Abstract
MicroRNAs (miRs) have emerged as potent regulators of pathways in physiological and disease contexts. This review focuses on the role of miRs in ageing of the cardiovascular system. Several miRs have been described to be regulated during ageing and some of these miRs are involved in the regulation of ageing‐related processes. We discuss the roles of miR‐34, miR‐217 and miR‐29, which are induced during ageing in the vasculature. The roles of miR‐34, miR‐29 (age‐induced) and miR‐18/19, which are decreased during ageing in the heart, are discussed as well. Furthermore, numerous miRs that play a role in diseases associated with ageing, like diabetes, atherosclerosis, hypertension, cardiac hypertrophy and atrial fibrillation, are also briefly discussed. miRs also serve as circulating biomarkers for cardiovascular ageing or ageing‐associated diseases. Finally, pharmacological modulation of ageing‐related miRs might become a promising strategy to combat cardiovascular ageing in a clinical setting.
Introduction
MicroRNAs (miRs) belong to the rapidly growing family of non‐coding RNAs with crucial and substantial regulatory functions in almost all cellular biological mechanisms. They act via RNA‐mediated gene silencing through RNA interference‐like pathways. The first microRNAs were described in the early 1990s regulating and timing larval development in the nematode Caenorhabditis elegans (Lee et al. 1993; Reinhart et al. 2000; Lee & Ambros, 2001). Since then, detailed insights have been gained into the biogenesis and regulation of microRNAs as well as specific roles in physiological regulatory mechanisms and dysregulation in pathophysiological conditions. To date, over 2500 miRs have been catalogued in humans in the latest release of the microRNA database miRBase, the majority of them by deep sequencing approaches and for many of these annotations, the functional importance has not yet been evaluated (Kozomara & Griffiths‐Jones, 2014).
MicroRNAs are small, ∼22 nucleotides in length, single stranded RNA molecules. By binding to their target mRNAs, miRs induce translational repression, mRNA deadenylation and mRNA decay (Huntzinger & Izaurralde, 2011). Predominantly, miR‐binding sites are located in the 3′ untranslated region (UTR) of their target mRNAs (Bartel, 2009). Target recognition is mainly driven by the miR ‘seed sequence’, a domain at the 5′ end reaching from nucleotide 2 to nucleotide 7 of the mature miR, and miRs with identical seed sequences belong to the same ‘miR family’ (Bartel, 2009). In the majority of target–miR interactions, the seed region binds perfectly complementarily, while the downstream nucleotides also contribute to base pairing but with varying numbers of mismatches (Fig. 1 A). The 3′ UTR of mRNAs can contain binding sites for different miRs and furthermore, due to the variations in binding of the nucleotides downstream of the seed region, one miR can target several hundred mRNAs (Fig. 1 A). Through these highly diversified binding interactions miRs can ‘fine tune’ entire signalling cascades and gene networks (Bartel, 2009).
Figure 1. MicroRNA binding and biogenesis .
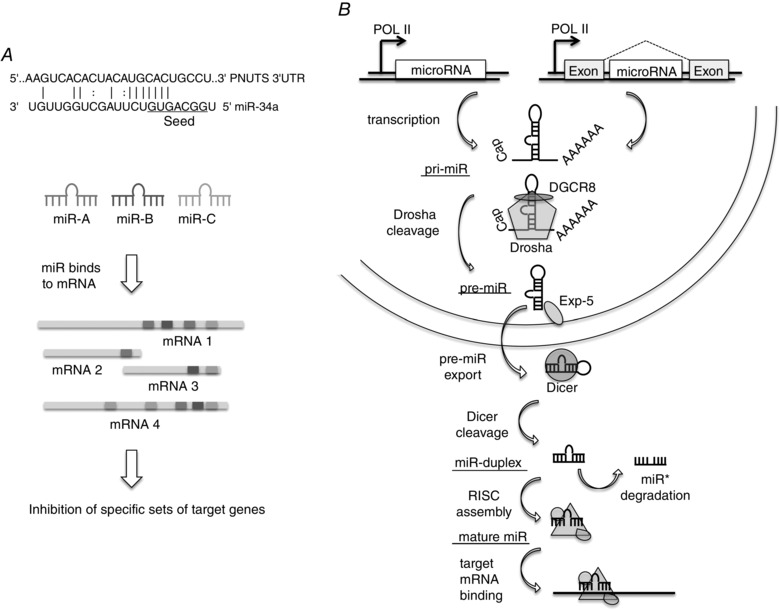
A, microRNAs (miRs) bind to target mRNAs mainly in a partially complementary fashion. Nucleotides 2–7 of the miR are referred to as a seed sequence and these nucleotides bind fully complementarily to the target sequence, whereas the other nucleotides of the miR only show interspersed complementarity as shown for miR‐34a and its target PNUTS (Boon et al. 2013). Different miRs (depicted by various grey tones) bind to a specific set of mRNA targets and these sets may be overlapping. One mRNA can be bound by multiple different miRs or even multiple times by the same miR. Each miR targets tens to hundreds of mRNAs, thereby coordinating complex regulatory networks. B, miRs are transcribed from the genome either from intergenic regions or within introns of genes. The primary miR transcript (pri‐miR) is processed by Drosha in association with DGCR8 (DiGeorge critical region 8), yielding a precursor miR (pre‐miR) that is exported by exportin 5 (Exp‐5). Pre‐miRs are cleaved by Dicer in the cytoplasm and one of the strands from the resulting miR duplex is incorporated in the RNA‐induced silencing complex (RISC) that binds to target mRNAs, thereby repressing expression levels.
MicroRNA biology
MicroRNAs encoded in the genomic DNA are mainly intergenic being located in the non‐coding regions between genes and transcribed by often unidentified promoters. However, specific miRs have been found to be transcribed from intronic regions (Lin et al. 2006; van Rooij et al. 2007). miRs are transcribed by RNA polymerase II (Pol II) and controlled by RNA Pol II‐associated transcription factors and epigenetic regulators as a long (>1 kb) primary RNA transcript (pri‐miR) (Lee et al. 2004). The pri‐miR forms a characteristic stem–loop structure and has the miR embedded in its stem (Fig. 1 B) (Ha & Kim, 2014). Each precursor miR contains two mature miRs, one in its 5′ and one in its 3′ strand (for example miR‐34a‐3p and miR‐34‐5p). In most cases, one is usually biologically more prevalent and is called the guide strand. The other is called the passenger strand or miR* and is mostly degraded. However, many examples also exist where the passenger strand is stable and functions as a miR as well.
