, respectively, originated within 5 Mb of a sperm DMR ( Table 2 A, B). Previously the DMR and regulated genes have been shown to exist within 2–5 Mb regions termed an epigenetic control region (ECR) [ 39 ], so a 5 Mb region was investigated. The genes proximal (i.e. up to 100 kb up and downstream) to the promoter associated DMRs identified previously in F3 generation vinclozolin lineage sperm were also investigated [ 10 , 38 ]. The correlation between the predicted targets of the altered miRNAs and 5′ halves (5′ UTR and 3′ UTR matching results only) and genes proximal to the DMRs was significant ( P value < 0.1) ( Table 3 ). However, the number of actual matches of predicted gene targets to DMR proximal genes was consistently lower than the number of expected random matches for the dysregulated miRNA, signifying a negative correlation ( Table S6 ). In contrast, the majority of actual matches observed for the putative 5′ halve targets (i.e. 5′ UTR and 3′ UTR only) was above the number of random matches, suggesting that 5′ halves possess the potential to post-transcriptionally regulate genes proximal to DMRs present in the sperm of F3 vinclozolin lineage rats ( Table S6 ).
Table 2.
proximity of differential expressed sncRNAs to DMRs
| DMRs (cell type) | No. of DMRs |
Distance from DMR (Mb)
|
|||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | ||
| (A) 5′ halves | |||||||
| F3 E13 PGC | 23 | 0 | 0 | 0 | 0 | 2 | 2 |
| F3 E16 PGC | 13 | 0 | 0 | 2 | 2 | 2 | 2 |
| F3 Sperm | 188 | 0 | 4 | 5 | 11 | 14 | 14 |
| (B) miRNA | |||||||
| F3 E13 PGC | 23 | 0 | 0 | 2 | 3 | 3 | 3 |
| F3 E16 PGC | 13 | 0 | 0 | 0 | 0 | 0 | 0 |
| F3 Sperm | 188 | 0 | 4 | 9 | 10 | 13 | 16 |
The genomic coordinates of the dysregulated (A) 5′ halves and (B) miRNA were compared to the locations of DMRs previously identified in F3 sperm, E13 PGCs, and E16 PGCs. The genes of sncRNA that were within 1–5 Mb of at least one of the DMR in its respective cell type were counted above. The sncRNA whose genes were contained within a DMR (0 Mb) were also counted
Table 3.
correlation between genes proximal to F3 sperm DMRs and predicted gene targets
| Distance from DMR (kb) | No. of proximal genes | miRNA–3′ UTR | 5′ halves–5′ UTR | 5′ halves–CDS | 5′ halves–3′ UTR |
|---|---|---|---|---|---|
| 0 | 65 | 14 a | 28 | 15 | 28 |
| 1 | 74 | 15 a | 34 | 19 | 29 |
| 2 | 83 | 16 a | 36 | 21 | 32 |
| 5 | 109 | 17 a | 40 | 30 | 37 |
| 10 | 129 | 20 a | 48 | 38 | 43 |
| 20 | 181 | 26 b | 66 | 52 | 56 |
| 50 | 313 | 40 b | 101 | 85 | 90 |
| 100 | 532 | 66 c | 150 | 141 | 139 |
| P -value | 0.011 | 0.064 | 0.66 | 0.061 | |
The genes up and downstream of the previously identified F3 sperm DMRs were compared to the predicted gene targets of the dysregulated miRNA and 5′ halves. P -values were determined by a Chi-square test.
a Two of the matching genes were predicted to be targeted by both up and down-regulated miRNA.
b Three of the matching genes were predicted to be targeted by both up and down-regulated miRNA.
c Four of the matching genes were predicted to be targeted by both up and down-regulated miRNA.
