Abstract
The adenovirus early region 1A (E1A) oncoprotein hijacks host cells via direct interactions with many key cellular proteins, such as KAT2B, also known as PCAF (p300/CBP associated factor). E1A binds the histone acetyltransferase (HAT) domain of KAT2B to repress its transcriptional activation. However, the molecular mechanism by which E1A inhibits the HAT activity is not known. Here we demonstrate that a short and relatively conserved N-terminal motif (cNM) in the intrinsically disordered E1A protein is crucial for KAT2B interaction, and inhibits its HAT activity through a direct competition with acetyl-CoA, but not its substrate histone H3. Molecular modeling together with a series of mutagenesis experiments suggests that the major helix of E1A cNM binds to a surface of the acetyl-CoA pocket of the KAT2B HAT domain. Moreover, transient expression of the cNM peptide is sufficient to inhibit KAT2B-specific H3 acetylation H3K14ac in vivo. Together, our data define an essential motif cNM in N-terminal E1A as an acetyl-CoA entry blocker that directly associates with the entrance of acetyl-CoA binding pocket to block the HAT domain access to its cofactor.
Keywords: acetyl coenzyme A (acetyl-CoA), acetyltransferase, adenovirus, enzyme inhibitor, structural model
Introduction
Adenoviruses, small DNA viruses with genomes of ∼35 kb have evolved to infect a broad range of animal species and human beings (1, 2). Human adenoviruses can be oncogenic in hamsters (3). Adenoviral E1A, encoded by a viral early region 1A, is essential for transcriptional regulation of the viruses, and reprogramming the host cells (4). Through alternative splicing, human adenoviruses produce two main E1A products, 13S and 12S, which share the same conserved region 1 (CR1),5 CR2, and CR4 motifs but not an extra CR3 motif only in 13S (5, 6).
E1A, as a molecular hub protein, regulates the host cells by interacting with multiple cellular proteins, including a variety of transcriptional coactivators, corepressors, and cell-cycle regulatory proteins (7–11). For example, E1A binds to pRB, the first E1A interaction protein ever identified, through CR1 and CR2 motifs to release E2F for transcriptional activation (12–16). E1A interaction with transcriptional coactivator p300 has been shown to associate with repression of many genes, especially those involved in terminal differentiation of cells (15, 17). The CR1 motif is essential to suppress p300/CBP-mediated p53 activation (18, 19). E1A also inhibits the p300/CBP-associated factor (KAT2B) through a direct interaction between the first 90 amino acids (aa) of E1A and KAT2B acetyltransferase domain (HAT), resulting in disruption of myogenesis, even though E1A does not appear to affect acetyltransferase activity (20, 21). On the contrary, two groups have shown that the CR2 and CR3 motifs but not the N-terminal region (1–76 aa) are indispensible in the inhibition of the acetylation activities of p300 and KAT2B, although the E1A N terminus dominates in binding to both proteins (22, 23).
KAT2B, also known as PCAF, and its ortholog KAT2A, known as GCN5, are GCN5-related N-acetyltransferases (GNAT) (24). They are conserved subunits of megadalton SAGA and ATAC complexes from yeast to human beings (25). Metazoan KAT2B and KAT2A have three conserved domains: the N-terminal extension region, the HAT domain, and the bromodomian (26). The HAT domain has a central globular core with a CoA binding pocket (27). KAT2A and −KAT2B are able to form dimeric states at least through their HAT domains (28). Current understanding of histone acetylation specificity is derived from the structures of Tetrahymena thermophila KAT2A (tKAT2A) with histone H3 or H4 peptides (29, 30). The H3 peptide with a conserved motif G-K-X-P binds the L-shaped cleft of the HAT domain close to the cofactor acetyl-CoA. Human KAT2B selectively acetylates N-terminal H3 peptides (19–27 aa) with a Km of ∼1 mm (31).
Molecular details about how E1A interacts with p300 through CR1 have been determined by nuclear magnetic resonance (32). However, the manner in which E1A interacts with KAT2B remains elusive. CR1 is essential for a direct binding to the HAT domain (21). However, mutations in the first 30 amino acids disrupt E1A binding to KAT2B (33). More intriguingly, clear discrepancies about E1A inhibition on KAT2B are found in vivo as described above. Here we defined all essential motifs in the E1A N terminus that interact with the HAT domain. We further demonstrate that E1A inhibits KAT2B acetylation by competition with acetyl-CoA, but not histone H3. Finally we have analyzed possible E1A binding surfaces on the HAT domain, and built a proposed E1A docking model.
Experimental Procedures
Protein Purifications and Peptides
His-tagged human KAT2B HAT domain (493–658 aa) was expressed and purified as described previously (28). GST-tagged E1A (1–126 and 39–126 aa) from human adenovirus 2 and GST-tagged Xenopus H3 (1–60 aa) were expressed using pGEX-6p-1 vector in BL21/DE3 Gold (Stratagene). Site-specific mutations were made using our modified QuikChange mutagenesis protocol (34). Overexpression of these proteins was induced with 0.25 mm isopropyl β-d-thiogalactopyranoside for 8 h at 25 °C. E1A and H3 proteins were first purified by glutathione Superflow-agarose (Pierce). Elutes were concentrated down using Centricons (Millipore) and exchanged into a buffer of 20 mm Tris, pH 8.0, 200 mm NaCl, and 5 mm β-mercaptoethanol. GST of the GST-H3 (1–60 aa) fusion protein was cleaved with PreScission protease and removed by SP Sepharose (GE Healthcare). Short E1A peptides and FITC or biotin labeling were chemically synthesized (Table 1).
TABLE 1.
E1A peptides and their sequences synthesized for KAT2B interaction
| Name | Serotype | Sequence | Region | Label |
|---|---|---|---|---|
| E1A2-13 | Adenovirus 2 | MRHIICHGGVITE | 1–13 | N-FITC |
| cNM | Adenovirus 2 | MRHIICHGGVITEEMAASLLDQLIEEVLADNLP | 1–33 | N-FITC |
| m21 | Adenovirus 2 | PSHFEPPTLHELYDLDVTAPE | 35–55 | N-FITC |
| CR1c | Adenovirus 2 | VTAPEDPNEEAVSQIFPESVMLAVQEGIDLFT | 51–82 | N-FITC |
| CR1 | Adenovirus 2 | PTLHELYDLDVTAPEDPNEEAVSQIFPESVMLAVQEG | 41–77 | N-FITC |
| E1A2s | Adenovirus 2 | GVITEEMAASLLDQLIEEVLAD | 9–30 | C-FITC |
| E1A12s | Adenovirus 12 | MTPLVLSYQEADDILEHLVDNFFNEVPS | 5–32 | C-biotin |
| E1A14-10 | Adenovirus 14 | MRHLRFLPQE | 1–10 | N-FITC |
| E1A14 | Adenovirus 14 | MRHLRFLPQEIISAETGNEILEFVVHALMGDDPE | 1–34 | C-biotin |
| E1A19 | Adenovirus 19 | MRHLRLLTSTVPLDMAALLLDDFVNTVLEDE | 1–31 | C-biotin |
| E1A3nat | Adenovirus 3 | MRHLRFLPQEIISSETGIEILEFVVNTLMGDD | 1–32 | NAa |
| E1A9nat | Adenovirus 9 | MRHLRLLPSTVPGELAVLMLEDFVDTVLEDE | 1–31 | NA |
| E1A12nat | Adenovirus 12 | MRTEMTPLVLSYQEADDILEHLVDNFFNEVPS | 1–32 | NA |
| E1A19nat | Adenovirus 19 | MRHLRLLTSTVPLDMAALLLDDFVNTVLEDE | 1–31 | NA |
a NA, not applicable.
