Abstract
Mutations in the c10orf2 gene encoding the human mitochondrial DNA replicative helicase Twinkle are linked to several rare genetic diseases characterized by mitochondrial defects. In this study, we have examined the catalytic activity of Twinkle helicase on model replication fork and DNA repair structures. Although Twinkle behaves as a traditional 5′ to 3′ helicase on conventional forked duplex substrates, the enzyme efficiently dissociates D-loop DNA substrates irrespective of whether it possesses a 5′ or 3′ single-stranded tailed invading strand. In contrast, we report for the first time that Twinkle branch-migrates an open-ended mobile three-stranded DNA structure with a strong 5′ to 3′ directionality preference. To determine how well Twinkle handles potential roadblocks to mtDNA replication, we tested the ability of the helicase to unwind substrates with site-specific oxidative DNA lesions or bound by the mitochondrial transcription factor A. Twinkle helicase is inhibited by DNA damage in a unique manner that is dependent on the type of oxidative lesion and the strand in which it resides. Novel single molecule FRET binding and unwinding assays show an interaction of the excluded strand with Twinkle as well as events corresponding to stepwise unwinding and annealing. TFAM inhibits Twinkle unwinding, suggesting other replisome proteins may be required for efficient removal. These studies shed new insight on the catalytic functions of Twinkle on the key DNA structures it would encounter during replication or possibly repair of the mitochondrial genome and how well it tolerates potential roadblocks to DNA unwinding.
Keywords: DNA helicase, DNA repair, DNA replication, genetic disease, mitochondria, Twinkle, branch migration, helicase
Introduction
The gene product of c10orf2, also known as Twinkle, is a DNA helicase required for the replication of human mitochondrial DNA (mtDNA) (1). Mutations in the c10orf2 gene encoding Twinkle helicase lead to mitochondrial deletions in post-mitotic tissues and are responsible for a number of hereditary disorders, including adult-onset progressive external ophthalmoplegia, hepatocerebral syndrome with mtDNA depletion syndrome, and infantile-onset spinocerebellar ataxia (2). Twinkle helicase collaborates with DNA polymerase γ (pol γA)2 and its associated processivity factor (pol γB) and the mitochondrial single-stranded DNA-binding protein as the minimally reconstituted mtDNA replisome (3). Although there has been much debate and interest in the mechanism(s) underlying mtDNA synthesis, it is generally thought, based on experimental evidence, that strand displacement DNA synthesis occurs for both the light and heavy guanine-rich strands of the circular double-stranded mitochondrial genome (4). Because there are two origins of replication for the heavy and light strands, a three-stranded displacement loop structure known as the D-loop was created. Displacement of DNA synthesis by pol γ is stimulated by the DNA unwinding activity catalyzed by Twinkle helicase.
A number of biophysical and biochemical studies have provided insight into the molecular and cellular functions of the mitochondrial replicative Twinkle DNA helicase. Twinkle is a member of the superfamily 4 DNA helicases, which includes the Escherichia coli DnaB replicative helicase as well as the bacteriophage T7 gene 4 helicase (5, 6). These helicases are known to form ring-like structures (7), and Twinkle's stoichiometry is that of a 6-subunit or 7-subunit architecture that can be modulated by solution components (8, 9). Twinkle catalyzes unwinding of duplex DNA with a 5′ to 3′ directionality fueled by the hydrolysis of nucleoside triphosphate (10–12). Twinkle can load onto single-stranded DNA circles (10, 12) or double-stranded DNA bubble structures (10), and it binds both single-stranded and double-stranded DNA (12). Twinkle anneals complementary single-stranded DNA (12), but the physiological significance of this biochemical activity remains to be understood. Although Twinkle plays a key role in separating strands at the mitochondrial replication fork, the enzyme is weakly active on G-quadruplex DNA structures (13), which are likely to form in the guanine-rich strand of the mitochondrial genome (13, 14). Presumably other proteins, including auxiliary DNA helicases (PIF1, DNA2, SUV3, and RECQ4) (15), may either assist or substitute for Twinkle in dealing with certain alternative DNA structures.
Despite the wealth of information that has been gained from molecular studies of Twinkle helicase, its precise role(s) in mtDNA replication remain to be fully characterized. Moreover, it is unclear whether Twinkle plays any role in mtDNA repair. In this work, Twinkle was tested on a series of DNA structures associated with the replication fork or DNA repair to ascertain the enzyme's substrate specificity. For the first time, we report Twinkle's activity on three-stranded DNA structures, including mobile and immobile D-loop substrates, as well open mobile three-stranded DNA structures designed to assay for putative branch migration activity of Twinkle and its directionality. In a second line of inquiry, we determined the effects of covalent oxidative DNA lesions or noncovalent protein-DNA complexes, both of which represent potential impediments to mitochondrial DNA metabolic processes, on Twinkle helicase activity. The results from these studies shed new insight on Twinkle's catalytic functions on key DNA structures it would encounter during the replication of the mitochondrial genome and potentially other processes, including DNA repair in the organelle.
Results
Twinkle Unwinding and Binding Activity on Replication Fork-associated DNA Structures
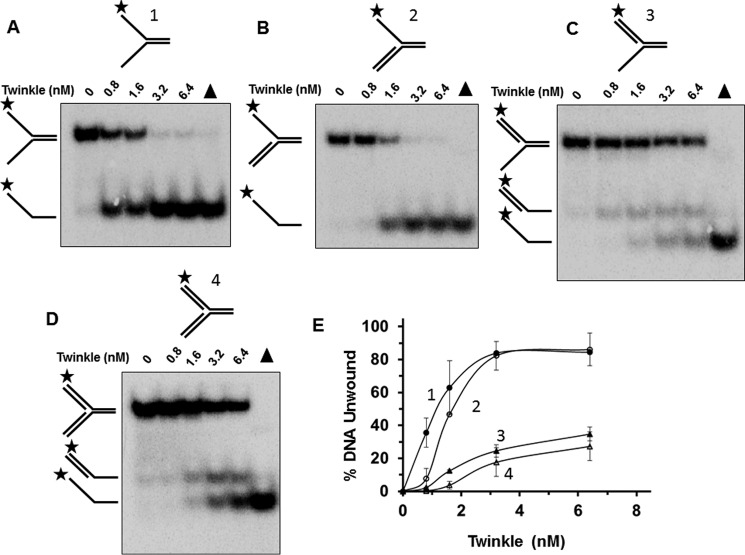
Although Twinkle DNA helicase is an essential player in mtDNA replication (1), its preferential activity on replication fork-associated structures has not been extensively examined. Therefore, we performed Twinkle protein titrations on a series of sequence-related DNA substrates that resemble DNA replication intermediates. Twinkle most efficiently unwound a forked duplex substrate with naked 5′ and 3′ single-stranded arms (substrate 1) (Fig. 1, A and E). A 5′ flap substrate representing an intermediate of strand displacement synthesis at the replication fork (substrate 2) was also unwound by Twinkle (Fig. 1B), but not to quite the same extent as the naked fork, particularly evident at a Twinkle concentration of 0.8 nm hexamer in which there was ∼7-fold difference in percent substrate unwound (Fig. 1E). Notably, Twinkle unwound the downstream 5′ flap oligonucleotide of the 5′ flap DNA substrate (Fig. 1B), consistent with its 5′ to 3′ directionality of unwinding (10, 12). Conversely, Twinkle showed relatively poor activity on a 3′ flap substrate (substrate 3) that lacks a pre-existing 5′ single-stranded tail (Fig. 1, C and E). In the case of the 3′ flap substrate, two minor radiolabeled products were observed, 3′ single strand tailed duplex and the single strand species (Fig. 1, C and E). Twinkle helicase activity on the synthetic replication fork structure with both 5′ and 3′ double-stranded arms (substrate 4) generated the same radiolabeled products as that for the 3′ flap substrate (Fig. 1D); unwound substrate was ∼4-fold less than that observed for the 5′ flap substrate at 1.6 nm Twinkle hexamer concentration (Fig. 1E), suggesting that the double strand character of the displaced strand negatively affects Twinkle unwinding.
FIGURE 1.
Twinkle unwinding activity on DNA replication structures. A–D, purified recombinant Twinkle helicase, designated in hexamer concentration, was incubated with the indicated replication fork DNA substrates 1–4 (0.5 nm) at 37 °C for 30 min as described under “Experimental Procedures.” Proteinase K-digested reaction products were resolved on non-denaturing 12% polyacrylamide gels. Representative gel images from at least three independent experiments are shown. Filled triangle, heat-denatured DNA substrate control. E, quantitative analysis of percent DNA substrate unwound from A to D. Fork with single-stranded 5′ and 3′ arms, filled circle; fork with 5′ single-stranded arm, open circle; fork with 3′ single strand arm, filled triangle; fork with double-stranded 5′ and 3′ arms, open triangle. Average values of at least three independent experiments with standard deviations indicated by error bars are shown.
Electrophoretic mobility shift assays (EMSA) were performed with the same replication fork-associated DNA substrates tested for helicase activity to evaluate DNA binding by Twinkle (Fig. 2). Here it was observed that DNA substrates with pre-existing single-stranded tails (naked fork (substrate 1), 3′ flap (substrate 3), 5′ flap (substrate 2)) were bound to a greater extent than the synthetic replication fork structure. A small fraction of the double-stranded replication fork substrate (substrate 4) was detectably bound by Twinkle at helicase protein concentrations of 3.2 nm or 6.4 nm. Interestingly, some binding of the 5′ flap or 3′ flap substrates was detected at 1.6 nm Twinkle, whereas this was not reproducibly observed for the forked duplex with both 5′ and 3′ single-stranded arms. The previously reported preferential binding of Twinkle to double-stranded DNA over single-stranded DNA (12) may contribute to the apparent difference; however, the poor binding of Twinkle to the synthetic replication fork suggests that single strand character in the fork substrate contributes to stable binding by Twinkle as detected by EMSA. These results suggest that for the replication fork structures, Twinkle preferentially binds those substrates with pre-existing single strand character; however, the preferential binding to 3′ flap substrate is not reflected by enhanced Twinkle helicase activity on that substrate.
FIGURE 2.
