Abstract
Diversity at pathogen genetic loci can be driven by host adaptive immune selection pressure and may reveal proteins important for parasite biology. Population-based genome sequencing of Plasmodium falciparum, the parasite responsible for the most severe form of malaria, has highlighted two related polymorphic genes called dblmsp and dblmsp2, which encode Duffy binding-like (DBL) domain-containing proteins located on the merozoite surface but whose function remains unknown. Using recombinant proteins and transgenic parasites, we show that DBLMSP and DBLMSP2 directly and avidly bind human IgM via their DBL domains. We used whole genome sequence data from over 400 African and Asian P. falciparum isolates to show that dblmsp and dblmsp2 exhibit extreme protein polymorphism in their DBL domain, with multiple variants of two major allelic classes present in every population tested. Despite this variability, the IgM binding function was retained across diverse sequence representatives. Although this interaction did not seem to have an effect on the ability of the parasite to invade red blood cells, binding of DBLMSP and DBLMSP2 to IgM inhibited the overall immunoreactivity of these proteins to IgG from patients who had been exposed to the parasite. This suggests that IgM binding might mask these proteins from the host humoral immune system.
Keywords: genetic polymorphism, host-pathogen interaction, parasite, plasmodium, protein-protein interaction, immunoglobulin M (IgM), merozoite
Introduction
Malaria is responsible for up to one million deaths annually and therefore remains one of the world's major health problems (1, 2). Among the different species of Plasmodium causing malaria in humans, Plasmodium falciparum is responsible for the highest burden of disease. The clinical symptoms of malaria are associated with the blood stage of the infection, when the merozoite stage of the parasite recognizes, invades, and develops within human erythrocytes (3). Because merozoites are directly exposed to circulating antibodies and passive immunization of infected children with purified immunoglobulins from clinically immune individuals reduces parasitemia (4), merozoite surface proteins are considered likely targets of host immunity and therefore potential vaccine candidates (5). Consistent with this, population-based P. falciparum genome sequence analysis (6) has revealed that genes encoding merozoite surface proteins are among the most polymorphic in the genome, with several exhibiting signatures of balancing selection, suggesting that host immune pressure maintains the presence of multiple distinct antigenic variants (7).
dblmsp and dblmsp2 (also known as msp3.4 and msp3.8, respectively) are two members of the P. falciparum msp3 family that comprises a cluster of eight paralogous genes on chromosome 10. The msp3 family encodes proteins, which are characterized by the presence of an NLR(K/N)(A/G/N) motif at their N terminus (8, 9), are secreted by the blood stage of the parasite, and located in both the parasitophorous vacuole and on the merozoite surface. Most proteins within this family also contain a C-terminal acidic and a coiled-coil region believed to be involved in oligomerization of the proteins (10), which form the SPAM (secreted polymorphic antigen associated with merozoites) domain (11). DBLMSP and DBLMSP2 are distinguished from the other MSP3 family members because they contain a Duffy binding-like (DBL)5 domain (8, 12). DBL domains are known to bind directly to host receptors and are present in other P. falciparum surface proteins including ligands involved in erythrocyte invasion such as EBA175 and EBA140 (13–15) and members of the PfEMP1 family, which are displayed on the surface of the infected erythrocyte (16). Sequencing of some African P. falciparum strains has shown that the genetic diversity in dblmsp and dblmsp2 is particularly high and concentrated in the region encoding the DBL domain (17–19). Clearly identifiable orthologs of dblmsp and dblmsp2 are not present in other Plasmodium species that infect humans, but the genome of the chimpanzee parasite Plasmodium reichenowi encodes a functional dblmsp gene, although dblmsp2 is a pseudogene (19). Despite these interesting features, the function of DBLMSP and DBLMSP2 and their role in the pathology of malaria are unknown.
Here, we used recombinant proteins produced in mammalian cells and a knock-out parasite line to show that the DBL domains of DBLMSP and DBLMSP2 from the 3D7 strain of P. falciparum bind avidly and directly to human IgM. Using population-based genome sequencing and bespoke assembly tools, we revealed widespread genetic diversity focused on the DBL domain, with two major allelic forms of both dblmsp and dblmsp2 observed in populations from Africa and Southeast Asia. Despite their diversity, binding of the DBL domains from different DBLMSP and DBLMSP2 sequence variants to human IgM was conserved, suggesting an important role in the parasite biology.
Results
DBLMSP and DBLMSP2 Bind Human IgM
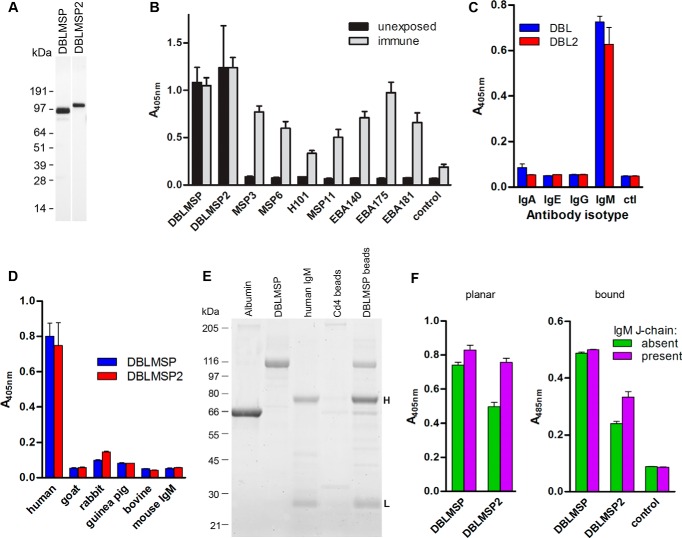
To gain insight into the functional role of DBLMSP and DBLMSP2 during malaria pathogenesis, we first expressed the entire coding region of both proteins from the P. falciparum 3D7 strain using a recently developed expression system based on mammalian cells, which has been shown to produce natively folded P. falciparum proteins (20). Both proteins were expressed (Fig. 1A), and their immunoreactivity to sera from malaria-exposed and unexposed control individuals was quantified. To our surprise, whereas other MSP3 family members (MSP3, MSP6, H101, and MSP11) and DBL-containing merozoite surface proteins (EBA140, EBA175, and EBA181) reacted, as expected, with only the exposed sera, DBLMSP and DBLMSP2 showed equally strong immunoreactivity to both the unexposed control and exposed sera (Fig. 1B). This suggested that recombinant DBLMSP and DBLMSP2 bound immunoglobulins present in normal serum from individuals without prior exposure to the malaria parasite. Indeed, when each protein was exposed to purified human immunoglobulin isotypes, strong binding was observed to IgM, but not to IgA, IgE, or IgG (Fig. 1C). This binding was specific to human IgM because they did not bind immunoglobulins from other mammalian species including goat, rabbit, guinea pig, and cow, or purified mouse IgM (Fig. 1D). To confirm these observations, we showed that DBLMSP-coated beads but not control beads incubated in normal human serum purified bands with masses there were consistent with the heavy and light chain of IgM (Fig. 1E), and their identities were subsequently confirmed by mass spectrometry.
FIGURE 1.
Recombinant P. falciparum DBLMSP and DBLMSP2 bind human IgM. A, anti-biotin Western blot showing expression of recombinant monobiotinylated DBLMSP and DBLMSP2 proteins from mammalian cells. B, DBLMSP and DBLMSP2 are immunoreactive with control, naïve human sera. Enzymatically monobiotinylated recombinant parasite proteins were immobilized on streptavidin-coated plates, and their immunoreactivity to pooled sera from either unexposed individuals (black bars) or immune Kenyan adults (gray bars) was tested. C, DBLMSP (blue) and DBLMSP2 (red) bind purified human IgM and no other immunoglobulin isotype, relative to no immunoglobulin control (ctl). D, DBLMSP and DBLMSP2 binding was restricted to human IgM, and did not bind sera from other species, including purified murine IgM. E, streptavidin-coated paramagnetic beads were coated in monobiotinylated His6-tagged recombinant DBLMSP or a control protein (Cd4-d3 + 4) and incubated in the presence of normal human serum. Following elution and resolution by SDS-PAGE, 80- and 25-kDa bands corresponding to the heavy (H) and light (L) chains of human IgM, respectively, were observed in the DBLMSP but not the negative control pulldown. Purified albumin, DBLMSP, and IgM protein are shown for comparison on the left. F, both DBLMSP and DBLMSP2 bound chimeric IgM containing human heavy chains regardless of whether it was antigen-bound (right panel) or not (left panel). Both DBLMSP and DBLMSP2 could bind IgM that contained or lacked a J-chain, although DBLMSP2 binding to IgM lacking a J-chain was reproducibly weaker. Negative control was the Cd4 tag alone. B–D and F represent one representative from two or more ELISA experiments; bars represent means ± S.D.; n = 3 replicate wells.
IgM adopts a flat, planar structure in solution but can change to a “staple” conformation upon antigen binding, thereby permitting interaction with other proteins such as C1q (21). To determine whether antigen bound to IgM influenced its interaction with DBLMSP and DBLMSP2, we made use of two transfectomas producing chimeric IgMs that recognize dinitrophenyl but differ by either the presence or absence of a murine J-chain. In these chimeric IgMs, the whole κ light chain and the variable region of the heavy chain are of murine origin, whereas the heavy chain Fc constant region is human (22). DBLMSP and DBLMSP2 bound IgM from both transfectomas, demonstrating that they interact with the human IgM heavy chain constant region, and this binding was independent of whether they were engaged with antigen or not. Binding of DBLMSP2 to IgMs lacking a J-chain, however, was consistently weaker (Fig. 1F).