Pulldown Assays
NTA-agarose affinity pulldown (NTA pulldown) was carried out using the His-tagged KAT2B HAT domain and Ni-NTA-agarose beads (Pierce). E1A peptides labeled with FITC were gently mixed with equal moles of KAT2B proteins and 10 μl the beads for 30 min at 4 °C in 1 ml of buffer of 20 mm Tris, pH 8.0, 50 mm NaCl, and 0.2% Triton X-100 unless indicated. The beads were collected by a low-speed centrifugation and washed 3 times using the same buffer to remove unspecific bindings. The proteins and bound peptides were separated on 18% Tricine SDS-PAGE gel and visualized under UV before being stained with Coomassie Blue. Similarly, for GST pulldown, GST-E1A fusion proteins were mixed with equal moles of the KAT2B HAT domain for 30 min at 4 °C in 1 ml of buffer of 20 mm Tris, pH 8.0, 250 mm NaCl, and 0.2% Triton X-100 unless indicated. The beads were collected and washed with the same buffer. The bound proteins were separated on 12% SDS-PAGE and stained with Coomassie Blue. Acetyl-CoA or CoA competition in NTA and GST pulldown experiments were examined with increasing concentrations of these cofactors during incubation.
Acetylation Assay in Vitro
The acetylation of histone H3 (1–60 aa, 5 μg) in GST fusion by 0.5 μg of the KAT2B HAT domain with 20 μm acetyl-CoA was set up in 25 μl of buffer of 50 mm Tris, pH 8.5, 10 mm butyric acid, 10% glycerol, 100 mm NaCl, and 0.1 mm EDTA for 30 min at 37 °C according to Chakravarti et al. (22). The resulting mixtures were separated on 15% SDS-PAGE and further blotted using anti-acetylated histone H3 antibody (06–599, Millipore). Half of the mixtures were used for another 15% SDS-PAGE and stained with Coomassie Blue as loading controls. The effects of E1A proteins (1–126 and 39–126 aa) and peptides (cNM and CR1) on histone acetylation were examined by mixing them with the KAT2B HAT domain at different molar ratios to acetyl-CoA as indicated and incubated at 37 °C for 3 min before acetyl-CoA was added. The mixture was again incubated at 37 °C for another 3 min for acetylation and stopped by 1× SDS sample buffer.
Enzymatic Kinetic Parameters
KAT2B HAT assay was carried out using the 5,5′-dithiobis(2-nitrobenzoic acid) method (35). 5,5′-Dithiobis(2-nitrobenzoic acid) reacts with the thiol group of CoA released from acetyl-CoA during acetylation to produce 2-nitro-5-thiobenzoate, which can be monitored at λ412 nm. The KAT2B HAT domain at 50 nm was set up in 100 μl of buffer containing 50 mm Tris, pH 8.0, 1 mm 5,5′-dithiobis(2-nitrobenzoic acid), 1 mm EDTA, and 200 μm acetyl-CoA with increasing H3 fragment (1–60 aa) as a substrate. Absorbance of each reaction at λ412 nm was recorded every 30 s for 30 min at 30 °C in a 96-well plate using POLARstar Omega. The kinetic parameters Km and Vmax were determined by non-linear curve fit to the Michaelis-Menten equation: v = Vmax[S]/(Km + [S]) in Origin 8 and kcat was calculated from equation Vmax = kcat [E].
Isothermal Titration Calorimetry
These ITC titration experiments were carried out at 25 °C using MicroCal iTC200 system (Marven) essentially following Freyer and Lewis (36). E1A peptides, acetyl-CoA, and CoA were dissolved in the same buffer of 20 mm Tris, pH 8.0, and 200 mm NaCl as the KAT2B HAT domain. The delay time was 60 s for the first 0.4-μl injection. The remaining titrations were completed with 2 μl of ligand per each injection for 20 intervals of 120 s. Reference power was 5 μcal/s. The stirring speed was 1,000 rpm. For the KAT2B and E1A titration, the best signal/noise ratio was obtained using 0.7 mm KAT2B protein in syringe and 50 μm E1A peptides in cell. The best signal/noise ratios for KAT2B and acetyl-CoA or CoA titrations were obtained with 50 μm HAT domain in the cell and 1 mm acetyl-CoA or CoA in the syringe. All ITC data were processed in Origin 7.0.
Homology Modeling and Docking
E1A sequences of all adenoviral serotypes were manually collected from GenBankTM for a global cNM conservation analysis using a web server PRALINE (37). The ab initial structures of the α-helical region in cNM of adenovirus serotype 2 E1A were built using a web server I-TASSER (38) and optimized by molecular operating environment (Chemical Computing Group Inc., Canada). Dockings of the cNM helical structures to the CoA-free KAT2B HAT domain (PDB code 4NSQ) were performed in ZDock server (39). The docking process was repeated three times using slightly different helical structures of cNM. The top 10 docking models were selected using anchor residues Leu20 and Leu23 of E1A2 and classified into three docking modes, which were further energy optimized using a Amber99 force field in the molecular operating environment, followed by the Flexible Peptide Docking server of Rosetta (40). The stability of these modeled complexes was finally examined by molecular dynamic simulation using a Amber99 force field in explicit solvent at 277 K. All structural graphics were prepared in PyMol (DeLano Scientific, LLC).
Co-immunoprecipitation
E1A wt and mut (Leu20 and Leu23 to Ala) were expressed in a C-terminal GFP fusion protein and HA tag. DNA fragments were inserted into EcoRI/SalI sites of our modified pLV-HA lentiviral vector (41). pCI-FLAG-KAT2B for the full-length KAT2B was purchased from Addgene and the HAT deletion mutant (ΔHAT, Δ493–658 aa) was made using our modified QuikChange mutagenesis protocol (34). The 293T cells in ф10 cm dishes at ∼80% confluence were co-transfected with E1A and KAT2B expression constructs using a PEI transfection protocol as described previously (42). Cells after 2 days of transfection were counted under the microscope and an equal number of cells were suspended in a buffer of 10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 1 mm DTT, 0.5 mm PMSF, and 1× protease inhibitor mixture for 10 min on ice. Cells were collected and lysed in a high-salt extraction buffer of 30 mm HEPES, pH 7.9, 20% glycerol, 0.6 m NaCl, 1% Nonidet P-40, 0.4 mm EDTA, 1.5 mm MgCl2, 1 mm DTT, 0.5 mm PMSF, and 1× protease inhibitor mixture for 30 min at 4 °C. 350 μg of whole cell extract was collected by centrifugation at 10,000 × g for 10 min and incubated with FLAG resin for 2 h at 4 °C in a buffer of 20 mm HEPES, pH 7.9, 10% glycerol, 0.2% Nonidet P-40, 0.2 mm EDTA, and 300 mm KCl. After an extensive wash with the same buffer, the bound proteins were eluted out with a FLAG peptide and analyzed in 8% SDS-PAGE, followed by Western blot analyses with anti-FLAG antibody (Proteintech, 20543-1-AP) and anti-HA antibody (Proteintech, 66006-1-IG).
Acetylation Inhibition Assay in Vivo
E1A-GFP-HA fusion proteins were overexpressed in 293T and HepG2 cells in a 6-well plate at 50% confluence infected with recombinant lentiviruses as described (41, 43). After 2 days of infection, the cells were selected in new medium containing 0.5 μg/ml of puromycin for 3 days. The total number of viable cells was counted under the microscope. The cells were collected and lysed with 1× SDS loading buffer and resolved in 12% SDS-PAGE, followed by Western blotting analyses with anti-H3K14ac antibody (PTM BioLabs, PTM-157), anti-HA antibody, and anti-β-Actin antibody (Proteintech, 66009-1-IG).
