Abstract
We describe an all-optical strategy for simultaneously manipulating and recording the activity of multiple neurons with cellular resolution in vivo. Concurrent two-photon optogenetic activation and calcium imaging is enabled by coexpression of a red-shifted opsin and a genetically encoded calcium indicator. A spatial light modulator allows tens of user-selected neurons to be targeted for spatiotemporally precise optogenetic activation, while simultaneous fast calcium imaging provides high-resolution network-wide readout of the manipulation with negligible optical crosstalk. Proof-of-principle experiments in mouse barrel cortex demonstrate interrogation of the same neuronal population during different behavioral states, and targeting of neuronal ensembles based on their functional signature. This approach extends the optogenetic toolkit beyond the specificity obtained with genetic or viral approaches, enabling high-throughput, flexible and long-term optical interrogation of functionally defined neural circuits with single-cell and single-spike resolution in the mammalian brain in vivo.
Introduction
Optogenetic actuators are rapidly revolutionizing experimental manipulations of neural activity, enabling activation and inactivation with millisecond precision at the spatial scale of populations of neurons1. At the same time, calcium indicators of activity in neurons now permit quantitative readout of neural activity from hundreds of neurons with cellular resolution2. A combination of these two experimental approaches would be highly desirable for manipulating and recording the activity of many neurons simultaneously at the spatial and temporal resolution at which they function in vivo (Fig. 1a). Two-photon excitation provides the optical sectioning and signal-to-noise required to achieve such a goal with single-cell and single-spike precision in both the imaging and the stimulation channels. However, two-photon calcium imaging and two-photon optogenetics have yet to be combined effectively (but see Ref. 3). Moreover, the ability to individually target multiple neurons using patterned photostimulation is crucial for generating and manipulating natural patterns of activity in vivo.
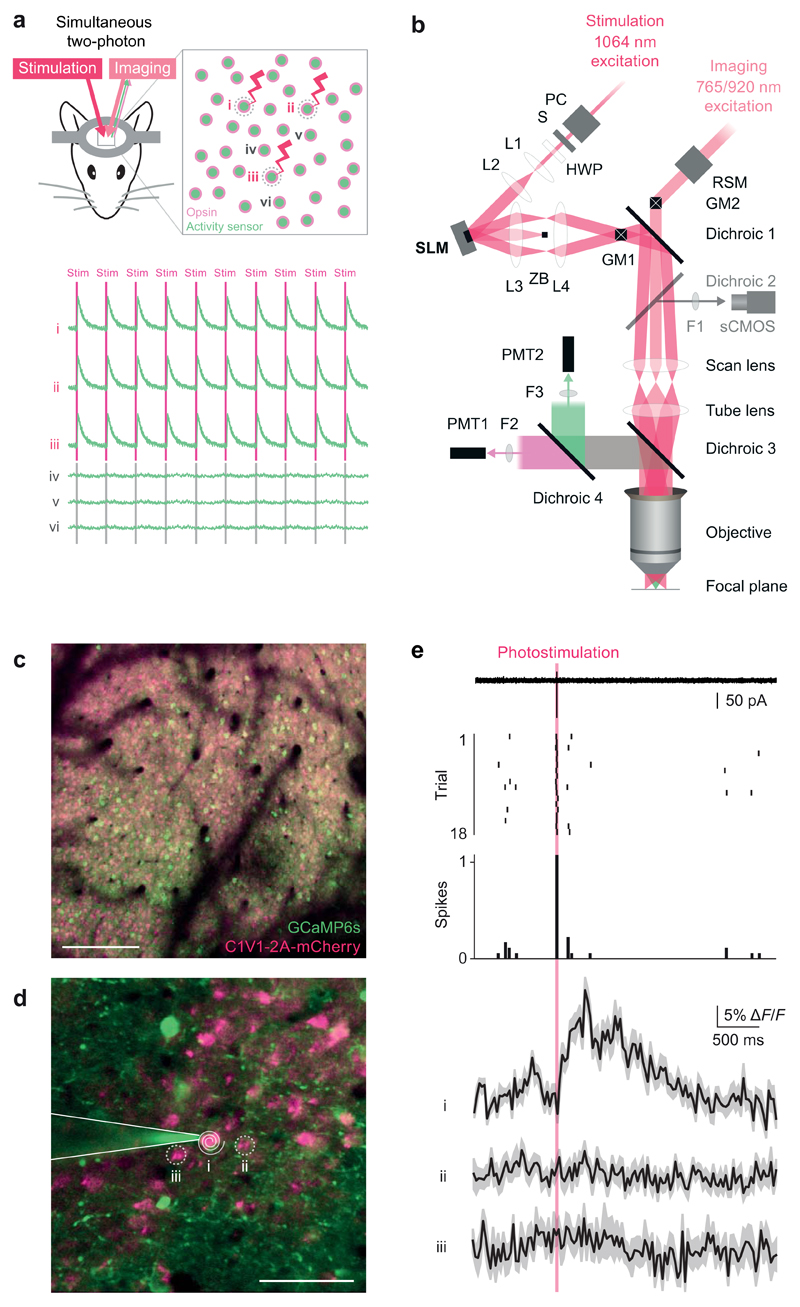
Figure 1. Single-cell two-photon optogenetic photostimulation and single action potential readout in vivo.
(a) Schematic illustration of the experimental goal. Successful interrogation of neural circuits at the resolution and precision at which they function requires a method with the specificity to simultaneously manipulate and record individual, user-selected neurons in the awake behaving animal. An all-optical approach to perform such experiments would be beneficial given the chronic and non-invasive nature in addition to the ability to localize neuron’s locations precisely in circuits where physical topology is critical. Top: Our approach utilizes a coexpression strategy to imbue neurons with read and write abilities: a calcium sensor generates an optical readout of activity while an opsin enables photostimulation. Bottom: The experimental goal would realize robust and reliable photostimulation in the user-selected neurons 1, 2, and 3 with sufficient resolution to avoid stimulating the immediately adjacent neurons 4, 5, and 6.