In the nucleus, the pri‐miR is processed by the nuclear RNase III Drosha and its essential co‐factor DGCR8 (named by its implication in a genetic disorder called DiGeorge syndrome) together forming the microprocessor complex (Lee et al. 2003; Denli et al. 2004; Gregory et al. 2004). Recently, it has been shown that methyltransferase‐like 3 (METTL3) methylates pri‐miRNAs, marking them for recognition and processing by DGCR8 (Alarcón et al. 2015). After binding of the microprocessor to the pri‐miR, the single‐stranded 5′ and 3′ arms are cut releasing the pre‐miR, a short hairpin‐like RNA with around 65 nucleotides in length. Deficiency of Drosha in the germline is embryonically lethal (Chong et al. 2010). Further, cardiac specific deletion of DGCR8 in mice leads to heart failure and dilated cardiomyopathy (DCM; Rao et al. 2009), highlighting the importance of miRs in cardiovascular homeostasis.
After nuclear processing by Drosha, the pre‐miR is exported from the nucleus for further maturation in the cytoplasm. In a complex with the protein exportin‐5 and GTP‐binding nuclear protein RanGTP the pre‐miR passes the nuclear core complex and is released into the cytoplasm (Bohnsack et al. 2004; Lund et al. 2004). After export, the pre‐miR is cleaved by Dicer, an RNase III‐type endonuclease, close to the terminal loop and a small RNA duplex containing miR/miR* is released (Bernstein et al. 2001; Hutvágner et al. 2001). Deletion of Dicer in the mouse germline is embryonically lethal (Bernstein et al. 2003). The cardiomyocyte specific deletion of Dicer also rapidly leads to DCM and heart failure (Chen et al. 2008).
Together with Argonaute proteins (human AGO1–4) the miR duplex forms the RNA‐induced silencing complex (RISC) which mediates all RNA‐silencing pathways (Liu et al. 2004). By removing the passenger strand the pre‐RISC turns into the mature RISC. The determination of the biologically active guide strand and the passenger strand, which is degraded quickly after release, is mainly dependent on the thermostability of the two ends of the RNA duplex (Schwarz et al. 2003).
Recently it has been shown that miRs may also act in the nucleus, even though miR‐loading into RISC only takes place in the cytoplasm (Gagnon et al. 2014). In this review we discuss the importance of miR‐mediated regulation of processes that are relevant for cardiovascular ageing. For an overview of miRs in cardiovascular biology in general, we refer to Quiat & Olson (2013), Boon & Dimmeler (2014), Greco et al. (2015), Schober et al. (2015) and Wronska et al. (2015).
Vascular ageing
Vascular ageing is characterized by detrimental effects in most cell types found in the vessel wall. These include changes in endothelial cells, smooth muscle cells and inflammatory cells. A few miRs have been described to regulate ageing‐related processes in these cells. One of the most described ageing‐induced miRs, miR‐34a, has been shown to regulate ageing‐related processes such as senescence in most of these cells (as well as in the heart, mentioned below). miR‐34a was first discovered in the context of cancer (He et al. 2007; Tarasov et al. 2007), where it was found to be induced by p53 and to regulate apoptosis (Hermeking, 2012). That miR‐34a is also important in vascular biology became clear with the study by Ito and colleagues (Ito et al. 2010) which showed that miR‐34a regulates the histone deacetylase silent mating‐type information regulation 2 homologue (SIRT1) in endothelial cells (see also Tabuchi et al. 2012). As SIRT1 acts as a longevity promoting factor (Haigis & Guarente, 2006), this mechanism contributes to the endothelial senescence‐inducing effects of miR‐34a. Ageing also induces miR‐34a expression in smooth muscle cells (Badi et al. 2014), where a reduction in SIRT1 likewise results in an increase in senescence and secretion of inflammatory factors. Like all miRs, miR‐34a also has multiple target genes and it is likely that additional target genes beyond SIRT1 are involved in inducing senescence and ageing in the vasculature (Fig. 2, Table 1).
Figure 2. Several miRs are involved in cardiac ageing and in vascular ageing .
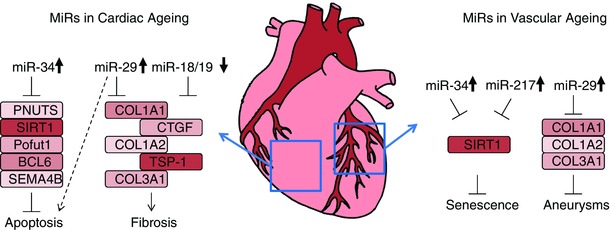
miRs that are involved in cardiac ageing include miR‐34, miR‐29, miR‐18 and miR‐19. miR‐34 is upregulated during ageing and induces apoptosis by inhibiting expression of anti‐apoptotic genes. miR‐29 is also upregulated and induces apoptosis, but counteracts fibrosis via suppression of extracellular matrix components. miR‐18 and miR‐19 are down‐regulated during ageing and normally inhibit pro‐fibrotic genes, thereby facilitating fibrosis. miR‐34, miR‐217 and miR‐29 are involved in vascular ageing. miR‐34 and miR‐217 are upregulated in endothelium and cause senescence via repression of SIRT1. miR‐29 is upregulated in smooth muscle cells, where it inhibits synthesis of extracellular matrix components, which contributes to aneurysm formation.
Table 1.