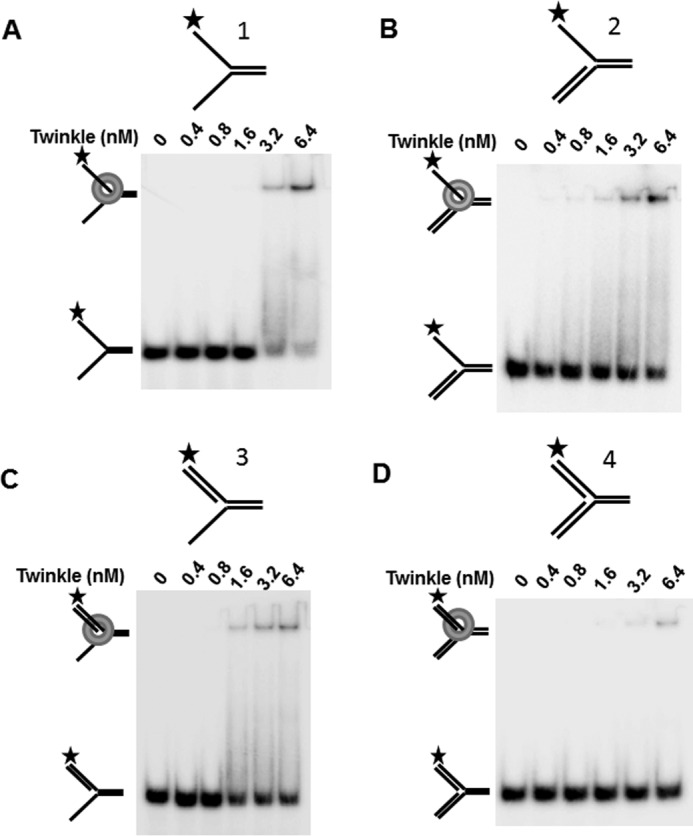
Twinkle DNA binding to replication fork structures. A–D, indicated concentration of Twinkle hexamer was incubated with the indicated replication fork DNA substrates 1–4 (0.5 nm) at 37 °C for 30 min as described under “Experimental Procedures.” DNA species from binding mixtures were resolved on non-denaturing 5% polyacrylamide gels. Representative images from EMSA of at least three independent experiments are shown.
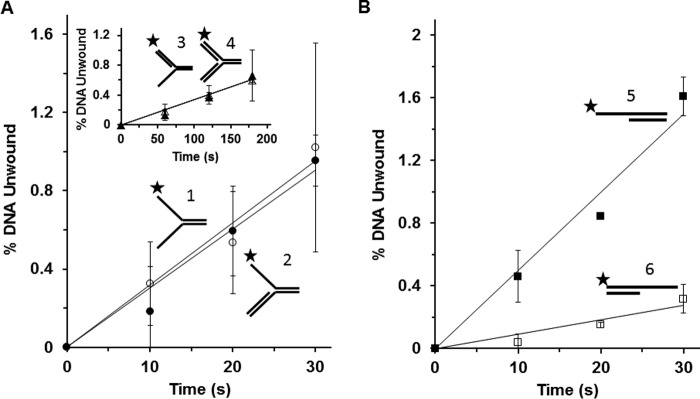
The observed differences in Twinkle helicase activity on the replication fork-associated DNA structures under multi-turnover conditions led us to perform experiments in which Twinkle was tested on these same substrates under single-turnover conditions. Under single-turnover conditions, Twinkle is incubated with a greater concentration of radiolabeled DNA substrate (2.5 nm) compared with the concentration of DNA substrate (0.5 nm) used for multi-turnover conditions; moreover, after pre-incubation of radiolabeled DNA substrate with Twinkle helicase, the reaction is initiated with the simultaneous addition of ATP and a large (100-fold) excess of unlabeled dT200 to limit any loading of Twinkle helicase molecules onto the radiolabeled substrate during the course of the incubation period. In control experiments, a 100-fold excess of dT200 in the helicase reaction resulted in a 95% reduction in Twinkle helicase activity on the forked duplex (substrate 1). Under these conditions, Twinkle helicase activity was determined as a function of time to obtain rates of DNA unwinding on the various substrates. As shown in Fig. 3A, Twinkle unwound the forked duplex substrate with 5′ and 3′ single-stranded arms (substrate 1) at a similar rate compared with the 5′ flap substrate (substrate 2), which were 10-fold greater than the 3′ flap substrate (substrate 3) or synthetic replication fork structure (substrate 4) (Fig. 3A, inset). The reduced but detectable unwinding of DNA substrates lacking a pre-existing 5′ single-stranded arm (synthetic replication fork or 3′ flap substrate) led us to examine Twinkle activity on simple 5′ tailed or 3′ tailed partial duplex substrates under single-turnover conditions. Here we observed that Twinkle unwound the 5′ tailed duplex substrate (substrate 5) at ∼5-fold greater rate than the 3′ tailed duplex (substrate 6) (Fig. 3B), consistent with its preferential 5′ to 3′ directionality of unwinding under multi-turnover conditions (10, 12).
FIGURE 3.
Single-turnover kinetics of Twinkle helicase activity on replication fork structures. Twinkle hexamer (3.2 nm) was pre-incubated with the indicated radiolabeled DNA substrate 1–6 (2.5 nm) prior to simultaneous addition of ATP and 100-fold excess of dT200, followed by incubation at specified time points as described under “Experimental Procedures” for single-turnover kinetic assays. Reaction products were resolved on native 12% polymacrylamide gels and analyzed. A, Twinkle (3.2 nm hexamer) unwinding kinetics on forked duplex substrate with single-stranded 5′ and 3′ arms (substrate 1) (filled circle) or 5′ flap substrate (substrate 2) (open circle). Inset, 3′ flap substrate (substrate 3) (filled triangle) or synthetic replication fork with duplex leading and lagging strand arms (substrate 4) (open triangle). B, Twinkle (6.4 nm hexamer) unwinding kinetics on 5′ single strand tailed duplex (substrate 5) (filled square) or 3′ single strand tailed duplex (substrate 6) (open square). Average values of at least three independent experiments with standard deviations indicated by error bars are shown.
Twinkle Catalytically Acts upon D-loop DNA Structures
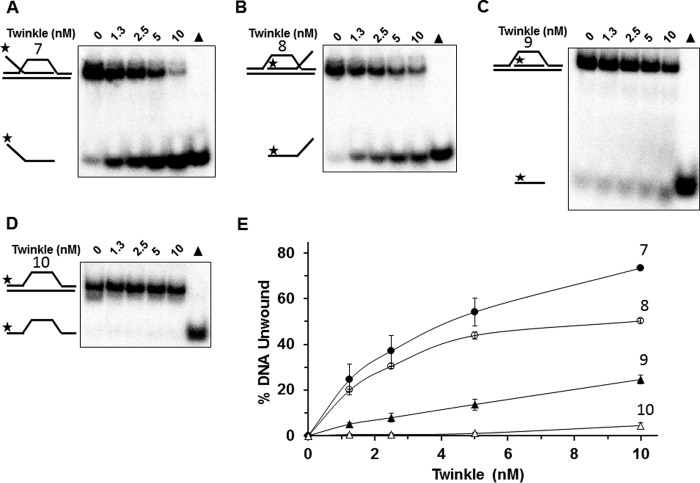
Although the precise mechanism of mtDNA replication is still debated, it is generally believed that a D-loop structure allows for effective and efficient replication of the heavy and light strands (16). In addition, the D-loop substrate represents a key early intermediate of homologous recombination (HR) repair (17). Thus, it is probable that Twinkle would encounter D-loop structures in the mitochondrial genome. However, up to this point, the catalytic activity of Twinkle has not been tested on D-loop structures. To address this, initial experiments were performed in which increasing concentrations of Twinkle were incubated with immobile three-stranded D-loop DNA substrates with a fixed noncomplementary 5′ ssDNA tail (substrate 7), 3′ ssDNA tail (substrate 8), or no tail altogether (substrate 9) as well as two-stranded bubble DNA substrate (substrate 10) for comparison. Twinkle was able to unwind the invading third strand of all three immobile D-loop DNA substrates (Fig. 4, A–C); however, Twinkle preferentially unwound the 5′ tailed immobile D-loop substrate followed closely by the 3′ tailed immobile D-loop substrate, and less efficiently by the no tail immobile D-loop substrate (Fig. 4E). The bubble DNA substrate was unwound very poorly by Twinkle throughout the protein titration (Fig. 4, D and E). Based on these results, we conclude that Twinkle is able to unwind immobile D-loop DNA substrates, with a preference toward those substrates that bear a noncomplementary 5′ or 3′ single-stranded tail as a component of the invading third strand.
FIGURE 4.
Twinkle helicase activity on immobile D-loop DNA substrates. A–D, indicated concentrations of Twinkle helicase were incubated with the specified immobile D-loop (substrates 7–9) or bubble (substrate 10) DNA substrates (0.5 nm) at 37 °C for 30 min as described under “Experimental Procedures.” Reaction products were resolved on non-denaturing 12% polyacrylamide gels. Representative gel images from at least three independent experiments are shown. E, quantitative analysis of percent DNA substrate unwound from A to D. Immobile D-loop with single-stranded 5′ tail (substrate 7), filled circle; immobile D-loop with single-stranded 3′ tail (substrate 8), open circle; immobile D-loop with flush invading strand (substrate 9), filled triangle; bubble DNA substrate (substrate 10), open triangle. Average values of at least three independent experiments with standard deviations indicated by error bars are shown.
We next sought to assess if Twinkle was capable of branch-migrating mobile three-stranded DNA substrates, because such structures would be physiologically relevant to its role during replication of the circular mitochondrial genome or potentially HR repair of mitochondrial DNA double strand breaks. We first tested Twinkle to branch-migrate mobile three-way junction DNA substrates that are designed to assess if the enzyme branch-migrates in a 5′ to 3′ (substrate 11) or 3′ to 5′ (substrate 13) direction. These substrates were previously used to characterize the 3′ to 5′ branch migration directionality catalyzed by the human recombinant RECQ1 helicase (18). We observed that the Twinkle enzyme preferentially acts to branch-migrate in the 5′ to 3′ direction (Fig. 5, A and C); however, a very low level of 3′ to 5′ branch migration was observed throughout the Twinkle titration (Fig. 5, B and C). Twinkle branch migration activity is dependent on ATP hydrolysis (Fig. 5D).
FIGURE 5.