(b) Optical layout of the SLM-based (spatial light modulator) two-photon patterned photostimulation, two-photon resonant-scanning, moving in vivo microscope. The photostimulation beam (300 fs, 5 W, 1064 nm pulsed laser, Fianium Ltd or 100 fs, 2.3 W, 1055 nm pulsed laser, Coherent) was controlled by a Pockels’ cell (PC, Conoptics Ltd.) and shutter (S, Vincent Associates), resized using a beam-expanding telescope (L1 [f = 50 mm] and L2 [f = 200 mm], plano-convex lenses, Thorlabs) to fill the SLM active area and polarization maximized for diffraction efficiency from the SLM with a half wave plate (HWP, WPH10M-1064, Thorlabs). SLM: reflective spatial light modulator, 7.68 mm × 7.68 mm active area, 512 × 512 pixels, (Boulder Nonlinear Systems). L3: 1” achromatic doublet, f = 400 or 250 mm. ZB: zero order block. L4: 2” achromatic doublet, f = 150 mm. GM1: Galvanometer mirror pair, 3 or 6 mm (Cambridge Technology, integrated into dual beam Ultima microscope by Bruker Corp. [formerly Prairie Technologies]). GM2: Galvanometer mirror pair, 6mm (Cambridge Technology, integrated by Bruker Corp.). The imaging beam path can also be scanned along the fast axis by a resonant scanning galvanometer mirror (RSM, Cambridge Technology, integrated by Bruker Corp.) relayed onto GM2. Dichroic 1: 1030 nm short pass (T1030SP, Chroma Technology). Dichroic 2: 660 nm long pass (660LP, Chroma). This dichroic is used to image SLM beamlets in widefield mode on the sCMOS camera only when dichroic 3 is removed. F1: 675/67 nm bandpass filter (Semrock). sCMOS camera: ORCA-Flash4.0 (Hamamatsu). Scan lens: f=75 mm. Tube lens f=180 mm. Dichroic 3: 700 nm long pass (T700lpxxr-xxt, Chroma). Dichroic 4: 575LP (HQ575dcxr, Chroma). F2: 525/70 nm bandpass filter (525/70m-2P, Chroma). F3: 607/45 nm bandpass filter (607/45m-2P, Chroma). PMT1: Multi-alkali photomultiplier tube (Hamamatsu). PMT2: GaAsP photomultiplier tube (Hamamatsu). Objective: 16X 0.8 NA (Nikon).
(c) A large field of view of neurons co-expressing GCaMP6s and C1V1 (scale bar, 100 μm).
(d) Inset from a large field of view (200 × 200 μm) for the experiment shown in (e) (scale bar, 50 μm). A two-photon targeted cell-attached patch clamp recording was obtained from neuron 1, which co-expressed GCaMP6s and C1V1. This neuron was targeted for optogenetic stimulation by a beamspot produced by the SLM that was then driven in a spiral pattern by the galvanometer mirrors (white spiral).
(e) Top: electrophysiological recording during photostimulation trials reveals reliable generation of single action potentials during the photostimulation period (red bar) as evidenced in the single sweep (from trial 2), raster plot, and peristimulus time histogram. Bottom: calcium imaging recordings obtained simultaneously showed a transient (mean ± SEM matching the amplitude and kinetics expected for a single action potential with GCaMP6s in the photostimulated neuron, while the nearby neurons showed no detectable transients. Similar results were seen in n = 3 neurons.
Previous implementations of simultaneous manipulation and readout of neural activity are associated with a range of limitations, including manipulation artifacts on the recording channel, inadvertent activation caused by the recording, or lack of spatial resolution. Solely electrophysiological approaches suffer from electrical artifacts in the recording channel during stimulation; the fact that recordings are invasive; and the inability to target large groups of neurons while maintaining single cell resolution. On the other hand, optical approaches have so far suffered from a similar drawback as one-photon actuation, which not only stimulates many cells simultaneously (even when purposefully minimized4), but can also generate a large optical artifact in the imaging channel due to spectral overlap of the actuator excitation and readout emission wavelengths. This results in data loss during the crucial photostimulation period as the imaging sensor must be either blanked or data discarded5–7. Promising approaches to differentiate the manipulation and recording wavelengths have been employed in C. elegans, though the red calcium indicator used does not provide single action potential resolution8. Additionally, using one-photon widefield excitation generates substantial autofluorescence throughout the tissue, which can generate a large optical artifact on the imaging channel, and moreover does not permit targetable manipulations with cellular resolution. Simultaneous two-photon holographic imaging and photostimulation methods relying on glutamate uncaging have only been employed in vitro9,10. Combined optical and electrophysiology approaches also show promise for specific applications11–13, though it can be difficult to ascertain which neurons are being directly activated and which are downstream of the activated neurons14,15.
Here, we take advantage of the superior optical sectioning afforded by in vivo two-photon microscopy16,17 to precisely activate multiple identified neurons while simultaneously performing high-speed calcium imaging with minimal crosstalk in mice in vivo. Independent manipulation of two femtosecond-pulsed laser beams enables precise control over the activated neurons and high resolution spatiotemporal recording of stimulated and nearby neurons. User-selected targeting of neurons for activation is performed with a programmable spatial light modulator18 (SLM), allowing the experimenter to photostimulate neurons based on their individual functional identities, not just genetic class. In combination with chronic window preparations19, our approach enables high-throughput probing of the same neural circuits in awake behaving animals over long timescales.
Results
Combining calcium imaging and single cell photostimulation in vivo
Our design goal was to be able to selectively activate groups of individually-selected neurons based on their functional identity, and then observe the response to stimulation in the selected neurons and the local network in an awake, behaving animal (Fig. 1a). We incorporated an SLM into a two-photon in vivo dual-beampath resonant-scanning microscope in order to provide simultaneous two-photon imaging and patterned optogenetic stimulation (Fig. 1b). We visualized neurons co-infected with GCaMP6s, an ‘ultrasensitive’ green-fluorescent genetically encoded calcium indicator20, and C1V1-2A-mCherry, a red-shifted opsin21 (Fig. 1c). We recorded high-speed (30 Hz) calcium imaging movies over a field of view of 200 × 200 µm using a resonant scanning system at 920 nm and photostimulated neurons via two-photon excitation at 1064 nm (Fig. 1d). We calibrated our all-optical approach by performing simultaneous cell-attached patch-clamp recordings in layer 2/3 of mouse somatosensory cortex (n = 3 experiments). The SLM was programmed to generate a single photostimulation spot, which was scanned in a spiral fashion22 over the neuronal cell body for 20 ms (see Methods). Single action potentials were reliably generated in the recorded neuron on every trial, resulting in large calcium transients in the photostimulated neuron, but not in neighboring neurons also expressing GCaMP6s and C1V1 (Fig. 1e). The spatial resolution of spiral photostimulation using 6 mm galvanometers (see Methods) was 12 µm laterally and ~24 µm axially, measured by incrementally shifting the photostimulation target (Supplementary Fig. 1). Action potential detection sensitivity was maintained even when imaging the entire 200 × 200 µm field of view, and imaging at this resolution did not result in photostimulation (Supplementary Fig. 2). In summary, we demonstrate that our approach is capable of simultaneous sensitive calcium imaging and precise two-photon optogenetic photostimulation in the same neurons in vivo.
