Abstract
Covalent modification of histones is a fundamental mechanism of regulated gene expression in eukaryotes, and interpretation of histone modifications is an essential feature of epigenetic control. Bromodomains are specialized binding modules that interact with acetylated histones, linking chromatin recognition to gene transcription. Because of their ability to function in a domain-specific fashion, selective disruption of bromodomain:acetylated histone interactions with chemical probes serves as a powerful means for understanding biological processes regulated by these chromatin adaptors. Here we describe the discovery and characterization of potent and selective small molecule inhibitors for the bromodomains of CREBBP/EP300 that engage their target in cellular assays. We use these tools to demonstrate a critical role for CREBBP/EP300 bromodomains in regulatory T cell biology. Because regulatory T cell recruitment to tumors is a major mechanism of immune evasion by cancer cells, our data highlight the importance of CREBBP/EP300 bromodomain inhibition as a novel, small molecule-based approach for cancer immunotherapy.
Keywords: acetylation; cellular immune response; chemical biology; chromatin regulation; forkhead box P3 (FOXP3); Treg, CBP, EP300, bromodomain, histone
Introduction
Regulatory T cells (Tregs)11 are essential for the maintenance of immunological self-tolerance, imposing a crucial check on the incidence of autoimmunity (1, 2). Tregs can also accumulate within tumors and are recruited from circulation by chemotactic mechanisms. Increased Treg loads are associated with poor prognosis in several cancer types, such as breast, ovarian, and gastric cancers (3–8). Intratumoral Tregs dampen effector T cell responses to tumor antigens, engendering an immunosuppressive microenvironment, and, as such, represent a major impediment to immune system clearance of tumor cells; specific inhibition of this subset therefore represents a powerful approach for bolstering anti-tumor immune responses, known as cancer immunotherapy (9–12).
Chromatin modifications govern diverse aspects of cellular function (13, 14). Lysine acetylation in histones and non-histone proteins (such as transcription factors) has been identified as a commonly utilized post-translational modification, allowing for widespread, dynamic, and reversible control of gene expression (15). Histone acetylation impacts gene transcription both by direct relaxation of chromatin architecture through alteration of DNA-histone interactions and by providing docking sites for recruitment and assembly of transcriptional complexes (16). Although histone acetyltransferase (HAT) enzymes deposit acetyl marks on histones, a group of conserved protein modules, bromodomains (BRDs), has evolved to bind acetyl lysine motifs, forging an essential link between chromatin modifications and transcriptional control (16–18).
The highly homologous, multifunctional mammalian proteins, CREBBP and EP300 (cAMP response element binding-protein binding protein and E1A binding protein 300, respectively; hereafter referred to as CBP/EP300) are especially important in this regard. CBP and EP300 each encode a single bromodomain and HAT activity, along with other protein interaction modules, and function as transcriptional co-activators (19, 20). Recent research underscores the importance of CBP/EP300 in Treg biology, because conditional deletion of either EP300 or CBP in mouse Tregs or inhibition of their HAT activity led to impaired Treg suppressive function and reduced tumor growth in murine cancer models (21, 22). In addition to evidence for the role of HAT activity, there is growing appreciation for the importance of the bromodomain in transactivation functions of CBP/EP300 (23). The conserved BRD fold provides a deep hydrophobic pocket suited to the binding of inhibitors, whereas diversity in surface and loop residues across BRD proteins allows for selective targeting by pharmacologic agents (23).
Here we describe the discovery and characterization of inhibitors of CBP/EP300 bromodomains. These compounds are potent, are highly selective over other bromodomains, and demonstrate target inhibition in cellular assays. A more detailed description of the optimization of this series is reported elsewhere (24). Employing these chemical probes to query bromodomain function, we find that inhibition of CBP/EP300 bromodomains in ex vivo differentiated human Tregs is sufficient to reduce FOXP3 expression, as well as mediators of Treg suppressive function, such as LAG-3, CTLA-4 and TIM-3. Our data reveal the importance of CBP/EP300 bromodomains in maintaining pro-tolerance programs in Tregs and point toward manipulation of CBP/EP300 bromodomain function as an approach to counter immunosuppression in cancer.