Twinkle helicase promotes branch migration of mobile three-way junction DNA structure preferentially in the 5′ to 3′ direction. A and B, indicated concentrations of Twinkle helicase hexamer were incubated with the specified DNA substrate (0.5 nm) at 37 °C for 30 min as described under “Experimental Procedures.” Reaction products were resolved on non-denaturing 12% polyacrylamide gels. Representative gel images from at least three independent experiments are shown. C, quantitative analysis of percent branch migration product in the 5′ to 3′ direction (substrate 11) (A, filled circle) or 3′ to 5′ direction (substrate 13) (B, open circle). Average values of at least three independent experiments with standard deviations indicated by error bars are shown. D, Twinkle-catalyzed branch migration is dependent on hydrolyzable ATP. Reaction mixtures containing 5 nm Twinkle and the 5′ to 3′ mobile three-way junction DNA substrate (substrate 11) (0.5 nm, A) were incubated and analyzed as described above. E, indicated concentrations of Twinkle helicase hexamer were incubated with a forked duplex DNA substrate (substrate 12) (0.5 nm) at 37 °C for 30 min as described under “Experimental Procedures.” The third oligonucleotide added to the reaction mixture requires helicase-catalyzed unwinding of the duplex region to anneal to one of the two unwound strands. Reaction products were resolved on non-denaturing 12% polyacrylamide gels. Representative gel images from at least three independent experiments are shown. F, quantitative analysis of percent helicase product. Average values of at least three independent experiments with standard deviations indicated by error bars are shown.
To address the relative contributions of Twinkle branch migration versus helicase activity on the branch-migrate mobile three-way junction DNA substrate, we tested for Twinkle helicase activity on a forked duplex DNA substrate (substrate 12) that was characterized by the same 63 base pair duplex found in the three-way junction substrate (substrate 11). Twinkle was able to unwind the forked duplex substrate, but not quite to the same extent as its branch migration activity on substrate 11 (Fig. 5, E and F), suggesting that both Twinkle branch migration and helicase activities may contribute to disruption of the three-way junction substrates used in the current study.
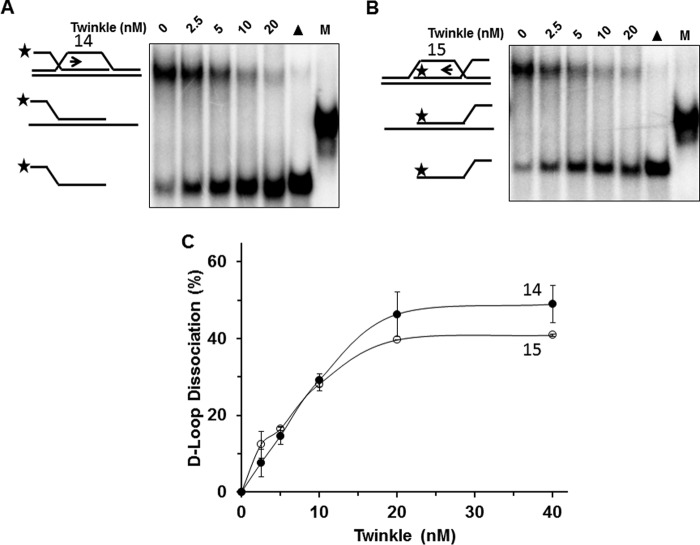
We then examined the ability of Twinkle to promote dissociation of mobile D-loop DNA substrates, which were previously shown to be acted upon by RECQ1 (18). In contrast to what was observed for the mobile three-way junction DNA substrates, Twinkle displayed a similar level of activity on the mobile D-loop substrates designed to test for 5′ to 3′ directionality (substrate 14) (Fig. 6, A and C) or 3′ to 5′ directionality (substrate 15) (Fig. 6, B and C). The difference in results between the two types of mobile DNA substrates (three-way junction versus D-loop) suggests that Twinkle recognizes and loads on to the mobile D-loop substrate in a manner distinct from the mobile three-way junction substrate. Moreover, the ability of Twinkle to efficiently unwind the invading strand with a 5′ or 3′ single-stranded tail in the immobile D-loop DNA substrates suggests that Twinkle recognizes the D-loop substrate irrespective of the third strand polarity and its helicase activity may contribute to dissociation of the mobile D-loop substrate.
FIGURE 6.
Twinkle helicase dissociates mobile D-loop DNA structures. A and B, indicated Twinkle helicase hexamer concentrations were incubated with the specified mobile D-loop DNA substrate (0.5 nm) at 37 °C for 30 min as described under “Experimental Procedures.” Reaction products were resolved on non-denaturing 12% polyacrylamide gels. Representative gel images from at least three independent experiments are shown. C, quantitative analysis of percent D-loop dissociation in the 5′ to 3′ direction (substrate 14) or 3′ to 5′ direction (substrate 15) (A, filled circle (substrate 14); B, open circle (substrate 15)). Average values of at least three independent experiments with standard deviations indicated by error bars are shown.
Effects of Oxidative DNA Lesions on Twinkle Helicase Activity
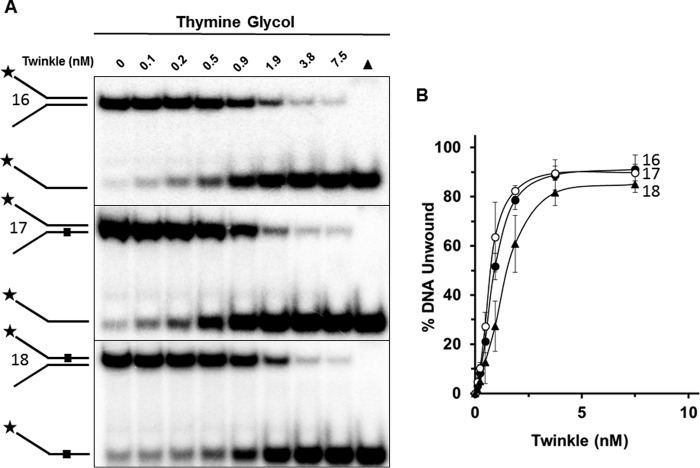
As the energy powerhouse organelles of the cell, mitochondria produce oxygen radicals at a significant rate as byproducts of oxidative phosphorylation and the electron transport chain (19). Persistence of these free radicals within the mitochondrial matrix can cause a variety of oxidative DNA lesions as well as damage to other macromolecules. A prominent form of oxidative DNA damage is Tg, which causes localized distortion of the DNA double helix (20) and blocks synthesis by a number of DNA polymerases (21). Given that Twinkle is likely to encounter Tg lesions during mtDNA replication, we set out to examine the effect of a single Tg residing in either the helicase-translocating or non-translocating strand within the double-stranded portion of a forked duplex DNA substrate that Twinkle efficiently unwinds. As shown in Fig. 7, Twinkle unwound the undamaged DNA substrate (substrate 16) and damaged DNA substrates in a protein concentration dependent manner; however, there were some apparent differences in the percent substrate unwound. Tg in the helicase translocating strand (substrate 18) inhibited Twinkle helicase activity by a maximal 2-fold at a Twinkle hexamer concentration of 0.9 nm, whereas there was no statistically significant effect of the non-translocating strand Tg (substrate 17) on Twinkle helicase activity throughout the protein titration.
FIGURE 7.
Twinkle is sensitive to a single thymine glycol in the helicase translocating strand. A, indicated Twinkle helicase hexamer concentrations were incubated with the specified thymine glycol containing DNA substrates (0.5 nm) at 37 °C for 30 min as described under “Experimental Procedures.” Reaction products were resolved on non-denaturing 12% polyacrylamide gels. Representative gel images from at least three independent experiments are shown. B, quantitative analysis of percent DNA substrate unwound from A. Undamaged substrate (substrate 16), filled circle; bottom (non-translocating) strand thymine glycol substrate (substrate 17), open circle; top (translocating) strand thymine glycol substrate (substrate 18), filled triangle. Average values of at least three independent experiments with standard deviations indicated by error bars are shown.
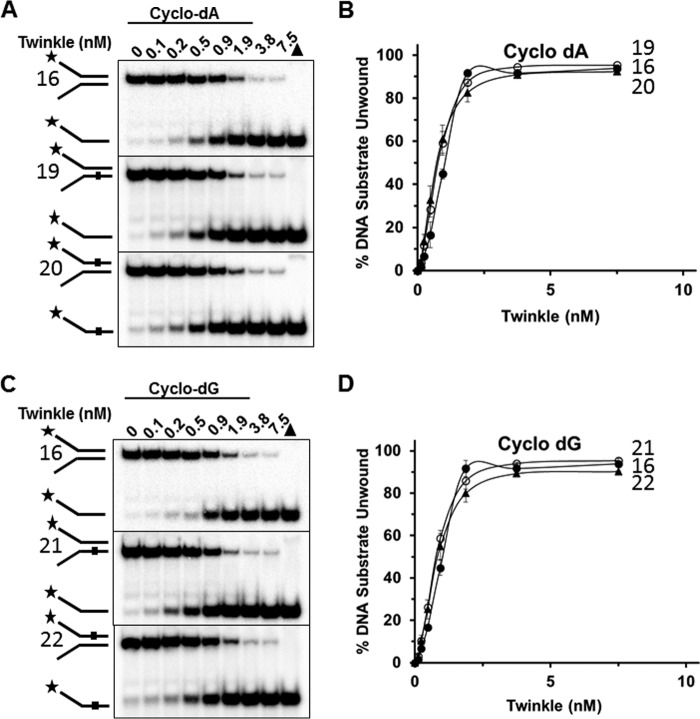
We then tested Twinkle helicase on a forked duplex harboring a cPu adduct, which distorts normal B-form double helical DNA by altering normal base stacking and helical twist (22, 23). Unlike the results from the experiments with the Tg forked duplex DNA substrates, there was no inhibitory effect of the either cyclo-dA (Fig. 8, A and B) or cyclo-dG (Fig. 8, C and D) in the translocating strand (substrate 20, substrate 22) or non-translocating strand (substrate 19, substrate 21), for that matter.
FIGURE 8.
Twinkle tolerates a single cyclopurine in the helicase translocating or non-translocating strands. A and C, indicated Twinkle helicase hexamer concentrations were incubated with the specified cyclo-dA (A) containing DNA substrates (substrates 19, 20) or cyclo-dG (C) containing DNA substrates (substrates 21 and 22) (0.5 nm) at 37 °C for 30 min as described under “Experimental Procedures.” Reaction products were resolved on non-denaturing 12% polyacrylamide gels. Representative gel images from at least three independent experiments are shown. B and D, quantitative analysis of percent DNA substrate unwound from A and C, respectively. Average values of at least three independent experiments with standard deviations indicated by error bars are shown. Undamaged substrate (substrate 16), filled circle; bottom (non-translocating) strand cPu substrate (substrates 19, 21), open circle; top (translocating) strand cPu substrate (substrates 20, 22), filled triangle.