An additional experiment examined the correlations with the transgenerational DMRs in the PGCs, which are the stem cells for the differentiated sperm cell. The previously identified DMRs in PGCs isolated from F3 generation vinclozolin lineage fetal testis at embryonic days E13 and E16 of development were analyzed [ 11 ]. Although the transgenerational DMRs in the E13 and E16 PGCs would not be directly relevant to the sperm DMRs, they are associated with the precursor cells of the sperm so correlations may be present. Initially, we investigated whether any of the transgenerationally altered miRNAs or 5′ halves originated near the E13 or E16 DMRs, but found very few correlations, relative to the sperm DMRs ( Table 2 A, B). Next, we compared the genes proximal to the DMRs and the predicted gene targets of the altered miRNA and 5′ halves. The genes proximal to the E13 PGC DMRs significantly correlated ( P value < 0.1) with the predicted 5′ halve targets, for the predictions from both the 5′ UTR and 3′ UTR-based matching ( Tables 4 A and S7A ). When we analyzed the E16 PGC DMRs, we again observed a significant correlation ( P value < 0.1) between the 5′ halves and proximal genes, this time just for the results of the 5′ UTR-based matching ( Tables 4 B and S7B ). There was a less significant correlation ( P value of 0.070 vs. 0.015 for the 5′ halves) between the predicted targets of the down-regulated miRNAs and E16 DMR proximal genes as well ( Tables 4 B and S7B ). Combined observations suggest an association between the transgenerationally altered DNA methylation regions in F3 generation male PGCs and sperm with the sperm-borne sncRNA expression.
Table 4.
correlation between genes proximal to F3 PGC DMRs and predicted gene targets
| Distance from DMR (kb) | No. of proximal genes | miRNA–3′ UTR | 5′ halves–5′ UTR | 5′ halves–CDS | 5′ halves–3′ UTR |
|---|---|---|---|---|---|
| (A) F3 E13 PGC | |||||
| 0 | 15 | 2 | 9 | 6 | 6 |
| 1 | 18 | 2 | 10 | 8 | 6 |
| 2 | 24 | 4 | 12 | 9 | 10 |
| 5 | 31 | 5 | 18 | 11 | 12 |
| 10 | 37 | 5 | 19 | 12 | 12 |
| 20 | 51 | 8 a | 24 | 16 | 19 |
| 50 | 94 | 15 a | 43 | 22 | 37 |
| 100 | 148 | 28 a | 67 | 38 | 62 |
| P -value | 0.97 | 6.79E−05 | 0.53 | 0.019 | |
| (B) F3 E16 PGC | |||||
| 0 | 7 | 2 | 5 | 3 | 2 |
| 1 | 9 | 3 | 5 | 3 | 4 |
| 2 | 9 | 3 | 5 | 3 | 4 |
| 5 | 16 | 6 | 8 | 4 | 6 |
| 10 | 22 | 9 | 12 | 5 | 8 |
| 20 | 32 | 9 | 16 | 8 | 8 |
| 50 | 57 | 11 | 23 | 15 | 15 |
| 100 | 93 | 19 | 38 | 23 | 26 |
| P -value | 0.070 | 0.015 | 0.90 | 0.97 | |
The genes up and downstream of the previously identified F3 E13 (A) and E16 (B) PGC DMRs were compared to the predicted gene targets of the dysregulated miRNA and 5′ halves. P -values were determined by a Chi-square test.
a One of the matching genes was predicted to be targeted by both up and down-regulated miRNA.
Discussion
Our data demonstrate that many sncRNAs were transgenerationally altered in the sperm of F3 generation vinclozolin lineage rats, tsRNAs in particular. The potential gene targets of both the altered miRNAs and 5′ halves were identified in order to determine whether the expression changes of these sncRNAs had any functional significance to the previously observed pathology of ancestral vinclozolin exposure.