An alternative method for generating single action potentials in cortical neurons in vivo, which is available on all commercially available two-photon microscopes, was utilized for testing in neurons expressing C1V1-2A-YFP via viral infection (Supplementary Fig. 3a). A spatiotemporal photostimulation pattern of a 10 × 10 grid of sites extending slightly beyond the borders of the neuron was chosen to ensure robust action potential generation in the recorded neuron. The photostimulation laser was directed to the target sites sequentially in raster fashion (0.1 ms/site, 0.1 ms inter-site interval, 20 ms total exposure time; Supplementary Fig. 3a), generating exactly one action potential with a latency of 17.3 ± 0.7 (mean ± SEM) ms, measured from photostimulation onset, in putative pyramidal neurons, in a similar fashion to recent work23. Increasing the total exposure time to 40 ms (0.2 ms/site, 0.2 ms inter-site interval) generated more action potentials at longer latencies due to the increased time it took to cover the cell soma (1.4 ± 0.09 action potentials, 33.2 ± 2.1 ms latency to first action potential, n = 4 neurons, 3 animals, 57 photostimulation trials, Supplementary Fig. 3b. Increasing the exposure time yet further to 120 ms (1 ms/site, 0.2 ms inter-site interval) generated a similar number of action potentials in the recorded neurons (1.4 ± 0.07 action potentials, 60 ± 2.6 ms latency, n = 9 neurons from 4 mice; 114 photostimulation trials; Supplementary Fig. 3b). Additional illumination did not result in more action potentials (perhaps due to opsin desensitization exhausting the restricted population of opsins available in the small photostimulation volume). For comparison, the same photostimulation paradigm was also tested on a fast-spiking, putative interneuron, which responded in a similar fashion, although with a shorter latency and more action potentials as expected from its intrinsic electrophysiological properties (3.8 ± 0.17 action potentials, 18.1 ± 1.3 ms latency, n = 1 neuron; 100 photostimulation trials; Supplementary Fig. 4).
The spatial resolution of photostimulation using the grid pattern with 3 mm galvanometers, measured by moving the pattern relative to the neuron, was 22 µm laterally and 67 µm axially (FWHM; Supplementary Fig. 3c). In total, we recorded from 19 neurons in five mice while photostimulating with a range of parameters and found a reliable strategy for optically generating action potentials in individual neurons with low jitter in vivo.
Simultaneous photostimulation of multiple selected neurons
We used the SLM to split the photostimulation laser into individually spatially targeted beamlets to activate multiple neurons simultaneously24. Animals virally infected with C1V1-2A-YFP or C1V1-2A-mCherry showed strong expression in neurons across large territories of cortical tissue, providing many targets to select for simultaneous photostimulation (Fig. 2a). Spiral scanning the beamlets as a group allowed more neurons to be targeted, as less power per beamlet is required (i.e. a high power density in a small beamspot is spatiotemporally multiplexed over a neuron, as opposed to an alternate strategy whereby much more power is directed on sample, but distributed over a larger area25). We confirmed that our photostimulation strategy generated action potentials via simultaneous two-photon targeted cell-attached recordings when just one of the selected ten targets was positioned over a neuron (Fig. 2b). Spiral scans lasting 11 or 16 ms reliably generated one action potential at similar latencies (1.2 ± 0.04 and 1.2 ± 0.05 action potentials, one or more action potentials on 98 ± 3 and 88 ± 4 % of trials, 26.4 ± 13.3 and 21.4 ± 3.3 ms latency, 18.8 ± 17.1 and 8.0 ± 4.0 ms jitter [defined as the standard deviation of the latency] respectively; 107 photostimulation trials in six neurons in four animals; Fig. 2c). Increasing the spiral duration increased the number of action potentials generated and latency (34 ms spiral: 1.5 ± 0.06 action potentials, one or more action potentials on 87 ± 8 % of trials, 34.9 ± 4.6 ms latency, 11.3 ± 3.3 ms jitter, 93 photostimulation trials in six neurons in four animals), presumably due to the increased photostimulation exposure time and the longer time it took to cover the cell body, respectively. Increasing the number of photostimulated neurons from ten to 20 reduced the laser power per spot, resulting in a slight increase in the latency (1.3 ± 0.06 and 1.2 ± 0.05 action potentials, one or more action potentials on 88 ± 6 and 100 ± 0 % of trials, 30.1 ± 8.5 and 35.9 ± 4.0 ms latency, 17.0 ± 11.9 and 5.6 ± 0.8 ms jitter for 16 ms and 34 ms spirals respectively; 70 photostimulation trials for six recorded neurons in three animals). In summary, we demonstrated optically generated action potentials with low jitter (comparable to previous in vitro work23,24,26) in 10 to 20 selected neurons, confirmed by recording from 11 neurons in seven animals. These experiments show that our approach can generate precisely timed action potentials in multiple, individually selected neurons using programmed patterns of photostimulation.
Figure 2. Precise photostimulation of multiple identified neurons in vivo.
(a) Left: An example field of view of somatosensory cortex layer 2/3 neurons expressing C1V1-2A-YFP (scale bar, 100 μm). Right: Protocol to target spiral photostimulation patterns to multiple locations using the SLM and galvanometer mirrors (scale bars, 100 μm).
(b) Left: Magnification of a, showing cell-attached patch-clamp recording configuration in which multiple locations were photostimulated while electrophysiologically confirming action potential generation in one of the targets. Middle: Raster of spike times around stimulus delivery over 10 trials (photostimulation, red bar). Right: Overlaid raw data showing low latency and jitter.
(c) Metrics evaluating performance for 10 and 20 spot photostimulation patterns and spiral photostimulations lasting 11, 16, or 34 ms. Error bars are SEM, n = six neurons, four mice.
Network readout during targeted multi-neuron stimulation
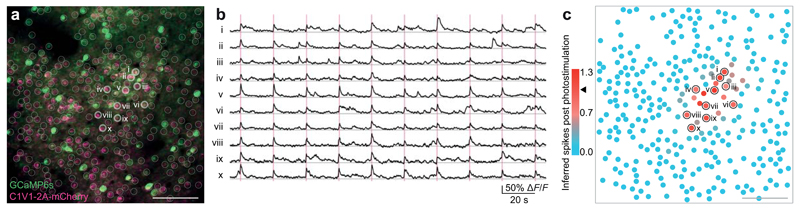
We combined the all-optical approach (Fig. 1) with the photostimulation of multiple specified neurons (Fig. 2) in order to investigate how precisely controlled photostimulation inputs are integrated by neural populations in vivo. We programmed the SLM to generate ten beamlets targeted to a cluster of ten neurons. While simultaneously imaging these and the surrounding 290 neurons at 30 Hz, we photostimulated the ten selected neurons (Fig. 3a). Individual target neurons showed strong and reliable responses to photostimulation (Fig. 3b-c). We performed this experiment in three mice. Only a small proportion of neighboring neurons responded (Supplementary Fig. 5), as expected from our and others’ previous work27,28. Some neurons expressed GCaMP6s very strongly, as observed previously20,29, and showed reduced responses to photostimulation (Supplementary Fig. 6a), though these neurons could be excluded from subsequent analysis. The response to photostimulation correlated slightly but not significantly with the expression of C1V1 (Supplementary Fig. 6b). In summary, we were able to optically invoke spatiotemporally precise action potentials in defined sets of neurons while simultaneously recording the responses of those neurons as well as many others in the same field of view.
