Abstract
Genetic evidence suggests that membranes rich in polyunsaturated fatty acids (PUFAs) act as supramolecular antioxidants that capture reactive oxygen species, thereby limiting damage to proteins. This process generates lipid fragmentation products including malondialdehyde (MDA), an archetypal marker of PUFA oxidation. We observed transient increases in levels of endogenous MDA in wounded Arabidopsis thaliana leaves, raising the possibility that MDA is metabolized. We developed a rigorous ion exchange method to purify enzymatically generated 13C- and 14C-MDA. Delivered as a volatile to intact plants, MDA was efficiently incorporated into lipids. Mass spectral and genetic analyses identified the major chloroplast galactolipid: α-linolenic acid (18:3)-7Z,10Z,13Z-hexadecatrienoic acid (16:3)-monogalactosyldiacylglycerol (18:3–16:3-MGDG) as an end-product of MDA incorporation. Consistent with this, the fad3-2 fad7-2 fad8 mutant that lacks tri-unsaturated fatty acids incorporated 14C-MDA into 18:2–16:2-MGDG. Saponification of 14C-labeled 18:3–16:3-MGDG revealed 84% of 14C-label in the acyl groups with the remaining 16% in the head group. 18:3–16:3-MGDG is enriched proximal to photosystem II and is likely a major in vivo source of MDA in photosynthetic tissues. We propose that nonenzymatically generated lipid fragments such as MDA are recycled back into plastidic galactolipids that, in their role as cell protectants, can again be fragmented into MDA.
Keywords: antioxidant, Arabidopsis thaliana, lipid oxidation, photosynthesis, polyunsaturated fatty acid (PUFA), reactive oxygen species (ROS), omega-3, photoprotection, singlet oxygen
Introduction
The susceptibility of fatty acids to oxidation by reactive oxygen species (ROS)3 increases with desaturation (1, 2), so that membranes rich in polyunsaturated fatty acids (PUFAs) are potentially vulnerable to oxidative damage. It is therefore remarkable that intra-organellar membranes in mitochondria and chloroplasts, organelles with highly oxidative metabolism, typically contain high percentages of PUFAs (3, 4). Both organelle types are active sites of ROS production (5, 6). For example, thylakoid membranes in chloroplasts are prone to photo-oxidative damage by singlet oxygen (1O2) that is produced through the interaction of excited (triplet-state) chlorophyll with oxygen (6–8). Moreover, the major site of 1O2 generation in chloroplasts, photosystem II (PSII), is surrounded by polyunsaturated fatty acid (PUFA)-rich lipids (9) and, strikingly, two-thirds of the FAs in thylakoids are triunsaturated fatty acids (TFAs), chiefly α-linolenic acid (10). These fatty acids are highly prone to oxidative fragmentation in vivo (11).
Cells counter the threat posed by 1O2 and other ROS with multiple protection mechanisms known to involve low molecular mass antioxidants. Among the best characterized of these cellular protectants are tocopherols (reviewed in Ref. 12). Indeed, mutants that lack tocopherol and plastochromanol are less protected against 1O2 and show elevated levels of nonenzymatic lipid oxidation (nLPO; 13, 14). Additionally, carotenoids both physically and chemically quench 1O2 (15). We proposed recently that a further layer of protection may also exist, one that uses PUFA-rich membranes as structural antioxidants. This hypothesis emerged from the fact that, even when grown under mild and relatively low light conditions, plants that lack TFAs displayed hallmark features of oxidative stress (16). TFAs act to protect proteins by absorbing ROS such as 1O2 and hydroxyl radicals produced under both healthy and stressful conditions (7, 11, 16).
The development of the supramolecular antioxidant hypothesis has been facilitated by the rigorous quantitative analysis of a PUFA fragmentation product, the 3-carbon dialdehyde malondialdehyde (MDA) that is generally considered to be harmful to the cells of humans (17, 18) and plants (19–21). MDA may also have activities in redox signaling (reviewed in Ref. 11). This compound, one of a plethora of PUFA oxidation products, is a robust marker of nonenzymatic lipid oxidation in animals (1) and in plants (e.g. 22). Furthermore, protocols exist for the quantitation of MDA by gas chromatography-mass spectrometry (GC/MS; 23). When combined with genetic analyses of mutants with reduced levels of TFAs, notably the Arabidopsis fad3-2 fad7-2 fad8 triple mutant (24), the use of such quantitation protocols has revealed that over 50% MDA in the leaves of Arabidopsis thaliana is derived from two oxidation-sensitive TFAs: α-linolenic acid (18:3) and 7Z,10Z,13Z-hexadecatrienoic acid (16:3; 16, 22).
The principal subcellular source of MDA in the leaves of A. thaliana is chloroplasts (organelles rich in 18:3 and 16:3 fatty acids), and the levels of MDA in wild-type chloroplasts were more than 2-fold higher than those in chloroplasts isolated from mutants lacking α-linolenic acid (25). These analyses highlight the fact that MDA is a relatively abundant molecule in leaves where its level is maintained in the nmol per gram dry weight range (16, 25). Such levels are typical of many primary metabolites in plant tissues e.g. (26). In the present study, and consistent with the literature (27), we found evidence for transient, light-dependent MDA accumulation in wounded leaves. These results led us to investigate the fate of isotope-labeled MDA in living plants, asking: can MDA be metabolized and, if so, which compounds are produced from this lipid damage-derived molecule?
Despite its simple composition (C3H4O2), isotopically labeled MDA of high purity is challenging to obtain. We therefore developed a rigorous method to purify MDA produced in a reaction catalyzed by an equine alcohol dehydrogenase (ADH; 28). The labeled MDA was purified on an anion exchange resin, fed as a volatile to plants, and traced to lipid pools by mass spectrometry. A major isotope-labeled 16:3- and 18:3-rich lipid was then identified. Based on this, we suggest that a potentially novel metabolic cycle ensures that carbon from damaged membranes is used to build new damage-prone membrane lipids that can again be re-utilized as ROS sinks.
Experimental Procedures
Plant Genotypes and Growth Conditions
Wild-type (WT) A. thaliana (Columbia), fad3-2 fad7-2 fad8 (24), and acs1 (29) were grown on soil at 22 °C for 5 weeks (metabolism studies) or ∼3.5 weeks (for wounding) under short day conditions (light: 100 μE m−2 s−1, 9 h light/15 h dark).
Reagents
Chemicals and analytic standards used were purchased from Sigma-Aldrich unless indicated. [1,2-14C]acetic acid sodium salt (14C-acetate) 100–120 mCi mmol−1 was purchased from Hartmann Analytics, Braunschweig, Germany. 1,3-[U-13C]propanediol, 99% 13C was obtained from Campro Scientific GmbH, Berlin, Germany, 1,3-[2-14C]propanediol 55 mCi mmol−1 from Hartmann Analytics.
MDA Quantification upon Wounding
Each biological replicate consisted of 3 leaves from 3 individual 3.5 week-old plants. 50% of the leaf surface from the tip was crushed with forceps. Plants were incubated in light (100 μE m−2 s−1) or the dark for 45 min and wounded at different time points prior to harvest into liquid nitrogen. MDA extraction was performed as described (25).
