Abstract
Positive strand RNA viruses replicate via a virally encoded RNA-dependent RNA polymerase (RdRP) that uses a unique palm domain active site closure mechanism to establish the canonical two-metal geometry needed for catalysis. This mechanism allows these viruses to evolutionarily fine-tune their replication fidelity to create an appropriate distribution of genetic variants known as a quasispecies. Prior work has shown that mutations in conserved motif A drastically alter RdRP fidelity, which can be either increased or decreased depending on the viral polymerase background. In the work presented here, we extend these studies to motif D, a region that forms the outer edge of the NTP entry channel where it may act as a nucleotide sensor to trigger active site closure. Crystallography, stopped-flow kinetics, quench-flow reactions, and infectious virus studies were used to characterize 15 engineered mutations in coxsackievirus B3 polymerase. Mutations that interfere with the transport of the metal A Mg2+ ion into the active site had only minor effects on RdRP function, but the stacking interaction between Phe364 and Pro357, which is absolutely conserved in enteroviral polymerases, was found to be critical for processive elongation and virus growth. Mutating Phe364 to tryptophan resulted in a genetically stable high fidelity virus variant with significantly reduced pathogenesis in mice. The data further illustrate the importance of the palm domain movement for RdRP active site closure and demonstrate that protein engineering can be used to alter viral polymerase function and attenuate virus growth and pathogenesis.
Keywords: plus-stranded RNA virus, protein engineering, vaccine development, viral polymerase, virology, polymerase fidelity
Introduction
The RNA-dependent RNA polymerases (RdRPs)2 from positive strand RNA viruses close their active sites for catalysis via a subtle NTP-induced conformational change within conserved motifs A and C (1). This palm domain-based closure mechanism differs from what is observed in other classes of replicative polymerases where the palm is fully structured prior to NTP binding, and the nascent base pair is delivered into the active site for catalysis via molecular motions originating in the finger domain (2, 3). Despite these different molecular motions, the structural end points are essentially equivalent, and the RdRPs retain the highly conserved polymerase active site geometry with aspartate residues and a magnesium-catalyzed two-metal reaction mechanism (4, 5). The origin of the palm-based movement in the viral RdRPs is likely the conserved molecular contact between the finger and thumb domains that stabilizes the protein structure at the expense of reducing finger domain flexibility (6).
One key characteristic of the viral RdRPs is their relatively low replication fidelity with mutation frequencies of 10−4–10−5 that result in a heterogeneous virus population often referred to as a quasispecies (7, 8). The population consensus sequence defines a particular virus and strain, but closer inspection of individual genomes reveals that they each contain a few random point mutations relative to the consensus. This pool of continually generated genetic diversity allows RNA viruses to rapidly adapt to different environments, enabling efficient replication in multiple cell types when infecting a host organism. The quasispecies population diversity is critically important for pathogenesis and virus growth, which can be attenuated in vivo by either decreasing diversity with high fidelity polymerase or increasing diversity with low fidelity polymerases (9–12).
The molecular interactions involved in the palm domain-based active site closure step led us to previously carry out mutagenesis studies showing that the fidelity of coxsackievirus B3 (CVB3) and poliovirus polymerases can be drastically altered by mutations within motif A (13). These structurally homologous enzymes differ 3–4-fold in processive elongation rates and nucleotide selectivity. Interestingly, mutations at structurally identical positions tend to increase the fidelity of the lower fidelity poliovirus polymerase but decrease the fidelity of the inherently higher fidelity CVB3 polymerase. Those data demonstrated that the RdRP active site closure mechanism provides a platform for the evolutionary fine-tuning of virus replication fidelity and suggested that protein engineering approaches to alter fidelity may be an avenue for developing live-attenuated virus vaccines.
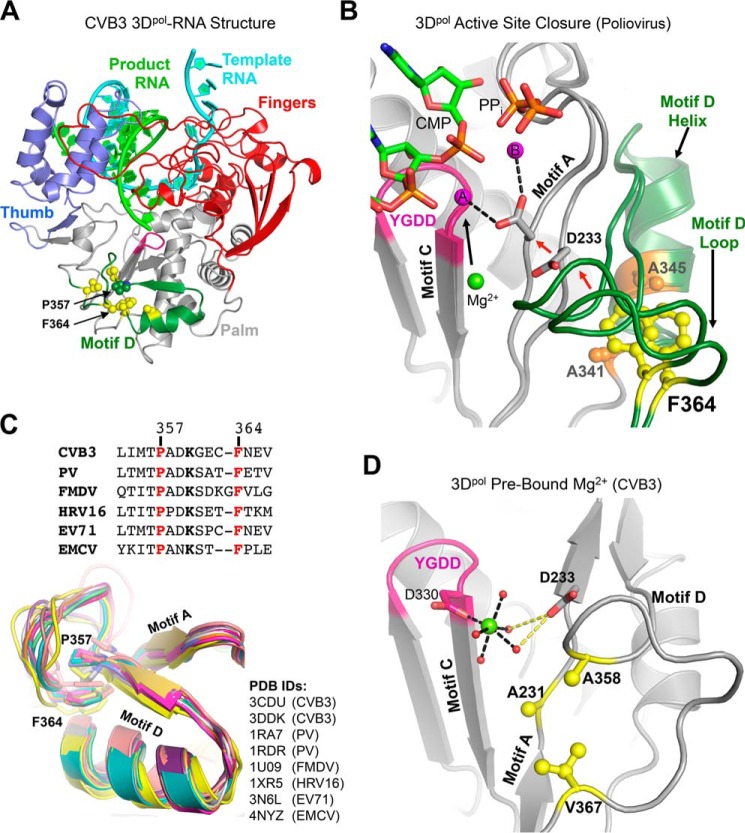
Multiple lines of experimental data have also identified motif D as an important regulator of RdRP function and as a reporter for active site conformational states. Motif D forms the outer rim of the NTP entry channel and is located immediately exterior to motif A (Fig. 1). NMR dynamics measurements using sparse 13C labeling show significant changes in local motion within motif D upon nucleotide binding (14), molecular dynamics trajectories suggest a role in NTP transport into the active site (15, 16), and kinetic isotope effects indicate that a conserved lysine in motif D is a proton donor during catalysis (17). Comparisons of open and closed active site conformations in RdRP crystal structures suggest an essentially rigid body movement of motif D that is tightly coupled to the movement of the adjacent motif A; there are only minor internal differences in how the motif is packed against the rest of the polymerase structure in the two states (Fig. 1B). However, Phe363 of poliovirus 3Dpol appears to undergo a sliding motion atop the motif D α-helix when comparing the open versus closed active site conformations (Fig. 1B). Structures of CVB3 polymerase have shown that certain motif A mutations that affect active site closure result in large conformational changes within motif D, including the displacement of Phe364, the structural equivalent of poliovirus 3Dpol Phe363, from its binding pocket (13). CVB3 elongation complex structures also show density for a polymerase-bound “metal A” Mg2+ ion that must be transported ≈5 Å into the catalytic center during active site closure where it would join the metal B ion that is delivered as part of the NTP-Mg2+ complex (1, 18). The extended hydration shell of the metal A ion and its likely path into the active site involve motif D, suggesting that the motif may play a role in controlling the dynamics of delivering Mg2+ for catalysis.
FIGURE 1.