Figure 3. Simultaneous fast calcium imaging and photostimulation of multiple neurons in vivo.
(a) A field of view of neurons in layer 2/3 of somatosensory cortex colabeled with C1V1-2A-mCherry and GCaMP6s (scale bar, 100 μm). Ten neurons were targeted for simultaneous photostimulation (white circles).
(b) Calcium transients from the ten photostimulated neurons showing the responses on all individual trials.
(c) Color-coded plot of the strength of photostimulation as measured by the sum of inferred spikes immediately post-stimulation. Stimulated neurons are circled in black. 300 total neurons, 120 trials, mean inferred action potentials post photostimulation = 1.0 ± 0.1 (mean ± SD, black arrow in legend).
Long-term reliability and stability of all-optical interrogation
We assessed the reliability and stability of multiple cell stimulation and readout. First, we photostimulated ten neurons (Supplementary Fig. 7a) and then found the same ten neurons one week later and photostimulated them again (Supplementary Fig. 7b; in three fields of view in three animals). We observed the same response to photostimulation on both days (Supplementary Fig. 7c). All neurons in all animals showed a highly significant coefficient of determination between their response on day 1 and day 8 (p < 0.0001, R2 = 0.7 to the identity line, Supplementary Fig. 7d).
All-optical interrogation in different behavioral states
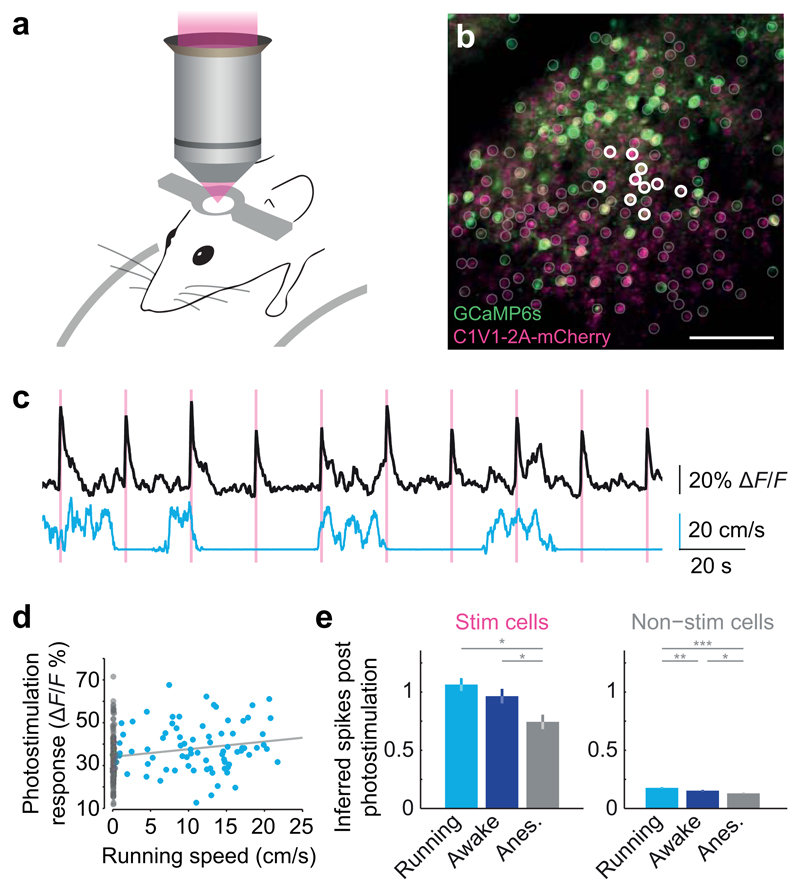
To demonstrate the power of our strategy for probing the functional properties of neural circuits in vivo we assessed how behavioral states modulate the responsiveness of neural circuits. We applied our approach to monitor target neurons and the local network response to photostimulation while mice were in different behavioral states: running, awake but stationary, or under light isoflurane anesthesia. Recent work has shown that locomotion is a salient modulator of neuronal activity in cortical circuits30–32. We performed experiments with awake and head-fixed mice allowed to run freely on a fixed-axis cylinder (Fig. 4a) while we repeatedly photostimulated ten target neurons (Fig. 4b). We simultaneously recorded the response to photostimulation and running speed (Fig. 4c). We found a significant correlation between mean amplitude of response to photostimulation and running speed (Spearman rho = 0.27, p < 0.0001; n = 22 experiments, 220 total stimulations in three mice; Fig. 4d). We performed the same patterned stimulation in the same field of view in the same mice while the animals were awake but not running, or lightly anesthetized (Fig 4e). We found a significant reduction of target neuron responses to photostimulation when animals were anesthetized (p < 0.001 one-way ANOVA; p < 0.001 running vs. anesthetized, p < 0.05 awake vs. anesthetized Tukey's test for multiple comparisons). There was a significant difference between all behavioral states for the responses of non-stimulated local network neurons (p < 0.0001 one-way ANOVA, p < 0.01 running vs. awake, p < 0.0001 running vs. anesthetized, p < 0.05 awake vs. anesthetized Tukey's test for multiple comparisons; n = three mice, 576 imaged neurons, 30 stimulated neurons, 280 trials distributed between behavioral states, minimum 20 trials per state per mouse). These results demonstrate the robustness of our approach for perturbing neuronal networks during ongoing activity and different behavioral states.
Figure 4. Behavioral state-dependence of network perturbations.
(a) Mice were head-fixed and allowed to run freely on a fixed-axis styrofoam treadmill.
(b) A field of view in which ten nearby neurons in layer 2/3 of mouse somatosensory cortex were chosen for photostimulation while they and the surrounding neurons across the field of view were simultaneously observed with high-speed calcium imaging.
(c) Top: mean calcium transient of all stimulated neurons in response to simultaneous photostimulation of ten neurons. Bottom: running speed.
(d) Correlation between mean perturbation of responses to photostimulation and running speed.
(e) Comparison of mean ± SD inferred spike responses to photostimulation during different behavioural states (n = 3 mice, 576 imaged neurons; Tukey’s test for multiple comparisons, * denotes p < 0.05).