The DBL Domain of DBLMSP and DBLMSP2 Binds Directly and Avidly to the IgM Cμ4 Domain
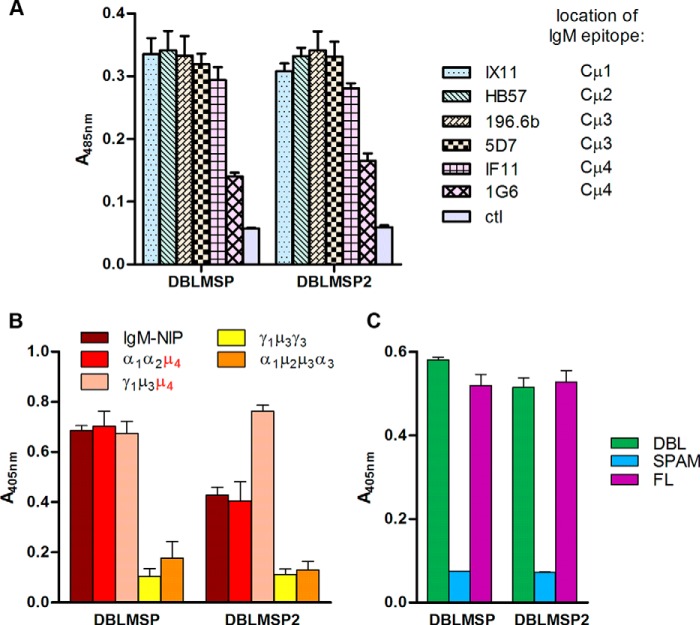
To identify the region on IgM that bound DBLMSP and DBLMSP2, we used mouse monoclonal antibodies that bind to specific domains of the human μ chain constant region (23, 24). Antibody 1G6, whose epitope is located within the Cμ4 domain, inhibited the interaction of DBLMSP and DBLMSP2 with human IgM, whereas antibodies IX11 and HB57, with epitopes located in the Cμ1 and Cμ2 domains, respectively, or the Cμ3-specific antibodies 196.6b and 5D7 did not (Fig. 2A). To confirm these observations, we also made use of chimeric recombinant antibodies in which domains of the human μ chain constant region replace domains from the human α or γ chain (25, 26). In all cases, binding of DBLMSP and DBLMSP2 proteins was observed only when the Cμ4 domain was present in the chimeric antibodies, and no binding was detected with domain-swapped antibodies containing only the Cμ2 or Cμ3 domains (Fig. 2B).
FIGURE 2.
The DBL domains directly interact with the Cμ4 domain of human IgM. A, monoclonal antibodies to human IgM with known domain-specific epitopes were used to map the DBLMSP and DBLMSP2 binding domain on human IgM heavy chain. Binding of both parasite proteins was inhibited by the Cμ4-specific 1G6 antibody but not by antibodies recognizing other domains or by the IF11 antibody, which is known to bind a different epitope on Cμ4. Negative control was immobilized biotinylated IgG. B, DBLMSP and DBLMSP2 bound the Cμ4 domain of human IgM. Systematic testing of a small panel of chimeric human immunoglobulins containing different combinations of heavy chain domains from either IgA, IgG, and IgM (numbered and labeled α, γ, and μ, respectively) showed DBLMSP and DBLMSP2 bound the Cμ4 domain only. C, the DBL domain of DBLMSP and DBLMSP2 bound human IgM. Fragments encompassing the DBL, but not the SPAM domain, of both DBLMSP and DBLMSP2 bound human IgM indistinguishably from the full-length (FL) protein. Bars represent means ± S.D. in all panels; n = 3 (A and C) or 4 (B) replicate wells.
To determine which regions of DBLMSP and DBLMSP2 were involved in IgM binding, we individually expressed either the DBL domain or the SPAM fragment. The SPAM fragments did not bind IgM, whereas the isolated DBL domains bound IgM as efficiently as the full-length proteins in this assay, indicating that the DBL domain encompassed all IgM binding activity (Fig. 2C).
DBLMSP and DBLMSP2 Form Multimers That Interact Avidly with Human IgM
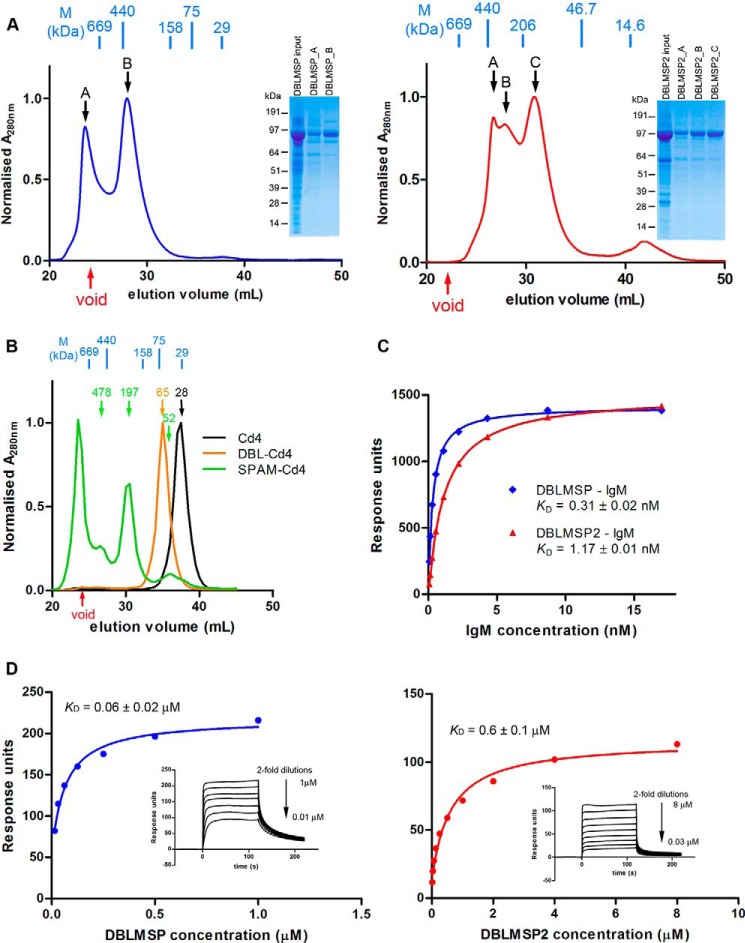
To determine whether DBLMSP and DBLMSP2 could, like MSP3, form oligomers, we first used size exclusion chromatography on each full-length protein. In both cases, polydisperse peaks were observed, suggesting the formation of higher order oligomers composed exclusively of the proteins of interest (Fig. 3A). We then resolved purified proteins encompassing the DBL domain and SPAM fragment by size exclusion chromatography. Although the DBL domain of DBLMSP2 eluted as a single monodisperse peak consistent with it being a monomer, the SPAM fragment elution profile was polydisperse, with peaks consistent with tetrameric and other higher order complexes and only a minor fraction consistent with a monomeric form (Fig. 3B). A similar analysis of DBLMSP gave comparable results, suggesting that both DBLMSP and DBLMSP2 are able to form higher order oligomers by interactions through their C terminus.
FIGURE 3.
DBLMSP and DBLMSP2 form oligomers and their DBL domain bind to human IgM with high affinity. A, size exclusion chromatography showing polydisperse profile of full-length DBLMSP (left panel, blue) and DBLMSP2 (right panel, red). The indicated fractions were resolved by denaturing SDS-PAGE gel, and the insets show that each peak is largely composed of the full-length protein. B, the SPAM fragment of DBLMSP2 induces oligomerization. Size exclusion chromatography showed purified SPAM-Cd4 (green) was polydisperse, containing oligomers of higher molecular mass relative to DBL-Cd4 (orange) and the Cd4 tag alone (black). The predicted monomeric molecular mass of SPAM-Cd4 is 52 kDa: the major peak lies within the void volume of the column used, whereas the other two peaks correspond to theoretical molecular masses of ∼480 and ∼200 kDa, respectively. Molecular markers shown in blue are thyroglobulin (669), ferritin (440), aldolase (158), conalbumin (75), and carbonic anhydrase (29). C, DBLMSP and DBLMSP2 bind directly and avidly to human IgM. Equilibrium binding analysis of full-length DBLMSP (blue) and DBLMSP2 (red) to human IgM, as determined by surface plasmon resonance. Biotinylated DBLMSP or DBLMSP2 were immobilized on a streptavidin-coated chip, and 2-fold serial dilutions of human IgM were used as analyte until equilibrium was reached. Reference-subtracted data were plotted, and the KD values estimated by fitting to a simple Langmuir binding isotherm. D, surface plasmon resonance was used to quantify the biophysical parameters of the monomeric DBL domains of DBLMSP and DBLMSP2 binding to human IgM. Increasing concentrations of purified DBL domains for both DBLMSP (blue, left panel) and DBLMSP2 (red, right panel) were injected as analytes over immobilized human IgM on a sensor chip until equilibrium had been reached (see inset). Reference-subtracted binding data were plotted as a function of injected DBL protein concentration, and the KD values were estimated by fitting to a simple (1:1 binding) Langmuir binding isotherm. Binding of both DBL domains showed clear evidence of saturation demonstrating the specificity of the interaction. The DBL domain of DBLMSP reproducibly bound with ∼10-fold higher affinity than DBLMSP2.
To quantify the interaction between IgM and both DBLMSPs and demonstrate that the proteins interacted directly, we used surface plasmon resonance. Binding of soluble, purified human IgM was tested against immobilized full-length DBLMSP or DBLMSP2, mimicking the orientation of the proteins that would be expected in vivo. IgM binding to both parasite proteins was clearly saturable, demonstrating the specificity of the interactions, with equilibrium binding constants (KD) of 0.3 and 1.1 nm for DBLMSP and DBLMSP2, respectively (Fig. 3C). Given the multimeric nature of both IgM and the two parasite proteins studied, these measurements reflect overall binding avidity and are not easily comparable with other monomeric protein interactions. We therefore immobilized human IgM on the chip and used the monomeric DBL domains as analytes. Using this approach, the affinity for human IgM was 200–550-fold weaker, reflecting the loss of avidity but nonetheless still strong, with an equilibrium binding affinity of 60 nm for DBLMSP versus 600 nm for DBLMSP2 (Fig. 3D). The physiological concentration of IgM in human serum is ∼1–2 mg/ml (1–2 μm), which is at least 3 orders of magnitude higher than the measured binding constants and implies that all serum-exposed DBLMSP and DBLMSP2 would be rapidly and irreversibly complexed with IgM.