GC/MS Analysis of Aldehydes
The detection of aldehydes was based on combined O-(2,3,4,5,6-pentafluorophenyl)methylhydroxylamine hydrochloride (PFBHA)- and N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA)-derivatization. Sodium tungstate (0.3 m, 100 μl) was added to 400 μl of crude or purified fractions and the samples centrifuged for 5 min at 1000 × g to precipitate protein. The supernatant was combined with 450 μl of buffer (0.21 m citric acid, 0.58 m Na2HPO4; pH 4). The pH was verified to be 4 and if necessary adjusted with 9 N H2SO4. For derivatization, 100 μl of PFBHA reagent (5 mg ml−1 in water) was added and incubated for 1 h at 22 °C. The PFBHA adducts were extracted by adding 0.5 ml of MeOH and 2 ml of hexane, and the organic phase acidified with 10 μl of 9 n H2SO4. The organic phase was recovered after centrifugation for 5 min at 1000 × g and brought to dryness with N2. BSTFA (50 μl) was added and silylation conducted for 30 min at 60 °C. Samples were injected directly into the GC/MS and analyzed in the positive chemical ionization mode (PCI) with selected ion monitoring (SIM) for the mass of MDA(PFBHA)2 (m/z 463) and 3-HPA(PFBHA)(Si(CH)3) (m/z 342). Splitless injection at 250 °C and carrier flow of 1.5 ml min−1. Initial oven temperature 60 °C for 2 min, followed by a ramp of 8 °C min−1 to 320 °C.
GC/MS Analysis of 1,3-Propanediol
Detection was based on boronate derivatization. The boronate derivative chosen was 4-(trifluoromethyl) phenylboronic acid (TMPBA) due to large adduct masses which facilitates MS analysis. Fractions of interest (10 μl) were mixed with 50 μl of 100 mm TMPBA in MeOH. Derivatization was allowed to proceed for 10 min at 25 °C. The adduct was extracted by adding 100 μl isooctane, mixing, and centrifugation at 1000 × g for 5 min at room temperature to separate phases. The organic phase was analyzed by GC/MS: splitless injection (240 °C) with methane for PCI, initial oven temperature 50 °C for 1 min, ramp of 40 °C min−1 until 280 °C; gas flow 1.5 liters min−1.
Generation of Isotope-labeled Malondialdehyde
Enzymatic generation of MDA from 1,3-propanediol using ADH was described earlier (28). 1,3-Propanediol was converted to MDA with recombinant equine alcohol dehydrogenase (Sigma-Aldrich; 0.5 units mg−1). 10-ml reactions in 50 mm sodium pyrophosphate buffer, pH 9 (reaction buffer, RB) were prepared containing 2 mm 1,3-propanediol, 2 mm NAD+, 2.4 units of lactate dehydrogenase, 20 mm sodium pyruvate, and 15 units of recombinant alcohol dehydrogenase. The reaction was evaluated by spectrophotometry at 266 nm against an equal enzymatic mix containing 20 mm pyrazole as inhibitor. MDA was partially purified on a weak anion exchange resin (Dowex Marathon WBA free base form, Sigma-Aldrich) which was used without strong acid activation and prepared by washing with RB. The column consisted of a poly-prep chromatography column (0.8 × 4 cm, Bio-Rad) filled with the resin (2 ml) and a syringe needle (Becton Dickinson AG (BD) Alschwil, Switzerland, Microlance 3 25-gauge 5/8, 0.5 × 16 mm gauge) plugged at the end to reduce and control the flow. The column was conditioned with 10 ml RB-buffer. Then the reaction mix was applied, and the column washed with 10 ml of RB-buffer. Bound MDA was eluted with 0.5 N NaCl in 10 ml of RB. Eluting fractions were collected in 24-well plates (Falcon Multiwell, BD Franklin Lakes). MDA was measured by spectrophotometry (Genesys 10S UV-Vis, Thermo Fisher Scientific) at 267 nm in 10 mm Na3PO4 buffer, pH 11 based on molar absorption coefficient (ε = 31500 m−1 cm−1 (30). Wavelength scanning of the combined MDA fractions was measured against a control run lacking 1,3-propanediol and MDA. Freshly generated MDA was used immediately.
Application of Volatile Malondialdehyde
Purified MDA ([12 C], [U-13 C], or [2-14 C]) was acidified with 4.5 N HCl. The acidified MDA (200 nmol/plant and one liter of air volume) was immediately pipetted onto cotton buds (3 buds per pot). The plants were kept during the application in transparent, sealed Plexiglas boxes (15 liters volume, 15 plants per box), or glass jars (1 liter volume, 1 plant).
Lipid Extraction from Arabidopsis Leaves
Lipids were extracted based on Ref. 31 with slight modifications. Leaves were submerged rapidly in 3 ml of 75 °C pre-heated isopropanol (+0.01% BHT) and heated for 15 min at 75 °C to prevent lipase activity. Samples were vortexed for 1 h after addition of 1.5 ml of CHCl3 and 0.6 ml of H2O. The organic phase was transferred to new tubes, and the leaves were extracted again with 4 ml of CHCl3/MeOH (2:1 + 0.01% butylated hydroxytoluene BHT). The combined extractions were washed by the addition of 1 m KCl (1 ml) and subsequent vortexing and centrifugation. The organic phase was then washed again with 2 ml of H2O. Samples were subsequently evaporated to dryness under N2 for analysis.
HPLC Fractionation of Polar Lipids
After evaporation of the solvent, lipids were resuspended in MeOH (1 ml). Polar lipids were fractionated on a reverse phase C18 column (XTerra Prep MSC18, 5 μm, 10 × 150 mm; Waters GmbH, Eschborn, Germany) with the following gradient: 1–100 min from 93% MeOH and H2O to 100% MeOH with a flow rate of 2 ml min−1. This was followed by a wash for 30 min with 100% MeOH, and a flow rate of 5 ml min−1 1. The obtained fractions were transferred into 15 ml of Flo-Scintillation mixture (PerkinElmer) and counted with Tri-Carb 2800TR (PerkinElmer) after letting the samples rest overnight in the dark. The fractions containing the peak of interest (18:3–16:3-MGDG for WT and 18:2–16:2-MGDG for fad3-2 fad7-2 fad8) were combined prior to scintillation counting (3 fractions), and only an aliquot used for the scintillation: the majority was used for further analysis.
Hydrolysis of Lipids
Total lipids or purified lipids were evaporated to dryness and resuspended in 5 ml of methanolic KOH (1 m). The saponification reaction was allowed to proceed overnight at room temperature. 10 ml NaCl 0.9% (w/v) was added and non-saponified lipids were removed by extracting 3 times with diethylether (5 ml). The aqueous phase was acidified with 600 μl HCl (4.5 m) and free fatty acids (FFAs) extracted with 3 × 5 ml hexane. The combined hexane fractions were washed again with H2O. The aqueous phase was mixed with 15 ml of scintillation mixture (Ultima Gold, PerkinElmer) and counted. FFAs were treated with 0.5 ml of NaClO 13% (w/v) for 1 h to bleach chlorophyll prior to scintillation counting in Insta-Fluor mixture (PerkinElmer). To establish counting efficiency samples were re-counted after addition of a known amounts of radioactivity.
UHPLC-QTOF-MS Analysis
Polar lipids were first separated from apolar lipids by resuspension of total dried lipid extracts in 1 ml of MeOH. The soluble lipids were then separated by HPLC as described above, monitored for UV absorbance, and the eluted peaks of interest were concentrated. A fraction (1 μl) of the concentrated HPLC fraction was separated and analyzed with an Acquity UPLC coupled to a quadrupole time-of-flight mass spectrometer (Waters) using negative electrospray ionization. Column: Acquity BEH C18, 50 × 1.0 mm i.d., 1.7-μm particle size. Solvents: A, water; B, methanol. Solvent gradient was 0–2 min from 85 to 100% MeOH, followed by 2 min of 100% MeOH and reequilibration for 1 min at 85% MeOH with a flow rate of 200 μl min−1. The high resolution mass spectrometer was operated over an 85–1200 Da mass range in MSE mode with alternating scans at low (4 eV) and high (10–30 eV ramp) collision energies. Internal calibration was obtained by constantly infusing a 400 ng/ml solution of the synthetic peptide leucine-enkephalin (m/z 554.2615) in the mass spectrometer.
