Abstract
The prominent role of voltage-gated sodium channel 1.7 (Nav1.7) in nociception was revealed by remarkable human clinical and genetic evidence. Development of potent and subtype-selective inhibitors of this ion channel is crucial for obtaining therapeutically useful analgesic compounds. Microproteins isolated from animal venoms have been identified as promising therapeutic leads for ion channels, because they naturally evolved to be potent ion channel blockers. Here, we report the engineering of highly potent and selective inhibitors of the Nav1.7 channel based on tarantula ceratotoxin-1 (CcoTx1). We utilized a combination of directed evolution, saturation mutagenesis, chemical modification, and rational drug design to obtain higher potency and selectivity to the Nav1.7 channel. The resulting microproteins are highly potent (IC50 to Nav1.7 of 2.5 nm) and selective. We achieved 80- and 20-fold selectivity over the closely related Nav1.2 and Nav1.6 channels, respectively, and the IC50 on skeletal (Nav1.4) and cardiac (Nav1.5) sodium channels is above 3000 nm. The lead molecules have the potential for future clinical development as novel therapeutics in the treatment of pain.
Keywords: directed evolution, pain, sodium channel, structure-function, toxin, ceratotoxin, Nav1.7, engineering
Introduction
Ion channels are an important class of targets for drugs with critical clinical need. Not only do channelopathies underlie a wide variety of human disorders, such as pain, cardiac arrhythmias, skeletal muscle myotonia, paralysis, epilepsy, cystic fibrosis, as well as other neurological diseases, but modifying ion channel function is a useful clinical intervention strategy in states such as pain, even when there is no known underlying defect in ion channel function. Pain represents a major global health issue and its management is still far from optimal. Consequently, there is a significant unmet clinical need for new analgesics with higher efficacy and reduced side effects (1).
Ample clinical and genetic evidence in humans has established the voltage-gated sodium channel 1.7 (Nav1.7) as a major player in peripheral pain pathways. Although loss-of-function mutations in Nav1.7 lead to complete inability to perceive pain (2, 3), gain-of-function of Nav1.7 results in debilitating painful disorders (4–7). Based on all available evidence, Nav1.7 is currently recognized as a prominent target for the development of new analgesic drugs, although recent evidence suggests additional involvement in regulating endogenous opioid systems (8, 9).
Nav1.7 belongs to the family of voltage-gated sodium channels that includes nine known members (Nav1.1–Nav1.9) with specific tissue localization and functional differences. These channels are vitally important for nerve conduction in the central and peripheral nervous system, skeletal muscle, and in the heart. Because they share the same domain structure and display overall amino acid sequence similarity, significant challenges exist in developing subtype-selective sodium channel inhibitors.
For these reasons, the search for novel Nav1.7 inhibitors has expanded from small molecules into biologics (10, 11). Venom toxins, in particular, have been identified as promising therapeutic leads, because they are generally more potent and selective than small molecules (12–14). Toxins from terrestrial and aquatic venomous species that influence sodium channel function fall broadly into two classes, according to their primary chemical composition: peptidic and non-peptidic compounds (15). Small peptide toxins (microproteins) are significant components of the complex biologically active mixtures present in animal venoms. Their distinct disulfide-bridged compact structure provides unique physicochemical stability and high resistance to proteolytic degradation (16, 17).
Among venomous animals, spiders represent by far the largest group of venomous species and are known to possess the most extensive array of microproteins with high potential therapeutic value (18). The remarkable preponderance of potent and selective ion channel modulators brought spider venom-derived peptides to the forefront of discovery for novel lead compounds to treat a variety of disorders (17). Although there are ∼73 identified spider microproteins with activity against vertebrate sodium channels, only some of these have been shown to have some activity toward Nav1.7, albeit their numbers are growing (19, 20). Several studies were undertaken recently with the aim of improving potency, selectivity, stability, and bioavailability of venom-derived lead peptide compounds targeting Nav1.7 (19, 21–23).
Here, we describe the generation of potent and selective molecules to block Nav1.7 with improved therapeutic properties. We selected ceratotoxin-1 (CcoTx1) (24), an inhibitor of neuronal sodium channels isolated from the venom of the tarantula Ceratogyrus cornuatus, as a starting point to create CcoTx1 variants. We applied a combination of directed evolution, saturation mutagenesis, structure-activity relationship, and chemical modification to create potent and selective microprotein inhibitors of the Nav1.7 sodium channel.
Experimental Procedures
Design of Phylogenetic Libraries and Production of Microproteins by Recombinant Expression
A total of 13 original libraries were designed following phylogenetic approach, with low to high diversities (2–4 mutations per clone) and analyzed before expression. Next, round of libraries were designed based on the best active clones from round 1. To generate the library, DNA shuffling was performed according to the method described previously (25). Briefly, degenerate primers incorporating a number of possible amino acids at specific positions representing active microproteins against the selected targets were generated. The primers are then introduced into an expression plasmid, and after expression and purification, microproteins are subjected to activity screening against the target. The best performing clones are then re-shuffled in a second evolution cycle, but then also tested against off-targets, thus obtaining data both on potency and selectivity.
Microproteins were cloned into in-house vector with T7 promoter OmpA for periplasmic expression, cellulose-binding domain as purification tag, tobacco etch virus protease splicing site, a small linker (GSGG), and the microprotein. They were expressed in Escherichia coli BL21 and induced with isopropyl 1-thio-β-d-galactopyranoside when the A600 ≥0.6, and expression was continued for additional 48 h at 26 °C before harvesting. The libraries were expressed in 24-well plates and centrifuged, and the supernatant was loaded into cellulose beads (Perloza MT100). The beads were washed with TBS, and tobacco etch virus protease was added to cleave the microproteins from the beads. Eluted samples were then run on a SDS-PAGE, and concentration was determined by SDS-PAGE staining. The proteins with good expression were selected, and their concentration was normalized and later tested for activity on sodium channels by electrophysiology. The most active microproteins were expressed in 500 ml of culture and further purified by size exclusion chromatography (AKTA Prime System). They were then characterized by SDS-PAGE, reverse phase chromatography (C8 column), and MALDI-TOF.
Production of Microproteins by Cell-free Expression
To allow for production of the microproteins without N-terminal methionine, as well as to facilitate their purification, the proteins were cloned as His6-SUMO11 fusions, which could be cleaved into the mature microprotein by SUMO proteinase (ULP) (26). Cell extract preparation, plasmid construction, and cell-free expression were carried out as described previously by Malakhov et al. and Zawada et al. (26, 27). Cell-free reactions were carried out in 48-well FlowerPlates (m2p labs) under shaking. Post cell-free expression, the microprotein fusions were purified via high throughput nickel-immobilized metal affinity chromatography using PhyTips (PhyNexus) according to the manufacturer's recommendations. The elution pools were buffer exchanged into 274 mm NaCl, 8 mm KCl, 10 mm HEPES, 3.8 mm CaCl2, 2 mm MgCl2, 20 mm glucose, 20 mm sucrose, pH 7.4, via desalting on Zeba Spin Desalting Plates (Thermo Fisher, catalog no. 89807) and subsequently treated with SUMO protease (Invitrogen, catalog no. 12588-018) overnight. SUMO protease and cleaved His6-SUMO were separated from the microprotein by scavenging chromatography on Q-Sepharose Fast Flow (GE Healthcare) by addition of Q-resin slurry to the microplates containing the desalted and SUMO-cleaved microprotein. Finally, the purified microprotein pools, recovered from the slurry supernatant, were adjusted to 137 mm NaCl, 4 mm KCl, 10 mm HEPES, 1.8 mm CaCl2, 1 mm MgCl2, 10 mm glucose, 10 mm sucrose, pH 7.4, by 2-fold dilution with 10 mm HEPES, pH 7.4, to match the buffer conditions of the QPatch electrophysiology assay.