dblmsp-deficient Merozoites Invade Erythrocytes Efficiently but Do Not Bind IgM
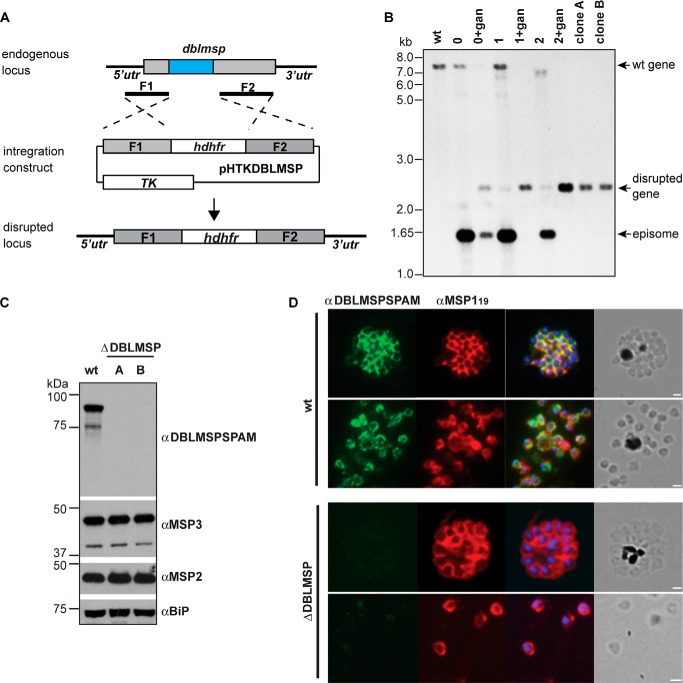
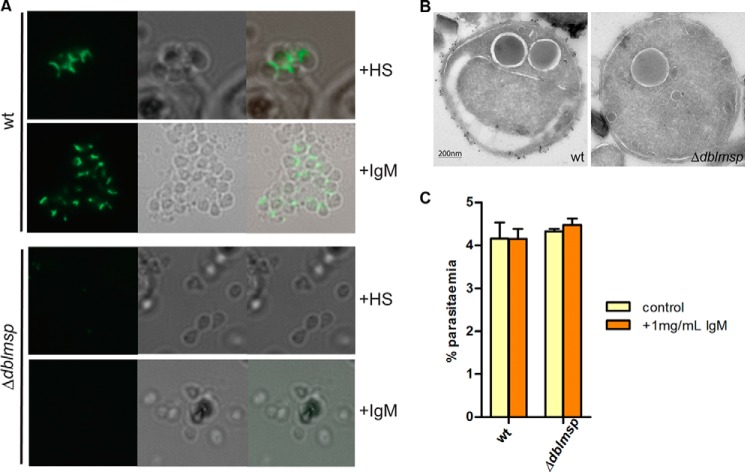
To demonstrate that native DBLMSP could bind human IgM, we generated Δdblmsp P. falciparum merozoites in which the dblmsp gene has been targeted (Fig. 4A) and confirmed this by Southern blotting (Fig. 4B). Using an antibody raised against the SPAM region of DBLMSP, we showed that the Δdblmsp parasites no longer expressed DBLMSP protein (Fig. 4C) and that it was absent from the surface of merozoites (Fig. 4D). We then used immunohistochemistry to test for the presence of human IgM at the surface of 3D7 wild-type or Δdblmsp parasites. In wild-type parasites, DBLMSP is expressed at the surface of merozoites in all schizonts, whereas DBLMSP2 is present in only a very small subset of schizonts (1% or less) (17). Following culture in the presence of purified human IgM or with human serum, IgM could be detected readily at the surface of wild-type but not dblmsp-deficient merozoites (Fig. 5A). These observations were further confirmed by immunoelectron microscopy of purified wild-type and knock-out merozoites incubated in the presence of purified human IgM, which showed regular punctate labeling at the surface of wild-type but not Δdblmsp merozoites (Fig. 5B). These data demonstrate that native DBLMSP binds IgM and is the sole IgM-binding protein at the surface of most merozoites. Δdblmsp knock-out mutants grown in the presence of medium containing a serum alternative that does not contain human IgM did not show any invasion phenotype when compared with the wild type. To determine whether the presence of human IgM on the merozoite surface had any effect on the ability of the wild-type parasite to invade red blood cells, we also performed invasion assays with parasites grown in either serum alternative alone or serum alternative supplemented with 1 mg/ml human IgM. No difference in invasion efficiency was observed, suggesting that the presence of human IgM on the merozoite surface does not affect erythrocyte invasion in vitro (Fig. 5C).
FIGURE 4.
Generation of dblmsp knock-out parasites. A, schematic representation of knock-out strategy for dblmsp. B, Southern blot on EcoRV/BglII-digested genomic DNA from wild-type and transgenic parasites: genomic DNA was extracted from P. falciparum 3D7 wild-type parasites, transgenic parasites in presence of WR99210 with or without ganciclovir, and clonal transfectants as indicated are the top of the blot. Bands corresponding to the endogenous gene, episome, and disrupted gene locus are indicated with arrows. Size markers are given on the left of the panel (in kb). C, Western blot analysis from the lysate of 1 × 106 schizonts prepared in SDS reducing sample buffer and incubated with the corresponding primary antibodies. DBLMSP could readily be detected in the lysate of wild-type parasites, but not of clones A and B. Sizes of molecular mass markers (kDa) are shown on the left of the panels. D, indirect immunofluorescence assay on thin smears of schizont and merozoites from wild-type and Δdblmsp parasites showing expression of DBLMSP (green, first panel) and MSP1 (red, second panel). The third panel depicts the overlay of both antibodies with the nuclear stain DAPI; the fourth panel shows the corresponding brightfield images. Scale bars represent 1 μm.
FIGURE 5.
Human IgM bind native DBLMSP protein. A, indirect immunofluorescence assay for detection of human IgM was conducted on live wild-type and Δdblmsp merozoites grown in either 20% human serum (+HS) or 0.125 mg/ml purified human IgM (+IgM). Alexa Fluor 488 goat anti-human IgM μ chain antibodies are depicted in green in panel 1. Panel 2 corresponds to the bright field image, and panel 3 shows the overlay. B, immunoelectron microscopy showing the presence of human IgM at the surface of wild-type but not Δdblmsp purified merozoites. Scale bar represents 200 nm. C, addition of 1 mg/ml purified human IgM to both wild-type and Δdblmsp parasites grown in Albumax did not affect their ability to infect red blood cells. Bars represent means ± S.D.; n = 3 replicate wells. A representative of four independent experiments is presented.
dblmsp and dblmsp2 Are Dimorphic
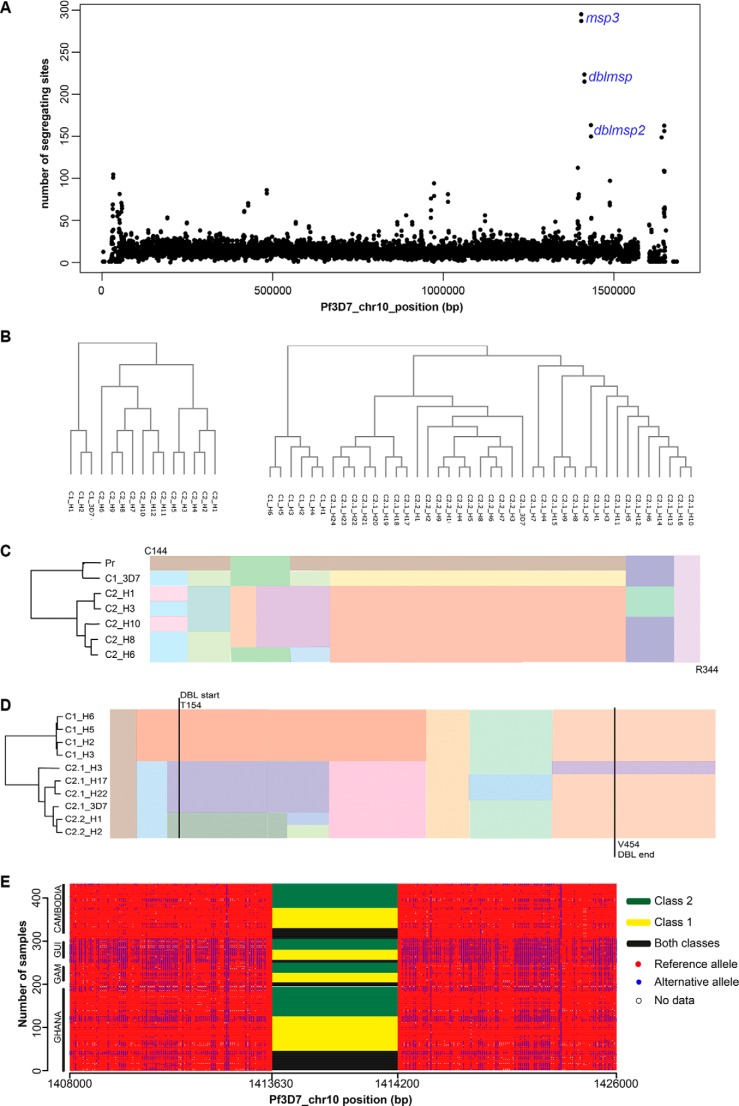
Our observations suggest that the DBLMSP and DBLMSP2 proteins from the 3D7 strain of P. falciparum parasite are able to bind human IgM. Sequencing of some P. falciparum isolates from Africa has, however, shown sequence variability in dblmsp and dblmsp2 (17–19). To further characterize the genetic diversity in these two genes, we looked at the sequence variability among field isolates from Africa and Southeast Asia. Characterizing and quantifying genetic architecture in field isolates using population-based genomic sequencing are challenging both because of their high degree of polymorphism and the appreciable levels of mixed infections. To circumvent these issues, we applied the Cortex de novo variation assembler (27, 28), which constructs a so-called de Bruijn graph representing the sequence and variation within the sample without the use of a reference genome. This method is agnostic to different types of genetic variation including indels and structural variants and is highly specific. Using this method, we generated biallelic variant calls across the P. falciparum genome using short-read Illumina sequence data from 434 field isolates from the Gambia, Ghana, Guinea, and Cambodia (minimum 50× coverage; mean read length, 74 bp) as reported elsewhere (17, 29, 30). Comparing genome sequences across P. falciparum isolates revealed striking regions of high sequence diversity, for example on chromosome 10 (Fig. 6A), and in particular around the MSP3 gene family. Using this approach, the highest peak was observed at msp3 whose two diverged haplotypes, within which recombination is reduced, have been very well studied in the literature and whose divergence precedes the P. reichenowi/P. falciparum split (31); however, we also observed extremely high peaks at dblmsp and dblmsp2.
FIGURE 6.
