Abstract
The viral vector-mediated overexpression of the defined transcription factors, Brn4/Pou3f4, Sox2, Klf4, and c-Myc (BSKM), could induce the direct conversion of somatic fibroblasts into induced neural stem cells (iNSCs). However, viral vectors may be randomly integrated into the host genome thereby increasing the risk for undesired genotoxicity, mutagenesis, and tumor formation. Here we describe the generation of integration-free iNSCs from mouse fibroblasts by non-viral episomal vectors containing BSKM. The episomal vector-derived iNSCs (e-iNSCs) closely resemble control NSCs, and iNSCs generated by retrovirus (r-iNSCs) in morphology, gene expression profile, epigenetic status, and self-renewal capacity. The e-iNSCs are functionally mature, as they could differentiate into all the neuronal cell types both in vitro and in vivo. Our study provides a novel concept for generating functional iNSCs using a non-viral, non-integrating, plasmid-based system that could facilitate their biomedical applicability.
Keywords: cell biology, cell therapy, neural stem cell (NSC), regenerative medicine, reprogramming
Introduction
Neural stem cells (NSCs),3 somatic stem cells of the brain and spinal cord, could differentiate into three major cell types (neurons, astrocytes, and oligodendrocytes) in the central nervous system (CNS) in response to relevant signaling pathways (1). A number of in vitro transplantation studies with disease animal models have demonstrated that the engrafted NSCs can functionally rescue disease-related phenotypes, proving their therapeutic potential (2–5). Thus, NSCs have long been considered as a potential source for replacing damaged cells and tissues often found in various neurodegenerative diseases (2–5). However, major concerns associated with these cells may preclude expediting advances into their therapeutic application. First, the accessibility of NSCs is largely restricted in vitro, due to their limited origins. Second, the autologous cell transplantation is not feasible using NSCs from their in vivo origins, and furthermore, allogeneic transplantation of NSCs may raise the potential for immune rejection. Finally, with current culture conditions, it is technically challenging to homogeneously maintain human NSCs in vitro (6, 7). Thus, NSC-like cells generated from easily accessible patient somatic cell types such as fibroblasts and blood cells could serve as an autologous source for therapeutic applications, thereby overcoming current obstacles to NSC-mediated translation research.
We and others have demonstrated the direct conversion of somatic fibroblasts into self-renewing and multipotent induced neural stem cells (iNSCs) or induced neural progenitor cells (iNPCs) by the forced expression of different sets of transcription factors (8–12). Recently, Thier et al. (12) have shown that the restricted expression of Oct4 by a tetracycline-dependent lentiviral vector, together with retrovirus-mediated overexpression of Sox2, Klf4, and c-Myc, could elicit the conversion of fibroblasts into iNSCs. Simultaneously, we have generated multipotent and self-renewing iNSCs from mouse fibroblasts by retrovirus-mediated overexpression of Brn4/Pou3f4, Sox2, Klf4, and c-Myc (BSKM) (9, 10). All the iNSCs generated in these studies closely resemble their in vivo counterparts in terms of morphology, gene expression profile, epigenetic status, and self-renewing capacity. They could also differentiate into neurons, astrocytes, and oligodendrocytes both in vitro and in vivo. Furthermore, in a recent preclinical study, iNSCs were shown to have therapeutic potential following transplantation into the spinal cord injury disease model (13).
Importantly, all these aforementioned studies have used either lentiviruses or retroviruses to deliver sets of transgenes into the starting cells. Although the transgenes become completely silenced in fully converted iNSCs, viral vectors can randomly integrate into the host genome and pose an increased risk for undesired integration-associated genotoxicity, insertional mutagenesis, and tumor formation due to uncontrolled reactivation of the transgenes. These drawbacks may pose significant challenges in the clinical translation of iNSCs, necessitating the development of a method for generating iNSCs without viral transduction.
The Epstein-Barr virus (EBV)-based vector containing oriP/EBNA-1 may be applied to this end for the following reasons. It can simply be transfected into cells without viral packing. After transfection, the vector can persist in the extra chromosomes within the nucleus of transfected cells as multicopy episomes without integrating into the host genome. After a certain time point, the vector can naturally be expelled from the initial transfected cells (14). Thus, the transfected cells are totally independent of episomal vector-derived transgenes. Recent studies have used this system as a reprogramming tool for generating integration-free induced pluripotent stem cells (iPSCs), rendering the oriP/EBNA-1 vector system potentially useful for the direct conversion technology (15, 16).
In this study, we employed episomal vectors to generate integration-free iNSCs. We found that a single transfection of episomal vectors containing BSKM was sufficient for generating iNSCs from mouse embryonic fibroblasts, albeit with a lower conversion efficiency than a viral system. The episomal vector-derived iNSCs (e-iNSCs) closely resemble control NSCs generated from the brain tissue as well as retrovirus-mediated iNSCs (r-iNSCs) in the morphology, global transcriptome, epigenetic features, and both in vitro and in vivo differentiation abilities. More importantly, e-iNSCs are indeed integration free. Therefore, our novel approach for generating integration-free iNSCs could expedite advances into their clinical translation.
Experimental Procedures
Ethics Statement
All mice used were bred and housed at the mouse facility of Konkuk University. All protocols in this study were approved by Institutional Animal Care and Use Committee (IACUC) of Konkuk University, and the methods were carried out in accordance with the approved guidelines.
Cell Culture
Mouse embryonic fibroblasts (MEFs) were derived from C3H mouse strain embryos at embryonic day 13.5 after carefully removing the head and all the internal organs including spinal cord. MEFs were maintained in DMEM (Biowest) containing 10% FBS (Biowest), 5 ml of penicillin/streptomycin/glutamine (Invitrogen), and 5 ml of MEM NEAA solution (Invitrogen) in 500 ml of MEF medium. The control NSCs and established iNSCs were maintained in NSC culturemedium: DMEM/F-12 supplemented with 10 ml of B27 supplements (Gibco), 10 ng/ml EGF (Peprotech), 10 ng/ml of bFGF (Peprotech), and 5 ml of penicillin/streptomycin/glutamine (Invitrogen) in 500 ml of NSC medium.
Generation of iNSCs
To generate e-iNSCs, 1 × 106 of MEFs were transfected using Amaxa P4 primary cell 4D-Nucleofector kit (Lonza) according to the manufacturer's instructions. Briefly, 1.5 μg of each episomal vector was mixed with 82 μl of P4 primary cell solution and 18 μl of supplement 1. The mixture of MEFs and episomal vectors was then transferred into NucleocuvetteTM Vessel and electroporated with CZ-167 program. The transfected cells were plated onto the gelatin-coated dish in MEF medium. Starting on the next day, the cells were cultured in NSC medium, which was replaced every other day with fresh medium until initial clusters were observed. To generate retroviral vector-mediated r-iNSCs, the MEFs were transduced with retroviral particles and cultured as previously described (9, 10). Briefly, 5 × 104 fibroblasts were plated onto the gelatin-coated 35-mm dish and incubated with ecotropic retroviruses for 48 h. After 48 h of incubation, the medium containing retroviral particles was replaced with NSC medium. To enrich the initial cluster of both e-iNSCs and r-iNSCs, non-reprogrammed fibroblasts or unwarranted cells were removed with a cell scraper as previously described (10). The initial iNSC clusters were observed around 4 weeks after initiation of reprogramming process. The clusters were maintained for 2–3 more days for maturation, and then passaged in a 1:1 ratio for the expansion and establishment of iNSCs. To establish the clonal iNSC lines, the iNSC bulk culture was stained with an antibody against the SSEA1, and SSEA1-positive single cells were sorted using BD FACSAriaTM (BD Biosciences) and plated onto laminin/poly-d-lysine-coated 96-well plates.
Gene Expression Analysis by RT-PCR and qPCR
Total RNA was isolated using the Hybrid-RTM kit (GeneAll), and 1 μg of total RNA was reverse transcribed into cDNA using the high capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's instructions. RT-PCR was performed using the GoTag green master mix (Promega). qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) on the ABI 7500 real-time PCR system (Applied Biosystems). ΔCt values were calculated by subtracting the Gapdh Ct value from that of target genes. Relative expression levels were calculated using the 2−ΔΔCt method. The sequence of primer sets was listed in Table 1.
TABLE 1.