Experimental Procedures
Chemical Synthesis
CPI098 (4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) is commercially available from Sigma-Aldrich and was used without further purification. Detailed descriptions of the chemical synthesis of CPI703, CPI644, CPI644-(−), and CPI571, along with descriptions of biochemical assays and crystallography are included in the supplemental text.
Thermal Shift Assay
All assays were carried out in 384-well plates. In a conical tube, CBP (4 mm) was combined with Sypro Orange (Life Technologies) to a final dye concentration of 5× in 50 mm Tris, 1 mm DTT, pH 8.5. The tube was centrifuged briefly to remove precipitate, and the protein:dye solution was then added to a black OptiplateTM plates (Greiner) and spun briefly (1 min, 900 × g), and then 23 ml of solution was transferred to either DMSO controls or fragments plated from 100 mm DMSO stocks into clear-bottomed Fluotrac200TM plates (Greiner) to a final compound concentration of 800 mm (0.8% v/v DMSO). Subsequently samples (15 ml) were transferred to LightCycler® 480 plates (Roche Diagnostics), spun (2 min, 900 × g), and analyzed on a Roche Lightcycler 480 II using a temperature gradient of 20–85 °C and a scanning rate of 1.2 °C/min. The midpoint of the melting transitions (Tm) were assessed using an application developed in-house measuring the first derivative of the rate of fluorescence change as a function of temperature. Compound-induced changes in the melting temperature, δTm, were calculated relative to DMSO controls within the same plate.
Crystallography
The co-crystal structures of CPI098 and CPI703 bound to the bromodomain of CBP were determined from crystals grown at 4 °C using the sitting drop technique. Crystals were grown using CBP protein at 24 mg/ml concentration stored in 20 mm HEPES, pH 7.5, 150 mm NaCl, and 1 mm tris-(2-carboxyethyl) phosphine hydrochloride that was equilibrated against either 0.2 m magnesium chloride, 0.1 m Bis-Tris, pH 6.5, and 27% PEG 3350 (CPI098) or 0.1 m Bicine:NaOH, pH 9.0, 20% (w/v) PEG 6000 (CPI703) for 5–20 days. Crystals were cryo-protected using a 50% mixture of paratone and paraffin oils before flash freezing in liquid nitrogen. The data were collected at the Advanced Photon Source on Beamline 22-ID-D (CPI098) or 21-ID-F (CPI703), and the structures were refined using refmac5 and the CCP4 suite of programs. The data collection and refinement statistics for CBP bound to CPI098 and CPI703 are detailed in supplemental Tables S3 and S4, respectively. The coordinates were deposited in the Protein Data Bank with accession codes: 4YK0 (CPI098) and 5DBM (CPI703).
ITC
Isothermal titration calorimetry (ITC) measurements were made on a MicroCal ITC200 instrument (Malvern Instruments). CBP was dialyzed overnight against 50 mm HEPES, pH 7.5, 150 mm NaCl. The solution was clarified by passage through a 0.22-μm Spin-X tube (Corning, Inc.), and the protein concentration was determined by absorbance at 280 nm measured with a NanoDrop instrument (Thermo Scientific). DMSO (0.2% v/v) was then added to the protein solution. Compounds were equilibrated in the calorimeter cell (15 μm, 200-μl volume, 25.0 °C, stirrer speed of 1000 rpm, 5 μcal/s reference power), and to this was added CBP (150 μm) in the same buffer as a series of sixteen 2.49-μl injections (4.98 s each) with an equilibration time of 120 s between each injection. The resulting data were fitted using the instrument's Origin 7.0 software.