Perhaps, the most striking vinclozolin-induced transgenerational effect on the sperm sncRNA transcriptome was the alteration of the majority of observed tsRNAs. Approximately 82% of the tsRNAs in the vinclozolin lineage F3 generation sperm were altered and ∼93% of the reads were attributed to 5′ halves. Comparing this to the other altered tsRNAs, which accounted for 81% (3′ halves), 65% (tRFs-5), and 49% (tRFs-3) of their respective species, we see that 5′ halves were dramatically altered in the transgenerational vinclozolin lineage sperm. This is especially noteworthy, given that 5′ halves are known to be highly enriched in mature sperm [ 24 ]. In the vinclozolin lineage sncRNA-Seq results, 5′ halves were the most abundant sncRNAs and accounted for ∼25% of all the annotated sncRNAs.
The gene categories that were significantly enriched in the lists of redundantly targeted genes for the down-regulated miRNA and the 5′ halve–5′ UTR targets were correlated to several of the transgenerational disease phenotypes. In the enrichment maps of both the altered miRNAs and 5′ halves, apoptosis was one of the largest clusters of enriched terms ( Fig. 3 a and b). An increase in spermatogenic cell apoptosis was observed previously in 90% of vinclozolin males, regardless of generation [ 1 , 2 ]. Gene categories and KEGG pathways related to the development and regulation of neurons were highly enriched within the lists of redundantly targeted genes for both the miRNAs and 5′ halves ( Fig. 3 a and b; Tables S4 and S5 ). Several of the phenotypes associated with vinclozolin-induced epigenetic transgenerational inheritance of disease involve the nervous system and brain development, despite no discernable morphological changes to the brain [ 2 , 12 , 40–42 ]. In a 2008 study by the Skinner lab, many neural-pathways were found to contain differentially expressed genes in the amygdala and hippocampus of F3 vinclozolin lineage male and female rats, including axon guidance, which was the only significant KEGG pathway identified for the down-regulated miRNA ( Table S4 ) [ 12 , 40 ]. In the same study, F3 generation vinclozolin male and female rats exhibited an increase and decrease in anxiety-like behaviors, respectively [ 40 ]. Other changes in behavior and the brain transcriptome of F3 vinclozolin rats have been observed in subsequent studies [ 12 , 41 , 42 ]. It is possible that aberrant gene targeting by the altered sperm miRNA and 5′ halves during early development could cause alterations in the developing brain and nervous system, ultimately manifesting as the transgenerational disease phenotypes described above.
Interestingly, an increase in 5′ halve expression has previously been linked to altered neuron survival and anxiety-related behaviors. Mice deficient in NSun2 ( NSun2−/− ), an RNA methyltransferase known to modify tRNAs, possessed elevated levels of 5′ halves in their skin and brain. This correlated with embryonic brain defects (of the cerebral cortex, hippocampus, and striatum) and, as was also seen in F3 generation vinclozolin lineage male rats, a reduction in anxiety-related behaviors [ 40 , 43 ]. NSun2−/− male mice are infertile as well, due to the arrest of spermatogenesis prior to the meiotic progression into the pachytene stage [ 44 ]. Therefore, it should come as no surprise that up-regulation of 5′ halves would correlate to the similar, albeit less severe, vinclozolin lineage transgenerational disease phenotype.
The ability of 5′ halves tsRNA to act as translational suppressors has been demonstrated in other organisms [ 29–32 ]. In one study, certain 5′ halves inhibited translation by displacing eIF4G/eIF4A from the 5′ end of mRNAs [ 29 ]. Based on the results of the RNAhybrid analysis, 5′ halves appear to preferentially bind to the 5′ UTRs of mRNAs, relative to the CDS and 3′ UTRs. The ability of 5′ halves to bind complementary sequences within mRNAs, and subsequently silence their expression has also been demonstrated [ 30 , 32 ]. Therefore, in some cases, the altered expression of 5′ halves could potentially regulate gene expression by binding to the 5′ UTRs and displacing cap-bound complexes and hindering translation of the target mRNA.