Production of Microproteins by Chemical Synthesis and Oxidative Refolding
Microprotein variants were chemically synthesized by Elimbio by standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) peptide synthesis protocols and delivered as HPLC-purified lyophilized product. Subsequently, lyophilized microproteins were solubilized in 100 mm Tris, pH 8.0, 150 mm NaCl at a concentration of about 0.5 mg/ml (∼125 μm). A mixture of reduced (G4705 Sigma) and oxidized glutathione (G4376 Sigma) (GSSG) was added for oxidative refolding to a final concentration of 2 mm each. After incubating the solution at 4 °C overnight, the pH was adjusted to pH ∼3 by adding TFA (roughly 1–2%). The refolded peptide was purified on a semi-preparative column Vydac 4.6-mm C18 using a gradient from 10 to 60% buffer B over 65 min (0.1% TFA; 90% acetonitrile). Fractions corresponding to the main peak were selected and tested for activity by electrophysiology. Large scale preparations were performed based on the described protocol by CPC Scientific. The molecular masses of the microproteins were as follows: CcoTx1, 4045.6 Da; 2670; 3970.7 Da; D1I, 3967.8 Da; D1Ia, 3968.8 Da; D1Za, 3964.8 Da; D1Z/M5Ia, 3946.7 Da; D1Z/M5I/R27N,a, 3904.7 Da; and D1Z/M5I/K18Y/R24K,a, 3953.7 Da. During the biophysical characterization of CcoTx1 variants, we noticed the presence of oxidized methionine; the oxidation increased over time and had a negative impact on the potency of the microproteins. For this reason, we replaced methionine at position 5 with isoleucine to remove the oxidation liability, resulting in D1Z/M5Ia variant. This substitution was selected based on the saturation mutagenesis data and had little impact on potency and selectivity. No oxidation of tryptophan residues was observed under tested conditions.
Crystallization and Structural Determination
Monoclonal antibody 6F1 was generated against the leading clone 2670 by standard hybridoma methods (28). Fab 6F1 was mixed 1:1 with 2670 to form the complex, and the complex was purified by size exclusion chromatography. The 6F1-Fab·2670 complex was concentrated to 7 mg/ml and mixed with reservoir solution at 1:1 volume ratio for crystallization by the hanging drop vapor diffusion method at 22 °C. Reservoir solution was 0.1 m zinc acetate, 0.08 m sodium cacodylate, pH 6.5, 14% polyethylene glycol 8000, and 20% glycerol. Crystals were directly flash-frozen in liquid nitrogen prior to data collection. X-ray diffraction data from crystals were collected at 100 K at the Advanced Light Source (ALS) beamline 5.0.2 at Lawrence Berkeley National Laboratory. The crystals belong to the space group P212121 with unit cell dimensions a = 97.32 Å, b = 98.44 Å, and c = 107.35 Å and diffracted to 1.75 Å (supplemental Table 1). All diffraction data were processed with DENZO and SCALEPACK (29). The structure of the 6F1-Fab·2670 complex was solved by molecular replacement using mouse IgG1 (PDB code 2VL5) and Hainantoxin-IV (PDB code 1NIY) as search models using PHASER (30). Model construction and rebuilding were performed using COOT (31). The structure of the 6F1-Fab·2670 complex was refined using REFMAC5 (32) in the CCP4 software suite (33), which reduced the R and Rfree values to 19.03 and 21.89%, respectively. The coordinates for the structure have been deposited in the PDB (PDB code 5EPM).
Electrophysiology
Ion channel currents were recorded using either an automated patch clamp system QPatch HT (Sophion, Biolin Scientific, Denmark) or a conventional patch clamp setup employing an EPC 10 patch clamp amplifier (HEKA Electronik GmbH, Germany).
QPatch Electrophysiology
Both single-hole and 10-hole QPlates with an integrated 250-μl waste container (QPlate 48L or 48X) were used enabling washes and application of multiple concentrations/compounds per well. Compounds were added to the cells with the eight pipettes via the QPlate integrated glass microfluidic pathways. For selectivity screens on ligand-gated channels, 5 μl of ligand was added for 3 s followed by washout (six times 5 μl of saline). Test compounds were typically preincubated for 345 s prior to agonist addition. The ligand-induced currents were acquired at 5 kHz for 5 s. During whole-cell recording, the membrane potential was held at −90 mV. The extracellular recording solution contained (in mm) 137 NaCl, 4 KCl, 10 HEPES, 1.8 CaCl2, 1 MgCl2, 10 glucose, 10 sucrose, adjusted to pH 7.4. The intracellular solution contained (in mm) 130 CsF, 10 NaCl, 5 EGTA, 10 HEPES, 4 Na2ATP, 0.1 Tris-GTP, adjusted to pH 7.2. All QPatch data were analyzed using Sophion's QPatch Assay software. Results are presented as means ± S.E.
Manual Patch Clamp Electrophysiology
Coverslips carrying cells were placed in a recording chamber and were constantly perfused at a rate of 0.5 ml/min with extracellular solution containing (in mm) 140 NaCl, 5 KCl, 2 CaCl2, 2 MgCl2, 10 HEPES, and 10 glucose (pH 7.4, 320 mosmol). Patch electrodes were pulled from borosilicate glass and fire-polished to 3–10-megohm tip resistance. The internal pipette solution (ATP-regenerating solution, pH 7.3, 300 mosmol) consisted of (in mm) 100 KCl, 2 MgCl2, 5 MgATP, 0.1 GTP, 15.5 phosphocreatine, 50 units/ml creatine phosphokinase, 10 1,2-bis(2-aminophenoxy)ethane-N,N,N,N-tetraacetic acid, tetrapotassium salt, and 10 HEPES. All recordings were made at room temperature (22–24 °C) using an EPC 10 patch clamp amplifier (HEKA Electronik GmbH, Germany). Cells were voltage-clamped at −90 mV, and control test responses were routinely recorded for a minimum of 10 min to ensure that the amplitude and kinetics of the response were stable. Data acquisition and analysis were performed with Pulse-PulseFit (HEKA Electronik GmbH, Germany) and IgorPro (WaveMetrics, Portland, OR). Compounds were applied to individual cells using a gravity-fed perfusion system. All reagents were purchased from Sigma unless otherwise noted. All data are expressed as means ± S.E.
Cell Culture
Cell lines recombinantly expressing particular ion channels were obtained from Pfizer's own cell line depository. Cells were thawed from frozen stocks and were grown in the appropriate culture media supplemented with required selection antibiotics in 75-cm2 Falcon culture flasks in an incubator at 37 °C and 6% CO2 with saturating humidity. The cells were used for electrophysiological recordings after 2–3 days culturing. For an automated patch clamp recording on the QPatch HT system, the cells were dissociated with enzymatic detachment solution Detachin (Genlantis, San Diego) and resuspended in serum-free medium supplemented with 25 mm HEPES and 1% penicillin/streptomycin (Mediatech, Manassas, VA). HEK293 cells stably expressing human Nav1.7 were grown in DMEM (catalog no. 10-01-CM, Mediatech, Manassas, VA), 10% FBS (JRScientific, Woodland, CA), 500 μg/ml G418 sulfate (Mediatech).
Results
Overview of the Microprotein Engineering Process
To engineer microproteins with high potency and selectivity toward Nav1.7, we utilized a combination of directed evolution (25), saturation mutagenesis (34), and chemical modification. We first established CcoTx1 as our starting molecule for directed evolution and developed a strategy for potency and selectivity screening. Next, we applied multiple cycles of directed evolution to generate molecules with better potency and specificity to Nav1.7. After each cycle, clones were tested on their activity toward Nav1.7 as well as Nav1.2 and Nav1.6 for selectivity. A careful evaluation of the amino acid sequences and the IC50 data resulted in the selection of variant 2670 as the molecule with the most improved characteristics. To explore the sequence space not sampled by the directed evolution, we performed an exhaustive saturation mutagenesis on the 2670 variant. We solved the structure of 2670 and analyzed the comprehensive mutagenesis data in the context of the structure. In addition to amino acid sequence variations, we also explored the effects of post-translational modifications such as C-terminal amidation and N-terminal pyroglutamate. Finally, we selected point mutations based on saturation mutagenesis data and combined them by utilizing the determined structure to yield the final candidates. Each step of this multipronged strategy is described in more detail below.