AlphaLISA
Inhibitory activity of compounds was determined by competition with the binding of purified His-FLAG-tagged bromodomains to H4-TetraAc-biotin peptide (New England Peptide) using AlphaLISA technology (PerkinElmer Life Sciences). Compound at varying concentrations were dispensed into 384-well Proxiplates (PerkinElmer Life Sciences) using Echo technology (Labcyte). For the assays, 0.5 μm His-FLAG-tagged CBP bromodomain (amino acids 1082–1197) was incubated with 0.003 μm H4-TetraAc-biotin for 20 min at room temperature in 1× reaction buffer (50 mm HEPES, pH 7.5, 1 mm tris(2-carboxyethyl)phosphine, 0.069 mm Brij-35, 150 mm NaCl, and 0.1 mg/ml BSA). Streptavidin acceptor beads and nickel donor beads (PerkinElmer Life Sciences) were added to 15 μg/ml with a Combi multidrop dispenser. Plates were sealed and incubated at 90 min in the dark at room temperature, and plates were read on an Envision plate reader (PerkinElmer Life Sciences) according to the manufacturer's instructions. Other bromodomains were assessed by AlphaLISA in a similar manner.
NanoBRET Cellular Assays
NanoBRET was carried out using the NanoBRETTM protein-protein interaction system (Promega) according to the manufacturer's instructions. Briefly, HEK293 cells were transiently co-transfected with a vector for histone H3.3-HaloTag and either a NanoLuc tagged CBP bromodomain, EP300 bromodomain, or full-length BRD4 expression construct. Transfected cells were plated in 96-well plates in the presence or absence of ligand and then treated with dose titrations of indicated compounds. The readings were performed on an Envision plate reader (PerkinElmer Life Sciences), and BRET readings were calculated by dividing the acceptor emission value (600 nm) by the donor emission value (460/50 nm).
Mesoscale Cellular Assay
Mesoscale was carried out according to the manufacturer's instructions (Meso Scale Discovery). CBP/EP300 double knock-out mouse embryonic fibroblasts (25) or HCT116 cells (compound treatment, 16 h) were used. Coat Standard MSD plates (catalog no. L15XA-3) with 30 μl/well capture antibody anti-histone (Millipore catalog no. MAB3422) at 4 μg/ml final concentration was used. The anti-H3K18ac (catalog no. 9675) and anti-histone H3 (catalog no. 4499) antibodies were obtained from Cell Signaling and used at 0.125 μg/ml. Data capture was performed using a MSD Sector Imager 2400 (Meso Scale Discovery).
Acetyltransferase Assay
EP300 acetyltransferase reactions were carried out in 50 mm Tris, pH 8, 100 mm NaCl, 1 mm DTT, 0.1 mm EDTA, 0.069 μm Brij-35, and 0.1 mg/ml BSA. A solution of EP300 (Active Motif) and 1:9 3H-labeled (PerkinElmer Life Sciences)/unlabeled (Sigma-Aldrich) acetyl-CoA was added to each well of a black 384-well plate (Greiner) containing 200 nl of compound (final concentration, 0.8% DMSO). Compounds were preincubated with enzyme and acetyl-CoA for 30 min. Reactions were initiated by the addition of biotinylated H3 (1–21) peptide (Anaspec). All enzymatic reactions were performed for 60 min at room temperature in a 25-μl assay volume. Final concentrations of reagents were 0.75 nm EP300, 1 μm peptide, and 2.3 μm acetyl-CoA. Reactions were terminated by the addition of 20 μl of 200 mm Tris, pH 8, 200 mm EDTA, 160 μm anacardic acid (Sigma Aldrich), transferred to a 384-well streptavidin FlashPlate (PerkinElmer Life Sciences), and counted by TopCount (PerkinElmer Life Sciences).
