Abstract
Mammalian actin-binding protein-1 (mAbp1) is an adaptor protein that binds actin and modulates scission during endocytosis. Recent studies suggest that mAbp1 impairs cell invasion; however, the mechanism for the inhibitory effects of mAbp1 remain unclear. We performed a yeast two-hybrid screen and identified the adaptor protein, FHL2, as a novel binding partner that interacts with the N-terminal actin depolymerizing factor homology domain (ADFH) domain of mAbp1. Here we report that depletion of mAbp1 or ectopic expression of the ADFH domain of mAbp1 increased Rho GTPase signaling and breast cancer cell invasion. Moreover, cell invasion induced by the ADFH domain of mAbp1 required the expression of FHL2. Taken together, our findings show that mAbp1 and FHL2 are novel binding partners that differentially regulate Rho GTPase signaling and MTLn3 breast cancer cell invasion.
Keywords: adaptor protein, cell invasion, cell migration, focal adhesion, Rho (Rho GTPase), FHL2, mAbp1
Introduction
Invasive migration of cancer cells is dependent on a dynamic actin cytoskeleton (1). Key regulators of the actin cytoskeleton include actin-binding proteins like cortactin, which have been implicated in invadopodia and podosome formation and invasive cell migration. Mammalian actin-binding protein-1 (mAbp1,2 Hip-55, DBNL) has significant structural homology to cortactin and binds actin with an N-terminal ADFH domain (2). Both mAbp1 and cortactin have identified roles during endocytosis and vesicle trafficking (3–5), and both proteins localize to lamellipodia, podosomes, and dorsal ruffles (4, 6, 7). However, in contrast to cortactin, mAbp1 impairs podosome formation and invasive migration of Src-transformed fibroblasts (8).
To determine how mAbp1 regulates cell invasion, we performed a yeast two-hybrid screen and identified the four-and-a-half LIM domain protein 2 (FHL2) as a novel mAbp1-binding partner that does not interact with cortactin. FHL2 (also known as DRAL or SLIM3) is a LIM domain protein consisting of four-and-a-half LIM domains. Each LIM domain contains two zinc finger loops that mediate protein-protein interactions. FHL2 interacts with >50 proteins (9) including integrins and focal adhesion kinase (FAK) (10, 11), actin (12), and the transcription factors estrogen receptor (ER), activator protein-1 (AP-1), and β-catenin (13–18). Interestingly, FHL2 is expressed at low levels in many tissues like breast, placenta, uterus, and the lungs (17, 19) and is up-regulated in cancer. For example, FHL2 expression is increased in many cancers including breast (20, 21), ovarian (22), prostate (23), lung (24), colon (25), brain (26), and skin cancers (27). In primary breast tumors, patients with high levels of FHL2 have a poorer survival rate (21), suggesting that FHL2 may be involved in cancer progression. Furthermore, overexpression of FHL2 in MDA-MB-231 human breast cancer cells enhances colony formation in soft agar, whereas knockdown reduces the ability to form colonies (20).
Here we report a novel interaction between the N-terminal ADFH domain of mAbp1 and FHL2 that modulates cell invasion. In the yeast, but not the mammalian form, the N terminus of Abp1 binds Arp2/3 (2, 28). We have now identified a novel binding partner that interacts with the N terminus of mAbp1. We found that depletion of mAbp1 or ectopic expression of the ADFH domain enhances cell invasion. Moreover the increased invasion observed with ectopic expression of the ADFH domain requires expression of FHL2. Our findings suggest that mAbp1 inhibits invasive migration of breast cancer cells at least in part through its interaction with FHL2 and the modulation of Rho GTPase signaling. These results identify a novel role for the interaction between mAbp1 and FHL2 in modulating MTLn3 breast cancer cell invasion.
Experimental Procedures
Reagents and Antibodies
Alexa Fluor 405 phalloidin and rhodamine phalloidin were purchased from Invitrogen. The following primary antibodies were used: rabbit anti-mAbp1 from the University of Wisconsin antibody facility as described previously (30), mouse anti-FHL2 clone F4B2-B11, (Santa Cruz, CA), mouse anti–β-actin clone AC-15 (Sigma), rabbit anti-GFP (Life Technologies), mouse anti-vinculin clone h-VIN1 (Sigma), rabbit anti-RhoA clone 67B9 (Cell Signaling Technology, Beverly, MA), mouse anti-myc clone 9E10 (Thermo Fisher, Waltham, MA), rabbit anti-His6 (Rockland Immunochemicals Inc, Limerick, PA), rabbit anti-tag RFP (Evrogen), mouse anti-β-tubulin (Cell Signaling Technology), and rabbit anti-Histone-3 (Cell Signaling Technology). Secondary antibodies used for immunofluorescence include Marina blue goat anti-mouse IgG (Invitrogen), Pacific blue goat anti-mouse (Invitrogen), rhodamine red-X goat anti-mouse IgG, TRITC donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA), and FITC donkey anti-rabbit IgG. Gamma glutathione-Sepharose beads were purchased from Amersham Biosciences. Secondary antibodies used for Odyssey Li-cor imaging of Western blots were mouse 680 (Invitrogen) and rabbit 800 (Rockland Immunochemicals). Rat tail collagen type I was used for collagen contraction assays (BD Biosciences). Gelatin from porcine skin (Sigma) was used for gelatin coating coverslips, and fibronectin was purified as described previously (31). EGF was acquired from Invitrogen.
Cell Culture
MTLn3 rat adenocarcinoma cells (provided by J. Condeelis and J. Segall, Albert Einstein College of Medicine, New York, NY) were cultured in α-minimum Eagle's medium supplemented with 5% FBS (HyClone, Logan, UT) and penicillin/streptomycin (Cellgro and Mediatech, Manassas, VA). MDA-MB-231 human mammary adenocarcinoma (provided by A. Rapraeger, University of Wisconsin, Madison, WI) were cultured in DMEM (Cellgro and Mediatech) supplemented with 10% FBS, 1% nonessential amino acids (Sigma), and penicillin/streptomycin (Cellgro and Mediatech). HEK 293 and PhxA cells (ATCC, Manassas, VA) were cultured in DMEM supplemented with 10% HI FBS (Hyclone), nonessential amino acids (Sigma), and penicillin/streptomycin (Cellgro and Mediatech).
Cell Transfections
Transient transfections of MTLn3 cells were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. In brief, 1 × 105 cells were cultured in a 6-well plate, and 2 μg of DNA and 3 μl of Lipofectamine 2000 reagent were used. Transfection mixtures incubated at room temperature in Opti-MEM media (Gibco for 30 min before adding to cells. Cells were incubated with the transfection mixture 4 h before washing 3× with PBS. Cells then were cultured by standard methods as described under “Cell Culture” above. HEK 293 transient transfections were performed by the standard calcium phosphate precipitation method. All cells, with the exception of HEK 293 (cultured at 10% CO2), were maintained at 5% CO2 at 37 °C.
DNA and siRNA Constructs
All constructs were generated by PCR amplification of product with cut sites in the primers as follows: full-length human mAbp1 and the ADFH, proline-rich region, or SH3 domains of mAbp1 were cloned into the pEGFP-C1 vector using the XhoI and BamHI cut sites. GFP-mAbp1-W415K was generated by site-directed mutagenesis of the wild-type construct. GST-m+Abp1 was cloned into pGex-4T1 with BamHI and XhoI cut sites. RFP-FHL2 was cloned into tagRFP-C1 vector with HindIII and BamHI cut sites. Myc-FHL2 was cloned into pcDNA3.1 myc vector with XhoI and EcoRI cut sites and the myc tag on the N terminus of FHL2. His-FHL2 was cloned into pTrcHisA vector with XhoI and EcoRI cut sites.
Retroviral Transduction of Cells
Stable knockdown MTLn3 lines were generated with the pSUPER.retro RNAi system per the manufacturer's instructions (OligoEngine, Seattle, WA). Briefly, to generate retrovirus for infection of MTLn3 cells, we plated 3 × 106 PhxA cells on a 10-cm dish. The PhxA cells were transfected with 12 μg of DNA using the calcium phosphate precipitation procedure. 24-h virus was discarded, and 4 ml of new media were added. The 48-h and 72-h retrovirus was harvested, filtered through a 0.2-μm syringe filter, and mixed with 1× Polybrene before adding to a 6-cm dish of 1 × 105 MTLn3 cells. Cells were incubated in retrovirus mixture for 6 h at 32 °C. Retroviral media was removed, and cells were recovered in standard media for 24 h before selection. Cells containing retrovirus were selected with 1 μg/ml puromycin (Sigma) for 5 days. Retroviral transduction of MDA-MB-231 cells was performed similarly with the exception that virus from 24, 48, 72, and 96 h was used for infection and virus-containing media remained on cells 24 h instead of 6. This longer infection was necessary for successful transduction of these cells.