Structural overview of picornaviral RdRP active site movements and motif D conservation. A, coxsackievirus B3 3Dpol elongation complex structure (Protein Data Bank code 4K4X) highlighting the location of motif D (green) within the palm domain and residues targeted for mutation (yellow spheres). B, close-up view of two conformations of the poliovirus 3Dpol elongation complex active site (Protein Data Bank codes 3OL6 and 3OL7) showing the concerted movements of motifs A and D that close the active site and reposition Asp233 and the prebound metal A Mg2+ ion (green sphere) to yield the two-metal ion (magenta) coordination geometry required for catalysis. Note the sliding movement of Phe364 atop Ala341 and Ala345 that are located on the motif D helix (CVB3 numbering used). C, sequence and structure alignment of the motif D loop from multiple picornaviruses showing the conservation of residues Pro357 and Phe364 (CVB3 numbering) that likely stabilize the loop conformation. The maximum likelihood structural superpositioning (34, 35) emphasizes how the proline-phenylalanine interaction stabilizes the conserved architecture of the motif D loop. D, close-up view of the open conformation CVB3 3Dpol active site (Protein Data Bank 4K4Z) showing the hydration shell around the prebound Mg2+ ion (green). Mg2+ coordination by waters (red spheres) and Asp330 is indicated by black dashes; Asp233 hydrogen bonds to two of the coordinating waters are shown as yellow dashes. Residues framing this hydration network that were targeted for mutagenesis are shown as yellow sticks. PV, poliovirus; FMDV, foot-and-mouth disease virus; EMCV, encephalomyocarditis virus; HRV, human rhinovirus; EV, enterovirus.
To further assess the role of motif D in controlling CVB3 polymerase rate and replication fidelity, we have carried out a biochemistry and virology study of CVB3 polymerase mutations that target two groups of structural interactions. First, mutations of highly conserved Phe364 (Fig. 1C) and its binding pocket slow processive elongation up to 7-fold in vitro and increase virus replication fidelity 2-fold in vivo, giving rise to the first genetically stable high fidelity CVB3 variant viruses. Second, mutations that disrupt the hydration network surrounding the bound metal A Mg2+ ion (Fig. 1D) also slow the polymerase and give rise to stable progeny viruses, but their effects on both rate and fidelity are fairly minor. These data provide further insights into the molecular mechanisms underlying viral RdRP active site closure and provide additional control points for engineering viral polymerase fidelity.
Experimental Procedures
Protein Expression and Purification
CVB3 polymerases were expressed in Escherichia coli from ubiquitin fusion constructs to generate the native N terminus required for activity (19) and purified as described previously (13). CVB3 mutant polymerases were generated using the QuikChange site-directed mutagenesis protocol and verified by DNA sequencing.
Crystallization and Structure Determination
CVB3 3Dpol crystals were grown at 16 °C by hanging drop vapor diffusion using NaCl and ammonium sulfate conditions (13). Crystals were harvested and frozen in liquid nitrogen, and diffraction data were collected at the Molecular Biology Consortium beamline 4.2.2 (Advanced Light Source, Berkeley, CA). Data were processed with XDS (20), and the structures were solved by molecular replacement using the wild type CVB3 3Dpol (Protein Data Bank code 3DDK) as the search model. Model building, refinement, and validation were performed with Coot (21), Phenix (22), and MolProbity (23), respectively, as distributed in the SBgrid suite (24).
Processive Elongation Kinetics
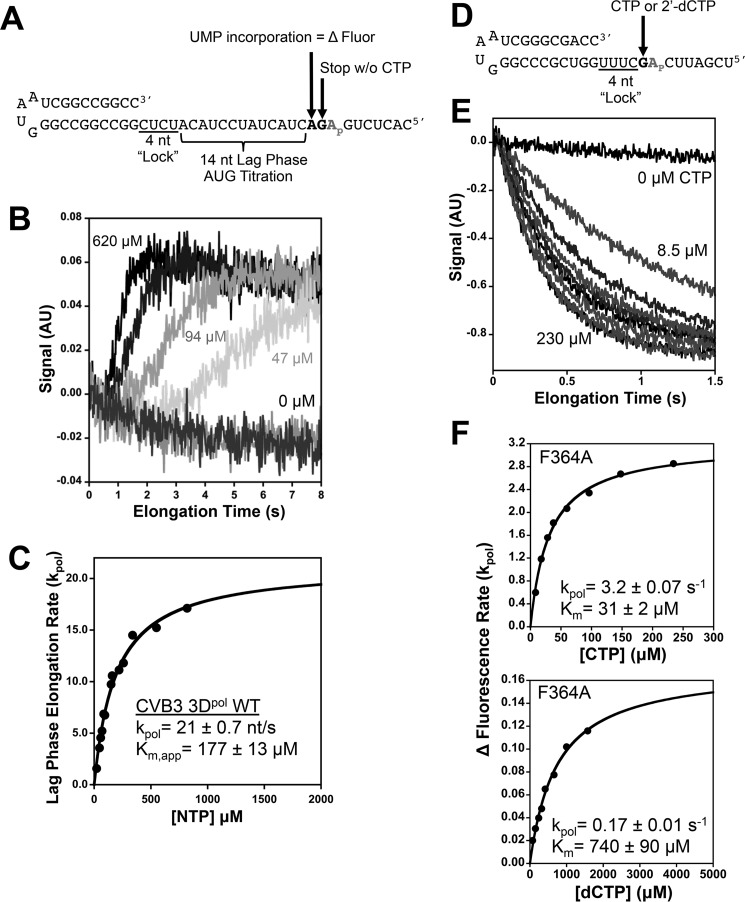
A synthetic hairpin primer-template RNA with an internal 2-aminopurine (2AP) in the template segment was used to measure processive elongation rates via a lag phase that represents elongation through 14 nucleotides (Fig. 3A) after which the 2AP is in the +2 position where it is fully unstacked from neighboring bases and exhibits maximal fluorescence (Fig. 3B). Kinetics experiments were performed with a Bio-Logic SFM-4000 titrating stopped-flow instrument equipped with a MOS-500 spectrometer. Fluorescence excitation was at 313 nm with a 10-nm bandwidth, and emission was detected using a Brightline 370/36 nm band pass filter (Semrock Inc., Rochester, NY). Preinitiated elongation complexes were generated with an excess of enzyme to drive RNA binding (7.5 μm 3Dpol, 5 μm snap cooled RNA, 60 μm each ATP and GTP), and after 10 min the samples were diluted to a final RNA concentration of 50 nm with SF buffer (25 mm HEPES, pH 7.0, 112.5 mm NaCl, 8 mm MgCl2, and 2 mm tris(2-carboxyethyl)phosphine). This was loaded into the stopped-flow instrument where the RNA concentration was further reduced to 25 nm during the stopped-flow experiments done at 30 °C.
FIGURE 3.
Processive elongation and single nucleotide incorporation measured by stopped-flow 2-aminopurine fluorescence. A, hairpin primer-template RNA on which preinitiated elongation complexes are assembled by only supplying GTP and ATP in the reaction, causing them to stall at a +4 product. Stopped-flow addition of ATP + GTP + UTP then results in rapid elongation to the +18 product where the complex stalls because CTP is not present. This translocates a unique 2-aminopurine base analog (Ap) into the +2 binding pocket on the polymerase where its fluorescence increases because it is fully unstacked from both neighboring bases. B, stopped-flow traces showing the shortening of the lag phase that reflects faster processive elongation as the NTP concentration is increased. C, curve fitting of the rates extracted from the lag phase versus NTP concentration allows the determination of processive elongation rates and Km values. D, structure of the hairpin primer-template RNA used for single nucleotide incorporation assays where preinitiation with ATP and GTP result in stalled elongation complexes with the template strand 2-aminopurine in the +2 position. E, stopped-flow traces demonstrating the single step quenching of fluorescence as 2-aminopurine is translocated from the +2 position to the +1 position. F, analysis of CVB3 3Dpol F364A single nucleotide turnover rates as a function of CTP (top) and 2′-dCTP (bottom) concentrations. AU, arbitrary units; nt, nucleotide(s).
Single Nucleotide Incorporation Assays
Stopped-flow experiments using a shorter hairpin primer-template RNA were used to determine the incorporation rates of a single cytosine opposite a templating guanosine positioned immediately downstream from the 2AP (Fig. 3D). Preinitiated elongation complexes were generated as in the processive elongation experiments but with the 2AP now prepositioned in the +2 site such that CTP incorporation and subsequent translocation will move the 2AP into the templating +1 site where the fluorescence is quenched due to stacking on the nascent duplex. These reactions were diluted to a final RNA concentration of 10 nm in the reaction cell, and maximal single nucleotide incorporation rates (kpol) and apparent Km values were determined by titrating CTP and analyzing the observed 2-aminopurine translocation rates (Fig. 3E). Nucleotide discrimination by the different mutant polymerases was assessed by comparing the incorporation of CTP with that of 2′-deoxy-CTP using the same assay and calculating the ratio of the CTP and dCTP catalytic efficiencies using the following relationship: Discrimination factor = (kpol/Km)CTP ÷ (kpol/Km)dCTP.