A correlation between the previously identified differential DMRs in the vinclozolin-induced transgenerational sperm with the altered sncRNA identified in this study demonstrated that ∼88% and ∼76% of the differentially expressed 5′ halves and miRNAs, respectively, originate within 5 Mb of a sperm DMR ( Table 2 A, B). Our correlation analysis also revealed that of the genes predicted to be targeted by the altered 5′ halves via their 5′ or 3′ UTRs, a significant number were within at least 100 kb of a sperm DMR ( Table 3 ). Previously, ECRs were identified in somatic tissues of vinclozolin lineage rats, spanning 2–5 Mb [ 39 ]. Based on these findings, it seems plausible that the altered 5′ halves are components of various ECRs, and that their differential expression is either due to the presence of the DMRs themselves, in response to other genes proximal to the DMRs, or a combination of both. In addition to sperm, a correlation between the differentially expressed sncRNAs and DMRs previously found in the E13 and E16 PGCs of vinclozolin lineage rats was made [ 11 ]. Although relatively fewer 5′ halves ( Table 2 A) and miRNAs ( Table 2 B) were expressed within 5 Mb of a PGC DMR, we did observe correlations between the gene targets of the sncRNAs and genes proximal to the DMRs. There were a significant number of putative 5′ halve gene targets that were within 100 kb of an E13 ( Table 4 A) or E16 ( Table 4 B) PGC DMR. A significant number of predicted gene targets of the down-regulated miRNA also matched genes proximal to the E16 PGC DMRs ( Table 4 B). It is possible that the altered expression of these miRNAs and 5′ halves during spermatogenesis is also influenced by dysregulation of the genes proximal to DMRs in PGCs. However, based on our observations, it seems that epigenetic alterations present at later stages of male germ cell development (i.e. sperm) play a potentially greater role in the differential expression of sperm-borne miRNA and 5′ halves. Future studies that assess the methylation and gene expression of vinclozolin lineage male germ cells, at several different stages of development, will be informative.
Observations show that ancestral exposure to vinclozolin causes transgenerational (i.e. F3 generation) alterations in the sncRNA expression levels in mature sperm. The altered miRNAs and 5′ halve tsRNA potential target genes were correlated to several of the previous observed vinclozolin transgenerational disease phenotypes. Interestingly, a dramatic up-regulation of 5′ halves was observed in the vinclozolin lineage sperm ( Table 1 ). In two other examples of epigenetic inheritance, the Kit and Sox9 paramutations, it was determined that the methyltransferase Dnmt2 was necessary for the transmission of the phenotype associated with the paramutation [ 26 ]. Dnmt2, like NSun2, confers stability to mature tRNA via cytosine-5 methylation and its loss affects 5′ halve production [ 43 , 45–47 ]. Because of this, tsRNAs, specifically 5′ halves, have been postulated to be effectors of epigenetic inheritance [ 26 , 48 ]. Our discovery that 5′ halves were abnormally enriched in transgenerational vinclozolin lineage sperm, and potentially correlated to the vinclozolin lineage transgenerational disease phenotype, provides additional evidence to the idea that sperm-borne 5′ halves act as epigenetic regulators. A new study in progress will determine whether similar observations involving an up-regulation of 5′ halves develop in another model of environmentally induced (e.g. DDT-induced) epigenetic transgenerational inheritance of disease. Overall, our findings highlight the potential importance of sperm-borne sncRNAs in the mechanism of vinclozolin-induced epigenetic transgenerational inheritance, as well as support the proposed role of 5′ halves as active epigenetic regulators during early development.