MicroRNAs that are regulated during ageing and/or ageing‐associated diseases
| MicroRNA | Associated disease | References |
|---|---|---|
| miR‐1 | Atrial fibrillation, mitochondrial function | (Girmatsion et al. 2009; Zhang et al. 2014) |
| miR‐133 | Cardiac hypertrophy | (Carè et al. 2007) |
| miR‐144 | Mitochondrial function | (Csiszar et al. 2014) |
| miR‐15 | Cardiac hypertrophy, atrial fibrillation | (Nishi et al. 2010; Tijsen et al. 2014) |
| miR‐18 | Cardiac fibrosis | (van Almen et al. 2011) |
| miR‐181 | Mitochondrial function, immunosenescence | (Das et al. 2012; Seeger et al. 2013) |
| miR‐19 | Cardiac fibrosis | (van Almen et al. 2011) |
| miR‐199 | Cardiac hypertrophy | (da Costa Martins et al. 2010) |
| miR‐208 | Cardiac hypertrophy | (van Rooij et al. 2007) |
| miR‐21 | Cardiac hypertrophy, cardiac fibrosis | (Thum et al. 2008; Patrick et al. 2010; Bang et al. 2014) |
| miR‐214 | Mitochondrial function | (el Azzouzi et al. 2013) |
| miR‐217 | Senescence | (Menghini et al. 2009) |
| miR‐22 | Cardiac fibrosis, senescence, cardiac hypertrophy | (Huang et al. 2013; Jazbutyte et al. 2013) |
| miR‐25 | Cardiac hypertrophy | (Dirkx et al. 2013) |
| miR‐26 | Atrial fibrillation | (Luo et al. 2013) |
| miR‐29 | Cardiac fibrosis, apoptosis, aortic aneurysms, atrial fibrillation | (van Rooij et al. 2008 b; Ye et al. 2010; Boon et al. 2011; Maegdefessel et al. 2012; Dawson et al. 2013; Abonnenc et al. 2013) |
| miR‐30 | Cardiac hypertrophy | (Wijnen et al. 2014) |
| miR‐328 | Atrial fibrillation | (Lu et al. 2010) |
| miR‐34 | Senescence, apoptosis, telomere attrition, cardiac hypertrophy | (Ito et al. 2010; Bernardo et al. 2012; Tabuchi et al. 2012; Boon et al. 2013; Yuan et al. 2014; Badi et al. 2014; Dimitrakopoulou et al. 2015) |
| miR‐378 | Cardiac hypertrophy | (Ganesan et al. 2013) |
| miR‐451 | Cardiac hypertrophy | (Kuwabara et al. 2015) |
| miR‐499 | Atrial fibrillation, mitochondrial function | (Wang et al. 2011; Ling et al. 2013 b) |
| miR‐92 | Endothelial dysfunction | (Chen et al. 2015) |
Other miRs that are involved in senescence and ageing of endothelial and smooth muscle cells include miR‐217 and miR‐29. The former was also shown to regulate the expression of SIRT1, thereby promoting endothelial ageing (Menghini et al. 2009). miR‐29 was found to be induced during ageing and play a role in aneurysm formation (Boon et al. 2011). Ageing is the major risk factor for the development of aneurysms, the pathological widening of large arteries, which greatly increases the risk of rupture of the artery with a very high mortality rate. miR‐29 regulates the expression of extracellular matrix proteins and thereby reduces the structural integrity of the vessel wall allowing aneurysm formation to take place (Boon et al. 2011; Maegdefessel et al. 2012).
Several other miRs have been described to be involved in disease processes that are associated with ageing; however, a direct role in ageing has not been firmly established. Many miRs were shown to affect atherosclerosis formation, which is reviewed elsewhere (Kumar et al. 2014; Menghini et al. 2014; Schober et al. 2015). Apoptosis in vascular cells, which is induced during ageing, is also regulated by miRs (Quintavalle et al. 2011). Ageing is a strong risk factor for arterial hypertension, partly via the β‐adrenergic system, which is regulated by miRs as well (reviewed in Bátkai & Thum, 2012; Ling et al. 2013 a). Furthermore, single nucleotide polymorphisms (SNPs) linked to arterial hypertension have been found in miR‐binding sites of genes of the renin–angiotensin system (Nossent et al. 2011). Finally, ageing is associated with an increase in diabetes mellitus and metabolic syndrome. The role of miRs in these age‐associated diseases is reviewed in Fernández‐Hernando et al. (2013), Paneni et al. (2013) and Beltrami et al. (2014).
One of the cellular mechanisms that causes ageing is oxidative stress. miR‐200 and miR‐210 have been described to regulate mitochondrial function and oxidative stress in the vasculature (for review, see Magenta et al. 2013). The oxidative stress response of endothelial cells includes expression changes in miRs as well, i.e. the upregulation of miR‐92a, which induces endothelial dysfunction (Chen et al. 2015). Interestingly, one can also exploit the cell‐type enriched expression patterns of miRs to specifically target certain cell types in the vasculature by including miR binding sites in the overexpression construct for miRs that are highly expressed in the tissue one does not want to target (so‐called detargeting). The endothelial‐enriched expression of miR‐126 was used to specifically detarget an adenoviral construct to overexpress p27 in vascular smooth muscle cells in the context of restenosis (Santulli et al. 2014 b).
Cardiac ageing
Ageing affects cardiac function in multiple manners. The most common age‐induced cardiac disease is diastolic dysfunction, also termed heart failure with preserved ejection fraction (Loffredo et al. 2014). This is caused by increased stiffness and fibrosis of the myocardium and is associated with endothelial dysfunction (Paulus & Tschöpe, 2013). Ageing‐induced changes in virtually all cell types in the heart contribute to these processes and several miRs were described to play a role in cardiac ageing.
In cardiomyocytes, the main ageing‐regulated miR that has been described is miR‐34. The miR‐34 family consists of miR‐34a, miR‐34b and miR‐34c. All these family members are induced during ageing (Boon et al. 2013). miR‐34a is the most highly expressed miR‐34 family member in cardiomyocytes and the increased miR‐34a expression in the aged heart is probably due to an increase in p53 signalling, known to be induced in ageing. miR‐34 family members induce apoptosis during ageing, but also after acute myocardial infarction (Bernardo et al. 2012; Boon et al. 2013). Next to SIRT1, miR‐34a directly inhibits the expression of several other target genes, including POFUT1, BCL6, SEMA4b (Bernardo et al. 2012) and PNUTS (Boon et al. 2013), thereby affecting cardiomyocyte apoptosis and heart function. Interestingly, miR‐34a is also induced in a genetic model for cardiac ageing (calstabin‐2 null mice) (Yuan et al. 2014) and integrated network analysis also confirmed a central role for miR‐34a in cardiac ageing (Dimitrakopoulou et al. 2015) (Fig. 2, Table 1).