Results
Changes in MDA Levels Following Wounding
In leaves, wounding causes both free (unbound) chlorophyll accumulation (32) and light-dependent singlet oxygen (1O2) generation (27, 32, 33). This could, in theory, lead to the accumulation of MDA following 1O2-dependent lipid fragmentation. To study MDA dynamics in adult-phase plants we moved a set of Arabidopsis plants into the dark or kept them in light. We then wounded expanded leaves (Fig. 1). In light-kept plants, MDA levels almost doubled 20 min after wounding from 8.6 nmol g−1 dry weight (D.W.) in control leaves to 15.4 nmol g−1 D.W. in wounded leaves. MDA levels started to decrease again at 45 min to 12.2 nmol g−1 D.W.
FIGURE 1.
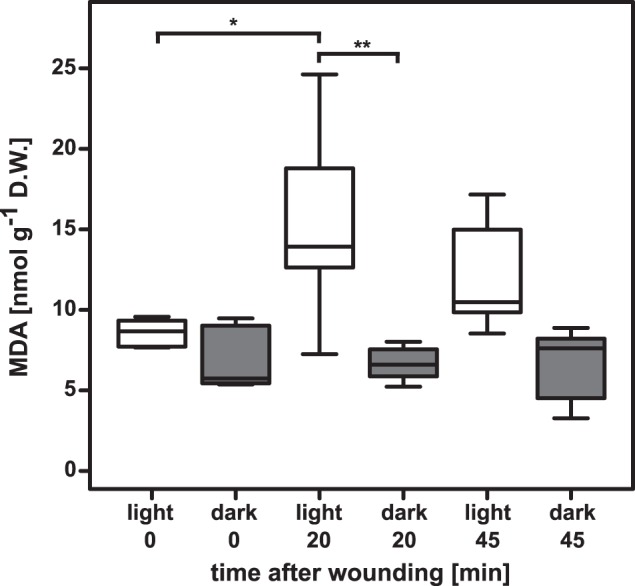
MDA levels change upon wounding in a light-dependent manner. MDA levels in leaves of wounded 3.5-week-old plants kept in the light or dark (n = 4–6). ANOVA analysis showed that the interaction between wounding and light was not significant (F2,26 = 3.175, p value = 0.05839, but was for the single factor light versus dark (F1,26 = 21.007, p value = 0.00010). Selected p values of 1:1 comparisons: increase of MDA in light-kept plants, 0 to 20 min after wounding; p value = 0.02924. Decrease in wounded light-kept plants from 20 min to 45 min; p value = 0.62643. Time point t = 0 min between white and dark kept plants; p value = 0.97111. In the dark MDA levels neither increased upon wounding nor showed any decrease at the later 45 min time point (p values = 0.99997 and 1.00000, respectively).
Conducting the same wounding procedure in dark-kept plants we observed a very different effect on MDA levels. First, MDA levels were slightly lower (7 nmol g−1 D.W.) in resting plants compared with the light kept plants. In the dark, MDA levels neither increased upon wounding nor showed any decrease at the later 45 min time point. To investigate possible MDA metabolism in light-kept leaves we developed a protocol to synthesize the isotope-labeled molecule.
Generation and Purification of Labeled MDA
MDA can be generated by acidification of 1,1,3,3-tetraethoxypropane (TEP) although this generates reactive species in addition to MDA (34). To maximize control over MDA generation we chose the enzymatic oxidation of 1,3-propanediol to MDA using equine ADH (28). The maximum conversion efficiency for the ADH reaction was reported to be up to 60%, leaving residual 1,3-propanediol as well as 3-hydroxypropanal (3-HPA) that must be removed if the MDA is used in a biological system. The highest efficiency we observed was between 40–50% conversion with 7.5 units mm−1 of 1,3-propanediol and a speed of about 400 pmol h−1 in the first 24 h. The maximum conversion was obtained after 2–3 days for 10 ml preparations containing 2 mm 1,3-propanediol. In our hands, the recombinant enzyme worked less efficiently than reported for the native horse ADH (28).
After synthesis at pH 9, MDA was bound to an anion exchange resin and washed prior to elution with NaCl. 2 ml of resin was capable of binding 5 μmol of MDA with 94 ± 6% of the loaded MDA being recovered. For further experiments, only fractions with an MDA concentration higher than 300 μm were pooled. The absorbance spectrum of combined eluted MDA fractions correlated with the reported MDA absorbance (30), showing a maximum at 267 nm. Wavelength scanning of 1,3-propanediol in Na3PO4 buffer did not show any absorbance in the MDA range. To summarize, from 20 μmol 1,3-propanediol we obtained 6 to 9 μmol of purified MDA, usually in a concentration between 1 and 2 mm.
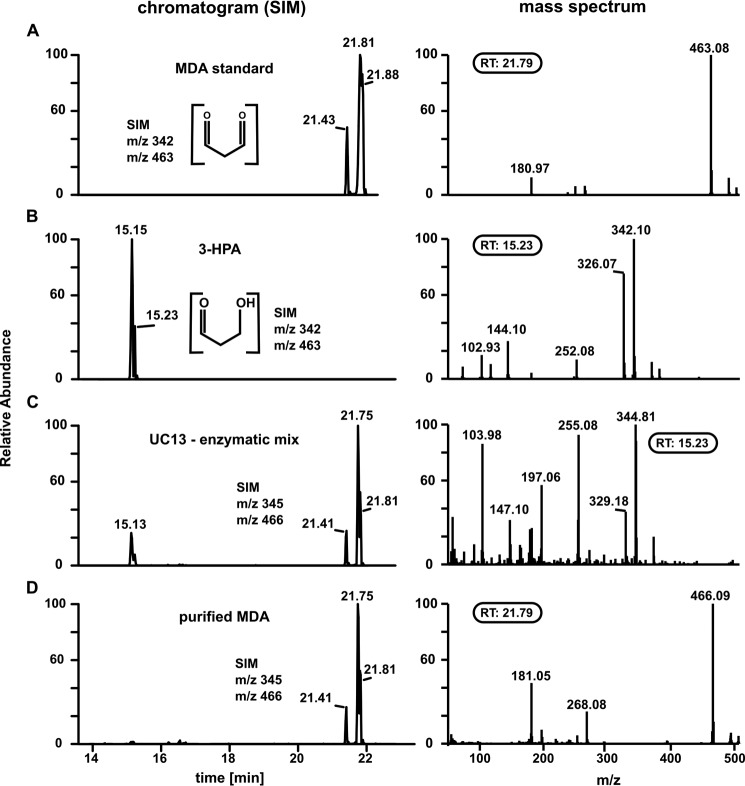
GC/MS analysis was used to examine MDA purity (Fig. 2). An MDA standard produced from tetrabutylammonium MDA showed two major peaks after derivatization with PFBHA according to the cis-or trans-confirmation of the PFBHA adduct (Fig. 2A). These peaks showed the expected mass of m/z 463, which derives from the bonding of one PFBHA molecule per aldehyde group of MDA (Fig. 2A, right panel). For 3-HPA we found a double peak with the expected mass of m/z 342 (Fig. 2B). 3-HPA binds to one molecule of PFBHA and the free hydroxy-group is further derivatized by the BSTFA adding an additional (Si(CH3)3)-group. When 1,3-[U-13C]propanediol was used as a substrate for ADH, the resulting masses for MDA and 3-HPA were shifted by +3 Da to m/z 466 and m/z 345, respectively and the mass spectrum of the first double peak eluting at 15.13 and 15.23 min (Fig. 2C, right panel) was similar to that of the 3-HPA standard (Fig. 2B). The major peak eluting between 21.41 and 21.81 min was found to be MDA (mass spectrum similar to Fig. 2D, right panel). The enzymatic mix was further purified by anion exchange to obtain MDA which was dominant in the eluate. We then ensured that 1,3-propanediol was not present in the purified MDA. 1,3-propanediol, which does not react with PFBHA, was derivatized with 4-(trifluoromethyl) phenylboronic acid (TMPBA) which forms cyclic structures with 1,3- and 1,2-diols (35). The structure and mass spectrum of the 1,3-propanediol:TMPBA adduct is shown in Fig. 3A. We loaded 1 mm 1,3-propanediol in reaction buffer (10 ml) onto the anion exchange column and proceeded similarly to the MDA purification procedure: The flow-through was collected, non-bound material was washed off with buffer, and bound compounds were eluted with 0.5 m NaCl. Importantly, no 1,3-propanediol was detected in this eluate (Fig. 3B).