Establishing CcoTx1 as Starting Molecule for Directed Evolution
To identify the best starting molecules for directed evolution, we tested the activity and expression in E. coli of 16 naturally occurring microproteins known to have activity against voltage-gated sodium channels. Because of the difficulty in expressing highly disulfide-linked microproteins, we explored several expression strategies, fusion proteins, and expression conditions. The most promising approach we found was to utilize the cellulose-binding domain as the fusion protein connected by GSGG linker at the N terminus of the venom-derived peptide. Four microproteins (CcoTx1, Huwentoxin-4, Hainantoxin-4, and Phrixotoxin-3) showed good expression, folding, and activity against Nav1.7 channel and were selected for initial directed evolution experiments (Fig. 1A).
FIGURE 1.
A, amino acid sequence alignment of microproteins included in the initial screening. CcoTx1 and three other molecules were selected for designing the initial libraries on which directed evolution was performed. B, schematic diagram of Nav1.7 with general domain structure. The locations of the four HA tag constructs (M1–M4) that retained a functional Nav1.7 channel are shown. M1 denotes insertion of the GSYPYDVPDYAGS sequence after residue 770, M2 after residue 829, M3 after residue 902, and M4 after residue 1540 (Uniprot Q15858). C, time course of Nav1.7 current block by 250 nm CcoTx1 in HEK293 cells transiently transfected with HA tag-modified Nav1.7 channels. HA tag insertion into the channel extracellular loops S1-S2/D2 (construct M1), S5-S6/D2 (construct M3), or S1-S2/D4 (construct M4) has no influence on CcoTx1 inhibition; however, HA tag insertion into the extracellular loop S3-S4/D2 (construct M2) abolishes the blocking activity of CcoTx1. The manual whole-cell patch clamp technique was used to record Nav1.7 currents. Nav1.7 currents were evoked by a 15-ms step depolarization to 0 mV every 10 s from a holding potential of −90 mV. Data are presented as normalized peak current amplitude versus time. Currents were normalized to the maximum amplitude of control peak current. The solid line indicates the time of compound application. D, amino acid sequence alignment of the S3-S4 region of domain 2 (D2) for several sodium channels (Nav1.1–Nav1.8). Main differences in Nav1.7 are highlighted.
Among the tested microproteins, CcoTx1 emerged as the most promising starting point due to its well behaved expression and good selectivity toward Nav1.4 and Nav1.5 channels. Selectivity against Nav1.4 and Nav1.5 is critical due to their predominant expression in skeletal (Nav1.4) and cardiac (Nav1.5) muscles. CcoTx1 was therefore used as the backbone for the construction of the initial phylogenetic libraries (see under “Experimental Procedures”).
Selectivity Counter-screening Strategy
In hope to better understand the molecular mechanisms of the interaction between CcoTx1 and Nav1.7 and guide the selection of additional channels for selectivity screening, we set out to identify the binding site of CcoTx1 on the Nav1.7 channel. To this end, we introduced HA tags (YPYDVPDYA sequence) at selected locations in the extracellular loops of the Nav1.7 channel (Fig. 1B) and tested channel activity in the presence and absence of CcoTx1. Out of the Nav1.7 constructs that retained activity, CcoTx1 was no longer able to inhibit the Nav1.7 construct M2, indicating that CcoTx1 likely interacts with the S3-S4 loop of domain DII (also called site 4). The same site has been identified also for other toxins (35–43).
Based on the local amino acid sequence differences in the DII domain, overall amino acid sequence conservation, as well as lack of selectivity of CcoTx1 toward Nav1.2 and Nav1.6, we selected these two channels for routine selectivity screening to assess the feasibility of developing subtype-selective Nav1.7 inhibitors.
Sodium channels subtypes Nav1.3, Nav1.8, and Nav1.9 have also been reported to be involved in pain signaling. Inhibition of these channels might be a desirable off-target effect,, and therefore we did not include these channels in routine selectivity screening. CcoTx1 showed only very modest inhibition of Nav1.4 and Nav1.5 in our initial experiments; therefore, these channels were only intermittently monitored for undesired activity. In addition to sodium channels, we also tested selectivity against an extensive panel of other ion channels (Table 1) to ensure that other classes of ion channels were not targeted by the engineered microproteins.
TABLE 1.
Broad selectivity screening of D1Ia
| Channel | Concentration | IC50 | % block | n |
|---|---|---|---|---|
| Nav1.7 | 0.1; 1; 10; 100 nm | 2 ± 0.3 nm | 33 | |
| Nav1.7 (monkey) | 0.1; 1; 10; 100 nm | 6 ± 1 nm | 10 | |
| Nav1.7 (dog) | 0.1; 1; 10; 100 nm | 3 ± 0.5 nm | 7 | |
| Nav1.7 (rat) | 0.1; 1; 10; 100 nm | 6 ± 1 nm | 11 | |
| Nav1.7 (mouse) | 0.1; 1; 10; 100 nm | 3 ± 1 nm | 5 | |
| Nav1.1 | 1; 10; 100; 500 nm | 60 ± 10 nm | 9 | |
| Nav1.1 (rat) | 0.1; 1; 10; 100 nm | 30 ± 8 nm | 8 | |
| Nav1.2 | 1; 10; 100; 500 nm | 50 ± 14 nm | 5 | |
| Nav1.3 | 0.1; 1; 10; 100 nm | 42 ± 5 nm | 8 | |
| Nav1.4 | 1000 nm | 21 ± 5 | 7 | |
| Nav1.5 | 1000 nm | >1 μm | 42 ± 7 | 4 |
| Nav1.6 | 1; 10; 100; 500 nm | 20 ± 2 nm | 3 | |
| Nav1.8+β1 | 1000 nm | >1 μm | 15 ± 8 | 10w |
| hERG | 1000 nm | −0.4 ± 4 | 3 | |
| Kv1.3 | 1000 nm | −8 ± 6 | 5 | |
| Kv1.5 | 1000 nm | −1 ± 2 | 2 | |
| Kir2.1 | 1000 nm | −4 ± 4 | 5 | |
| Kv4.3/KChIP2.2 | 10; 100; 500; 1000 nm | 180 ± 30 nm | 6 | |
| KvLQT1/minK | 1000 nm | 13 ± 3 | 5 | |
| KCNQ2/3 | 1000 nm | 1 ± 5 | 4 | |
| KCNQ3/5 | 1000 nm | 1 ± 2 | 8w | |
| KCNQ4 | 1000 nm | 0 ± 2 | 7w | |
| BK1 | 1000 nm | −5 ± 8 | 5 | |
| SK2 | 1000 nm | 5 ± 3 | 4 | |
| SK3 | 1000 nm | 8 ± 4 | 4 | |
| IK1 | 1000 nm | 3 ± 4 | 8w | |
| Kv1.1/1.2 | 1000 nm | 2 ± 3 | 3 | |
| Cav1.2 | 1000 nm | 41 ± 4 | 5 | |
| Cav2.2 | 1000 nm | 18 ± 6 | 3 | |
| HCN2 | 1000 nm | −0.3 ± 2 | 4 | |
| HCN4 | 1000 nm | −3 | 1 | |
| ASIC1a | 1000 nm | 17 ± 6 | 8 | |
| ASIC3 | 1000 nm | −12 ± 2 | 3 | |
| α7nAchR | 1000 nm | −25 ± 9 | 3 | |
| P2X2 TT | 1000 nm | 10 ± 12 | 4 | |
| TRPA1 | 1000 nm | 36 ± 5 | 8w | |
| TRPM8 | 1000 nm | 8 ± 5 | 2 | |
| TRPV1 | 1000 nm | 9 ± 5 | 10w | |
| TRPV6 | 1000 nm | −7 ± 6 | 4 |
Generation of a Potent Inhibitor to Nav1.7 by Directed Evolution
To improve potency and selectivity of CcoTx1, we applied three rounds of directed evolution. During the first round, we generated several protein libraries based on CcoTx1 with diversity introduced based on three other molecules (Huwentoxin-4, Hainantoxin-4, and Phrixotoxin-3). More than five thousand microproteins from the initial libraries were expressed and purified in high throughput format and tested for Nav1.7 channel inhibition activity at a single concentration of 100 nm using an automated patch clamp system (QPatch). Overall, 184 microproteins from the first cycle of directed evolution (round 1) showed activity comparable with or better than wild-type CcoTx1. IC50 values were then determined for the active microproteins in dose titration studies using QPatch. Finally, microproteins with confirmed potency to Nav1.7 were tested for selectivity to other voltage-gated sodium channels.