The most efficient rat mAbp1 shRNA target sequences were at nucleotides: 1334 sense (GATCCCCGTGGCTTCCTTGTTGCCTATTCAAGAGATAGGCAACAAGGAAGCCACTTTTTA) and 1334 antisense (AGCTTAAAAAGTGGCTTCCTTGTTGCCTATCTCTTGAATAGGCAACAAGGAAGCCACGGG) and 1464 sense (GATCCCCATGCTTACCGCAGCCTCTATTCAAGAGATAGAGGCTGCGGTAAGCATTTTTTA) and 1464 antisense (AGCTTAAAAAATGCTTACCGCAGCCTCTATCTCTTGAATAGAGGCTGCGGTAAGCATGGG). To generate an FHL2 knockdown in MTLn3 cells, 12 targets were attempted with the most efficient knockdown targeted at start nucleotide 152 with the primers 152 sense (GATCCCCGCAAGGACTTGTCCTACAATTCAAGAGATTGTAGGACAAGTCCTTGCTTTTTA) and 152 antisense (AGCTTAAAAAGCAAGGACTTGTCCTACAATCTCTTGAATTGTAGGACAAGTCCTTGCGGG). MDA-MB-231 stableknockdowns were generated with primers targeting nucleotide 1748: sense (GATCCCCGTGAACGTAGAGAATTGTTTTCAAGAGAAACAATTCTCTACGTTCACTTTTT) and 1748 antisense (AGCTAAAAAGTGAACGTAGAGAATTGTTTCTCTTGAAAACAATTCTCTACGTTCACGGG).
Collagen Contraction
Collagen contraction experiments were performed as follows. Desired volume of collagen was mixed 1:1 with 150 mm HEPES (Cellgro) in 2× PBS for a stock concentration of 4.5 mg/ml collagen and incubated on ice. 1 × 106 MDA-MB-231 cells were counted and resuspended in 656 μl of media. 444 μl of collagen stock was mixed with cells for a final collagen concentration of 2 mg/ml and immediately transferred to 1 well of a 6-well plate. The plate was rocked to spread collagen across the entire surface then cultured at 37 °C for 1 h to polymerize. When polymerized, 2 ml of media were added to well, and collagen was loosened from the well with gentle rocking. Cells were cultured, and images were taken at 24 h and 48 h. Percent contraction was quantified on metamorph imaging software.
Immunoblotting and Immunoprecipitation
Cells were scraped into lysis buffer (20 mm HEPES, 50 mm KCl, 1 mm EDTA, 1% Nonidet P-40, 0.2 mm PMSF, 1 μg/ml pepstatin, 2 μg/ml aprotinin, and 1 μg/ml leupeptin) on ice and clarified by centrifugation. Protein concentrations were determined with a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. Equal protein levels were denatured in SDS and loaded on 4–20% SDS-polyacrylamide gradient gels. Separated proteins were transferred to nitrocellulose, probed with the appropriate antibodies, and imaged with an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). For GFP immunoprecipitation experiments, transiently transfected HEK 293 cells were cultured to ∼80% confluency on 10-cm plates, washed once with PBS, and lysed in lysis buffer. Lysates were clarified by centrifugation and incubated with 3 μg of rabbit-anti-GFP antibody. Immune complexes were captured on Gammabind G-Sepharose beads (GE Healthcare), washed three times in lysis buffer, and analyzed by immunoblotting.
Rho Pulldown Assay
Rho GTPase pulldown assays were performed with Rhotekin-RBD Protein GST beads (Cytoskeleton, Inc.) according the manufacturer's instructions with some modifications. In brief, 3 × 106 MTLn3 cells were cultured on 15-cm dishes, serum-starved for 24 h, then treated with 5 nm EGF for 5 min. Cells were immediately lysed on ice in Rho lysis buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 100 mm NaCl, 1% Triton X-100, and 1% PMSF, leupeptin, and aprotinin) and centrifuged at 14,000 rpm for 5 min for clarification. A sample of lysate was removed for Western blot loading control, and the remainder of lysate was placed in a tube containing 60 μg of Rhotekin-RBD beads to capture active Rho. Tubes were incubated for 30 min at 4 °C before centrifuging for 1 min at 1000 rpm and washed 3× with wash buffer (25 mm Tris, pH 7.5, 30 mm MgCl2, 40 mm NaCl, and 1% PMSF, leupeptin, and aprotinin). Meanwhile, the positive and negative control lysates were incubated at room temperature for 15 min with 15 mm EDTA and 0.2 mm GTPγS or 1 mm GDP, respectively. GTPγS is a non-hydrolysable GTP analog so that active Rho is pulled down from cell lysates. GDP inactivates Rho and blocks its binding to rhotekin RBD beads. MgCl2 (60 mm final concentration) was added to stop all reactions, and lysates were incubated on rhotekin beads and washed as described above. All samples were run on SDS-PAGE. The amount of Rho pulled down was calculated by densitometry analysis of Western blots. The ratio of active Rho pulled down to input Rho was calculated first. Then the ratio of active Rho pulled down per condition was normalized to control shRNA cells.
Cell Fractionation
Cell fractionation was performed as previously described (32). In brief, cells were scraped from confluent 10-cm dishes on ice into 1 ml of ice-cold PBS. Cells were briefly centrifuged, and supernatant was removed. Cells were then resuspended in 900 μl of 0.1% Nonidet P-40 in PBS and titrated 3×. 300 μl of lysate was removed as the whole cell lysate. Remaining lysate was briefly centrifuged, and supernatant was removed as the cytosolic fraction. Nuclear pellet was resuspended in 180 μl of SDS buffer. Samples were loaded on SDS page and analyzed on Western blot.
Immunofluorescence
Coverslips were acid-washed, ethanol-sterilized, coated with 2 mg/ml gelatin, and subsequently coated with 10 μg/ml fibronectin overnight at 4 °C. 3.0 × 104 cells were plated in α-minimum Eagle's medium on coverslips for 6 h at 37 °C and 5% CO2. Cells were fixed with 3.7% formaldehyde for 10 min, quenched with 0.15 m glycine for 10 min, permeabilized with 0.2% Triton X-100 for 10 min, washed with PBS 3 times, and blocked with 5% goat serum for 1 h or overnight at 4 °C. Cells were incubated with primary antibody and/or rhodamine phalloidin for minimum of 1 h, washed, then incubated with fluorophore-conjugated secondary antibodies for 1 h.
Cells were imaged either with a Zeiss Observer Z.1 inverted microscope or with a Nikon Eclipse TE300 inverted microscope. On the Zeiss Observer Z.1 inverted microscope, images were captured with a Coolsnap ES2 camera or with an Evolve 512 Delta EM-CCD camera for confocal mode (Photometrics, Huntington Beach, CA) using a 63× oil/NA 1.4 Plan Apo immersion objective (Zeiss Immersol 518F oil) and processed with Zen (blue edition) software (Carl Zeiss). Images from the Nikon Eclipse TE300 were captured with a Coolsnap ES2 camera using a 60× oil/NA 1.4 Plan Apo oil immersion objective and processed with Zen imaging software.
Invasion Assays
Cells were serum-starved for 24 h, and 2 × 105 cells were seeded into top chamber of 8-μm pore boyden chambers coated with Matrigel (BD Biosystems). Serum-starving medium for MTLn3 cells contained 1% FBS and antibiotics. Serum-starving medium for other cell types contained 0.2% fatty acid free BSA and no serum. Cells were allowed to invade for 16 or 48 h as indicated toward serum-containing media. Cells were removed from the top chamber, and cells remaining on the membrane were stained with a Hema-3 stain kit (Thermo Fisher Scientific). Cells were counted from 12 representative 20× field of view images per condition from at least three independent experiments. All images were acquired with an Olympus 1X-70 inverted microscope (Olympus America, Melville, NY) using a 20×/NA 0.4 objective. Images were processed with Metavue imaging software v6.2 (Universal Imaging, Downingtown, PA).
Statistical Analyses
Statistical analyses used were unpaired t test or one-way analysis of variance (ANOVA) with Tukey post tests. p < 0.05 was considered significant.
