Abstract
The green odor of plants is characterized by green leaf volatiles (GLVs) composed of C6 compounds. GLVs are biosynthesized from polyunsaturated fatty acids in thylakoid membranes by a series of enzymes. A representative member of GLVs (E)-2-hexenal, known as the leaf aldehyde, has been assumed to be produced by isomerization from (Z)-3-hexenal in the biosynthesis pathway; however, the enzyme has not yet been identified. In this study, we purified the (Z)-3:(E)-2-hexenal isomerase (HI) from paprika fruits and showed that various plant species have homologous HIs. Purified HI is a homotrimeric protein of 110 kDa composed of 35-kDa subunits and shows high activity at acidic and neutral pH values. Phylogenetic analysis showed that HIs belong to the cupin superfamily, and at least three catalytic amino acids (His, Lys, Tyr) are conserved in HIs of various plant species. Enzymatic isomerization of (Z)-3-hexenal in the presence of deuterium oxide resulted in the introduction of deuterium at the C4 position of (E)-2-hexenal, and a suicide substrate 3-hexyn-1-al inhibited HI irreversibly, suggesting that the catalytic mode of HI is a keto-enol tautomerism reaction mode mediated by a catalytic His residue. The gene expression of HIs in Solanaceae plants was enhanced in specific developmental stages and by wounding treatment. Transgenic tomato plants overexpressing paprika HI accumulated (E)-2-hexenal in contrast to wild-type tomato plants mainly accumulating (Z)-3-hexenal, suggesting that HI plays a key role in the production of (E)-2-hexenal in planta.
Keywords: enzyme mechanism, enzyme purification, lipid peroxidation, plant biochemistry, polyunsaturated fatty acid (PUFA)
Introduction
Plants emit various volatile organic compounds, such as terpenes, volatile plant hormones, and fatty acid derivatives. Abiotic and/or biotic stresses often stimulate the emission of volatile organic compounds (1). In response to wounding, the emission of C6 compounds has been widely observed in many plants. This emission is a fast bioprocess, with that of (Z)-3-hexenal being observed within a few minutes after wounding (2, 3) followed by (E)-2-hexenal (3). C6 compounds such as n-hexanal, (Z)-3-hexenal, (E)-2-hexenal, and the corresponding alcohol and ester derivatives are collectively named green leaf volatiles (GLVs)2 because they have a characteristic green leaf odor. Recent investigations of the biological effects of GLVs suggested that GLVs contribute to protection against the invasion of fungi and insects due to their pathological effects (4–6). In addition, GLVs may be involved in abiotic stress response; a GLV with α,β-unsaturated carbonyl bonds (E)-2-hexenal has been shown to be a signal chemical inducing abiotic stress-associated gene expression (7).
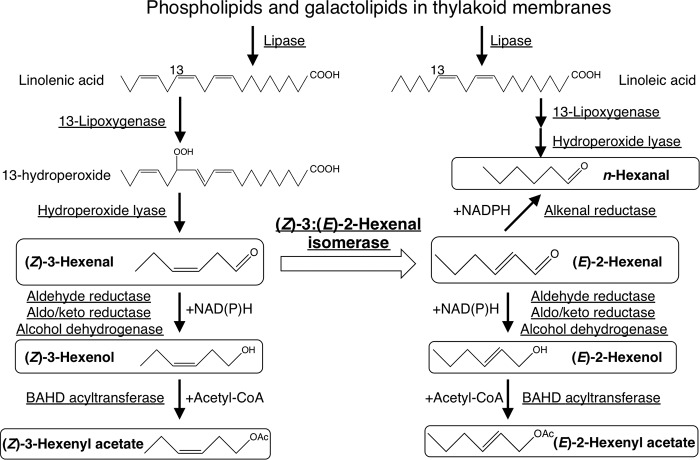
GLVs are produced from thylakoid membrane-bound polyunsaturated fatty acids in chloroplasts by a series of enzymes (Fig. 1). Briefly, linolenic acid released from lipase from thylakoid membranes is peroxidized by 13-lipoxygenase (8) and then cleaved by hydroperoxide lyase to produce (Z)-3-hexenal in chloroplasts (9, 10). Branching from (Z)-3-hexenal to (E)-2-hexenal then forms unsaturated GLVs in two series, i.e. (Z)-3 and (E)-2, based on the position of the unsaturated bond in their structures. (Z)-3-Hexenal and (E)-2-hexenal are subsequently reduced to alcohols by aldehyde reductases, aldo/keto reductases (11), and alcohol dehydrogenases (12). Alcohol forms of GLVs are further converted to ester forms by a BAHD acyltransferase (2). A saturated form of GLV, n-hexanal, is produced through the oxidation of linoleic acid mediated by 13-lipoxygenase and hydroperoxide lyase along with the hydrogenation of the C-C double bond in (E)-2-hexenal by alkenal reductase (13).
FIGURE 1.
GLV biosynthesis pathway in higher plants. Major GLV species are enclosed by round-shaped squares. Enzymes catalyzing each reaction are underlined. Isomerization step catalyzed by HI is shown by a white arrow. Enzymatic reactions for alcohol- and acetate-forms formation need cofactors.
Among GLV species, (E)-2-hexenal was identified by Curtius and Franzen (14) at an early stage in GLV research history and named the leaf aldehyde (originally named Blätteraldehyd in German). Despite this early discovery, enzymes involved in (E)-2-hexenal production have remained unknown, although it has been assumed to be catalyzed by an isomerase (15, 16). In this study we identified (Z)-3:(E)-2-hexenal isomerase (HI) in various plant species and identified the common catalytic amino acids needed for isomerase activity by analysis of the enzymatic mechanism of isomerization. Overexpression of paprika (Capsicum annuum) HI (CaHI) in tomato plants led to a drastic change in hexenal composition in both leaves and fruits, indicating that this HI is a critical enzyme determining green odor in planta.
Experimental Procedures
Materials
Paprika (C. annuum L.), Arabidopsis (Arabidopsis thaliana, Columbia-0), tomato (Solanum lycopersicum, cv. Micro-Tom), potato (Solanum tuberosum cv. Sassy), tobacco (Nicotiana benthamiana), alfalfa (Medicago sativa), cucumber (Cucumis sativus L.), and rice (Oryza sativa, cv. Nipponbare) were sown on Jiffy-7 peat pellets (Sakata Seed Co., Yokohama, Japan) and kept at 4 °C for 3 days in the dark. The plants were then transferred to the conditions of a 14-h-light (80 μmol of photons m−2 s−1)/10-h dark cycle at 23 °C (for Arabidopsis, tomato, potato, tobacco, alfalfa, cucumber, and rice) or 28 °C (for paprika). Wounding treatment to leaves was performed with nipping by tweezers. (Z)-3-Hexenal was obtained from Bedoukian Research Inc. (Danbury, CT). Other chemicals of research grade were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan). (Z)-3-Nonenal and (Z,Z)-3,6-nonadienal were obtained by oxidation with the Dess-Martin periodinane (Wako Pure Chemical) from (Z)-3-nonen-1-ol and (Z,Z)-3,6-nonadienol, respectively (Tokyo Chemical Industry, Tokyo, Japan).