Twinkle Dynamically Interacts with Both DNA Strands
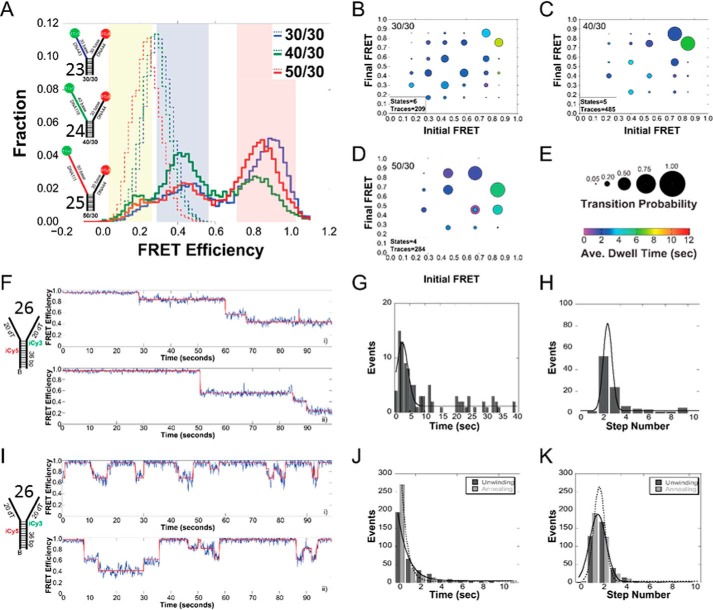
To test for a direct interaction between the helicase and the nontranslocating strand of the model DNA fork, single molecule FRET (smFRET) experiments were performed as described previously (24, 25). Three FRET-labeled DNA fork substrates (30/30 (substrate 23), 40/30 (substrate 24), 50/30 (substrate 25)) were tested that had 30 bases (dT) on the 5′ arm of the fork where the helicase loads, and increasing 30, 40, and 50 bases (dT) on the 3′ excluded arm of the fork. Histograms for the immobilized DNA forks alone produced a single stable low-FRET peak corresponding to the termini of the fork arms not being in close proximity (Fig. 9A). Increasingly longer 3′ fork arms correspond to lower FRET signals as expected.
FIGURE 9.
Histograms and ExPRT plots of Twinkle bound to DNA forks. A, histogram of the FRET signals from the 30/30 DNA fork substrate (substrate 23) (blue), 40/30 DNA fork substrate (substrate 24) (green), and 50/30 DNA fork substrate (substrate 25) (red), DNA fork substrates alone (dashed lines) and after addition of 25 nm Twinkle (solid lines). The length of the 5′-translocating strand was kept constant at 30 (dT) nucleotides, and the length of the 3′ excluded strand was varied from 30 to 50 (dT) nucleotides. Yellow, blue, and red regions indicate low, medium, and high FRET states, respectively. ExPRT plots for Twinkle bound to the 30/30 (B), 40/30 (C), and 50/30 (D) DNA forks. Each marker on the ExPRT plot represents a transition from the initial FRET state on the x axis to the final FRET state on the y axis. E, size of the marker corresponds to the fraction of analyzed traces that exhibit that particular transition (transition probability), and the color represents the dwell time (seconds) of the initial state. Representative traces from smFRET experiments showing undirectional unwinding (F) or alternating unwinding and reannealing (I) for substrate 26. The calculated FRET signal is shown in blue, and the fit to ideal states is overlaid in red. A global quantification of the unwinding time or number of steps for unidirectional unwinding (G and H) or alternating unwinding and reannealing (J and K). For multistate smFRET traces, the individual unwinding (dark gray) and annealing (light gray) times and steps are quantified individually. The lines correspond to fits to a Gaussian equation.
When Twinkle was added to each of these forks, there was a concomitant shift to higher FRET states for all three substrates in the histogram (Fig. 9A). On the 30/30 substrate, Twinkle's interaction with the fork results in a predominantly high-FRET population with additional smaller medium FRET populations. The 30/50 fork produced a nearly identical histogram, but the 30/40 substrate had larger low- and medium-FRET populations with a smaller high-FRET population. In the case of all three substrates, we see a significant high-FRET population, which has previously been shown to correspond to a wrapping interaction where the excluded strand wraps around the outer surface of the helicase (24, 25).3 The presence of multiple populations within the histogram data indicates that there are multiple FRET states sampled by each model fork substrate upon Twinkle binding. However, the histogram alone does not reveal any information concerning possible dynamic transitioning between these states.
To probe the dynamics of the interaction, we generated ExPRT plots (Fig. 9, B–D) for each data set. The novel ExPRT analysis quantifies all unique transitions, explicit probabilities, and dwell times on a single plot for all smFRET traces containing two or more transitions. Each marker represents a single transition from an initial FRET state on the x axis to a final FRET state on the y axis. The size of the marker corresponds to the fraction of traces analyzed that exhibited that specific transition, and the color of the marker corresponds to the average dwell time spent in the initial state before the transition (Fig. 9E). The number of FRET states decreases (6 to 5 to 4) as the length of the excluded strand increases, correlating with greater stabilization of the excluded strand interaction along its binding path(s). Despite this difference, there are also similarities among the three ExPRT plots. For example, there is significantly greater stability of the excluded strand interaction corresponding to the highest FRET state in each ExPRT plot as the length of the 3′ strand increases. Markers indicating the highest initial FRET state have longer dwell times, mostly between 5 and 8 s. All other markers representing transitions between alternative FRET states, even those that return to the highest FRET state, have shorter dwell times between 1 and 4 s. Another similarity exists between the 30/30 (Fig. 9B) and 40/30 (Fig. 9C) datasets in which the largest markers, which are the most commonly seen transitions, occur between adjacent FRET states. This is consistent with multiple points of contact between the excluded strand and the exterior surface of the helicase, where in most cases there is a “stepping” in the disruption or formation of these contacts such that most transitions are occurring between adjacent states. Transitions occurring between neighboring states are indicative of wrapping the excluded strand around the hexamer to form the most stably bound high FRET state. The “peeling off” of this strand, where adjacent contacts are disrupted one at a time, leads to the less stable lower FRET states. The ExPRT plot for Twinkle on the 50/30 fork (Fig. 9D) shows that the highest initial FRET state can transition into either the second or third highest final FRET states with roughly equal probability. This may be a consequence of the longer excluded strand arm of the fork being able to occupy distinct pathways on the external surface of the helicase.
DNA Unwinding and Reannealing by Twinkle Measured by smFRET
To directly monitor the specific DNA unwinding and reannealing of DNA by Twinkle, we again utilized smFRET. The biotinylated fork DNA substrate was altered to include internal Cy3 and Cy5 dyes 6 bp apart in the duplex region adjacent to the fork arms (substrate 26). Upon addition of ATP, Twinkle displayed single molecule FRET traces consistent with either alternating unwinding/reannealing or unidirectional stepwise unwinding at a 2:1 ratio, respectively, over the acquisition time (100 s) (Fig. 9, F and I). It should be noted that the time frame of our experiments is limited by the lifetime of the fluorescent dyes and that events that take place on the time scale of minutes are lost. Therefore, if an unwinding event takes tens of seconds to occur, a subsequent rewinding event would likely not be captured. The traces shown in Fig. 9, F and I, also demonstrate that Twinkle does not always unwind the fork substrate completely. This could be a result of the biotin-streptavidin junction at the base of the duplex that could act as a block to the helicase as shown previously for the NS3 helicase (27) and the T7 gp41–61 helicase-primase (28).
To be certain that stepwise changes in FRET corresponded to the action of Twinkle, smFRET experiments were compared using ATP or the nonhydrolyzable analog AMP-PNP. Upon addition of AMP-PNP, ∼2.5 and 4% of observed molecules display clear unwinding/reannealing and unwinding events, respectively. When ATP is added, ∼17 and 8% of the molecules display clear unwinding/reannealing and unwinding events, respectively. These results are consistent with a previous bulk study that showed Twinkle annealing activity can take place in the presence of a non-hydrolyzable nucleotide analogue or in the absence of nucleotide, but rates of annealing increased when a hydrolyzable nucleotide was added (12).
Using smFRET, we can quantify the steps taken by Twinkle for unwinding or reannealing of the fork as well as the time it takes for these events to occur. A quantification of the unwinding or reannealing times for all traces shows that the mean unwinding time is 4-fold greater when unidirectionally unwinding than when molecules are alternating between unwinding and reannealing (Fig. 9, G versus J). Unidirectional unwinding also tended to have a greater number of steps (2.3 ± 0.7) for the progression on our fork substrate compared with alternating unwinding/reannealing events where the average step numbers were 1.7 ± 1.0 and 1.4 ± 0.8, respectively, for each. The cumulative smFRET data suggest that Twinkle on its own may occasionally release its grip on the translocating strand and allow the DNA to reanneal. However, maintaining contact with the excluded strand or reengagement on the translocating strand can reverse the regression to reactivate unwinding in a forward direction.
Effect of Protein-DNA Complexes on Twinkle Unwinding
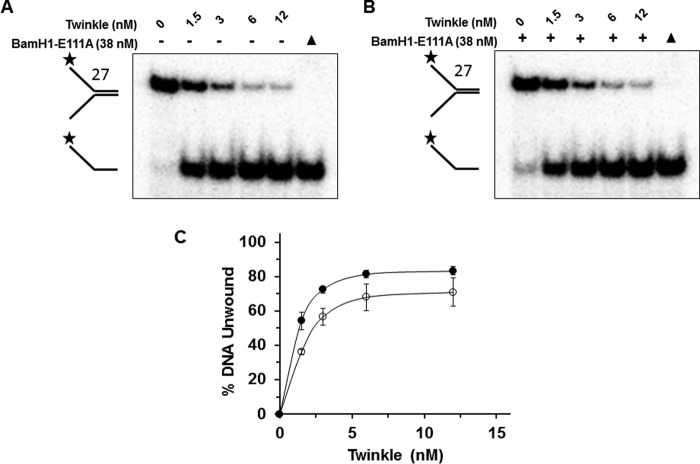
Recent experimental results from biochemical and biological assays suggest that certain DNA helicases may use their motor ATPase function to displace proteins bound to DNA (29). Although the mitochondrial genome is devoid of histones, the mitochondrial transcription factor A (TFAM) is highly abundant, and it is estimated that the pool of TFAM protein would be sufficient to entirely coat the double-stranded DNA of the mitochondrial genome (30). TFAM is known to play an important role in transcriptional regulation and mtDNA organization (31), so we thought to test whether Twinkle possesses the ability to displace proteins such as TFAM bound to duplex DNA. Initially, we tested whether the catalytically inactive BamHI-E111A restriction endonuclease bound to forked duplex substrate with the cognate palindromic BamHI recognition sequence (substrate 27) could inhibit Twinkle-catalyzed unwinding of the DNA substrate. Previously, we had shown that BamHI-E111A bound to the forked duplex substrate potently blocks DNA unwinding of the substrate by the 5′ to 3′ FANCJ helicase (32). Using a saturating concentration of BamHI-E111A (38 nm) that we previously determined bound nearly all the forked duplex substrate (32), Twinkle (12 nm) efficiently unwound the BamHI-E111A-bound forked duplex to an extent only slightly less (∼8%) than that observed in reactions in which BamHI-E111A was omitted from the reaction mixture (Fig. 10A). Increasing concentrations of Twinkle displayed only a modestly reduced level of helicase activity on the BamHI-E111A-bound forked duplex compared with the protein-free forked duplex at lower concentrations of Twinkle (1.5–6 nm) (Fig. 10, B–D). A reduction of ∼1.5-fold was observed at a Twinkle hexamer concentration of 1.5 nm.