One of the criteria for selecting CcoTx1 as a starting point was its good selectivity to Nav1.4 and Nav1.5. We therefore continued to monitor the lack of activity toward Nav1.4 and Nav1.5 but utilized the closely related Nav1.2 and Nav1.6 channels for stringent selectivity screening. The activity of the selected variants on Nav1.7, Nav1.2, and Nav1.6 from round 1 are shown in Fig. 2, A and B (black dots). Compounds with increased potency to Nav1.7 and decreased potency to Nav1.2 and Nav1.6 were selected for a second cycle of directed evolution. Phylogenetic libraries were built based on the best clones, and the screening process was repeated as described in round 1. Rounds 1 and 2 (Fig. 2, A and B, black and blue dots) produced a limited number of more potent and selective clones. Therefore, we decided to introduce additional diversity at selected positions in the library (round 3; see under “Experimental Procedures” for details). This appeared to help to gain additional potency (Fig. 2, A and B, red dots) and suggested that introducing additional diversity not represented in the phylogenetic tree might be beneficial.
FIGURE 2.
A, potency scatter plot (IC50 Nav1. 7 versus IC50 counter-screening channel Nav1.2) of molecules generated by directed evolution experiments. B, same plot as in A, but with Nav1.6 as counter-screening channel. The starting molecule (CcoTx1) and the molecule selected for saturation mutagenesis and optimization (2670) are depicted in yellow. Increased potency and selectivity are highlighted by black arrows. The three rounds of directed evolution are color-coded black (round 1), blue (round 2), and red (round 3). C–F, certain amino acids that increase selectivity are preferentially found in some positions. Position-specific potency scatter plots are shown (Nav1.7 versus Nav1.2), where each compound's potency on the two channels is represented with a letter corresponding to the amino acid at that specific position. Positions 5, 12, and 20 (C and D) show good separation between amino acids, suggesting clear selectivity preferences. The plots indicate that a methionine achieves better selectivity than tryptophan at position 5; glutamate achieves better selectivity than lysine at position 12, and arginine achieves better selectivity than tyrosine at position 20. In contrast, at position 32 (F), no strong separation is observed between aspartate and lysine.
Surprisingly, analysis of potency and selectivity data on a per position basis (Fig. 2, C–F) showed that, even in the context of multiple mutations per molecule, selectivity trends and preferences of some amino acids could be observed. For example at position 5, methionine showed better selectivity toward Nav1.7 over Nav1.2 relative to tryptophan (Fig. 2C). Similar observations could also be made for glutamine at position 12 (Fig. 2D) and arginine at position 20 (Fig. 2E). Not all positions showed preferences as exemplified by position 32 where neither aspartate nor lysine can be associated with high selectivity (Fig. 2F). A comprehensive analysis of amino acids at all positions was not possible at this stage due to the biased sampling of the sequence space by the phylogenetic shuffling approach and limitations on the number of potency and selectivity measurements. Hence, we selected one of the best molecules from round 3 of the directed evolution approach (named 2670) and exhaustively explored the sequence space by single amino acid substitution saturation mutagenesis.
Optimization of 2670 Properties by Saturation Mutagenesis
To further improve potency and selectivity to Nav1.7, we decided to switch to a semi-rational approach based on saturation mutagenesis and structure-activity relationship. To take full advantage of the saturation mutagenesis, we switched to a cell-free expression system that allowed more control of the expression and folding conditions and resulted in a higher percentage of properly folded and active variants (see “Experimental Procedures”).
When designing the strategy for saturation mutagenesis, we avoided modifying or introducing additional cysteines to retain the proper disulfide structure of these microproteins. More specifically, we mutated all of the 27 non-cysteine amino acids to all the other 18 amino acids for a total of 486 compounds. About 93% (453 out of 486) of the designed compounds expressed at a workable concentration (>10 nm) in a 96-well format and were tested for activity on Nav1.7 using an automated patch clamp system QPatch (Fig. 3). Approximately 60% (274 compounds) of the molecules showed IC50 of 100 nm or better against Nav1.7 channel suggesting good structural robustness of the CcoTx1 scaffold.
FIGURE 3.
Results of saturation mutagenesis are expressed as a potency table against Nav1.7 for all single mutant variants screened. Potency (IC50 in nm) is color-coded from red to blue. The color scheme is selected such that the IC50 improvement is red, worsening is blue, and white is no change with respect to initial compound 2670. Light gray shows cysteines that were not mutated in the study. Dark gray shows positions with no data due to low or no expression. Positions with mainly blue cells (such as positions Met-5, Phe-6, Ser-23, Trp-28, Lys-30, and Trp-31) are likely to form the interface with the channel, although positions with mainly white cells (Asp-10, Glu-12, Asn-13, and Lys-15) are likely to be less involved in the interaction. Finally, positions with red cells (Asp-1, Ser-8, and Ser-25) represent mutations that improve the potency of the molecule. The top of the table shows the amino acid sequence of the 2670 microprotein together with the connectivity of the disulfide bridges.
Analysis of the mutants showed that 83 compounds had improved potency compared with 2670 (better than 25 nm), and 14 of these compounds had potency better than 12.5 nm. The substitutions in these 14 compounds were distributed among five positions as follows: Asp-1 (substitution to Tyr, Phe, Ile, and Thr), Gly-4 (substitutions to positively charged residues Arg, Lys, and His), Ser-8 (Phe and Arg), Val-21 (Ile), and Ser-25 (Trp, Tyr, Arg, and Lys). The fact that several pairs of chemically similar amino acids show up in this list (Tyr/Phe for Asp-1, Arg/Lys for Gly-4, Tyr/Trp and Arg/Lys for Ser-25) is an indicator of the robustness of the generated data. The activity table (Fig. 3) indicates that substitutions in the C-terminal region of the molecule, especially positions 23, 28, and 30–33, are much less tolerated, whereas mutations in the central part of the amino acid sequence, positions 10–15, are well tolerated. Other positions where substitutions are not very well tolerated are at positions 3, 5, 6, 20, and 21.
Compounds with good potency to Nav1.7 (IC50 <100 nm), were counter-screened against sodium channels Nav1.2 and Nav1.6 using an automated patch clamp system QPatch (supplemental Fig. 1, A and B). Overall, most compounds presented a reduced potency to Nav1.2 and Nav1.6. Specifically, the IC50 value for Nav1.2 ranged from 23.6 to 1460 nm with an average IC50 value of 275 nm, almost twice the Nav1.2 IC50 value for 2670 (146 nm). Similarly for Nav1.6, the IC50 value ranged from 15.5 to 891 nm with a population average of 128 nm slightly higher than the Nav1.6 IC50 value of 2670 (92 nm). We found 39 compounds with better potency to Nav1.2 and there were 79 with better potency to Nav1.6.
We also evaluated these data in terms of fold selectivity rather than IC50 values (supplemental Fig. 2, A and B) and compared this to the starting 2670 microprotein. The fold selectivity was calculated by dividing the IC50 for Nav1.2 or Nav1.6 by the IC50 on Nav1.7. Through this analysis we identified 63 compounds with improved selectivity for Nav1.7 over Nav1.2. Out of these compounds, 21 were better by a factor of 2 or more. Similarly, there were 108 compounds with improved selectivity for Nav1.7 over Nav1.6 with 24 having selectivity improved by a factor of 2 or more. Out of these 24, 14 have also improved selectivity to Nav1.7 over Nav1.2 channel.