Genome sequencing of 434 P. falciparum isolates reveals dblmsp and dblmsp2 are dimorphic and divergent. A, the number of segregating sites across chromosome 10 from 434 P. falciparum isolates sequenced from the Gambia, Ghana, Guinea, and Cambodia, plotted in half-overlapping 500-bp windows showing isolated regions of high sequence diversity. B, cluster dendrogram of the 15 dblmsp haplotypes (left) and 41 dblmsp2 haplotypes (right) identified in the study. Both trees were obtained using ClustalW. C, a schematic representation of the amino acid multiple sequence alignment of six representative exemplar haplotypes for the DBL domain of DBLMSP between residues Cys144 and Arg344, and the P. reichenowi ortholog. Note the long non-recombining region has two alternate allelic forms (orange and yellow), of which the yellow is relatively close to the P. reichenowi sequence (shown in brown). A phylogenetic tree indicating the relationships of the sequences built by clustering haplotypes based on a distance matrix (where distance corresponds to the number of SNP and indel differences) is shown on the left. D, a schematic representation of the amino acid multiple sequence alignment of 10 representative exemplar haplotypes for the DBL domain of DBLMSP2 between residues Glu102 and Gly550. Note the long non-recombining region has two alternate allelic forms (orange for class 1 and purple/dark green for subclasses 2.1/2.2, respectively). A phylogenetic tree indicating the relationships of the sequences built by clustering haplotypes based on a distance matrix (where distance corresponds to the number of SNP and indel differences) is shown on the left. E, haplotype structure in the region of DBLMSP; rows are samples, and columns are polymorphic sites. Outside the DBL domain, the reference allele is shown in red, the alternate allele is in blue, and missing data are in white. Within the DBL domain, the class of allele found in each sample is shown: class1 only (yellow), class 2 only (green), or both present (black). Samples are split by country and then sorted first by DBLMSP type and then by haplotype structure outside the DBL domain. Note that the x axis is not drawn to scale.
After close examination of the variant calls within dblmsp and dblmsp2, we found a large number of overlapping variants. Because Cortex only calls biallelic variants, samples with multiple infections, which contain more than two alleles at a single site, will return no value. Because few sites were multiallelic in all samples, we took the union of all sites in the 434 samples and removed redundancy; this union contained the vast majority of polymorphic sites. The overlapping alleles in dblmsp and dblmsp2 were long (up to ∼1 kb) and shared sequence. As a result of this, together with the unknown multiplicity of infection and lack of phasing information with the short Illumina reads, we took a conservative approach and did not attempt to genotype the implied long multiallelic variants. Applying these approaches, we established that dblmsp and dblmsp2 each contained one major cluster of long non-recombining haplotypes (15 haplotypes in dblmsp and 41 in dblmsp2), with differing lengths (∼570 bp in dblmsp and 1350 bp in dblmsp2) and all overlapping the region encoding the DBL domain. In dblmsp, these 15 haplotypes grouped into two major allelic forms that we termed class 1 (3 haplotypes) and 2 (12 haplotypes) (Fig. 6, B and C). In dblmsp2, we found two major allelic forms, class 1 (6 haplotypes) and class 2 (35 haplotypes), which we subdivided into two subclasses (c2.1 and c2.2) on the basis of multiple sequence alignments (Fig. 6, B and D). To quantify the divergence between haplotypes, we designated d(c1,c2) as the mean number of SNP and indel differences between classes c1 and c2. For dblmsp, d(c1,c1) = 1.77 and d(c2,c2) = 22.5, whereas d(c1,c2) = 185. Similarly for dblmsp2, d(c1,c1) = 2.9, d(c2.1,c2.1) = 34.2, d(c2.2,c2.2) = 7.0, and d(c2.1, c2.2) = 61, whereas d(c1, c2.1) = 239 and d(c1,c2.2) = 194, demonstrating that for both genes, the two main allelic classes were deeply diverged.
To get an estimate of population frequencies, we used the alignments to find sequence markers characteristic of the main classes and then queried the de Bruijn graph for all 434 samples to see which samples showed evidence of class 1 and/or class 2 haplotypes. Using this approach, we were able to find evidence for a haplotype from either dimorphic form even in samples where no call had been made. Our findings confirmed that both dimorphic forms co-existed at high frequencies in each gene and all four countries (Fig. 6E). Haplotype classes in all 434 samples were clearly identified in the region encoding the DBL domain, with SNP and indel sites present on either side. Outside the DBL domain, a strong long range linkage disequilibrium structure was observed, but all four countries showed different patterns. Within the DBL domain, however, samples from all countries showed both allelic classes at appreciable frequencies, a powerful signal of balancing selection. Although this had been shown previously in Africa, to our knowledge this is the first time it has been demonstrated in Southeast Asia. In the case of dblmsp, one of the two haplotype classes was very close to the P. reichenowi sequence (Fig. 6C) and is a strong example of balancing selection. Until further samples of P. reichenowi are sequenced, we will not know whether the selection pressure is present in P. falciparum only or whether this is a trans-species polymorphism (32). Interestingly, the population counts for haplotype classes in dblmsp showed much higher levels of mixed infection than expected (Fig. 6E), from which we inferred the existence of a dblmsp paralog, as has been suggested before (19). In conclusion, the lack of recombination within the class-defining region of the DBL domain and the long range linkage disequilibrium observed on either side of this region are striking and provide convincing evidence for strong selection pressure on the DBL domains of DBLMSP and DBLMSP2, suggesting they play an important role in parasite biology.
Most Naturally Occurring DBL Domain Variants Can Bind Human IgM
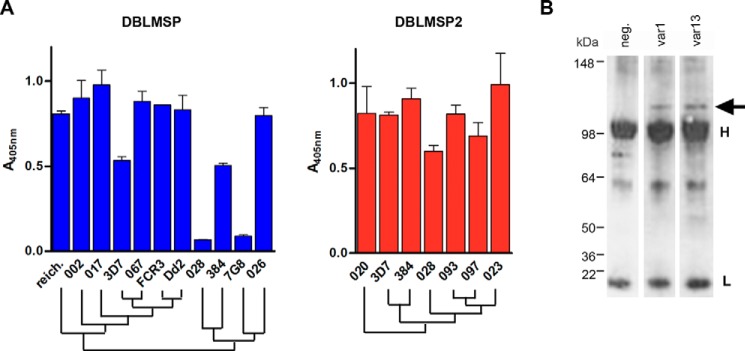
Because the genetic variability within dblmsp and dblmsp2 is concentrated on the region encoding the DBL domain, we tested whether the ability to bind IgM was retained across DBL sequence variants representing the major allelic forms for both genes found in our and others' analyses (18, 19). For DBLMSP, a total of 10 DBL variants from 3 P. falciparum laboratory strains: 7G8, Dd2, and FCR3; 6 field isolates; and the P. reichenowi ortholog sharing between 59 and 99% sequence identity were selected (supplemental Fig. S1). Except for the DBL domains from 7G8 and 028, IgM binding was conserved across all variants, including the most divergent (384 and P. reichenowi) (Fig. 7A). Similarly for DBLMSP2, seven representative DBL variants with sequence identities ranging from 62 to 98% were selected (supplemental Fig. S1). Although variant 082 repeatedly did not produce protein, all the other six bound human IgM (Fig. 7A). Interestingly, whereas the 7G8 and 028 DBLMSP DBL domains could not bind human IgM, their shared DBLMSP2 DBL sequence was able to do so.
FIGURE 7.
Binding to human IgM is conserved across DBLMSP and DBLMSP2 variants. A, IgM binding to representative DBL domains from 11 DBLMSP (blue) and seven DBLMSP2 (red) was conserved. Only the DBLMSP DBL domains from the laboratory strain 7G8 and isolate 028 did not show binding. A representative from two experiments is shown in each panel. Bars represent means ± S.D.; n = 3 replicate wells. reich, P. reichenowi. B, Western blot showing that DBLMSP is associated to human IgM that were purified from long term cultures of IT4 var1 or var13 parasite; the negative (neg) control corresponds to culture medium without parasites. Note that the anti-rabbit secondary antibody used in this experiment cross-reacts with the light (25 kDa) and heavy chains (50 kDa) of human IgG.
To further confirm that binding to IgM was conserved among sequence variants, we performed IgM pulldown from long term parasite culture supernatants from the var1 and var13 substrains of the IT4 strain (whose dblmsp sequence is identical to that of FCR3) grown in the presence of human serum. Following SDS-PAGE and incubation with an anti-DBLMSP antibody, a band corresponding to the DBLMSP protein was identified in the immunoprecipitates from parasite cultures but not from the negative control supernatant, in which no parasite was grown (Fig. 7B). In summary, despite their high level of sequence polymorphism, binding of the DBL domains to human IgM was conserved across most of the DBLMSP and DBLMSP2 DBL domain variants tested.
Binding of Human IgM to Their DBL Domains Masks DBLMSP and DBLMSP2 from Host Antibodies
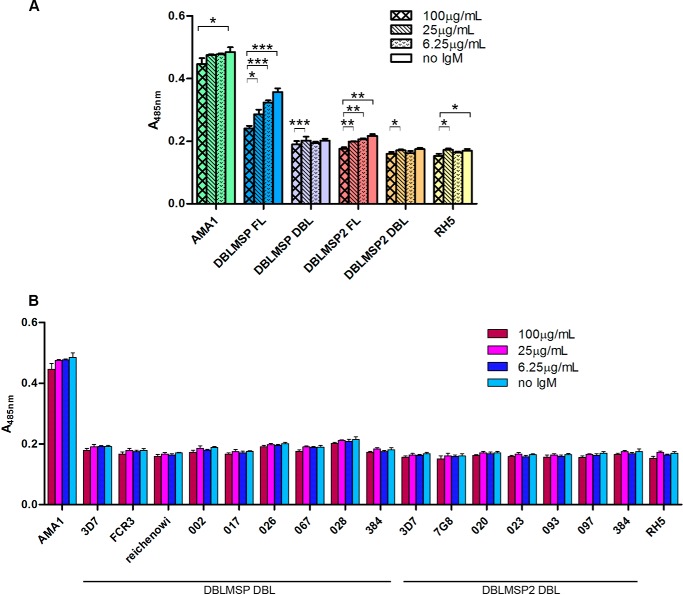
The fact that the DBL domains of DBLMSP and DBLMSP2 retain IgM binding despite their extraordinary sequence diversity suggests that DBLMSP and DBLMSP2 are targeted by the host immune system and that IgM binding promotes parasite survival. To determine whether individuals regularly exposed to P. falciparum mount an immune response to DBLMSP and DBLMSP2, we compared the immunoreactivity of the full-length proteins and DBL domains of both DBLMSP and DBLMSP2 from the 3D7 strain to purified IgG from Malawian immune adults (33). As comparators, we used AMA1 and RH5, which have previously been shown to be highly and weakly immunoreactive, respectively, to antibodies in sera from Kenyan, Malian, and Senegalese immune adults (20, 34–36). In the absence of human IgM, the immunoreactivity of the full-length 3D7 isoform of DBLMSP was intermediate, being weaker than the immunoreactivity of AMA1 but significantly higher than that of RH5. Full-length DBLMSP2, however, was only very slightly immunoreactive when compared with RH5 (Fig. 8A). Consistent with the large number of variants observed in P. falciparum populations, almost no binding of purified hyperimmune IgG was observed to either of the two DBL domains. To confirm this was not merely because the sera were collected from individuals who had not been exposed to the 3D7 variants used in this assay, we also tested the immunoreactivity of other DBL variants for DBLMSP and DBLMSP2 and confirmed that they were generally not immunoreactive (Fig. 8B).