Results
E1A cNM Is a Major Binding Ligand to the KAT2B HAT Domain
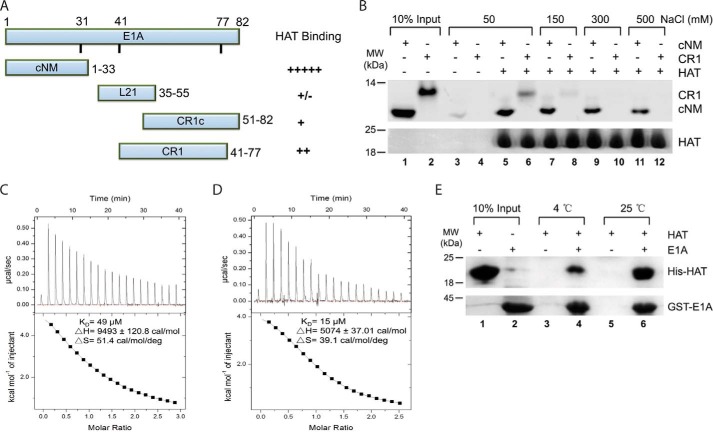
Because the E1A N-terminal region (1–90 aa) is sufficient for KAT2B binding (21), we first asked which parts of it are essential. A series of peptides of E1A2 (E1A from serotype 2 adenovirus) were chemically synthesized, some of which were labeled with the fluorescent probe FITC (Table 1). We examined the interactions of these peptides with 6× His-tagged KAT2B HAT domain (493–658 aa) using NTA affinity pulldown assay (NTA pulldown) (Fig. 1A). The peptide with the highest affinity was found to be the conserved N-terminal motif (cNM, 1–33 aa). CR1 (41–77 aa) bound, but with a lower affinity than cNM. The peptide CR1c (51–82 aa) bound significantly more weakly than CR1, suggesting that the N-terminal portion of CR1 is important for its interaction with the HAT domain. The linker peptide L21 (35–55 aa) appeared to be dispensable.
FIGURE 1.
E1A cNM plays a dominant role in binding to the KAT2B HAT domain. A, interactions between E1A N-terminal peptides and the KAT2B HAT domain quantified in NTA pulldown experiments. The peptides that cover different E1A regions are indicated on the left. Semi-quantitative summary from at least three pulldown experiments is shown on the right. The pulldown experiments were performed with 1.5 μm HAT domain and equal moles of E1A peptides labeled with FITC at 4 °C. The bound peptides were visualized by UV fluorescence and estimated by measuring the band intensity using ImageJ. B, ion strength dependence of the E1A and KAT2B interactions. The NTA pulldown experiment was performed with 1.5 μm KAT2B HAT domain and 1 μm E1A peptides in the same condition and buffers with different NaCl concentrations as indicated. The upper half is the bound peptides visualized under UV. The amounts of inputs are indicated in lanes 1 and 2. Any possible unspecific binding was detected with the E1A peptides mixed with NTA beads in the lowest salt buffer (lanes 3 and 4). The lower half is the KAT2B protein used for each reaction stained with Coomassie Blue. C and D, ITC titrations of E1A cNM (C) and E1A (1–126 aa) (D) with the KAT2B HAT domain. Raw titration curves are at the top and thermodynamic parameters derived from global fitting are shown at the bottom. E, the KAT2B and E1A interaction is affected by temperature. The GST pulldown experiment was performed with 3.0 μm GST-E1A (1–126 aa) fusion protein incubated with 3.0 μm KAT2B HAT domain and 10 μl of GST-agarose beads at either 4 or 25 °C. The amounts of KAT2B and E1A proteins used for each reaction are indicated in lanes 1 and 2.
We examined the dependence of cNM and CR1 interactions with the HAT domain upon salt concentrations (Fig. 1B). Although both peptides bound to the HAT domain in the buffer containing 50 mm NaCl (lanes 3–6), cNM, but not CR1, retained an interaction with a significant affinity when NTA pulldowns were performed at higher salt concentrations (lanes 7–12), suggesting that cNM is a dominant motif in E1A for the interaction with the HAT domain.
We next determined the binding affinity of the cNM peptide with the KAT2B HAT domain using isothermal titration calorimetry (ITC). We measured a binding affinity of Kd = 49 μm (Fig. 1C). In comparison, an E1A fragment (1–126 aa) with a GST fusion bound the HAT domain at Kd = 15 μm, indicating other motifs also contribute to the E1A binding to the HAT domain (Fig. 1D). More importantly, large positive ΔH and ΔS values measured for the E1A and KAT2B interaction suggested that their interaction is driven mainly by interface hydrophobicity, but not hydrogen bonds, and thus the binding should be more favored at higher temperature. To test this possibility, we incubated the E1A and KAT2B mixture for an additional 30 min at 25 °C before performing NTA pulldowns (Fig. 1E). Compared with all steps done at 4 °C, E1A indeed bound the HAT domain with a significantly greater affinity under these conditions (lane 6 versus 4). A hydrophobic interaction between E1A and KAT2B is also consistent with the resistance to increasing ionic strength for cNM and the HAT domain pulldowns(Fig. 1B).
E1A Competes with Acetyl-CoA for Inhibition
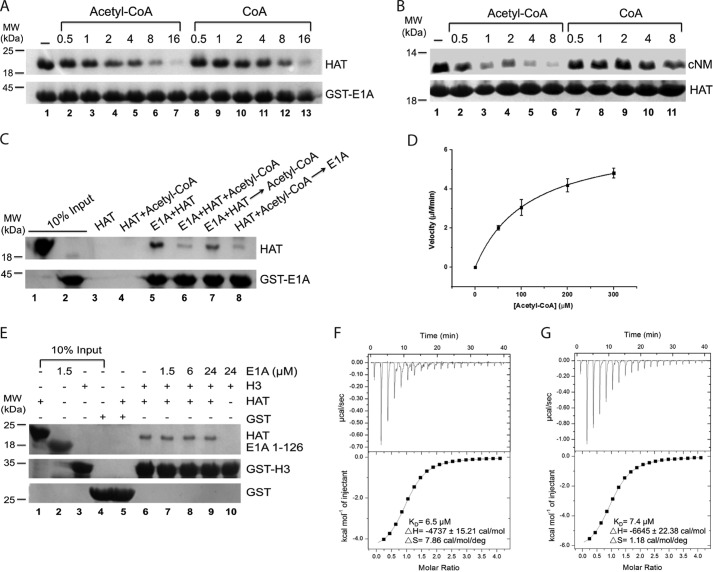
Having identified a key section of E1A in binding with the HAT domain, we next asked how E1A inhibits its acetyltransferase activity. We tested the possibility of direct competition, whereby E1A either competes off substrates or interferes with acetyl-CoA binding. We used a GST-tagged E1A (1–126 aa) fusion protein as bait to pull down the His-tagged KAT2B HAT domain in the presence of increasing concentrations of the cofactor (Fig. 2A). We observed that the amount of KAT2B associated with E1A gradually reduced with increasing concentrations of acetyl-CoA, and was almost absent in the 16× acetyl-CoA (lanes 1–7). Similarly, CoA had a lower capacity to inhibit the KAT2B and E1A interaction (lanes 8–13). In a reciprocal NTA pulldown experiment using the His-tagged KAT2B HAT domain, the cNM peptide interaction was severely weakened by acetyl-CoA, and by CoA to a certain extent (Fig. 2B). The competition was also observed using the human KAT2A HAT domain (data not shown). These experiments supported the possibility that E1A is a direct competitor with acetyl-CoA for the HAT domain.