Optogenetic manipulation targeted to functional ensembles
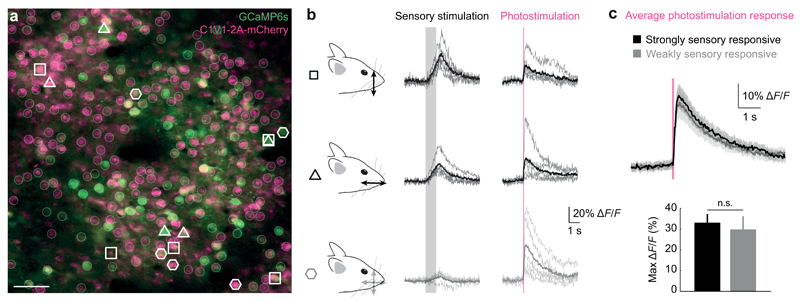
Our approach allowed us to target different groups of neurons within the population based on their individual functional properties. To achieve this goal, we first performed intrinsic imaging to identify barrel C2 in somatosensory cortex (Fig. 5a). Next, two-photon imaging of the responses of individual neurons to sensory stimulation within the barrel revealed neurons that responded strongly or weakly to particular stimulation orientations (Fig. 5b). We targeted photostimulation to five neurons within each of the three groups while the animal was awake and running on a styrofoam treadmill. The response to photostimulation was not significantly different between the strongly sensory-responsive (both groups) and weakly sensory-responsive groups (max ΔF/F response to photostimulation: 33 ± 4 % and 30 ± 6 %, p = 0.55, Mann-Whitney test; n = 25 and 15 strongly and weakly sensory responsive neurons, respectively, in two fields of view; Fig. 6c). This experiment highlights the utility of our approach for activating functionally defined ensembles of neurons in the awake, behaving animal.
Figure 5. Targeting neurons for optogenetic manipulation based on their individual functional signatures in vivo.
(a) A field of view (scale bar, 50 µm) showing neurons co-expressing GCaMP6s (green) and C1V1-2A-mCherry (purple) in the C2 barrel of mouse somatosensory cortex.
(b) Groups of individually identified neurons were selected for photostimulation based on their response to dorsal-ventral and rostro-caudal whisker stimulation (open symbols). Five neurons that responded differently or not at all to sensory stimulation (gray shading) were simultaneously photostimulated (red line).
(c) Simultaneous photostimulation of five neurons responding strongly to a given sensory stimulation or weakly to both stimuli revealed similar responses (mean ± SEM) to photostimulation (n = 1 mouse).
Discussion
Probing neural circuits at the spatial and temporal resolution at which they function is crucial to understanding how populations of neurons work together to drive behavior. Optical approaches provide a means to directly address these questions in a minimally invasive manner. We have developed an all-optical strategy for activating and recording the same neurons with cellular resolution in mice in vivo using a dual two-photon optogenetic and calcium imaging approach. The strategy relies on viral coexpression of a genetically encoded activity sensor (GCaMP6s) and a red-shifted opsin (C1V1). A spatial light modulator enables targeting tens of user-selected neurons for temporally and spatially precise, simultaneous optogenetic activation using 1064 nm excitation. In parallel, simultaneous fast calcium imaging with single action potential resolution provides readout of the manipulation as well as its effect on hundreds of other neurons. The ability to target neurons based on their functional signature, rather than simply their genetic identity, will enable one to replay and manipulate natural patterns of activity in neural circuits1.
We calibrated the reliability and temporal precision of both activation and readout of activity using cell-attached targeted patch clamp recordings from identified neurons. The combination of indicator sensitivity and lack of spectral overlap between the optogenetic excitation and GCaMP6s emission wavelengths minimizes the stimulation artifact. The optical nature of the approach combined with a chronic window preparation provides the flexibility to select individual neurons for stimulation while the animal is awake and behaving under the microscope over weeks to months, and the ability to target activation of neurons which are normally active during a behavioral task. We demonstrate proof-of-principle experiments by activating groups of layer 2/3 pyramidal neurons in mouse barrel cortex in defined spatiotemporal patterns during behavior. Our method enables high-throughput, flexible, and long-term optical interrogation of neural circuits in the mammalian brain in vivo.
This strategy provides a significant advance over previous approaches in which neurons were defined either anatomically or genetically, as it allows targeting of individual neurons based on their specific functional properties, as characterized by activity measured during observation-only imaging experiments. Such single neuron precision, simultaneous stimulation and readout experiments will open up new avenues for optogenetic experiments.
Previous methods for single neuron precision optogenetic manipulation relied on raster or spiral scanning methods to spatiotemporally multiplex a small excitation beamspot over a cell body22–24,33–35 or holographically ‘painting’ the cell body combined with temporal focusing to provide optical sectioning3,26. All of these approaches have thus far only been demonstrated in vitro, aside from our previous proof-of-principle experiments23 showing that the two-photon optogenetic excitation strategy could work in vivo and recent work that came out while this work was in review3. Other work has shown that temporal focusing combined with general phase contrast enables action potential generation via two-photon excitation of channelrhodopsin deep in brain slices36.
Our approach, combining SLM-based beam splitting with spiral scanning, enables simultaneous addressing of a large population of neurons in vivo. There is a tradeoff between the number of neurons that can be simultaneously addressed versus how quickly a simultaneous stimulation pattern can be performed, dictated by the power and time required for a given spatiotemporal multiplexing strategy. In alternative holographic and/or temporal focusing based approaches, simultaneous photostimulations can be achieved more quickly, at the cost of addressing fewer neurons per ‘shot’. The advantage of such alternative strategies may be more efficient shaping of the temporal profile of the response pattern. Our strategy attempts to maximize the number of neurons per stimulation while minimizing photostimulation time by splitting the beam and using a fast scanning strategy to photostimulate neurons. Thus, while all of these technologies are currently under active development, our approach is well-suited for generation of synchronous activity among ensembles of neurons, which can be performed at high rates in a sequence. We expect that the underlying biological question at hand will dictate the choice between these different approaches in vivo.
Our all-optical, dual two-photon approach combining fast readout with high-speed calcium imaging of the most recent ‘ultrasensitive’ calcium indicator20 drastically reduces the crosstalk problem observed in previous all-optical approaches relying on one-photon excitation5–7. These approaches required blocking the readout during photostimulation, resulting in loss of crucial data. Additionally, the ability to directly observe neuronal locations and target neurons with single-cell precision removes any confusion about which cells are directly stimulated and which are downstream14,15, which has been a problem for other simultaneous readout and manipulation strategies relying on the combination of electrophysiology and optogenetics, such as optrodes11–13. Further development of new molecular tools37, including characterization of their two-photon excitation, will enable their integration into the in vivo approach presented here. In addition, advances in faster and more complex imaging methods38–40 and analysis41, can also be incorporated into the readout side of the approach.