FIGURE 8.
IgM binding to full-length DBLMSP and DBLMSP2 masks them from the host immune system. A, normalized amounts of full-length (FL) DBLMSP, DBLMSP2, RH5, the entire ectodomains of AMA1, or the DBL domain of both DBLMSP and DBLMSP2 expressed as β-lactamase-tagged pentamers were precomplexed with varying concentrations of human IgM, and the immunoreactivity of the complexes to purified IgG from Malawian adults quantified by β-lactamase substrate hydrolysis. The immunoreactivity of full-length DBLMSP and DBLMSP2 decreased with increasing concentrations of IgM. The AMA1 and RH5 proteins were used as exemplars of high and low immunoreactivity, respectively, and their immunoreactivity, as well as those of the DBL domains, was only marginally affected by the presence of IgM. *, 0.02 < p ≤ 0.05; **, 0.01 < p ≤ 0.02; ***, p < 0.01; two-tailed paired t test. Shown is one representative of three experiments. Bars represent means ± S.D.; n = 3 replicate wells; all proteins were the 3D7 strain sequences. B, DBL domains from a representative panel of DBLMSP and DBLMSP2 variants were expressed as β-lactamase pentamers and precomplexed with varying concentrations of human IgM before incubating with purified immobilized IgG from Malawian adults. The immunoreactivity of all the DBL domains was very low, irrespective of the IgM concentration. The entire ectodomains of AMA1 and RH5 from the 3D7 strain were used as exemplars of high and low immunoreactivity, respectively. Bars represent means ± S.D.; n = 3 replicate wells.
Non-immune human IgM is also known to bind to some P. falciparum PfEMP1 variants expressed at the surface of infected erythrocytes, which are associated with rosetting or placental malaria (24, 37). Although the function of IgM binding in rosetting remains unclear, association to the var2CSA variant responsible for placental malaria has been linked to masking of the parasite from antigen-specific IgG (38, 39). To determine whether binding of IgM to the DBL domains of DBLMSP and DBLMSP2 could have a steric immunological “masking” effect, we precomplexed each of the two parasite proteins with varying concentrations of purified human IgM before incubating them with purified hyperimmune IgG. The presence of IgM significantly reduced the overall immunoreactivity of both full-length proteins in a dose-dependent manner, an effect that was not observed for AMA1 and RH5 (Fig. 8A). Interestingly, the presence of IgM had almost no effect on the already very limited immunoreactivity of the DBL domains. This suggests that binding of IgM to DBLMSP and DBLMSP2 masks antibody epitopes that are located in other regions of the proteins.
Discussion
The deeply diverged lineages preserved in dblmsp and dblmsp2 across P. falciparum populations suggests that they are under extremely strong selection and must have important biological functions. One possibility is that they are direct targets of the host immune response and are therefore potential targets for therapeutic intervention, but their function has remained unknown. Here, we have demonstrated that the DBL domains in both proteins bind directly and avidly to host IgM and that this function is conserved between the different sequence variants identified in P. falciparum isolates.
The population-based sequence analysis of dblmsp and dblmsp2 reported here extends earlier findings that both genes exhibit a strong allelic dimorphism with limited interallelic recombination across parasite isolates from all populations sampled. Strikingly, we have now shown that both allelic classes in dblmsp and dblmsp2 are present at relatively high frequency in populations from West Africa (Ghana, the Gambia, and Guinea) and Southeast Asia (Cambodia), with recombination absent (or selected against) within a specific block of the DBL domain. The pattern of balancing selection preserving two deeply diverged haplotypes with reduced recombination has previously been seen in msp3, which lacks a DBL domain. We would draw a distinction between this pattern, and the known dimorphism at EBA175, which is simply a single indel, rather than a long preserved haplotype. This unusual genetic architecture raises the question as to how this dimorphism arose and is maintained within the population. Following our demonstration that the DBL domain can bind host IgM, one possibility is that there is a selective advantage to binding the two major IgM species that either contain or lack a J-chain. Dimorphism, however, is also observed in genes encoding other merozoite surface proteins, suggesting selection by a broader dichotomous feature within the immune system of the host population.
Despite the large number of polymorphisms within the DBL domains of both DBLMSP and DBLMSP2, the ability to bind the constant region of human IgM was broadly conserved across representative sequence variants, suggesting that this interaction plays an important role in the parasite biology. Using the known crystal structure of the DBL domain for DBLMSP2 (9) to model the DBL domains of DBLMSP and DBLMSP2 used in our functional analysis, we observed that the majority of the differences between IgM binders and non-binders are located in helices h1, h2a, and h4 (supplemental Fig. S2). These helices could therefore be potential binding sites for human IgM. Although DBLMSP is highly expressed in all mature schizonts in the parasitophorous vacuole and located on the merozoite surface, DBLMSP2 is only expressed in a small percentage of schizonts (17, 40). One suggestion could therefore be that DBLMSP2 is expressed at the surface of the minority of merozoites committed to gametocytogenesis. One interesting observation was that although all tested DBLMSP2 variants were able to bind IgM, some DBLMSP variants did not. Although this may indicate that dblmsp function—or its ability to bind IgM—is dispensable, which may be consistent with an earlier finding that some dblmsp sequences from field isolates contained premature stop codons (18, 19), 3 of 14 cloned P. falciparum lines were found to contain multiple sequences for dblmsp (19). These results, combined with our observation of a higher proportion than expected of mixed haplotypes at the dblmsp locus, suggest the existence of functional paralogous dblmsp genes that can substitute in some isolates and strains.
Our finding that DBLMSP binds to IgM with a KD almost 4 orders of magnitude lower than physiological plasma IgM concentrations demonstrates that DBLMSP would rapidly and irreversibly be saturated with host IgM once merozoites are exposed to host blood following schizont rupture. Consequently, plasma-exposed DBLMSP should really be considered as a DBLMSP-IgM complex: something that must be taken into account when investigating the role of these proteins in the blood stages of the parasite. Previous research has suggested a role for DBLMSP in erythrocyte invasion, supported by the presence of a DBL domain, which is shared by other invasion ligands and its localization to the surface of merozoites (9, 40, 41), through its interaction with MSP1 (42, 43). However, addition of anti-DBLMSP antibodies at very high concentration (20 mg/ml) only had a modest 25% decrease on invasion (41), and deletion of the dblmsp gene in the 3D7 laboratory strain did not affect the invasion efficiency of the parasite in our study and others (41). The presence of human IgM in the parasite culture did not either affect the ability of wild-type or Δdblmsp parasites to invade red blood cells, suggesting that DBLMSP is not essentially required for parasite invasion in vitro.
DBL domains, in addition to binding erythrocyte receptors from the glycophorin family, are known to mediate IgM binding in some PfEMP1 variants that are displayed on the surface of infected erythrocytes (37, 44–46). This IgM binding, which is involved in adhesion of infected erythrocytes (47, 48), has recently been suggested to cluster PfEMP1 on the erythrocyte surface thereby increasing their avidity for host receptors (49, 50). Alternatively, PfEMP1-mediated IgM binding could also be implicated in masking from immune IgG, thereby acting as a steric shield from the host adaptive immune system (38). The conservation of IgM binding to DBLMSP, despite high levels of sequence polymorphism, suggests it might be an important immune evasion strategy for the parasite. Consistent with this, a comparative immunoreactivity analysis performed with purified IgG from Malawian adults revealed that the DBL variants of DBLMSP and DBLMSP2 are poorly immunogenic and that binding to human IgM further decreases the overall immunoreactivity to the full-length proteins. This masking, however, did not seem to affect the accessibility of IgGs to other merozoite surface proteins because surface labeling of PfMSP1 from 3D7 parasites grown in the presence or absence of human IgM looked similar by immunofluorescence analysis (data not shown). Binding of host IgM to DBLMSP and DBLMSP2 might therefore mask specific domains that are important for the parasite biology on these two proteins but whose function remains unknown.
In conclusion, we have extended the genetic characterization of two P. falciparum genes that are under strong balancing selection and shown that the proteins both bind directly and with high affinity to host IgM and thereby shield the parasite from the host adaptive immune response. Given that both infected erythrocytes and merozoites are able to bind host IgM, IgM-binding proteins could be a general immunoprotective mechanism used by P. falciparum and possibly other parasites and pathogens.
Experimental Procedures
Ethics Statement
Use of erythrocytes and serum from human donors for P. falciparum culture was approved by the National Health Service Cambridgeshire 4 Research Ethics Committee. All subjects provided written informed consent. The use of animals to raise antisera was performed according to UK Home Office governmental regulations and in accordance with European directive 2010/63/EU regarding the use of laboratory animals. Research was approved by the Sanger Institute Animal Welfare and Ethical Review Board.
P. falciparum Population Sequence Analysis
434 samples from Gambian, Ghanaian, Guinean, and Cambodian origin sequenced by the MalariaGEN consortium underwent standard QC protocols as previously documented (29) and were sequenced to a depth of at least 50× with Illumina reads (mean read length, 74 bp) and PCR-based library preparation. Samples underwent Cortex assembly using the “Independent workflow” as previously described (28) using a k-mer of 31. All variants called within dblmsp and dblmsp2 were collated and clustered into groups separated by at least 31 bp, revealing 15 different haplotypes overlapping the DBL domain in dblmsp and 41 in dblmsp2. The GenBankTM accession numbers are KM577604–KM577618 for dblmsp and KM589600–KM589640 for dblmsp2.