Results
mAbp1 Inhibits Invasion of Breast Cancer Cells
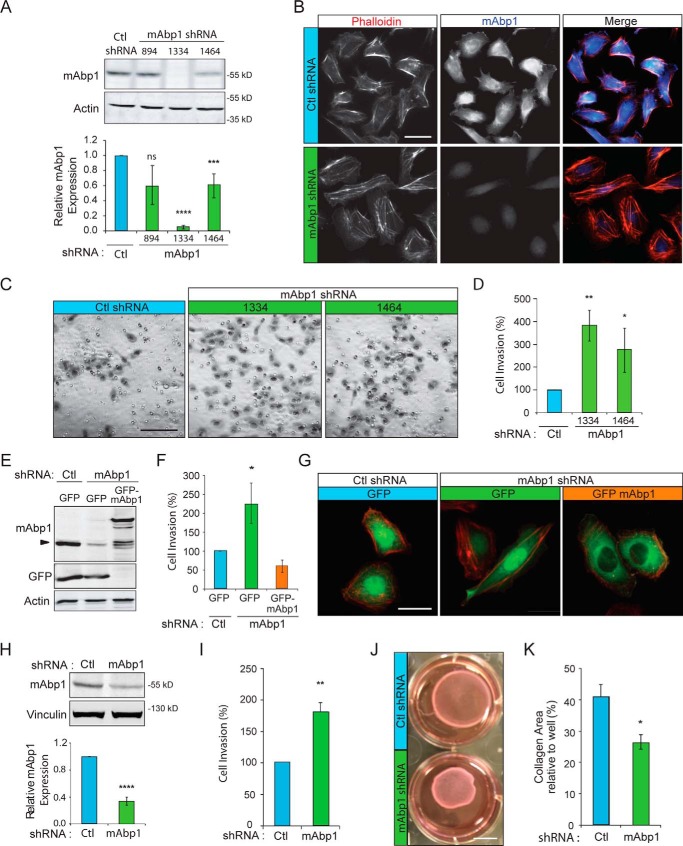
We previously reported that mAbp1 impairs the migration of Src-transformed NIH 3T3 cells (8). To investigate whether mAbp1 also negatively regulates invasion of breast cancer cells, we depleted mAbp1 in MTLn3 and MDA-MB-231 breast cancer cell lines using retroviral shRNA (Fig. 1, A, B, and H). Cell invasion through Matrigel-coated boyden chambers was enhanced 2–4-fold in mAbp1-deficient breast cancer cell lines (Fig. 1, C, D, and I). This enhanced invasion was rescued to control levels with exogenous transient expression of GFP-mAbp1 in mAbp1-deficient MTLn3 cells (Fig. 1F). GFP-tagged mAbp1 localized to the cytoplasm and perinuclear regions (Fig. 1G) similar to endogenous mAbp1 as detected by immunofluorescence (Fig. 1B). Taken together, these findings identify an inhibitory role for mAbp1 during invasive migration of breast cancer cell lines.
FIGURE 1.
mAbp1 inhibits invasion of MTLn3 and MDA-MB-231 breast cancer cells. A, control and mAbp1 shRNA stable lines were generated by retroviral transduction of MTLn3 breast cancer cells; target nucleotide sites are noted. Western blot of cell lysates was probed for endogenous mAbp1 and actin as a loading control. Densitometry analysis of mAbp1 expression relative to actin and normalized to control is shown. n = 8; ***, p = 0.0010; ****, p < 0.0001. ns, not significant. B, mAbp1 immunofluorescence and rhodamine phalloidin (actin) staining of fixed control and 1334 shRNA cells cultured on gelatin and fibronectin-coated coverslips. Endogenous mAbp1 localizes to cytoplasm, perinuclear region, and in lamellipodia. Scale bar = 20 μm. C, invasive migration of control or mAbp1 knockdown lines (1334 and 1464). Cells were serum-starved for 24 h, then seeded in Matrigel-coated boyden chambers and allowed to invade toward serum-containing media for 16 h. Invaded cells were stained, imaged with 20× objective, and counted. Scale bar = 100 μm. D, quantification of cell invasion. Percentage of cells invaded relative to control from 12 images taken at 20× magnification. Four independent experiments. **, p = 0.0068; *, p = 0.0462. E, expression of GFP or GFP-mAbp1 in control or mAbp1 shRNA MTLn3 cells. Shown in a Western blot probed for mAbp1, GFP, and actin as a loading control. F, cells transiently transfected with GFP or GFP-mAbp1 were serum-starved for 24 h after transfection, then seeded on Matrigel-coated boyden chambers and allowed to invade toward serum for 16 h. Percentage of cells invaded relative to control from 12 images taken at 20× magnification. Data are from three independent experiments. *, p = 0.0116. G, fluorescent images of GFP or GFP-mAbp1 expression in transiently transfected control or mAbp1 shRNA cells, fixed, and stained with rhodamine phalloidin (actin). GFP-mAbp1 localizes to focal adhesions similar to endogenous mAbp1. Scale bar = 20 μm. H, stable control and mAbp1 knockdown MDA-MB-231 cell lines were generated by retroviral transduction of shRNA. Shown is a Western blot of cell lysates probed for endogenous mAbp1 and vinculin as a loading control. Densitometry analysis of mAbp1 levels relative to control is shown. n = 9; ****, p < 0.0001. I, quantification of invasion between control or mAbp1 shRNA MDA-MB-231 cells through Matrigel-coated boyden chambers. Cells were serum-starved 24 h, seeded on Matrigel, and allowed to invade toward serum media for 48 h. Percentage of cells invaded relative to control from 12 images were taken at 20× magnification. The experiment was performed in quadruplicate. **, p = 0.0069. J, control and mAbp1 shRNA MDA-MB-231 cells were cultured in 1 mg/ml collagen and floated in media to allow for collagen contraction to occur for 48 h. Scale bar = 1 cm. K, quantification of the percentage of contracted collagen area relative to total area. n = 4; *, p = 0.0164. Error bars represent the S.E.
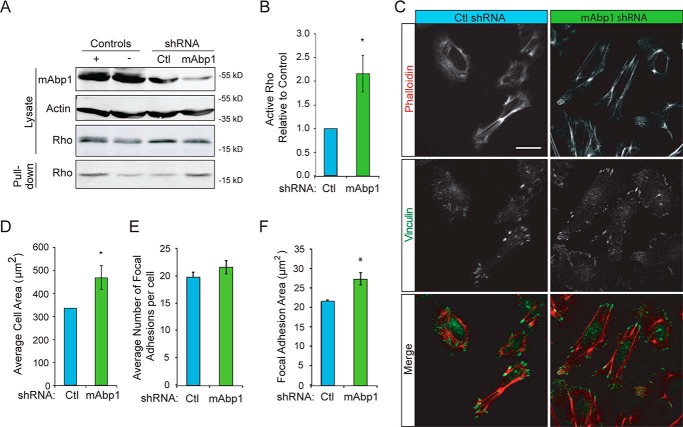
Invasive migration is dependent on Rho GTPase activityand subsequent cell contractility. To determine how mAbp1 impairs cell invasion, we used MDA-MD-231 cells that grow well in three-dimensional collagen gels to examine how mAbp1 expression modulates cell contraction. We found that in accordance with increased invasion, depletion of mAbp1 increased the contraction of collagen gels, indicating that mAbp1-deficient cells have enhanced cell contractility and force application in three-dimensional collagen (Fig. 1, J, and K). Likewise, mAbp1-deficient cells had higher RhoA activity (Fig. 2, A and B). Cell forces are applied to the extracellular matrix via adhesion sites known as focal adhesions. We found that mAbp1-deficient cells had larger focal adhesions and were more spread on two-dimensional surfaces (Fig. 2, C–F). Taken together, these results suggest that mAbp1 negatively regulates the invasion of breast cancer cells, at least in part through the modulation of Rho GTPase signaling.
FIGURE 2.
mAbp1-deficient breast cancer cells have more Rho activity and cell area. A, Rho pulldown assay. MTLn3 control and mAbp1 shRNA breast cancer cells were serum-starved 24 h then treated with 5 nm EGF for 5 min. Cell lysates were applied to Rhotekin binding beads for 30 min to capture active Rho. Western blots of cell lysate input and active Rho were pulled down. Blots were probed for mAbp1, actin, and RhoA. B, quantification of active Rho pulled down from control or mAbp1 shRNA cells. The amount of active RhoA was normalized to control; n = 4; *, p = 0.0236. C, representative images of cells cultured on fibronectin-coated coverslips and stained with rhodamine phalloidin and immunofluorescent labeling of vinculin. Scale bar = 20 μm. D, quantification of the average two-dimensional cell spread area. n > 50 cells per experiment. *, p = 0.0401. E, average number of focal adhesions per cell in MTLn3 control or mAbp1 shRNA cells. n > 50 cells per experiment. F, quantification of average focal adhesion area in control or mAbp1 shRNA cells; n > 40 cells per experiment. *, p = 0.0179. All experiments were performed in triplicate. Error bars represent the S.E.