Crude Extract Preparation and HI Activity Measurement
Plant material was homogenized with two volumes of 50 mm Hepes-NaOH, pH 7.0, on ice. After centrifugation at 10,000 × g for 10 min at 4 °C, the supernatant was used as a crude extract. Crude extract (up to 500 μl, depending on the activity) was incubated in 1 ml of reaction mixture containing 10 mm (Z)-3-hexenal in 50 mm Hepes-NaOH, pH 7.0, for 30 min at 25 °C. The reaction was stopped and derivatized with 2,4-dinitrophenylhydrazine by the addition of 25 μl of 20 mm 2,4-dinitrophenylhydrazine in acetonitrile and 20 μl of HCOOH. After 10 min, dinitrophenylhydrazone (DNP) derivatives were extracted with 300 μl of n-hexane. After centrifugation, 150 μl of the hexane layer was recovered and dried in vacuo. The residue was dissolved in 200 μl of acetonitrile and filtered through a Cosmonice Filter (Nacalai Tesque, Kyoto, Japan), after which 10-μl aliquots were subjected to HPLC as described previously (17).
Purification of CaHI
Paprika pericarp (640 g) was homogenized with 1.5 volumes of 50 mm Hepes-NaOH, pH 7.0. Debris was removed by filtration through two layers of gauze, and the filtrate was centrifuged at 10,000 × g for 10 min. Ammonium sulfate was added to the supernatant to 30% saturation followed by centrifugation at 10,000 × g for 10 min. After lipids floating on the supernatant were removed by filtration through two layers of gauze, the supernatant was applied to a phenyl-Sepharose column (40 ml, GE Healthcare) equilibrated with 30% saturated ammonium sulfate in 50 mm potassium phosphate buffer (K-PB), pH 7.0. Proteins were eluted with 50 mm K-PB, pH 7.0, and the eluent was applied directly to a hydroxyapatite (Nacalai Tesque) column equilibrated with 50 mm K-PB, pH 7.0. After washing with 250 mm K-PB, pH 7.0, HI activity was eluted with 250 mm potassium phosphate, pH 7.0, containing 1% Triton X-100. Nine volumes of 20 mm Hepes-NaOH, pH 7.0, containing 0.1% β-d-dodecyl maltoside were added to the fraction containing activity, which was then applied to a Mono Q mini column (total volume 2 ml, GE Healthcare). After washing with 20 mm Hepes-NaOH, pH 7.0, containing 0.1% β-d-dodecyl maltoside, proteins were eluted with 20 mm Hepes-NaOH, pH 7.0, containing 0.1% β-d-dodecyl maltoside and 1 m NaCl. The enzyme solution was concentrated by ultrafiltration using Centricon-30 (Merck Millipore) and diluted by 9 volumes of 20 mm Hepes-NaOH, pH 7.0, containing 0.1% β-d-dodecyl maltoside. The proteins were subjected to anion exchange column chromatography twice (Mono Q, 5/5, and subsequent MonoQ1.6/5, SMART System, GE Healthcare) and fractionated with a gradient of 0–0.6 M NaCl in 20 mm Hepes-NaOH, pH 7.0, containing 0.1% β-d-dodecylmaltoside. All procedures were done below 4 °C. Protein concentration was determined by the method of Bradford (18).
Internal Amino Acid Sequencing of Purified Enzyme
Purified paprika enzyme (2 μg) dissolved in 1 ml of 70% HCOOH was chemically fragmented with 1 mg of BrCN at room temperature overnight in the dark. Fragmented peptides recovered by centrifugal evaporation were dissolved in SDS-sample buffer and then separated by Tricine-SDS-PAGE. After electrophoresis, the separated peptides were blotted onto a PVDF membrane with a semidry blotting system (Atto Corp., Tokyo, Japan), and peptides were visualized by CBB R-250 staining. Stained bands were excised from the membrane, and their amino acid sequences were determined with a protein sequencer (Procise Model 492, Applied Biosystems, Foster City, CA).
Cloning of CaHI and Quantitative RT-PCR (qRT-PCR)
A similarity search of internal amino acid sequences was performed against a Solanaceae-specific database (SolCyc Home). To confirm that Ca08g14620, including both internal amino acid sequences, was identical to CaHI, the open reading frame (ORF) of Ca08g14620 was cloned as follows. Total RNA was isolated from paprika fruits with an RNeasy Plant Mini Kit (Qiagen), and cDNA was then synthesized with ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan). To obtain the ORF of Ca08g14620, PCR was performed with ExTaq DNA polymerase (Takara Bio Inc., Shiga, Japan) using primers CaHI BamHI 5′ (5′-GGATCCATGGATTTAATATTGGCATCG-3′, the additional restriction site is underlined) and CaHI SalI 3′ (5′-GTCGACTTAAGGTGGGGCAATGACTGC-3′). The PCR product was cloned into the TA Cloning vector pMD19 and then sequenced with a BigDye® Terminator v3.1 Cycle Sequencing kit for confirmation. qRT-PCR was performed with Thunderbird SYBR Green qPCR Mix (TOYOBO) and a LightCycler Nano System (Roche Applied Science) using template cDNA prepared as described above. Primers used for qRT-PCR are shown in supplemental Table S1. For analysis of relative transcript levels, internal standard mRNA (a list is shown in supplemental Table S1) was used in all qRT-PCR experiments, and the expression levels of genes of interest were normalized to that of the internal standard by subtraction of the cycle threshold (CT) value of the internal standard from the CT value of the gene of interest.
Phylogenetic Analysis
Alignment and phylogenetic tree analyses were performed using the software MEGA 5 (19). A list of protein sequences used for analysis is shown in supplemental Table S2.
Heterogeneous Expression of HIs
The ORF of HI was amplified by PCR using the primers shown in supplemental Table S3 and then subcloned into pMD19 (Takara Bio). After confirming DNA sequence, each HI gene was inserted into pColdProS2 vector (Takara Bio), and Escherichia coli strain BL21 (DE3) was transformed with the resultant expression plasmid. E. coli harboring the expression vector was grown with shaking at 37 °C in LB broth with 50 μg/ml ampicillin to midlogarithmic phase. Expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside to 0.1 mm, and cultures were grown for a further 16 h at 15 °C and harvested by centrifugation. The pellet was washed twice with phosphate-buffered saline (PBS) and then resuspended with 400 μl of PBS. After disruption of the cells by sonication (40 watts, 10 s, three times) on ice, recombinant HI protein in the soluble fraction was purified on a His SpinTrap column (GE Healthcare) according to the manufacturer's instructions. An electrophoretically homogenous HI fraction was used for enzyme assays. Point mutation of recombinant CaHI (rCaHI) was performed with a PrimeSTAR Mutagenesis Basal kit (Takara Bio) using the primers shown in supplemental Table S4.
3-Hexyn-1-al Synthesis and Measurement of Its Inhibitory Activity against HI
3-Hexyn-1-al was chemically synthesized from 3-hexyn-1-ol (Tokyo Chemical Industry) by oxidation with the Dess-Martin periodinane (Wako Pure Chemical) reagent according to the method of Wavrin and Viala (20). To confirm its purity, synthesized 3-hexyn-1-al was derivatized with 2,4-dinitrophenylhydrazine as described above and then analyzed by LC-MS and NMR. Synthesized 3-hexyn-1-al was incubated with purified rCaHI for 4 h on ice, and free 3-hexyn-1-al was then removed by ultrafiltration using Vivaspin 500–30K (GE Healthcare). Residual HI activity was measured by standard assay as described above.