Recombinant Protein Production
All P. falciparum proteins were produced recombinantly by transient transfection of HEK293E cells (51) essentially as described (20) except Py MSP119 (52). Briefly, chemically synthesized genes were codon-optimized for mammalian expression and cloned in frame with an exogenous signal peptide (53), and potential N-linked glycosylation sequons were mutated to prevent inappropriate glycosylation of Plasmodium proteins. All recombinant proteins contain a C-terminal Cd4d3 + 4 tag (54) followed by either an enzymatically biotinylatable sequence, a His6 tag, or a pentamerization sequence followed by β-lactamase (55). Monobiotinylated proteins were produced by cotransfecting with a plasmid encoding secreted BirA (55, 56). Proteins were processed and purified using His-Trap purification columns (GE Healthcare) as previously described (57, 58).
Protein Purification from Human Serum or Parasite Culture Supernatant
Streptavidin-coated paramagnetic beads (100 μl, 1 μm diameter; Life Technologies) were saturated with 20 μg of enzymatically monobiotinylated DBLMSP or a Cd4 tag-alone control, isolated with a magnet, and washed three times with PBS before incubating with 1 ml of filtered human serum (Sigma) for one h at 4 °C. Beads were washed four times with 1 ml of PBS and eluted with 200 μl of 1% SDS. 20 μl were resolved by SDS-PAGE under reducing conditions and stained with SYPRO Orange (Sigma), and the gel image captured on a Typhoon 9400 phosphorimaging device (GE Healthcare).
Anti-human IgM agarose beads (Sigma) were incubated with long term parasite culture supernatants from the IT4 var1 and var13 strains grown in the presence of human serum or control culture medium without parasite for 1 week at 37 °C. After five washes in PBS, the beads were resuspended in loading buffer in the presence or absence of DTT, and eluates were blotted onto nitrocellulose membranes (Amersham Biosciences Protran) followed by blocking in PBS, 0.1% Tween 20, 5% nonfat milk powder) and incubated for 1 h with a rabbit anti-full-length DBLMSP antibody at a 1:100 dilution. After further washes, the membrane was incubated with an anti-rabbit HRP-conjugated IgG secondary antibody (1:1000; Sigma) and developed using 3′,3′-diaminobenzidine (DAKO) according to the manufacturer's instructions.
Parasite Culture and Transfection
P. falciparum parasites (3D7 strain) were grown in vitro in RPMI 1640 medium containing Albumax II or 10% human serum as described previously (59) and transfected using standard protocols (60). Transfected parasite cultures were selected with 10 nm WR99210 (kind gift of Jacobus Pharmaceuticals) and 10 μm ganciclovir (Sigma-Aldrich) and cloned by limiting dilution.
dblmsp Knock-out Construct Design and Characterization
For gene disruption via double homologous recombination in P. falciparum, a 520-bp 5′ fragment (F1 region) and a 659-bp 3′ fragment (F2 region) comprising ORF and UTR sequences of dblmsp were amplified using primers 348F1for (5′-AATAAACCGCGGCACATTTAATTAAGGTTGTATTTACTG-3′, SacIIsite underlined), 348F1rev (5′-TTATTTAGATCTGACATTTTTAATACCCTTACAAAAATTTTC-3′; BglII site underlined), 348F2for (5′-TTATTTATCGATAGTGTTAGGGATTCTAGTAATCTAGATCAACG-3′; ClaI site underlined), and 348F2rev (5′-TTTATTCCTAGGCATATCTTCTGTCAAACCCTTAAAAATAGTTTC-3′; AvrII site underlined). Amplification products were digested using SacII + BglII (F1 flank) and ClaI + AvrII (F2 flank) and cloned into pHTK vector (60), resulting in construct pHTKDBLMSP.
For Southern blotting, the genomic DNA from P. falciparum 3D7 wild-type parasites, transgenic parasites in presence of WR99210 with or without ganciclovir, and clonal transfectants were digested with EcoRV and BglII, separated on a 0.7% agarose gel, and transferred onto a nitrocellulose membrane using standard techniques. A radioactive probe was generated from the 520-bp F1 flank in construct pHTKDBLMSP using random priming (Decaprime II; Ambion). Hybridization was carried out at 62 °C overnight before stringent washes and autoradiography.
For Western blot analysis, total lysate from 1 × 106 schizonts was prepared in SDS reducing sample buffer and boiled at 95 °C for 5 min. The lysates were separated on precast 10% Bis Tris NuPAGE polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes by electroblotting. The membranes were blocked and incubated with rabbit anti-DBLMSPSPAM (1:2000), rabbit anti-MSP3 (1:2000), mouse anti-MSP2 (1:1000) (61), and rat anti-BiP antibodies (1:1000). Bound antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (Bio-Rad).
For immunofluorescence analysis, thin smears of schizont and merozoites from wild-type and Δdblmsp parasites were incubated with anti-DBLMSPSPAM (1:2000) and anti-MSP119 (clone 1E1, 1:3000) antibodies diluted in 3% BSA/PBS and incubated for 1 h at room temperature before washes and incubation with secondary Alexa Fluor-labeled (488 and 594; Invitrogen) antibodies at 1:5000.
Human IgM Binding to Merozoites
To detect IgM binding to merozoites, parasites were tightly synchronized before late stage schizonts were purified using 70% Percoll, put back into culture containing RPMI + Albumax II with the addition of 20% human serum or 0.125 mg/ml purified human IgM (Sigma). IgM was detected using an Alexa Fluor 488 goat anti-human IgM μ chain antibody (Molecular Probes; preadsorbed against human IgG) at 1:1000. The slides were viewed on a Zeiss Axioplan 2 imaging system with Plan Apochromat 100×/1.4 oil immersion objective. Images were captured using Axiovision 4.6.3 software and edited using Adobe Photoshop.
For immunogold labeling, merozoites were fixed in 4% paraformaldehyde in 0.1 m phosphate buffer at pH 7.4 for 1 h at room temperature, rinsed three times in buffer, and infiltrated with 1% and then 10% gelatin before immersing in 2.3 m sucrose in phosphate buffer overnight at 4 °C for cryoprotection. Frozen samples were prepared by mounting onto aluminum pins and rapidly immersing in liquid nitrogen in preparation for ultrathin 80 nm sectioning on a Leica EM FC6 ultramicrotome. Ultra thin sections were labeled as per Tokuyasu (62), with a rabbit anti-human IgM antiserum (Abcam) diluted 1:25, and detected with 10-nm protein A gold. Imaging was performed on an FEI 120kV Spirit Biotwin with a Tietz F4.15 CCD camera.
Cell Culture of Transfectomas Secreting Anti-dinitrophenyl Human IgM
Transfectomas expressing anti-dinitrophenyl IgM antibodies containing the human constant heavy chains either with (Xp) or without (Gp) the mouse J-chain (22) were a kind gift of Prof. Marc Shulman. Transfectomas were grown in DMEM supplemented with 10% fetal bovine serum, 0.6 mg/ml G418, and 0.02% β-mercaptoethanol, and supernatants were harvested and filtered before use in ELISA and AVEXIS experiments.
ELISA and Primary Antibodies
Biotinylated recombinant P. falciparum proteins were normalized and immobilized on streptavidin-coated plates, preblocked with HBS and 0.1% Tween 20 (HBST), 2% BSA for 30 min. After washing with HBST, serial dilutions of either pooled human sera from 10 malaria-exposed or malaria-naïve individuals, transfectoma cell culture supernatants, or purified primary antibodies were incubated for 90 min. The plates were again washed with HBST, and appropriate alkaline phosphatase-conjugated secondary antibodies were incubated for 1 h. The plates were washed with HBST and once with HBS before adding p-nitrophenyl at 1 mg/ml, and absorbance at 405 nm was quantified on either a PHERAstar Plus or FLUOstar Optima plate reader (BMG Labtech). For domain mapping experiments involving chimeric human antibodies, an HRP-conjugated anti-human secondary antibody was used. Following PBS washes, tetramethylbenzidine substrate was added, and absorbance was quantified at 450 nm. All procedures were performed at room temperature. Primary antibodies used were: anti-Cd4 OX68 (1:1000; AbD Serotec); purified human IgA, IgE, IgM (3 μg/ml; Sigma), or IgG (3 μg/ml; Bethyl Laboratories); human, goat, rabbit, guinea pig, or bovine serum (1:1000; Sigma); purified mouse IgM (10 μg/ml MEM-150; Abcam); or domain-swapped human antibodies at 25 nm (25, 26). All alkaline-phosphatase-conjugated secondary antibodies were from Sigma, except anti-rabbit immunoglobulins (Jackson ImmunoResearch).
Mapping DBLMSP and DBLMSP2 Binding Site on Human IgM
Domain-specific monoclonal antibodies that bind the human constant μ chain (23) were incubated for 90 min with either biotinylated human IgM or biotinylated human IgG (used as a control) before washing and adding pentamerized, β-lactamase-tagged DBLMSP or DBLMSP2 and incubating for a further hour. After washes, the β-lactamase substrate nitrocefin was added at 125 μg/ml, and colorimetric turnover was measured by absorbance reading at 485 nm. Antibodies were biotinylated using EZ-link Sulfo-NHS-LC-biotin (Pierce). Construction of the homology model for the DBL domain of DBLMSP and DBLMSP2 was done using the Phyre2 engine.
Surface Plasmon Resonance Analysis
Surface plasmon resonance analysis was performed on a BIAcore T100 instrument at 37 °C in HBS-EP buffer, using streptavidin-coated sensor chips (GE Healthcare) essentially as described (63). Briefly, 300 response units of the biotinylated Cd4 tag was used as a reference, and molar equivalents of full-length biotinylated DBLMSP or DBLMSP2 were immobilized in query flow cells. Increasing concentrations of purified human IgM were injected for 30 min at 10 μl/min until equilibrium had been reached, and binding was quantified from reference-subtracted sensorgrams. The surface was regenerated after each cycle with either 2 m NaCl for 60 s for DBLMSP, or 10 mm glycine HCl, pH 3.0, for 20 s for DBLMSP2, and duplicate injections of the same concentration of IgM in each experiment showed no loss of activity between each cycle. For the binding analysis using the DBL domains only, 500 response units of biotinylated human IgG were immobilized as a reference, and a molar equivalent of biotinylated human IgM was immobilized in the query flow cell. Each purified DBL domain was resolved by gel filtration on a Superdex 200 Tricorn 10/600 column to remove any protein aggregates that might interfere with the kinetic measurements. Increasing concentrations of DBL domains were injected at 20 μl/min for 2 min in each cycle until equilibrium had been reached. The surface was regenerated after each cycle with 2 m NaCl for 60 s, with no loss of activity. Data analysis was performed using BIAcore analysis software.