The Inhibitory Effects of mAbp1 on Cell Invasion Requires the C-terminal SH3 Domain
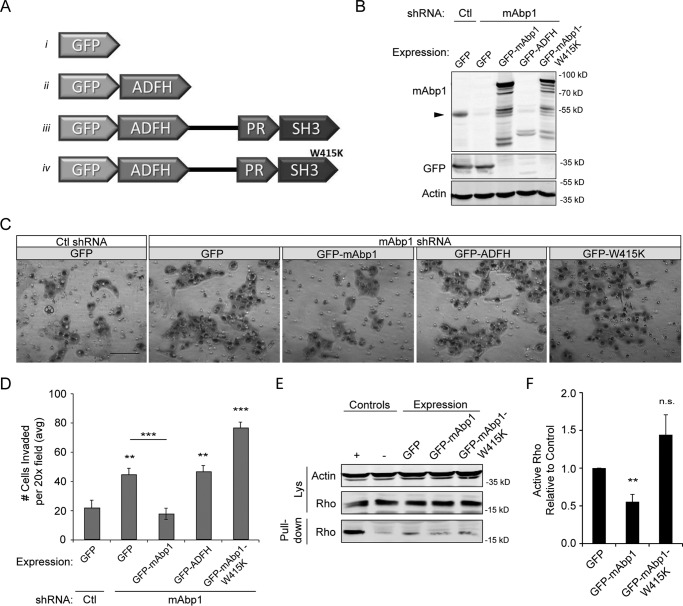
Mammalian Abp1 is an adaptor protein that binds to the actin cytoskeleton through its N-terminal ADFH domain and to dynamin (2, 33, 34), WIP (7), and other proteins through its C-terminal SH3 domain (Fig. 3A). To determine whether the SH3 domain is necessary for the inhibitory effects of mAbp1 on invasion, we expressed either full-length mAbp1, the ADFH domain alone, or an SH3 domain mutant (mAbp1-W415K) in mAbp1-deficient MTLn3 cells and measured invasion through Matrigel (Fig. 3, A–D). mAbp1-depleted cells had increased invasion, and expression of WT GFP-mAbp1 rescued this effect. Interestingly, expression of the GFP-mAbp1-W415K or the GFP-ADFH domain alone was not sufficient for rescue (Fig. 3, B–D). A caveat is that the expression levels of the ADFH domain were low in the rescue lines as shown by immunostaining (Fig. 3B). Regardless, these results suggest that a functional SH3 domain is necessary for the inhibitory effects of mAbp1 on invasion. We next asked if a functional SH3 domain was required for mAbp1 inhibition of RhoA activity. We overexpressed GFP, GFP-mAbp1, or GFP-mAbp1-W415K in MTLn3 cells and pulled down active Rho from lysates (Fig. 3, E and F). We found that RhoA activity was decreased with mAbp1 overexpression but not with mAbp1-W415K, suggesting that the SH3 domain interactions are required for the regulation of Rho activity by mAbp1.
FIGURE 3.
mAbp1 regulates MTLn3 cell invasion. A, schematic of GFP constructs with the ADFH domain alone, WT mAbp1 and mAbp1-W415K (SH3 domain mutant). B, control or mAbp1 shRNA MTLn3 cells were transiently transfected with GFP constructs, and lysates were probed for mAbp1, GFP, and actin as a loading control. The Western blot was cut to immunostain GFP and mAbp1 separately; however, GFP staining of all constructs can be seen in Fig. 4D. The arrow denotes endogenous mAbp1 expression in control and mAbp1 shRNA cells. C, transiently transfected MTLn3 cells were serum-starved for 24 h, seeded in Matrigel coated boyden chambers, and allowed to invade toward serum-containing media for 16 h. Representative images of invaded cells at 20× magnification. Scale bar = 100 μm. D, quantification of invasion of transiently transfected control or mAbp1-deficient cells. Average number of cells invaded from 12 images taken at 20× magnification. n = 4. **, p = 0.0067; ***, p = 0.0010; **, p = 0.0080; ***, p = 0.0003. E, MTLn3 cells overexpressing GFP, GFP-mAbp1, or GFP-mAbp1-W415K were serum-starved then treated with 5 nm EGF for 5 min. Active Rho was pulled down with Rhotekin binding beads and probed for RhoA and actin as a loading control. Overexpression of mAbp1, but not the SH3 mutant mAbp1-W415K, reduced RhoA activity in mAbp-deficient cells. F, quantification of active RhoA pulled down by densitometry analysis. The amount of active RhoA was normalized to control; n = 3. **, p = 0.0056. ns, not significant. Error bars represent S.E.
The N-terminal ADFH Domain of mAbp1 Interacts with FHL2
To determine how mAbp1 inhibits invasive migration, we performed a yeast two-hybrid screen with full-length human mAbp1 and identified several novel binding partners (Table 1). One protein of particular interest was FHL2, as it has been implicated in breast cancer progression. To confirm the interaction between mAbp1 and FHL2, we performed in vitro GST pulldown assays (Fig. 4A). Beads coated with GST alone did not pull down His-tagged FHL2, while beads coated with GST-mAbp1 interacted with His-FHL2. To confirm the interaction in mammalian cells, we performed a co-immunoprecipitation experiment by pulling down endogenous FHL2 and probing for mAbp1 (Fig. 4B). In addition, we expressed GFP or GFP-mAbp1 with myc-FHL2 in HEK 293 cells and immunoprecipitated GFP; myc-FHL2 interacted with GFP-mAbp1 but not GFP alone (Fig. 4C).
TABLE 1.
Yeast two-hybrid screen of human mAbp1 protein interactions
| Gene | Protein name | Function | References |
|---|---|---|---|
| ATP1B3 | ATPase, Na+/K+ transporting, β3 polypeptide | Non catalytic component of Na+/K+ ATPase transporter | 50, 51 |
| FHL2 | Four-and-a-half LIM domains 2, DRAL, SLIM-3 | Translates cell signaling pathways to transcriptional regulation. Focal adhesions. | 37, 39, 52, 53 |
| GCC2 | GRIP and coiled-coil domain containing 2, GCC185, Golgin | Peripheral membrane protein localized to trans-Golgi network. Vesicular transport between endosomes and Golgi. | 54, 55 |
| IDH3A | Isocitrate dehydrogenase | Metabolism | 56 |
| LGALS2 | Lectin galactoside-binding soluble 2 | Mutant confers risk for myocardial infarction. T-cell apoptosis. | 57, 58 |
| PTS | 6-Pyruvoyltetrahydropterin synthase | Metabolism | 59, 60 |
| RBL1 | Retinoblastoma-like 1, p107 | Regulator of cell division cycle. May have tumor suppressor activities. | 61, 62 |
| REPS1 | RALBP1-associated Eps domain csontaining 1, RalBP1 | Interacts with intersectin1, endophilin, and amphiphysin. Endocytosis. | 63 |
FIGURE 4.
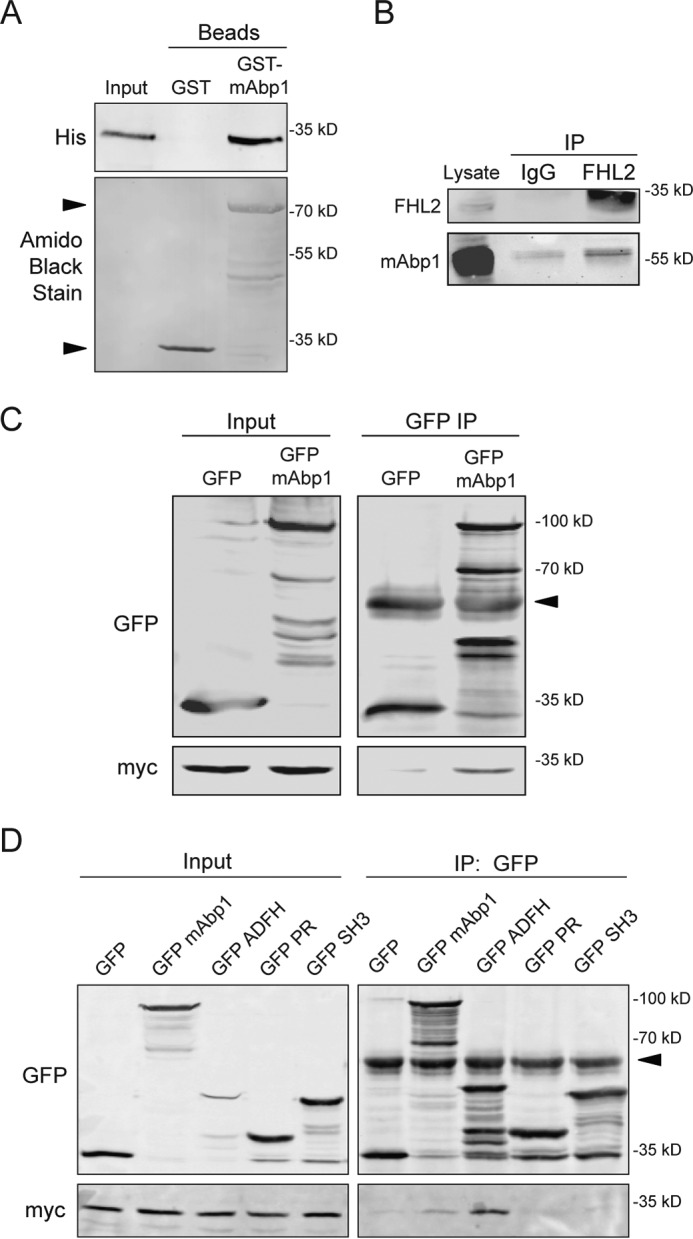
mAbp1 interacts with FHL2 through its ADFH domain. A, an in vitroGST pulldown assay was performed with purified His-tagged FHL2 and GST beads or GST-tagged mAbp1 beads. His-tagged FHL2 interacted with GST-mAbp1 but not GST beads alone. His-tagged FHL2 was probed using anti-His antibody. The presence of GST and GST-mAbp1 (arrow) was analyzed by staining the blot with Amido Black stain (lower panel). B, co-immunoprecipitation (IP) of endogenous FHL2 and mAbp1 from MTLn3 cell lysate. FHL2 was immunoprecipitated with anti-FHL2 but not with IgG alone, and co-immunoprecipitated mAbp1 was probed by Western blot. C, co-immunoprecipitation of mAbp1 and FHL2 overexpression in mammalian cells. GFP-mAbp1 and myc-FHL2 were transiently overexpressed in HEK-293 cells. Cells were lysed and incubated with GFP antibody overnight, then harvested with gamma-G-Sepharose beads. myc-FHL2 did not pull down with GFP alone but did interact with GFP-mAbp1. The blot was probed for GFP and myc. The arrow indicates IgG band. D, the ADFH domain of mAbp1 interacts with FHL2. GFP constructs containing full-length mAbp1, the ADFH domain, the proline-rich region, or the SH3 domain alone were transiently expressed with myc-FHL2 in HEK 293 cells. GFP antibody was used to pull down GFP fusion proteins, and immunoblotting was performed to detect co-immunoprecipitated myc-FHL2. The arrow indicates IgG band. All experiments were performed in triplicate.