Bromo Dot Cellular Assays
U2OS-C413 cells express the bromodomain of CBP in the context of the BRD9 protein. Specifically, amino acid residues 114–253 of BRD9 (UniProt Q9H8M2) have been replaced by amino acids 1064–1205 of CBP (UniProt Q92793). This chimeric protein is expressed in fusion with a C-terminally located ZsGreen (ZsG) fluorescent tag in a tetracycline-inducible manner. For the bromo dot assay, 5000 cells/well were seeded in 384-well imaging plates in the presence of 2 μg/ml doxycycline for 16 h. Compounds diluted in fresh medium were added to the cells for 2 h at 37 °C. The cells were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Images of cells were acquired using ImageXpress Micro (Molecular Devices) and processed with the Transfluor Module of MetaXpress software. Foci per nucleus values were obtained from four adjacent images in each well, and there were two technical replicates for each compound concentration. Dose-response curves were generated by plotting these numbers for each compound concentration, and EC50 values were calculated by a four-parameter non-linear regression model in GraphPad Prism. The average values from DMSO-treated wells were used to constrain the bottom of the curve.
Human T Cell Cultures
Leukopak samples were procured from the Biological Specialty Corporation (Colmar, PA), and peripheral blood mononuclear cells were isolated by Ficoll (GE Biosciences) density gradient centrifugation. Naive CD4+ CD45RA+ T cells were isolated from peripheral blood mononuclear cells to a purity >98% using Miltenyi naive human T cell isolation kits (130-094-131; Miltenyi Biotech). Isolated cells were cultured at 106 cells/ml under Treg-polarizing conditions, using human T-activator CD3/CD28 Dynabeads® (11132D; Invitrogen), human TGFβ at 10 ng/ml and human IL-2 at 10 units/ml (100-B and 202-IL, respectively; R&D Biosystems). For unpolarized TH0 cultures, isolated cells were cultured with Dynabeads alone, without the addition of exogenous cytokines.
FACS Antibodies
Tregs were stained for CTLA-4 (46-1529-42; eBioscience), CD25 (12-0259-42; eBioscience), LAG-3 (17-2239-42; eBioscience), PD-1 (47-2799-42; eBioscience), TIM3 (25-3109-42; eBioscience), and FOXP3 (12-4777-42; eBioscience). For FOXP3 intracellular staining, cells were fixed and permeabilized using the Foxp3/transcription factor staining kit (00-5523-00; eBioscience). In addition to staining for CTLA-4, LAG-3, PD-1, and TIM-3, CD8 T cells were also fixed and permeabilized using the Foxp3/transcription factor staining kit and stained for Perforin (11-9994-42; eBioscience) and Granzyme B (561142; BD Biosciences). The cells and markers were quantified on the BD FACSCantoTM II flow cytometry analyzer (BD Bioscience), and data were analyzed using FlowJo Software, gating on live cells for all FACS plots shown.
Lentiviral shRNA Knockdown of CBP and EP300
Naïve human T cells were cultured under Treg-inducing conditions as described above and at ∼16 h post-activation were infected with lentivirus harboring shRNAs specific for either EP300 or CBP (three independent hairpins per protein were cloned into pLKO.1-based lentiviral vectors (25) (see supplemental Table S2). Lentiviral supernatants were added to T cells in the presence of 8 μg/ml Sequa-brene (S2667-1VL; Sigma) followed by spin infection at 2100 rpm, 90′ at 30 °C. Transduced cells were selected by the addition of 1 μg/ml puromycin after 24 h; infection rates were monitored by measuring GFP fluorescence.
Global Expression Profiling
Naïve CD4+ T cells were treated with 4 μm CPI703 or DMSO under Treg polarizing conditions (described above) for 4 days. Total RNA was prepared by using a Qiagen RNeasy Plus mini kit (with columns for elimination of genomic DNA) according to the manufacturer's protocol. Samples were processed and hybridized on Affymetrix exon arrays, and the data were acquired at ALMAC Diagnostics. CEL files were analyzed with the Affymetrix Expression Console software.