Primers for RT-PCR and qPCR
| Gene name | Accession number | Sequence | Annealing temperature |
|---|---|---|---|
| Pax6 | NM_ 001244198 | 5′-CAAGTTCCCGGGAGTGAACC-3′ | 60 °C |
| 5′-TCCACATAGTCATTGGCAGA-3′ | |||
| Sox1 | NM_ 009233 | 5′-GTGCCCCTGACGCACATCTA-3′ | 60 °C |
| 5′-GAGCGGCCTTTTATCGAGAG-3′ | |||
| Sox2 | NM_ 011443 | 5′-ACGGCCATTAACGGCACACT-3′ | 60 °C |
| 5′-TTTTGCACCCCTCCCAATTC-3′ | |||
| Nestin | NM_ 016701 | 5′-TCCTGGTCCTCAGGGGAAGA-3′ | 60 °C |
| 5′-TCCACGAGAGATACCACAGG-3′ | |||
| Olig2 | NM_ 016967 | 5′-ACCACCACGTGTCGGCTATG-3′ | 60 °C |
| 5′-TGGTCCAGCTCCCCTTCTTG-3′ | |||
| Blbp | NM_ 021272 | 5′-GGATGGCAAGATGGTCGTGA-3′ | 60 °C |
| 5′-TGGGACTCCAGGAAACCAAG-3′ | |||
| Mash1 | NM_ 008553 | 5′-CAGAGGAACAAGAGCTGCTG-3′ | 60 °C |
| 5′-GATCTGCTGCCATCCTGCTT-3′ | |||
| Col1a1 | NM_007742 | 5′-CCCTGCCTGCTTCGTGTAAA-3′ | 60 °C |
| 5′-TCGTCTGTTTCCAGGGTTGG-3′ | |||
| Thy1 | NM_009382 | 5′-TTCCCTCTCCCTCCTCCAAGC-3′ | 60 °C |
| 5′-TCGAGGGCTCCTGTTTCTCCTT-3′ | |||
| Pdgfrß | NM_001146268 | 5′-CAGGACCTCTGGCTGAAGCA-3′ | 60 °C |
| 5′-TCTGGGAGGCAGAAGGGAGAT-3′ | |||
| Acta2 | NM_007392 | 5′-GAAGCTGGGCCCTCCATCGT-3′ | 60 °C |
| 5′-AACTGGAGGCGCTGATCCACA-3′ | |||
| Gapdh | NM_008084 | 5′-CCAATGTGTCCGTCGTGGAT-3′ | 60 °C |
| 5′-TGCCTGCTTCACCACCTTCT-3′ | |||
| pCXLE-Brn4 | 5′-CCAATGTGTCCGTCGTGGAT-3′ | 60 °C | |
| 5′-TGCCTGCTTCACCACCTTCT-3′ | |||
| pCXLE-Sox2 | 5′-TATGGGCACAGCAGCTCCAT-3′ | 60 °C | |
| 5′-GCAGAATTCGCCCTTTACGC-3′ | |||
| pCXLE-Klf4 | 5′-GTGGTACCTCACCCTTACCGAGTC-3′ | 60 °C | |
| 5′-TGCAGCTGGCTCAGCTTAGAA-3′ | |||
| pCXLE-cMyc | 5′-GTGGTACCTCACCCTTACCGAGTC-3′ | 60 °C | |
| 5′-TGGTGGGCACAGGATTCACT-3′ | |||
| oriP/EBNA-1 | 5′-CATCATCATCCGGGTCTCCA-3′ | 60 °C | |
| 5′-CAGTGCTTGGGCCTTCTCCT-3′ |
Whole Genome Expression Analysis
Total RNA samples were prepared from MEFs, e-iNSCs, r-iNSCs, and control NSCs using the RNeasy Mini Kit (Qiagen). Microarray was carried out according to the manufacturer's instructions (Affymetrix). Briefly, 100 ng of total RNA per samples was used to generate double-stranded cDNA using a T7-oligo(dT) primer (GeneAtlasTM 3′ IVT Express Kit). Biotin-labeled cRNA was synthesized from the double-stranded cDNA and fragmented after purification. 7.5 μg of fragmented and labeled cRNA was hybridized for 16 h at 45 °C on AffymetrixGeneChip arrays (Mouse Genome 430 PM Array Strip). The chips were then washed and stained in the Affymetrix Fluidics Station, and fluorescence was detected using the Affymetrix Imaging Station. Raw data were background corrected and subsequently normalized using the Partek Express Affymetrix Edition under the Robust Microarray Analysis algorithm. Among 45,141 total annotated genes, 2,198 genes that were differentially expressed by at least 4-fold between MEFs and control NSCs were selected for analysis. Heat maps and hierarchical clustering of samples were generated by using the MeV software. Original data were uploaded to the Gene Expression Omnibus database (accession number GSE67319).
Flow Cytometry
For flow cytometry analysis, cells were dissociated with trypsin, washed once with PBS, and resuspended with FACS buffer (PBS containing 5% FBS). 1 × 106 cells were incubated with FITC-conjugated antibody raised against anti-SSEA1 (Santa Cruz Biotechnology, 1:10) for 15 min at 4 °C. Cells were washed once with FACS buffer and resuspended with PBS for analysis using a BD FACSAriaTM (BD Biosciences).
Karyotyping
The confluent monolayer of iNSCs cultured in a 25T flask was treated with 10 μg/ml of colcemid (Gibco) for 4 h to arrest cells in the metaphase state. The cells were gently washed three times with 0.075 m KCl hypotonic solution (Merck) and 1% sodium citrate (Merck), and then treated with hypotonic solution at 37 °C for 25 min. The cells were dissociated by shaking the flask and then collected into a 15-ml conical tube. After centrifugation at 1500 rpm for 10 min, the supernatant was removed, and the cell pellet was washed three times with fixative solution (methane: acetic acid = 3:1, Merck). The cell pellet was suspended with fixative solution and dropped on the cold wet slide. The slide was treated with trypsin, and then stained with Giemsa (Sigma). The slide was observed under the light microscope.
DNA Methylation Analysis
To determine the DNA methylation status of iNSCs, genomic DNA was treated with sodium bisulfite to convert all unmethylated cytosine residues into uracil residues using EpiTect Bisulfite Kit (Qiagen) according to the manufacturer's instructions. Briefly, PCR amplifications were performed using SuperTaq polymerase (Ambion) in a total volume of 25 μl and a protocol of a total of 40 cycles of denaturation at 94 °C for 30 s, annealing at the appropriate temperature for each target region for 30 s, extension at 72 °C for 30 s with a 1st denaturation at 94 °C for 5 min, and a final extension at 72 °C for 10 min. Primer sequences and annealing temperatures were described in our previous study (9, 13) and listed in Table 2. For each primer set, 3 μl of product from the first round of PCR was used in the second round of PCR as template. The amplified products were verified by electrophoresis on 1% agarose gel. PCR products were subcloned using the PCR 2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions. Reconstructed plasmids were purified using the QIAprep Spin Miniprep Kit (Qiagen) and individual clones were sequenced (Macrogen, Korea). Clones were analyzed using QUMA software.
TABLE 2.
Primers used for DNA methylation analysis
| Gene name | Sequence | Annealing temperature |
|---|---|---|
| Nestin enhancer 1st | 5′-GATCCCAGTGTGGTGGTACG-3′ | 45 °C |
| 5′-GGCTTCAGCTCCGTCTCCAT-3′ | ||
| Nestin enhancer 2nd | 5′-GTGTGGTGGTACGGGAAATC-3′ | 60 °C |
| 5′-GAGAAGGACGGGAGCAGAG-3′ | ||
| Col1a11st | 5′-GTTAGGTAGTTTTGATTGGTTGG-3′ | 55 °C |
| 5′-ACAATAACCCCTAAAAAAAACAAAAA-3′ | ||
| Col1a12nd | 5′-TGGTATAAAAGGGGTTTAGGTTAGT-3′ | 60 °C |
| 5′-ACAATAACCCCTAAAAAAAACAAAAA-3′ |
Immunocytochemistry
The cells were fixed with 4% paraformaldehyde (Sigma) for 20 min at room temperature, and then blocked with Dulbecco's PBS (Biowest) containing 0.3% Triton X-100 (Sigma) and 5% FBS (Biowest) for 2 h at room temperature.