Quench-flow Kinetics
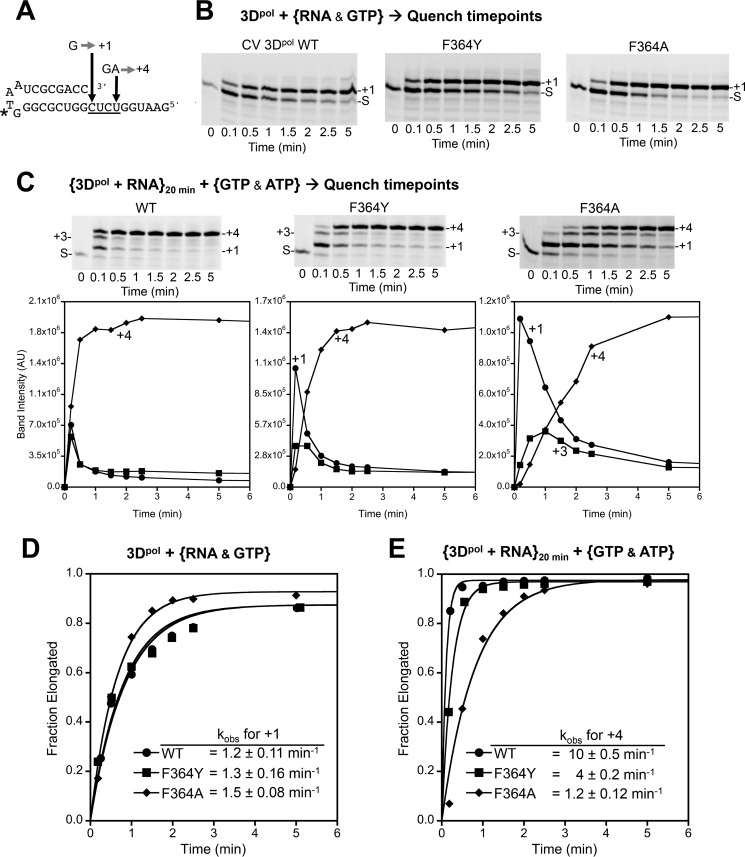
Rates of elongation complex formation were determined using a LI-COR Biosciences IRDye800-labeled hairpin primer-template RNA that allows formation of a +1 product in the presence of GTP or a +4 product in the presence of GTP and ATP (Fig. 6A). Final concentrations were 5 μm 3Dpol, 0.5 μm RNA, and 40 μm each NTP in SF buffer at room temperature. Time point samples were quenched with SF buffer containing NaCl and EDTA at final postquench concentrations of 300 and 25 mm, respectively. For the +1 product formation reactions, the RNA and GTP were mixed simultaneously with 3Dpol in initiation reactions that are largely limited by the RNA binding kinetics. To eliminate RNA binding as a rate-limiting step, a second set of reactions were done wherein the RNA and 3Dpol were preincubated for 20 min prior to mixing with GTP and ATP to trigger elongation to a defined +4 product. These reactions were set up in 40 μl volumes, from which 2 μl samples were removed and quenched for every time point. All quench reaction products were analyzed by denaturing PAGE (15% acrylamide and 7 m urea), imaged on a LI-COR Biosciences Odyssey imager, and quantified with ImageStudio (LI-COR Biosciences).
FIGURE 6.
Perturbing the Pro357-Phe364 interaction affects both pre- and postcatalysis active site motions. A, sequence of the hairpin primer-template RNA used in elongation reactions where incubation with only GTP results in formation of a +1 product, whereas incubation with GTP and ATP will yield a +4 product. * denotes location of LI-COR Biosciences IRDye800 attachment. B, +1 product formation by wild type and mutant polymerases observed by denaturing PAGE followed by infrared imaging of the IRDye label. S marks starting RNA material position. C, +4 product formation in a reaction where 3Dpol and RNAwere preincubated to minimize effects from the slow RNA binding step prior to initiation. The plots of band intensities underscore the processive elongation defects of the F364A mutant where there is significant buildup of the intermediate +1 product as compared with wild type. D and E, single exponential rates for +1 and +4 product formation reactions shown in B and C. AU, arbitrary units.
Generation of Virus Stocks and Infections
All variants were constructed using the QuikChange XL site-directed mutagenesis kit (Stratagene) and the CVB3-Nancy infectious cDNA. 4 μg of in vitro transcribed infectious RNA were electroporated into 6 × 106 HeLa cells. To determine the cytopathic effect, 500 μl of these virus stocks were used to infect fresh HeLa cell monolayers for three more passages. For each passage, virus was harvested by two freeze-thaw cycles. Two independent stocks were generated for each virus. For mutagen assays, HeLa cells were pretreated for 1 h with different concentrations of ribavirin or 5-fluorouracil and infected at a multiplicity of infection of 0.01 with passage 3 virus. 48 h postinfection, virus titers were determined by TCID50. 10-Fold serial dilutions of virus were prepared in 96-well round bottom plates in DMEM. Dilutions were performed in octuplicate, and 100 μl of the dilution were transferred to 104 cells plated in 100 μl of DMEM and 10% newborn calf serum. After 5 days, living cell monolayers were colored by crystal violet. TCID50 values were determined by the method of Reed and Muensch (36).
Virus Replication Kinetics
For one-step growth kinetics, HeLa cells were infected at a multiplicity of infection of 10 and frozen at different time points after infection. For quantitative RT-PCR analysis, total RNA from infected cells was extracted by TRIzol reagent (Invitrogen) and purified. The TaqMan RNA-to-Ct one-step RT-PCR kit (Applied Biosystems) was used to quantify viral RNA. Each 25-μl reaction contained 5 μl of RNA, 100 μm each primer (forward, 5′-GATCGCATATGGTGATGATGTGA-3′; reverse, 5′-AGCTTCAGCGAGTAAAGATGCA-3′, and 25 pmol of probe 5′-[6-Fam]CGCATCGTACCCATGG-3′ in an ABI 7000 machine. Reverse transcription was performed at 50 °C for 30 min and 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. A standard curve was generated using in vitro transcribed genomic RNA.
Sequencing
Viral RNA from passage 3 virus stocks was extracted and RT-PCR -amplified using the primers sets 878Forward and 2141Rev covering part of the structural protein coding region. The resulting PCR products were TOPO TA cloned (Invitrogen) and sequenced, and an 1168-nucleotide sequence was analyzed per clone using Lasergene software (DNAStar Inc.). The mutation frequency was calculated using the total mutations identified per population over the total number of nucleotides sequenced for that population multiplied by 104. For each population, the number of clones presenting no mutations, one, two, three, etc. were quantified and used for statistical testing by Mann-Whitney U test. The number of clones analyzed per population were as follows: A358V, 95 clones; A358T, 96 clones; V367I, 89 clones; V367L, 82 clones; A231T, 89 clones; A231V, 95 clones; F364W, 81 clones; F364Y, 85 clones; and wild type, 84 clones.
Protection Studies
Mice were housed in the Institut Pasteur animal facilities in BSL-2 conditions with water and food supplied ad libitum and handled in accordance with institutional guidelines for animal welfare. All studies were carried out in 5-week-old BALB/c male mice obtained from Charles River Breeding Laboratories. Mice were infected intraperitoneally with 105 TCID50 in 0.20 ml of PBS. Tissue-specific viral titers were determined by TCID50 assay after harvesting whole organs that were homogenized in PBS using a Precellys 24 tissue homogenizer (Bertin Technologies).