To determine which mAbp1 domain interacts with FHL2, we expressed either the GFP-tagged ADFH domain, proline-rich region, or the SH3 domain alone, along with His-FHL2 and performed co-immunoprecipitation experiments (Fig. 4D). Surprisingly, we found that FHL2 interacted with the ADFH domain but not the SH3 or proline-rich region regions of mAbp1. Moreover, the interaction between FHL2 and the ADFH domain was more robust than FHL2 interaction with full-length mAbp1. This result suggests that conformation of full-length mAbp1 may modulate FHL2 binding. Although GFP-ADFH domain expression was low relative to full-length GFP-mAbp1 (as shown by immunofluorescence), immunoprecipitation of GFP enriched GFP-ADFH to comparable levels (Fig. 4D). Taken together, these findings suggest that FHL2 interacts directly with the ADFH domain of mAbp1. Interestingly, the ADFH domain of mAbp1 is not conserved in cortactin, thus providing a key distinction between the binding partners of mAbp1 and cortactin. It is an intriguing idea that FHL2 interactions may play a role in the differential effects of mAbp1 and cortactin on cell invasion.
FHL2 Is Necessary for Invasive Migration of Breast Cancer Cells
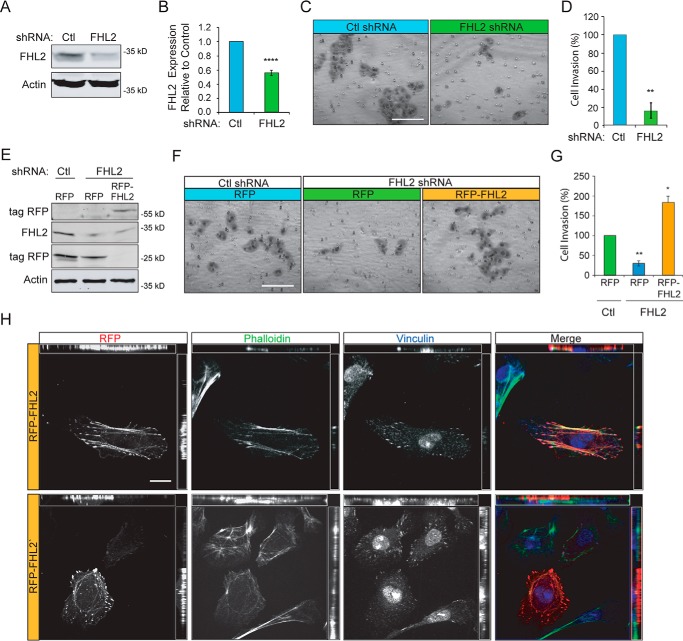
FHL2 expression is up-regulated in breast cancer (20, 21); however, the role of FHL2 during breast cancer cell invasion is not known. To determine how FHL2 affects invasive migration of breast cancer cells, we depleted FHL2 in MTLn3 cells using retroviral shRNA (Fig. 5, A and B) and found that even with a partial knockdown, invasive migration was significantly impaired (Fig. 5, C and D). To confirm the role of FHL2 during invasive migration, FHL2-deficient cell invasion was rescued with transient exogenous expression of RFP-FHL2 (Fig. 5, E–G). Consistent with previous reports, exogenous RFP-FHL2 localized at focal adhesions and along stress fibers in MTLn3 cells (Fig. 5H).
FIGURE 5.
FHL2 mediates invasive migration of MTLn3 breast cancer cells. A, control and FHL2 shRNA stable lines were generated by retroviral transduction. Immunoblots were probed for FHL2 and actin as a loading control. B, quantification of FHL2 knockdown in stable shRNA lines. n = 5; ****, p < 0.0001. C, control or FHL2 shRNA cells were serum-starved 24 h, seeded in Matrigel coated boyden chambers, and allowed to invade toward serum-containing media for 16 h. Representative images of invaded cells on a membrane at 20× magnification. Scale bar = 100 μm. D, quantification of cell invasion. Percentage of cell invasion relative to control from 12 images taken at 20× magnification and normalized to control. Data are from three independent experiments. **, p = 0.0044. E, exogenous FHL2 expression rescues invasion. RFP or RFP-FHL2 were transiently expressed in control or FHL2 shRNA MTLn3 cell lines. Lysates were probed for RFP, FHL2, and actin as a loading control. F, representative images of invasion of FHL2-deficient cells transiently expressing RFP or RFP-FHL2. Cells were serum-starved 24 h and invaded through Matrigel toward serum media for 16 h. Scale bar = 100 μm. G, percentage of cell invasion relative to control from 12 images taken at 20× magnification. Data are from three independent experiments. *, p = 0.0297; **, p = 0.0040. H, representative images of RFP-FHL2 in FHL2 shRNA cells labeled with Alexa 488 phalloidin (green) and vinculin immunostaining (blue). RFP-FHL2 localizes to focal adhesions and stress fibers and occasionally in the nucleus similar to endogenous FHL2. Scale bar = 20 μm.
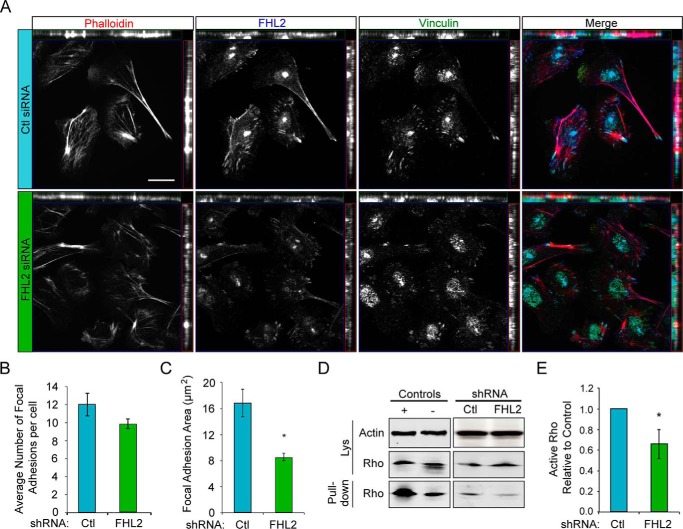
To determine if FHL2 modulates focal adhesions, we imaged focal adhesions and stress fibers in control and FHL2-deficient cells. Similar to exogenous RFP-FHL2 expression, endogenous immunofluorescence of FHL2 was localized to focal adhesions and stress fibers (Fig. 6A). We found that FHL2-deficient MTLn3 cells had reduced focal adhesion area, although the total numbers of focal adhesions were not significantly different (Fig. 6, B and C). Accordingly, we also found that FHL2-deficient cells had reduced RhoA activity by pulldown assay in the presence of EGF stimulation (Fig. 6, D and E). These findings are consistent with the reported roles for FHL2 in heart muscle and contractility of the sarcomere (35), wound healing (36), and the invasion of glioblastoma cells (25, 26). These findings suggest that FHL2 is necessary for efficient invasion of MTLn3 breast cancer cells, likely in part through its effects on Rho GTPase activity.
FIGURE 6.
FHL2 modulates focal adhesions and Rho activation. A, endogenous FHL2 localizes to focal adhesions, along some stress fibers and the cytoplasm. Control or FHL2 shRNA MTLn3 cells were cultured on fibronectin-coated coverslips and stained with rhodamine phalloidin (actin) and immunolabeled for FHL2 and vinculin. Scale bar = 20 μm. B, average focal adhesion number in MTLn3 control or FHL2 shRNA cells. n > 50 cells per experiment. C, average focal adhesion area in control or FHL2 shRNA cells; n > 40 focal adhesions per experiment; *, p = 0.0190. D, FHL2 mediates RhoA activation. Control and FHL2 shRNA cells were serum-starved 24 h and treated with 5 nm EGF for 5 min. Lysates were incubated with Rhotekin beads to pull down active Rho. RhoA levels were detected by immunoblotting. E, quantification of active RhoA pulled down by densitometry with normalization to control. Active RhoA was reduced in FHL2 deficient cells. n = 8. *, p = 0.0314. All experiments were performed in triplicate. Error bars represent S.E.