The cells were then incubated with primary antibodies at 4 °C for 16 h, washed three times with Dulbecco's PBS, and then incubated with appropriate fluorescence-conjugated secondary antibody for 2 h at room temperature in the dark. Nuclei were stained with Hoechst 33342 (Sigma). Primary antibodies used for immunofluorescence are as follows: mouse anti-Nestin (Millipore, 1:200), goat anti-Sox2 (Santa Cruz Biotechnology, 1:200), mouse anti-SSEA1 (Santa Cruz Biotechnology, 1:100), rabbit anti-Olig2 (Millipore, 1:200), mouse anti-Tuj1 (Covance, 1:500), rabbit anti-GFAP (DAKO, 1:500), and rat anti-MBP (Abcam, 1:100).
In Vitro Differentiation
For differentiation into neurons, iNSCs were plated onto laminin/polylysine-coated dishes at 2.5 × 104 cells per cm2 in NSC medium. The next day, medium was replaced with neural differentiation medium: DMEM/F-12 supplemented with 2 ml of B27 supplements (Gibco), 1 ml of penicillin/streptomycin/glutamine (Invitrogen), and 10 ng/ml of bFGF (Peprotech) in 100 ml of neural differentiation medium. On day 4 of differentiation, the medium was changed into neural differentiation medium containing 200 mm ascorbic acid (Sigma) without growth factors for 8–10 more days. For differentiation into astrocytes, iNSCs were cultured in DMEM/F-12 supplemented with 10% FBS and 1% penicillin/streptomycin/glutamine on gelatin-coated dishes for 5 days. Finally, for differentiation into oligodendrocytes, iNSCs were plated onto laminin/polylysine-coated dishes at 2.5 × 104 cells per cm2 in NSC medium. The next day, the medium was replaced with oligodendrocyte differentiation medium: DMEM/F-12 supplemented with 2 ml of B27 supplements, 1 ml of penicillin/streptomycin/glutamine, 10 ng/ml of bFGF, and 10 ng/ml of PDGF (Sigma) in 100 ml of oligodendrocyte differentiation medium. On day 4 of differentiation, the medium was changed into oligodendrocyte differentiation medium containing 30 ng/ml of T3 (Sigma) and 200 mm ascorbic acid for another 4 days. The differentiation medium was replaced with fresh medium every other day.
Electrophysiological Recordings
The conventional whole cell patch clamp technique was performed to record membrane currents and potential (Em) using an EPC8 (HEKA, Mahone Bay, Nova Scotia, Canada) patch clamp amplifier with a BNC-2111 interface (National Instrument, Austin, TX). A coverslip with the adherent cells was placed into the recording chamber and perfused with normal Tyrode solution containing the 143 mm NaCl, 5.4 mm KCl, 0.33 mm NaH2PO4, 1.8 mm CaCl2, 0.5 mm MgCl2, 5 mm HEPES, and 11 mm glucose, and adjusted to pH 7.4 with NaOH. Patch pipettes were pulled from borosilicate capillary tubes (Clark Electromedical Instruments, Pangbourne, UK) using a puller (PP-83; Narishige, Tokyo, Japan) and filled with an internal solution containing 140 mm KCl, 5 mm NaCl, 5 mm MgATP, 10 mm HEPES, 0.5 mm GTP, and 10 mm EGTA, and adjusted to pH 7.2 with KOH. Whole cell Na+ currents and K+ currents were elicited by depolarizing voltage steps in 10-mV increments between −70 and +50 mV (100-ms duration and 3-s intervals) from a holding potential of −80 mV. Em was recorded under current-clamp mode and the action potentials were evoked with current injections (from −20 to 580 pA with 20-ms duration and 3-s intervals). Data were digitized at a sampling rate of 10 kHz, and low-pass filtered at 1 kHz. All chemicals were purchased from Sigma.
Transplantation
Postnatal day 2 (P2) Sprague-Dawley rat pups were used as recipients. Transplantation was performed under deep hypothermia and with the rats positioned in a stereotaxic frame. The iNSCs (1 × 106 cells in 5 μl culture medium) were transplanted into the cortex region (coordinates: 0 mm anteroposterior (AP), ±1.5 mm mediolateral (ML) from bregma, −1.5 to −2.0 mm dorsoventral (DV) from the surface of the brain), and injections were performed using a 10-μl siliconized Hamilton syringe with a pulled, beveled glass pipette tip (80–90 μm inner diameter) connected to a syringe pump (Pump 11 Elite Nanomite Syringe Pump; Harvard Apparatus) at a rate of 1 μl/min. The glass pipette was kept in place for an additional 5 min to prevent leakage upon withdrawal. After injection, Cyclosporin A (CipolInjTM; Chongkundang Pharmaceutical) was administered at 10 mg/kg−1 day−1 subcutaneously until the animals were sacrificed for the analysis. 4 weeks after transplantation, the animals were anesthetized by inhalation of isoflurane (Forane; Choongwae Pharma) and transcardially perfused first with cold normal saline (0.9% NaCl) followed by 4% paraformaldehyde in 0.1 m PBS (pH 7.4). The brains were dissected, fixed overnight at 4 °C, cryoprotected in 30% sucrose in 0.1 m PBS, embedded in M-1 embedding matrix (Thermo Scientific), and coronally sectioned at 10 μm thickness using a cryostat (Leica Microsystems).
Statistical Analysis
Data are presented as mean ± S.D. For direct comparisons, a two-tailed Student's t test was used to calculate p values. p < 0.05 was considered as significant.
Results
Single Transfection of Episomal Vectors Is Sufficient for Generating iNSCs
We have previously shown that the retroviral infection of the defined set of transcription factors Brn4/Pou3f4, Sox2, Klf4, and c-Myc (BSKM) could directly convert fibroblasts into iNSCs (9, 10). Although the retrovirus-mediated iNSCs (r-iNSCs) exhibited molecular and cellular characteristics similar to those of control NSCs from brain tissue, the integration of exogenous factors in the iNSC genome might preclude the clinical translation of iNSCs. Thus, in the current study, we tried to generate integration-free iNSCs using a non-viral vector system. Based on our previous studies (9, 10), the generation of iNSCs normally takes about 4–5 weeks upon retroviral introduction of BSKM. Thus, we first tested whether the episomal vector system could sufficiently deliver and maintain the reprogramming factors. For this, we first transfected an episomal vector encoding mCherry into MEFs through nucleofection, and then monitored its expression by both fluorescence microscopy and fluorescence-activated cell sorting (FACS) over a period of 4 weeks (supplemental Fig. S1, A and B). mCherry was expressed in ∼43% cells on day 5 post-transfection. Although the number of mCherry-positive cells continuously decreased upon subsequent culture, mCherry expression was maintained even after 4 weeks of transfection (6.31 ± 0.43%, supplemental Fig. S1, A and B). The non-transfected MEFs did not yield mCherry-positive cells (0.33 ± 0.08%, supplemental Fig. S1, A and B). Overall, these data indicate that our episomal vector system might be adequate for introducing and maintaining the reprogramming factors for generating iNSCs, and that the episome-mediated expression of exogenous factors becomes gradually extinct with the progression of in vitro culture.
To generate iNSCs from the MEFs, we next transfected episomal vectors encoding BSKM into MEFs (Fig. 1A and also see the method), which are free from contamination with any neuronal cell types, as evidenced by the absence of immunoreactive signals for both NSC markers (Sox2, Nestin, and Olig2) and a neuronal marker (Tuj1) in our starting cell population (supplemental Fig. S1C). On day 2 post-transfection, the transfected MEFs were cultured in NSC culture medium to support the growth of iNSCs. At 4 weeks post-transfection, we observed the first iNSC clusters displaying typical NSC-like morphology (Fig. 1B). In contrast, we did not observe any NSC-like colonies when the cells were transfected with an episomal vector containing mCherry and cultured under an identical culture condition for the entire period (Fig. 1B). Upon further passaging, we were able to establish three lines of e-iNSCs displaying morphology and marker expression patterns similar to those of control NSCs from brain tissue (Fig. 1, B–E). Of three lines established, one e-iNSC line was further characterized throughout this study. Taken together, this data indicates that a single transfection of non-viral episomal vectors are sufficient for generating iNSCs.
FIGURE 1.