Results
Structural Characterization of Motif D Mutants
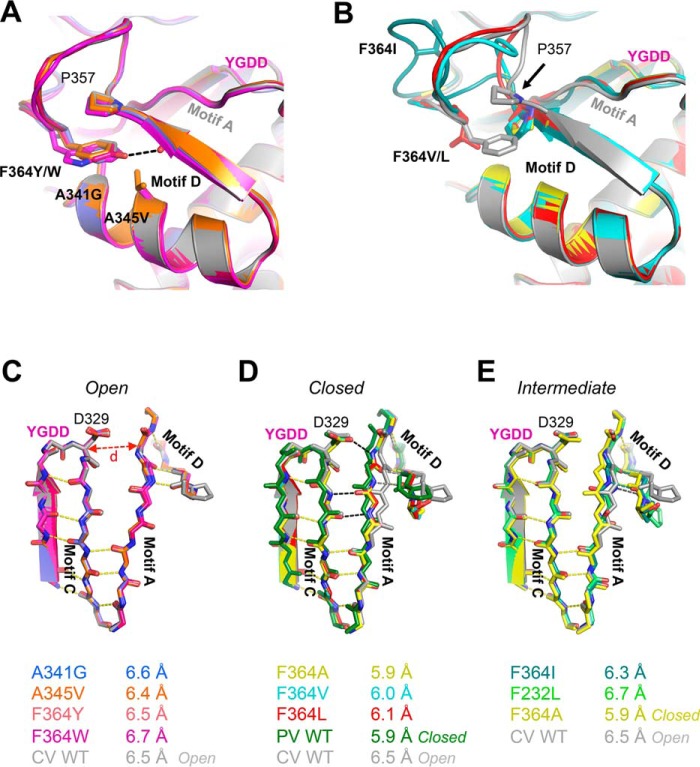
Crystal structures at 1.8–2.6-Å resolution were determined for eight motif D mutants (Table 1) using previously established ammonium sulfate and sodium chloride crystallization conditions (13). Superpositioning of the mutant and wild type 3Dpol structures clearly delineates them into two groups, one with the native conformations for the motif D loop and active site (Fig. 2A) and the other with various motif D loop distortions that are accompanied by a movement of Pro357 and closure of the active site (Fig. 2B). These structures reveal that removal of Phe364 from its binding pocket can drive structural changes in motif A and lead to a closed 3Dpol RdRP active site in the absence of RNA and a nucleotide triphosphate.
TABLE 1.
Data collection and refinement statistics
r.m.s., root mean square.
| Mutation | F364Y | F364W | F364A | F364V | F364L | F364I | A341G | A345V |
|---|---|---|---|---|---|---|---|---|
| Protein Data Bank code | 4ZPA | 4ZPB | 4ZP6 | 4ZP7 | 4ZP8 | 4ZP9 | 4ZPC | 4ZPD |
| Data collection | ||||||||
| Space group | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 |
| Cell dimensions | ||||||||
| a, b, c (Å) | 73.94, 73.94, 287.11 | 74.61, 74.61, 289.43 | 74.23, 74.23, 288.95 | 74.57, 74.57, 289.83 | 74.22, 74.22, 289.45 | 74.39, 74.39, 288.25 | 74.29, 74.29, 288.15 | 74.45, 74.45, 289.64 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 52.3–2.67 (2.76–2.67)a | 51.9–1.8 (1.86–1.8) | 52.5–1.65 (1.71–1.65) | 52.7–1.9 (1.97–1.9) | 51.8–1.9 (1.96–1.9) | 52.6–1.8 (1.86–1.8) | 52.5–1.6 (1.65–1.6) | 51.9–1.8 (1.86–1.8) |
| Rmerge | 0.1495 (0.776) | 0.09355 (0.756) | 0.0501 (1.09) | 0.1131 (0.7139) | 0.149 (1.404) | 0.1281 (1.47) | 0.107 (2.084) | 0.1035 (1.082) |
| CC½b | 0.996 (0.801) | 0.999 (0.831) | 1 (0.236) | 0.998 (0.785) | 0.998 (0.487) | 0.999 (0.577) | 0.999 (0.207) | 0.999 (0.608) |
| CC*b | 0.999 (0.943) | 1 (0.953) | 1 (0.618) | 1 (0.938) | 0.999 (0.809) | 1 (0.855) | 1 (0.585) | 1 (0.87) |
| Mean I/σI | 18.22 (2.91) | 20.28 (2.87) | 31.26 (0.76) | 16.31 (2.56) | 12.88 (1.36) | 17.04 (1.57) | 14.88 (0.68) | 19.84 (1.80) |
| Completeness (%) | 98.0 (100) | 100 (98.0) | 85.0 (43.0) | 100 (98.0) | 100 (96.0) | 100 (99.0) | 0.98 (0.89) | 100 (97.0) |
| Multiplicity | 13.1 (11.0) | 13.6 (12.5) | 11.0 (3.1) | 13.2 (10.7) | 13.0 (10.1) | 11.5 (10.6) | 11.3 (7.5) | 13.4 (10.2) |
| Refinement | ||||||||
| Resolution (Å) | 52–2.67 (2.76–2.67) | 51.9–1.8 (1.86–1.8) | 52.5–1.65 (1.71–1.65) | 52.7–1.9 (1.97–1.9) | 51.8–1.9 (1.96–1.9) | 52.6–1.8 (1.86–1.8) | 52.5–1.6 (1.65–1.6) | 51.9–1.8 (1.86–1.8) |
| Total reflections | 311,358 | 1,052,872 | 920,933 | 863,216 | 851,533 | 873,625 | 1,224,064 | 1,035,067 |
| Unique reflections | 23,856 | 77,189 | 83,791 | 65,517 | 65,409 | 76,295 | 108,427 | 76,995 |
| Rwork/Rfree (%) | 20.5/22.9 | 18.8/22.7 | 18.9/22.0 | 18.8/22.0 | 20.0/23.3 | 19.6/23.3 | 21.3/23.0 | 20.5/22.9 |
| No. atoms | ||||||||
| Protein | 3,692 | 3,693 | 3,625 | 3,643 | 3,630 | 3,687 | 3,689 | 3,692 |
| Water | 85 | 654 | 548 | 577 | 567 | 686 | 522 | 437 |
| B-factors | ||||||||
| Protein | 26.7 | 22.2 | 23.8 | 24.2 | 27.8 | 24.6 | 26.6 | 26.7 |
| Water | 34.6 | 33.3 | 34.3 | 34.3 | 37.6 | 35.3 | 35.0 | 34.6 |
| r.m.s. deviations | ||||||||
| Bond lengths (Å) | 0.007 | 0.008 | 0.007 | 0.007 | 0.007 | 0.007 | 0.008 | 0.007 |
| Bond angles (°) | 0.87 | 0.84 | 0.87 | 0.83 | 0.82 | 0.83 | 0.85 | 0.87 |
| Ramachandran | ||||||||
| Favored (%) | 98 | 98 | 98 | 97 | 98 | 98 | 98 | 98 |
| Outliers (%) | 0.22 | 0 | 0 | 0 | 0 | 0 | 0 | 0.22 |
a Values in parentheses are for highest resolution shell.
b See Ref. 37.
FIGURE 2.
Crystal structures of CVB3 3Dpol motif D mutants. A, mutations that do not alter the wild type conformation of the motif D loop include A341G (blue), A345V (orange), F364Y (pink), and F364W (magenta). The F364Y mutation may stabilize this conformation via a new hydrogen bond to the backbone carbonyl of Met355 (black dashed line), and F364W may reinforce the interaction with Pro357 by providing a larger aromatic surface area. YGDD marks the active site β-turn within motif C. B, mutation of Phe364 to small hydrophobic and β-branched residues precludes the stacking interaction with Pro357, inducing a downward movement of Pro357. F364A is shown in yellow, F364V is in cyan, F364L is in red, and F364I is in teal. C–E, mutations to Phe364 result in a closing of the active site in the absence of RNA and NTPs as shown in these top views of the active site motifs. C, the open conformation seen with A341G, A345V, and the aromatic F364Y and F364W mutations matches the structure of the wild type 3Dpol (gray). The open conformation is characterized by partial hydrogen bonding (yellow dashes) between motifs A and C and an ≈6.5-Å distance between Asp329 Cα in Motif C and Tyr234 Cα in motif A. D, The F364A, F364L, and F364V mutant structures show closed active sites characterized by an ≈0.5-Å movement of motif A toward motif C that is stabilized by the formation of three new hydrogen bonds (black dashes). The hydrogen bonds between motifs A and D are maintained (gray dashes), demonstrating the concerted movement of motifs A and D to close the active site. The open wild type CVB3 (gray) and closed poliovirus (PV) (green) conformation structures are shown for comparison. E, the F364I mutant (teal with nitrogen atoms in blue and oxygen atoms in red) is in a partially closed state where the hydrogen bonding network of the closed active site has not fully formed. The open wild type (gray), closed F364A (yellow), and F232L (green) structures are shown for comparison.