Our ability to read out and manipulate activity in the same population of neurons provides us with the unprecedented opportunity to target optogenetic manipulation based on the functional signature of the neurons. Previously, optogenetic manipulations could be targeted based on genetic and/or anatomical identity alone42, or on c-fos expression43, which only provides a very indirect readout of activity and lacks temporal precision. Here we provide proof-of-principle experiments showing that we can use a detailed functional characterization of the response of the neurons to different sensory stimuli in order to enable targeting of specific ensembles in the population. This will allow optogenetic manipulation to be targeted with far greater precision than previously possible, enabling investigation of the neural code in only those neurons within a population exhibiting a specific functional signature during sensory processing or defined behaviors. Future work using this approach should provide fundamental new insights into information processing in neural circuits in vivo.
Online Methods
All surgical procedures were carried out under licence from the UK Home Office in accordance with the Animal (Scientific Procedures) Act 1986. C57/BL6 mice (mus musculus) approximately 4 weeks to 3 months old of both genders were used without randomization. Animals were anesthetized with an intraperitoneal injection of a ketamine/xylazine mixture (0.1/0.01 mg/g body weight) or with isoflurane during surgical procedures.
Viral injections, headplate and chronic window installation
Craniotomies were drilled (NSK UK Ltd.) over barrel cortex (right hemisphere, 2 mm posterior from and 3.5 mm lateral of bregma, 0.5 mm diameter). Calibrated injection pipettes bevelled to a sharp point (inner diameter ~15 µm) were coupled to a hydraulic injection apparatus driven by a syringe pump (Harvard Apparatus). Virus (AAV2-CaMKIIa-C1V1(E162T)-p2A-EYFP, AAVdj-CaMKIIa-C1V1(E162T)-TS-P2A-mCherry-WPRE1, or AAV1-Syn-GCaMP6s-WPRE-SV40) was front-loaded into the injection pipette and injected at a rate of 0.1 µL min-1 into layer 2/3 (~300 µm deep). Animals that received two separate injections were first injected with 1 µL of virus and then the scalp incision closed with Vetbond (3M). Subsequent injections, also 1 µL, were performed through the same craniotomy site that was still visible 24 days later. Animals singly injected received 1 µL of virus. Co-injected animals were injected with a mixture containing 0.1 µL GCaMP6s virus and 0.9 µL C1V1-2A-mCherry virus.
For chronic experiments, the scalp was removed bilaterally from the midline to the temporalis muscles and a metal headplate with 5 mm circular imaging well fixed to the skull overlying somatosensory cortex with dental cement (Super-Bond C&B, Sun-Medical) before virus injection. Intrinsic imaging2 was performed to further localize the injection site to the C2 barrel in certain experiments. The C2 whisker was threaded into a trimmed glass pipette glued to a piezoeletric actuator (Physik Instrumente). The actuator was programmed to oscillate at 10 Hz for 4 seconds with a 20 second interstimulus interval. The brain was perfused with sterile external solution (150 mM NaCl, 2.5 mM KCl, 10 mM HEPES, 2 mM CaCl2, 1 mM MgCl2) to make the skull semi-transparent and imaged with a CCD camera (Pike F-032b, Allied Vision Technologies) while a red LED illuminated the surface. Increase in reflectance from the brain indicated the barrel, which could be localized relative to the blood vessel pattern as visualized with a green LED. After virus injection (see above), a 3 mm circular craniotomy was performed during which any bleeding was washed away with sterile external solution or stanched with Sugi-sponges (Sugi, Kettenbach). Cranial windows composed of a 3 mm circular glass coverslip cemented to a 2 mm square glass coverslip (UQG Optics) with UV-curable optical cement (NOR-61, Norland Optical Adhesive), were press-fit into the craniotomy, sealed to the skull by a thin layer of cyanoacrylate (VetBond), and fixed in place by dental cement.
Following all surgical procedures, animals recovered for at least 7 days with post-operative administration of 0.7 µL/mL Rimadyl and 4 µL/mL Baytril in drinking water.
Titration of calcium indicator expression
The sensitivity of the readout in our all-optical approach depended heavily on the expression of GCaMP6s. To assess stability of calcium indicator expression, we injected animals with GCaMP6s virus at three viral titers: 3.22e13 genomes/ml stock and lower titer solutions of 3.22e12 and 3.22e11 genomes/ml diluted in a buffer solution (20 mM Tris, pH 8.0, 140 mM NaCl, 0.001% pluronic F-68). 100 nL of virus was injected at 0.1 µL/min into L2/3 barrel cortex (2 mm posterior from and 3.5 mm lateral of bregma, 300 µm below dura). We divided 18 adult (P60 – P80) C57/BL6 mice of both sexes into 3 cohorts of 6 mice each and injected each cohort with one of the above dilutions. Each 6 animal viral-titer cohort was then divided such that, within each cohort, 2 animals could be used for each of 3 time points: 1 week, 2 week and 3 week post-injection. At each of these time points an acute craniotomy was performed to enable two-photon imaging (see below) under isoflurane anesthesia (5% for induction, 1.5% for surgery and 0.5% for imaging). Two-photon images tiling the infection site were taken at 10 µm intervals from the cortical surface to a maximum depth of 500 µm (depending on the extent of expression). Acute imaging was used in order to minimize potential confounding artifacts introduced by chronic window implantation. The crucial parameter was a reduction in virus titer to 10% of its original level (i.e. 1012 genomes/ml), which resulted in stable expression as long as 12 weeks post-injection (Supplementary Fig. 8). Next, we confirmed single action potential resolution by performing two-photon targeted cell-attached recordings while imaging at 60 Hz (n = 4 neurons; Supplementary Fig. 9a). We found that the number of spikes in a burst occurring within a 250 ms window could be reliably reported by integrating the results of the deconvolution algorithm3 (Supplementary Fig. 9b). Integrating the result of this inference over a 433 ms window resulted in the best performance as defined by the strength of the correlation between the true number of spikes and the inferred number of spikes (Supplementary Fig. 9c). We also quantified the hit rate (94 ± 3 %) and false positive rate (7 ± 6 %) on 3 neurons as in ref. 16, not including one held-out neuron on which we based the classifier (Supplementary Fig. 9d).
The need to reduce the titer of the GCaMP6s-producing virus meant we could dilute it directly with the virus expressing C1V1-2A-mCherry (see above). After injecting this mixture of viruses into the animals during a single surgical procedure, a chronic window was installed to provide long-term optical access to the cortical tissue of interest. After five weeks, we observed co-labeling of neurons with both GCaMP6s and C1V1-2A-mCherry, which were easily spectrally separated based on emission wavelength.
Cell-attached targeted patch-clamp recordings
Headplate installation was performed as described above. Two-photon targeted patch-clamp recordings4 were obtained using glass pipettes pulled from borosilicate glass (~5 MΩ pipette resistance) and filled with external solution. Signals were amplified on a MultiClamp 700a (Molecular Devices), filtered with a Bessel filter above 4 kHz, and recorded at 20 kHz with custom software (‘PackIO’5) written in LabView (National Instruments).