The ADFH Domain of mAbp1 Increases Invasive Migration, and This Effect Requires FHL2
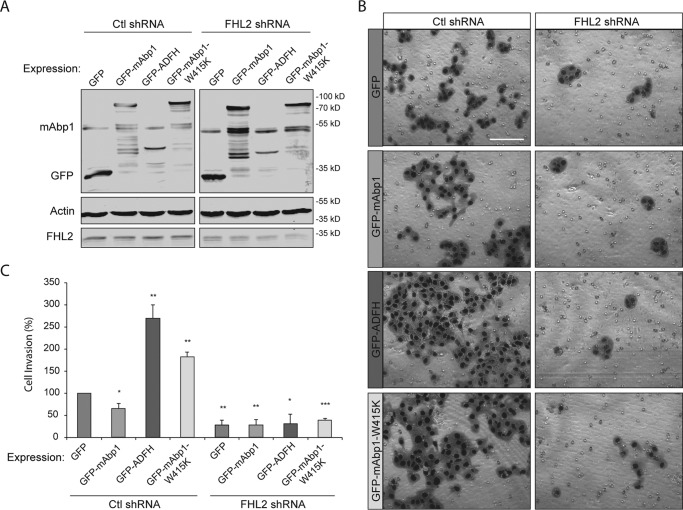
To determine whether mAbp1 affects invasion through its interaction with FHL2, we overexpressed WT mAbp1, ADFH domain alone, and mAbp1-W415K and assessed invasion through Matrigel in control and FHL2-deficient cells (Fig. 7). As expected, overexpression of full-length GFP-mAbp1 in wild-type cells impaired invasive migration. By contrast, ectopic expression of the ADFH domain alone dramatically increased invasive migration of MTLn3 cells (Fig. 7, B and C), suggesting that the ADFH domain may block the inhibitory effects of endogenous mAbp1 on invasion. In support of this idea, we found that overexpression of the SH3 domain mutant (GFP-mAbp1-W415K) also enhanced invasive migration. Our results confirm the hypothesis that mAbp1 binds FHL2 through the ADFH domain and inhibits FHL2 invasive activity via its SH3 domain or its interactions. Although the enhanced invasion of full-length mAbp1-W415K-expressing cells was not as dramatic as expression of the ADFH domain alone, this difference may be due to reduced levels of FHL2 interaction with full-length mAbp1 in comparison with the ADFH domain alone (Fig. 4D).
FIGURE 7.
FHL2 modulates invasion induced by the ADFH domain of mAbp1. A, MTLn3 lysates from control or FHL2 shRNA cells overexpressing GFP, GFP-mAbp1, GFP-ADFH, or GFP-mAbp1-W415K and probed for GFP, endogenous mAbp1, FHL2, and actin by immunoblotting. B, cells were serum-starved 24 h, seeded on Matrigel invasion chambers, and allowed to invade toward serum-containing media for 16 h. Representative images of invaded cells. Scale bar = 100 μm. C, quantification of cell invasion. Shown is the percentage of cells invaded relative to control from 12 images taken at 20× magnification. Data are from independent experiments. **, p = 0.0067; **, p = 0.0080; ***, p < 0.0030.
To determine how the interaction between mAbp1 and FHL2 modulates invasive cell migration, we determined if expression of FHL2 was necessary for the invasive migration induced by expression of the ADFH domain of mAbp1. We found that FHL2 expression was required for the induction of invasive migration with ectopic expression of the ADFH domain or W415K-mAbp1 mutant (Fig. 7, B and C). However, a potential caveat is that invasive migration may be dependent on FHL2 expression independent of mAbp1 effects. Taken together, these findings support the idea that mAbp1 sequesters FHL2 and impairs FHL2-Rho signaling to affect cell invasion. Furthermore, these results suggest that the inhibitory effect of mAbp1 on invasion requires the SH3 domain and its specific binding partners.
Discussion
Here we report a novel interaction between mAbp1 and the multifunctional adaptor protein FHL2. We show that mAbp1 impairs MTLn3 cell invasion and Rho GTPase signaling, while FHL2 increases MTLn3 invasion and RhoA activity. We show that mAbp1 interacts with FHL2 through its N-terminal ADFH domain. Our data suggest that mAbp1 regulates cell invasion at least in part through its effects on FHL2 and Rho GTPase activation. In support of this idea, expression of the ADFH domain of mAbp1, lacking the inhibitory SH3 domain, is sufficient to induce invasive cell migration, and this effect requires the expression of FHL2.
FHL2 is an adaptor protein that has known functions at focal adhesions and also may function as a transcriptional cofactor in the nucleus. The interaction between mAbp1 and FHL2 is particularly interesting as FHL2 is up-regulated during invasive migration of metastatic osteosarcoma cells, colon cancer, and glioblastomas (25, 26, 38) and has been implicated in migration during wound closure (20, 21, 25, 36). In colon cancer, FHL2 promotes epithelial to mesenchymal transition by suppressing E-cadherin and promoting vimentin and MMP-9 expression (25, 39), while in vitro, FHL2 has been shown to alter transactivation of key breast cancer related genes (13, 17, 18). Although FHL2 expression can alter the invasive capacity of cancer cells, it remains unclear whether FHL2 mediates its effects on invasion through focal adhesions or its transcription cofactor activity. Our findings suggest that FHL2 and mAbp1 regulate cell invasion through effects on focal adhesions and the activation of Rho GTPase signaling.
Cell motility and invasion are dependent on the tight regulation of Rho-ROCK signaling and cell contractility (40–43). Inhibition of Rho or ROCK impairs breast cancer invasion (44) and metastasis in vivo (45). Our work suggests that the actin-binding protein mAbp1 and FHL2 play opposing roles in modulating RhoA activity and cell invasion. We found that mAbp1 inhibits cellular contraction of collagen and inhibits RhoA activity while FHL2 facilitates RhoA activation. Not surprisingly, the activity of RhoA correlated with the invasive potential of mAbp1- and FHL2-depleted breast cancer cell lines. Future work will be needed to determine whether FHL2 and its interaction with mAbp1 regulate actomyosin contraction and invasive migration in three-dimensional collagen gels or within tissues in vivo.
Our findings suggest that the SH3 domain of mAbp1 is necessary for the inhibitory effects of mAbp1 on cell invasion. Moreover, our results suggest that mAbp1 may inhibit invasion by sequestering FHL2 and that this inhibitory effect requires the SH3 domain of mAbp1. Interestingly, we found that FHL2 interacted more strongly with the ADFH domain of mAbp1 compared to full-length mAbp1, suggesting that the conformation of full-length mAbp1 may regulate the interaction between mAbp1 and FHL2. Overexpression of the ADFH domain in cells with endogenous mAbp1 resulted in enhanced invasion dependent on FHL2 expression, suggesting that the ADFH domain was sufficient to overcome the inhibitory effects of full-length mAbp1 on cell invasion. An increased affinity of the ADFH domain for FHL2 may be sufficient to block endogenous mAbp1 binding to FHL2 thereby modulating FHL2 function and Rho GTPase signaling.
These results suggest that the mAbp1-FHL2 interaction may be regulated by conformational changes in mAbp1 potentially through phosphorylation, additional protein interactions, SH3 domain modifications, or protease activity. Recently, putative phosphorylation sites have been identified by mass spectroscopy of His-tagged mAbp1 in HEK 293 cells (46). Serine 99 and threonine 114 are both located in the ADFH domain and are attractive targets that may alter FHL2 binding.
mAbp1 serine 269 is also an interesting potential regulatory target since phosphorylation of mAbp1 serine 269 by the maternal embryonic leucine zipper kinase (MELK) promotes tumor growth and invasion of MCF-7 breast cancer cells. Our work is the first to suggest that endogenous mAbp1 functions as an inhibitor of cancer cell invasion. It will be interesting to determine whether phosphorylation of serine 269 affects the interaction between mAbp1 and FHL2 and cell invasion (47). Furthermore, it is intriguing that mAbp1 serine 269 is close to the calpain-2 cleavage sites of mAbp1 (7, 30). Calpain-2 activity is required for several cancer lines including breast cancer (29, 30, 48, 49). Calpain-2 cleavage of mAbp1 would effectively separate the N-terminal ADFH domain from the regulatory C-terminal SH3 domain and thus could alter cell invasion, possibly through ADFH interactions with FHL2. These possible modes of regulation of the mAbp1-FHL2 interaction remain to be tested and would provide further insight into potential targets that modulate the invasive migration of cancer cells.