Structures that retained the wild type conformation of the motif D loop and an open active site include F364Y, F364W, A341G, and A345V, all of which feature conservative amino acid substitutions (Fig. 2C). Conversely, the more radical F364A, F364V, and F364L mutations resulted in structures with closed active sites (Fig. 2D) as well as motif D loop distortions and heterogeneity such that the loop itself could not be well modeled into the electron density maps. The F364I structure has a partially closed active site (Fig. 2E) that may be due to the NaCl crystallization condition; this crystal form has a Na+ ion in the metal A binding site that interacts with Asp233 via a water molecule (13), and this interaction may hamper the movement of Asp233 and motif A that is necessary for active site closure. These structures show that the Phe364 binding pocket is able to accommodate every planar aromatic amino acid with minimal structural changes in motif D. The tyrosine and tryptophan mutations may in fact strengthen this conformation as the F364Y side chain hydroxyl forms a new hydrogen bond with the backbone carbonyl of Met355, and the F364W mutant provides a larger aromatic surface area for the interaction (Fig. 2A). In contrast, small hydrophobic amino acids in place of Phe364 do not insert into the pocket, and as a result Pro357 drops down, collapsing the pocket and shifting motif A toward motif C in a way that effectively closes the active site in the absence of a bound NTP. Additionally, several structures showed weak density for residues 19–21 in the index finger that comprise the +2 nucleotide binding pocket in the 3Dpol-RNA complex (18), indicating that this region of CVB3 polymerase is flexible in the absence of bound RNA.
Elongation Rates
Processive elongation rates were determined by stopped-flow fluorescence using a template strand 2AP in a synthetic hairpin primer-template RNA (Fig. 3, A–C). Stalled elongation complexes were first assembled at high concentration to drive polymerase-RNA binding and then diluted to 25 nm final concentration as they were mixed with varying concentrations of NTPs in the stopped-flow instrument. NTP solutions contained ATP:UTP:GTP at a 6:1:1 mole ratio that represents their approximate in vivo ratios (25, 26), and the reported Km values reflect the ATP concentration in that mixture. Elongation proceeds for precisely 14 nucleotides after which there is a clear increase in 2AP fluorescence as the base analog enters the +2 nucleotide pocket and loses stacking interactions with neighboring nucleotides that previously quenched fluorescence. The result is a data trace with a lag phase that reflects the time needed to incorporate 14 nucleotides on the heteropolymeric template (Fig. 3B) and from which we can elucidate maximal elongation rates (kpol) and NTP Km values (Fig. 3C) using methods previously developed for 5′-fluorescein-labeled RNA templates (27).
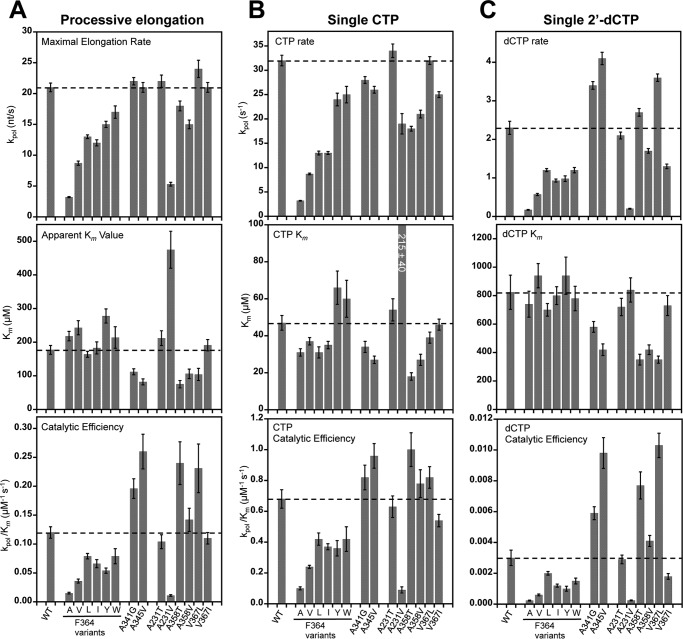
The Phe364 mutations have relatively modest effects on the NTP Km values but slow elongation rates up to almost 7-fold with effects that are largely in proportion to the size of the replacement residue (Fig. 4A). This suggests that the mutants are primarily deficient in the active site conformational changes required for catalysis. Based on these data and the crystal structures discussed above, we hypothesize that the various mutations slow elongation rates because they alter the interactions with the Phe364 binding pocket to favor the open or the closed conformation, either of which would result in a higher energy barrier to the repeated cycling of the active site during processive elongation. The tyrosine and tryptophan mutations at Phe364 have stronger interactions with the binding pocket that favor the open conformation, whereas the alanine, valine, leucine, and isoleucine mutations all compromise the interaction with Pro357 and favor the closed state. In contrast to the Phe364 mutations, the A341G and A345V mutations located in the helix upon which Phe364 packs have essentially no effect on rate but did lower NTP Km values 2-fold. We suspect these two mutations allow for greater flexibility of the motif D loop while neither compromising nor reinforcing the Pro357-Phe364 stacking interaction, and this results in slightly more efficient initial NTP binding during elongation.
FIGURE 4.
Summary of processive and single nucleotide elongation data. A, plots of processive elongation rates (kpol; top), Km values (middle), and catalytic efficiencies (kpol/Km; bottom) for all mutations in this study. Dashed horizontal lines indicate WT values, and error bars reflect the S.E. obtained from curve fitting the data. For the catalytic efficiencies, these errors are propagated as root mean square fractional errors. B and C, analogous plots for single nucleotide turnover rates with CTP and 2′-dCTP substrates. nt, nucleotides.
The second set of mutations were designed to influence the movement of the metal A Mg2+ ion into the polymerase active site for catalysis. These also slowed processive elongation, but the effects were quite small. A231T and V367I have kpol and Km values and catalytic efficiencies (kpol/Km) nearly identical to those of the wild type enzyme, whereas the A358T, A358V, and V367L mutants have similar rates but lower NTP Km values. The exception was a strong effect from the A231V mutation that points to the importance of a polar environment in this region of the polymerase. A231V displays a nearly 4-fold reduction in rate and nearly 3-fold increase in Km, making it the least efficient polymerase examined. In contrast, A231T, which effectively replaces one of the A231V methyl groups with a hydroxyl, restored essentially wild type behavior. Our interpretation of these data is that adding the valine with two methyl groups disrupts the transport of the hydrated metal ion either into the prebound metal A site or between the prebound site and the catalytic center.
Single Nucleotide Incorporation and Discrimination
Single nucleotide turnover rates for cytosine incorporation followed by translocation were measured by the quenching of 2-aminopurine fluorescence as the base analog moves from the unstacked +2 position into the +1 position where it is stacked on the priming duplex (Fig. 3, D–F). Rapid mixing stopped-flow data show single exponential rates of fluorescence loss, and analysis of these rates yields a single nucleotide turnover rate constant (kpol) and affinity (Km) for CTP. The general trends observed in processive elongation assays hold true in the single CMP incorporation experiments with the set of Phe364 variants being 1.3–10-fold slower than the wild type enzyme but retaining similar NTP Km values (Fig. 4B). The A341G and A345V mutations have slightly higher catalytic efficiencies due to lower Km values despite modestly slower CTP incorporation rates. Among the mutations targeting divalent ion binding, A231T and V367I have rates and Km values nearly identical to wild type 3Dpol, whereas the A358T, A358V, and V367L mutants display modestly slower rates and lower Km values. Again, the A231V mutant has the greatest effect with an ≈2-fold decrease in rate and ≈5-fold increase in Km that together reduced catalytic efficiency almost 10-fold.