Imaging and photostimulation
Two-photon imaging was performed by raster scanning a femtosecond-pulsed laser beam (Chameleon Ultra II, Coherent) via standard or resonant galvanometer raster scanning with a moving in vivo microscope (Bruker Corporation, formerly Prairie Technologies). A 16x/0.8-NA objective (Nikon) was used for all experiments. GCaMP6s was imaged with excitation wavelength of 920 nm during calcium imaging and mCherry was imaged at 765 nm.
In longitudinal experiments, the same field of view could be found over weeks based on the blood vessel pattern observed under the chronic window. The chronic window allowed visualization of a 2 × 2 mm area of cortex, while the maximum field of view allowed by two-photon imaging was 832 × 832 µm. Thus, it was straightforward to localize the same general area based on large blood vessels visualized through the eyepieces of the microscope. Afterwards, images at high resolution obtained via two-photon imaging could be precisely matched to images from previous sessions based on blood vessels and any uniquely shaped processes or strongly labeled neurons. Micrometer-scale resolution was confirmed by comparing two high resolution images such as in Fig. 4a-b.
A reflective spatial light modulator (SLM, 7.68 mm × 7.68 mm active area, 512 × 512 pixels, optimized for 1064 nm, Meadowlark Optics/Boulder Nonlinear Systems) was coupled to the microscope using a lightpath similar to that for an in vitro microscope6, but with the significant difference that the entire microscope, including SLM and beam expansion telescope, moves relative to the sample. A computer running BNS_DVI Version 1.3 (Meadowlark Optics/Boulder Nonlinear Systems) displayed the phase mask on the SLM via a NVIDIA GeForce GTX 660 Ti graphics card and DVI cable connected to the SLM driver electronics (Meadowlark Optics/Boulder Nonlinear Systems). The manufacturer supplied lookup table converted pixel values to voltages driving the liquid crystal active area of the SLM. The zero order beam was blocked at the focus of L3 with a small piece of foil glued to a coverslip (Fig. 1b).
The excitation source for the photostimulation path was a femtosecond pulsed laser fixed at 1064 nm (total output 5 watts, pulse width 250-400 fs, Fianium Ltd) or 1055 nm (total output 2.3 watts, pulse width 100 fs, Coherent). The Gerchberg-Saxton (GS) algorithm7 was used to calculate phase masks to be displayed on the SLM that would result in the desired pattern with individual beams on each targeted neuron. We found that diffraction efficiency to targets situated at the periphery of the field of view, which required large angles to be generated by the SLM, was lower than for targets at the center of the field of view with the standard GS algorithm. This effect was likely due to the increasing impact of interpixel crosstalk in our SLM as diffracting to large angles requires higher resolution phase masks (smaller diffracting features) that increase chromatic aberration and are more prone to interpixel crosstalk due to the larger variation in neighboring pixel values. A linear weighting of desired spot intensity prior to the GS algorithm was sufficient to counteract this effect and resulted in sufficiently homogeneous power distribution (<6% standard deviation) among targets. The positioning of these beams relative to the two-photon images of the neurons was registered using a widefield sCMOS camera (ORCA-Flash4.0, Hamamatsu), which imaged the sample plane via the objective and tube lens, while photostimulating a homogeneously fluorescent sample. Alternatively, the photostimulation beams could be registered by burning a homogeneously fluorescent sample that could be visualized by the two-photon imaging laser. Spiral photostimulation patterns (3 rotations, 20 µm diameter) were generated by moving all beamlets simultaneously with a pair of galvanometer mirrors conjugate to the SLM plane. Power on sample for photostimulation using grid pattern ranged from 28-150 mW, generally 100 mW (for all experiments in Supplementary Fig. 3). Power on sample for spiral photostimulation experiments ranged from 20-80 mW. Power distribution was even across beamlets generated by the SLM (59 ± 3 mW, n = 3 patterns) as measured on sample by loading phase masks and blocking all but a single beamlet near the focus of L3 (see Fig. 1b.).
Sensory stimulation and behavior
Animals were gradually acclimatized to head-fixation if necessary and trained to sit passively in the apparatus. Animals generally ran freely on the styrofoam treadmill while running speed was measured with a rotary encoder on the axle. Whisker stimulation, which occurred under isoflurane anesthesia, consisted of either one second long, 10 Hz stimulation with a one-dimensional piezoelectric actuator (Physik Instrumente) or one second long, random, white-noise filtered stimuli with a two-dimensional piezoelectric actuator (Noliac) onto which a capillary tube was glued as a guide for an individual whisker.
Analysis
Movement correction was performed on all calcium imaging movies using a discrete Fourier transform (DFT) based algorithm8. This algorithm computes the cross-correlation between two images via selective upsampling with a matrix-multiply DFT, thus increasing speed and reducing memory requirements. Contours defining neuronal somata were selected by hand using both GCaMP and C1V1 channels. Spike inference was performed with a fast nonnegative deconvolution algorithm, for which the only required parameter is the imaging time step3. Analyses were performed with MATLAB and ImageJ; statistical tests were performed with Prism or MATLAB. All values are mean ± SEM unless otherwise noted.
Supplementary Material
Acknowledgments
We thank M. Pachitariu, C. Stringer, S. Turaga, M. London, and N. Pettit for helpful discussion, analysis routines and software; C. Wilms, C. Schmidt-Hieber, A. Roth, B. Clark, I. Bianco, S. Smith, B. Judkewitz and D. Peterka for discussion and comments on the manuscript, the staff at Bruker Corporation (formerly Prairie Technologies) for enabling customization of the microscope, M. Lochrie at the Stanford Neuroscience Gene Vector and Virus Core (Grant P30 NS069375-01A1) for advice on use of AAVdj, and K. Deisseroth (Stanford University) for plasmids and access to AAVdj virus. This work was supported by grants from the Wellcome Trust, the Gatsby Charitable Foundation, the European Commission (Marie Curie IIF), the European Molecular Biology Organization, the Medical Research Council, and the European Research Council.
Footnotes
Statement of Competing Financial Interests
All authors declare that there are no competing financial interests
Author Contributions
A.M.P., L.E.R., and H.W.P. performed experiments and analyzed data. A.M.P., L.E.R., H.W.P., and M.H. designed the study and wrote the paper.
Contributor Information
Adam M. Packer, Wolfson Institute for Biomedical Research, University College London, Gower Street, London WC1E 6BT, UK; Department of Neuroscience, Physiology and Pharmacology, University College London, Gower Street, London WC1E 6BT, UK
Lloyd E. Russell, Wolfson Institute for Biomedical Research, University College London, Gower Street, London WC1E 6BT, UK; Department of Neuroscience, Physiology and Pharmacology, University College London, Gower Street, London WC1E 6BT, UK
Henry W.P. Dalgleish, Wolfson Institute for Biomedical Research, University College London, Gower Street, London WC1E 6BT, UK; Department of Neuroscience, Physiology and Pharmacology, University College London, Gower Street, London WC1E 6BT, UK
Michael Häusser, Wolfson Institute for Biomedical Research, University College London, Gower Street, London WC1E 6BT, UK; Department of Neuroscience, Physiology and Pharmacology, University College London, Gower Street, London WC1E 6BT, UK.