FIGURE 2.
Purity of final MDA eluate. Left column, chromatograms; right column, mass spectra (PCI mode). A, MDA generated from tetrabutylammonium malondialdehyde salt. The double peak results from either cis-or trans-formation of the adduct. B, purified 3-HPA. Peak eluting at 15.23 min is 3-HPA, the other one at 15.15 a derivatization agent artifact. C, products generated by enzymatic conversion of [U -13C]propanediol. Both MDA and 3-HPA were observed showing the expected m/z +3 mass shift. D, 13C-MDA purified by anion-exchange chromatography. Samples were derivatized using PFHBA.
FIGURE 3.
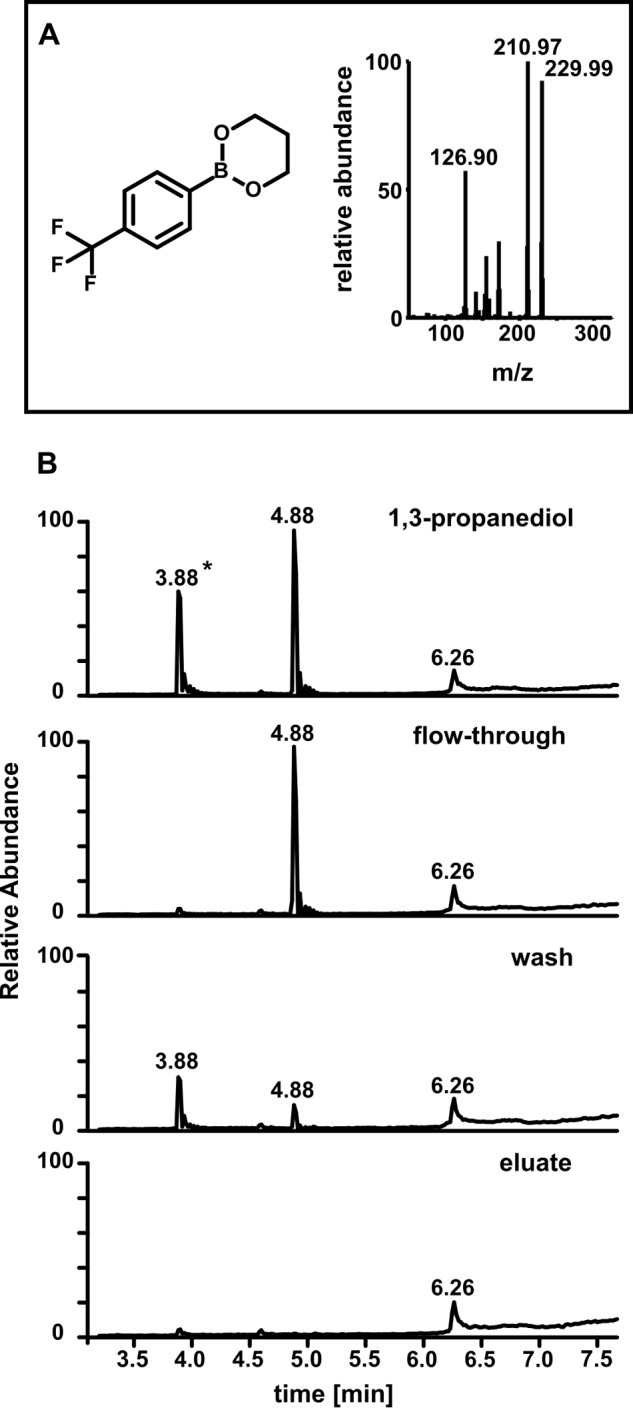
Analysis of 1,3-propanediol in anion exchange fractions by GC/MS. A, chemical structure and mass spectrum of the 1,3-propanediol:TMPBA adduct (PCI mode). B, 1,3-propanediol presence in different anion exchange fractions. Total ion currents were monitored. Starting material: 1,3-propanediol (1 mm) in 50 mm pyrophosphate buffer, pH 9. Flow-through after the mix was loaded on a weak basic anion exchange column, wash of the column with pH 9 pyrophosphate buffer; eluate, combined fractions eluted with 0.5 N NaCl; *, contaminant.
Incorporation of 14C-MDA-derived Radioactivity into Plant Lipids
[U-13C]MDA was used to establish an uptake procedure for volatile MDA into living plants. Exposure of single plants to 25 nmol of volatilized 13C-MDA in a 1 liter air volume doubled the resting level of MDA in leaves. Extracted MDA displayed two MDA mother ions, one for the endogenous MDA and a new one with a mass shift of 3 Da according to three incorporated heavy-labeled carbons. To test the hypothesis that exogenous MDA might be incorporated into lipids, plants were incubated with 14C-MDA [0.45 nCi nmol−1] for 24 h at which time total lipids were extracted. The extracted lipids dissolved in diethylether were loaded onto silica columns.
Scintillation counting showed 50–70% of the total counts being present in the water soluble fraction or remained bound in the tissue, whereas 30–50% of the 14C was recovered in apolar lipid-containing extracts. These findings recapitulated similar observations from 14C-MDA feeding experiments in mice (36). After feeding of mice with 14C-MDA, 20% of the radioactivity was found 4 h later in lipids. Preliminary thin-layer chromatography (TLC) analysis of lipid extracts confirmed radioactivity in discrete molecular species.
Identification of an MDA End-point Metabolite
To identify and purify the major lipid species showing 14C incorporation, we separated polar lipids by reverse-phase HPLC, monitoring the eluent at 200–500 nm (37) (Fig. 4A). Quantitative scintillation counting of the fractions (Fig. 4B) revealed a major radioactive peak with a retention time of about 32 min (Fig. 4B), correlating with the observation from the TLC on which we found high radiation concentrated in one band. The hypothesis that the major radioactive peak observed in HPLC might contain TFAs was tested using the fad3-2 fad7-2 fad8 mutant (24). fad3-2 fad7-2 fad8 plants were exposed for 24 h to 14C-MDA and polar lipids analyzed by HPLC and scintillation counting (Fig. 5). As judged by the UV-absorbance profile, the polar lipid composition of the mutants (Fig. 5A) was strikingly different compared with that of the WT (Fig. 4A). The fad3-2 fad7-2 fad8 mutant lacked the major radioactive peak observed at 32 min in the WT, but contained a new major peak at 45 min (Fig. 5A, black box).
FIGURE 4.