FIGURE 10.
Twinkle displaces BamHI-E111A bound to forked duplex DNA substrate and unwinds the forked duplex. A and B, indicated concentrations of Twinkle (hexamer) were incubated at 37 °C for 30 min with a BamHI forked duplex substrate (substrate 27) (0.5 nm) in the absence (A) or presence (B) of catalytically inactive BamHI-E111A restriction endonuclease (38 nm) as described under “Experimental Procedures.” Proteinase K-digested products were electrophoresed on non-denaturing 12% polyacrylamide gels. Representative gel image from at least three independent experiments is shown. C, quantitative analysis of percent DNA substrate unwound from A and B. Naked forked duplex, filled circles; BamHI-E111A-bound forked duplex, open circles. Average values of at least three independent experiments with standard deviations indicated by error bars are shown.
We then set out to test the effect of TFAM, a factor known to bind its promoter-associated target sequence with a high affinity (33), for its effect on Twinkle helicase activity. The forked duplex substrate (substrate 28) used for these studies harbored a light strand promoter-specific recognition sequence in which TFAM is reported to bind in the low nanomolar range. TFAM bound the forked duplex in a protein concentration-dependent manner, as demonstrated by EMSA (Fig. 11A). Twinkle (23 nm hexamer) unwound 60% of the forked duplex when TFAM was omitted. TFAM inhibited unwinding of the forked duplex DNA substrate by Twinkle (23 nm) in a TFAM protein concentration-dependent manner (Fig. 11, B and C). Only 16% of the substrate was unwound when 86 nm TFAM was used in the pre-binding step prior to Twinkle addition (∼25% control activity). Complete inhibition of Twinkle helicase activity was observed at TFAM concentrations of 171 or 342 nm.
FIGURE 11.
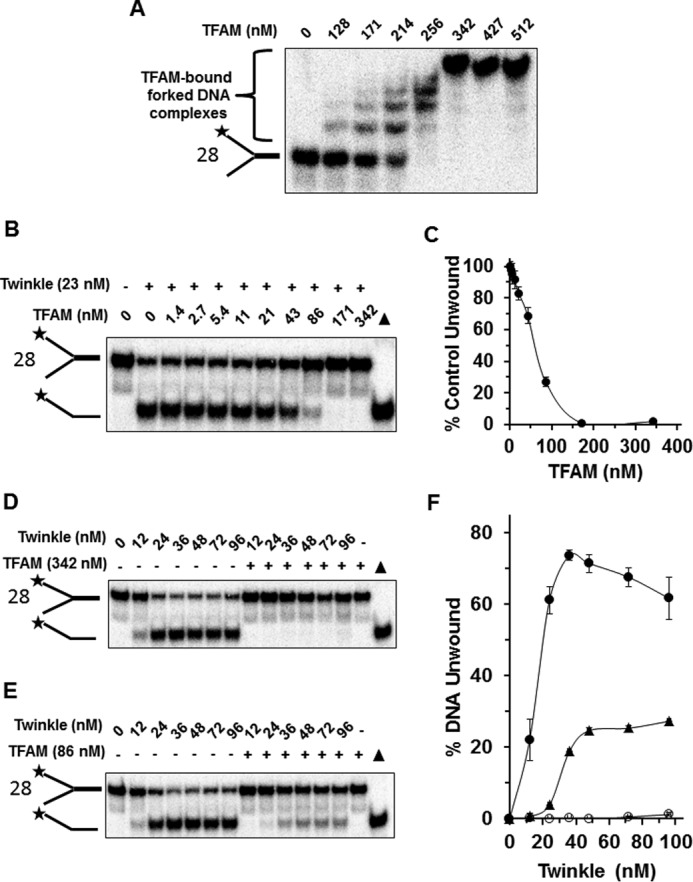
TFAM bound to forked duplex DNA substrate inhibits DNA unwinding by Twinkle helicase. A, TFAM binding to forked duplex. The indicated concentration of TFAM was incubated with the TFAM forked duplex DNA substrate (substrate 28) (0.5 nm) at 24 °C for 30 min as described under “Experimental Procedures.” DNA species from binding mixtures were resolved on non-denaturing 5% polyacrylamide gels. Representative image from EMSA of at least three independent experiments is shown. B, Twinkle helicase (23 nm hexamer) was incubated at 37 °C for 30 min with a forked duplex substrate (0.5 nm) that had been pre-bound with the indicated concentrations of TFAM as described under “Experimental Procedures.” Proteinase K-digested products were electrophoresed on non-denaturing 12% polyacrylamide gels. Representative gel image from at least three independent experiments is shown. C, quantitative analysis of forked duplex DNA substrate unwound (percent control activity compared with the naked forked duplex in which TFAM was omitted from pre-binding step) from experiments as conducted in B. D and E, indicated concentrations of Twinkle hexamer were incubated at 37 °C for 30 min with forked duplex DNA substrate (0.5 nm) that had been pre-incubated in the absence or presence of 342 nm TFAM (D) or 86 nm TFAM (E). Proteinase K-digested products were electrophoresed on non-denaturing 12% polyacrylamide gels. Representative gel image from at least three independent experiments is shown. F, quantitative analysis of forked duplex DNA substrate unwound from experiments as conducted in D and E. Naked forked duplex, filled circles; TFAM (342 nm)-bound forked duplex, open circles; TFAM (86 nm)-bound forked duplex, filled triangles. Average values of at least three independent experiments with standard deviations indicated by error bars are shown.
We next asked whether increasing Twinkle concentration was able to overcome TFAM inhibition (Fig. 11, D–F). For initial experiments, the forked duplex substrate was pre-incubated with 342 nm TFAM, a concentration in which all the DNA substrate was bound according to the EMSA results (Fig. 11A). Here, Twinkle unwound the forked duplex in a protein concentration-dependent manner; however, TFAM (342 nm) completed inhibited Twinkle helicase activity throughout the Twinkle titration (Fig. 11, D and F). We next used a lower TFAM concentration of 86 nm and repeated the Twinkle titration. Nearly 30% of the substrate was unwound at the highest concentration tested, 96 nm Twinkle hexamer. Nonetheless, there was significantly less substrate unwound when TFAM (86 nm) was present compared with reactions in which TFAM was omitted throughout the Twinkle titration range (Fig. 11, E and F). Based on these results, we conclude that TFAM inhibits Twinkle helicase activity on the simple forked duplex substrate in a profound manner.
Discussion
In this study, we have undertaken a biochemical analysis of the ATP-dependent catalytic strand separation activities of the purified human recombinant mitochondrial DNA helicase Twinkle. This work has provided new insight to Twinkle's apparent DNA substrate specificity, its newly discovered branch migration activity, and the ability of the enzyme to tolerate roadblocks it is likely to encounter during mitochondrial genome replication. The key conclusion from the Twinkle helicase assays with replication fork-associated structures is that Twinkle preferentially unwinds DNA substrates with pre-existing 5′ and 3′ single-stranded tails but is also quite active on a 5′ flap substrate, a key intermediate of strand displacement synthesis during DNA replication or DNA repair synthesis. These results are consistent with previous findings from Falkenberg and co-workers (11). The robust activity of Twinkle on a 5′ flap substrate was made even more apparent under single-turnover conditions, in which the rate of Twinkle-catalyzed DNA unwinding was comparable with the forked duplex with single-stranded 5′ and 3′ arms. The residual activity of Twinkle on a 3′ flap or synthetic replication fork structure may reflect some thermal breathing at the junction or blunt duplex end to allow Twinkle loading; however, we cannot dismiss a mechanism in which a small fraction of Twinkle helicase molecules bind to 3′ single-stranded tails or duplex arms and disrupts base pairs allowing Twinkle to load onto the opposite strand enabling it to translocate with 5′ to 3′ directionality and unwind the entire duplex. This may also be a factor under single-turnover conditions for a simple 3′ ssDNA tailed duplex in which a low rate of Twinkle helicase activity was observed. Although previous experimental data from fluorescence anisotropy titrations performed by Patel and co-workers (12) demonstrated that Twinkle binds a 20-mer duplex DNA fragment with 2-fold greater affinity than a 20-mer single-stranded oligonucleotide, our EMSA data show that a greater fraction of forked DNA structures with one or both single-stranded arms is bound by Twinkle compared with the level of Twinkle binding to synthetic replication fork structure with duplex leading and lagging strand arms. Presumably, Twinkle binding to the duplex regions of the 3′-tailed substrate, 3′ flap substrate, or synthetic replication fork may contribute to the modest level of unwinding observed for these substrates.
In addition to conventional replication fork-associated structures, we determined that Twinkle catalytically dissociates the invading strand of immobile or mobile D-loop DNA structures irrespective of the single strand polarity of the third strand with similar preferential activity; however, the extent of dissociated invading strand with a free 5′ ssDNA end was slightly greater at the highest Twinkle concentrations tested. Nonetheless, the experimental data suggest that unlike the replication fork-associated structures, Twinkle does not require an obligatory 5′ single-stranded loading tail to efficiently unwind the D-loop substrates. Twinkle's ability to unwind the invading strand of the immobile D-loop substrates in which the invading strand has a non-base-paired 5′ ssDNA tail or a non-base-paired 3′ ssDNA tail (Fig. 4, A, B and E) suggests that the traditionally defined 5′ to 3′ DNA helicase is able to recognize the three-stranded junction irrespective of the tail directionality status and initiate unwinding at the junction to release the invading strand. In contrast to what was observed for the immobile D-loop substrates with an invading third strand that has single strand character, Twinkle displays relatively poor helicase activity on the immobile D-loop substrate that has a “flush” third strand, i.e. one in which there is no pre-existing single strand character (Fig. 4, C and E). This result suggests that Twinkle loading at either the upstream nick or downstream nick to initiate unwinding of the third strand occurs rather inefficiently.