FIGURE 2.
Acetyl-CoA and CoA compete E1A off for KAT2B binding. A, competition of acetyl-CoA and CoA with E1A for KAT2B. The GST pulldown experiment was carried out with 1.5 μm GST-E1A (1–126 aa) mixed with 3 μm KAT2B HAT domain (lane 1) and increasing amounts of acetyl-CoA (lanes 2–7) or CoA (lanes 8–13). The ratios of acetyl-CoA or CoA to KAT2B are indicated on the top. The lower gel indicates the GST-E1A protein used in each lane. B, competition of acetyl-CoA and CoA with the cNM motif. The NTA pulldown experiment was carried out with 1.5 μm His-tagged KAT2B HAT domain mixed with 1 μm cNM (lane 1) and increasing amounts of acetyl-CoA (lanes 2–6) or CoA (lanes 7–11). The ratios of acetyl-CoA or CoA to cNM are indicated on the top. The lower gel indicates the KAT2B protein used in each lane. C, mutual competition between E1A and acetyl-CoA in KAT2B binding. GST-E1A (1–126 aa, 1.5 μm) was directly incubated with 1.5 μm KAT2B (lane 5), or together with (lane 6), 10 min prior to (lane 7), or 10 min after (lane 8) the addition of 3 μm acetyl-CoA. KAT2B or KAT2B and the acetyl-CoA mixture were incubated with GST beads for possible unspecific bindings (lanes 3 and 4). The amounts of KAT2B and E1A proteins used in each lane are shown in lanes 1 and 2. D, enzymatic kinetics of the KAT2B HAT domain on histone H3. Experiment was set up using 50, 100, 200, and 300 μm H3 (1–60 aa). y axis indicates velocity of KAT2B acetylation on each concentration of H3. E, competition of E1A with histone H3 peptide for the KAT2B HAT domain. GST-H3 (1–60 aa, 1.5 μm) was mixed with 1.5 μm KAT2B protein and a concentration gradient of E1A (1–126 aa) (lanes 6–9). KAT2B (1.5 μm) was incubated with 1.5 μm GST (lane 5) and 24 μm E1A (1–126 aa) was incubated with 1.5 μm H3 (lane 10) as negative controls. The amounts of KAT2B, E1A, H3, and GST used in each lane are shown in lanes 1–4. The bound KAT2B is shown on the top gel. GST-E1A and GST are shown on the middle and bottom gels. F and G, ITC titrations of acetyl-CoA (E) and CoA (F) to the KAT2B HAT domain. Titrations were carried out using 50 μm KAT2B protein with 1 mm acetyl-CoA or CoA at 25 °C.
We then asked whether E1A and acetyl-CoA competition was conditional. To answer this question, we incubated E1A or acetyl-CoA with the HAT domain for 10 min before adding an equal amount of their competitors in a GST pulldown experiment (Fig. 2C). E1A gained a significant ability to compete with acetyl-CoA when it was preincubated with KAT2B (lane 7 versus 8). However, preincubation of acetyl-CoA with KAT2B did not improve its competition (lane 6 versus 8). Collectively these data suggested that acetyl-CoA and E1A are competitive, and E1A is a weaker competitor for binding to the HAT domain.
To examine the possibility of competition with any HAT substrates, we purified an N-terminal fragment of Xenopus histone protein H3 (1–60 aa), which was readily soluble and stable in low salt buffers compared with the full-length H3. The acetylation kinetic parameters (Km = 116.69 ± 2.76 μm, kcat = 133.20 ± 1.28 min−1) showed that this N-terminal H3 fragment could be an efficient substrate of the KAT2B HAT domain (Fig. 2D). E1A did not inhibit the H3 and KAT2B interaction even though 16× E1A was used in our pulldown experiment (Fig. 2E). All these data suggested that cNM inhibits KAT2B activity mainly through removal of acetyl-CoA, but not the KAT2B substrate H3, by competition.
We next hypothesized that the ability of acetyl-CoA and CoA in E1A competition might be a result of their stronger binding affinities with the KAT2B HAT domain. We measured their dissociation constants by isothermal titration calorimetry, finding a Kd of 6.5 μm for acetyl-CoA and 7.4 μm for CoA, nearly 8 times stronger than that for cNM (Fig. 2, F and G). Interestingly, negative enthalpy and positive entropy in both reactions suggest that acetyl-CoA and CoA binding are driven by hydrogen bond formation and hydrophobic interactions. We note that the entropy change for acetyl-CoA binding is nearly 7 times larger than that for CoA, suggesting the acetyl group contributes the large hydrophobicity for binding to the HAT domain.
The Hydrophobic Nature of the Interface between E1A and the HAT Domain
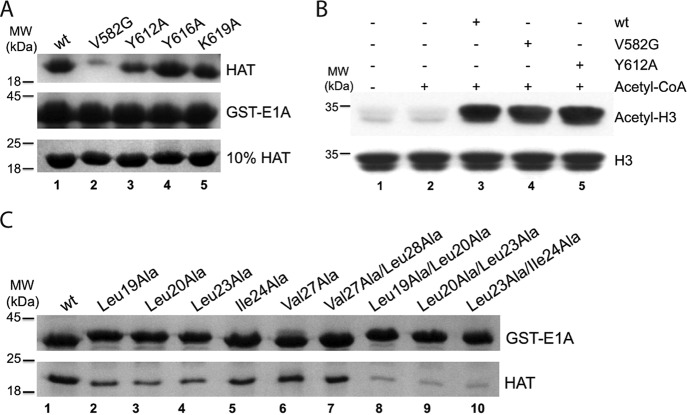
Acetyl-CoA is bound into a cavity formed by the surface of loop β5α4 (609–614 aa) and α4, and further fixed in place by another loop β4α3 (577–585 aa) in the KAT2B HAT domain (27). The competitive relationship of acetyl-CoA and E1A suggested that E1A might bind to the same surface area on KAT2B. To test this possibility, we introduced mutations into the surface around the CoA binding site to examine whether these would affect KAT2B binding in GST-E1A pulldown assays. Compared with wt, the Y612A mutation clearly weakened, and the V582G mutation almost abolished the interaction of KAT2B with E1A, whereas other two mutations (Y616A and K619A) had no effect (Fig. 3A). None of these protein surface mutations affect KAT2B acetyltransferase activity and therefore presumably did not change the overall conformation of the HAT domain (Fig. 3B). Together, these mutagenesis experiments supported the contention that the protein surface around the CoA pocket of KAT2B is important for E1A binding.
FIGURE 3.
Key residues in KAT2B and E1A interaction. A, disruptive mutations in the KAT2B HAT domain. The GST-E1A (1–126 aa) pulldown experiment was performed with 3.0 μm KAT2B HAT domains (wt and mutants). The bound KAT2B proteins are shown on the top gel and inputs are on the bottom gel. GST-E1A loading controls are shown on the middle gel. B, acetyltransferase activity of KAT2B mutants. H3 alone (lane 1), and further mixed with 20 μm acetyl-CoA without KAT2B (lane 2) were loaded as controls. C, disruptive E1A mutations for KAT2B interaction. GST pulldown experiment was performed with equal amounts of GST-E1A (1–126 aa) fusion protein and its mutants bound on GST beads (upper band) to pulldown the KAT2B HAT domain (lower band).