Treg Suppression Assay
Human Tregs were differentiated in vitro (as described above) for 4 days. Additional IL-2 (at 10 units/ml) was added to the cultures on day 2, and FOXP3 expression was checked on day 4 (∼80% FOXP3 positive). The cells were taken off Dynabead stimulation, washed, and counted. Naïve T cells were labeled with carboxyfluorescein succinimidyl ester (C34554; Life Technologies) using the manufacturer's protocol. Co-cultures of naïve T cells and Tregs were set up at a 1:1 ratio. Human T-Activator CD3/CD28 Dynabeads® were added at a 1:4 ratio of beads to cells.
Deep ChIP Sequencing (ChIP-seq) and RNA Sequencing (RNA-seq)
Naïve human CD4+ T cells were treated with 4 μm CPI703 or DMSO under Treg polarizing conditions (described above) for 4 days. For ChIP-seq, the cells were pelleted, fixed in 1% formaldehyde for 10 min, lysed, and sonicated. Chromatin samples were precleared with protein A Dynabeads (Life Technologies) and incubated overnight at 4 °C with anti-H3K18 Ac (9675; Cell Signaling), anti-H3K27 Ac (ab4729; Abcam), and anti-H3K4 Me3 (ab8580; Abcam). Chromatin-antibody complexes were precipitated using protein A Dynabeads followed by washes in radioimmune precipitation assay buffer and Tris/EDTA. Samples were digested with RNase A and treated with proteinase K and 10% SDS, followed by cross-link reversal at 65 °C. DNA was purified using MinElute PCR purification kits (Qiagen). DNA libraries for ChIP-seq were prepared using Ovation Ultralow DR multiplex system kits (0330-32; NuGEN) followed by Illumina sequencing at the MIT BioMicro Center. For RNA-seq, RNA was isolated using Qiagen RNeasy Plus mini kits and sequenced at Ocean Ridge Biosciences (Palm Beach Gardens, FL).
Luminex Cytokine Assays
Cytokines were quantified from 72-h cell supernatants using Luminex multiplex assays (HTH17MAG-14K-12; Millipore), as per the manufacturer's protocol.
Cell Viability
Cell viability was assessed using CellTitreGlo®, which detects any change in the number of viable cells based on quantitation of ATP (G7572; Promega). Absolute live cell numbers were determined by trypan blue staining followed by analysis using the Countess automated cell counter (Invitrogen).
Real Time RT-PCR
RNA was purified from cells using an RNeasy Plus mini kit (Qiagen) according to the manufacturer's protocol. First strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen). Quantitative real time PCR was performed using FastStart Universal Probe master mix (Roche) and TaqMan probes (FOXP3, LAG3, HAVCR2 (TIM3), PDCD1 (PD1), PRF1, GRZMB, and EOMES) on the Stratagene MxPro3005p. Glucose-6-phosphate dehydrogenase was used as housekeeping gene (05-046-246-001; Roche).
Western Blotting
Cell pellets were resuspended in radioimmune precipitation assay buffer containing complete protease inhibitor mixture (04693116001; Roche), incubated on ice for 15 min, and spun at 12,000 × g. Supernatants thus obtained were boiled in loading buffer for 10 min at 95 °C and loaded on 4–20% Tris-glycine mini gels (EC6028BOX; Novex) followed by overnight transfer at 35 V onto PVDF membranes. The membranes were blocked with 5% milk in TBS, 0.1% Tween. Rabbit anti-human EP300 (sc-584; Santa Cruz) and rabbit anti-human CBP (sc-369; Santa Cruz) were used to detect EP300 and CBP, respectively, with goat anti-rabbit:HRP as secondary antibody. Blots were developed using the Pierce ECL Western blotting substrate.
Global Expression Profiling Data Analysis
The gene level RMA algorithm was applied, with the core set of probe sets and the annotation file Huex-1_0-st-v2_na31_hg19 provided by Affymetrix. The data were exported using the function “Export Probe Set Results (pivot table) with Annotations to TXT.” The RMA algorithm provides data in log space. The replicate profiles were averaged, and each probe set was displayed on scatter plot as a point. Gene set enrichment analysis (GSEA) was done using the GseaPreranked tool in GSEA with the following parameters: Score_scheme, Weighted; Make_sets, TRUE; Mode, Max_probe; Gene set collection, C7; Include_only_symbols, TRUE; Set_min, 15; Nperm, 1000; Rnd_seed, timestamp. The gene expression data are available at the Gene Expression Omnibus with accession no. GSE66596.