In contrast, Twinkle branch-migrates mobile three-way DNA structures with a strong 5′ to 3′ directionality preference. To our knowledge, this and another study (34) published while ours was under revision are the first reported demonstrations that Twinkle possesses DNA branch migration activity of a D-loop, a DNA substrate the enzyme is likely to encounter during mtDNA replication. Collectively, the in vitro studies suggest that Twinkle may be capable of acting upon D-loop substrates by a combination of its branch migration and helicase activity, the latter not restricted to D-loops with pre-existing 5′ single-stranded DNA tails. These findings are pertinent to the finding that Twinkle (and pol γ) are highly enriched in the D-loop region and may be relevant to reloading of the replisome components at the end the mitochondrial D-loop (35).
Consistent with previous work showing that Twinkle is an unwinding and annealing helicase (12), we now show at the single molecule level that during stepwise unwinding Twinkle will regress and anneal duplex DNA more than half the time. Our smFRET fork binding assays show that multiple binding paths and contacts on the helicase exterior are sampled by the non-translocating strand. It is possible that some or all of these binding interactions could play a role in Twinkle's unwinding and annealing activities.
The repetitive release and re-engagement of the DNA strand may be important in processing D-loops and three-stranded junctions. The low efficiency in DNA unwinding could be enhanced through binding of mitochondrial SSB, which is known to stimulate Twinkle helicase activity (11). Alternatively, or in addition, the coupled action of DNA synthesis by mtDNA polymerase γ with Twinkle unwinding may help to stabilize the unwound strands, similar to what was reported for the bacteriophage T7 DNA polymerase and helicase in which polymerase rapidly traps single-stranded bases arising from helicase action at the fork (36).
We also assessed Twinkle's ability to unwind past potential mitochondrial genomic roadblocks, either covalent DNA lesions or static protein-DNA complexes. Given that damage to the mitochondrial genome is believed to occur readily in the oxidative environment of the mitochondrial matrix (37), we assayed Twinkle helicase activity on DNA substrates with a site- and strand-specific cPu or Tg, two structurally distinct oxidative lesions. Tg is characterized by the introduction of two hydroxyl groups to the pyrimidine at positions C5 and C6, causing a significant localized change to B-form DNA and the base to assume an extrahelical position that is more accessible to solvent and other molecules (20). In contrast, the cPu DNA lesion is characterized by the cyclization of deoxyguanine or deoxyadenine such that a second glycosidic bond is made between the base and the corresponding sugar residue, resulting in perturbation of normal helix twist and abnormal base pair stacking (22, 23). A single Tg residing in the translocating strand inhibited Twinkle helicase, whereas the sequence-related hexameric DnaB replication fork helicase of E. coli was unaffected by the Tg lesion in either the translocating or non-translocating strand (38). In contrast to what was observed for Tg, the cPu lesion did not affect Twinkle helicase activity, suggesting that the Twinkle multimeric (hexameric/heptameric) ring (8, 9), which has a variable central channel diameter of 130–160 Å, is sufficiently large to accommodate the single-stranded DNA with the cPu lesion inside the ring.
Although the cPu or Tg lesion in the non-translocating strand did not affect Twinkle helicase activity, smFRET measurements of DNA binding strongly suggest that Twinkle makes multiple points of contact between the excluded strand and the exterior surface of the helicase. The number of smFRET states and transitions for Twinkle binding the excluded strand are much greater than that seen for SsoMCM or EcDnaB (24)3; however, the dwell times in each state are similar. This implies that Twinkle does not necessarily have greater dynamics for binding the excluded strand; rather instead, it samples more exterior binding paths, especially for the 30/30 fork. In contrast, Twinkle binding to DNA is similar to both EcDnaB and SsoMCM in that the largest dwell times occur at the highest FRET states, and the probabilities for transitions between the two highest FRET states are greater than the lower less abundant FRET states. This implies that the most stable surface-excluded strand interaction occurs when the non-translocating strand is tightly bound to the helicase exterior via multiple points of contact. Previously published biochemical data suggest that the sequence- and structure-related T7 gene 4 helicase and EcDnaB make contact with the displaced strand during DNA unwinding (39–41). Like Twinkle, EcDnaB, which also forms a ring-like structure (42, 43), fully tolerates the cPu lesion in the translocating or non-translocating strands (44). The differences between cPu and Tg on DNA structure are significant. A cPu alters sugar pucker that affects base-stacking interactions. In contrast, a Tg negatively affects the aromatic character and planarity of the base and causes it to assume an extrahelical residence, resulting in a significant localized structural change to DNA deviating from the B-form, which is likely to underlie the inhibition of Twinkle helicase activity.
Aside from DNA damage, DNA-protein complexes may affect mitochondrial replication. TFAM coating mtDNA may not only sterically prevent binding of replication, transcription, and DNA repair factors but may also exert more indirect effects by wrapping around duplex DNA and altering the DNA helix writhe and packaging (31). Up to this point, there were no reports indicating whether Twinkle uses its motor ATPase function to displace protein bound to duplex DNA. Our results show that TFAM can potently block Twinkle duplex unwinding. Interestingly, Twinkle was able to largely overcome the helicase inhibition imposed by BamHI-E111A bound to the DNA substrate, despite the high affinity of the restriction endonuclease for the cognate recognition sequence (Kd = 2.95E-11 m). We surmise that the unique interaction of TFAM binding specifically as well as non-specifically to the DNA substrate and also its effect on DNA conformation play a significant role in Twinkle helicase inhibition. In addition, the number of TFAM molecules bound to the forked duplex substrate is likely to contribute to the inhibitory effect on Twinkle helicase activity. In a physiological setting, it seems reasonable to propose that Twinkle operating in the context of mitochondrial replication or DNA repair may require other proteins to facilitate efficient removal of TFAM bound to the mitochondrial genomic DNA.
Electron microscopy and agarose gel analyses have provided evidence that human heart mtDNA contains multimeric junctional DNA complexes, including four-way and three-way junctions typically associated with replication forks or intermediates of HR repair (45). Overexpression of Twinkle was shown to increase the presence of Holliday junction type structures in heart mtDNA of transgenic mice (45). These findings coupled with observations that the major strand exchange recombination protein Rad51 and the related HR proteins Rad51C and Xrcc3 are found in human mitochondria (46) suggest that HR repair of mitochondrial double strand breaks is likely to be important for maintaining genome stability in the organelle. Supporting this idea, a deficiency in Rad51 or Rad59 impairs the repair of a targeted mitochondrial double strand break in S. cerevisiae (47). Moreover, a demonstrated role of Rad51 to help cells replicate their mtDNA during conditions of replication stress suggests that HR is important to maintain DNA synthesis (48).
These observations led us to test whether Twinkle is capable of acting upon three-stranded DNA molecules that represent model D-loop structural intermediates that occur early in HR repair as a consequence of invasion of a single strand of a double strand break into the recipient duplex. Twinkle was capable of dissociating mobile or immobile D-loop DNA substrates to release the invading third strand, irrespective of the third strand's directionality. Thus, Twinkle uses elements of the D-loop structure itself, rather than simply recognizing the single strand tail of the invading strand, to act upon it. If the latter case were true, one would have expected that the substrate with only the 5′ single-stranded invading strand would have been preferred by Twinkle; however, this was not the case as the D-loop structures with an invading strand characterized by a 3′ single-stranded element were good substrates for Twinkle.
Although Twinkle did not display a strong directionality preference for dissociation of the D-loop substrates, a comparison of its catalytic strand separation activity on mobile three-way junction DNA structures designed to assess for branch migration directionality demonstrated that Twinkle preferentially catalyzes branch migration in the 5′ to 3′ direction. The demonstrated ability of Twinkle to catalyze branch migration in vitro is likely to be relevant to the observation that Twinkle overexpression increases the abundance of Holliday junction structures in vivo (45). Moreover, Twinkle 5′ to 3′ branch migration activity may also play a role in facilitating mitochondrial genome replication past a leading strand template lesion that blocks DNA synthesis (Fig. 12). Template switching enables the leading strand to be replicated past the lesion; the recombinant DNA molecule can then be acted on by Twinkle loading at the single-stranded gap onto the replication fork leading strand template to restore the parental leading and lagging strand duplexes. Further studies that address this proposed mechanism of action by Twinkle in a biological setting will be informative to explain perhaps an unappreciated role of Twinkle to deal with mitochondrial genome replication stress. For example, recent work demonstrated that Twinkle overexpression suppresses oxidative induced replication stalling in cardiomyocyte mitochondria, thereby reducing mtDNA point mutations and rearrangements (49).
FIGURE 12.
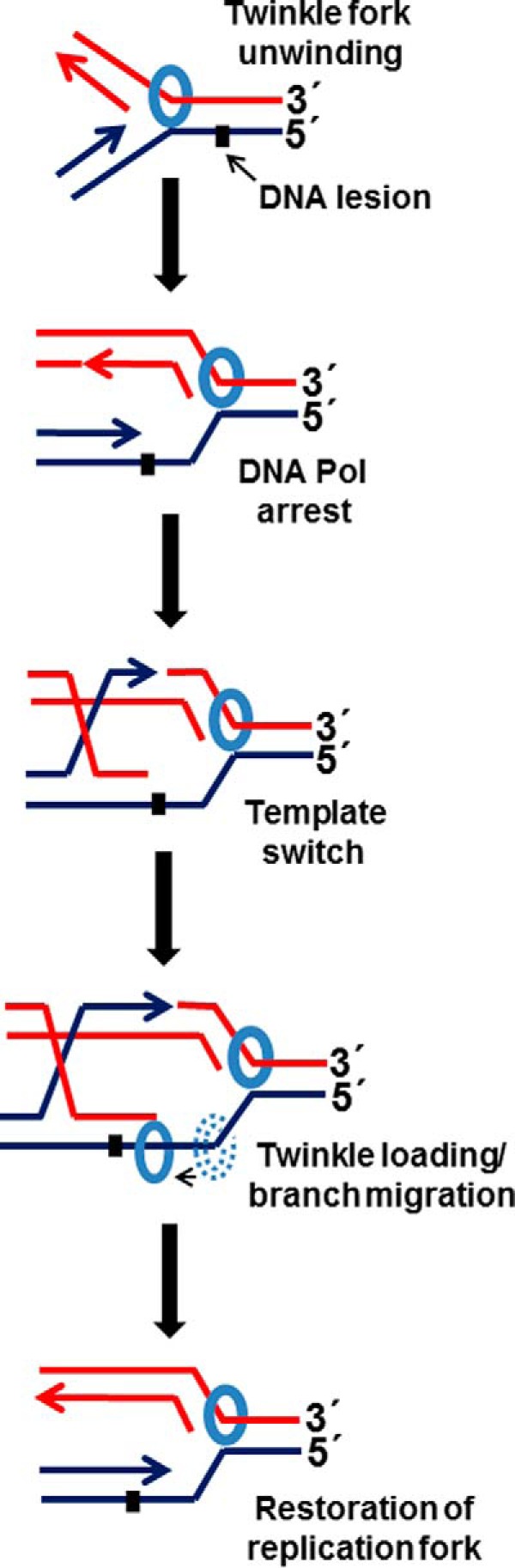
Proposed model for Twinkle branch migration activity at a stalled mitochondrial DNA replication fork. See text for details.