Effects of Post-translational Modification on Potency and Selectivity of D1I
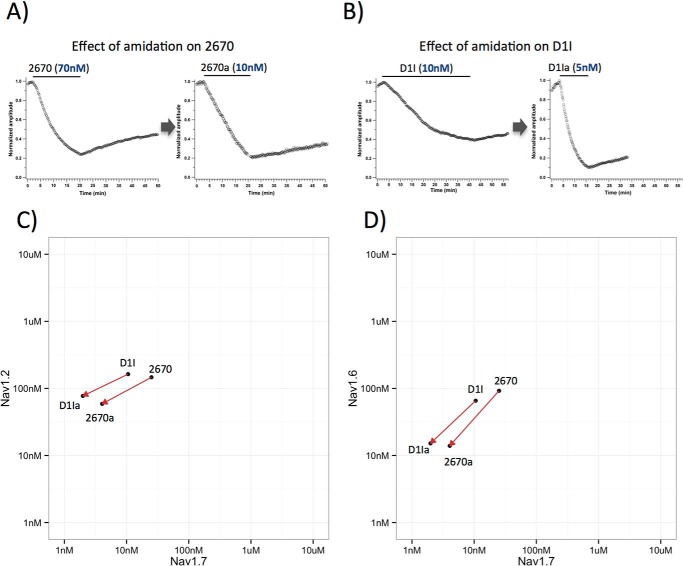
Before starting to combine single amino acid substitutions to further improve potency of the lead molecule, we investigated chemical modifications known to affect the activity of naturally occurring microproteins. Amidation of the C terminus is known to affect activity for several of the microproteins isolated from spider and snake venoms (35). Our saturation mutagenesis data show that the C terminus of 2670 does not tolerate many substitutions and likely interacts directly with the Nav1.7 channel. In contrast, extension of the C terminus with additional amino acids did not significantly affect its activity (data not shown). It was therefore unclear whether C-terminal amidation of lead compounds would have any effect on potency and selectivity. To investigate the effect of amidation on CcoTx1-like microproteins, we turned to chemical synthesis followed by refolding to generate an amidated 2670 (denoted as 2670a). In addition we selected a molecule named D1I from the saturation mutagenesis study. D1I is a variant with a better activity and selectivity profile and is identical to 2670 except that the aspartate at the first position is replaced by isoleucine. Its amidated version is referred to as D1Ia. The addition of a C-terminal amidation to both 2670 and D1I significantly increased their potency to Nav1.7 by a factor of 6.2 (from 25.1 ± 1.3 to 4.1 ± 1.7 nm) and 5.3 (from 10.6 ± 1.8 to 2.0 ± 0.2 nm), respectively (Fig. 4, A and B). When we introduced C-terminal amidation in other compounds of the CcoTx1-like family (data not shown), we also noticed a similar boost in potency by a factor of 5–10.
FIGURE 4.
A and B, time course of Nav1.7 current block by the non-amidated and amidated variants of compounds 2670 and D1I in HEK293 cells stably expressing the human Nav1.7 channel. The manual whole-cell patch clamp technique was used to record Nav1.7 currents. Nav1.7 currents were evoked by a 15-ms step depolarization to 0 mV every 10 s from a holding potential of −90 mV. Currents were normalized to the maximum amplitude of control peak current. Data presented as normalized peak current amplitude versus time. The time of compound application is indicated by solid line. C and D, effect of amidation for 2670 and D1I illustrated in selectivity plots. Amidation increases potency for both compounds on Nav1.7 but also on Nav1.2 and Nav1.6. Improvement in potency is accompanied by selectivity benefits against Nav1.2 but not Nav1.6.
We next asked the question whether the effect of the C-terminal amidation is specific for Nav1.7 or whether it translates in a similar manner to other Nav channels. Potency for Nav1.2 increases upon amidation by a factor of 2.5 for 2670 (from 146.3 ± 9.9 to 58.8 ± 9.4 nm) and by a factor of 2.1 for D1I (from 162.9 ± 52 nm to 77.3 ± 8.6 nm). Potency for Nav1.6 increases by a factor of 6.6 for 2670 (from 92.1 ± 6.8 to 13.9 ± 2.6 nm) and 4.3 for D1I (from 65.5 ± 9.2 to 15.1 ± 1.0 nm). From these data, it appears that the C-terminal amidation of these variants not only gives a significant potency improvement but also provides selectivity benefits against Nav1.2 but not Nav1.6 channels (Fig. 4, C and D).
We also introduced N-terminal pyroglutamate, as this modification was reported to enhance binding strength to the sodium channel of Huwentoxin-IV (44), a microprotein in the same family as CcoTx1 (35). CcoTx1, like other site 4 microproteins (binding to S3-S4 linker of domain II) (36–38), can be effectively dislodged from its binding site on the Nav1.7 channel by strong depolarization (23, 44). In our studies, 5 nm D1Ia induced almost complete block of Nav1.7 current after a 20-min application; however, a subsequent positive depolarizing pulse to +50 mV for 1 min has reversed the blocking effect of D1Ia by 83 ± 10% (n = 5) (supplemental Fig. 3A). In contrast, in the same experiment inhibition of Nav1.7 induced by the variant containing N-terminal pyroglutamate was reversed by positive depolarizing pulse by only 56 ± 20% (n = 5) (supplemental Fig. 3B) indicating a moderate increase in microprotein binding strength. Additionally, both the selectivity and potency of N-terminal pyroglutamate variant for Nav1.7 channel remained similar to D1Ia so we decided to continue with the N-terminally modified variant (D1Za).
Broad Selectivity Screening
To check the selectivity more broadly, we tested one of the lead compounds (D1Ia) against a large panel of channels, including various sodium channels (Nav1.1, Nav1.2, Nav1.3, Nav1.4, Nav1.5, Nav1.6, Nav1.7, and Nav1.8), Nav1.7 from different species (human, monkey, dog, rat, and mouse), potassium channels (KCNQ2/3, KCNQ3/5, KCNQ4, Kir2.1, Kir3.1/3.4, Kir6.2/SUR1, Kir6.2/SUR2A, Kv1.1, Kv1.2, Kv1.4, Kv1.5, Kv3.4, Kv4.2/KChiP2.2, Kv4.3/KChiP2.2, and KvLQT/mink and BK, SK1, SK2, SK3, and SK4), calcium channels (Cav1.2, Cav2.1, and Cav3.2), purinergic receptor channels (P2X1, P2X2, P2X3, P2X4, and P2X7), transient receptor potential channels (TRPA1, TRPM8, TRPV1, and TRPV6), hyperpolarization-activated cyclic nucleotide-gated channels (HCN1, HCN2, and HCN4), GABA receptor ligand-gated ion channels (GABA-A1 and GABA-A5), chloride channels (CFTR and CLC-1), acid-sensing ion channels (ASIC1a and ASIC3a), and serotonin receptor ligand-gated ion channels 5-HT3A. This broad selectivity screen was performed on an automated patch clamp system QPatch by using established voltage-gated or ligand application protocols for particular channel types (ChanTest.com). The data shown in Table 1 represent a list of ion channels on which the lead compound D1Ia was tested. The D1Ia concentration used in testing, the calculated IC50 value or % of block (− denotes potentiation), and the number of replicates (w denotes use of 10 holes wells on QPatch plates) are shown for each channel. D1Ia up to the highest concentration tested (1 μm) was without effect on HERG channels. D1Ia generally showed little activity on most channels tested even at a high concentration of 1 μm with the exception of limited activity on Kv4.3 (IC50 180 nm), Cav1.2 (41% block at 1 μm), and hTRPA1 (36% block at 1 μm) channels.
Structure of 2670
We first attempted to combine the best mutants from the saturation mutagenesis and created a panel of double mutants in the hope of further improving the potency and selectivity. To our surprise, the double mutants were not significantly better than the single substitutions (supplemental Fig. 4). To better understand the saturation mutagenesis data in the context of the three-dimensional structure of these microproteins and to improve our ability to combine mutations in a more productive way, we decided to solve the crystal structure of variant 2670.