Generation of Polyclonal Antibodies
A polyclonal antibody against the SPAM fragment of DBLMSP was generated. Briefly, a 619-bp DNA product (corresponding to amino acids 439–633 of DBLMSP) was amplified from 3D7 genomic DNA using primers 348ForPET30 (5′-GGTATTGAGGGTCGCAAAGAATTTAAAGATAACGTTACACTTCTTAAAGC-3′) and 348revbPET30 (5′-AGAGGAGAGTTAGAGCCTTAACTTTGTTGTTGATTACTGAGTTCTTTTTCC-3′) and cloned into pET-30 Xa/LIC expression vector (Novagen). Expression plasmids were sequenced before transformation into Escherichia coli BL21(DE3)pLysS, and expression was induced with 1 mm isopropyl β-d-thiogalactopyranoside for 4 h at 30 °C. Recombinant DBLMSPSPAM protein was purified using nickel-nitrilotriacetic acid affinity chromatography under non-denaturing conditions. Purified rDBLMSPSPAM was used for antibody production in rabbits (Harlan Laboratories Inc.) following standard procedures. Rabbit antibodies were subsequently affinity-purified using rDBLMSPSPAM coupled to CNBr-activated Sepharose 4B (GE Healthcare) and IgG-selected using protein G-Sepharose (Sigma-Aldrich).
Polyclonal antibodies against the full-length 3D7 form of DBLMSP were raised in rabbits (Cambridge Research Biochemicals). The full-length, His-tagged protein was produced in mammalian cells, purified by nickel-nitrilotriacetic acid chromatography using HisTrap column (GE Healthcare) and injected every 2 weeks over an 11-week period with the first injection in complete Freund's adjuvant, and the subsequent five injections in incomplete adjuvant. Rabbit antibodies were subsequently affinity-purified using HiTrap protein G columns (GE Healthcare).
Immunoreactivity Analysis
Pentamerized, normalized β-lactamase-tagged parasite proteins were incubated with serial dilutions of purified human IgM for 90 min before being transferred to 20 μg/ml purified IgG from Malawian adults (33) and immobilized on protein G-coated microtiter plates (Pierce). After 60 min, the plates were washed and incubated with nitrocefin at 125 μg/ml. The incubations were performed at room temperature, and absorbance was read at 485 nm as described above.
Author Contributions
C. C. identified and characterized the interactions between human IgM and recombinant DBLMSP proteins, performed the binding and immunoreactivity studies on DBL variants, and wrote the manuscript. Z. I., S. C. M., A. M., and D. P. K. collated the samples, designed, and analyzed the data corresponding to the genetic analysis of field isolates. E. K. and A. A. H. designed, performed, and analyzed the data relating to the generation and analysis of Δdblmsp and wild-type parasites. A. J. P. performed the IgM purification from human serum. G. K., D. G., and G. D. purified the merozoites and performed the immunoelectron microscopy experiments. L. Y. B. and J. C. R. performed and analyzed parasite culture experiments. S. C. M. and R. J. P. performed the mapping experiment on human IgM and the immunoprecipitation of IgM from parasite culture supernatant and built the homology model. G. J. W. conceived and coordinated the study and wrote the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Marc Shulman and Alister Craig for kindly providing the transfectomas and purified IgG from Malawian adults, respectively. Our thanks also go to Britta Urban, Magnus Manske, and Antoine Claessens for expert help and advice, as well as Katy Lloyd and Nadia Cross for technical help. This publication uses sequencing data from the MalariaGEN Community Project on P. falciparum population genomics as described in Ref. 29, and we thank Lucas Amenga-Etego, David Conway, Alfred Amambua-Ngwa, Victor Mobegi, and Rick Fairhurst for graciously allowing us to use data on samples that they have contributed to the project.
This work was supported by Wellcome Trust Grant 098051 (to C.C., L. Y. B., A. J. P., D. G., G. D., D. P. K., J. C. R., and G. J. W.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1 and S2.
- DBL
- Duffy binding-like
- EBA
- erythrocyte-binding antigen
- MSP
- merozoite surface protein.
References
- 1. Murray C. J., Rosenfeld L. C., Lim S. S., Andrews K. G., Foreman K. J., Haring D., Fullman N., Naghavi M., Lozano R., and Lopez A. D. (2012) Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet 379, 413–431 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization (2012) World Malaria Report 2012, World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Cowman A. F., Berry D., and Baum J. (2012) The cellular and molecular basis for malaria parasite invasion of the human red blood cell. J. Cell Biol. 198, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen S., McGregor I. A., and Carrington S. (1961) γ-Globulin and acquired immunity to human malaria. Nature 192, 733–737 [DOI] [PubMed] [Google Scholar]
- 5. Richards J. S., and Beeson J. G. (2009) The future for blood-stage vaccines against malaria. Immunol. Cell Biol. 87, 377–390 [DOI] [PubMed] [Google Scholar]
- 6. Manske M., Miotto O., Campino S., Auburn S., Almagro-Garcia J., Maslen G., O'Brien J., Djimdé A., Doumbo O., Zongo I., Ouedraogo J. B., Michon P., Mueller I., Siba P., Nzila, A. et al. (2012) Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487, 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weedall G. D., and Conway D. J. (2010) Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol. 26, 363–369 [DOI] [PubMed] [Google Scholar]
- 8. Singh S., Soe S., Weisman S., Barnwell J. W., Pérignon J. L., and Druilhe P. (2009) A conserved multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum. PLoS One 4, e5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hodder A. N., Czabotar P. E., Uboldi A. D., Clarke O. B., Lin C. S., Healer J., Smith B. J., and Cowman A. F. (2012) Insights into Duffy binding-like domains through the crystal structure and function of the merozoite surface protein MSPDBL2 from Plasmodium falciparum. J. Biol. Chem. 287, 32922–32939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gondeau C., Corradin G., Heitz F., Le Peuch C., Balbo A., Schuck P., and Kajava A. V. (2009) The C-terminal domain of Plasmodium falciparum merozoite surface protein 3 self-assembles into α-helical coiled coil tetramer. Mol. Biochem. Parasitol. 165, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McColl D. J., Silva A., Foley M., Kun J. F., Favaloro J. M., Thompson J. K., Marshall V. M., Coppel R. L., Kemp D. J., and Anders R. F. (1994) Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 68, 53–67 [DOI] [PubMed] [Google Scholar]
- 12. Wickramarachchi T., Cabrera A. L., Sinha D., Dhawan S., Chandran T., Devi Y. S., Kono M., Spielmann T., Gilberger T. W., Chauhan V. S., and Mohmmed A. (2009) A novel Plasmodium falciparum erythrocyte binding protein associated with the merozoite surface, PfDBLMSP. Int. J. Parasitol. 39, 763–773 [DOI] [PubMed] [Google Scholar]
- 13. Orlandi P. A., Klotz F. W., and Haynes J. D. (1992) A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum, recognizes the terminal Neu5Ac(α2–3)Gal-sequences of glycophorin A. J. Cell Biol. 116, 901–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maier A. G., Duraisingh M. T., Reeder J. C., Patel S. S., Kazura J. W., Zimmerman P. A., and Cowman A. F. (2003) Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 9, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sim B. K., Chitnis C. E., Wasniowska K., Hadley T. J., and Miller L. H. (1994) Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264, 1941–1944 [DOI] [PubMed] [Google Scholar]
- 16. Kraemer S. M., and Smith J. D. (2006) A family affair: var genes, PfEMP1 binding, and malaria disease. Curr. Opin. Microbiol. 9, 374–380 [DOI] [PubMed] [Google Scholar]
- 17. Amambua-Ngwa A., Tetteh K. K., Manske M., Gomez-Escobar N., Stewart L. B., Deerhake M. E., Cheeseman I. H., Newbold C. I., Holder A. A., Knuepfer E., Janha O., Jallow M., Campino S., Macinnis B., Kwiatkowski D. P., and Conway D. J. (2012) Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 8, e1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ochola L. I., Tetteh K. K., Stewart L. B., Riitho V., Marsh K., and Conway D. J. (2010) Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol. Biol. Evol. 27, 2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tetteh K. K., Stewart L. B., Ochola L. I., Amambua-Ngwa A., Thomas A. W., Marsh K., Weedall G. D., and Conway D. J. (2009) Prospective identification of malaria parasite genes under balancing selection. PLoS One 4, e5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crosnier C., Wanaguru M., McDade B., Osier F. H., Marsh K., Rayner J. C., and Wright G. J. (2013) A library of functional recombinant cell surface and secreted Plasmodium falciparum merozoite proteins. Mol. Cell. Proteomics 12, 3976–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. MacKenzie M. R., Creevy N., and Heh M. (1971) The interaction of human IgM and C1q. J. Immunol. 106, 65–68 [PubMed] [Google Scholar]
- 22. Collins C., Tsui F. W., and Shulman M. J. (2002) Differential activation of human and guinea pig complement by pentameric and hexameric IgM. Eur. J. Immunol. 32, 1802–1810 [DOI] [PubMed] [Google Scholar]
- 23. Rudich S. M., Mihaesco E., Winchester R., and Mongini P. K. (1987) Analysis of the domain specificity of various murine anti-human IgM monoclonal antibodies differing in human B lymphocyte signaling activity. Mol. Immunol. 24, 809–820 [DOI] [PubMed] [Google Scholar]
- 24. Ghumra A., Semblat J. P., McIntosh R. S., Raza A., Rasmussen I. B., Braathen R., Johansen F. E., Sandlie I., Mongini P. K., Rowe J. A., and Pleass R. J. (2008) Identification of residues in the Cmu4 domain of polymeric IgM essential for interaction with Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1). J. Immunol. 181, 1988–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Braathen R., Sorensen V., Brandtzaeg P., Sandlie I., and Johansen F. E. (2002) The carboxyl-terminal domains of IgA and IgM direct isotype-specific polymerization and interaction with the polymeric immunoglobulin receptor. J. Biol. Chem. 277, 42755–42762 [DOI] [PubMed] [Google Scholar]