References
- 1.Packer AM, Roska B, Hausser M. Targeting neurons and photons for optogenetics. Nat Neurosci. 2013;16:805–815. doi: 10.1038/nn.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grienberger C, Konnerth A. Imaging calcium in neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Rickgauer JP, Deisseroth K, Tank DW. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat Neurosci. 2014 doi: 10.1038/nn.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber D, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fajardo O, Zhu P, Friedrich RW. Control of a specific motor program by a small brain area in zebrafish. Front Neural Circuits. 2013;7:67. doi: 10.3389/fncir.2013.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson NR, Runyan CA, Wang FL, Sur M. Division and subtraction by distinct cortical inhibitory networks in vivo. Nature. 2012;488:343–348. doi: 10.1038/nature11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang YP, Oertner TG. Optical induction of synaptic plasticity using a light-sensitive channel. Nat Methods. 2007;4:139–141. doi: 10.1038/nmeth988. [DOI] [PubMed] [Google Scholar]
- 8.Akerboom J, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anselmi F, Ventalon C, Begue A, Ogden D, Emiliani V. Three-dimensional imaging and photostimulation by remote-focusing and holographic light patterning. Proc Natl Acad Sci U S A. 2011;108:19504–19509. doi: 10.1073/pnas.1109111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dal Maschio M, et al. Simultaneous two-photon imaging and photo-stimulation with structured light illumination. Opt Express. 2010;18:18720–18731. doi: 10.1364/OE.18.018720. [DOI] [PubMed] [Google Scholar]
- 11.LeChasseur Y, et al. A microprobe for parallel optical and electrical recordings from single neurons in vivo. Nat Methods. 2011;8:319–325. doi: 10.1038/nmeth.1572. [DOI] [PubMed] [Google Scholar]
- 12.Anikeeva P, et al. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat Neurosci. 2012;15:163–170. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Royer S, et al. Multi-array silicon probes with integrated optical fibers: light-assisted perturbation and recording of local neural circuits in the behaving animal. Eur J Neurosci. 2010;31:2279–2291. doi: 10.1111/j.1460-9568.2010.07250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lima SQ, Hromadka T, Znamenskiy P, Zador AM. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS One. 2009;4:e6099. doi: 10.1371/journal.pone.0006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kravitz AV, Owen SF, Kreitzer AC. Optogenetic identification of striatal projection neuron subtypes during in vivo recordings. Brain Res. 2013;1511:21–32. doi: 10.1016/j.brainres.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- 17.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 18.Nikolenko V, et al. SLM Microscopy: Scanless Two-Photon Imaging and Photostimulation with Spatial Light Modulators. Front Neural Circuits. 2008;2:5. doi: 10.3389/neuro.04.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtmaat A, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci U S A. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prakash R, et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Packer AM, et al. Two-photon optogenetics of dendritic spines and neural circuits. Nat Methods. 2012;9:1202–1205. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peron S, Svoboda K. From cudgel to scalpel: toward precise neural control with optogenetics. Nat Methods. 2011;8:30–34. doi: 10.1038/nmeth.f.325. [DOI] [PubMed] [Google Scholar]
- 26.Papagiakoumou E, et al. Scanless two-photon excitation of channelrhodopsin-2. Nat Methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwan AC, Dan Y. Dissection of cortical microcircuits by single-neuron stimulation in vivo. Curr Biol. 2012;22:1459–1467. doi: 10.1016/j.cub.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.London M, Roth A, Beeren L, Hausser M, Latham PE. Sensitivity to perturbations in vivo implies high noise and suggests rate coding in cortex. Nature. 2010;466:123–127. doi: 10.1038/nature09086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Y, et al. A cortical circuit for gain control by behavioral state. Cell. 2014;156:1139–1152. doi: 10.1016/j.cell.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niell CM, Stryker MP. Modulation of visual responses by behavioral state in mouse visual cortex. Neuron. 2010;65:472–479. doi: 10.1016/j.neuron.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider DM, Nelson A, Mooney R. A synaptic and circuit basis for corollary discharge in the auditory cortex. Nature. 2014;513:189–194. doi: 10.1038/nature13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu P, et al. Optogenetic Dissection of Neuronal Circuits in Zebrafish using Viral Gene Transfer and the Tet System. Front Neural Circuits. 2009;3:21. doi: 10.3389/neuro.04.021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc Natl Acad Sci U S A. 2010;107:11981–11986. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Losonczy A, Zemelman BV, Vaziri A, Magee JC. Network mechanisms of theta related neuronal activity in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2010;13:967–972. doi: 10.1038/nn.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papagiakoumou E, et al. Functional patterned multiphoton excitation deep inside scattering tissue. Nature Photonics. 2013;7:274–278. [Google Scholar]
- 37.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahrens MB, et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prevedel R, et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat Meth. 2014;11:727–730. doi: 10.1038/nmeth.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quirin S, Jackson J, Peterka DS, Yuste R. Simultaneous imaging of neural activity in three dimensions. Front Neural Circuits. 2014;8:29. doi: 10.3389/fncir.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freeman J, et al. Mapping brain activity at scale with cluster computing. Nat Meth. 2014 doi: 10.1038/nmeth.3041. advance online publication. [DOI] [PubMed] [Google Scholar]
- 42.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Grimm D, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grinvald A, Lieke E, Frostig RD, Gilbert CD, Wiesel TN. Functional architecture of cortex revealed by optical imaging of intrinsic signals. Nature. 1986;324:361–364. doi: 10.1038/324361a0. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein JT, et al. Fast nonnegative deconvolution for spike train inference from population calcium imaging. J Neurophysiol. 2010;104:3691–3704. doi: 10.1152/jn.01073.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- 5.Packer AM. Understanding the nervous system as an information processing machine: dense, nonspecific, canonical microcircuit architecture of inhibition in neocortex and a neural circuit for angular velocity computation. PhD thesis; Columbia University: 2011. [Google Scholar]
- 6.Nikolenko V, et al. SLM Microscopy: Scanless Two-Photon Imaging and Photostimulation with Spatial Light Modulators. Front Neural Circuits. 2008;2:5. doi: 10.3389/neuro.04.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerchberg RW, a. S WO. A practical algorithm for the determination of the phase from image and diffraction plane pictures. Optik. 1972;35 [Google Scholar]
- 8.Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008;33:156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.