1H NMR Analysis
All NMR experiments were performed on a JEOL JNM-AL300 (300 MHz at 1H, JEOL Ltd., Tokyo, Japan), and chemical shifts were assigned relative to the solvent signal. Hexenal-DNP derivatives were dissolved in chloroform-d2 and added to tubes having a diameter of 5 mm.
Production of Transgenic Tomato Overexpressing CaHI
A DNA fragment of CaHI amplified with primers CaHI NdeI 5′ (5′-CATATGGATTTAATATTGGCATCG-3′) and CaHI SalI 3′ was digested with NdeI and SalI and ligated into the pRI101-AN vector (Takara Bio Inc.) to construct a 35Spromoter-driven expression plasmid. After Agrobacterium tumefaciens (strain C58C1Rifr) was transformed by pRI101-AN::35S::CaHI, cotyledons of tomato plants (cv. Micro-Tom) were transformed by Agrobacterium-mediated transformation with kanamycin resistance as a selectable marker (21). For confirmation of insertion of 35S-driven CaHI and the neomycin phosphotransferase II gene in regenerated transgenic tomato plants, genome PCR was performed using the primers 35S-F (5′-TCGCCGTAAAGACTGGCGAACA-3′) and CaHI SalI 3′. Candidates were further examined by qRT-PCR and volatile analysis to establish transgenic lines. Finally, transgenic lines showing stable seed production were used for analysis.
Volatile Analysis
Harvested tomato materials were placed in a Falcon tube (15 ml) with three stainless steel beads (5 mm inner diameter), and the tube was then sealed tightly with Parafilm. After samples were frozen in liquid N2, the tissues were completely disrupted by vigorous vortexing. When volatiles in intact tissues were analyzed, 1 ml of saturated CaCl2 solution was added to inactivate the enzymes. An SPME fiber (50/30 μm DVB/Carboxen/PDMS, Supelco, Bellefonte, PA) was exposed to the headspace of the tube for 30 min (for leaf) or 60 min (for fruit) at 25 °C. The fiber was inserted into the insertion port of a GC-MS (QP-5050, Shimadzu, Kyoto, Japan) equipped with a 0.25-μm × 30-m Stabiliwax column (Restek, Bellefonte, PA). The column temperature was programmed as follows: 40 °C for 1 min, increasing by 15 °C min−1 to 180 °C for 1 min (22). The carrier gas (He) was delivered at a flow rate of 1 ml min−1. Splitless injection with a sampling time of 1 min was used. The fiber was held in the injection port for 1 min to remove all compounds fully from the matrix. The temperatures of the injector and interface were 200 °C and 230 °C, respectively. The mass detector was operated in electron impact mode with ionization energy of 70 eV. To identify compounds, retention indices and MS profiles of corresponding authentic specimens were used.
Homology Modeling
Three-dimensional structures of HI proteins were deduced by homology modeling using SWISS-MODEL (23–25). The amino acid sequences of HIs were used to search for templates in the database. The templates were selected based on the Global Model Quality Estimation values and the QMEAN scores (26). Templates and the sequence identity between the HIs and their corresponding templates are shown in the figures. The protein structures were analyzed with Swiss PdbViewer (27).
Accession Number
The full sequences of the CaHI and alfalfa HI (MsHI) have been deposited at the DDBJ data bank with accession numbers LC146479 and LC146718, respectively.
Results
Purification of CaHI from Red Paprika
At the start of this study, we screened plant materials for high HI activity. We first assayed activity in red bell pepper (C. annuum L.) fruits because bell pepper showed a large change in leaf aldehyde composition during ripening; (Z)-3-hexenal was the predominant GLV in green bell pepper, whereas (E)-2-hexenal became the predominant GLV during the change in fruit color from green to red (28). This phenomenon suggested that abundant HI activity occurs in ripening bell pepper. As expected, we detected activity in red pepper fruits, and another bell pepper variant, red paprika, showed the most abundant activity (supplemental Fig. S1). We detected no isomerase activities in Arabidopsis leaves, tobacco leaves, or tomato fruits. We accordingly used red paprika as a source for purification of HI.
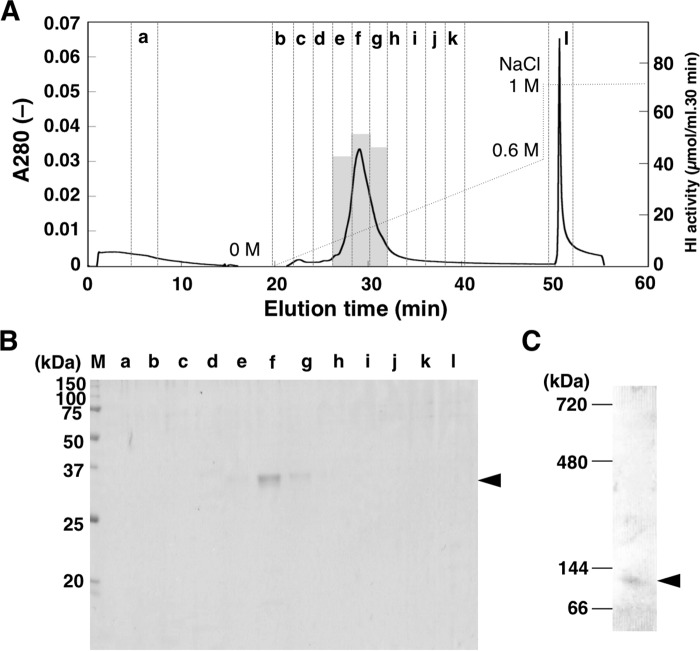
CaHI was purified from pericarp of red paprika fruits by successive column chromatography steps (Table 1). At the final step of purification, fractions containing activity matched elution peaks of proteins (Fig. 2A). SDS-PAGE analysis showed that each fraction contained a single protein of 35 kDa (Fig. 2B), suggesting that CaHI was purified to homogeneity. Native CaHI is a trimetric protein, given that the molecular mass of nondenatured isomerase was estimated by BN-PAGE to be 110 kDa (Fig. 2C). Isomerization of hexenal by purified CaHI is unidirectional given that CaHI did not convert (E)-2-hexenal to (Z)-3-hexenal (supplemental Fig. S2, A and B). (Z)-3-Nonenal and (Z,Z)-3,6-nonadienal, which are substrates for other fatty acid-derived volatiles (E)-2-nonenal (29) and (E,Z)-2,6-nonadienal (30), respectively, were also isomerized (supplemental Fig. S2, C and D). Kinetic parameters against (Z)-3-hexenal are shown in Table 2. CaHI showed high activity at acidic to neutral pH (supplemental Fig. S3A).
TABLE 1.