MicroRNAs that are present in cardiac fibroblasts and are regulated during ageing appear to have a link to fibrosis, which is known to be induced during ageing. miR‐18 and miR‐19, which are expressed from the same primary cluster, miR‐17‐92, are reduced in aged mouse hearts (van Almen et al. 2011). Connective tissue growth factor (CTGF) and thrombospondin‐1 (TSP‐1) are the main pro‐fibrotic targets of these miRs and a reduction of miR‐18 and miR‐19 during ageing contributes to the increased expression of CTGF and TSP‐1, resulting in increased fibrosis and a decline in heart function. Interestingly, the fibrosis‐inhibiting miR‐29 is also increased in the heart during ageing (Boon et al. 2013) and even though exogenous miR‐29 reduces fibrosis (van Rooij et al. 2008 b; Abonnenc et al. 2013), the endogenous induction does not seem to be able to prevent fibrosis during ageing. A possible mechanism could be the contribution of miR‐29 to apoptosis of cardiomyocytes during ageing, since inhibition of miR‐29 was described as preventing ischaemia‐induced cardiomyocyte apoptosis (Ye et al. 2010). Finally, miR‐22 has also been shown to be induced during ageing in the mouse heart, where it induces fibroblast migration and senescence that contribute to fibrosis in ageing (Jazbutyte et al. 2013).
Several other miRs have been described to play a role in processes related to cardiac ageing or ageing in general. We will briefly discuss these miRs here. miRs that are involved in heart failure in general are reviewed in Tritsch et al. (2013) and Zhuo et al. (2014). Further, the contribution of miRs to clinical management of heart failure was recently described (Sardu et al. 2014). Control of cardiac hypertrophy and fibrosis, which are also induced during ageing, by miRs was shown for miR‐133 (Carè et al. 2007), miR‐21 (Thum et al. 2008; Patrick et al. 2010; Bang et al. 2014), miR‐208 (van Rooij et al. 2007), miR‐15 (Tijsen et al. 2014), miR‐25 (Dirkx et al. 2013), miR‐199 (da Costa Martins et al. 2010), miR‐22 (Huang et al. 2013), miR‐451 (Kuwabara et al. 2015), miR‐378 (Ganesan et al. 2013) and miR‐30 (Wijnen et al. 2014). Ageing also induces the prevalence of atrial fibrillation and several miRs have been described in the context of atrial fibrillation: miR‐1 (Girmatsion et al. 2009), miR‐26 (Luo et al. 2013), miR‐29 (Dawson et al. 2013), miR‐328 (Lu et al. 2010) and miR‐499 (Ling et al. 2013 b), reviewed in Santulli et al. (2014 a). Increased oxidative stress in the heart and impaired mitochondrial functional are hallmarks of cardiac ageing. Several miRs have been identified to control mitochondrial function and oxidative stress in the heart that may contribute to cardiac ageing: miR‐1 (Zhang et al. 2014), miR‐144 (Csiszar et al. 2014), miR‐199 and miR‐214 (el Azzouzi et al. 2013), miR‐181 (Das et al. 2012), miR‐499 (Wang et al. 2011) and miR‐15 (Nishi et al. 2010).
Clinical outlook
The development of miRNA therapeutics has gained considerable momentum over the past years. Inhibition of miRs with anti‐miRs that sterically block the specific miR seems most promising and has been pioneered by anti‐miRs against miR‐122 to treat hepatitis C (Lanford et al. 2010). Phase II clinical trials using these anti‐miRs (called miravirsen) are very promising (Janssen et al. 2013). There are no clinical trials reported to date with anti‐miRs targeting cardiovascular disease, but inhibition of many miRs was shown to be therapeutically beneficial in mouse models (van Rooij et al. 2008 a) or even in large animal models (Hinkel et al. 2013). Several of the ageing‐related miRs discussed in this review may also prove promising therapeutic targets. For instance, inhibition of miR‐34a in the myocardium may prevent or even ameliorate age‐induced cardiac dysfunction. One should, however, be very careful in choosing delivery strategies for the anti‐miRs, as many miRs are expressed in a variety of cell types and may have dissimilar roles in different cells. The pro‐apoptotic miR‐34a would be an interesting target to prevent cardiomyocyte apoptosis, but inhibition of miR‐34a may simultaneously induce tumorigenesis (Hermeking, 2010). In fact, delivery of miR‐34a mimics is being developed as treatment for liver cancer (clinical trial number NCT01829971).
Even though anti‐miR chemistries appeared to be safe and well‐tolerated in the clinical trials performed so far, a recent study showed that the phosphorothioate‐modified RNA backbone used in anti‐miRs can induce platelet aggregation via activation of glycoprotein VI on platelets (Flierl et al. 2015). However, most anti‐miRs are only 16 nucleotides long and therefore too small to facilitate glycoprotein VI dimerization and subsequent platelet aggregation (Flierl et al. 2015).
Finally, miRs may serve as biomarkers of cardiovascular ageing. Many miRs have been proposed as biomarkers for cardiovascular disease (Fichtlscherer et al. 2011; Zampetaki et al. 2012; Watson et al. 2015), but only a few circulating miRs reflect cardiovascular ageing. Reduced expression of miR‐181c in the peripheral blood was shown to be associated with ageing and chronic heart failure as well as immunosenescence (Seeger et al. 2013), circulating miR‐34a was shown to correlate with age in mice (Li et al. 2011), and plasma levels of miR‐21 are increased with ageing (Olivieri et al. 2012). Furthermore, several reports describe differential levels of circulating miRs in centenarians, compared to young control subjects (Noren Hooten et al. 2010; Gombar et al. 2012; Serna et al. 2012; Meder et al. 2014). As these miRs are not cardiovascular specific, the levels do not necessarily reflect cardiovascular disease but may help in risk stratification as well as in monitoring disease progression. For example, miRs found in blood plasma can function as biomarkers for diabetes mellitus (Guay & Regazzi, 2013). Circulating miRs that specifically reflect cardiovascular ageing or might emerge to be direct therapeutic targets are still elusive.
Additional information
Competing interests
The authors have no conflicts of interest to declare.
Funding
Work in R.A.B.’s laboratory is funded by the Hessian Ministry of Higher Education, Research and the Arts, funding reference number: III L 4‐518/17.004 (2013).