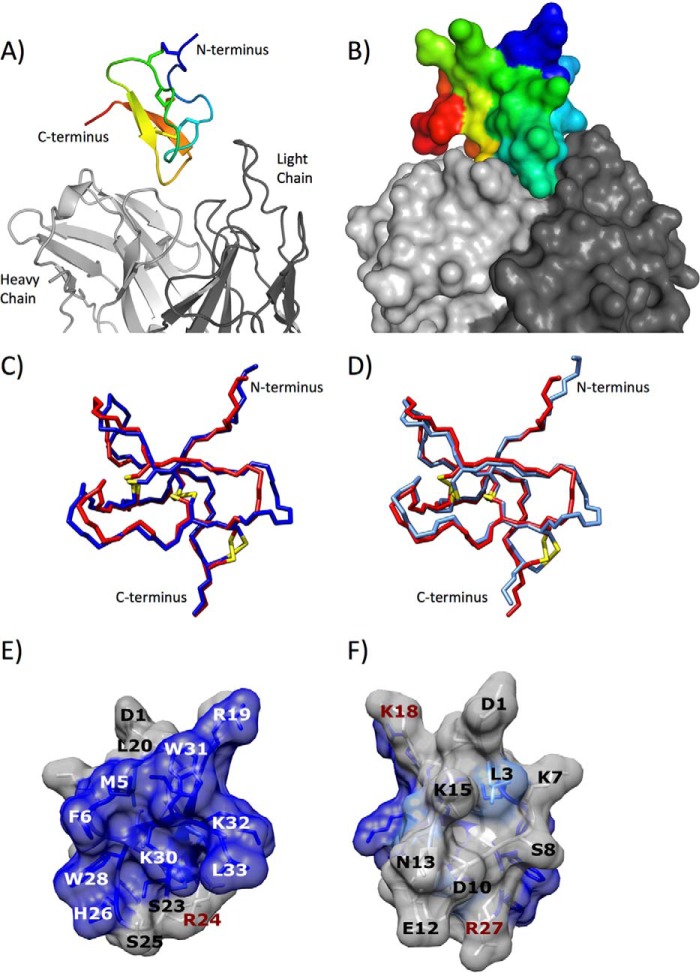
Venom peptides of the same family have been difficult to crystallize, and the currently known structures of Huwenoxin-IV, Hanatoxin-I, Hainantoxin-IV, SGTx1, and GrTx (42, 45–49) were determined by NMR techniques. We utilized a different approach and generated a high affinity antibody with a low dissociation rate and obtained a high resolution x-ray crystal structure of 2670 in complex with the Fab fragment of this antibody (supplemental Table 1). The structure of the 2670 variant in complex with the Fab fragment is shown in Fig. 5, A and B. 2670 adopts an inhibitor cystine knot motif common to inhibitory peptides (50). The backbone of 2670 superposed to Hainantoxin-IV and Huwentoxin-IV is shown in Fig. 5, C and D, and resulted in ∼0.6 and ∼0.9 Å r.m.s.d. for 31 Cα atoms, respectively. Although the backbones of the three cysteine knot molecules are similar, the overall shape of these small proteins is highly impacted by even a small number of side chain substitutions (supplemental Fig. 5).
FIGURE 5.
A, ribbon representation of the crystal structure of variant 2670 in complex with 6F1 Fab fragment. 2670 is colored in rainbow gradient going from N terminus (blue) to C terminus (red). 6F1 Fab fragment is shown in gray, with only part of the structure visible. B, surface representation of the complex in the same orientation and color-coding as in A. C, backbone representation of variant 2670 (red) superposed to Huwentoxin-IV (blue) resulting in an r.m.s.d. of 0.9 Å. D, backbone representation of variant 2670 (red) superposed to Hainantoxin-IV (light blue) resulting in an r.m.s.d. of 0.6Å. Cysteine disulfide bridges are shown in stick representation in both C and D. E and F, functional epitope (blue and cyan) of 2670 against Nav1.7 channel mapped on crystal structure of 2670. Functional epitope is defined as all positions for which the fraction of substitutions worsening potency by a factor 2 or more is larger than a threshold (blue, at least 75%; cyan, 50–75%).
Combining Mutations and Selection of Final Molecules
The structure of 2670 allowed a more refined interpretation of the saturation mutagenesis data for the inhibition of Nav1.7. We first identified the residues that make significant contributions to the binding to Nav1.7 by calculating the percentage of substitutions with potency decreased by a factor of 2 or more relative to 2670. Eleven positions (residues 5–6, 19–20, 23, 26, 28, and 30–33) have at least 75% of substitutions with decreased potency and were mapped on the structure of 2670 (Fig. 5, E and F). An additional four positions (residues 3, 11, 14, and 21) that have 50–75% of substitutions with decreased potency are also shown. The data clearly show a well defined interaction patch defining the likely orientation of the interaction with Nav1.7. The interaction region is composed of several hydrophobic residues (Met-5, Phe-6, Trp-28, Trp-31, and Leu-33), as well as other polar or positively charged residues (Arg-19, His-26, Lys-30, and Lys-32). Leu-20 and Ser-23 likely play a structural role and might not contribute directly to the binding. Comparison of 2670 interacting residues and previously published data on Hainantoxin-IV (41) and Huwentoxin-IV (42) is shown in supplemental Fig. 6.
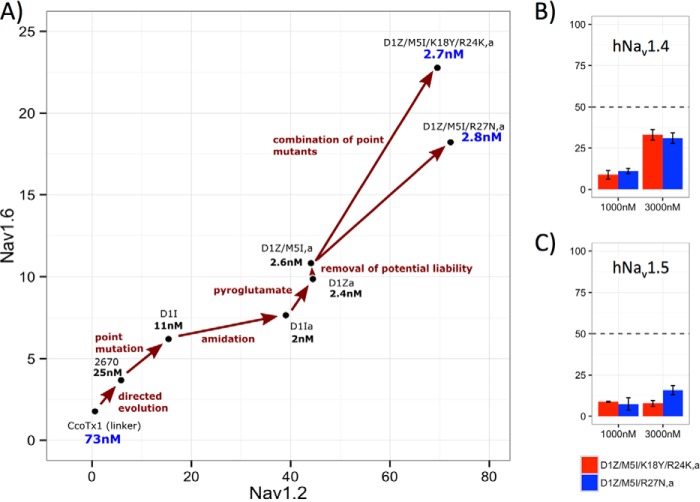
Next, we identified residues that had a significant impact on selectivity but were not part of the main Nav1.7 interaction interface (positions 18, 24, and 27) (Figs. 3 and 5, E and F). We further hypothesized that due to the small size of the CcoTx1, combining single mutants that are in close proximity might produce unexpected results, so we combined mutants that were on the opposite sides of the molecule. With this approach, we selected (K18Y/R24K) and (R27N) as potential mutants to test in combination with the lead amidated D1Z/M5I microprotein (D1Z/M5Ia). We found that addition of K18Y/R24K or R27N provided significant selectivity improvement without loss in Nav1.7 potency (Fig. 6A).
FIGURE 6.
A, evolution of compounds starting from CcoTx1 up to the rationally engineered multiple mutants (top-right corner). Selectivity to Nav1.2 and Nav1.6 is reported on the axis and potency is illustrated in nm on the plot. Effect of D1Z/M5I/K18Y/R24K,a and D1Z/M5I/R27N,a on human (B) Nav1.4 and (C) Nav1.5. The microprotein activity was tested at a concentration of 1000 and 3000 nm and percent inhibition was measured. Even at 3000 nm neither microprotein reached 50% inhibition on either channel, giving better than 1000-fold selectivity (hNav1.7 potency of these microproteins is ∼2.7 nm; A).
Our entire protein engineering process to obtain selective and potent Nav1.7 microproteins is summarized in Fig. 6A. Our most selective compound has an IC50 to Nav1.7 of 2.5 nm, which is an ∼30-fold improvement over the parental CcoTx1 molecule. The selectivity of the closely related channels Nav1.2 and Nav1.6 is 80- and 20-fold, respectively (IC50 on Nav1.2 of 186–200 nm and IC50 on Nav1.6 of 50–60 nm). The final molecules were also tested against the Nav1.4 and Nav1.5 channels expressed in skeletal and cardiac muscles (Fig. 6, B and C). The final molecules show great selectivity against these critical channels, where half-maximal inhibition was not achieved even at 3000 nm concentration, giving us more than 1000-fold selectivity against Nav1.4 and Nav1.5.