Summary of purification of CaHI from red paprika
| Total protein | Total activity | Specific activity | Purification | Yield | |
|---|---|---|---|---|---|
| mg | nmol/min | nmol/min mg | -Fold | % | |
| Crude extracta | 16,800 | 647,000 | 38.3 | 1.0 | 100 |
| Phenyl-Sepharose | 866 | 130,000 | 150 | 3.92 | 20.1 |
| Hydroxyapatite | 153 | 98,700 | 646 | 16.9 | 15.3 |
| Mono Q5/5 | 0.80b | 10,000 | 12,600 | 328 | 1.56 |
| Mono Q1.6/5 | 0.012b | 3,840 | 320,000 | 8,350 | 0.59 |
a Prepared from 640 g of red paprika pericarp.
b Protein concentration was estimated using EA 280 (0.1% protein) = 1.
FIGURE 2.
Purification of CaHI from paprika pericarp. A, elution profile of CaHI in the final step of purification. HI activity in fractions containing proteins was measured by standard assay. Activity is shown by a gray bar. B, protein profile in each fraction (small letters are as in panel A; M, molecular marker) was analyzed by SDS-PAGE. C, molecular mass of native HI was determined by BN-PAGE. Arrowheads indicate bands of purified CaHI.
TABLE 2.
Kinetic parameters of purified CaHI and recombinant HIs
Ca-, C. annuum; Sl-, S. lycopersicum; St-, S. tuberosum; Ms-, M. sativa; Cs-, C. sativus; Os-, O. sativa; r-, recombinant.
| kcat | Km | kcat/Km | |
|---|---|---|---|
| s−1 | mm | s−1 mm−1 | |
| CaHI | 760 | 0.73 | 1040 |
| rCaHI | 521 ± 13.0 | 1.78 ± 0.06 | 293 ± 2.60 |
| rSlHI1 | 264 ± 18.1 | 0.20 ± 0.04 | 1349 ± 126 |
| rStHI1 | 27.7 ± 1.75 | 0.33 ± 0.03 | 83.7 ± 1.08 |
| rStHI2 | 159 ± 13.9 | 0.72 ± 0.10 | 224 ± 13.7 |
| rMsHI | 308 ± 13.9 | 0.32 ± 0.04 | 990 ± 79.1 |
| rCsHI1 | 951 ± 66.2 | 0.56 ± 0.08 | 1749 ± 172 |
| rOsHI1 | 43.5 ± 2.23 | 1.23 ± 0.12 | 35.8 ± 1.61 |
Cloning of CaHI
To clone the cDNA encoding CaHI, we determined the internal amino acid sequences. After the purified protein was chemically cleaved by BrCN, polypeptide fragments were separated by Tricine-SDS-PAGE and blotted onto PVDF membranes (supplemental Fig. S4A). Among the blotted peptides, amino acid sequences of two major polypeptides could be determined (supplemental Fig. S4B). A homology search of these sequences using a Solanaceae-specific database indicated that the amino acid sequence of an unknown protein encoded by Ca08g14620 contained both sequences. To confirm that this unknown protein was identical to CaHI, the ORF of the unknown protein was cloned into pColdProS2 plasmid and then produced as a recombinant protein (supplemental Fig. S5A). The purified recombinant protein in soluble form showed high HI activity (supplemental Fig. S5B), indicating that Ca08g14620 encoded CaHI. rCaHI, like native CaHI, showed high activity at weakly acidic and neutral pH values (supplemental Fig. S3 B).
HI Belongs to the Cupin Superfamily
BLAST search using the amino acid sequence of CaHI indicated that it is a member of the cupin superfamily. This superfamily is a large protein family containing diverse functional proteins such as storage proteins and various enzymes (31). A subfamily including CaHI is located near germin and a germin-like protein family including 11S globulin and vicilin (Fig. 3). CaHI and highly homologous proteins in Solanaceae species such as tomato and potato are included in a clade named Solanaceae HI, and another clade closely related to HI is named Solanaceae HI-like. We produced proteins belonging to clade HI and HI-like as recombinant proteins and assayed their activity. Proteins belonging to clade HI showed activity, but one representative (SlHI-like 1 from tomato) belonging to clade HI-like did not (supplemental Fig. S5C). To investigate the physiological role of HIs, their gene expression levels at different developmental stages were analyzed. Expression levels of HIs in paprika and potato showed developmental stage-specific expression, being extremely high in ripe fruits and sprouts, respectively (supplemental Fig. S6, A and B). In contrast, expression levels of tomato HIs were consistently low (supplemental Fig. S5C), consistent with our inability to detect HI activity in tomato leaves and fruits (supplemental Fig. S1).
FIGURE 3.
Phylogenic tree of HIs. Proteins having catalytic HKY and homologous proteins but not having catalytic HKY are named as HI and HI-like, respectively. Left, HI and HI-like clades (shown by a dotted circle) belong to germin and germin-like protein family in the cupin superfamily. Right, enlarged view of HI and HI-like clades. Recombinant proteins showing HI activity or no activity are marked with ○ or −, respectively. Numbers in parentheses indicate identity against CaHI. Values at the nodes indicate percentage of bootstrap support (of 1000 bootstrap replicates).
Determination of Amino Acid Residues Essential for Enzymatic Activity
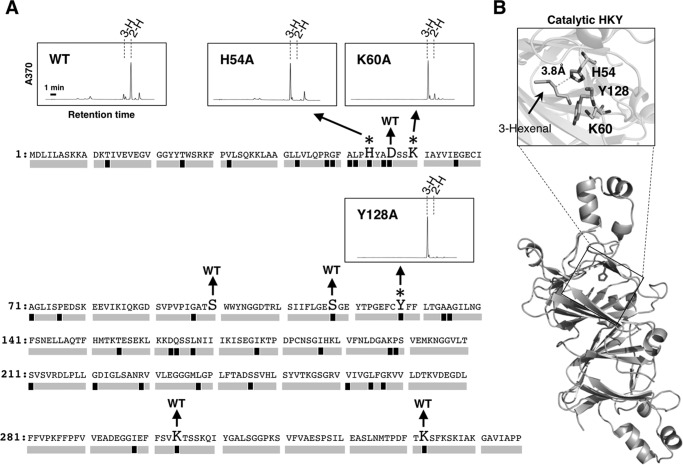
To identify amino acid residues essential for the isomerization reaction, we considered the action mode of bacterial β-hydroxydecanoyl thiolester dehydrase (32) in which a His residue plays a critical role in the isomerization reaction. Among 357 amino acid residues, 38 differed among proteins belonging to Solanaceae HI and HI-like clades, and only His-54 was conserved in all proteins in the HI clade but not in the HI-like clade, suggesting this His as a candidate catalytic amino acid in HIs. To test this hypothesis, we produced point-mutated rCaHI(H54A). As expected, rCaHI(H54A) showed no activity (Fig. 4A), indicating that His-54 plays a critical role in enzymatic reaction. By homology modeling to deduce other catalytic amino acids, Lys-60 and Tyr-128 were found to be candidates due to their locations near His-54 (Fig. 4B). We accordingly produced rCaHI(K60A) and rCaHI(Y128A) and found that these point mutations caused loss of isomerase activity (Fig. 4A). These results suggested that these three amino acid residues (named catalytic HKY) form a catalytic site in HI.
FIGURE 4.