Biographies
Timon Seeger received his MD from the School of Medicine, University of Heidelberg, Germany and finished his internal medicine residency at the University Hospital of Frankfurt, Germany. During this time he pursued his research at Professor Stefanie Dimmeler's Institute for Cardiovascular Regeneration, Centre of Molecular Medicine, University of Frankfurt, studying the role of microRNAs in cardiac diseases and ageing as well as cardiac regeneration. In 2014 he started a German Research Association funded postdoctoral fellowship at Professor Joseph Wu's Cardiovascular Institute at Stanford University focusing on molecular dysregulations in cardiac diseases using iPSC derived cardiomyocytes.

Reinier Boon received his Master's degree in Medical Biology and PhD in vascular biology from the University of Amsterdam, the Netherlands. After receiving his PhD degree, he moved to the Institute for Cardiovascular Regeneration in Frankfurt, Germany, where he started postdoctoral work in the laboratory of Professor Dimmeler. Dr Boon has been a group leader in the Institute for Cardiovascular Regeneration in Frankfurt since 2011 and studies the role of non‐coding RNA in cardiovascular ageing.
References
- Abonnenc M, Nabeebaccus AA, Mayr U, Barallobre‐Barreiro J, Dong X, Cuello F, Sur S, Drozdov I, Langley SR, Lu R, Stathopoulou K, Didangelos A, Yin X, Zimmermann W‐H, Shah AM, Zampetaki A & Mayr M (2013). Extracellular matrix secretion by cardiac fibroblasts: role of microRNA‐29b and microRNA‐30c. Circ Res 113, 1138–1147. [DOI] [PubMed] [Google Scholar]
- Alarcón CR, Lee H, Goodarzi H, Halberg N & Tavazoie SF (2015). N 6‐methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badi I, Burba I, Ruggeri C, Zeni F, Bertolotti M, Scopece A, Pompilio G & Raucci A (2014). MicroRNA‐34a induces vascular smooth muscle cells senescence by SIRT1 downregulation and promotes the expression of age‐associated pro‐inflammatory secretory factors. J Gerontol A Biol Sci Med Sci 70, 1304–1311. [DOI] [PubMed] [Google Scholar]
- Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, Ponimaskin E, Schmiedl A, Yin X, Mayr M, Halder R, Fischer A, Engelhardt S, Wei Y, Schober A, Fiedler J & Thum T (2014). Cardiac fibroblast‐derived microRNA passenger strand‐enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest 124, 2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bátkai S & Thum T (2012). MicroRNAs in hypertension: mechanisms and therapeutic targets. Curr Hypertens Rep 14, 79–87. [DOI] [PubMed] [Google Scholar]
- Beltrami C, Angelini TG & Emanueli C (2014). Noncoding RNAs in diabetes vascular complications. J Mol Cell Cardiol 89, 42–50. [DOI] [PubMed] [Google Scholar]
- Bernardo BC, Gao X‐M, Winbanks CE, Boey EJH, Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du X‐J, Lin RCY & McMullen JR (2012). Therapeutic inhibition of the miR‐34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci U S A 109, 17615–17620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM & Hannon GJ (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV & Hannon GJ (2003). Dicer is essential for mouse development. Nat Genet 35, 215–217. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K & Gorlich D (2004). Exportin 5 is a RanGTP‐dependent dsRNA‐binding protein that mediates nuclear export of pre‐miRNAs. RNA 10, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon RA & Dimmeler S (2014). MicroRNAs in myocardial infarction. Nat Rev Cardiol 12, 135–142. [DOI] [PubMed] [Google Scholar]
- Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Tréguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Müller OJ, Potente M, Zeiher AM, Hermeking H & Dimmeler S (2013). MicroRNA‐34a regulates cardiac ageing and function. Nature 495, 107–110. [DOI] [PubMed] [Google Scholar]
- Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, Vinciguerra M, Rosenthal N, Sciacca S, Pilato M, van Heijningen P, Essers J, Brandes RP, Zeiher AM & Dimmeler S (2011). MicroRNA‐29 in aortic dilation: implications for aneurysm formation. Circ Res 109, 1115–1119. [DOI] [PubMed] [Google Scholar]
- Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Høydal M, Autore C, Russo MA, Dorn GW 2nd, Ellingsen O, Ruiz‐Lozano P, Peterson KL, Croce CM, Peschle C & Condorelli G (2007). MicroRNA‐133 controls cardiac hypertrophy. Nat Med 13, 613–618. [DOI] [PubMed] [Google Scholar]
- Chen J‐F, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ & Wang D‐Z (2008). Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A 105, 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wen L, Martin M, Hsu C‐Y, Fang L, Lin F‐M, Lin T‐Y, Geary MJ, Geary GG, Zhao Y, Johnson DA, Chen J‐W, Lin S‐J, Chien S, Huang H‐D, Miller YI, Huang P‐H & Shyy JY‐J (2015). Oxidative stress activates endothelial innate immunity via sterol regulatory element binding protein 2 (SREBP2) transactivation of microRNA‐92a. Circulation 131, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MMW, Zhang G, Cheloufi S, Neubert TA, Hannon GJ & Littman DR (2010). Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev 24, 1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csiszar A, Gautam T, Sosnowska D, Tarantini S, Banki E, Tucsek Z, Toth P, Losonczy G, Koller A, Reglodi D, Giles CB, Wren JD, Sonntag WE & Ungvari Z (2014). Caloric restriction confers persistent anti‐oxidative, pro‐angiogenic, and anti‐inflammatory effects and promotes anti‐aging miRNA expression profile in cerebromicrovascular endothelial cells of aged rats. Am J Physiol Heart Circ Physiol 307, H292–H306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa Martins PA, Salic K, Gladka MM, Armand A‐S, Leptidis S, el Azzouzi H, Hansen A, Coenen‐de Roo CJ, Bierhuizen MF, van der Nagel R, van Kuik J, de Weger R, de Bruin A, Condorelli G, Arbones ML, Eschenhagen T & De Windt LJ (2010). MicroRNA‐199b targets the nuclear kinase Dyrk1a in an auto‐amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol 12, 1220–1227. [DOI] [PubMed] [Google Scholar]