Methods
Animal Studies and Breeding
Female and male rats of an outbred strain Hsd:Sprague Dawley (Harlan) at 70–100 days of age were fed ad lib with a standard rat diet and ad lib tap water for drinking. To obtain time-pregnant females, the female rats in proestrus were pairmated with male rats. The sperm-positive (Day 0) rats were monitored for diestrus and body weight. On Days 8 through 14 of gestation [ 36 ], the six different females per group were administered daily intraperitoneal injections of vinclozolin (100 mg/kg BW/day) or dimethyl sulfoxide (vehicle). The vinclozolin was obtained from Chem Service Inc. (West Chester PA, USA) and was injected in a 200 µl DMSO/sesame oil vehicle as previously described [ 4 ]. Treatment lineages are designated “control” or “vinclozolin” lineages. The gestating female rats treated were designated as the F0 generation. The offspring of the F0 generation rats were the F1 generation. Nonlittermate females and males aged 70–90 days from F1 generation of control, or vinclozolin lineages were bred to obtain F2 generation offspring. The F2 generation rats were bred to obtain F3 generation offspring. As epigenetic transgenerational inheritance involves parent of origin germline epigenetic information, the optimal phenotype is obtained through breeding within the lineage, so the control and vinclozolin lineages used intrabreeding within the lineage to obtain the F3 generation. No sibling or cousin breeding was used to avoid any inbreeding artifacts. Only the F0 generation gestating female was directly treated transiently with vinclozolin. The control and vinclozolin lineages were housed in the same room and racks with lighting, food and water as previously described [ 36–38 ]. The epididymal sperm was collected from 1-year-old adult males and three pools of three males ( n = 9) used for the analysis, with all males being from different litters for both the control and vinclozolin lineages. All experimental protocols for the procedures with rats were preapproved by the Washington State University Animal Care and Use Committee (IACUC approval # 02568).
RNA Isolation
The F3 generation vinclozolin and control lineage male epididymal sperm were collected and processed as previously described and stored at −70°C until use [ 10 ]. The total RNA was isolated using the mirVana miRNA Isolation Kit (Life Technologies) following the manufacturer’s instructions with modifications at the lysis stage. In brief, after addition of lysis buffer, the frozen sperm pellets were homogenized at low settings for 90 s, followed by a 5-min incubation at 65°C. Samples were then placed on ice, and the default protocol was resumed. For quality control, RNA integrity numbers (RIN) were obtained by RNA 6000 Nano chips run on an Agilent 2100 Bioanalyzer (Agilent). A RIN of 2–4 indicates good sperm RNA quality.
Sequencing
Prior to library preparation, total sperm RNA samples were enriched for small RNAs using the protocol provided in the Ion RNA-Seq Kit v2 (Life Technologies). Small RNA-enriched samples were used for small RNA library preparation, using the same kit, and barcoded with Ion Xpress Barcode Adapters (Life Technologies). Quality control was performed using Agilent High Sensitivity chips (Agilent). Libraries were loaded onto the same Ion PI chip via the Ion PI Template OT2 200 v3 and Ion PI Sequencing 200 v3 kits, and sequenced on an Ion Proton Sequencer (Life Technologies).
Bioinformatics
The sncRNA-Seq data were annotated as follows: reads shorter than 15 nt were discarded. The remaining reads were matched to known rat sncRNA, consisting of mature miRNA (miRBase, release 21), tRNA (Genomic tRNA Database, rn5), piRNA (piRNABank), rRNA (ENSEMBL, release 76), and mitochondrial RNA (ENSEMBL, release 76) using Sequery (0–2 mismatches allowed) [ 34 , 49–52 ]. Unmatched reads were matched to mouse testis [ 53 ] and sperm endo-siRNA (M.K.S., in preparation) with Sequery (0 mismatches allowed). The remaining unmatched reads were aligned to the rat genome (rn5) via Bowtie (settings -n 2 -k 3 –best -S –al -q) [ 54 ]. Aligned reads were matched to the genomic coordinates of known rat mature miRNA, tRNA, rRNA, and mitochondrial RNA (same databases used previously). Read counts were obtained by in-house Python scripts. Unnormalized read counts were used for differential expression analysis via DESeq2 ( Padj ≤ 0.1) [ 55 ].
Reads initially matched to tRNA were extracted and matched to the 5′ and 3′ halves of the full length tRNA (split at the 3′ end of the anticodon), via Sequery (0–2 mismatches allowed) [ 52 ]. Reads ≥27 nt and ≤26 nt were referred to as halves and tRFs, respectively, and further classified as 5′ halves, tRF-5s, 3′ halves, and tRF-3s based on whether the read aligned to the 5′ or 3′ half of the tRNA ( Fig. 1 d).