Author Contributions
L. R. B. contributed to the intellectual design of experiments, execution, and analysis of experiments and wrote the manuscript. D. B. contributed to the design of experiments, execution of the yeast two-hybrid experiments, and GST pulldown assays. S. D. O. contributed confocal immunofluorescent images. A. H. contributed to the intellectual design and editing of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 CA009135 (NCI Cancer Biology; to L. B.) and R01 CA085862 (NCI; to A. H.). The authors declare no competing or financial interests. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- mAbp1
- mammalian actin-binding protein-1
- FHL2
- four-and-one-half LIM domain protein-2
- ADFH
- actin depolymerization factor homology
- SH3
- Src homology 3
- TRITC
- tetramethylrhodamine isothiocyanate
- RFP
- red fluorescent protein
- ROCK
- Rho-associated protein kinase
- GTPγS
- guanosine 5′-O-(thiotriphosphate).
References
- 1. Yamaguchi H., and Condeelis J. (2007) Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 1773, 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu W., and Stamnes M. (2006) The actin-depolymerizing factor homology and charged/helical domains of drebrin and mAbp1 direct membrane binding and localization via distinct interactions with actin. J. Biol. Chem. 281, 11826–11833 [DOI] [PubMed] [Google Scholar]
- 3. Connert S., Wienand S., Thiel C., Krikunova M., Glyvuk N., Tsytsyura Y., Hilfiker-Kleiner D., Bartsch J. W., Klingauf J., and Wienands J. (2006) SH3P7/mAbp1 deficiency leads to tissue and behavioral abnormalities and impaired vesicle transport. EMBO J. 25, 1611–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kessels M. M., Engqvist-Goldstein A. E., and Drubin D. G. (2000) Association of mouse actin-binding protein 1 (mAbp1/SH3P7), an Src kinase target, with dynamic regions of the cortical actin cytoskeleton in response to Rac1 activation. Mol. Biol. Cell 11, 393–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao H., Orth J. D., Chen J., Weller S. G., Heuser J. E., and McNiven M. A. (2003) Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol. Cell. Biol. 23, 2162–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu H., and Parsons J. T. (1993) Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J. Cell Biol. 120, 1417–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cortesio C. L., Perrin B. J., Bennin D. A., and Huttenlocher A. (2010) Actin-binding protein-1 interacts with WASp-interacting protein to regulate growth factor-induced dorsal ruffle formation. Mol. Biol. Cell 21, 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boateng L. R., Cortesio C. L., and Huttenlocher A. (2012) Src-mediated phosphorylation of mammalian Abp1 (DBNL) regulates podosome rosette formation in transformed fibroblasts. J. Cell Sci. 125, 1329–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johannessen M., Møller S., Hansen T., Moens U., and Van Ghelue M. (2006) The multifunctional roles of the four-and-a-half-LIM only protein FHL2. CMLS 63, 268–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Samson T., Smyth N., Janetzky S., Wendler O., Müller J. M., Schüle R., von der Mark H., von der Mark K., and Wixler V. (2004) The LIM-only proteins FHL2 and FHL3 interact with α- and β-subunits of the muscle α7β1 integrin receptor. J. Biol. Chem. 279, 28641–28652 [DOI] [PubMed] [Google Scholar]
- 11. Wixler V., Geerts D., Laplantine E., Westhoff D., Smyth N., Aumailley M., Sonnenberg A., and Paulsson M. (2000) The LIM-only protein DRAL/FHL2 binds to the cytoplasmic domain of several α and β integrin chains and is recruited to adhesion complexes. J. Biol. Chem. 275, 33669–33678 [DOI] [PubMed] [Google Scholar]
- 12. Coghill I. D., Brown S., Cottle D. L., McGrath M. J., Robinson P. A., Nandurkar H. H., Dyson J. M., and Mitchell C. A. (2003) FHL3 is an actin-binding protein that regulates α-actinin-mediated actin bundling: FHL3 localizes to actin stress fibers and enhances cell spreading and stress fiber disassembly. J. Biol. Chem. 278, 24139–24152 [DOI] [PubMed] [Google Scholar]
- 13. Kobayashi S., Shibata H., Yokota K., Suda N., Murai A., Kurihara I., Saito I., and Saruta T. (2004) FHL2, UBC9, and PIAS1 are novel estrogen receptor α-interacting proteins. Endocr. Res. 30, 617–621 [DOI] [PubMed] [Google Scholar]
- 14. Labalette C., Renard C. A., Neuveut C., Buendia M. A., and Wei Y. (2004) Interaction and functional cooperation between the LIM protein FHL2, CBP/p300, and β-catenin. Mol. Cell. Biol. 24, 10689–10702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin B., Schneider R., Janetzky S., Waibler Z., Pandur P., Kühl M., Behrens J., von der Mark K., Starzinski-Powitz A., and Wixler V. (2002) The LIM-only protein FHL2 interacts with β-catenin and promotes differentiation of mouse myoblasts. J. Cell Biol. 159, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morlon A., and Sassone-Corsi P. (2003) The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc. Natl. Acad. Sci. U.S.A. 100, 3977–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan J., Zhu J., Zhong H., Lu Q., Huang C., and Ye Q. (2003) BRCA1 interacts with FHL2 and enhances FHL2 transactivation function. FEBS Lett. 553, 183–189 [DOI] [PubMed] [Google Scholar]
- 18. Xiong Z., Ding L., Sun J., Cao J., Lin J., Lu Z., Liu Y., Huang C., and Ye Q. (2010) Synergistic repression of estrogen receptor transcriptional activity by FHL2 and Smad4 in breast cancer cells. IUBMB Life 62, 669–676 [DOI] [PubMed] [Google Scholar]
- 19. Chan K. K., Tsui S. K., Lee S. M., Luk S. C., Liew C. C., Fung K. P., Waye M. M., and Lee C. Y. (1998) Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene 210, 345–350 [DOI] [PubMed] [Google Scholar]
- 20. Martin B. T., Kleiber K., Wixler V., Raab M., Zimmer B., Kaufmann M., and Strebhardt K. (2007) FHL2 regulates cell cycle-dependent and doxorubicin-induced p21Cip1/Waf1 expression in breast cancer cells. Cell Cycle 6, 1779–1788 [DOI] [PubMed] [Google Scholar]
- 21. Gabriel B., Fischer D. C., Orlowska-Volk M., zur Hausen A., Schüle R., Müller J. M., and Hasenburg A. (2006) Expression of the transcriptional coregulator FHL2 in human breast cancer: a clinicopathologic study. J. Soc. Gynecol. Investig. 13, 69–75 [DOI] [PubMed] [Google Scholar]
- 22. Gabriel B., Mildenberger S., Weisser C. W., Metzger E., Gitsch G., Schüle R., and Müller J. M. (2004) Focal adhesion kinase interacts with the transcriptional coactivator FHL2 and both are overexpressed in epithelial ovarian cancer. Anticancer Res. 24, 921–927 [PubMed] [Google Scholar]
- 23. Kinoshita M., Nakagawa T., Shimizu A., and Katsuoka Y. (2005) Differently regulated androgen receptor transcriptional complex in prostate cancer compared with normal prostate. Int. J. Urol. 12, 390–397 [DOI] [PubMed] [Google Scholar]
- 24. Borczuk A. C., Shah L., Pearson G. D., Walter K. L., Wang L., Austin J. H., Friedman R. A., and Powell C. A. (2004) Molecular signatures in biopsy specimens of lung cancer. Am. J. Resp. Crit. Care Med. 170, 167–174 [DOI] [PubMed] [Google Scholar]
- 25. Zhang W., Jiang B., Guo Z., Sardet C., Zou B., Lam C. S., Li J., He M., Lan H. Y., Pang R., Hung I. F., Tan V. P., Wang J., and Wong B. C. (2010) Four-and-a-half LIM protein 2 promotes invasive potential and epithelial-mesenchymal transition in colon cancer. Carcinogenesis 31, 1220–1229 [DOI] [PubMed] [Google Scholar]
- 26. Li M., Wang J., Ng S. S., Chan C. Y., Chen A. C., Xia H. P., Yew D. T., Wong B. C., Chen Z., Kung H. F., and Lin M. C. (2008) The four-and-a-half-LIM protein 2 (FHL2) is overexpressed in gliomas and associated with oncogenic activities. Glia 56, 1328–1338 [DOI] [PubMed] [Google Scholar]
- 27. Westphal P., Mauch C., Florin A., Czerwitzki J., Olligschläger N., Wodtke C., Schüle R., Büttner R., and Friedrichs N. (2015) Enhanced FHL2 and TGF-β1 expression is associated with invasive growth and poor survival in malignant melanomas. Am. J. Clin. Path. 143, 248–256 [DOI] [PubMed] [Google Scholar]