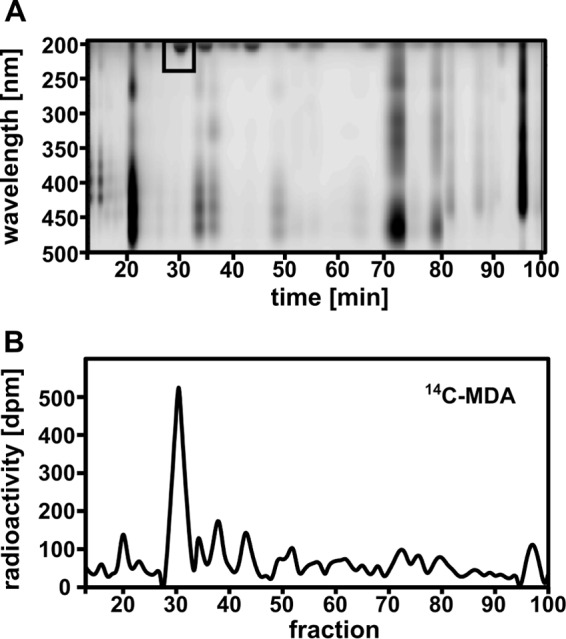
HPLC separation of lipid extracts from WT plants exposed to 14C-MDA for 24 h. A, chromatogram of polar leaf lipids monitored by light absorption (200–500 nm). B, radiation distribution in HPLC-fractions measured by liquid scintillation. (Pool of leaves from 4 plants, 300,000 dpm/plant l−1; SA: 0.67 nCi nmol−1).
FIGURE 5.
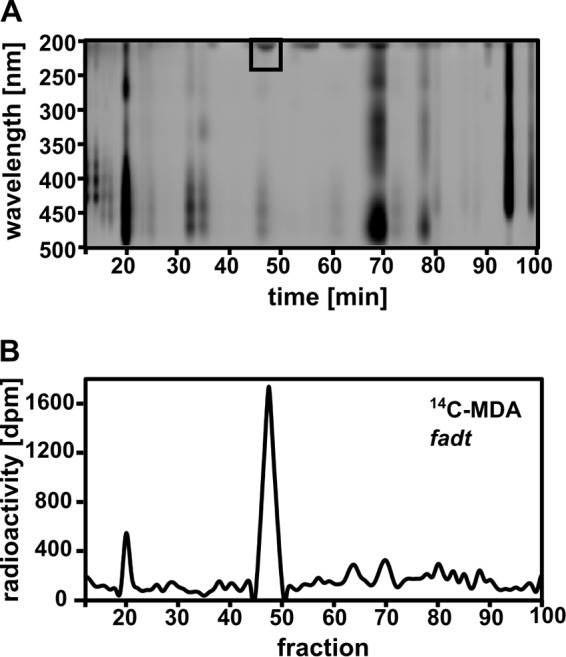
HPLC separation of lipid extracts from fad3-2 fad7-2 fad8 plants exposed to 14C-MDA for 24 h. A, polar leaf lipids from 14 C-MDA treated fad3-2 fad7-2 fad8 (fadt) plants, separated by HPLC and monitored by light absorption. B, radiation distribution in HPLC-fractions measured by liquid scintillation. (Pool of leaves from 5 plants, 400,000 dpm/plant l−1; SA: 0.9 nCi nmol−1).
To identify the major 14C-containing lipid obtained after treating plants with 14C-MDA, the fraction of interest was isolated from an identical HPLC separation using 12C-MDA. Lipid extracts first separated by HPLC were analyzed by UHPLC-QTOF-MS (Fig. 6). One major peak eluted at 2.25 min after some early eluting putative breakdown products (Fig. 6A). Analyzing the high resolution fragmentation spectrum of the major peak we concluded that the molecule was 18:3–16:3-monogalactosyldiacylglycerol (18:3–16:3-MGDG, Fig. 6, B and C). The molecule gave a parent ion at m/z 745.4888 (corresponding formula C43H69O10) and a formate adduct at m/z 791.4941. The fragmentation peaks at m/z 249.1857 (C16H25O2) and m/z 277.2171 (C18H29O2) represented linolenic acid (18:3) and hexadecatrienoic acid (16:3) moieties, respectively. This result required genetic verification.
FIGURE 6.
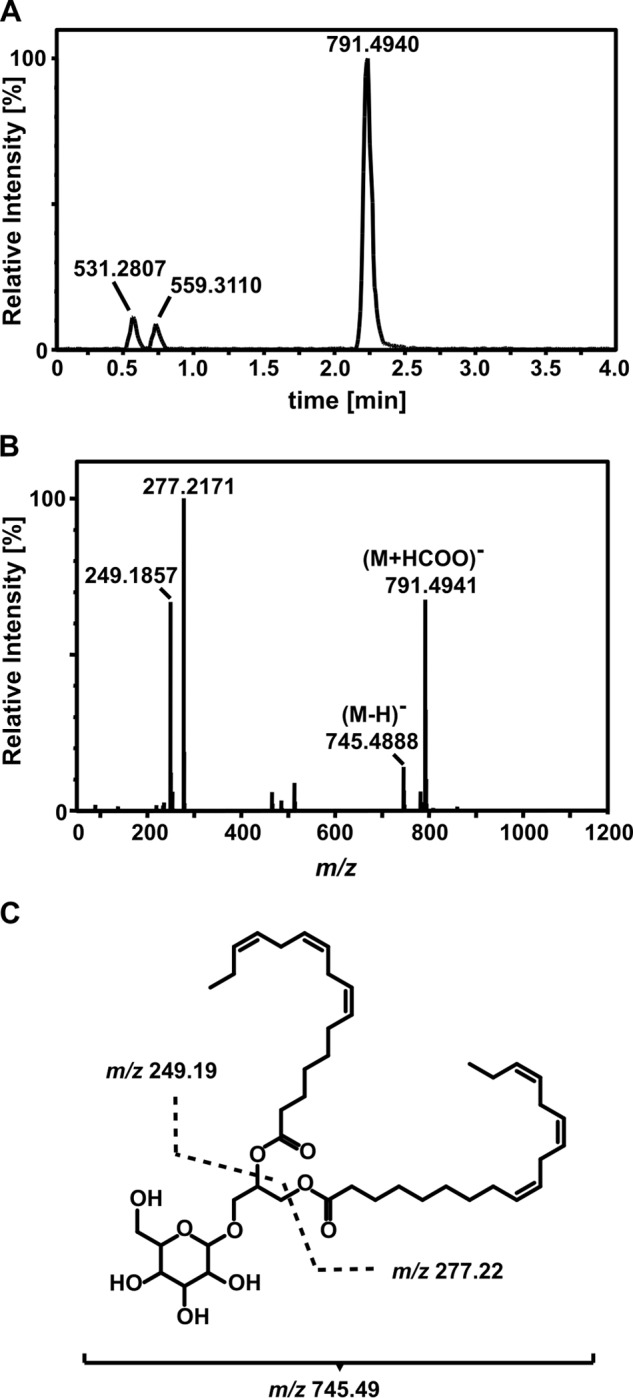
Identification of 18:3–16:3-MGDG as an end-point MDA metabolite. A, UHPLC-QTOF-MS analysis of the major product of 14C-MDA incorporation extracted from plants exposed to 12C-MDA (4 plants combined) and pre-separated by HPLC. B, mass spectrum of the peak at 2.25 min room temperature. C, chemical structure of the molecule and the fragments identified by MS.
UHPLC-QTOF-MS confirmed that the corresponding 14C-MDA incorporation peak in fad3-2 fad7-2 fad8 lipids was not 18:3–16:3-MGDG, since its retention time (2.4 min) and MS/MS spectrum differed. The parent ion at m/z 749.5204 (C43H73O10) and fragment ions at m/z 251.2016 (C16H27O2) and m/z 279.2329 (C18H31O2), indicating hexadecadienoic acid (16:2) and linoleic acid (18:2), respectively, revealed that the molecule was 18:2–16:2-MGDG. This result supported the previous finding (Fig. 4) that a major MDA-derived metabolite in WT leaves was 18:3–16:3-MGDG. Quantitative analysis after saponification (38) of the 14C-labeled 18:3–16:3-MGDG revealed that 84 ± 3.3% of the 14C-counts in lipids were present in the FA-moiety with the rest in the head-group.