Discussion
Microproteins from venoms are an obvious choice for the development of ion channel blockers as they naturally evolved to modulate ion channels. The main drawback with natural occurring microproteins, however, is that they are usually not specific to the therapeutic targets of interest. To improve specificity and potency, we utilized a combination of directed evolution and saturation mutagenesis coupled with structural activity relationship design to engineer highly potent and selective microproteins against Nav1.7.
The directed evolution strategy has been successfully applied in the past to evolve enzymes with high specificity (25, 51) and served as our initial approach. In a typical directed evolution screening, high throughput assays are necessary to explore libraries of large diversity. Although high throughput electrophysiology has come a long way in the last decade, the throughput of these functional assays cannot match the throughput of display technologies such as phage or yeast display.
To gain potency and selectivity, we took advantage of phylogenetic libraries that allow exploration of an optimized amino acid sequence space and can sample amino acid sequences with higher functional probability. This approach allowed us to explore multiple mutations simultaneously but at a price of exploring an amino acid sequence landscape that is relatively close to the natural microproteins.
This limitation led us to further improve the evolved microproteins by adding saturation mutagenesis. Interestingly, the results from saturation mutagenesis show a great tolerability of the CcoTx1 fold to mutations. Out of all the microproteins designed, 93% express well in a high throughput cell-free expression system, with no optimization for the individual compounds. Almost two-thirds of the tested compounds inhibited Nav1.7 with IC50 of 100 nm or better. Saturation mutagenesis, when compared with phylogenetic exploration of the sequence space, has the advantage to exhaustively cover all of the possible single-amino acid substitutions, but only one at a time. We solved the crystal structure of one of the CcoTx1 variants developed in our study and utilized the structure-activity relationship analysis to identify mutations that were additive and combined these to generate microproteins with improved properties.
In summary, we utilized naturally occurring microprotein CcoTx1, which has good activity against the established pain target Nav1.7. One of the critical properties to select CcoTx1 as a starting point to engineer potent analgesics was its low activity against Nav1.4 and Nav1.5 channels. We further evolved CcoTx1 to be more active and selective by combination of semi-rational approaches and achieved high potency against Nav1.7 (2.7 nm) with more than 1000-fold selectivity against Nav1.4/Nav1.5 and 80/20-fold selectivity against some of the most closely related Nav1.2 and Nav1.6 channels. The lead molecules have the potential for future clinical development as promising therapeutics for the treatment of pain.
Author Contributions
V. B., A. C., O. B., and M. G. C. performed the directed evolution experiments. A. R. S., R. H., and A. G. performed the cell free expression experiments. G. Z., A. H. M., and A. S. performed the electrophysiology experiments. A. Rossi and P. S. performed chemical modification and refolding experiments. A. Rossi analyzed and led the interpretation of the IC50 data. P. S. and M. R. performed the crystallography experiments. P. S., A. S., D. F., A. Rossi, A. Rajpal, D. L. S., O. B., and J. P. conceived and coordinated the studies. A. Rossi, P. S., and A. S. wrote the paper.
Supplementary Material
Acknowledgment
We thank Santiago Farias for MS analysis.
All authors were employees of Pfizer, Amunix, and Sutro Biopharma at the time of the study.
The atomic coordinates and structure factors (code 5EPM) have been deposited in the Protein Data Bank (http://wwpdb.org/).

This article contains supplemental Tables S1 and S2 and Figs. S1–S6.
- SUMO
- small ubiquitin-like modifier
- r.m.s.d.
- root mean square deviation
- PDB
- Protein Data Bank.
References
- 1. Goldberg D. S., and McGee S. J. (2011) Pain as a global public health priority. BMC Public Health 11, 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cox J. J., Reimann F., Nicholas A. K., Thornton G., Roberts E., Springell K., Karbani G., Jafri H., Mannan J., Raashid Y., Al-Gazali L., Hamamy H., Valente E. M., Gorman S., Williams R., et al. (2006) An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldberg Y. P., MacFarlane J., MacDonald M. L., Thompson J., Dube M. P., Mattice M., Fraser R., Young C., Hossain S., Pape T., Payne B., Radomski C., Donaldson G., Ives E., Cox J., et al. (2007) Loss-of-function mutations in the Nav1.7 gene underlie congenital indifference to pain in multiple human populations. Clin. Genet. 71, 311–319 [DOI] [PubMed] [Google Scholar]
- 4. Yang Y., Wang Y., Li S., Xu Z., Li H., Ma L., Fan J., Bu D., Liu B., Fan Z., Wu G., Jin J., Ding B., Zhu X., and Shen Y. (2004) Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J. Med. Genet. 41, 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fertleman C. R., Baker M. D., Parker K. A., Moffatt S., Elmslie F. V., Abrahamsen B., Ostman J., Klugbauer N., Wood J. N., Gardiner R. M., and Rees M. (2006) SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron 52, 767–774 [DOI] [PubMed] [Google Scholar]
- 6. Estacion M., Han C., Choi J. S., Hoeijmakers J. G., Lauria G., Drenth J. P., Gerrits M. M., Dib-Hajj S. D., Faber C. G., Merkies I. S., and Waxman S. G. (2011) Intra- and interfamily phenotypic diversity in pain syndromes associated with a gain-of-function variant of NaV1.7. Mol. Pain 7, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Faber C. G., Hoeijmakers J. G., Ahn H. S., Cheng X., Han C., Choi J. S., Estacion M., Lauria G., Vanhoutte E. K., Gerrits M. M., Dib-Hajj S., Drenth J. P., Waxman S. G., and Merkies I. S. (2012) Gain-of-function Nav1.7 mutations in idiopathic small fiber neuropathy. Ann. Neurol 71, 26–39 [DOI] [PubMed] [Google Scholar]
- 8. King G. F., and Vetter I. (2014) No gain, no pain: NaV1.7 as an analgesic target. ACS Chem. Neurosci. 5, 749–751 [DOI] [PubMed] [Google Scholar]
- 9. Minett M. S., Pereira V., Sikandar S., Matsuyama A., Lolignier S., Kanellopoulos A. H., Mancini F., Iannetti G. D., Bogdanov Y. D., Santana-Varela S., Millet Q., Baskozos G., MacAllister R., Cox J. J., Zhao J., and Wood J. N. (2015) Endogenous opioids contribute to insensitivity to pain in humans and mice lacking sodium channel Nav1.7. Nat. Commun. 6, 8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chessell I. P., Dudley A., and Billinton A. (2012) Biologics: the next generation of analgesic drugs? Drug Discov. Today 17, 875–879 [DOI] [PubMed] [Google Scholar]
- 11. Lee J. H., Park C. K., Chen G., Han Q., Xie R. G., Liu T., Ji R. R., and Lee S. Y. (2014) A monoclonal antibody that targets a NaV1.7 channel voltage sensor for pain and itch relief. Cell 157, 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Harvey A. L. (2014) Toxins and drug discovery. Toxicon 92, 193–200 [DOI] [PubMed] [Google Scholar]
- 13. Yang S., Xiao Y., Kang D., Liu J., Li Y., Undheim E. A., Klint J. K., Rong M., Lai R., and King G. F. (2013) Discovery of a selective NaV1.7 inhibitor from centipede venom with analgesic efficacy exceeding morphine in rodent pain models. Proc. Natl. Acad. Sci. U.S.A. 110, 17534–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Craik D. J., Fairlie D. P., Liras S., and Price D. (2013) The future of peptide-based drugs. Chem. Biol. Drug Des. 81, 136–147 [DOI] [PubMed] [Google Scholar]