Determination of catalytic amino acids of HI. A, effect of point mutation on HI activity. Amino acids written in large print indicate point-muted amino acids, and chromatograms of the point-muted proteins completely losing activity are shown. Asterisks suggest essential amino acids to show the HI activity, and point-mutated amino acids with no effect on the HI activity are indicated by WT. Black boxes under the amino acid sequence of CaHI indicate amino acids conserved in all proteins belonging to Solanaceae HI clade but not in HI-like clade. Gray boxes show amino acids not conserved in Solanaceae HI clade or conserved both HI and HI-like clades. 3-H and 2-H in chromatograms indicate peaks of (Z)-3- and (E)-2-hexenal-DNPs, respectively. B, homology modeling of CaHI to deduce catalytic amino acids. Catalytic amino acids locate in the same pocket and near substrate (Z)-3-hexenal. PDB ID of template protein, identity between CaHI and template protein, and QMEAN score are 2e9q, 23%, and 0.62, respectively.
HIs Are Distributed among Various Plant Species
Given that (E)-2-hexenal has been detected in various plant species other than Solanaceae (9), the presence of other types of HI was expected. To identify other HIs, we searched in other plant species for proteins showing lower similarity but conserving the catalytic HKY, finding candidates in alfalfa, cucumber, and rice sequences (alignment is shown in supplemental Fig. S7). We produced recombinant HIs and assayed their activity. All recombinant proteins having catalytic HKY showed HI activity (supplemental Fig. S8), confirming that HIs are widely distributed among various plant species. Phylogenetic tree analysis showed that these miscellaneous HIs form a branched clade from Solanaceae HI, and HI-like proteins having no HKY catalytic amino acids are present as in the case of Solanaceae HI (Fig. 3). HIs of monocotyledonous plants (OsHIs) are located far from those of dicotyledonous plants.
Catalysis Mode of HI
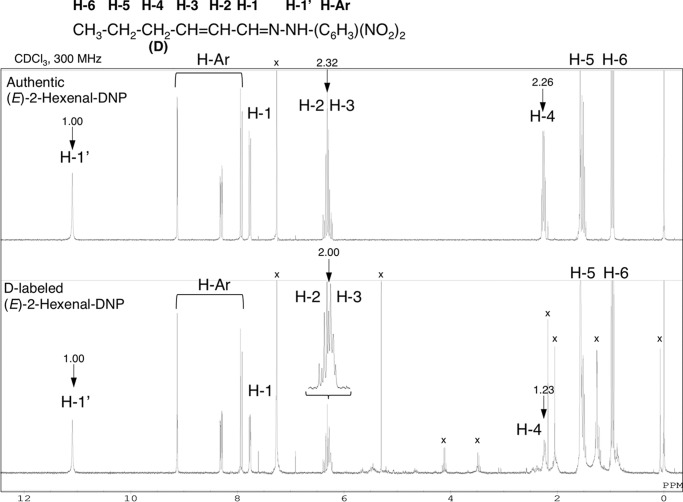
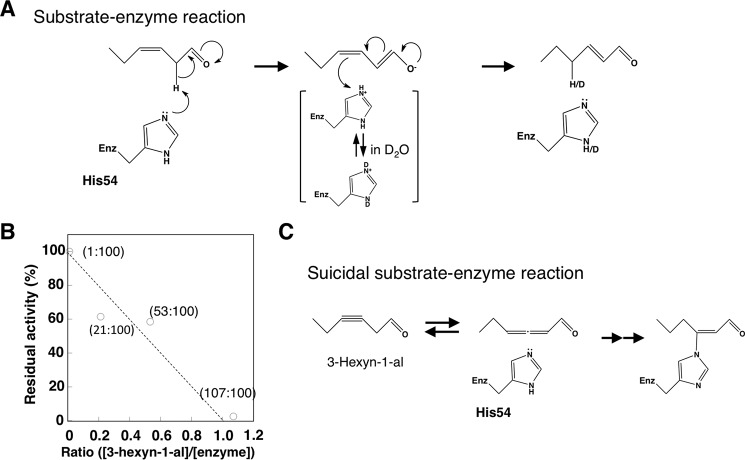
To investigate the catalytic mechanism of HI, (Z)-3-hexenal was incubated with rCaHI in the presence of D2O as a solvent, and the enzymatic product was analyzed by 1H NMR to identify the positions and geometry of the C-C double bond in 2-hexenal. 1H NMR spectra of authentic (E)-2-hexenal-DNP and enzymatic D-labeled 2-hexenal are shown in Fig. 5. The signals H-2 (6.35 ppm, m, 1H) and H-3 (6.27 ppm, m, 1H) were identified as the protons on the double bond, and the coupling constants between H-2 and H-3 (J = 15.6 Hz) were determined to be E. By comparison of the authentic and enzymatic products, deuterium was introduced mainly at the C4 position (H-4) given that the integrated area of H-4 in enzymatically produced hexenal-DNP was half that in authentic 2-hexenal-DNP. Incorporation of deuterium in C4 position (H-4) was also supported by disturbed 1H signals of C3 position (H-3) in D-labeled (E)-2-hexenal-DNP (Fig. 5) due to substituted deuterium of H-4 position. These results indicate that rCaHI isomerized (Z)-3-hexenal to (E)-2-hexenal by abstraction of H+ at the C2 position and subsequent H+ donation at the C4 position in a His-54-mediated keto-enol tautomerism reaction mode (a plausible catalytic mechanism is shown in Fig. 6A). To further investigate the catalytic mechanism, we produced point-mutated rCaHI (K60R) and rCaHI (Y120F) to evaluate the roles of the ϵ-NH2 group in the Lys-60 and OH group in Tyr-128, respectively (supplemental Fig. S9A). rCaHI (K60R) completely lost HI activity, but rCaHI (Y128F) retained the activity (supplemental Fig. S9B), suggesting that Lys-60 was involved in the catalysis mechanism.
FIGURE 5.
1H NMR spectra of authentic (upper) and D-labeled (lower) (E)-2-hexenal-DNPs. The letters indicate the position of protons and their corresponding signals. Arrows indicate integrated values of signals (that of H-1′ is set to 1). Solvent signals are indicated by ×. Ar, aromatic ring.
FIGURE 6.
Hypothetical catalytic mechanism of HI. A, His-mediated isomerization from (Z)-3-hexenal to (E)-2-hexenal catalyzed by HI. In this scheme γ-nitrogen of His is depicted as a representative example of the catalyst. B, inhibition of rCaHI activity by suicidal substrate, 3-hexyn-1-al. Numbers in parentheses indicate pmol of 3-hexyn-1-al and enzyme used for analysis, respectively. Activity without 3-hexyn-1-al was defined as 100% activity. Theoretically stoichiometric relationship between enzyme and suicidal substrate is indicated by a dotted line. C, suicidal substrate inhibitory mechanism of HI. Also in this scheme, γ-nitrogen of His is depicted as a representative example.