We next examined possible critical sites in E1A for KAT2B binding. As demonstrated above, cNM is the dominant motif in KAT2B binding; we therefore generated a series of E1A mutants within this motif for GST pulldown experiments. The mutations (Leu19, Leu20, and Leu23 to Ala) clearly reduced E1A binding affinity for KAT2B, whereas other mutations (Ile24, Val27, and Leu28 to Ala) were less effective (Fig. 3C, lanes 1–7). Moreover, double mutations (Leu19, Leu20, Leu23, and Ile24 to Ala) markedly suppressed the E1A and KAT2B interaction (Fig. 3C, lanes 8–10). All these indicated that E1A binds KAT2B through hydrophobic interfaces mainly formed by Leu20 and Leu23, with involvement of Leu19 and Ile24 consistent with the large positive entropy changes observed in the ITC experiments (Fig. 1, C and D).
Docking Model of the E1A and KAT2B Complex
To understand better the molecular mechanisms of the E1A and KAT2B interactions, we attempted to crystallize the KAT2B HAT domain in complex with E1A using a variety of cNM peptides as listed in Table 1. We solved several co-crystal structures of E1A and HAT domain complexes, or crystal structures of the HAT domain soaked with peptides, but could not find any electron density of bound E1A peptides. We then resorted to molecular modeling and docking.
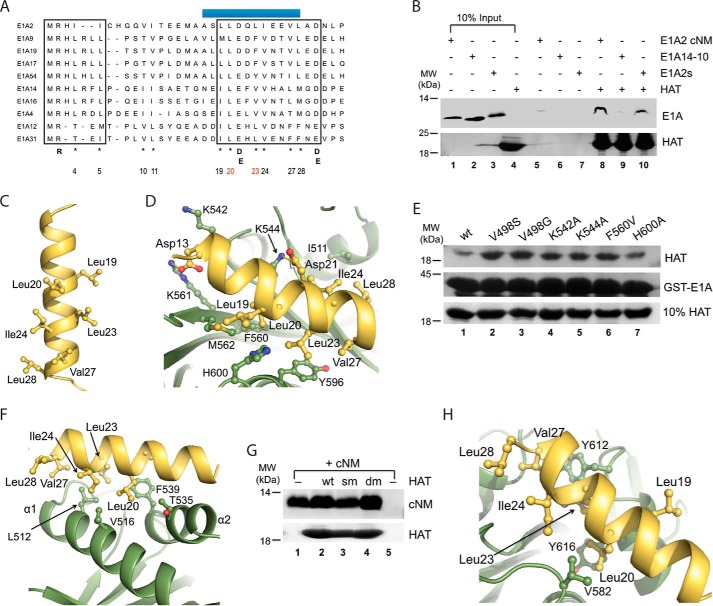
The E1A cNM motif contains a short amphipathic helix but its sequence conservation is much lower than CR1, CR2, CR3, and CR4 (6, 11). To characterize this motif, we collected 11 unique E1A sequences with 31–90% identity from all adenoviral serotypes (sequence identity of 18–90%, and E1A40 was removed because its identity to others is below 10%) for a multiple sequence alignment using the program PRALINE, which predicted an invariable α-helix in this motif (Fig. 4A). With slight adjustments, an additional N-terminal loop (1–7 aa) was also predicted. The loop is characterized by the presence of an invariable residue Arg2, which is critical for E1A function in targeting p300/CBP activities (44). The helical region of cNM contains three relatively conserved hydrophobic patches (19–20, 23–24, and 27–28 in E1A2). Interestingly, two acidic residues Asp/Glu (21, 29) are also highly conserved.
FIGURE 4.
Dock cNM to the KAT2B HAT domain. A, alignment of unique E1A cNM sequences from different adenovirus serotypes. Conserved regions are boxed. A predicted short α-helix is highlighted with a blue bar on the top. The conserved residues are indicated below the sequences. Highly conserved hydrophobic residues are labeled with an asterisk and numbered according to E1A2, among which two residues are colored in red for their key roles in KAT2B interaction. B, conserved submotifs of cNM to interact with KAT2B. The NTA pulldown was set up with 3 μm HAT domain and 1.5 μm E1A14-10 (1–10 aa) or E1A2s (α-helical region) peptides. Possible unspecific bindings of these E1A peptides to NTA beads were examined in lanes 5–7. The top was the gel visualized with UV and the bottom was the gel stained with Coomassie Blue. The amounts of the E1A peptides and KAT2B are shown in lanes 1–4. C, a modeled structure of the α-helical region of E1A2 cNM. Important residues contributed to its hydrophobic surface are shown in sticks and further labeled. D, F, and H, docking complex models of KAT2B and E1A2 α-helix. The key residues of KAT2B that are involved in hydrophobic interactions and hydrogen bonds are shown in sticks and labeled with single-letter codes, whereas the key residues in E1A are shown in sticks and labeled with three-letter codes. Hydrogen bonds are indicated with yellow broken lines. E, effects of mutations of KAT2B in E1A interaction. The GST pulldown experiment was performed with 3.0 μm wt and mutated KAT2B HAT domains (loading controls in the bottom gel) and equal moles of GST-E1A (1–126 aa, loading controls in the middle gel). G, effects of mutations on KAT2B dimeric interfaces for E1A interaction. The NTA pulldown experiment was set up with 3.0 μm wt and mutated KAT2B HAT domains and equal moles of E1A cNM peptide. The bound peptide on beads is shown in the top gel. The KAT2B loading control is shown in the bottom gel. The amount of E1A peptide used for each lane is shown in lane 1 and unspecific binding of the peptide to the beads is shown in lane 5. wt, KAT2B (493–658 aa); sm, KAT2B (493–658 aa, L512D/V516D/F539A); dm, KAT2B (496–658 aa, L512D/V516D/F539A).
To investigate contributions of these two submotifs in cNM to the HAT domain binding, we examined two peptide fragments that cover the first 10–13 amino acids (peptides E1A2-13 and E1A14-10) and the helical region (peptide E1A2s, 9–30 aa) of E1A (Table 1) in a NTA pulldown experiment. The E1A2s peptide was shown to interact strongly with the HAT domain, whereas the E1A14-10 peptide was only barely detectable (Fig. 4B).
Because the helical region of cNM is dominant in the HAT binding, we next modeled the α-helix of E1A2, resulting in a putative structure with all hydrophobic patches aligned on one side (Fig. 4C). We then docked the α-helix to the crystal structure of the KAT2B HAT domain using ZDock and optimized in molecular operating environment and Rosetta. We selected the top 10 models from 2000 complexes, which could be classified into three groups (Fig. 4, D–H). In 4 of 10 models, E1A was docked to a HAT domain surface with both hydrophobic and charge-charge interactions (Fig. 4D). Although this appeared to be the best model, mutation of all critical residues in the interface, including Val498, Lys542, LysK544, Phe560, and His600, did not disrupt the E1A and KAT2B interaction in this pulldown experiment (Fig. 4E).
In 3 of 10 models, E1A was docked to a hydrophobic surface formed by the α1 and α2 helices of the HAT domain, the dominant dimer interface of the HAT domain (Fig. 4F). We also examined this model using two KAT2B HAT mutants in the dimer interface in an NTA pulldown assay (Fig. 4G). A HAT domain with triple mutations of L512A/M513A/V516A (sm) and a shortened KAT2B HAT domain with triple mutations of L512D/V516D/F539A (dm), which are defective in dimerization (28), retained similar binding affinity with cNM, apparently excluding this model of the HAT dimer interface.