In 2′-dCTP utilization experiments, the wild type rate and Km decrease 14- and 17-fold, respectively, as compared with the CTP values. Among the mutant polymerases, the effects on rate are generally much stronger than the effects on NTP binding (Fig. 4C). One interesting observation is that the mutations at the base of the Phe364 binding pocket, A341G and A345V, increase dCTP incorporation rates while decreasing the CTP rates, suggesting that these mutations may be low fidelity polymerases that are more prone to nucleotide misincorporation.
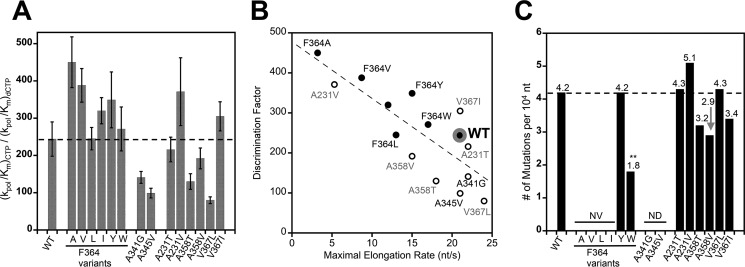
As an indirect measure of polymerase fidelity we can calculate nucleotide discrimination factors as the ratio of the catalytic efficiencies for CTP and 2′-dCTP incorporation, i.e. (kpol/Km)CTP ÷ (kpol/Km)dCTP. This is shown in Fig. 5A and additionally illustrated by plotting the discrimination factors versus maximal processive elongation rates in Fig. 5B. We have previously demonstrated a correlation between elongation rate and the nucleotide discrimination factor for mutations in motif A where high discrimination factors are predictive of viruses with higher replication fidelity (13). In the data presented here, all the Phe364 variants have higher discrimination factors than wild type, whereas the A341G and A345V mutations in the Phe364 binding pocket show decreased discrimination. The mutations targeting the metal A site show modest effects that increase and decrease nucleotide discrimination with the strongest effects being from A358T and V367L mutations whose location in the structure (Fig. 1D) suggests that they may affect the entry of a metal ion into the prebound metal A site.
FIGURE 5.
Nucleotide discrimination and deep sequencing-based fidelity of 3Dpol variants. A, CTP versus 2′-dCTP discrimination factors calculated as the ratio of their catalytic efficiencies. Error bars reflect root mean square propagation of experimental errors from Fig. 4. B, correlation plot of maximal elongation rates based on processive elongation (Fig. 4A) versus the nucleotide discrimination factors. Phe364 mutations are shown as closed symbols, and wild type is marked with a large gray ring. C, mutation frequencies of the variant polymerases based on molecular clone sequencing of progeny virus genomes following infection in tissue culture. Between 81 and 96 clones (94,000 and 112,000 nucleotides) were sequenced per population. **, p < 0.01 by two-tailed Mann-Whitney U test; n = 165; all other values not significant, p > 0.05. NV denotes non-viable viruses, and ND denotes not determined. nt, nucleotides.
Mechanism of Phe364 Mutant Effects on Active Site Motions
To further investigate the mechanism behind the slowed elongation rates of the Phe364 mutants, we examined the behavior of F364Y and F364A via rapid quench reactions. These allow us to directly visualize the RNA products formed as opposed to indirectly assessing polymerase turnover via RNA translocation state-dependent 2-aminopurine fluorescence. F364A was chosen because it has a preclosed active site, which may increase the incorporation rate of the first nucleotide. F364Y was chosen because it forms a new side chain hydrogen bond that may stabilize the open conformation and thus slow the active site closure step. First, a short primer-template hairpin RNA (Fig. 6A) was used to measure RNA binding followed by one guanosine incorporation step in a single reaction where 3Dpol is combined with a premixed RNA + GTP solution and the kinetics are rate-limited by the RNA binding step. Both mutants readily initiate with rates and yields that are comparable with the wild type enzyme (Fig. 6, B and D). Second, we examined processive elongation by 4 nucleotides in a reaction where 3Dpol was first prebound to the same RNA, and this 3Dpol-RNA complex was then mixed with both GTP and ATP (40 μm final concentration for each) in a quench-flow instrument. All three polymerases showed a rapid loss of starting material within 6 s, but the two mutants clearly have slower processive elongation kinetics than wild type based on slowed accumulation of +1 and +3 intermediate species and the time needed to produce the final +4 product (Fig. 6, C and E). Notably and despite the structural observation that it has a preclosed active site, F364A is slower than F364Y both at incorporating the first nucleotide and at elongation over multiple catalytic cycles. The overall effects on processive elongation were quantitated by analyzing the buildup for the final +4 species over time, yielding single exponential rates of 10 ± 0.5, 4 ± 0.2, and 1.2 ± 0.1 per minute for the wild type, F364Y, and F364A polymerases, respectively. Note that in this experiment we do not see significant buildup of the +2 species after ATP addition because GTP has an ≈35-fold lower Km than ATP (28), and as a result the +2 species quickly incorporates guanosine to generate the +3 species, which then more slowly progresses through an adenosine addition to yield the final +4 species.
Infectious Virus Studies
Several of the mutant polymerases were incorporated into virus genomic RNA and transfected into HeLa cells to see whether they could support virus replication and yield enough progeny virus for deep sequencing to obtain mutation frequencies. Among the Phe364 mutations, only the tyrosine and tryptophan variants were recovered. The lack of progeny virus from isoleucine, valine, and alanine mutations point to a strong requirement for a planar amino acid at residue 364. Mutation frequencies calculated from molecular clones (29) showed that the F364Y mutant retained wild type fidelity despite having a higher 2′-hydroxyl discrimination factor in the biochemical assays. In contrast, the F364W mutation had a dramatic effect on mutation frequency, which went down more than 2-fold from 4.2 to 1.8 mutations per 10 kb synthesized (Fig. 5C). This is the first genetically stable high fidelity CVB3 polymerase variant we have isolated. The mutants targeting the metal A site all supported virus growth with final titers within 1 log of the wild type virus, but their one-step growth curves showed growth delays of 4–6 h (data not shown). These mutations resulted in small non-significant changes in fidelity.
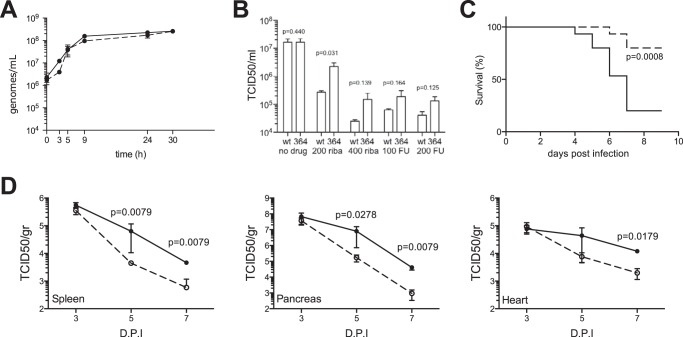
The strong high fidelity effects of the F364W mutation led us to further assess its effects on nucleotide analog drug resistance in tissue culture and on tissue tropism and pathogenesis in mice. First, both wild type and F364W virus presented the same RNA synthesis profiles in one-step growth curves (Fig. 7A). The retention of the mutation at the last time point was confirmed by sequencing. The mutant virus exhibited resistance to both ribavirin and 5-fluorouracil (Fig. 7B) with about 10-fold higher titers than wild type on ribavirin and ≈3-fold higher titers on 5-fluorouracil. The mutant virus showed significantly reduced virulence in vivo with 12 of 15 mice surviving after 9 days versus 3 of 15 for the wild type (Fig. 7C). Consistent with this, tissue samples showed that late stage growth of the F364W virus is attenuated by about 1 log in spleen, pancreas, and heart (Fig. 7D). Thus, the F364W mutation significantly attenuated coxsackievirus B3 growth in mice.