- 28. Goode B. L., Rodal A. A., Barnes G., and Drubin D. G. (2001) Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J. Cell Biol. 153, 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jang H. S., Lal S., and Greenwood J. A. (2010) Calpain 2 is required for glioblastoma cell invasion: regulation of matrix metalloproteinase 2. Neurochem. Res. 35, 1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mamoune A., Luo J. H., Lauffenburger D. A., and Wells A. (2003) Calpain-2 as a target for limiting prostate cancer invasion. Cancer Res. 63, 4632–4640 [PubMed] [Google Scholar]
- 31. Ruoslahti E., Hayman E. G., Pierschbacher M., and Engvall E. (1982) Fibronectin: purification, immunochemical properties, and biological activities. Methods Enzymol. 82, 803–831 [DOI] [PubMed] [Google Scholar]
- 32. Suzuki K., Bose P., Leong-Quong R. Y., Fujita D. J., and Riabowol K. (2010) REAP: a two-minute fractionation method. BMC Res. Notes. 3, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kessels M. M., Engqvist-Goldstein A. E., Drubin D. G., and Qualmann B. (2001) Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J. Cell Biol. 153, 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He K., Xing R., Yan X., Tian A., Zhang M., Yuan J., Lv Z., Fang X., Li Z., and Zhang Y. (2015) Mammalian actin-binding protein 1/HIP-55 is essential for the scission of clathrin-coated pits by regulating dynamin-actin interaction. FASEB J. 29, 2495–2503 [DOI] [PubMed] [Google Scholar]
- 35. Lange S., Auerbach D., McLoughlin P., Perriard E., Schäfer B. W., Perriard J. C., and Ehler E. (2002) Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J. Cell Sci. 115, 4925–4936 [DOI] [PubMed] [Google Scholar]
- 36. Wixler V., Hirner S., Müller J. M., Gullotti L., Will C., Kirfel J., Günther T., Schneider H., Bosserhoff A., Schorle H., Park J., Schüle R., and Buettner R. (2007) Deficiency in the LIM-only protein Fhl2 impairs skin wound healing. J. Cell Biol. 177, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Müller J. M., Metzger E., Greschik H., Bosserhoff A.-K., Mercep L., Buettner R., and Schüle R. (2002) The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J. 21, 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brun J., Dieudonné F. X., Marty C., Müller J., Schüle R., Patiño-García A., Lecanda F., Fromigué O., and Marie P. J. (2013) FHL2 silencing reduces Wnt signaling and osteosarcoma tumorigenesis in vitro and in vivo. PloS ONE 8, e55034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang W., Wang J., Zou B., Sardet C., Li J., Lam C. S., Ng L., Pang R., Hung I. F., Tan V. P., Jiang B., and Wong B. C. (2011) Four and a half LIM protein 2 (FHL2) negatively regulates the transcription of E-cadherin through interaction with Snail1. Eur. J. Cancer 47, 121–130 [DOI] [PubMed] [Google Scholar]
- 40. Yoshioka K., Nakamori S., and Itoh K. (1999) Overexpression of small GTP-binding protein RhoA promotes invasion of tumor cells. Cancer Res. 59, 2004–2010 [PubMed] [Google Scholar]
- 41. Itoh K., Yoshioka K., Akedo H., Uehata M., Ishizaki T., and Narumiya S. (1999) An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat. Med. 5, 221–225 [DOI] [PubMed] [Google Scholar]
- 42. Sahai E., and Marshall C. J. (2003) Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5, 711–719 [DOI] [PubMed] [Google Scholar]
- 43. Wyckoff J. B., Pinner S. E., Gschmeissner S., Condeelis J. S., and Sahai E. (2006) ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 16, 1515–1523 [DOI] [PubMed] [Google Scholar]
- 44. Wu D., Asiedu M., and Wei Q. (2009) Myosin-interacting guanine exchange factor (MyoGEF) regulates the invasion activity of MDA-MB-231 breast cancer cells through activation of RhoA and RhoC. Oncogene 28, 2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu J., Yue P., Artym V. V., Mueller S. C., and Guo W. (2009) The role of the exocyst in matrix metalloproteinase secretion and actin dynamics during tumor cell invadopodia formation. Mol. Biol. Cell 20, 3763–3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu N., Sun N., Gao X., and Li Z. (2014) Phosphosite mapping of HIP-55 protein in mammalian cells. Int. J. Mol. Sci. 15, 4903–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chung S., Suzuki H., Miyamoto T., Takamatsu N., Tatsuguchi A., Ueda K., Kijima K., Nakamura Y., and Matsuo Y. (2012) Development of an orally administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget 3, 1629–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cortesio C. L., Chan K. T., Perrin B. J., Burton N. O., Zhang S., Zhang Z. Y., and Huttenlocher A. (2008) Calpain 2 and PTP1B function in a novel pathway with Src to regulate invadopodia dynamics and breast cancer cell invasion. J. Cell Biol. 180, 957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carragher N. O., Walker S. M., Scott Carragher L. A., Harris F., Sawyer T. K., Brunton V. G., Ozanne B. W., and Frame M. C. (2006) Calpain 2 and Src dependence distinguishes mesenchymal and amoeboid modes of tumour cell invasion: a link to integrin function. Oncogene 25, 5726–5740 [DOI] [PubMed] [Google Scholar]
- 50. Malik N., Canfield V., Sanchez-Watts G., Watts A. G., Scherer S., Beatty B. G., Gros P., and Levenson R. (1998) Structural organization and chromosomal localization of the human Na, K-ATPase β3 subunit gene and pseudogene. Mamm. Genome 9, 136–143 [DOI] [PubMed] [Google Scholar]
- 51. Malik N., Canfield V. A., Beckers M. C., Gros P., and Levenson R. (1996) Identification of the mammalian Na,K-ATPase β3 subunit. J. Biol. Chem. 271, 22754–22758 [DOI] [PubMed] [Google Scholar]
- 52. Morgan M. J., and Madgwick A. J. (1996) Slim defines a novel family of LIM-proteins expressed in skeletal muscle. Biochem. Biophys. Res. Commun. 225, 632–638 [DOI] [PubMed] [Google Scholar]
- 53. Wei Y., Renard C. A., Labalette C., Wu Y., Lévy L., Neuveut C., Prieur X., Flajolet M., Prigent S., and Buendia M. A. (2003) Identification of the LIM protein FHL2 as a coactivator of β-catenin. J. Biol. Chem. 278, 5188–5194 [DOI] [PubMed] [Google Scholar]
- 54. Luke M. R., Kjer-Nielsen L., Brown D. L., Stow J. L., and Gleeson P. A. (2003) GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 278, 4216–4226 [DOI] [PubMed] [Google Scholar]
- 55. Derby M. C., Lieu Z. Z., Brown D., Stow J. L., Goud B., and Gleeson P. A. (2007) The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic 8, 758–773 [DOI] [PubMed] [Google Scholar]
- 56. Kim Y.-O., Oh I.-U., Park H.-S., Jeng J., Song B. J., Huh T.-L. (1995) Characterization of a cDNA clone for human NAD(+)-specific isocitrate dehydrogenase α-subunit and structural comparison with its isoenzymes from different species. Biochem. J. 308, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ozaki K., Inoue K., Sato H., Iida A., Ohnishi Y., Sekine A., Sato H., Odashiro K., Nobuyoshi M., Hori M., Nakamura Y., and Tanaka T. (2004) Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-α secretion in vitro. Nature 429, 72–75 [DOI] [PubMed] [Google Scholar]
- 58. Sturm A., Lensch M., André S., Kaltner H., Wiedenmann B., Rosewicz S., Dignass A. U., and Gabius H. J. (2004) Human galectin-2: novel indicator of T cell apoptosis with distinct profile of caspase activation. J. Immunol. 173, 3825–3837 [DOI] [PubMed] [Google Scholar]
- 59. Blau N., Scherer-Oppliger T., Baumer A., Riegel M., Matasovic A., Schinzel A., Jaeken J., and Thöny B. (2000) Isolated central form of tetrahydrobiopterin deficiency associated with hemizygosity on chromosome 11q and a mutant allele of PTPS. Hum. Mutat. 16, 54–60 [DOI] [PubMed] [Google Scholar]
- 60. Thöny B., Leimbacher W., Bürgisser D., and Heizmann C.W. (1992) Human 6-pyruvoyltetrahydropterin synthase: cDNA cloning and heterologous expression of the recombinant enzyme. Biochem. Biophys. Res. Commun. 189, 1437–1443 [DOI] [PubMed] [Google Scholar]
- 61. Zhu L., van den Heuvel S., Helin K., Fattaey A., Ewen M., Livingston D., Dyson N., Harlow E. (1993) Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 7, 1111–1125 [DOI] [PubMed] [Google Scholar]
- 62. O'Connor R. J., Schaley J. E., Feeney G., and Hearing P. (2001) The p107 tumor suppressor induces stable E2F DNA binding to repress target promoters. Oncogene 20, 1882–1891 [DOI] [PubMed] [Google Scholar]
- 63. Dergai O., Novokhatska O., Dergai M., Skrypkina I., Tsyba L., Moreau J., and Rynditch A. (2010) Intersectin 1 forms complexes with SGIP1 and Reps1 in clathrin-coated pits. Biochem. Biophys. Res. Commun. 402, 408–413 [DOI] [PubMed] [Google Scholar]