Experimental Procedures
Recombinant Proteins
Recombinant human Twinkle protein (gene product of c10orf2) with a C-terminal hexahistidine tag was expressed in bacterial cells and purified, and protein concentration was determined by extinction coefficient as described previously (supplemental Fig. S1) (12). Recombinant human mitochondrial transcription factor A (residues 43–246) was expressed with an N-terminal His-tagged maltose-binding protein fusion and was purified untagged as described previously (supplemental Fig. S1) (50). The catalytically inactive BamHI-E111A restriction endonuclease, used in a previous study (32), was provided by New England Biolabs (Ipswich, MA).
DNA Substrates
The oligonucleotides (Loftstrand Labs, Rockville, MD) used for preparation of the various radiolabeled DNA substrates are shown in supplemental Table S1 and are referred to in the following preparation of DNA substrates, shown in supplemental Fig. S2 and S3. Note that the asterisks denoted on DNA structures in the figures indicate 32P label at the 5′ end of the corresponding oligonucleotide in the DNA substrate. The replication fork-associated DNA structures with single-stranded or double-stranded 5′ and/or 3′ arms were prepared as described previously (51). The immobile D-loop substrates were prepared as described (52). The mobile three-way junction and mobile D-loop DNA substrates were prepared by sequential annealing as described (18). The forked duplex DNA substrates with either thymine glycol (Tg) (38) or cyclopurine (cPu) (44) in the helicase translocating or non-translocating strands were prepared as described. The forked duplex substrate harboring a BamHI restriction endonuclease recognition site was prepared as described (32). This same procedure was used to prepare the forked duplex substrate harboring a TFAM- binding site derived from the light strand promoter (5′-tgtgttagttggggggtgactgttaaa-3′) (53) using the appropriate oligonucleotides shown in supplemental Table S1.
DNA Helicase Assays
Helicase reaction mixtures (20 μl) for multiturnover reaction conditions contained 10 fmol of the indicated forked duplex or immobile D-loop DNA substrate (0.5 nm DNA substrate concentration) and the concentration(s) of Twinkle hexamer specified in the figure legend. Reaction conditions were as described previously (54) except for the inclusion of 25 mm KCl and the additional presence throughout the incubation period of a 100-fold excess of oligonucleotide of the same sequence as the labeled strand in the partial duplex substrate to serve as a displaced strand trap (12). Reaction mixtures were incubated at 37 °C for the specified time periods. Helicase reactions were terminated by addition of 20 μl of Stop buffer containing final concentrations of 0.3% SDS, 9 mm EDTA, 0.02% bromphenol blue, 0.02% xylene cyanol, and 500 μg/ml proteinase K. Proteinase K digestion was performed at 37 °C for 15 min, and products were resolved on nondenaturing 12% polyacrylamide (19:1 acrylamide/bisacrylamide) gels (51). Single-turnover reaction conditions were the same as those for multiturnover conditions with the following exceptions: 1) reaction mixtures contained 50 fmol of DNA substrate (2.5 nm DNA substrate concentration); 2) the specified concentration of Twinkle (indicated in figure legend) was pre-incubated with the DNA substrate for 5 min at 24 °C prior to simultaneous addition of ATP and a 100-fold excess of dT200, incubation for the specified time periods at 37 °C, followed by quench and proteinase K digestion. In control experiments, a 100-fold excess of dT200 in the helicase reaction resulted in a 95% reduction in Twinkle helicase activity (supplemental Fig. S4). Gels were scanned by PhosphorImager and quantitated as described previously (51).
Electrophoretic Mobility Shift Assays
DNA binding mixtures (20 μl) contained 10 fmol of the indicated forked duplex substrate (0.5 nm DNA substrate concentration) and the concentration(s) of Twinkle hexamer or TFAM specified in the figure legends. DNA binding incubation conditions were the same as those for helicase assays except that ATP was omitted, and binding mixtures were incubated at 24 °C for 30 min. After incubation, 4 μl of loading dye (74% glycerol, 0.01% xylene cyanol, 0.01% bromphenol blue) was added to each mixture, and samples were loaded onto native 8% (19:1 acrylamide/bisacrylamide) polyacrylamide gels and electrophoresed at 200 V for 2.5 h at 4 °C using 0.5× TBE as the running buffer. The resolved radiolabeled species were visualized using a PhosphorImager and analyzed with ImageQuant software (GE Healthcare).
DNA Branch Migration Assays
Twinkle DNA branch migration assays with the mobile three-way junction DNA substrates or mobile D-loop substrates were performed in the same manner as helicase assays described above except that the 100-fold excess of oligonucleotide of the same sequence as the labeled strand in the partial duplex substrate was omitted from the reaction mixtures. Products were resolved and quantitated as described above.
Protein Displacement/Helicase Assays
The forked duplex DNA substrate (0.5 nm) harboring either the BamHI-E111A cognate restriction endonuclease recognition site or the TFAM-binding site was incubated with 38 nm BamHI-E111A or the indicated concentration of TFAM, respectively, at 24 °C in Twinkle helicase reaction buffer containing ATP as described above for 15 min, followed by addition of Twinkle helicase. The reaction mixtures (20 μl) were then incubated at 37 °C for 15 min. Reactions were terminated by addition of 20 μl of Stop buffer containing 500 μg/ml proteinase K as described above. Reactions were further incubated at 37 °C for 15 min, and products were resolved on nondenaturing 12% (19:1 acrylamide/bisacrylamide) gels in 1× TBE. Gels were scanned by PhosphorImager and quantitated as described previously (51).
Single Molecule FRET-based DNA Binding and Unwinding Measurements
The oligonucleotides (IDT Corp., Coralville, IA) used to create the fluorescently labeled DNA substrates are listed in supplemental Table S1. The DNA substrates were immobilized on a pegylated quartz slide via biotin-streptavidin interactions (55). The smFRET buffer conditions were 20 mm Tris-HCl (pH 7.5), 1 mm DTT, 5 mm AMP-PNP, and 10 mm MgCl2 as well as an oxygen-scavenging system (1 mg/ml glucose oxidase, 0.4% (w/v) d-glucose, 0.04 mg/ml catalase, and 2 mm Trolox). The concentration of Twinkle used in the assays was 25 nm. Unwinding assays were initiated by addition of 5 mm ATP (final concentration). Data were collected on a prism-based total internal reflection microscope at 10 frames/s for 5 or more regions, with each region containing 50–250 single molecules. A 532-nm diode laser was used to excite the Cy3 fluorophore. The resulting Cy3 and Cy5 fluorescence signals were separated using a 610-nm dichroic long pass mirror, a 580/40 bandpass filter, a 660-nm long pass filter, and then imaged by an EM-CCD iXon camera (Andor, Belfast, UK) (56). Regions of signal intensity were fit to two-dimensional gaussians to identify single molecule FRET pairs, and subsequently corrected for thermal drift and local background intensity (57). FRET efficiency, Eapp, was calculated using Equation 1,
 |
where IA and ID are the intensities of the acceptor and donor signals, respectively.
Single Molecule FRET Data Analysis for the Fork Binding and Unwinding Assays
Traces were stitched together when the experimental conditions were identical, and the resulting trace was fit to ideal states using the vbFRET software package (26). The number of FRET states was determined by fitting the stitched trace to the maximum number of states where the states are Eapp = 0.1 away from one another, and the variation of one state does not overlap with another. Once fit to ideal states, traces were then unstitched and analyzed utilizing the ExPRT (Explicit Probability and Rate Transition) analysis program. The ExPRT plot analysis produces transition plots from ideal traces where a marker's position corresponds to a given transition where the initial state corresponds to the x axis position, and the final state corresponds to the y axis position. The size of the marker corresponds to the fraction of all traces that contain that particular transition, and the color of the marker corresponds to the dwell time of an initial state preceding the transition. The collected dwell times for each transition are fit to a single and double exponential survival curve. If the R2 value of the double exponential fit was greater than 0.970 and increased from the R2 value of the single exponential fit by more than 0.015, then the rates from the double exponential fit were used to create a concentric marker with two colors, each representing a dwell time based on the two rates. Otherwise, the dwell time was defined as the average of all measured dwell times for a given transition. Only dwell times that were both preceded and followed by transitions were taken into account in this analysis.
For the unwinding assay, each individual trace was fit to ideal states using vbFRET software. Unwinding and rewinding events were then identified, and the number of steps and the time for unwinding or rewinding to occur were collected. The unwinding time is defined as starting from the point in which there is a transition from the high FRET state and concludes when the lowest FRET state is reached. Similarly, the rewinding time is defined at that point when the lowest FRET state is departed until the highest FRET state is reached. Histograms were generated from these data and fit with a standard gaussian equation.
Author Contributions
I. K., J. D. C., S. K. B., J. A. S., and R. M. B. designed, performed, and analyzed the experiments shown in Figs. 1–8 and 10. S. M. C. and M. A. T. designed, performed, and analyzed the experiments shown in Fig. 9. E. Y. and M. G. D. provided purified recombinant TFAM. I. K. and R. M. B. wrote the paper. All authors reviewed the results, provided critical comments and input, proofread, and approved the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Laboratory of Molecular Gerontology (NIA, National Institutes of Health) for helpful discussion. We are grateful to Becky Kucera and Dr. Jurate Bitinaite (New England Biolabs) for the BamHI-E111A-purified protein. We thank Sanford Leuba (University of Pittsburgh) for continued access to the smFRET instrumentation.
This work was supported by National Institutes of Health, NIA, University of Pittsburgh, Baylor University, Research Scholar Grant RSG-11-049-01-DMC (to M. A. T.) from the American Cancer Society, and National Institutes of Health Grant GM100021 (to M. G.-D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1 and Figs. S1–S4.
S. M. Carney, H. N. McFarland, S. H. Leuba, and M. A. Trakselis, submitted for publication.
- pol
- polymerase
- cPu
- cyclopurine
- D-loop
- displacement loop
- HR
- homologous recombination
- TFAM
- mitochondrial transcription factor A
- Tg
- thymine glycol.
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- smFRET
- single molecule FRET
- ssDNA
- single-stranded DNA.