In one model, E1A was directly docked to the surface of acetyl-CoA binding pocket (Fig. 4H). Although this was only found in 1 of 10 models, this was fully supported by our biochemical data discussed above (Figs. 2 and 3). The remaining 2 of the top 10 models included unreasonable contacts between E1A and KAT2B (data not shown).
cNM Inhibits the KAT2B HAT Activity Both in Vitro and in Vivo
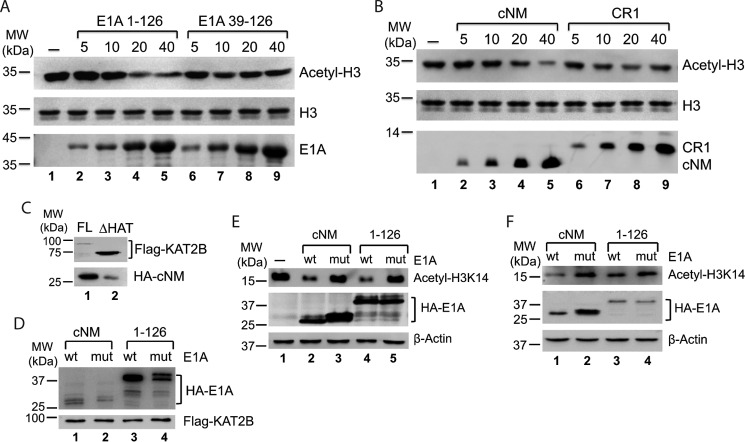
From the data above, we defined the essential regions of E1A in interactions with KAT2B in vitro. We wanted to know whether these motifs were sufficient to inhibit KAT2B HAT activity. Acetylation of histone H3 by KAT2B could be clearly inhibited by the E1A (1–126 aa) fragment. However, an E1A fragment lacking cNM (39–126 aa) was barely able to inhibit KAT2B activity (Fig. 5A). A further experiment using the cNM and CR1 peptides showed that cNM indeed has an ability to inhibit HAT activity, whereas CR1 is not significant (Fig. 5B). All these in vitro data suggested that cNM but not CR1 is a dominant motif in E1A in the inhibition of KAT2B activity.
FIGURE 5.
E1A inhibits KAT2B HAT activity. A and B, cNM is essential for E1A to inhibit KAT2B HAT activity in vitro. Acetylation reactions were set up by mixing 0.5 μg of KAT2B and a concentration gradient of E1A before 5 μm acetyl-CoA was added. The acetyl-lysine was detected by using anti-acetyl-H3 antibody (the top gel). The loading controls of H3 and E1A are shown in the middle and bottom gels. H3 acetylation without E1A is shown in lane 1. C and D, E1A cNM interacts with the KAT2B HAT domain in vivo (C) and their interaction can be affected by Leu20/Leu23 mutations in E1A (D). FLAG-KAT2B full-length (FL, lane 1) and HAT deletion (ΔHAT, lane 2) were expressed with cNM or E1A (1–126 aa) in 293T cells and co-immunoprecipitated with FLAG gel. All bound fractions were examined in Western blot using anti-HA and anti-FLAG antibodies. E and F, E1A inhibits KAT2B-specific histone H3 acetylation in 293T (E) and HepG2 (F) cells. The cells were infected with recombinant lentiviruses to express E1A proteins and selected under a pressure of puromycin for 3 days. A lentivirus produced from an empty vector pLV-HA was used for a negative control (E, lane 1). Whole cell extracts were analyzed in Western blot using antibodies against H3K14 acetylation (anti-H3K14ac, top gel), HA tag (anti-HA, middle gel), and β-actin (anti-β-actin, bottom gel). β-Actin was used as a loading control.
We next examined whether this inhibition also occurred in vivo. A HAT deletion mutation markedly reduced E1A binding to KAT2B, suggesting the HAT domain is indeed dominant in the E1A interaction (Fig. 5C). On the other hand a cNM double mutant (Leu20 and Leu23 to Ala) had a clearly weaker interaction with KAT2B than wt, as did the E1A (1–126 aa) fragment (Fig. 5D). All the data further supported our observation that E1A and KAT2B interact through the HAT domain and cNM. Consistently, overexpression of E1A fragments inhibited KAT2B-specific histone acetylation, H3K14, whereas double mutants did not have any effects, which was observed in both 293T and HepG2 cell lines (Fig. 5, E and F). Therefore, we conclude that E1A directly interacts with the HAT domain using the cNM motif to inhibit the KAT2B HAT activity.
Discussion
Adenovirus E1A is a key regulator for transcription of viral genes (45). E1A is also an intrinsically disordered protein that serves as a hub for a variety of host cell proteins (7, 11, 15, 46). The molecular mechanism by which E1A inhibits KAT2B activity still remains unclear. Here we show that cNM, a relatively conserved motif located in the N-terminal region of E1A, is essential for binding the HAT domain and inhibiting its HAT activity.
E1A cNM Is a Dominant Binder
Reid et al. (21) have demonstrated that E1A interacts with p300 and KAT2B through its N-terminal region (1–90 aa). CR1 (50–80 aa) is a conserved motif in this region (6, 11). Alanine mutations in the CR1 region have been shown to effectively prevent E1A from binding to p300 (47), consistent with a solution structure of E1A in complex with the p300 CH3 domain (32). Alanine mutations of 6 amino acids within CR1 also prevent E1A from binding to KAT2B (21). However, we have found that CR1, binding of which is mediated by charge-charge interactions binds more weakly to KAT2B compared with cNM (Fig. 1).
Intriguingly, the region of the first 1–50 aa is not conserved except for a short α-helix (6, 11). However, our analyses reveal the cNM motif in this region, containing an invariable α-helix and a small submotif (1–7 aa) (Fig. 4A). It has been shown that a completely conserved arginine in this 7-residue submotif is critical for E1A to inhibit p300 activity (16, 47). However, compared with the α-helical motif, this 7-residue motif has a significantly weaker interaction with KAT2B (Fig. 4B). Therefore, the cNM motif, and in particular its α-helix, is an essential binding site for the E1A and KAT2B interaction.
cNM Competes with Acetyl-CoA for the KAT2B HAT Domain
Acetyl-CoA is an essential cofactor for HAT activity. The binding affinity of CoA for KAT2A was previously determined to be 5.1 μm, similar to acetyl-CoA (Kd of 8.5 μm) using equilibrium dialysis and fluorescence anisotropy (48). CoA may be able to stabilize KAT2A as acetyl-CoA because the tKAT2A HAT domain can form a stable complex with CoA and H3 peptide (30). Both acetyl-CoA and CoA bind the KAT2B HAT domain with Kd of 6.5–7.4 μm determined in our ITC experiments (Fig. 2, F and G), and both are able to compete with E1A for binding to the KAT2B HAT domain (Fig. 2, A and B). Acetyl-CoA is a stronger competitor, which cannot be simply explained by their binding constants. One possibility is that acetyl-CoA is better stabilized by more hydrophobic interactions because acetyl-CoA has a 7× larger entropy change than CoA in interactions with KAT2B (Fig. 2, F and G).
The long E1A fragment (1–126 aa) has a Kd of 15 μm, about 3 times stronger than cNM (Fig. 1, C and D). The full-length E1A may have a similar or even higher affinity than acetyl-CoA and CoA because it has been shown to interact with KAT2B through additional motifs, such as CR3 (49, 50). Moreover, our ITC experiments have shown that the E1A and KAT2B interaction has a large positive entropy change, indicating that E1A may undergo some conformational changes to stabilize their complex. Therefore, E1A and acetyl-CoA may be equivalent for competitive binding to the HAT domain (Fig. 2C). In contrast, the N-terminal tail of histone H3 binds significantly more weakly because the Kd of the H3 peptide and KAT2A/KAT2B complex is about 100 μm in the presence of acetyl-CoA (51). Consistent with this, a direct interaction between KAT2B and H3 was readily detected in our pulldown experiment only when using low stringency buffers (50–100 mm NaCl) (data not shown). Moreover, H3 seemed incapable to compete with E1A for the KAT2B HAT domain (Fig. 2E).