3-Hexyn-1-al Acts as a Suicidal Substrate for HI
As suggested by studies of bacterial isomerases (32, 33), an analogous compound having a C-C triple bond can behave as a suicidal substrate in an isomerization reaction by binding irreversibly to the catalytic His. Accordingly, we prepared 3-hexyn-1-al, a triple bond-containing analog of (Z)-3-hexenal, and tested its inhibitory activity. As shown in Fig. 6B, 3-hexyn-1-al inhibited rCaHI stoichiometrically. The removal of non-bound 3-hexyn-1-al by ultrafiltration after HI and 3-hexyn-1-al incubation did not restore activity, showing that inhibition by 3-hexyn-1-al was irreversible. This finding showed that plant HI shares a similar enzymatic property with bacterial isomerases that are irreversibly inhibited by suicidal substrates (a plausible inhibitory mechanism is shown in Fig. 6C).
Wounding Treatment-induced HI Gene Expression
(E)-2-Hexenal is assumed to play a protective role in wounding response because it shows antifungal activity (5, 34), leading to a hypothesis that HI genes are induced by wound treatment. To test this hypothesis, induction of HI gene expression in wounded paprika and tomato leaves was investigated. Expression of SlHI1, CaHI, and PINII genes, known as typical wound-inducible genes (35), was enhanced by wounding treatment (Fig. 7), suggesting that HIs may be regulated at the transcriptional level in response to wounding, leading to enhanced (E)-2-hexenal production.
FIGURE 7.
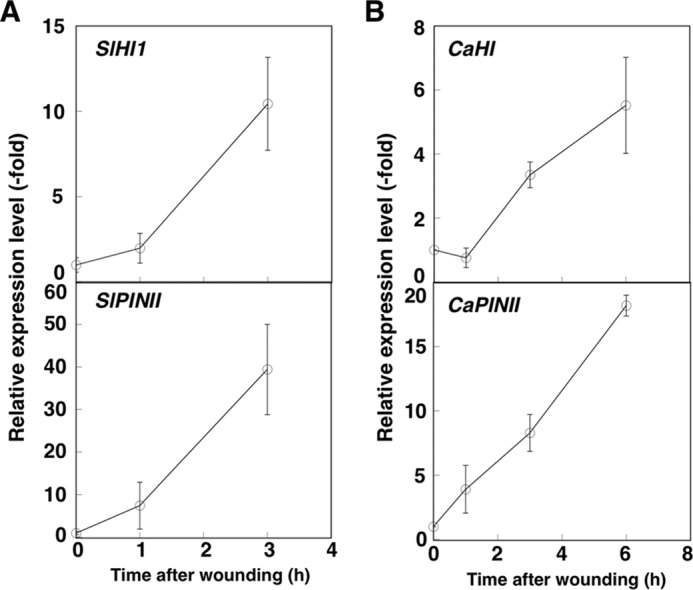
Induction of HIs and PINII by wounding treatment. After leaves of tomato (A) and paprika (B) were wounded by tweezers, expressed genes were quantified by qRT-PCR. The relative expression level of the zero-time sample against the internal standard gene (the actin genes SlACT and CaACT for tomato and paprika samples, respectively) was set to 1. Data are the means ± S.E. (n = 3).
Overexpression of CaHI in Transgenic Tomatoes Drastically Changes the (E)-2-Hexenal Production in Planta
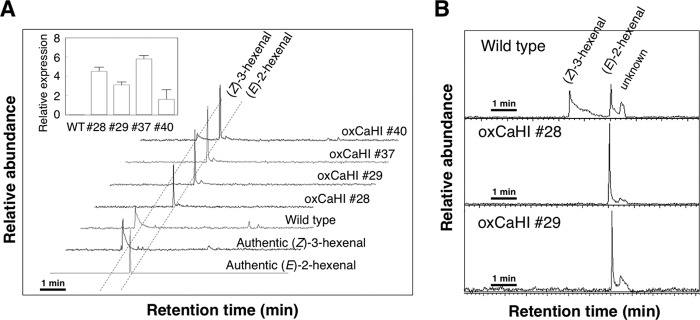
(Z)-3-Hexenal is known to be the most abundant volatile in tomato fruits and is thus recognized as a source of tomato-like flavor (36). To assess the biological impact of HI on hexenal composition, we produced transgenic tomato plants overexpressing CaHI and assayed their hexenal composition. In both leaves and fruits of wild-type tomatoes, (Z)-3-hexenal was detected as the main hexenal, as reported by previous authors (Fig. 8), suggesting that the very low expression of inherited tomato HI genes does not contribute to production of (E)-2-hexenal. In contrast, overexpression of CaHI in transgenic tomato plants resulted in a drastic change in (Z)-3/(E)-2-hexenal proportion in leaves (Fig. 8A), corresponding to the much higher expression of CaHI introduced exogenously (Fig. 8A, inset). In fruits, the wild-type contained (Z)-3-hexenal as the main hexenal (85% ± 5.8%), but (E)-2-hexenal was the sole hexenal in transgenic plants (Fig. 8B). These results suggest that the CaHI transgene CaHI functions effectively and thus determines hexenal composition in planta.
FIGURE 8.
Overexpression of CaHI drastically changed (Z)-3-hexenal and (E)-2-hexenal composition in transgenic tomatoes. A, (Z)-3- and (E)-2-hexenal analysis in wild-type and CaHI-overexpressing tomato (oxCaHI) leaves. Volatiles were collected by SPME and then analyzed by GC-MS. SIM (Selected ion monitoring, m/z = 98) chromatograms to detect (Z)-3- and (E)-2-hexenals are shown. Inset, confirmation of enhanced expression of CaHI in transgenic tomatoes by qRT-PCR (the actin gene SlACT was used as an internal standard). Numbers indicate transgenic tomato lines showing high CaHI expression. B, SIM chromatogram (m/z = 98) of volatiles from ripe tomato fruits.
Discussion
In this study we identified HIs responsible for (E)-2-hexenal production in various plant species and showed evidence that plants produce (E)-2-hexenal by enzymatic reaction. Expression analysis of HIs indicated that HI levels remained low except under several physiological conditions such as wounding and at specific developmental stages (Fig. 7 and supplemental Fig. S6), suggesting that the physiological roles of (E)-2-hexenal are limited to certain conditions. Previous reports suggested that (E)-2-hexenal as well as (Z)-3-hexenal were wounding-responsive volatiles, given that they showed antibiotic and defense gene-inducing activities (4, 5, 34). In response to wounding, the production of GLVs including both (Z)-3- and (E)-2-hexenals is an early event, a reflexive response that starts within a few minutes after wounding and leads to the presence of large amounts of detectable GLVs after 10 min (3). (Z)-3- and (E)-2-hexenal production is mediated by a series of enzymes including lipase, 13-lipoxygenase, hydroperoxide lyase, and HI (Fig. 1), and these enzymes need no cofactors for their reactions. This biochemical property would facilitate the rapid hexenal burst after wounding. At the transcriptional level, expression of HIs was low except for a specific developmental stage (supplemental Fig. S6) and was induced by wounding treatment over a period of hours (Fig. 7). Given that emission of (E)-2-hexenal continues for hours after wounding (3), this transcriptional regulation of HIs in wounding response might support this long term emission.
The site of production of (Z)-3-hexenal are chloroplasts because both 13-lipoxygenase and hydroperoxide lyase are chloroplastic proteins, whereas cytosol is likely to be a subsequent production site of (E)-2-hexenal judged from the amino acid sequence of HI indicating HIs as cytosolic proteins; that is, native CaHI includes N-terminal amino acid sequence (supplemental Fig. S4B), and computational protein localization predictors also indicated that no transit signature sequence was found in C-terminal region.