- 15. Gilchrist J., Olivera B. M., and Bosmans F. (2014) Animal toxins influence voltage-gated sodium channel function. Handb. Exp. Pharmacol. 221, 203–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pineda S. S., Undheim E. A., Rupasinghe D. B., Ikonomopoulou M. P., and King G. F. (2014) Spider venomics: implications for drug discovery. Future Med. Chem. 6, 1699–1714 [DOI] [PubMed] [Google Scholar]
- 17. Saez N. J., Senff S., Jensen J. E., Er S. Y., Herzig V., Rash L. D., and King G. F. (2010) Spider-venom peptides as therapeutics. Toxins 2, 2851–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wood D. L., Miljenović T., Cai S., Raven R. J., Kaas Q., Escoubas P., Herzig V., Wilson D., and King G. F. (2009) ArachnoServer: a database of protein toxins from spiders. BMC Genomics 10, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chow C. Y., Cristofori-Armstrong B., Undheim E. A., King G. F., and Rash L. D. (2015) Three peptide modulators of the human voltage-gated sodium channel 1.7, an important analgesic target, from the venom of an Australian tarantula. Toxins 7, 2494–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klint J. K., Smith J. J., Vetter I., Rupasinghe D. B., Er S. Y., Senff S., Herzig V., Mobli M., Lewis R. J., Bosmans F., and King G. F. (2015) Seven novel modulators of the analgesic target NaV 1.7 uncovered using a high-throughput venom-based discovery approach. Br. J. Pharmacol. 172, 2445–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murray J. K., Ligutti J., Liu D., Zou A., Poppe L., Li H., Andrews K. L., Moyer B. D., McDonough S. I., Favreau P., Stöcklin R., and Miranda L. P. (2015) Engineering potent and selective analogues of GpTx-1, a tarantula venom peptide antagonist of the Na(V)1.7 sodium channel. J. Med. Chem. 58, 2299–2314 [DOI] [PubMed] [Google Scholar]
- 22. Revell J. D., Lund P. E., Linley J. E., Metcalfe J., Burmeister N., Sridharan S., Jones C., Jermutus L., and Bednarek M. A. (2013) Potency optimization of Huwentoxin-IV on hNav1.7: a neurotoxin TTX-S sodium-channel antagonist from the venom of the Chinese bird-eating spider Selenocosmia huwena. Peptides 44, 40–46 [DOI] [PubMed] [Google Scholar]
- 23. Park J. H., Carlin K. P., Wu G., Ilyin V. I., Musza L. L., Blake P. R., and Kyle D. J. (2014) Studies examining the relationship between the chemical structure of protoxin II and its activity on voltage-gated sodium channels. J. Med. Chem. 57, 6623–6631 [DOI] [PubMed] [Google Scholar]
- 24. Bosmans F., Rash L., Zhu S., Diochot S., Lazdunski M., Escoubas P., and Tytgat J. (2006) Four novel tarantula toxins as selective modulators of voltage-gated sodium channel subtypes. Mol. Pharmacol. 69, 419–429 [DOI] [PubMed] [Google Scholar]
- 25. Crameri A., Raillard S. A., Bermudez E., and Stemmer W. P. (1998) DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391, 288–291 [DOI] [PubMed] [Google Scholar]
- 26. Malakhov M. P., Mattern M. R., Malakhova O. A., Drinker M., Weeks S. D., and Butt T. R. (2004) SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics 5, 75–86 [DOI] [PubMed] [Google Scholar]
- 27. Zawada J. F., Yin G., Steiner A. R., Yang J., Naresh A., Roy S. M., Gold D. S., Heinsohn H. G., and Murray C. J. (2011) Microscale to manufacturing scale-up of cell-free cytokine production–a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 108, 1570–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Köhler G., and Milstein C. (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 [DOI] [PubMed] [Google Scholar]
- 29. Otwinoswski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33. Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 34. Tripathi A., and Varadarajan R. (2014) Residue specific contributions to stability and activity inferred from saturation mutagenesis and deep sequencing. Curr. Opin. Struct. Biol. 24, 63–71 [DOI] [PubMed] [Google Scholar]
- 35. Klint J. K., Senff S., Rupasinghe D. B., Er S. Y., Herzig V., Nicholson G. M., and King G. F. (2012) Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads. Toxicon 60, 478–491 [DOI] [PubMed] [Google Scholar]
- 36. Sokolov S., Kraus R. L., Scheuer T., and Catterall W. A. (2008) Inhibition of sodium channel gating by trapping the domain II voltage sensor with protoxin II. Mol. Pharmacol. 73, 1020–1028 [DOI] [PubMed] [Google Scholar]
- 37. Nicholson G. M. (2007) Insect-selective spider toxins targeting voltage-gated sodium channels. Toxicon 49, 490–512 [DOI] [PubMed] [Google Scholar]
- 38. Xiao Y., Bingham J. P., Zhu W., Moczydlowski E., Liang S., and Cummins T. R. (2008) Tarantula huwentoxin-IV inhibits neuronal sodium channels by binding to receptor site 4 and trapping the domain ii voltage sensor in the closed configuration. J. Biol. Chem. 283, 27300–27313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Billen B., Bosmans F., and Tytgat J. (2008) Animal peptides targeting voltage-activated sodium channels. Curr. Pharm. Des. 14, 2492–2502 [DOI] [PubMed] [Google Scholar]
- 40. Bosmans F., and Swartz K. J. (2010) Targeting voltage sensors in sodium channels with spider toxins. Trends Pharmacol. Sci. 31, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cai T., Luo J., Meng E., Ding J., Liang S., Wang S., and Liu Z. (2015) Mapping the interaction site for the tarantula toxin hainantoxin-IV (β-TRTX-Hn2a) in the voltage sensor module of domain II of voltage-gated sodium channels. Peptides 68, 148–156 [DOI] [PubMed] [Google Scholar]
- 42. Minassian N. A., Gibbs A., Shih A. Y., Liu Y., Neff R. A., Sutton S. W., Mirzadegan T., Connor J., Fellows R., Husovsky M., Nelson S., Hunter M. J., Flinspach M., and Wickenden A. D. (2013) Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin huwentoxin-IV (mu-TRTX-Hh2a). J. Biol. Chem. 288, 22707–22720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ogata N., and Ohishi Y. (2002) Molecular diversity of structure and function of the voltage-gated Na+ channels. Jpn. J. Pharmacol. 88, 365–377 [DOI] [PubMed] [Google Scholar]
- 44. Rong M., Duan Z., Chen J., Li J., Xiao Y., and Liang S. (2013) Native pyroglutamation of huwentoxin-IV: a post-translational modification that increases the trapping ability to the sodium channel. PLoS ONE 8, e65984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee C. W., Kim S., Roh S. H., Endoh H., Kodera Y., Maeda T., Kohno T., Wang J. M., Swartz K. J., and Kim J. I. (2004) Solution structure and functional characterization of SGTx1, a modifier of Kv2.1 channel gating. Biochemistry 43, 890–897 [DOI] [PubMed] [Google Scholar]
- 46. Li D., Xiao Y., Xu X., Xiong X., Lu S., Liu Z., Zhu Q., Wang M., Gu X., and Liang S. (2004) Structure-activity relationships of hainantoxin-IV and structure determination of active and inactive sodium channel blockers. J. Biol. Chem. 279, 37734–37740 [DOI] [PubMed] [Google Scholar]
- 47. Peng K., Shu Q., Liu Z., and Liang S. (2002) Function and solution structure of huwentoxin-IV, a potent neuronal tetrodotoxin (TTX)-sensitive sodium channel antagonist from Chinese bird spider Selenocosmia huwena. J. Biol. Chem. 277, 47564–47571 [DOI] [PubMed] [Google Scholar]
- 48. Takahashi H., Kim J. I., Min H. J., Sato K., Swartz K. J., and Shimada I. (2000) Solution structure of hanatoxin1, a gating modifier of voltage-dependent K+ channels: common surface features of gating modifier toxins. J. Mol. Biol. 297, 771–780 [DOI] [PubMed] [Google Scholar]
- 49. Takeuchi K., Park E., Lee C., Kim J., Takahashi H., Swartz K., and Shimada I. (2002) Solution structure of ω-grammotoxin SIA, a gating modifier of P/Q and N-type Ca2+ channel. J. Mol. Biol. 321, 517–526 [DOI] [PubMed] [Google Scholar]
- 50. Pallaghy P. K., Nielsen K. J., Craik D. J., and Norton R. S. (1994) A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 3, 1833–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Packer M. S., and Liu D. R. (2015) Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.