Direct conversion of fibroblasts into iNSCs using episomal vectors. A, schematic illustration depicting the procedure for the direct conversion of somatic cells into iNSCs using episomal vectors. B, the morphology of the first iNSC clusters at 4 weeks after transfection, established iNSCs at passage 10, and control NSCs. Scale bars, 100 μm. C, expression of NSC and fibroblast markers in e-iNSCs and r-iNSCs analyzed by qPCR. Expression levels are normalized to those in MEFs. Error bars indicate the standard deviation of triplicate values. D and E, immunofluorescence microscopy images of e-iNSCs and r-iNSCs using antibodies against Sox2/Nestin (D) and Olig2/SSEA1. E, control NSCs were used as a positive control. Scale bars, 100 μm.
Gradual Conversion into iNSCs Using Episomal Vectors
Although we had thoroughly characterized r-iNSCs in our previous studies (9, 10), the mechanism underlying the direct conversion of somatic cells into iNSCs still remained elusive.
Thus, to understand the direct conversion process into iNSCs, we analyzed the conversion kinetics during the early phase of neural induction. To this end, we introduced the reprogramming mixture and analyzed the expression pattern of both fibroblast and NSC markers in a time course manner. We were unable to observe the activation of endogenous NSC markers up to 3 weeks of transfection. However, all NSC markers were eventually up-regulated at 4 weeks of transfection, although the level of each marker was still lower than that of control NSCs. In contrast to the slow activation of NSC markers, the fibroblast markers were suppressed within just 1 week of transfection (supplemental Fig. S2A).
We also investigated the cellular and molecular events that occur in the maturation phase of e-iNSC generation. For this, we performed several experiments including proliferation analysis, gene expression analysis, FACS analysis, immunocytochemistry, and DNA methylation analyses using e-iNSCs from distinct passages (passages 5 and 20). We first checked the proliferation rate of e-iNSCs to evaluate their self-renewal capacity (supplemental Fig. S2B). Despite the NSC-like morphology, e-iNSCs at passage 1 still displayed a relatively slower proliferation than control NSCs (supplemental Fig. S2B). However, it became dramatically enhanced upon further passaging and e-iNSCs at passage 5 already exhibited the proliferation rate comparable with that of control NSCs as well as iNSCs at passage 20, indicating that the acquisition of self-renewal capacity is a gradual process (supplemental Fig. S2B).
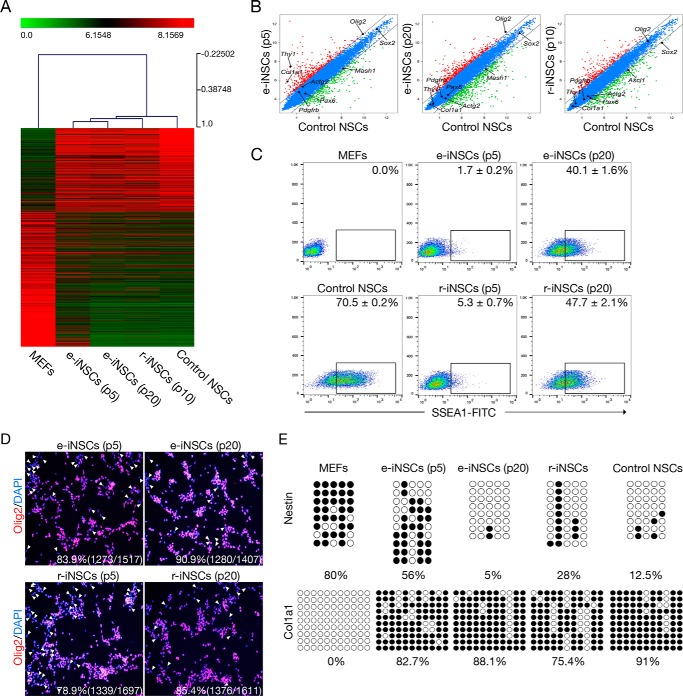
Next, we monitored the transcriptional changes during the maturation phase of e-iNSC generation by comparing the gene expression patterns of NSC and fibroblast markers using qPCR. In contrast to early passage e-iNSCs (passage 5), which showed the poor activation of the endogenous NSC program and incomplete suppression of fibroblast markers, late-passage e-iNSCs (passages 10, 15, and 20) exhibited significantly reprogrammed patterns for both NSC and fibroblast markers in a time-dependent manner (supplemental Fig. S2, C and D). To evaluate the reprogramming status of e-iNSCs in high resolution, we analyzed the global gene expression profile of the e-iNSCs using microarray. Heat map analysis and pairwise scatter plots showed the genome-wide conversion of fibroblasts into an NSC-like state. Specifically, a set of genes not expressed in MEFs became activated in e-iNSCs. Most of the genes in this set were related to the NSC transcriptional program. Conversely, a set of genes highly expressed in MEFs became silenced in e-iNSCs. Those genes were associated with the donor cell-specific program. Clustering analysis revealed that e-iNSCs were clustered together with r-iNSCs and control NSCs, but clearly distinct from the starting cells, MEFs. Interestingly, late passage e-iNSCs were more closely clustered with control NSCs than early passage e-iNSCs (Fig. 2, A and B). This could be due to the poor activation of the endogenous NSC program and incomplete silencing of the somatic program at early passages. These data also support the notions that direct conversion is a gradual process in which the donor cell-specific program becomes gradually repressed, whereas the NSC program becomes gradually activated over subsequent passaging.
FIGURE 2.
A gradual conversion process into iNSCs. A, heat map representing the global gene expression profile of MEFs, e-iNSCs (passages 5 and 20), r-iNSCs (passage 10), and control NSCs. Genes that are more than 4-fold differentially expressed between MEFs and control NSCs are represented. The color bar at the top of the heat map indicates gene expression in log2 scale. Red and green colors represent higher and lower expression levels, respectively. B, pairwise scatter plot analysis of the global gene expression profiles of MEFs, e-iNSCs (passages 5 and 20), r-iNSCs (passage 10), and control NSCs. The black lines delineate the boundaries of 2-fold difference in gene expression levels. C, FACS analysis of early (p5) and late passage (p20) of both e-iNSCs and r-iNSCs using antibody against SSEA1. D, immunofluorescence microscopy images of early (p5) and late (p20) passage of both e-iNSCs and r-iNSCs using antibody against Olig2. The Olig2-negative cells are indicated by arrowheads. E, DNA methylation status of second intron of Nestin and promoter region of Col1a1 in MEFs, e-iNSCs (passage 5 and 20), r-iNSCs (passage 10), and control NSCs was assessed by bisulfite sequencing PCR. Open and filled circles represent unmethylated and methylated CpGs, respectively.
We next performed FACS analysis for checking the number of SSEA1-positive population. As in our previous studies (10, 17), only 2–5% of iNSCs from the early passage was SSEA1-positive (Fig. 2C), although they already showed the homogenous morphology (data not shown). To our surprise, the numbers of SSEA1-positive cells were gradually increased upon serial passaging and more than 40% of both e-iNSCs and r-iNSCs at p20 became SSEA1-positive (Fig. 2C). Because SSEA1 is not a genuine marker for iNSCs due to its overlapping expression in iPSCs (18), we next performed an additional immunocytochemistry using an antibody against Olig2 (Fig. 2D). Similar to SSEA1 expression, the number of Olig2-positive population from both e-iNSCs and r-iNSCs was significantly increased upon serial passaging.
Finally, to understand this gradual process on the epigenetic level, we next checked the DNA methylation status of both Nestin, a NSC marker (19), and Col1a1, a fibroblast marker (13). Notably, the promoter region of Col1a1 was dramatically de novo methylated even in the early passage e-iNSCs to the level similar to control NSCs (Fig. 2E). This high level of DNA methylation on the Col1a1 locus was stably maintained for the long-term culture of e-iNSCs, suggesting that the induced neural stemness by the transient transfection but not by viral system is quite stable.