HIs showed characteristic expression at specific developmental stages. In the case of paprika, higher expression of CaHI was observed in ripe fruits (supplemental Fig. S6A). This expression might contribute to producing (E)-2-hexenal as an antifungus volatile as in strawberry (37). In potato, higher expression of StHI1 and StHI2 was observed in sprouts (supplemental Fig. S6B). Because in transgenic potato, depletion of hydroperoxide lyase to decrease GLV contents caused an increase in aphid performance (38), higher expression of StHIs may promote the production of (E)-2-hexenal as an insect repellent to protect sprouts from pests (39).
The isomerization catalyzed by HI is likely to be the keto-enol tautomerism reaction mode (Fig. 6A), similar to that of the keto-enol tautomerism-mediated isomerization of the double bond of fatty acid derivatives, which has been well studied in bacterial fatty acid metabolism. In the case of β-hydroxydecanoyl thioester dehydrase, which catalyzes the reaction of double bond isomerization on 10-carbon thioesters of acyl carrier protein in the biosynthesis of unsaturated fatty acids under anaerobic conditions (40), the catalytic mode is also the keto-enol tautomerism mediated by a catalytic His, and a specific suicidal substrate, 3-decynoyl-N-acetylcysteamine, inactivated the enzyme by irreversible binding to the catalytic His (32). Also in HIs, a specific suicidal substrate, 3-hexyn-1-al, completely inhibited HI activities irreversibly (Fig. 6), suggesting that the His residue plays a critical role in catalytic function, plausibly the migration of H+. The importance of two other catalytic amino acids (Lys-60 and Tyr-128) was also shown by complete loss of HI activity of rCaHI(K60A) and rCaHI(Y128A) (Fig. 4). In the case of Lys-60, basic amino acid substitution with Arg (K60R) resulted in the complete loss of activity (supplemental Fig. S9), suggesting that the protonated ϵ-NH2 group in Lys-60 under neutral and acidic pH conditions was essential to the keto-enol tautomerism as a proton donor. On the other hand, Tyr-128 might contribute in forming the substrate binding pocket, given that the OH group in Tyr-128 was apparently not involved in the catalysis, as shown by the retained activity of rCaHI (Y128F).
Identification of quantitative trait loci that affect the volatile emissions of tomato fruits has been studied, because tomato breeders wish to combine good flavor with high fruit firmness, long shelf life, and high disease resistance (41). However, conventional breeding for sensory quality has been severely limited (42). For this reason, genetic manipulation is applied for improvement of tomato flavor. Changes in expression levels of alcohol dehydrogenase (43), fatty acid desaturase (44), and linalool synthase (45) resulted in changes in composition of volatiles. Among tomato volatiles, (Z)-3-hexenal is the main flavor volatile determining tomato flavor, thus that (Z)-3-hexenal is assigned as a source of tomato-like flavor (36). As shown in this study (Fig. 8), both leaves and fruits of overexpressing CaHI plants contained (E)-2-hexenal as the main green odor but not (Z)-3-hexenal. Alonso et al. (42) found that (E)-2-hexenal is one of the major contributors determining sensory differences among traditional and hybrid tomato types, suggesting that gene manipulation of HI is a candidate approach with high potential in molecular breeding for tomato flavor.
In conclusion, we identified HIs in various plant species, and our identification of HIs allows the completion of the scheme of GLV biosynthesis in plants. HIs share a small number of catalytic amino acids, and catalytic His is plausibly responsible for the keto-enol tautomerism involved in isomerization of (Z)-3-hexenal to (E)-2-hexenal. Higher expression of the gene encoding HI in transgenic tomatoes led to enhanced (E)-2-hexenal production, suggesting that HI plays a key role in the production of (E)-2-hexenal in planta.
Author Contributions
Y. Y. designed the study. M. K, and Y. Y. performed the research, M. K. and H. T. analyzed the chemical data, M. M. analyzed the molecular biological data, Y. S. supervised the research, and M. K. and Y. Y. wrote the paper.
Supplementary Material
Acknowledgments
Tomato seeds (Micro-Tom, TOMJPF00001) were provided by University of Tsukuba, Gene Research Center, through the National Bio-Resource Project (NBRP) of the Japan Agency for Research and Development (AMED), Japan.
This work was supported by JSPS (Japan Society for the Promotion of Science) KAKENHI Grant 15K07325 (to Y. Y.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables 1–4 and Figs. S1–S9.
- GLV
- green leaf volatile
- HI
- (Z)-3:(E)-2-hexenal isomerase
- DNP
- 2,4-dinitrophenylhydrazone
- CaHI
- Capsicum annuum HI
- rCaHI
- recombinant CaHI
- K-PB
- potassium phosphate buffer
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- qRT
- quantitative real time
- OsHI
- O. sativa HI.
References
- 1. Niinemets U. (2010) Mild versus severe stress and BVOCs: thresholds, priming, and consequences. Trends Plant Sci. 15, 145–153 [DOI] [PubMed] [Google Scholar]
- 2. D'Auria J. C., Pichersky E., Schaub A., Hansel A., and Gershenzon J. (2007) Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J. 49, 194–207 [DOI] [PubMed] [Google Scholar]
- 3. Fall R., Karl T., Hansel A., Jordan A., and Lindinger W. (1999) Volatile organic compounds emitted after leaf wounding: on-line analysis by proton-transfer-reaction mass spectrometry. J. Geophys. Res. 104, 15963–15974 [Google Scholar]
- 4. Kishimoto K., Matsui K., Ozawa R., and Takabayashi J. (2005) Volatile C6-aldehyde and allo-ocimene activate defense genes and induce resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol. 46, 1093–1102 [DOI] [PubMed] [Google Scholar]
- 5. Kishimoto K., Matsui K., Ozawa R., and Takabayashi J. (2006) Components of C6-aldehyde-induced resistance in Arabidopsis thaliana against a necrotrophic fungal pathogen, Botrytis cinerea. Plant Sci. 170, 715–723 [Google Scholar]
- 6. Scala A., Allmann S., Mirabella R., Haring M. A., and Schuurink R. C. (2013) Green leaf volatiles: a plant's multifunctional weapon against herbivores and pathogens. Int. J. Mol. Sci. 14, 17781–17811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamauchi Y., Kunishima M., Mizutani M., and Sugimoto Y. (2015) Reactive short-chain leaf volatiles act as powerful inducers of abiotic stress-related gene expression. Sci. Rep. 5, 8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen G., Hackett R., Walker D., Taylor A., Lin Z., and Grierson D. (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 136, 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hatanaka A., Kajiwara T., and Sekiya J. (1987) Biosynthetic pathway for C6-aldehydes formation from linolenic acid in green leaves. Chem. Phys. Lipids 44, 341–361 [Google Scholar]