Acetate as a Potential MDA Metabolite
14C-acetate is readily incorporated in MGDG in 18:3 plants (39, 40). To examine whether MDA might travel through acetate as a metabolic intermediate, we analyzed 14C-MDA incorporation into lipids in the acetyl-CoA synthetase (ACS) mutant acs1 (29). ACS uses acetate, ATP and CoA to generate acetyl-CoA which can be used for FA synthesis and a 90% decrease of 14C-acetate incorporation into FAs relative to the WT was reported for acs1 plants (29). We compared 14C-MDA incorporation in mutant and WT plants after 6 h. As reported, incorporation of 14C-acetate into FAs was found to be strongly reduced in acs1 plants compared with the WT. However, the acs1 mutation did not affect 14C-MDA incorporation into FAs (Fig. 7).
FIGURE 7.
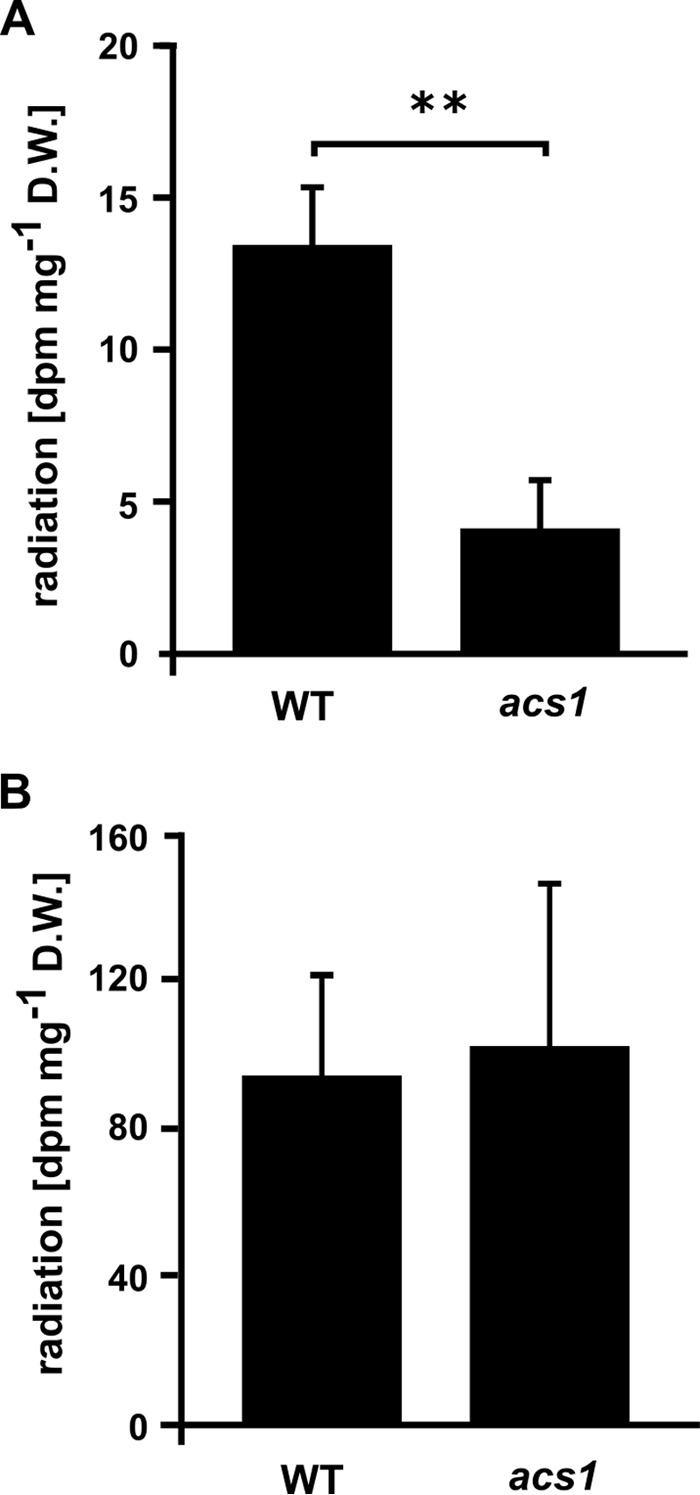
14C-acetate and 14C-MDA incorporation into lipids of WT and acs1 plants. A, 14C-acetate incorporation in WT and acs1 plants. B, 14C-MDA incorporation in WT and acs1 plants. For both treatments total lipids were saponified following 6 h exposure of plants to the volatile tracer. The FA-containing phase was extracted and analyzed by scintillation counting; counts shown as disintegrations per minute (dpm) per mg dry weight (D.W.) plant tissue. 5 biological replicates per genotype and treatment were used (14C-acetate: 100,000 dpm/plant l−1; SA: 0.225 nCi nmol−1, 14C-MDA: 400,000 dpm/plant l−1; SA: 0.9 nCi nmol−1). **, p < 0.001.
Discussion
Most MDA in healthy leaf tissue originates from the fragmentation of chloroplast PUFAs (19, 25) and this is likely to involve 1O2-induced fatty acid chain fragmentation (19, 25), a process that is light-dependent (11). Consistent with this, we found that leaves that had been wounded in the light produced more MDA than those wounded in the dark and we observed that MDA increases after wounding were transient. We set out to find major metabolites in which carbon was recruited from exogenous lipid fragments, basing our strategy on the use of high purity isotope-labeled MDA.
MDA is commonly obtained from its tetrabutylammonium salt or from the acid hydrolysis of 1,1,3,3-tetraethoxypropane (TEP). Neither of these methods is useful for the generation of high purity isotope-labeled MDA. For example, the generation of MDA from TEP produces other reactive side-products such as β-ethoxy-acrolein and β-methoxy-acrolein (34). Both of these aldehydes might be toxic or have biological activity, potentially interfering with the analysis of MDA metabolism. We therefore used enzymatic conversion of 1,3-propanediol to MDA (28). Summerfield and Tappel (28) purified the MDA that they generated enzymatically by acidifying to pH 3 to make MDA volatile. The MDA was driven into the gas phase by heating to 50 °C, then captured by condensing the vapor at −78 °C. However, this method is suboptimal for MDA purification due to the danger of generating artifacts through heating (41). In the present study we used the fact that MDA is charged above pH 4.5 to trap MDA on anion exchange columns, while the unbound alcohols were eluted. To prevent loss of volatile MDA a high salt concentration was chosen over acidification for the elution of MDA from the column. The purification technique we developed is simple, versatile and permitted MDA to be isolated in millimolar concentrations. It should prove useful in further studies employing MDA.
MDA was taken up and rapidly turned over in Arabidopsis leaves, findings that recapitulated similar observations from 14C-MDA feeding experiments in rodents (36, 42, 43). These earlier experiments in animals suggested that MDA was metabolized through the TCA cycle (43) or might be used in fatty acid (FA) assembly (36). Our results clearly show MDA incorporation into FAs in plants.
At its metabolic entry point, MDA could be either reduced or oxidized (41). If this is the case then both pathways would produce unstable and potentially short-lived intermediates. Yamauchi et al. (44) characterized several plant enzymes capable of reducing reactive carbonyls. Such enzymes would likely produce 3-hydroxypropanal (3-HPA) from MDA. However, we were unable to detect this aldehyde in leaves. The alternative is oxidative metabolism where ADHs have been proposed to catalyze MDA oxidation to malonate semialdehyde (MSA; 36). We note that (methyl)malonate semialdehyde oxidative decarboxylases that might catalyze the direct conversion of MSA to acetyl-CoA (45, 46) might exist in Arabidopsis. This led us to synthesize MSA using conversion from ethyl 3,3-diethoxypropanoate (47) and we used it as a standard for GC/MS analysis, but we were unable to detect MSA in leaves. In vitro, MSA is unstable and quickly decomposes to acetaldehyde and CO2. Moreover, if acetaldehyde was generated from MDA it might be converted through ADH action to acetate. However, the fact that the acs1 mutant showed WT levels of MDA incorporation suggests that acetate may not be a major intermediate in MDA metabolism in leaves or that a metabolic bypass exists. How MDA enters FA synthesis pathways is still unclear but in the present work we unequivocally identified a product of its incorporation into lipids.