We also checked the DNA methylational changes on the Nestin regulatory region. To our surprise, the early passage iNSCs still exhibited the relatively high level of DNA methylation on the Nestin enhancer and this residual methylation was eventually erased upon further passaging, resulting the complete DNA demethylation at passage 20 (Fig. 2E). All together, our data indicates that the initial phase of direct conversion toward an iNSC state (supplemental Fig. S2) as well as the maturation process of iNSCs occur gradually (Fig. 2 and supplemental Fig. S2, C and D).
e-iNSCs Are Functionally Comparable with Control NSCs
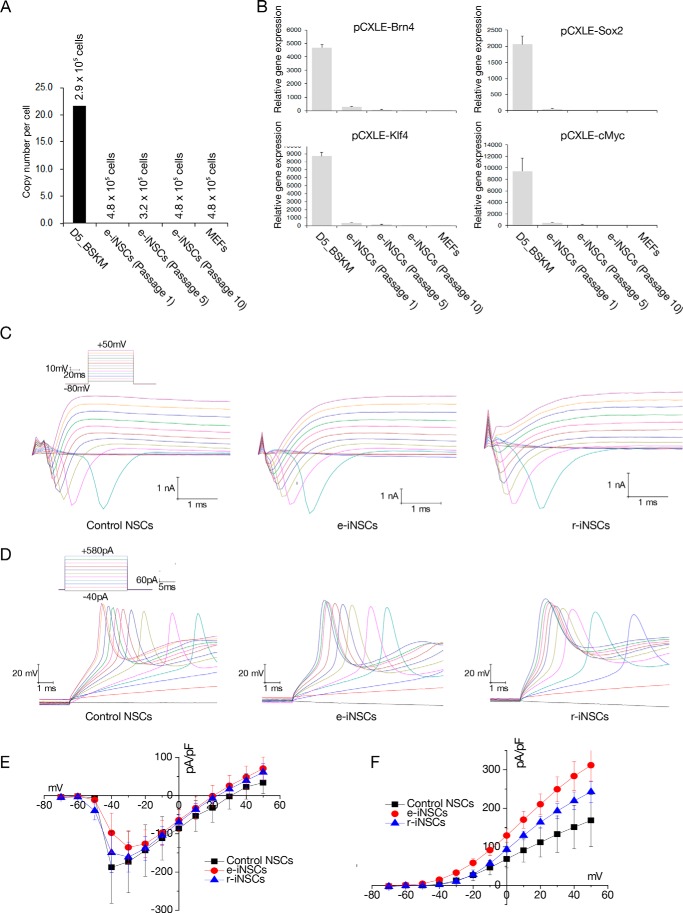
We next investigated the potential integration of episomal vectors in the established e-iNSCs by assessing the copy number of vector sequences in these cells. qPCR analysis revealed about 22 copies of the vector sequences per cell on day 5 post-transfection (Fig. 3A). However, we were unable to detect any copies of vector sequences once the e-iNSCs became the dominant population (at passage 1); we detected fewer than 0.02 copies of the vector sequences at passage 10, which was comparable with the copy number of non-transfected MEFs (Fig. 3A). As expected, we were unable to detect the expression of transgenes in the established e-iNSCs (Fig. 3B). Thus, e-iNSCs are indeed free from vector integration into the host genome.
FIGURE 3.
Electrophysiological properties of e-iNSCs. A, the copy numbers of episomal vectors in e-iNSCs (passages 1, 5, and 10). Numbers indicate the number of estimated cell numbers in each cell. MEFs on day 5 post-transfection were used as a positive control. B, the episomal vector-mediated transgene expression in the established e-iNSCs (passages 1, 5, and 10) was examined by qPCR. The expression levels are normalized to those of MEFs. Error bars indicate standard derivation of duplicate values. C, representative voltage-clamp recordings in response to increasing voltage pulses, of which protocols are shown as the inset, from neurons derived from control NSCs, e-iNSCs, and r-iNSCs, respectively, after 14–16 days of differentiation. D, representative current-clamp recordings showing typical action potentials in response to increasing current pulses, of which protocols are shown as the inset, from neurons derived from control NSCs, e-iNSCs, and r-iNSCs, respectively. E and F, summary of the current-voltage (I-V) relationships for the voltage-gated Na+ (E) and K+ currents (F) in neurons derived from control NSCs (n = 3), e-iNSCs (n = 11), and r-iNSCs (n = 9).
To estimate the functionality of these integration-free e-iNSCs, we first differentiated e-iNSCs into neurons for 2 weeks and then performed electrophysiology recordings. Similar to neurons from control NSCs and r-iNSCs, neurons from e-iNSCs generated typical voltage-gated Na+ and K+ currents in response to step depolarizing voltage pulses (Fig. 3C). Accordingly, in response to the depolarizing current injections, neurons derived from e-iNSCs (n = 11) generated action potentials similar to neurons from control NSCs and r-iNSCs (Fig. 3D). Moreover, the amplitudes and current-voltage (I-V) relationships of the Na+ and K+ currents from all the cell types were comparable (Fig. 3, E and F). Therefore, our data indicate that the electrophysiological properties of the neurons derived from e-iNSCs are comparable with those of r-iNSCs and control NSCs.
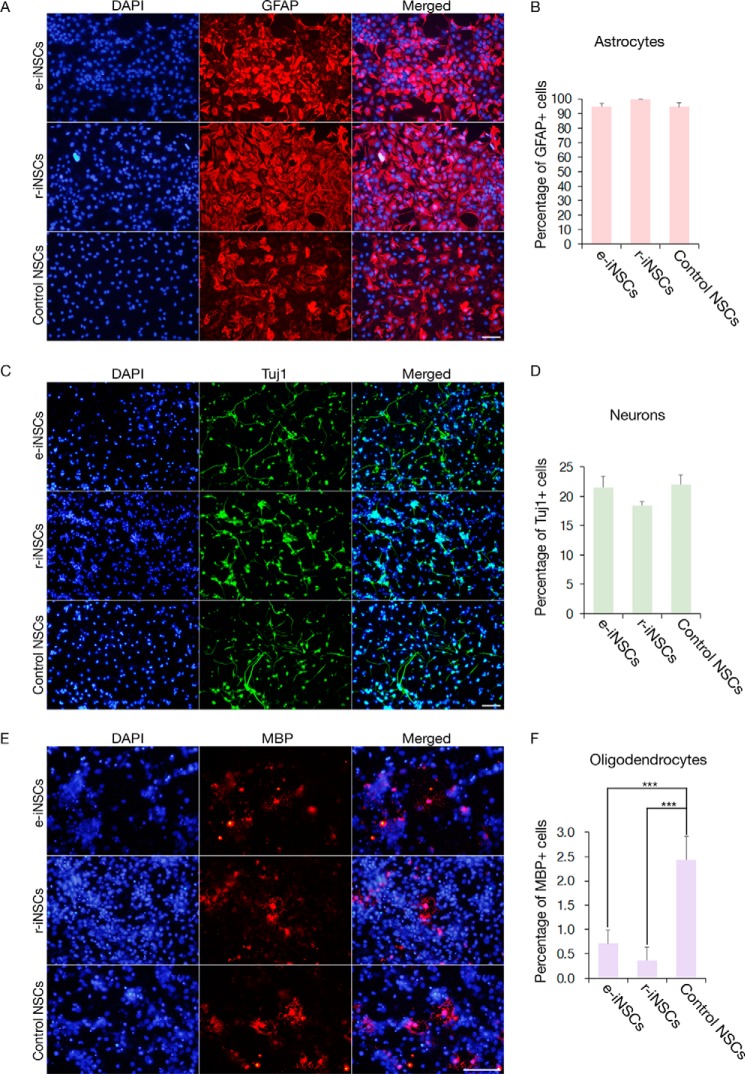
We next assessed the in vitro functionality of e-iNSCs by inducing their differentiation into astrocytes, neurons, and oligodendrocytes. The e-iNSCs could differentiate into astrocytes and neurons as determined by GFAP and Tuj1 staining, respectively, with similar efficiency as r-iNSCs and control NSCs (Fig. 4, A–D). Furthermore, the prolonged in vitro culture of e-iNSCs up to passage 40 did not affect the in vitro differentiation potential of e-iNSCs into neuronal and glial cell types (supplemental Fig. S3). However, consistent with a previous study (9), only a small number of e-iNSCs differentiated into MBP-positive oligodendrocytes (Fig. 4, E and F). The efficiency of this conversion was similar to that of r-iNSCs but significantly lower than that of control NSCs. Nevertheless, these results indicate that e-iNSCs could maintain in vitro functionality similar to control NSCs even in the absence of exogenous reprogramming factors.
FIGURE 4.
In vitro differentiation potential of iNSCs. A, C, and E, differentiation potential of e-iNSCs and r-iNSCs into astrocytes (A), neurons (C), and oligodendrocytes (E), as shown by immunostaining with antibodies against GFAP, Tuj1, and MBP, respectively. B, D, and F, the efficiency of differentiation into astrocytes (B), neurons (D), and oligodendrocytes (E) was compared by quantifying the numbers of GFAP, Tuj1, and MBP positive cells, respectively. Control NSCs were used as a positive control for determining the differentiation efficiency. Scale bars, 100 μm. ***, p < 0.001.