References
- 1. Rajala N., Gerhold J. M., Martinsson P., Klymov A., and Spelbrink J. N. (2014) Replication factors transiently associate with mtDNA at the mitochondrial inner membrane to facilitate replication. Nucleic Acids Res. 42, 952–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Copeland W. C. (2012) Defects in mitochondrial DNA replication and human disease. Crit. Rev. Biochem. Mol. Biol. 47, 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Korhonen J. A., Pham X. H., Pellegrini M., and Falkenberg M. (2004) Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 23, 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McKinney E. A., and Oliveira M. T. (2013) Replicating animal mitochondrial DNA. Genet. Mol. Biol. 36, 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shutt T. E., and Gray M. W. (2006) Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 62, 588–599 [DOI] [PubMed] [Google Scholar]
- 6. Ziebarth T. D., Farr C. L., and Kaguni L. S. (2007) Modular architecture of the hexameric human mitochondrial DNA helicase. J. Mol. Biol. 367, 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singleton M. R., Dillingham M. S., and Wigley D. B. (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 8. Fernández-Millán P., Lázaro M., Cansız-Arda Ş., Gerhold J. M., Rajala N., Schmitz C. A., Silva-Espiña C., Gil D., Bernadó P., Valle M., Spelbrink J. N., and Solà M. (2015) The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Res. 43, 4284–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ziebarth T. D., Gonzalez-Soltero R., Makowska-Grzyska M. M., Núñez-Ramírez R., Carazo J. M., and Kaguni L. S. (2010) Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. J. Biol. Chem. 285, 14639–14647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jemt E., Farge G., Bäckström S., Holmlund T., Gustafsson C. M., and Falkenberg M. (2011) The mitochondrial DNA helicase TWINKLE can assemble on a closed circular template and support initiation of DNA synthesis. Nucleic Acids Res. 39, 9238–9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korhonen J. A., Gaspari M., and Falkenberg M. (2003) TWINKLE Has 5′ → 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 278, 48627–48632 [DOI] [PubMed] [Google Scholar]
- 12. Sen D., Nandakumar D., Tang G. Q., and Patel S. S. (2012) The human mitochondrial DNA helicase TWINKLE is both an unwinding and an annealing helicase. J. Biol. Chem. 287, 14545–14556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bharti S. K., Sommers J. A., Zhou J., Kaplan D. L., Spelbrink J. N., Mergny J. L., and Brosh R. M. Jr. (2014) DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J. Biol. Chem. 289, 29975–29993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong D. W., Pereira F., Barrett S. P., Kolesar J. E., Cao K., Damas J., Yatsunyk L. A., Johnson F. B., and Kaufman B. A. (2014) Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics 15, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding L., and Liu Y. (2015) Borrowing nuclear DNA helicases to protect mitochondrial DNA. Int. J. Mol. Sci. 16, 10870–10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wanrooij S., and Falkenberg M. (2010) The human mitochondrial replication fork in health and disease. Biochim. Biophys. Acta 1797, 1378–1388 [DOI] [PubMed] [Google Scholar]
- 17. Kowalczykowski S. C. (2015) An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bugreev D. V., Brosh R. M. Jr., and Mazin A. V. (2008) RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 283, 20231–20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zorov D. B., Juhaszova M., and Sollott S. J. (2014) Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 94, 909–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kung H. C., and Bolton P. H. (1997) Structure of a duplex DNA containing a thymine glycol residue in solution. J. Biol. Chem. 272, 9227–9236 [DOI] [PubMed] [Google Scholar]
- 21. Berquist B. R., and Wilson D. M. 3rd. (2012) Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 327, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang H., Das R. S., Basu A. K., and Stone M. P. (2011) Structure of (5′S)-8,5′-cyclo-2′-deoxyguanosine in DNA. J. Am. Chem. Soc. 133, 20357–20368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zaliznyak T., Lukin M., and de los Santos C. (2012) Structure and stability of duplex DNA containing (5′S)-5′,8-cyclo-2′-deoxyadenosine: an oxidatively generated lesion repaired by NER. Chem. Res. Toxicol. 25, 2103–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graham B. W., Schauer G. D., Leuba S. H., and Trakselis M. A. (2011) Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res. 39, 6585–6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rothenberg E., Trakselis M. A., Bell S. D., and Ha T. (2007) MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J. Biol. Chem. 282, 34229–34234 [DOI] [PubMed] [Google Scholar]
- 26. Bronson J. E., Fei J., Hofman J. M., Gonzalez R. L. Jr., and Wiggins C. H. (2009) Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 97, 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Myong S., Bruno M. M., Pyle A. M., and Ha T. (2007) Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 317, 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee W., Jose D., Phelps C., Marcus A. H., and von Hippel P. H. (2013) A single-molecule view of the assembly pathway, subunit stoichiometry, and unwinding activity of the bacteriophage T4 primosome (helicase-primase) complex. Biochemistry 52, 3157–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brosh R. M., Jr. (2013) DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13, 542–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ngo H. B., Lovely G. A., Phillips R., and Chan D. C. (2014) Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 5, 3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bogenhagen D. F. (2012) Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta 1819, 914–920 [DOI] [PubMed] [Google Scholar]
- 32. Sommers J. A., Banerjee T., Hinds T., Wan B., Wold M. S., Lei M., and Brosh R. M. Jr. (2014) Novel function of the Fanconi anemia group J or RECQ1 helicase to disrupt protein-DNA complexes in a replication protein A-stimulated manner. J. Biol. Chem. 289, 19928–19941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Campbell C. T., Kolesar J. E., and Kaufman B. A. (2012) Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim. Biophys. Acta 1819, 921–929 [DOI] [PubMed] [Google Scholar]
- 34. Sen D., Patel G., and Patel S. S. (2016) Homologous DNA strand exchange activity of the human mitochondrial DNA helicase TWINKLE. Nucleic Acids Res. 44, 4200–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jemt E., Persson Ö., Shi Y., Mehmedovic M., Uhler J. P., DávilaLópez M., Freyer C., Gustafsson C. M., Samuelsson T., and Falkenberg M. (2015) Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 43, 9262–9275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stano N. M., Jeong Y. J., Donmez I., Tummalapalli P., Levin M. K., and Patel S. S. (2005) DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature 435, 370–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muftuoglu M., Mori M. P., and de Souza-Pinto N. C. (2014) Formation and repair of oxidative damage in the mitochondrial DNA. Mitochondrion 17, 164–181 [DOI] [PubMed] [Google Scholar]
- 38. Suhasini A. N., Sommers J. A., Mason A. C., Voloshin O. N., Camerini-Otero R. D., Wold M. S., and Brosh R. M. Jr. (2009) FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J. Biol. Chem. 284, 18458–18470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Donmez I., and Patel S. S. (2006) Mechanisms of a ring shaped helicase. Nucleic Acids Res. 34, 4216–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Manhart C. M., and McHenry C. S. (2015) Identification of subunit binding positions on a model fork and displacements that occur during sequential assembly of the Escherichia coli primosome. J. Biol. Chem. 290, 10828–10839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Satapathy A. K., Kulczyk A. W., Ghosh S., van Oijen A. M., and Richardson C. C. (2011) Coupling dTTP hydrolysis with DNA unwinding by the DNA helicase of bacteriophage T7. J. Biol. Chem. 286, 34468–34478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. San Martin C., Radermacher M., Wolpensinger B., Engel A., Miles C. S., Dixon N. E., and Carazo J. M. (1998) Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure 6, 501–509 [DOI] [PubMed] [Google Scholar]
- 43. Yang S., Yu X., VanLoock M. S., Jezewska M. J., Bujalowski W., and Egelman E. H. (2002) Flexibility of the rings: structural asymmetry in the DnaB hexameric helicase. J. Mol. Biol. 321, 839–849 [DOI] [PubMed] [Google Scholar]
- 44. Khan I., Suhasini A. N., Banerjee T., Sommers J. A., Kaplan D. L., Kuper J., Kisker C., and Brosh R. M. Jr. (2014) Impact of age-associated cyclopurine lesions on DNA repair helicases. PLoS ONE 9, e113293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pohjoismäki J. L., Goffart S., Tyynismaa H., Willcox S., Ide T., Kang D., Suomalainen A., Karhunen P. J., Griffith J. D., Holt I. J., and Jacobs H. T. (2009) Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J. Biol. Chem. 284, 21446–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sage J. M., Gildemeister O. S., and Knight K. L. (2010) Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J. Biol. Chem. 285, 18984–18990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stein A., Kalifa L., and Sia E. A. (2015) Members of the RAD52 epistasis group contribute to mitochondrial homologous recombination and double strand break repair in Saccharomyces cerevisiae. PLoS Genet. 11, e1005664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sage J. M., and Knight K. L. (2013) Human Rad51 promotes mitochondrial DNA synthesis under conditions of increased replication stress. Mitochondrion 13, 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pohjoismäki J. L., Williams S. L., Boettger T., Goffart S., Kim J., Suomalainen A., Moraes C. T., and Braun T. (2013) Overexpression of Twinkle-helicase protects cardiomyocytes from genotoxic stress caused by reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 110, 19408–19413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yakubovskaya E., Guja K. E., Eng E. T., Choi W. S., Mejia E., Beglov D., Lukin M., Kozakov D., and Garcia-Diaz M. (2014) Organization of the human mitochondrial transcription initiation complex. Nucleic Acids Res. 42, 4100–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brosh R. M. Jr., Waheed J., and Sommers J. A. (2002) Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 277, 23236–23245 [DOI] [PubMed] [Google Scholar]
- 52. Gupta R., Sharma S., Sommers J. A., Jin Z., Cantor S. B., and Brosh R. M. Jr. (2005) Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 280, 25450–25460 [DOI] [PubMed] [Google Scholar]
- 53. Wang K. Z., Zhu J., Dagda R. K., Uechi G., Cherra S. J. 3rd, Gusdon A. M., Balasubramani M., and Chu C. T. (2014) ERK-mediated phosphorylation of TFAM downregulates mitochondrial transcription: implications for Parkinson's disease. Mitochondrion 17, 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Farge G., Holmlund T., Khvorostova J., Rofougaran R., Hofer A., and Falkenberg M. (2008) The N-terminal domain of TWINKLE contributes to single-stranded DNA binding and DNA helicase activities. Nucleic Acids Res. 36, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Roy R., Hohng S., and Ha T. (2008) A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fagerburg M. V., and Leuba S. H. (2011) in DNA Nanotechnology, Methods and Protocols (Zuccheri G., and Sumorí B., eds) pp. 273–290, Humana Press Inc., Totowa, NJ [Google Scholar]
- 57. Rasband W. S. (2009) ImageJ, National Institutes of Health, Bethesda [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.