FIGURE 7.
F364W increases resistance to RNA mutagens and is attenuated in vivo. A, one-step replication curves of wild type (solid line) and F364W (dashed line) viruses. HeLa cells were infected at a multiplicity of infection of 10, and at the times indicated the total viral RNA genomes were quantified by quantitative RT-PCR. B, the F364W virus exhibits resistance to nucleoside analogs in Vero cells treated with either ribavirin or 5-fluorouracil. Histograms represent mean values and S.E.; p values based on unpaired, two-tailed t test are indicated; n = 3. C, survival curve of mice (15 per group) infected with either wild type (solid line) or F364W (dashed line) virus monitored over 14 days, the first 9 days of which are shown. p value is indicated based on log rank Mantel-Cox test. D, tissue-specific viral titers (TCID50/g of organ) in mice infected with either wild type (solid line) or F364W (dashed line) virus harvested at 3, 5, and 7 days postinfection. Mean values with error bars reflecting the standard error of the mean are shown; p values determined by two-tailed Mann-Whitney U test are indicated; n = 5.
Discussion
Building upon our previous work identifying structure-function relationships in engineered fidelity variant picornaviral polymerases (9, 13), here we focused our attention on the conformational flexibility of motif D and its impact on CVB3 3Dpol kinetics, fidelity, and structure. The initial structures of poliovirus 3Dpol elongation complexes showed that RdRP active site closure involves a concerted rigid body movement of motif A and the loop portion of motif D that establishes antiparallel β-sheet backbone hydrogen bonding between motifs A and C. This process fully structures the canonical polymerase palm domain active site by repositioning the essential Asp233 in motif A to coordinate both divalent metals needed for catalysis (1). Point mutations within motif A can have profound effects on polymerase elongation rate and replication fidelity, which can be either increased or decreased depending on the specific viral polymerase being studied (13). An alternate conformation of the loop portion of motif D was then captured in a crystal structure of the low fidelity CVB3 3Dpol F232L variant (13). This alternate motif D loop conformation involved Phe364 moving from its binding pocket under Pro357 to instead bind near the base of the thumb domain, and it showed that the motif D loop could be restructured independently of motif A. This is consistent with the conformational heterogeneity within motif D that has been observed by NMR (14) and in molecular dynamics simulations (15) where data indicate that motions in this region of the polymerase are sensitive to NTP binding. Sequence and structure alignments of picornaviral polymerases further show that the original conformation is highly conserved and seemingly stabilized by a stacking interaction between Phe364 and Pro357 (Fig. 1C), suggesting that the interactions of these two residues may be important for RdRP function by controlling the motif D loop conformation.
In this study, we examined whether perturbations of this interaction could impact loop flexibility and reveal aspects of the functional role motif D plays during active site closure. We targeted residue Phe364 with mutagenesis to small and β-branched hydrophobic residues (Ala, Val, Leu, and Ile) and with more conservative changes to tyrosine and tryptophan, the other two planar aromatic amino acids. We also mutated Ala341 and Ala345, two residues that form a relatively flat floor at the bottom of the hydrophobic pocket into which Phe364 is inserted. In a second set of mutations, we targeted the active site metal A ion by changes to residues Ala231, Ala358, and Val367 that may impact the transport of magnesium into the active site for catalysis by virtue of being indirectly involved in the extended ion hydration shell (18). Crystallography, stopped-flow kinetics, rapid quench reactions, and infectious virus studies were used to characterize the effects of the mutants on 3Dpol structure and biochemistry and on coxsackievirus B3 growth and pathogenesis.
The data point to Phe364 being a key control point for both processive replication rate and fidelity. Mutations of this residue slow the polymerase elongation rate (kpol) with a continuum of effects ranging from 1.2- to 7-fold, whereas the Km for NTP remains largely unchanged, suggesting that residue 364 plays a role in the dynamics of nucleotide reorientation and active site closure that occurs after the initial NTP binding event. The crystal structures show that a planar amino acid (Phe, Tyr, or Trp) is needed at residue 364 to maintain the active site in an open conformation. Mutations to Ile, Ala, Val, and Leu all show Pro357 dropping down toward the Phe364 binding pocket to result in either a partially or a fully closed active site. Furthermore, Tyr and Trp were the only Phe364 mutations that supported virus growth, indicating that stabilizing the loop conformation and/or the default open active site is essential for proper 3Dpol function during infection. Sequencing of the progeny virus population further revealed that F364Y does not affect replication fidelity, but F364W increased fidelity 2.3-fold, resulting in a very strong high fidelity variant making only 1.8 mutations per 10 kb synthesized. This is the first isolation of a genetically stable high fidelity variant of CVB3, an enzyme that already has fairly high fidelity in comparison with the poliovirus RdRP. Combined with our previous studies of motif A mutations that result in low fidelity variants (9, 13), we have now recovered viable CVB3 isolates with 3Dpol variants whose mutation frequencies vary over a 6-fold range from 1.8 to 11.2 mutations per 10 kb of RNA synthesized.
The details of the 3Dpol catalytic cycle have been studied in great detail to reveal five major steps: NTP binding, a precatalysis conformational change that reorients the NTP into the active site, catalysis, a postcatalysis conformational change, and finally translocation of the RNA to reset the active site (30). The precatalysis reorientation step is rate-limiting and a major fidelity checkpoint, resulting in a strong correlation between elongation rate and replication fidelity. The structures of picornaviral elongation complexes show that the molecular rearrangement that takes place during the active site closure step is the inward movement of motif A to fully fold the core palm domain β-sheet and create a canonical replicative polymerase active site (Fig. 1B) (1, 18). Our prior studies of 3Dpol motif A mutations showed a strong correlation between replication fidelity obtained from deep sequencing and the presence of an NTP 2′-hydroxyl group based on the CTP-versus-dCTP discrimination assay (13). Those findings suggested that proper hydroxyl positioning and the resulting hydrogen bonding network emanating from the 2′-OH play a major role in driving active site closure for catalysis. However, such a 2′-OH correlation does not appear to hold for the set of motif D mutants described in this work. For example, the F364Y and F364W mutants both have slightly elevated discrimination factors, but only the tryptophan has a significant fidelity effect in an infectious virus context. Among the mutants targeting the metal A site, the biochemical data would predict lowered fidelity, but the virus-based observation is one of slightly higher fidelity variants. These new data do retain the in vitro biochemical observation that faster polymerases have reduced 2′-OH discrimination (Fig. 5B), but for motif D mutants these effects are no longer clearly predictive of change to in vivo virus mutation frequencies.
The kinetic data also indicate that the cycling of the active site and motif D from an open to closed conformation is important for the first round of catalysis, supporting molecular dynamics data arguing that motif D plays a role in nucleotide transport into the active site (31). Wild type 3Dpol and the F364Y mutant have open active sites and comparable rates for the first nucleotide incorporation event based on the observed disappearance of starting material, but the F364A mutant is significantly slower despite having a preclosed active site that might be expected to accelerate incorporation of the first nucleotide (Fig. 6C). Thus, we conclude that the active site must be cycled to the open state to enable initial NTP binding, and the NTP is then repositioned to trigger active site closure and catalysis. Subsequent incorporation steps, i.e. processive elongation, are slowed in both F364Y and F364A mutants because they stabilize the open or the closed state, respectively, and either effect will reduce the rate at which the polymerase progresses through the full catalytic cycle. The molecular origin of the lower in vivo fidelity observed for F364W variant virus is not clear from the structures or the biochemical data, and the in vitro biochemical behavior of this mutant 3Dpol is very similar to that of the non-fidelity variant F364Y (Fig. 4). The fidelity effect is likely due to differences in motif D dynamics that alter the efficiency of NTP delivery into the active site or the coupling between motifs A and D, and elucidating the fine details of these motions will likely require atomic level dynamics data from NMR and computational approaches.