E1A Inhibition
The E1A region (1–76 aa) has been shown to be important for p300 binding, but it inhibits p300 acetylation activity only to a certain extent (14, 22, 33). Although the critical region in E1A to interact with KAT2A/2B is found to be the first 29 amino acids, it is not sufficient to inhibit the acetylation activity in vitro of KAT2A/2B (52). Additional evidence indicates that E1A does not directly affect the HAT activity in vitro (21). However, our data here strongly suggest that E1A is able to inhibit HAT activity both in vitro and in vivo (Fig. 5). In our in vitro experiments, its inhibition effect could be further enhanced by preincubation of E1A and KAT2B before adding acetyl-CoA. Indeed, when reactions were performed by simply mixing cNM with acetyl-CoA and KAT2B for a long time, the inhibition of cNM was clearly reduced. In contrast, E1A (1–126 aa) did not appear to be affected by preincubation treatment (data not shown). One rationale is that E1A cNM binds KAT2B more weakly than acetyl-CoA. The longer E1A fragments or the full-length E1A could be a stronger competitor. E1A cNM appeared to interact specifically with the KAT2B HAT domain (Fig. 5C). Endogenous KAT2B HAT activity could be significantly inhibited by overexpression of E1A (Fig. 5, E and F). A longer E1A fragment (1–126 aa) resulted in stronger inhibition than cNM. Similarly, complete inhibition of E1A on p300 appears to require all E1A motifs (22). Furthermore, stable interaction of CR1 with the p300/CBP CH3 domain (32) may result in the formation of a tri-component inhibitory complex in vivo together with p300 and KAT2B, which also interact through the CH3 domain (20).
A Molecular Model of E1A and KAT2B Interaction
We made a great effort to determine crystal structures of the E1A and KAT2B association by screening E1A peptides of cNM and CR1 regions of various lengths and sequences (Table 1). But we failed to generate any well ordered electron density for bound peptides on KAT2A or the KAT2B HAT domains, probably due to low binding affinity of cNM peptides. The E1A (1–126 aa) fragment and the KAT2B HAT domain complex could not be maintained in size exclusion chromatography (data not shown). We therefore turned to molecular modeling of possible complexes of the KAT2B HAT domain and E1A, because the α-helical region of cNM is a dominant submotif of E1A in binding to KAT2B.
cNM was predicted to have an amphipathic α-helical region (6, 11). Mutation of its hydrophobic residues significantly reduced E1A binding to p300, KAT2A, and KAT2B (33). An Leu23 and Ile24 double mutant of E1A5 12S no longer bound KAT2B (50). Our theoretical model of this helix from E1A2 suggested that three pairs of hydrophobic residues form a continuous hydrophobic surface on one side (Fig. 4C). Our biochemical and modeling studies further defined that Leu19, Leu20, and Leu23 and Ile24 but not Val27 and Leu28 residues are essential in the E1A and KAT2B interaction (Fig. 3C). Although the first model among the top 10 models was dominant and apparently best in that cNM was docked directly to a hydrophobic surface of the HAT domain, it was excluded by our mutagenesis experiment (Fig. 4, D and E). So was the KAT2B dimeric interface (Fig. 4, F and G). The model of cNM to the acetyl-CoA binding site suggested the possibility of blocking acetyl-CoA entrance is a plausible mechanism for the E1A inhibition of KAT2B (Fig. 4H).
In summary, we have defined the short N-terminal conserved region cNM as a key motif involved in E1A for KAT2B interaction. All the data presented here suggest that cNM inhibits the KAT2B activity through a direct competition with cofactor acetyl-CoA but not substrate H3. The strategy taken by adenoviruses as a cofactor entry blocker can be possibly used to develop new KAT2B inhibitors to treat relevant diseases, for example, neuroinflammatory Alzheimer disease.
Author Contributions
S. S., S. Z., J. L., and C. G. conducted all experiments. K. L. performed the docking and model analyses of E1A and PCAF complexes. Y. C. performed the co-immunoprecipitation experiments. A. H., Z. J., and R. C. designed and analyzed experiments. Q. J. and X. Y. joined in formulation of experimental designs and data analyses. S. S. wrote a draft of the manuscript.
Acknowledgments
We thank Dr. Karolin Luger (Colorado State University) for Xenopus histone expression constructs. We also thank Dr. David M. J. Lilley (University of Dundee) for critical revision of our manuscript.
This work was supported by the National Science Foundation of China Grants 31170685 and 90919036 (to A. H.), 81361120386, 31570751, and 31270809 (to R. C.) and Project 985 (0660ZK1022). The authors declare that they have no conflicts of interest with the contents of this article.
- CR1
- conserved region 1
- cNM
- conserved N-terminal motif
- HAT
- histone acetyltransferase
- ITC
- isothermal titration calorimetry
- KAT2B
- K-lysine acetyltransferase 2A
- KAT2B
- K-lysine acetyltransferase 2B
- NTA
- nitrilotriacetic acid
- PCAF
- p300/CBP associated factor
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- CBP
- cAMP-response element-binding protein.