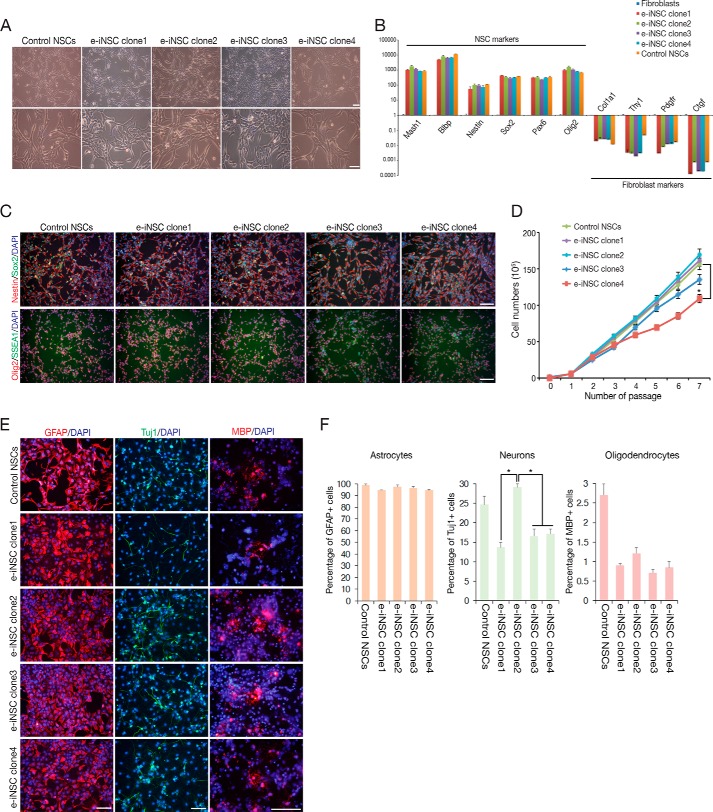
As we showed in our previous studies (9, 10) as well as in the current study (Fig. 2, B–D), the conversion process into iNSCs using either episomal vectors or retroviruses is a gradual process and thus, the initial iNSCs might be comprised of distinct populations of cells with various degrees of reprogramming. Indeed, we previously showed that the SSEA1-positive early iNSC population exhibits the mosaic expression of Olig2, indicating that iNSCs from early passage still represent a heterogeneous population (10). Thus, to better understand the heterogeneity of e-iNSCs, we established 4 clonal lines of e-iNSCs by single cell sorting and determined their reprogramming status (Fig. 5). All clonal lines exhibited the morphology, proliferation rate, and gene expression pattern similar to control NSCs (Fig. 5, A–D). However, their in vitro differentiation potentials were somewhat different among the clonal lines (Fig. 5, E and F). Although all clonal lines showed similar differentiation potential into glial cell types such as astrocytes and oligodendrocytes, they showed distinct differentiation patterns into neurons, indicating that the initial e-iNSCs are indeed a heterogeneous population comprised of functionally distinct subpopulations.
FIGURE 5.
Characterization of clonal e-iNSC lines. A, the morphology of established clonal e-iNSC lines after single cell sorting. B, expression of NSC and fibroblast markers in clonal e-iNSC lines analyzed by qPCR. Expression levels are normalized to those in MEFs. Error bars indicate the standard deviation of triplicate values. C, immunofluorescence microscopy images of clonal e-iNSC lines using antibodies against Sox2/Nestin and Olig2/SSEA1. D, proliferation of clonal e-iNSC lines. 1 × 105 cells were passaged every 2 days on the wells of 12-well plates. *, p < 0.05. E, in vitro differentiation potential of clonal e-iNSC lines into astrocytes, neurons, and oligodendrocytes, as shown by immunostaining with antibodies against GFAP, Tuj1, and MBP, respectively. F, the efficiency of differentiation into astrocytes, neurons, and oligodendrocytes was examined by quantifying the numbers of GFAP, Tuj1, and MBP positive cells against nuclear staining. Control NSCs were used as a positive control for determining the differentiation efficiency. Scale bars, 100 μm. *, p < 0.05.
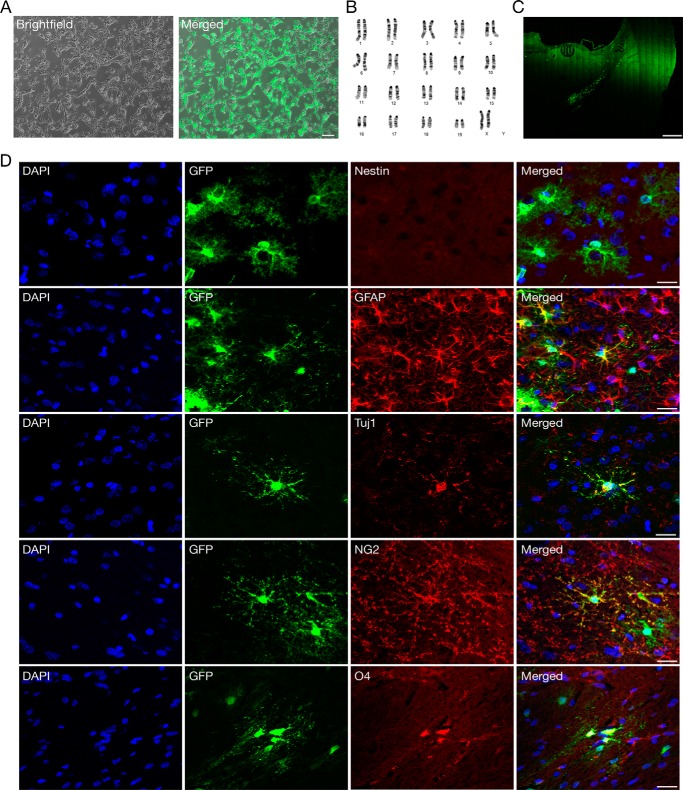
Finally, to examine the in vivo multipotency of iNSCs, we labeled karyotypically normal e-iNSCs with green fluorescent protein (GFP) (Fig. 6, A and B). After 4 weeks of transplantation of e-iNSCs into the cortical region of the brain, we were able to observe the survival and migrating engrafted e-iNSCs (Fig. 6C). As in our previous transplantation studies (13, 20), here we found no obvious evidence for tumor formation in any of the transplanted rats (data not shown). To further investigate the fate of the engrafted e-iNSCs, we checked for the presence of Nestin, a progenitor marker, but found no GFP-positive cells expressing Nestin, indicating that the engrafted e-iNSCs had lost their stem cell identity (Fig. 6D). Instead, the engrafted e-iNSCs had differentiated into all the neuronal cell types such as neurons (GFP+/Tuj1+), astrocytes (GFP+/GFAP+), and oligodendrocytes (GFP+/NG2+ or GFP+/O4+) (Fig. 6D). Finally, e-iNSCs are as functionally mature as control NSCs, as evidenced by their in vitro and in vivo differentiation capacity.
FIGURE 6.
In vivo differentiation of engrafted e-iNSCs. A, e-iNSCs were labeled with GFP before transplantation. Scale bars, 100 μm. B, karyotypic analysis of e-iNSCs. C, integration and migration of engrafted iNSCs. GFP-labeled iNSCs were injected into the cortical region of the brain and analyzed at 4 weeks after transplantation. Scale bars, 500 μm. D, the transplanted e-iNSCs showed differentiation into neurons (GFP+/Tuj1+), astrocytes (GFP+/GFAP+), and oligodendrocytes (GFP+/NG2+ or GFP+/O4+), as determined by immunohistochemistry. The transplanted iNSCs lost their stem cell identity, as shown by the absence of Nestin+ cells at 4 weeks after transplantation. Scale bars, 20 μm.
Discussion
Recent studies have described the generation of integration-free iNPCs using different sets of reprogramming factors with small molecules (21, 22). Wang et al. (21) used six factors (OCT4, SOX2, SV40LT, KLF4, and MIR302-367) in the presence of five small molecules (CHIR99021, PD0325901, A83-01, thiazovivin, and DMH1) to generate iNPCs from human urine cells. More recently, Lu et al. (22) reported that the introduction of four iPSC factors by Sendai virus, with the combined treatment of leukemia inhibitory factor, SB431542, and CHIR99021, could lead to the conversion of both human and monkey fibroblasts into iNPCs. Although both iNPCs are integration free, they were generated by factor combinations that could readily facilitate the generation of iPSCs (23). Furthermore, we previously showed that short-term exposure of fibroblasts in NSC medium containing both bFGF and EGF could dramatically boost iPSC generation due to the enhanced proliferation of the starting cells (24). Therefore, the possibility that iNPCs may have differentiated from an intermediate pluripotent cell state could not be excluded. Moreover, both iNPCs could not be maintained without the assistance of different combinations of small molecules, indicating that the cellular identity of the iNPCs may not be stable.