The emergence of the F364W mutations as a genetically stable high fidelity variant is attributed to two distinct effects. First, the sole tryptophan codon is UGG, and any single base mutation of this codon will result in small or charged amino acids (i.e. Gly, Ser, Cys, Leu, and Arg) that likely cannot support virus replication based on our virus growth results. Thus, the Trp364 variant virus effectively includes a genetic poison pill that minimizes its reversion potential. Second, at the level of a functional polymerase, the tryptophan creates a larger surface for the sliding motion that takes place relative to the motif D helix (Fig. 1B), but it does so by nonspecific hydrophobic interactions that do not strongly favor either the open or the closed conformation of the active site. Consequently, it has a small effect on the elongation rate, reducing it from 20 to 17 nucleotides/s, and remains fast enough to support virus growth. This is in stark contrast to two known high fidelity variants that were first identified in poliovirus but are not functional in coxsackievirus (9, 13): 3Dpol G64S was originally selected as a ribavirin-resistant poliovirus (32), and in CVB3 it is slightly higher fidelity than wild type based on in vitro polymerase assays, but the mutation reduced virus replication by almost 100-fold, and it reverted to wild type after only three passages. Similarly, K359R is a viable high fidelity motif D mutant in poliovirus (33), but the structurally equivalent K360R slows the CVB3 3Dpol elongation rate by ≈35-fold, and it does not support virus growth.
A long term goal of understanding structure-function relationships in the viral RdRPs is to use such information to attenuate in vivo virus growth in ways that can lead to suitable vaccine strains. Prior studies with poliovirus and coxsackievirus have shown that either increasing or decreasing RdRP fidelity can attenuate growth in vivo. One potential advantage of doing this via a protein engineering approach is the identification of specific mutations that can retain function but are unlikely to arise by virus-based adaptation pathways. Such mutations may also be less likely to revert by the same pathways. In this study, we used a combination of structure-based protein engineering and virology to identify 3Dpol F364W as a new high fidelity variant of CVB3 that significantly attenuates virus growth in mice. The parental phenylalanine residue is highly conserved among picornaviruses, and a tryptophan is never observed, suggesting that the nuances of codon usage have prevented this fully functional polymerase variant from appearing in any enterovirus, and likewise this limits the reversion potential of the variant. Our findings show that motif D and the conserved structural interactions with Phe364 provide a powerful control point for engineering polymerase fidelity as a tool for attenuating virus growth.
Author Contributions
S. M. conducted the biochemistry and structural biology experiments, analyzed the results, and wrote the relevant parts of the paper with O. B. P. A. B. conducted the experiments relating to the metal A binding site. S. B. and G. M. conducted the virology studies and wrote those sections of the paper together with V. M. S. M. and O. B. P. conceived the idea for the project.
Acknowledgments
We thank G. Campagnola for helpful discussions and experiment assistance and J. Nix for beamline support.
This work was supported by National Institutes of Health Grant R01 AI059130 (to O. B. P.) and a LabEx Integrative Biology of Emerging Infectious Diseases program grant (to M. V.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- RdRP
- RNA-dependent RNA polymerase
- 3Dpol
- picornaviral RNA-dependent-RNA polymerase
- CV
- coxsackievirus
- TCID50
- 50% tissue culture infective dose
- 2AP
- 2-aminopurine.
References
- 1. Gong P., and Peersen O. B. (2010) Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kornberg R. D. (2007) The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 12955–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Steitz T. A. (2009) The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr. Opin. Struct. Biol. 19, 683–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steitz T. A. (1998) A mechanism for all polymerases. Nature 391, 231–232 [DOI] [PubMed] [Google Scholar]
- 5. Ferrer-Orta C., Ferrero D., and Verdaguer N. (2015) RNA-dependent RNA polymerases of picornaviruses: from the structure to regulatory mechanisms. Viruses 7, 4438–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thompson A. A., Albertini R. A., and Peersen O. B. (2007) Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J. Mol. Biol. 366, 1459–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andino R., and Domingo E. (2015) Viral quasispecies. Virology 479–480, 46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lauring A. S., Frydman J., and Andino R. (2013) The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 11, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gnädig N. F., Beaucourt S., Campagnola G., Bordería A. V., Sanz-Ramos M., Gong P., Blanc H., Peersen O. B., and Vignuzzi M. (2012) Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. U.S.A. 109, E2294–E2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Korboukh V. K., Lee C. A., Acevedo A., Vignuzzi M., Xiao Y., Arnold J. J., Hemperly S., Graci J. D., August A., Andino R., and Cameron C. E. (2014) RNA virus population diversity, an optimum for maximal fitness and virulence. J. Biol. Chem. 289, 29531–29544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pfeiffer J. K., and Kirkegaard K. (2005) Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 1, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stapleford K. A., Rozen-Gagnon K., Das P. K., Saul S., Poirier E. Z., Blanc H., Vidalain P. O., Merits A., and Vignuzzi M. (2015) Viral polymerase-helicase complexes regulate replication fidelity to overcome intracellular nucleotide depletion. J. Virol. 89, 11233–11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campagnola G., McDonald S., Beaucourt S., Vignuzzi M., and Peersen O. B. (2015) Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J. Virol. 89, 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang X., Smidansky E. D., Maksimchuk K. R., Lum D., Welch J. L., Arnold J. J., Cameron C. E., and Boehr D. D. (2012) Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 20, 1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moustafa I. M., Shen H., Morton B., Colina C. M., and Cameron C. E. (2011) Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 410, 159–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moustafa I. M., Korboukh V. K., Arnold J. J., Smidansky E. D., Marcotte L. L., Gohara D. W., Yang X., Sánchez-Farrán M. A., Filman D., Maranas J. K., Boehr D. D., Hogle J. M., Colina C. M., and Cameron C. E. (2014) Structural dynamics as a contributor to error-prone replication by an RNA-dependent RNA polymerase. J. Biol. Chem. 289, 36229–36248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Götte M., Konigsberg W., and Cameron C. E. (2009) Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 16, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gong P., Kortus M. G., Nix J. C., Davis R. E., and Peersen O. B. (2013) Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PLoS One 8, e60272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gohara D. W., Ha C. S., Kumar S., Ghosh B., Arnold J. J., Wisniewski T. J., and Cameron C. E. (1999) Production of “authentic” poliovirus RNA-dependent RNA polymerase (3Dpol) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17, 128–138 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., and Sliz P. (2013) Collaboration gets the most out of software. eLife 2, e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 26. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., and Kim B. (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gong P., Campagnola G., and Peersen O. B. (2009) A quantitative stopped-flow fluorescence assay for measuring polymerase elongation rates. Anal. Biochem. 391, 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnold J. J., and Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43, 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Beaucourt S., Borderia A. V., Coffey L. L., Gnadig N. F., Sanz-Ramos M., Beeharry Y., and Vignuzzi M. (2011) Isolation of fidelity variants of RNA viruses and characterization of virus mutation frequency. J. Vis. Exp. 2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ng K. K., Arnold J. J., and Cameron C. E. (2008) Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 320, 137–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen H., Sun H., and Li G. (2012) What is the role of motif D in the nucleotide incorporation catalyzed by the RNA-dependent RNA polymerase from poliovirus? PLoS Comput. Biol. 8, e1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfeiffer J. K., and Kirkegaard K. (2003) A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U.S.A. 100, 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weeks S. A., Lee C. A., Zhao Y., Smidansky E. D., August A., Arnold J. J., and Cameron C. E. (2012) A Polymerase mechanism-based strategy for viral attenuation and vaccine development. J. Biol. Chem. 287, 31618–31622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Theobald D. L., and Wuttke D. S. (2006) THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics 22, 2171–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Theobald D. L., and Wuttke D. S. (2008) Accurate structural correlations from maximum likelihood superpositions. PLoS Comput. Biol. 4, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reed L. J. and Muench H. (1938) A simple method for estimating fifty per cent endpoints. Am. J. Hyg. 27, 493–497 [Google Scholar]
- 37. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]