To identify major products of MDA metabolism we concentrated on plants that had been exposed for 24 h to 14C-MDA. A major radioactive species derived from MDA was identified as 18:3–16:3-MGDG. MGDG is one of the most abundant lipids on earth and is also the dominant lipid species in leaves of wild-type Arabidopsis (>40% of total lipids (4). The thylakoid membranes of this plant are highly enriched in 16:3- and 18:3-containing MGDGs (10). Here, we found (Fig. 1) ∼10 nmol per g dry weight MDA in 3.5-week-old leaves. MDA levels in older leaves are reported to be higher (16, 25). This suggests that MDA levels may increase upon full leaf expansion. These levels are low relative to the large MGDG pools in leaves, so, in our experiments, only a fraction of MGDG is likely to be subject to oxidative fragmentation after wounding.
MGDGs play important structural and functional roles, for example in stabilizing photosystem II (PSII; 48) and MGDG deficiency causes reduced photoprotection (49). Several of the lipids tightly associated with the PSII reaction center are MGDGs (9, 50). They separate PS II from the antenna and subunits and provide an environment that facilitates turnover of the D1 subunit (9). TFAs, which are enriched in thylakoid galactolipids, are necessary for recovery from photoinhibition (10). Genetic work on MGDG itself has revealed important functions in chloroplast function and survival. On the one hand an MGDG synthase 1-deficient mutant (mgd1–2) in Arabidopsis causes a dwarf-albino phenotype and is incapable of correctly assembling subunit-proteins for PS II (51). Also, treatment with the inhibitor galvestine-1 led to a reduced MGDG content which was accompanied by impaired chloroplasts development and fewer thylakoids (52). On the other hand, overexpression of a rice MGDG synthase in tobacco strongly increased tolerance to salt stress (53). Here, we propose that MGDGs surrounding PS II, a major site of 1O2 formation (6), might provide a buffer to protect proteins from damage by 1O2 and/or by hydroxyl radicals in plastids. Some effects of either reducing or increasing MGDG synthesis may be related to ROS capture and protein protection by this lipid.
In summary, it is intriguing that a major end-point metabolite of MDA metabolism (18:3–16:3-MGDG) represents the likely substrate for production of MDA. Here, we propose that MDA resulting from omega-3 fatty acid fragmentation serves as an intermediate in an important lipid repair cycle that may be widespread in nature. Photosynthesis predates the origin of eukaryotes, and PUFAs including triunsaturated fatty acids are present in the thylakoids in cyanobacteria (reviewed in 54). It is therefore possible that the PUFA/MDA cycle we have begun to delineate has its origins deep in time. Furthermore, analogous cycles may occur in other biological contexts where ROS generation is associated with high levels of membrane PUFAs.
Author Contributions
E. S. S., O. S., and G. G. performed experiments; E. S. S., G. G., and E. E. F. analyzed data; E. E. F. and E. S. S. wrote the manuscript.
Acknowledgments
We thank Y. Poirier (University of Lausanne) for critical comments. S. Vollenweider (Givaudan) provided 3-HPA.
This work was supported with a National Center of Competence in Research “Plant Survival” grant and Swiss National Science Foundation Grants 3100A0-122441 and 31003A-138235. The authors declare that they have no conflicts of interest with the contents of this article.
- ROS
- reactive oxygen species
- PUFA
- polyunsaturated fatty acid
- MDA
- malondialdehyde
- TFA
- triunsaturated fatty acid
- MSA
- malonate semialdehyde.
References
- 1. Liu J., Yeo H. C., Doniger S. J., and Ames B. N. (1997) Assay of aldehydes from lipid peroxidation: gas chromatography-mass spectrometry compared to thiobarbituric acid. Anal. Biochem. 245, 161–166 [DOI] [PubMed] [Google Scholar]
- 2. Frankel E. N. (2012) Lipid Oxidation, Oily Press [Google Scholar]
- 3. Block M. A., Dorne A., Joyard J., and Douce R. (1983) Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from Spinach. J. Biol. Chem. 258, 13281–13286 [PubMed] [Google Scholar]
- 4. Li-Beisson Y., Shorrosh B., Beisson F., Andersson M. X., Arondel V., Bates P. D., Baud S., Bird D., Debono A., Durrett T. P., Franke R. B., Graham I. A., Katayama K., Kelly A. A., Larson T., Markham J. E., Miquel M., Molina I., Nishida I., Rowland O., Samuels L., Schmid K. M., Wada H., Welti R., Xu C., Zallot R., and Ohlrogge J. (2010) Acyl-lipid metabolism. The Arabidopsis Book 8, e0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blokhina O., and Fagerstedt K. V (2010) Reactive oxygen species and nitric oxide in plant mitochondria: origin and redundant regulatory systems. Physiol. Plant. 138, 447–62 [DOI] [PubMed] [Google Scholar]
- 6. Krieger-Liszkay A. (2005) Singlet oxygen production in photosynthesis. J. Exp. Botany 56, 337–346 [DOI] [PubMed] [Google Scholar]
- 7. Zoeller M., Stingl N., Krischke M., Fekete A., Waller F., Berger S., and Mueller M. J. (2012) Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiol. 160, 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Triantaphylidès C., Krischke M., Hoeberichts F. A., Ksas B., Gresser G., Havaux M., Van Breusegem F., and Mueller M. J. (2008) Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 148, 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., and Saenger W. (2009) Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 10. Vijayan P., and Browse J. (2002) Photoinhibition in mutants of Arabidopsis deficient in thylakoid unsaturation. Plant Physiol. 129, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farmer E. E., and Mueller M. J. (2013) ROS-Mediated Lipid Peroxidation and RES-Activated Signaling. Annu. Rev. Plant Biol. 64, 429–450 [DOI] [PubMed] [Google Scholar]
- 12. Maeda H., and DellaPenna D. (2007) Tocopherol functions in photosynthetic organisms. Curr. Opin. Plant Biol. 10, 260–265 [DOI] [PubMed] [Google Scholar]
- 13. Sattler S. E., Mène-Saffrané L., Farmer E. E., Krischke M., Mueller M. J., and DellaPenna D. (2006) Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell. 18, 3706–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rastogi A., Yadav D. K., Szymańska R., Kruk J., Sedlářová M., and Pospíšil P. (2014) Singlet oxygen scavenging activity of tocopherol and plastochromanol in Arabidopsis thaliana: relevance to photooxidative stress. Plant. Cell Environ. 37, 392–401 [DOI] [PubMed] [Google Scholar]
- 15. Ramel F., Birtic S., Cuiné S., Triantaphylidès C., Ravanat J.-L., and Havaux M. (2012) Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 158, 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mène-Saffrané L., Dubugnon L., Chételat A., Stolz S., Gouhier-Darimont C., and Farmer E. E. (2009) Nonenzymatic oxidation of trienoic fatty acids contributes to reactive oxygen species management in Arabidopsis. J. Biol. Chem. 284, 1702–1708 [DOI] [PubMed] [Google Scholar]
- 17. Marnett L. J. (2002) Oxy radicals, lipid peroxidation and DNA damage. Toxicology. 181–182, 219–222 [DOI] [PubMed] [Google Scholar]
- 18. Refsgaard H. H., Tsai L., and Stadtman E. R. (2000) Modifications of proteins by polyunsaturated fatty acid peroxidation products. Proc. Natl. Acad. Sci. U.S.A. 97, 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mano J. (2012) Reactive carbonyl species: their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 59, 90–97 [DOI] [PubMed] [Google Scholar]