In the current study, we were able to generate iNSCs by the single transfection of episomal vectors encoding four NSC factors (Brn4/Pou3f4, Sox2, Klf4, and c-Myc), which could not generate iPSCs (9). The generated e-iNSCs were very similar to control NSCs and r-iNSCs in terms of morphology, global gene expression pattern, epigenetic status, and functionality as determined by patch clamp recordings and both in vitro and in vivo differentiation. Furthermore, e-iNSCs do not rely on either continuous expression of exogenous factors or treatment with multiple small molecules for maintaining their stemness, indicating that our conversion method induces the stable and solid NSC state on the somatic cells, unlike other reported methods. The conversion efficiency of fibroblasts into an NSC state using episomal vectors is, however, relatively lower than that of retroviral vector-mediated conversion potentially due to the relatively low transfection efficiency of episomal vectors (15, 25). Thus, further optimization is necessary for enhancing the reprogramming efficiency, for instance, by using small molecules as in previous iPSC studies.
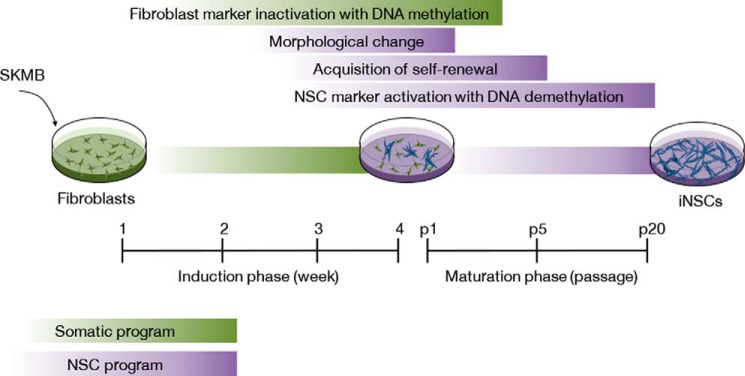
To understand the underlying mechanism of iNSC generation, in the current study we performed several experiments for exploring the conversion kinetics and found that the direct conversion process into an iNSC state is gradual. Furthermore, we also noticed that the following cellular and molecular events occur in a sequential manner during the induction and maturation phases of neural stemness, 1) inactivation of fibroblast marker (within 1 week of transfection) with de novo methylation, 2) acquisition of NSC-like morphology (about 4 weeks of transfection), 3) initiation of endogenous NSC marker activation (after 4 weeks of transfection), 4) acquisition of self-renewal capacity (about passage 5), and finally, 5) further fine tuning of NSC transcriptional program (passage 15–20) with DNA demethylation on NSC marker (Fig. 7).
FIGURE 7.
Direct conversion kinetics into an iNSC state. Schematic diagram depicting the procedure for the direct conversion of fibroblasts into iNSCs.
In line with our previous study (10), we also found that the initial e-iNSCs are a heterogeneous population comprised of functionally distinct subpopulations (Fig. 5). There are a few possible reasons explaining the heterogeneity of e-iNSCs. First, the transfection procedure could cause the heterogeneity. Because the transfection efficiency into each single fibroblast might be different, each single cell therefore might contain the different copy numbers of episomal vector. The secondary reprogramming system (26) using the transgenic fibroblast line where all reprogramming factors are integrated into the identical integration sites with same copy numbers would be very helpful to understand this heterogeneous nature of iNSCs. Second, the purity of the starting population might also cause the heterogeneity. In our previous studies as well as the current study, we mainly used fibroblasts that are actually heterogeneous. Thus, using more specified somatic cell types such as blood cells and hepatocytes would be helpful to explain this heterogeneity in e-iNSCs. Third, the gradual reprogramming process could also cause the heterogeneity of e-iNSCs. An depth study on the heterogeneity of iNSCs will be required for unveiling the underlying mechanism of direct conversion process.
Direct cellular conversion is a relatively simple and efficient procedure that is associated with a low risk for tumor formation. The technology may be an alternative to iPSCs, but some potential outcomes must be addressed before translating it to the clinic. These may stem from the use of both oncogenes for producing certain cell types (9, 12, 27, 28) and a viral system for integrating exogenous reprogramming factors into the somatic cell genome (9–11, 28–40). For example, insertional mutagenesis may occur and lead to the continuous expression of exogenous reprogramming factors in directly converted cells, potentially affecting cellular functionality (41). Furthermore, it might also lead to tumor formation, as evidenced by the development of T-cell acute leukemia-like syndrome in the two patients who received integrating retroviruses in a gene therapy trial for the treatment of X-linked severe combined immunodeficiency disease (X-SCID) (42, 43). Taken together, these findings clearly suggest that e-iNSCs might be a safer cell source than r-iNSCs as well as NSCs derived from pluripotent stem cells. A recent study demonstrated the derivation of iNSCs from blood cells (44). As blood cells are one of the easily accessible autologous cell sources, further studies will aim to assess whether our episomal vector system could affect the direct conversion of human blood cells, and other human somatic cells, into iNSCs.
Author Contributions
S. M. K. performed most of the experiments. J. K. and J. K. H. analyzed the in vivo differentiation potential of the iNSCs. S. W. P., H. P., and Y. M. B. performed electrophysiology recordings. J. K., K. T. L., H. H., and I. L. performed the microarray analysis. K. K. performed the DNA methylation analysis. T. H. K., J. H. Y., and K. P. K. constructed the episomal vectors. K. P. K., H. T. L., and H. R. S. edited the manuscript. D. W. H. conceived the project and wrote the manuscript.
Supplementary Material
Acknowledgments
We appreciate all members of the Han laboratory for their helpful discussions.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), supported by Ministry of Education, Science and Technology Grant 2011-0013885, and Korea Health Technology R&D Project, Ministry of Health & Welfare Grant A120392. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S3.
- NSC
- neural stem cell
- iNSC
- induced neural stem cell
- iNPC
- induced neural progenitor cell
- iPSC
- induced pluripotent stem cell
- e-iNSC
- episomal vector-derived iNSC
- MEF
- mouse embryonic fibroblast
- qPCR
- quantitative PCR
- r-iNSC
- retrovirus-mediated iNSC.