- 10. Howe G. A., Lee G. I., Itoh A., Li L., and DeRocher A. E. (2000) Cytochrome P450-dependent metabolism of oxylipins in tomato: cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 123, 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamauchi Y., Hasegawa A., Taninaka A., Mizutani M., and Sugimoto Y. (2011) NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 286, 6999–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bate N. J., Riley J. C. M., Thompson J. E., and Rothstein S. J. (1998) Quantitative and qualitative difference in C6-volatile production from the lipoxygenase pathway in an alcohol dehydrogenase mutant of Arabidopsis thaliana. Physiol. Plant. 104, 97–104 [Google Scholar]
- 13. Mano J., Torii Y., Hayashi S., Takimoto K., Matsui K., Nakamura K., Inzé D., Babiychuk E., Kushnir S., and Asada K. (2002) The NADPH:quinone oxidoreductase P1-ζ-crystallin in Arabidopsis catalyzes the α,β-hydrogenation of 2-alkenals: detoxication of the lipid peroxide-derived reactive aldehydes. Plant Cell Physiol. 43, 1445–1455 [DOI] [PubMed] [Google Scholar]
- 14. Curtius T., and Franzen H. (1912) Über die chemischen Bestandteile grüner Pflanzen: Über den Blätteraldehyd. Ann. Chem. 390, 89–121 [Google Scholar]
- 15. Phillips D. R., Matthew J. A., Reynolds J., and Fenwick G. R. (1979) Partial purification and properties of a cis-3:trans-2-enal isomerase from cucumber fruit. Phytochemistry 18, 401–404 [Google Scholar]
- 16. Noordermeer M. A., Veldink G. A., and Vliegenthart J. F. (1999) Alfalfa contains substantial 9-hydroperoxide lyase activity and a 3Z:2E-enal isomerase. FEBS Lett. 443, 201–204 [DOI] [PubMed] [Google Scholar]
- 17. Yamauchi Y., Hasegawa A., Mizutani M., and Sugimoto Y. (2012) Chloroplastic NADPH-dependent alkenal/one oxidoreductase contributes to the detoxification of reactive carbonyls produced under oxidative stress. FEBS Lett. 586, 1208–1213 [DOI] [PubMed] [Google Scholar]
- 18. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 19. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wavrin L., and Viala J. (2002) Clean and efficient oxidation of homoallylic and homopropargylic alcohols into β,γ-unsaturated aldehydes by the Dess-Martin periodinane. Synthesis 3, 326–330 [Google Scholar]
- 21. Sun H.-J., Uchii S., Watanabe S., and Ezura H. (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 47, 426–431 [DOI] [PubMed] [Google Scholar]
- 22. Matsui K., Sugimoto K., Mano J., Ozawa R., and Takabayashi J. (2012) Differential metabolisms of green leaf volatiles in injured and intact parts of a wounded leaf meet distinct ecophysiological requirements. PLoS ONE 7, e36433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 24. Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., and Schwede T. (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protocols 4, 1–13 [DOI] [PubMed] [Google Scholar]
- 25. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benkert P., Künzli M., and Schwede T. (2009) QMEAN server for protein model quality. Nucleic Acids Res. 37, W510–W514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guex N., and Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 28. Luning P. A., Carey A. T., Roozen J. P., and Wichers H. J. (1995) Characterization and occurrence of lipoxygenase in bell peppers at different ripening stages in relation to the formation of volatile flavor compounds. J. Agric. Food Chem. 43, 1493–1500 [Google Scholar]
- 29. Galliard T., Phillips D. R., and Reynolds J. (1976) The formation of cis-3-nonenal, trans-2-nonenal and hexenal from linoleic acid hydroperoxide isomers by a hydroperoxide cleavage enzyme system in cucumber ( Cucumis sativus) fruits. Biochim. Biophys. Acta 441, 181–192 [DOI] [PubMed] [Google Scholar]
- 30. Hatanaka A., Kajiwara T., and Harada T. (1975) Biosynthetic pathway of cucumber alcohol: trans-2,cis-6-nonadienol via cis-3,cis-6-nonadienal. Phytochemistry 14, 2589–2592 [Google Scholar]
- 31. Dunwell J. M., Purvis A., and Khuri S. (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65, 7–17 [DOI] [PubMed] [Google Scholar]
- 32. Leesong M., Henderson B. S., Gillig J. R., Schwab J. M., and Smith J. L. (1996) Structure of a dehydratase isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure 4, 253–264 [DOI] [PubMed] [Google Scholar]
- 33. Endo K., Helmkamp G. M. Jr., and Bloch K. (1970) Mode of inhibition of β-hydroxydecanoyl thioester dehydrase by 3-decynoyl-N-acetylcysteamine. J. Biol. Chem. 245, 4293–4296 [PubMed] [Google Scholar]
- 34. Croft K.., Juttner F., and Slusarenko A. J. (1993) Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv. phaseolicola. Plant Physiol. 101, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ryan C. A. (1990) Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu. Rev. Phytopathol. 28, 425–449 [Google Scholar]
- 36. Tandon K. S., Baldwin E. A., and Shewfelt R. L. (2000) Aroma perception of individual volatile compounds in fresh tomatoes (Lycopersicon esculentum, Mill.) as affected by the medium of evaluation. Postharvest Biol. Technol. 20, 261–268 [Google Scholar]
- 37. Myung K., Hamilton-Kemp T. R., and Archbold D. D. (2006) Biosynthesis of trans-2-hexenal in response to wounding in strawberry fruit. J. Agric. Food Chem. 54, 1442–1448 [DOI] [PubMed] [Google Scholar]
- 38. Vancanneyt G., Sanz C., Farmaki T., Paneque M., Ortego F., Castañera P., and Sánchez-Serrano J. J. (2001) Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc. Natl. Acad. Sci. U.S.A. 98, 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Moraes C. M., Mescher M. C., and Tumlinson J. H. (2001) Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature 410, 577–580 [DOI] [PubMed] [Google Scholar]
- 40. Bloch K. (1971) 15β-Hydroxydecanoyl thioester dehydrase. The Enzymes, 3rd Ed, Vol. 5, pp. 441–464, Academic Press, NY [Google Scholar]
- 41. Mathieu S., Cin V. D., Fei Z., Li H., Bliss P., Taylor M. G., Klee H. J., and Tieman D. M. (2009) Flavour compounds in tomato fruits: identification of loci and potential pathways affecting volatile composition. J. Exp. Bot. 60, 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alonso A., García-Aliaga R., García-Martínez S., Ruiz J. J., and Carbonell-Barrachina A. A. (2009) Characterization of Spanish tomatoes using aroma composition and discriminant analysis. Food Sci. Tech. Int. 15, 47–55 [Google Scholar]
- 43. Speirs J., Lee E., Holt K., Yong-Duk K., Steele Scott N., Loveys B., and Schuch W. (1998) Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols. Plant Physiol. 117, 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang C., Xing J., Chin C.-K., Ho C.-T., and Martin C. E. (2001) Modification of fatty acids changes the flavor volatiles in tomato leaves. Phytochemistry 58, 227–232 [DOI] [PubMed] [Google Scholar]
- 45. Lewinsohn E., Schalechet F., Wilkinson J., Matsui K., Tadmor Y., Nam K.-H., Amar O., Lastochkin E., Larkov O., Ravid U., Hiatt W., Gepstein S., and Pichersky E. (2001) Enhanced levels of the aroma and flavor compound S-linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiol. 127, 1256–1265 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.