References
- 1. Green M. (1970) Oncogenic viruses. Annu. Rev. Biochem. 39, 701–756 [DOI] [PubMed] [Google Scholar]
- 2. Sprengel J., Schmitz B., Heuss-Neitzel D., Zock C., and Doerfler W. (1994) Nucleotide sequence of human adenovirus type 12 DNA: comparative functional analysis. J. Virol. 68, 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trentin J. J., Van Hoosier G. L. Jr., and Samper L. (1968) The oncogenicity of human adenoviruses in hamsters. Proc. Soc. Exp. Biol. Med. 127, 683–689 [DOI] [PubMed] [Google Scholar]
- 4. Nevins J. R., Ginsberg H. S., Blanchard J. M., Wilson M. C., and Darnell J. E. Jr. (1979) Regulation of the primary expression of the early adenovirus transcription units. J. Virol. 32, 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimelman D., Miller J. S., Porter D., and Roberts B. E. (1985) E1a regions of the human adenoviruses and of the highly oncogenic simian adenovirus 7 are closely related. J. Virol. 53, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Avvakumov N., Wheeler R., D'Halluin J. C., and Mymryk J. S. (2002) Comparative sequence analysis of the largest E1A proteins of human and simian adenoviruses. J. Virol. 76, 7968–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berk A. J. (2005) Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24, 7673–7685 [DOI] [PubMed] [Google Scholar]
- 8. Flint J., and Shenk T. (1997) Viral transactivating proteins. Annu. Rev. Genet. 31, 177–212 [DOI] [PubMed] [Google Scholar]
- 9. Gallimore P. H., and Turnell A. S. (2001) Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene 20, 7824–7835 [DOI] [PubMed] [Google Scholar]
- 10. Bayley S. T., and Mymryk J. S. (1994) Adenovirus e1a proteins and transformation (review). Int. J. Oncol. 5, 425–444 [DOI] [PubMed] [Google Scholar]
- 11. Pelka P., Ablack J. N., Fonseca G. J., Yousef A. F., and Mymryk J. S. (2008) Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 82, 7252–7263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dyson N., and Harlow E. (1992) Adenovirus E1A targets key regulators of cell proliferation. Cancer Surv. 12, 161–195 [PubMed] [Google Scholar]
- 13. Moran E. (1993) DNA tumor virus transforming proteins and the cell cycle. Curr. Opin. Genet. Dev. 3, 63–70 [DOI] [PubMed] [Google Scholar]
- 14. Wang H. G., Rikitake Y., Carter M. C., Yaciuk P., Abraham S. E., Zerler B., and Moran E. (1993) Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol. 67, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frisch S. M., and Mymryk J. S. (2002) Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 3, 441–452 [DOI] [PubMed] [Google Scholar]
- 16. Ferrari R., Pellegrini M., Horwitz G. A., Xie W., Berk A. J., and Kurdistani S. K. (2008) Epigenetic reprogramming by adenovirus E1A. Science 321, 1086–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lang S. E., and Hearing P. (2003) The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22, 2836–2841 [DOI] [PubMed] [Google Scholar]
- 18. Lill N. L., Grossman S. R., Ginsberg D., DeCaprio J., and Livingston D. M. (1997) Binding and modulation of p53 by p300/CBP coactivators. Nature 387, 823–827 [DOI] [PubMed] [Google Scholar]
- 19. O'Connor M. J., Zimmermann H., Nielsen S., Bernard H. U., and Kouzarides T. (1999) Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J. Virol. 73, 3574–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang X. J., Ogryzko V. V., Nishikawa J., Howard B. H., and Nakatani Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382, 319–324 [DOI] [PubMed] [Google Scholar]
- 21. Reid J. L., Bannister A. J., Zegerman P., Martínez-Balbás M. A., and Kouzarides T. (1998) E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 17, 4469–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chakravarti D., Ogryzko V., Kao H. Y., Nash A., Chen H., Nakatani Y., and Evans R. M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96, 393–403 [DOI] [PubMed] [Google Scholar]
- 23. Hamamori Y., Sartorelli V., Ogryzko V., Puri P. L., Wu H. Y., Wang J. Y., Nakatani Y., and Kedes L. (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96, 405–413 [DOI] [PubMed] [Google Scholar]
- 24. Nagy Z., and Tora L. (2007) Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26, 5341–5357 [DOI] [PubMed] [Google Scholar]
- 25. Spedale G., Timmers H. T., and Pijnappel W. W. (2012) ATAC-king the complexity of SAGA during evolution. Genes Dev. 26, 527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang X. J. (2004) The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32, 959–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clements A., Rojas J. R., Trievel R. C., Wang L., Berger S. L., and Marmorstein R. (1999) Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 18, 3521–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi S., Lin J., Cai Y., Yu J., Hong H., Ji K., Downey J. S., Lu X., Chen R., Han J., and Han A. (2014) Dimeric structure of p300/CBP associated factor. BMC Struct. Biol. 14, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rojas J. R., Trievel R. C., Zhou J., Mo Y., Li X., Berger S. L., Allis C. D., and Marmorstein R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98 [DOI] [PubMed] [Google Scholar]
- 30. Clements A., Poux A. N., Lo W. S., Pillus L., Berger S. L., and Marmorstein R. (2003) Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol. Cell 12, 461–473 [DOI] [PubMed] [Google Scholar]
- 31. Trievel R. C., Li F. Y., and Marmorstein R. (2000) Application of a fluorescent histone acetyltransferase assay to probe the substrate specificity of the human p300/CBP-associated factor. Anal. Biochem. 287, 319–328 [DOI] [PubMed] [Google Scholar]
- 32. Ferreon J. C., Martinez-Yamout M. A., Dyson H. J., and Wright P. E. (2009) Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc. Natl. Acad. Sci. U.S.A. 106, 13260–13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rasti M., Grand R. J., Mymryk J. S., Gallimore P. H., and Turnell A. S. (2005) Recruitment of CBP/p300, TATA-binding protein, and S8 to distinct regions at the N terminus of adenovirus E1A. J. Virol. 79, 5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mao Y., Lin J., Zhou A., Ji K., Downey J. S., Chen R., and Han A. (2011) Quikgene: a gene synthesis method integrated with ligation-free cloning. Anal. Biochem. 415, 21–26 [DOI] [PubMed] [Google Scholar]
- 35. Ellman G. L. (1959) Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 36. Freyer M. W., and Lewis E. A. (2008) Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 84, 79–113 [DOI] [PubMed] [Google Scholar]
- 37. Simossis V. A., and Heringa J. (2005) PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 33, W289–W294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pierce B. G., Wiehe K., Hwang H., Kim B. H., Vreven T., and Weng Z. (2014) ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. London N., Raveh B., Cohen E., Fathi G., and Schueler-Furman O. (2011) Rosetta FlexPepDock web server: high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 39, W249–W253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu X., Lu X., Liu R., Ai N., Cao Z., Li Y., Liu J., Yu B., Liu K., Wang H., Zhou C., Wang Y., Han A., Ding F., and Chen R. (2014) Histone cross-talk connects protein phosphatase 1α (PP1α) and histone deacetylase (HDAC) pathways to regulate the functional transition of bromodomain-containing 4 (BRD4) for inducible gene expression. J. Biol. Chem. 289, 23154–23167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen R., Liu M., Li H., Xue Y., Ramey W. N., He N., Ai N., Luo H., Zhu Y., Zhou N., and Zhou Q. (2008) PP2B and PP1α cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 22, 1356–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ai N., Hu X., Ding F., Yu B., Wang H., Lu X., Zhang K., Li Y., Han A., Lin W., Liu R., and Chen R. (2011) Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 39, 9592–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Horwitz G. A., Zhang K., McBrian M. A., Grunstein M., Kurdistani S. K., and Berk A. J. (2008) Adenovirus small E1A alters global patterns of histone modification. Science 321, 1084–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Flint J., and Shenk T. (1989) Adenovirus E1A protein paradigm viral transactivator. Annu. Rev. Genet. 23, 141–161 [DOI] [PubMed] [Google Scholar]
- 46. Ferreon A. C., Ferreon J. C., Wright P. E., and Deniz A. A. (2013) Modulation of allostery by protein intrinsic disorder. Nature 498, 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ferrari R., Gou D., Jawdekar G., Johnson S. A., Nava M., Su T., Yousef A. F., Zemke N. R., Pellegrini M., Kurdistani S. K., and Berk A. J. (2014) Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe 16, 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tanner K. G., Langer M. R., Kim Y., and Denu J. M. (2000) Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J. Biol. Chem. 275, 22048–22055 [DOI] [PubMed] [Google Scholar]
- 49. Ablack J. N., Cohen M., Thillainadesan G., Fonseca G. J., Pelka P., Torchia J., and Mymryk J. S. (2012) Cellular GCN5 is a novel regulator of human adenovirus E1A-conserved region 3 transactivation. J. Virol. 86, 8198–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pelka P., Ablack J. N., Shuen M., Yousef A. F., Rasti M., Grand R. J., Turnell A. S., and Mymryk J. S. (2009) Identification of a second independent binding site for the pCAF acetyltransferase in adenovirus E1A. Virology 391, 90–98 [DOI] [PubMed] [Google Scholar]
- 51. Tanner K. G., Langer M. R., and Denu J. M. (2000) Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 39, 11961–11969 [DOI] [PubMed] [Google Scholar]
- 52. Shuen M., Avvakumov N., Walfish P. G., Brandl C. J., and Mymryk J. S. (2002) The adenovirus E1A protein targets the SAGA but not the ADA transcriptional regulatory complex through multiple independent domains. J. Biol. Chem. 277, 30844–30851 [DOI] [PubMed] [Google Scholar]