- 20. Yamauchi Y., Furutera A., Seki K., Toyoda Y., Tanaka K., and Sugimoto Y. (2008) Malondialdehyde generated from peroxidized linolenic acid causes protein modification in heat-stressed plants. Plant Physiol. Biochem. 46, 786–793 [DOI] [PubMed] [Google Scholar]
- 21. Biswas M. S., and Mano J. (2015) Lipid peroxide-derived short-chain carbonyls mediate hydrogen peroxide-induced and salt-induced programmed cell death in plants. Plant Physiol. 168, 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weber H., Chételat A., Reymond P., and Farmer E. E. (2004) Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 37, 877–888 [DOI] [PubMed] [Google Scholar]
- 23. Yeo H. C., Helbock H. J., Chyu D. W., and Ames B. N. (1994) Assay of malondialdehyde in biological fluids by gas chromatography-mass spectrometry. Anal. Biochem. 220, 391–396 [DOI] [PubMed] [Google Scholar]
- 24. McConn M., and Browse J. (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell. 8, 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmid-Siegert E., Loscos J., and Farmer E. E. (2012) Inducible malondialdehyde pools in zones of cell proliferation and developing tissues in Arabidopsis. J. Biol. Chem. 287, 8954–8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roessner U., Luedemann A., Brust D., Fiehn O., Linke T., Willmitzer L., and Fernie A. (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell. 13, 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pospíšil P., and Prasad A. (2014) Formation of singlet oxygen and protection against its oxidative damage in Photosystem II under abiotic stress. J. Photochem. Photobiol. B Biol. 137, 39–48 [DOI] [PubMed] [Google Scholar]
- 28. Summerfield F. W., and Tappel A. L. (1978) Enzymatic synthesis of malonaldehyde. Biochem. Biophys. Res. Commun. 82, 547–552 [DOI] [PubMed] [Google Scholar]
- 29. Lin M., and Oliver D. J. (2008) The role of acetyl-coenzyme a synthetase in Arabidopsis. Plant Physiol. 147, 1822–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Esterbauer H., Schaur R. J., and Zollner H. (1991) Chemistry and bio-chemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 11, 81–128 [DOI] [PubMed] [Google Scholar]
- 31. Pattada S., Roth M. R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X., and Devaiah S. P. (2006) Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dα1 knockout mutant. Phytochemistry 67, 1907–1924 [DOI] [PubMed] [Google Scholar]
- 32. Kariola T., Brader G., Li J., and Palva E. T. (2005) Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell. 17, 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Flors C., Fryer M. J., Waring J., Reeder B., Bechtold U., Mullineaux P. M., Nonell S., Wilson M. T., and Baker N. R. (2006) Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 57, 1725–1734 [DOI] [PubMed] [Google Scholar]
- 34. Marnett L. J., and Tuttle M. A. (1980) Comparison of the mutagenicities of malondialdehyde and the side products formed during its chemical synthesis. Cancer Res. 40, 276–282 [PubMed] [Google Scholar]
- 35. Brooks C., and Maclean I. (1971) Cyclic n-butylboronates as derivatives of polar bifunctional groups for gas chromatography and mass spectrometry. J. Chromatogr. 9, 193–204 [Google Scholar]
- 36. Marnett L. J., Buck J., Tuttle M. M. A., Basu A. K. A., and Bull A. W. (1985) Distribution and oxidation of malondialdeyhde in mice. Prostaglandins 30, 241–254 [DOI] [PubMed] [Google Scholar]
- 37. Holte L. L., van Kuijk F. J., and Dratz E. A. (1990) Preparative high-performance liquid chromatography purification of polyunsaturated phospholipids and characterization using ultraviolet derivative spectroscopy. Anal. Biochem. 188, 136–141 [DOI] [PubMed] [Google Scholar]
- 38. Behal R. H., Lin M., Back S., and Oliver D. J. (2002) Role of acetyl-coenzyme A synthetase in leaves of Arabidopsis thaliana. Arch. Biochem. Biophys. 402, 259–267 [DOI] [PubMed] [Google Scholar]
- 39. Roughan P. (1970) Turnover of the glycerolipids of pumpkin leaves. Biochem. J. 117, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Slack C. R., and Roughan P. G. (1975) The kinetics of incorporation in vivo of (14C)acetate and (14C)carbon dioxide into the fatty acids of glycerolipids in developing leaves. Biochem. J. 152, 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Janero D. R. (1990) Malondialdehyde and thiobarbituric acid-reactivity as diagnostic indices of lipid peroxidation and peroxidative tissue injury. Free Radic. Biol. Med. 9, 515–540 [DOI] [PubMed] [Google Scholar]
- 42. Draper H. H., McGirr L. G., and Hadley M. (1986) The metabolism of malondialdehyde. Lipids. 21, 305–307 [DOI] [PubMed] [Google Scholar]
- 43. Siu G. M., and Draper H. H. (1982) Metabolism of malonaldehyde in vivo and in vitro. Lipids 17, 349–355 [DOI] [PubMed] [Google Scholar]
- 44. Yamauchi Y., Hasegawa A., Taninaka A., Mizutani M., and Sugimoto Y. (2011) NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 286, 6999–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lloyd D. (1965) The purification and properties of malonic semialdehyde oxidative decarboxylase from Prototheca zopfii. Biochem. J. 96, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scholem R. D., and Brown G. K. (1983) Metabolism of malonic semialdehyde in man. Biochem. J. 216, 81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson T. (1963) Water-soluble organic acids. The Organic Constituents of Higher Plants: Their Chemistry and Interrelationships, Vol. 40, pps. 36–44, American Chemical Society [Google Scholar]
- 48. Wada H., and Murata N. (2010) The Role of Glycolipids in Photosynthesis in Lipids in Photosynthesis Essential and Regulatory Functions, pp. 265–282 [Google Scholar]
- 49. Aronsson H., Schöttler M., Kelly A., Sundqvist C., Dörmann P., Karim S., and Jarvis P. (2008) Monogalactosyldiacylglycerol deficiency in Arabidopsis affects pigment composition in the prolamellar body and impairs thylakoid membrane energization and photoprotection in leaves. Plant Physiol. 148, 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loll B., Kern J., Saenger W., Zouni A., and Biesiadka J. (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 51. Kobayashi K., Narise T., Sonoike K., Hashimoto H., Sato N., Kondo M., Nishimura M., Sato M., Toyooka K., Sugimoto K., Wada H., Masuda T., and Ohta H. (2013) Role of galactolipid biosynthesis in coordinated development of photosynthetic complexes and thylakoid membranes during chloroplast biogenesis in Arabidopsis. Plant J. 73, 250–261 [DOI] [PubMed] [Google Scholar]
- 52. Botté C. Y., Deligny M., Roccia A., Bonneau A.-L., Saïdani N., Hardré H., Aci S., Yamaryo-Botté Y., Jouhet J., Dubots E., Loizeau K., Bastien O., Bréhélin L., Joyard J., Cintrat J.-C., Falconet D., Block M. A., Rousseau B., Lopez R., and Maréchal E. (2011) Chemical inhibitors of monogalactosyldiacylglycerol synthases in Arabidopsis thaliana. Nat. Chem. Biol. 7, 834–842 [DOI] [PubMed] [Google Scholar]
- 53. Wang S., Uddin M. I., Tanaka K., Yin L., Shi Z., Qi Y., Mano J., Matsui K., Shimomura N., Sakaki T., Deng X., and Zhang S. (2014) Maintenance of chloroplast structure and function by overexpression of the OsMGD gene leads to enhanced salt tolerance in tobacco. Plant Physiol. 165, 1144–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Russell N. J., and Nichols D. S. (1999) Polyunsaturated fatty acids in marine bacteria–a dogma rewritten. Microbiology 145, 767–779 [DOI] [PubMed] [Google Scholar]