- Das S, Ferlito M, Kent OA, Fox‐Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E & Steenbergen C (2012). Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res 110, 1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson K, Wakili R, Ordög B, Clauss S, Chen Y, Iwasaki Y, Voigt N, Qi XY, Sinner MF, Dobrev D, Kääb S & Nattel S (2013). MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 127, 1466–1475, 1475e1–e28. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BBJ, Plasterk RHA, Ketting RF & Hannon GJ (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235. [DOI] [PubMed] [Google Scholar]
- Dimitrakopoulou K, Vrahatis AG & Bezerianos A (2015). Integromics network meta‐analysis on cardiac aging offers robust multi‐layer modular signatures and reveals micronome synergism. BMC Genomics 16, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirkx E, Gladka MM, Philippen LE, Armand AS, Kinet V, Leptidis S, El Azzouzi H, Salic K, Bourajjaj M, da Silva GJ, Olieslagers S, van der Nagel R, de Weger R, Bitsch N, Kisters N, Seyen S, Morikawa Y, Chanoine C, Heymans S, Volders PG, Thum T, Dimmeler S, Cserjesi P, Eschenhagen T, da Costa Martins PA & De Windt LJ (2013). Nfat and miR‐25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat Cell Biol 15, 1282–1293. [DOI] [PubMed] [Google Scholar]
- el Azzouzi H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K, McClellan EA, Poels E, Sluimer JC, van den Hoogenhof MM, Armand AS, Yin X, Langley S, Bourajjaj M, Olieslagers S, Krishnan J, Vooijs M, Kurihara H, Stubbs A, Pinto YM, Krek W, Mayr M, da Costa Martins PA, Schrauwen P & De Windt LJ (2013). The hypoxia‐inducible microRNA cluster miR‐199a∼214 targets myocardial PPARδ and impairs mitochondrial fatty acid oxidation. Cell Metab 18, 341–354. [DOI] [PubMed] [Google Scholar]
- Fernández‐Hernando C, Ramírez CM, Goedeke L & Suárez Y (2013). MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol 33, 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtlscherer S, Zeiher AM & Dimmeler S (2011). Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol 31, 2383–2390. [DOI] [PubMed] [Google Scholar]
- Flierl U, Nero TL, Lim B, Arthur JF, Yao Y, Jung SM, Gitz E, Pollitt AY, Zaldivia MTK, Jandrot‐Perrus M, Schäfer A, Nieswandt B, Andrews RK, Parker MW, Gardiner EE & Peter K (2015). Phosphorothioate backbone modifications of nucleotide‐based drugs are potent platelet activators. J Exp Med 212, 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Chu Y, Janowski BA & Corey DR (2014). RNAi factors are present and active in human cell nuclei. Cell Rep 6, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, Leierseder S, Loyer X, Giacca M, Zentilin L, Thum T, Laggerbauer B & Engelhardt S (2013). MiR‐378 controls cardiac hypertrophy by combined repression of mitogen‐activated protein kinase pathway factors. Circulation 127, 2097–2106. [DOI] [PubMed] [Google Scholar]
- Girmatsion Z, Biliczki P, Bonauer A, Wimmer‐Greinecker G, Scherer M, Moritz A, Bukowska A, Goette A, Nattel S, Hohnloser SH & Ehrlich JR (2009). Changes in microRNA‐1 expression and IK1 up‐regulation in human atrial fibrillation. Heart Rhythm 6, 1802–1809. [DOI] [PubMed] [Google Scholar]
- Gombar S, Jung HJ, Dong F, Calder B, Atzmon G, Barzilai N, Tian X‐L, Pothof J, Hoeijmakers JHJ, Campisi J, Vijg J & Suh Y (2012). Comprehensive microRNA profiling in B‐cells of human centenarians by massively parallel sequencing. BMC Genomics 13, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S, Gorospe M & Martelli F (2015). Noncoding RNA in age‐related cardiovascular diseases. J Mol Cell Cardiol 83, 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan K‐P, Amuthan G, Chendrimada T, Doratotaj B, Cooch N & Shiekhattar R (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240. [DOI] [PubMed] [Google Scholar]
- Guay C & Regazzi R (2013). Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 9, 513–521. [DOI] [PubMed] [Google Scholar]
- Ha M & Kim VN (2014). Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524. [DOI] [PubMed] [Google Scholar]
- Haigis MC & Guarente LP (2006). Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev 20, 2913–2921. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA & Hannon GJ (2007). A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H (2010). The miR‐34 family in cancer and apoptosis. Cell Death Differ 17, 193–199. [DOI] [PubMed] [Google Scholar]
- Hermeking H (2012). MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 12, 613–626. [DOI] [PubMed] [Google Scholar]
- Hinkel R, Penzkofer D, Zühlke S, Fischer A, Husada W, Xu Q‐F, Baloch E, van Rooij E, Zeiher AM, Kupatt C & Dimmeler S (2013). Inhibition of microRNA‐92a protects against ischemia/reperfusion injury in a large‐animal model. Circulation 128, 1066–1075. [DOI] [PubMed] [Google Scholar]
- Huang Z‐P, Chen J, Seok HY, Zhang Z, Kataoka M, Hu X & Wang D‐Z (2013). MicroRNA‐22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res 112, 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E & Izaurralde E (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12, 99–110. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T & Zamore PD (2001). A cellular function for the RNA‐interference enzyme Dicer in the maturation of the let‐7 small temporal RNA. Science 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Ito T, Yagi S & Yamakuchi M (2010). MicroRNA‐34a regulation of endothelial senescence. Biochem Biophys Res Commun 398, 735–740. [DOI] [PubMed] [Google Scholar]
- Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez‐Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA & Hodges MR (2013). Treatment of HCV infection by targeting microRNA. N Engl J Med 368, 1685–1694. [DOI] [PubMed] [Google Scholar]