Transgenerationally altered miRNA gene targets were predicted using RNAhybrid (settings -n 50 -m 50000 -c -d 2.29,0.18 -p 0.05 -e -20) and miRanda (default settings; score ≥ 140 and energy ≤−20) against rat 3′ UTR sequences (ENSEMBL, release 76), discarding predictions not made by both programs [ 34–37 ]. Dysregulated 5′ halve gene targets were predicted using RNAhybrid (settings -n 50 -m 50000 -c -d xi, θ -p 0.01 -e -20) against rat 5′ UTR, CDS, and 3′ UTR sequences (ENSEMBL, release 76) using xi,θ values (3.07,0.28), (3.00,0.27), and (2.85,0.25), respectively. The xi,θ values were obtained using RNAcalibrate and known 5′ halve sequences (tRFdb) or mature miRNA (miRBase, release 21) [ 35 , 49 , 56 ].
GO term enrichment analysis was performed using DAVID and the categories GOTERM_BP_FAT, GOTERM_MF_FAT, GOTERM_CC_FAT, and KEGG pathways [ 57 ]. Benjamini score ≤0.05 was used as the significance cutoff.
The significance of the number of observed matches between the predicted gene targets and genes proximal to DMRs ( Tables 2 and 3 , Supp. Table 5 ) was determined using a Chi-square test. The observed number of matches was compared to the expected number of random matches, defined as the number of proximal genes multiplied by the percentage of all known rat coding genes targeted by the dysregulated sncRNA. The adjusted “ Padj ” values are from DESeq2, the differential expression analysis program which adjusts the P -values to account for multiple testing using the Benjamini–Hochberg procedure, keeping the amount of Type I errors under control. The significance threshold was set at 0.1 for the DESeq2 differential expression and Chi-square analyses.
Conflict of interest: None declared.
Supplementary Material
Supplementary Data
Acknowledgements
This work was supported, in part, by grants from the NIH (ES012974 to M.K.S.) and the Templeton Foundation (PID: 50183 to M.K.S. and W.Y.). Sequencing was conducted in the Single Cell Genomics Core supported, in part, by a NIH Grant (P20RR18751). MKS and WY conceived the study; AS conducted the experiments; AS, WY and MKS analyzed the data; AS, WY and MKS wrote the manuscript. MKS and WY contributed equally to the study. Correspondence and requests for materials should be addressed to skinner@wsu.edu or wyan@medicine.nevada.edu. The authors declare no competing financial interests.
Supplementary data
Supplementary data are available at EnvEpig online.
References
- 1. Anway MD, Cupp AS, Uzumcu M, et al. . Epigenetic transgenerational actions of endocrine disruptors and male fertility . Science 2005. ; 308 : 1466 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anway MD, Leathers C, Skinner MK . Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease . Endocrinology 2006. ; 147 : 5515 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nilsson EE, Skinner MK . Environmentally induced epigenetic transgenerational inheritance of disease susceptibility . Transl Res 2015. ; 165 : 12 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yan W . Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance . Mol Cell Endocrinol 2014. ; 398 : 24 – 30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruner-Tran KL, Osteen KG . Developmental exposure to TCDD reduces fertility and negatively affects pregnancy outcomes across multiple generations . Reprod Toxicol 2011. ; 31 : 344 – 50 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manikkam M, Guerrero-Bosagna C, Tracey R, et al. . Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures . PLoS One 2012. ; 7 : e31901 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manikkam M, Tracey R, Guerrero-Bosagna C, et al. . Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of adult-onset disease and sperm epimutations . PLoS One 2013. ; 8 : e55387 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rissman EF, Adli M . Minireview: transgenerational epigenetic inheritance: focus on endocrine disrupting compounds . Endocrinology 2014. ; 155 : 2770 – 80 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manikkam M, Haque MM, Guerrero-Bosagna C, et al. . Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult onset disease through the female germline . PLoS One 2014. ; 9 : e102091 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guerrero-Bosagna C, Settles M, Lucker B, et al. . Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome . PLoS One 2010. ; 5 : e13100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skinner M, Guerrero-Bosagna C, Haque MM, et al. . Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and subsequent germline . PLoS One 2013. ; 8 : e66318 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skinner MK, Savenkova M, Zhang B, et al. . Gene bionetworks involved in epigenetic transgenerational inheritance of altered mate preference: environmental epigenetics and evolutionary biology . BMC Genomics 2014. ; 16 : 337 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casas E, Vavouri T . Sperm epigenomics: challenges and opportunities . Front Genet 2014. ; 5 : 330 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sendler E, Johnson GD, Mao S, et al. . Stability, delivery and functions of human sperm RNAs at fertilization . Nucleic Acids Res 2013. ; 41 : 4104 – 17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krawetz SA, Kruger A, Lalancette C, et al. . A survey of small RNAs in human sperm . Hum Reprod 2011. ; 26 : 3401 – 12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ostermeier GC, Miller D, Huntriss JD, et al. . Reproductive biology: delivering spermatozoan RNA to the oocyte . Nature 2004. ; 429 : 154 . [DOI] [PubMed] [Google Scholar]
- 17. Jodar M, Selvaraju S, Sendler E, et al. . The presence, role and clinical use of spermatozoal RNAs . Hum Reprod Update 2013. ; 19 : 604 – 24 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wagner KD, Wagner N, Ghanbarian H, et al. . RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse . Dev Cell 2008. ; 14 : 962 – 9 . [DOI] [PubMed] [Google Scholar]
- 19. Rassoulzadegan M, Grandjean V, Gounon P, et al. . RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse . Nature 2006. ; 441 : 469 – 74 . [DOI] [PubMed] [Google Scholar]
- 20. Yuan S, Oliver D, Schuster A, et al. . Breeding scheme and maternal small RNAs affect the efficiency of transgenerational inheritance of a paramutation in mice . Sci Rep 2015. ; 5 : 9266 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gapp K, Jawaid A, Sarkies P, et al. . Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice . Nat Neurosci 2014. ; 17 : 667 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ortogero N, Schuster AS, Oliver DK, et al. . A novel class of somatic small RNAs similar to germ cell pachytene PIWI-interacting small RNAs . J Biol Chem 2014. ; 289 : 32824 – 34 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ro S, Ma HY, Park C, et al. . The mitochondrial genome encodes abundant small noncoding RNAs . Cell Res. 2013. ; 23 : 759 – 74 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng H, Shi J, Zhang Y, et al. . A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm . Cell Res 2012. ; 22 : 1609 – 12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gebetsberger J, Polacek N . Slicing tRNAs to boost functional ncRNA diversity . RNA Biol 2013. ; 10 : 1798 – 806 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiani J, Grandjean V, Liebers R, et al. . RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2 . PLoS Genet 2013. ; 9 : e1003498 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function . Cell 2004. ; 116 : 281 – 97 . [DOI] [PubMed] [Google Scholar]
- 28. Kumar P, Anaya J, Mudunuri SB, et al. . Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets . BMC Biol 2014. ; 12 : 78 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ivanov P, Emara MM, Villen J, et al. . Angiogenin-induced tRNA fragments inhibit translation initiation . Mol Cell 2011. ; 43 : 613 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Elbarbary RA, Takaku H, Uchiumi N, et al. . Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA . PLoS One 2009. ; 4 : e5908 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamasaki S, Ivanov P, Hu GF, et al. . Angiogenin cleaves tRNA and promotes stress-induced translational repression . J Cell Biol 2009. ; 185 : 35 – 42 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Q, Lee I, Ren J, et al. . Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection . Mol Ther 2013. ; 21 : 368 – 79 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sobala A, Hutvagner G . Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells . RNA Biol 2013. ; 10 : 553 – 63 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Flicek P, Amode MR, Barrell D, et al. . Ensembl 2014 . Nucleic Acids Res 2014. ; 42 ( Database issue ): D749 – 55 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rehmsmeier M, Steffen P, Hochsmann M, et al. . Fast and effective prediction of microRNA/target duplexes . RNA 2004. ; 10 : 1507 – 17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kruger J, Rehmsmeier M . RNAhybrid: microRNA target prediction easy, fast and flexible . Nucleic Acids Res 2006. ; 34(Web Server issue) : W451 – 4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. John B, Enright AJ, Aravin A, et al. . Human MicroRNA targets . PLoS Biol 2004. ; 2 : e363 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skinner MK, Guerrero-Bosagna C, Haque MM . Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations . Epigenetics 2015. ; 10 : 762 – 71 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Skinner MK, Manikkam M, Haque MM, et al. . Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions . Genome Biol 2012. ; 13 : R91 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skinner MK, Anway MD, Savenkova MI, et al. . Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior . PLoS One 2008. ; 3 : e3745 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gillette R, Miller-Crews I, Skinner MK, et al. . Distinct actions of ancestral vinclozolin and juvenile stress on neural gene expression in the male rat . Front Genet 2015. ; 6 : 56 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crews D, Gillette R, Scarpino SV, et al. . Epigenetic transgenerational inheritance of altered stress responses . Proc Natl Acad Sci USA 2012. ; 109 : 9143 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blanco S, Dietmann S, Flores JV, et al. . Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders . EMBO J 2014. ; 33 : 2020 – 39 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hussain S, Tuorto F, Menon S, et al. . The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation . Mol Cell Biol 2013. ; 33 : 1561 – 70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schaefer M, Pollex T, Hanna K, et al. . RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage . Genes Dev 2010. ; 24 : 1590 – 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tuorto F, Liebers R, Musch T, et al. . RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis . Nat Struct Mol Biol 2012. ; 19 : 900 – 5 . [DOI] [PubMed] [Google Scholar]
- 47. Hussain S, Sajini AA, Blanco S, et al. . NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs . Cell Rep 2013. ; 4 : 255 – 61 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kiani J, Rassoulzadegan M. A. load of small RNAs in the sperm—how many bits of hereditary information? Cell Res 2013. ; 23 : 18 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kozomara A, Griffiths-Jones S . miRBase: annotating high confidence microRNAs using deep sequencing data . Nucleic Acids Res 2014. ; 42 ( Database issue ): D68 – 73 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chan PP, Lowe TM . GtRNAdb: a database of transfer RNA genes detected in genomic sequence . Nucleic Acids Res 2009. ; 37 ( Database issue ): D93 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sai Lakshmi S, Agrawal S . piRNABank: a web resource on classified and clustered Piwi-interacting RNAs . Nucleic Acids Res 2008. ; 36 ( Database issue ): D173 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ortogero N, Hennig GW, Langille C, et al. . Computer-assisted annotation of murine Sertoli cell small RNA transcriptome . Biol Reprod 2013. ; 88 : 3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Song R, Hennig GW, Wu Q, et al. . Male germ cells express abundant endogenous siRNAs . Proc Natl Acad Sci USA 2011. ; 108 : 13159 – 64 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Langmead B, Trapnell C, Pop M, et al. . Ultrafast and memory-efficient alignment of short DNA sequences to the human genome . Genome Biol 2009. ; 10 : R25 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Love MI, Huber W, Anders S . Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 . Genome Biol 2014. ; 15 : 550 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kumar P, Mudunuri SB, Anaya J, et al. . tRFdb: a database for transfer RNA fragments . Nucleic Acids Res 2015. ; 43 ( Database issue ): D141 – 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dennis G, Jr, Sherman BT, Hosack DA, et al. . DAVID: Database for Annotation, Visualization, and Integrated Discovery . Genome Biol 2003. ; 4 : P3 . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data