- 26. Sørensen V., Rasmussen I. B., Sundvold V., Michaelsen T. E., and Sandlie I. (2000) Structural requirements for incorporation of J chain into human IgM and IgA. Int. Immunol. 12, 19–27 [DOI] [PubMed] [Google Scholar]
- 27. Iqbal Z., Caccamo M., Turner I., Flicek P., and McVean G. (2012) De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat. Genet. 44, 226–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iqbal Z., Turner I., and McVean G. (2013) High-throughput microbial population genomics using the Cortex variation assembler. Bioinformatics 29, 275–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miotto O., Almagro-Garcia J., Manske M., Macinnis B., Campino S., Rockett K. A., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J. M., Duong S., Nguon C., Chuor C. M., Saunders D., et al. (2013) Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 45, 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mobegi V. A., Duffy C. W., Amambua-Ngwa A., Loua K. M., Laman E., Nwakanma D. C., MacInnis B., Aspeling-Jones H., Murray L., Clark T. G., Kwiatkowski D. P., and Conway D. J. (2014) Genome-wide analysis of selection on the malaria parasite Plasmodium falciparum in West African populations of differing infection endemicity. Mol. Biol. Evol. 31, 1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Okenu D. M., Thomas A. W., and Conway D. J. (2000) Allelic lineages of the merozoite surface protein 3 gene in Plasmodium reichenowi and Plasmodium falciparum. Mol. Biochem. Parasitol. 109, 185–188 [DOI] [PubMed] [Google Scholar]
- 32. Ségurel L., Thompson E. E., Flutre T., Lovstad J., Venkat A., Margulis S. W., Moyse J., Ross S., Gamble K., Sella G., Ober C., and Przeworski M. (2012) The ABO blood group is a trans-species polymorphism in primates. Proc. Natl. Acad. Sci. U.S.A. 109, 18493–18498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taylor T. E., Molyneux M. E., Wirima J. J., Borgstein A., Goldring J. D., and Hommel M. (1992) Intravenous immunoglobulin in the treatment of paediatric cerebral malaria. Clin. Exp. Immunol. 90, 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Douglas A. D., Williams A. R., Illingworth J. J., Kamuyu G., Biswas S., Goodman A. L., Wyllie D. H., Crosnier C., Miura K., Wright G. J., Long C. A., Osier F. H., Marsh K., Turner A. V., Hill A. V., and Draper S. J. (2011) The blood-stage malaria antigen PfRH5 is susceptible to vaccine-inducible cross-strain neutralizing antibody. Nat. Commun. 2, 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patel S. D., Ahouidi A. D., Bei A. K., Dieye T. N., Mboup S., Harrison S. C., and Duraisingh M. T. (2013) Plasmodium falciparum merozoite surface antigen, PfRH5, elicits detectable levels of invasion-inhibiting antibodies in humans. J. Infect. Dis. 208, 1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tran T. M., Ongoiba A., Coursen J., Crosnier C., Diouf A., Huang C. Y., Li S., Doumbo S., Doumtabe D., Kone Y., Bathily A., Dia S., Niangaly M., Dara C., Sangala J., et al. (2014) Naturally acquired antibodies specific for Plasmodium falciparum reticulocyte-binding protein homologue 5 inhibit parasite growth and predict protection from malaria. J. Infect. Dis. 209, 789–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Semblat J. P., Raza A., Kyes S. A., and Rowe J. A. (2006) Identification of Plasmodium falciparum var1CSA and var2CSA domains that bind IgM natural antibodies. Mol. Biochem. Parasitol. 146, 192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barfod L., Dalgaard M. B., Pleman S. T., Ofori M. F., Pleass R. J., and Hviid L. (2011) Evasion of immunity to Plasmodium falciparum malaria by IgM masking of protective IgG epitopes in infected erythrocyte surface-exposed PfEMP1. Proc. Natl. Acad. Sci. U.S.A. 108, 12485–12490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Czajkowsky D. M., Salanti A., Ditlev S. B., Shao Z., Ghumra A., Rowe J. A., and Pleass R. J. (2010) IgM, Fc mu Rs, and malarial immune evasion. J. Immunol. 184, 4597–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tetteh K. K., Osier F. H., Salanti A., Kamuyu G., Drought L., Failly M., Martin C., Marsh K., and Conway D. J. (2013) Analysis of antibodies to newly described Plasmodium falciparum merozoite antigens supports MSPDBL2 as a predicted target of naturally acquired immunity. Infect. Immun. 81, 3835–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sakamoto H., Takeo S., Maier A. G., Sattabongkot J., Cowman A. F., and Tsuboi T. (2012) Antibodies against a Plasmodium falciparum antigen PfMSPDBL1 inhibit merozoite invasion into human erythrocytes. Vaccine 30, 1972–1980 [DOI] [PubMed] [Google Scholar]
- 42. Lin C. S., Uboldi A. D., Epp C., Bujard H., Tsuboi T., Czabotar P. E., and Cowman A. F. (2016) Multiple Plasmodium falciparum merozoite surface protein 1 complexes mediate merozoite binding to human erythrocytes. J. Biol. Chem. 291, 7703–7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin C. S., Uboldi A. D., Marapana D., Czabotar P. E., Epp C., Bujard H., Taylor N. L., Perugini M. A., Hodder A. N., and Cowman A. F. (2014) The merozoite surface protein 1 complex is a platform for binding to human erythrocytes by Plasmodium falciparum. J. Biol. Chem. 289, 25655–25669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rowe J. A., Shafi J., Kai O. K., Marsh K., and Raza A. (2002) Nonimmune IgM, but not IgG binds to the surface of Plasmodium falciparum-infected erythrocytes and correlates with rosetting and severe malaria. Am. J. Trop. Med. Hyg. 66, 692–699 [DOI] [PubMed] [Google Scholar]
- 45. Jeppesen A., Ditlev S. B., Soroka V., Stevenson L., Turner L., Dzikowski R., Hviid L., and Barfod L. (2015) Multiple Plasmodium falciparum erythrocyte membrane protein 1 variants per genome can bind IgM via its Fc fragment Fcμ. Infect. Immun. 83, 3972–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pleass R. J., Moore S. C., Stevenson L., and Hviid L. (2016) Immunoglobulin M: restrainer of inflammation and mediator of immune evasion by Plasmodium falciparum malaria. Trends Parasitol. 32, 108–119 [DOI] [PubMed] [Google Scholar]
- 47. Scholander C., Treutiger C. J., Hultenby K., and Wahlgren M. (1996) Novel fibrillar structure confers adhesive property to malaria-infected erythrocytes. Nat. Med. 2, 204–208 [DOI] [PubMed] [Google Scholar]
- 48. Rasti N., Namusoke F., Chêne A., Chen Q., Staalsoe T., Klinkert M. Q., Mirembe F., Kironde F., and Wahlgren M. (2006) Nonimmune immunoglobulin binding and multiple adhesion characterize Plasmodium falciparum-infected erythrocytes of placental origin. Proc. Natl. Acad. Sci. U.S.A. 103, 13795–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Akhouri R. R., Goel S., Furusho H., Skoglund U., and Wahlgren M. (2016) Architecture of human IgM in complex with P. falciparum erythrocyte membrane protein 1. Cell Reports 14, 723–736 [DOI] [PubMed] [Google Scholar]
- 50. Stevenson L., Laursen E., Cowan G. J., Bandoh B., Barfod L., Cavanagh D. R., Andersen G. R., and Hviid L. (2015) α2-Macroglobulin can crosslink multiple Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) molecules and may facilitate adhesion of parasitized erythrocytes. PLoS Pathogens 11, e1005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Durocher Y., Perret S., and Kamen A. (2002) High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ling I. T., Ogun S. A., and Holder A. A. (1994) Immunization against malaria with a recombinant protein. Parasite Immunol. 16, 63–67 [DOI] [PubMed] [Google Scholar]
- 53. Crosnier C., Staudt N., and Wright G. J. (2010) A rapid and scalable method for selecting recombinant mouse monoclonal antibodies. BMC Biol. 8, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brown M. H., and Barclay A. N. (1994) Expression of immunoglobulin and scavenger receptor superfamily domains as chimeric proteins with domains 3 and 4 of CD4 for ligand analysis. Protein Eng. 7, 515–521 [DOI] [PubMed] [Google Scholar]
- 55. Bushell K. M., Söllner C., Schuster-Boeckler B., Bateman A., and Wright G. J. (2008) Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 18, 622–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun Y., Gallagher-Jones M., Barker C., and Wright G. J. (2012) A benchmarked protein microarray-based platform for the identification of novel low-affinity extracellular protein interactions. Anal. Biochem. 424, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bartholdson S. J., Bustamante L. Y., Crosnier C., Johnson S., Lea S., Rayner J. C., and Wright G. J. (2012) Semaphorin-7A is an erythrocyte receptor for P. falciparum merozoite-specific TRAP homolog, MTRAP. PLoS Pathogens 8, e1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kerr J. S., and Wright G. J. (2012) Avidity-based extracellular interaction screening (AVEXIS) for the scalable detection of low-affinity extracellular receptor-ligand interactions. J. Vis. Exp. 61, e3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Trager W., and Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 60. Duraisingh M. T., Triglia T., and Cowman A. F. (2002) Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int. J. Parasitol. 32, 81–89 [DOI] [PubMed] [Google Scholar]
- 61. Gerold P., Schofield L., Blackman M. J., Holder A. A., and Schwarz R. T. (1996) Structural analysis of the glycosyl-phosphatidylinositol membrane anchor of the merozoite surface proteins-1 and -2 of Plasmodium falciparum. Mol. Biochem. Parasitol 75, 131–143 [DOI] [PubMed] [Google Scholar]
- 62. Tokuyasu K. (1997) Immunocytochemistry on ultrathin cryosections. In Cells: A Laboratory Manual: Subcellular Localization of Genes and Their Products (Specer D. L., and Leinwand G. R., eds) pp 131.131–131.127, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 63. Crosnier C., Bustamante L. Y., Bartholdson S. J., Bei A. K., Theron M., Uchikawa M., Mboup S., Ndir O., Kwiatkowski D. P., Duraisingh M. T., Rayner J. C., and Wright G. J. (2011) Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature 480, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.