References
- 1. Gage F. H. (2000) Mammalian neural stem cells. Science 287, 1433–1438 [DOI] [PubMed] [Google Scholar]
- 2. Blurton-Jones M., Kitazawa M., Martinez-Coria H., Castello N. A., Müller F. J., Loring J. F., Yamasaki T. R., Poon W. W., Green K. N., and LaFerla F. M. (2009) Neural stem cells improve cognition via BDNF in a transgenic model of Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 106, 13594–13599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xuan A. G., Luo M., Ji W. D., and Long D. H. (2009) Effects of engrafted neural stem cells in Alzheimer's disease rats. Neurosci. Lett. 450, 167–171 [DOI] [PubMed] [Google Scholar]
- 4. Martino G., and Pluchino S. (2006) The therapeutic potential of neural stem cells. Nat. Rev. Neurosci. 7, 395–406 [DOI] [PubMed] [Google Scholar]
- 5. Ourednik J., Ourednik V., Lynch W. P., Schachner M., and Snyder E. Y. (2002) Neural stem cells display an inherent mechanism for rescuing dysfunctional neurons. Nat. Biotechnol. 20, 1103–1110 [DOI] [PubMed] [Google Scholar]
- 6. Jakel R. J., Schneider B. L., and Svendsen C. N. (2004) Using human neural stem cells to model neurological disease. Nat. Rev. Genetics 5, 136–144 [DOI] [PubMed] [Google Scholar]
- 7. Xu X., Warrington A. E., Bieber A. J., and Rodriguez M. (2011) Enhancing CNS repair in neurological disease: challenges arising from neurodegeneration and rewiring of the network. CNS Drugs 25, 555–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cassady J. P., D'Alessio A. C., Sarkar S., Dani V. S., Fan Z. P., Ganz K., Roessler R., Sur M., Young R. A., and Jaenisch R. (2014) Direct lineage conversion of adult mouse liver cells and B lymphocytes to neural stem cells. Stem Cell Reports 3, 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ring K. L., Tong L. M., Balestra M. E., Javier R., Andrews-Zwilling Y., Li G., Walker D., Zhang W. R., Kreitzer A. C., and Huang Y. (2012) Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11, 100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim S. M., Flaßkamp H., Hermann A., Araúzo-Bravo M. J., Lee S. C., Lee S. H., Seo E. H., Lee S. H., Storch A., Lee H. T., Schöler H. R., Tapia N., and Han D. W. (2014) Direct conversion of mouse fibroblasts into induced neural stem cells. Nat. Protoc. 9, 871–881 [DOI] [PubMed] [Google Scholar]
- 11. Lujan E., Chanda S., Ahlenius H., Südhof T. C., and Wernig M. (2012) Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. U.S.A. 109, 2527–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thier M., Wörsdörfer P., Lakes Y. B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nöthen M. M., Brüstle O., and Edenhofer F. (2012) Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10, 473–479 [DOI] [PubMed] [Google Scholar]
- 13. Hong J. Y., Lee S. H., Lee S. C., Kim J. W., Kim K. P., Kim S. M., Tapia N., Lim K. T., Kim J., Ahn H. S., Ko K., Shin C. Y., Lee H. T., Schöler H. R., Hyun J. K., and Han D. W. (2014) Therapeutic potential of induced neural stem cells for spinal cord injury. J. Biol. Chem. 289, 32512–32525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Craenenbroeck K., Vanhoenacker P., and Haegeman G. (2000) Episomal vectors for gene expression in mammalian cells. Eur. J. Biochem. 267, 5665–5678 [DOI] [PubMed] [Google Scholar]
- 15. Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I. I., and Thomson J. A. (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K., Shibata T., Kunisada T., Takahashi M., Takahashi J., Saji H., and Yamanaka S. (2011) A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 [DOI] [PubMed] [Google Scholar]
- 17. Kim S. M., Lim K. T., Kwak T. H., Lee S. C., Im J. H., Hali S., In Hwang S., Kim D., Hwang J., Kim K. P., Chung H. J., Kim J. B., Ko K., Chung H. M., Lee H. T., Schöler H. R., and Han D. W. (2016) Induced neural stem cells from distinct genetic backgrounds exhibit different reprogramming status. Stem Cell Res. 16, 460–468 [DOI] [PubMed] [Google Scholar]
- 18. Kim J. B., Sebastiano V., Wu G., Araúzo-Bravo M. J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., Meyer J., Hübner K., Bernemann C., Ortmeier C., Zenke M., Fleischmann B. K., et al. (2009) Oct4-induced pluripotency in adult neural stem cells. Cell 136, 411–419 [DOI] [PubMed] [Google Scholar]
- 19. Han D. W., Do J. T., Araúzo-Bravo M. J., Lee S. H., Meissner A., Lee H. T., Jaenisch R., and Schöler H. R. (2009) Epigenetic hierarchy governing Nestin expression. Stem Cells 27, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 20. Hemmer K., Zhang M., van Wüllen T., Sakalem M., Tapia N., Baumuratov A., Kaltschmidt C., Kaltschmidt B., Schöler H. R., Zhang W., and Schwamborn J. C. (2014) Induced neural stem cells achieve long-term survival and functional integration in the adult mouse brain. Stem Cell Reports 3, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang L., Wang L., Huang W., Su H., Xue Y., Su Z., Liao B., Wang H., Bao X., Qin D., He J., Wu W., So K. F., Pan G., and Pei D. (2013) Generation of integration-free neural progenitor cells from cells in human urine. Nat. Methods 10, 84–89 [DOI] [PubMed] [Google Scholar]
- 22. Lu J., Liu H., Huang C. T., Chen H., Du Z., Liu Y., Sherafat M. A., and Zhang S. C. (2013) Generation of integration-free and region-specific neural progenitors from primate fibroblasts. Cell Reports 3, 1580–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H. S., Hao E., Hayek A., and Ding S. (2009) A chemical platform for improved induction of human iPSCs. Nat. Methods 6, 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han D. W., Greber B., Wu G., Tapia N., Araúzo-Bravo M. J., Ko K., Bernemann C., Stehling M., and Schöler H. R. (2011) Direct reprogramming of fibroblasts into epiblast stem cells. Nat. Cell Biol. 13, 66–71 [DOI] [PubMed] [Google Scholar]
- 25. Schlaeger T. M., Daheron L., Brickler T. R., Entwisle S., Chan K., Cianci A., DeVine A., Ettenger A., Fitzgerald K., Godfrey M., Gupta D., McPherson J., Malwadkar P., Gupta M., Bell B., et al. (2015) A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 33, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wernig M., Lengner C. J., Hanna J., Lodato M. A., Steine E., Foreman R., Staerk J., Markoulaki S., and Jaenisch R. (2008) A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 26, 916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Du Y., Wang J., Jia J., Song N., Xiang C., Xu J., Hou Z., Su X., Liu B., Jiang T., Zhao D., Sun Y., Shu J., Guo Q., Yin M., Sun D., et al. (2014) Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 14, 394–403 [DOI] [PubMed] [Google Scholar]
- 28. Huang P., Zhang L., Gao Y., He Z., Yao D., Wu Z., Cen J., Chen X., Liu C., Hu Y., Lai D., Hu Z., Chen L., Zhang Y., Cheng X., et al. (2014) Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14, 370–384 [DOI] [PubMed] [Google Scholar]
- 29. Buganim Y., Itskovich E., Hu Y. C., Cheng A. W., Ganz K., Sarkar S., Fu D., Welstead G. G., Page D. C., and Jaenisch R. (2012) Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell 11, 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caiazzo M., Dell'Anno M. T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T. D., Menegon A., Roncaglia P., Colciago G., Russo G., Carninci P., Pezzoli G., Gainetdinov R. R., Gustincich S., et al. (2011) Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227 [DOI] [PubMed] [Google Scholar]
- 31. Efe J. A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G., Chen J., and Ding S. (2011) Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13, 215–222 [DOI] [PubMed] [Google Scholar]
- 32. Ginsberg M., James D., Ding B. S., Nolan D., Geng F., Butler J. M., Schachterle W., Pulijaal V. R., Mathew S., Chasen S. T., Xiang J., Rosenwaks Z., Shido K., Elemento O., Rabbany S. Y., and Rafii S. (2012) Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFβ suppression. Cell 151, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., and Hui L. (2011) Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 [DOI] [PubMed] [Google Scholar]
- 34. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., and Srivastava D. (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Najm F. J., Lager A. M., Zaremba A., Wyatt K., Caprariello A. V., Factor D. C., Karl R. T., Maeda T., Miller R. H., and Tesar P. J. (2013) Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat. Biotechnol. 31, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., Citri A., Sebastiano V., Marro S., Südhof T. C., and Wernig M. (2011) Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sekiya S., and Suzuki A. (2011) Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 [DOI] [PubMed] [Google Scholar]
- 38. Son E. Y., Ichida J. K., Wainger B. J., Toma J. S., Rafuse V. F., Woolf C. J., and Eggan K. (2011) Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., and Wernig M. (2010) Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang N., Zuchero J. B., Ahlenius H., Marro S., Ng Y. H., Vierbuchen T., Hawkins J. S., Geissler R., Barres B. A., and Wernig M. (2013) Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 31, 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., and Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 42. Hacein-Bey-Abina S., von Kalle C., Schmidt M., Le Deist F., Wulffraat N., McIntyre E., Radford I., Villeval J. L., Fraser C. C., Cavazzana-Calvo M., and Fischer A. (2003) A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348, 255–256 [DOI] [PubMed] [Google Scholar]
- 43. Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M. P., Wulffraat N., Leboulch P., Lim A., Osborne C. S., Pawliuk R., Morillon E., Sorensen R., Forster A., Fraser P., Cohen J. I., de Saint Basile G., et al. (2003) LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419 [DOI] [PubMed] [Google Scholar]
- 44. Lee J. H., Mitchell R. R., McNicol J. D., Shapovalova Z., Laronde S., Tanasijevic B., Milsom C., Casado F., Fiebig-Comyn A., Collins T. J., Singh K. K., and Bhatia M. (2015) Single transcription factor conversion of human blood fate to NPCs with CNS and PNS developmental capacity. Cell Rep. 11, 1367–1376 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.