- Jazbutyte V, Fiedler J, Kneitz S, Galuppo P, Just A, Holzmann A, Bauersachs J & Thum T (2013). MicroRNA‐22 increases senescence and activates cardiac fibroblasts in the aging heart. Age (Dordr) 35, 747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A & Griffiths‐Jones S (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42, D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kim CW, Simmons RD & Jo H (2014). Role of flow‐sensitive microRNAs in endothelial dysfunction and atherosclerosis: mechanosensitive athero‐miRs. Arterioscler Thromb Vasc Biol 34, 2206–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara Y, Horie T, Baba O, Watanabe S, Nishiga M, Usami S, Izuhara M, Nakao T, Nishino T, Otsu K, Kita T, Kimura T & Ono K (2015). MicroRNA‐451 exacerbates lipotoxicity in cardiac myocytes and high‐fat diet‐induced cardiac hypertrophy in mice through suppression of the LKB1/AMPK pathway. Circ Res 116, 279–288. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt‐Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S & Orum H (2010). Therapeutic silencing of MicroRNA‐122 in primates with chronic hepatitis C virus infection. Science 327, 198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC & Ambros V (2001). An extensive class of small RNAs in Caenorhabditis elegans . Science 294, 862–864. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL & Ambros V (1993). The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14 . Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S & Kim VN (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom K‐H, Lee S, Baek SH & Kim VN (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23, 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Khanna A, Li N & Wang E (2011). Circulatory miR34a as an RNAbased, noninvasive biomarker for brain aging. Aging (Albany NY) 3, 985–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S‐L, Miller JD & Ying S‐Y (2006). Intronic microRNA (miRNA). J Biomed Biotechnol 2006, 26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S, Nanhwan M, Qian J, Kodakandla M, Castillo AC, Thomas B, Liu H & Ye Y (2013. a). Modulation of microRNAs in hypertension‐induced arterial remodeling through the β1 and β3‐adrenoreceptor pathways. J Mol Cell Cardiol 65, 127–136. [DOI] [PubMed] [Google Scholar]
- Ling T‐Y, Wang X‐L, Chai Q, Lau T‐W, Koestler CM, Park SJ, Daly RC, Greason KL, Jen J, Wu L‐Q, Shen W‐F, Shen W‐K, Cha Y‐M & Lee H‐C (2013. b). Regulation of the SK3 channel by microRNA‐499 – potential role in atrial fibrillation. Heart Rhythm 10, 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas F V, Marsden CG, Thomson JM, Song J‐J, Hammond SM, Joshua‐Tor L & Hannon GJ (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441. [DOI] [PubMed] [Google Scholar]
- Loffredo FS, Nikolova AP, Pancoast JR & Lee RT (2014). Heart failure with preserved ejection fraction: molecular pathways of the aging myocardium. Circ Res 115, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, Zhang Y, Shan H, Luo X, Bai Y, Sun L, Song W, Xu C, Wang Z & Yang B (2010). MicroRNA‐328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 122, 2378–2387. [DOI] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE & Kutay U (2004). Nuclear export of microRNA precursors. Science 303, 95–98. [DOI] [PubMed] [Google Scholar]
- Luo X, Pan Z, Shan H, Xiao J, Sun X, Wang N, Lin H, Xiao L, Maguy A, Qi XY, Li Y, Gao X, Dong D, Zhang Y, Bai Y, Ai J, Sun L, Lu H, Luo XY, Wang Z, Lu Y, Yang B & Nattel S (2013). MicroRNA‐26 governs profibrillatory inward‐rectifier potassium current changes in atrial fibrillation. J Clin Invest 123, 1939–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegdefessel L, Azuma J, Toh R, Merk DR, Deng A, Chin JT, Raaz U, Schoelmerich AM, Raiesdana A, Leeper NJ, McConnell MV, Dalman RL, Spin JM & Tsao PS (2012). Inhibition of microRNA‐29b reduces murine abdominal aortic aneurysm development. J Clin Invest 122, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magenta A, Greco S, Gaetano C & Martelli F (2013). Oxidative stress and microRNAs in vascular diseases. Int J Mol Sci 14, 17319–17346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder B, Backes C, Haas J, Leidinger P, Stähler C, Großmann T, Vogel B, Frese K, Giannitsis E, Katus HA, Meese E & Keller A (2014). Influence of the confounding factors age and sex on microRNA profiles from peripheral blood. Clin Chem 60, 1200–1208. [DOI] [PubMed] [Google Scholar]
- Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, Vasa‐Nicotera M, Ippoliti A, Novelli G, Melino G, Lauro R & Federici M (2009). MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 120, 1524–1532. [DOI] [PubMed] [Google Scholar]
- Menghini R, Stöhr R & Federici M (2014). MicroRNAs in vascular aging and atherosclerosis. Ageing Res Rev 17, 68–78. [DOI] [PubMed] [Google Scholar]
- Nishi H, Ono K, Iwanaga Y, Horie T, Nagao K, Takemura G, Kinoshita M, Kuwabara Y, Mori RT, Hasegawa K, Kita T & Kimura T (2010). MicroRNA‐15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J Biol Chem 285, 4920–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB & Evans MK (2010). microRNA expression patterns reveal differential expression of target genes with age. PLoS One 5, e10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossent AY, Hansen JL, Doggen C, Quax PHA, Sheikh SP & Rosendaal FR (2011). SNPs in microRNA binding sites in 3′‐UTRs of RAAS genes influence arterial blood pressure and risk of myocardial infarction. Am J Hypertens 24, 999–1006. [DOI] [PubMed] [Google Scholar]
- Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, Rippo MR, Galeazzi R, Abbatecola AM, Marcheselli F, Monti D, Ostan R, Cevenini E, Antonicelli R, Franceschi C & Procopio AD (2012). Age‐related differences in the expression of circulating microRNAs: miR‐21 as a new circulating marker of inflammaging. Mech Ageing Dev 133, 675–685. [DOI] [PubMed] [Google Scholar]
- Paneni F, Beckman JA, Creager MA & Cosentino F (2013). Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part I. Eur Heart J 34, 2436–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E & Olson EN (2010). Stress‐dependent cardiac remodeling occurs in the absence of microRNA‐21 in mice. J Clin Invest 120, 3912–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus WJ & Tschöpe C (2013). A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J Am Coll Cardiol 62, 263–271. [DOI] [PubMed] [Google Scholar]
- Quiat D & Olson EN (2013). MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest 123, 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintavalle C, Garofalo M, Croce CM & Condorelli G (2011). ‘ApoptomiRs’ in vascular cells: their role in physiological and pathological angiogenesis. Vascul Pharmacol 55, 87–91. [DOI] [PubMed] [Google Scholar]
- Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, Lodish HF & Blelloch R (2009). Loss of cardiac microRNA‐mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 105, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR & Ruvkun G (2000). The 21‐nucleotide let‐7 RNA regulates developmental timing in Caenorhabditis elegans . Nature 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Santulli G, Iaccarino G, De Luca N, Trimarco B & Condorelli G (2014. a). Atrial fibrillation and microRNAs. Front Physiol 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Wronska A, Uryu K, Diacovo TG, Gao M, Marx SO, Kitajewski J, Chilton JM, Akat KM, Tuschl T, Marks AR & Totary‐Jain H (2014. b). A selective microRNA‐based strategy inhibits restenosis while preserving endothelial function. J Clin Invest 124, 4102–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardu C, Marfella R, Santulli G & Paolisso G (2014). Functional role of miRNA in cardiac resynchronization therapy. Pharmacogenomics 15, 1159–1168. [DOI] [PubMed] [Google Scholar]
- Schober A, Nazari‐Jahantigh M & Weber C (2015). MicroRNA‐mediated mechanisms of the cellular stress response in atherosclerosis. Nat Rev Cardiol 12, 361–374. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N & Zamore PD (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208. [DOI] [PubMed] [Google Scholar]
- Seeger T, Haffez F, Fischer A, Koehl U, Leistner DM, Seeger FH, Boon RA, Zeiher AM & Dimmeler S (2013). Immunosenescence‐associated microRNAs in age and heart failure. Eur J Heart Fail 15, 385–393. [DOI] [PubMed] [Google Scholar]
- Serna E, Gambini J, Borras C, Abdelaziz KM, Mohammed K, Belenguer A, Sanchis P, Avellana JA, Rodriguez‐Mañas L & Viña J (2012). Centenarians, but not octogenarians, up‐regulate the expression of microRNAs. Sci Rep 2, 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T, Satoh M, Itoh T & Nakamura M (2012). MicroRNA‐34a regulates the longevity‐associated protein SIRT1 in coronary artery disease: effect of statins on SIRT1 and microRNA‐34a expression. Clin Sci (Lond) 123, 161–171. [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G & Hermeking H (2007). Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR‐34a is a p53 target that induces apoptosis and G1‐arrest. Cell Cycle 6, 1586–1593. [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J & Engelhardt S (2008). MicroRNA‐21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984. [DOI] [PubMed] [Google Scholar]
- Tijsen AJ, van der Made I, van den Hoogenhof MM, Wijnen WJ, van Deel ED, de Groot NE, Alekseev S, Fluiter K, Schroen B, Goumans M‐J, van der Velden J, Duncker DJ, Pinto YM & Creemers EE (2014). The microRNA‐15 family inhibits the TGFβ‐pathway in the heart. Cardiovasc Res 104, 61–71. [DOI] [PubMed] [Google Scholar]
- Tritsch E, Mallat Y, Lefebvre F, Diguet N, Escoubet B, Blanc J, De Windt LJ, Catalucci D, Vandecasteele G, Li Z & Mericskay M (2013). An SRF/miR‐1 axis regulates NCX1 and annexin A5 protein levels in the normal and failing heart. Cardiovasc Res 98, 372–380. [DOI] [PubMed] [Google Scholar]
- Van Almen GC, Verhesen W, van Leeuwen REW, van de Vrie M, Eurlings C, Schellings MWM, Swinnen M, Cleutjens JPM, van Zandvoort MAMJ, Heymans S & Schroen B (2011). MicroRNA‐18 and microRNA‐19 regulate CTGF and TSP‐1 expression in age‐related heart failure. Aging Cell, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rooij E, Marshall WS & Olson EN (2008. a). Toward MicroRNA‐based therapeutics for heart disease: the sense in antisense. Circ Res 103, 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J & Olson EN (2007). Control of stress‐dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579. [DOI] [PubMed] [Google Scholar]
- Van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA & Olson EN (2008. b). Dysregulation of microRNAs after myocardial infarction reveals a role of miR‐29 in cardiac fibrosis. Proc Natl Acad Sci U S A 105, 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J‐X, Jiao J‐Q, Li Q, Long B, Wang K, Liu J‐P, Li Y‐R & Li P‐F (2011). miR‐499 regulates mitochondrial dynamics by targeting calcineurin and dynamin‐related protein‐1. Nat Med 17, 71–78. [DOI] [PubMed] [Google Scholar]
- Watson CJ, Gupta SK, O'Connell E, Thum S, Glezeva N, Fendrich J, Gallagher J, Ledwidge M, Grote‐Levi L, McDonald K & Thum T (2015). MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail 17, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen WJ, van der Made I, van den Oever S, Hiller M, de Boer BA, Picavet DI, Chatzispyrou IA, Houtkooper RH, Tijsen AJ, Hagoort J, van Veen H, Everts V, Ruijter JM, Pinto YM & Creemers EE (2014). Cardiomyocyte‐specific miRNA‐30c over‐expression causes dilated cardiomyopathy. PLoS One 9, e96290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wronska A, Kurkowska‐Jastrzebska I & Santulli G (2015). Application of microRNAs in diagnosis and treatment of cardiovascular disease. Acta Physiol (Oxf) 213, 60–83. [DOI] [PubMed] [Google Scholar]
- Ye Y, Hu Z, Lin Y, Zhang C & Perez‐Polo JR (2010). Downregulation of microRNA‐29 by antisense inhibitors and a PPAR‐γ agonist protects against myocardial ischaemia–reperfusion injury. Cardiovasc Res 87, 535–544. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Chen Z, Santulli G, Gu L, Yang Z‐G, Yuan Z‐Q, Zhao Y‐T, Xin H‐B, Deng K‐Y, Wang S‐Q & Ji G (2014). Functional role of Calstabin2 in age‐related cardiac alterations. Sci Rep 4, 7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard J‐M, Mayr A, Weger S, Schett G, Shah A, Boulanger CM, Willeit J, Chowienczyk PJ, Kiechl S & Mayr M (2012). Prospective study on circulating microRNAs and risk of myocardial infarction. J Am Coll Cardiol 60, 290–299. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y & Fu X‐D (2014). MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo R, Fu S, Li S, Yao M, Lv D, Xu T & Bei Y (2014). Desregulated microRNAs in aging‐related heart failure. Front Genet 5, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]