Immunoprecipitation of FOXP3 and Acetyl-FOXP3 Detection
Human Tregs were differentiated in the presence of 4 μm CPI703 for 36 h or 4 days. Whole cell extracts were prepared using complete lysis buffer (40010; Active Motif) and FOXP3 was immunoprecipitated using anti-Foxp3 (14-4777-82; eBioscience) directly coupled to NHS-activated magnetic beads (88826; Thermo Fisher; per manufacturer's protocol), followed by immunoblotting with a pan-acetyl Lys antibody (Ac-K2–100; 9814S; Cell Signaling) or anti-Foxp3. MG132 was added at 20 μm to reduce proteasome-induced FOXP3 degradation ∼6 h before preparation of whole cell extracts. Band densities were quantified using ImageJ software.
Results
Identification of Selective Small Molecule Inhibitors of CBP/EP300 Bromodomains
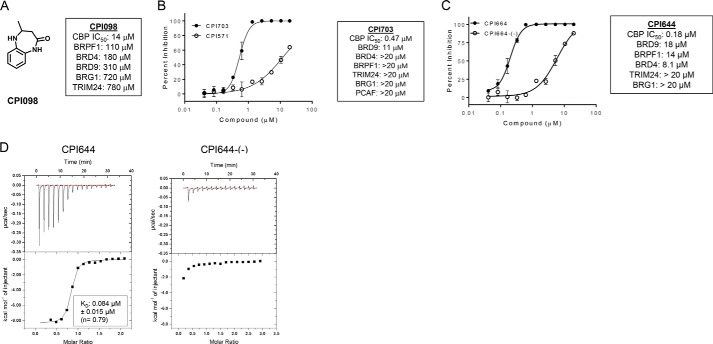
To identify small molecule ligands for CBP/EP300 bromodomains, we screened a library of ∼1200 fragments by differential scanning fluorimetry. Compounds that shifted the melting temperature of the CBP bromodomain by at least 1 °C and demonstrated a dose-response in the melting temperature shift when titrated were considered hits. Compounds were further validated by 15N heteronuclear single quantum coherence NMR, ITC, and a peptide competition AlphaLISA assay (data not shown). These studies identified 4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one(CPI098) as a 14 μm inhibitor of CBP by AlphaLISA that displayed promising selectivity over other bromodomains tested, including 13-fold selectivity over the BRD4 bromodomain 1 (BD1) (Fig. 1A).
FIGURE 1.
Biochemical description and characterization of CBP/EP300 probe molecules. A, structure of CPI098 and summary table indicating potency against other bromodomains. B, CBP inhibition by CPI703 and CPI571. Representative AlphaLISA data are shown for each compound (duplicate, ± S.E.). Across multiple replicates CPI703 (closed circles) inhibits with an IC50 = 0.47 ± 0.07 μm (n = 4), and CPI571 (open circles) has an IC50 = 12.2 ± 0.4 μm (n = 3) (values ± S.E.). C, CBP inhibition by CPI644 and CPI644-(−). Representative AlphaLISA data is shown for each compound (duplicate, ± S.E.). Across multiple replicates CPI644 inhibits with IC50 = 0.18 ± 0.06 μm (n = 5) and CPI644-(−) (open circles) with IC50 = 6.0 ± 0.6 μm (n = 3) (values ± S.E.). D, ITC analysis with CPI644 or CPI644-(−) and the bromodomain of CBP. A binding stoichiometrically of n = 0.79 and a KD of 0.084 μm was observed for CPI644, although no detectable binding was observed for CPI644-(−).
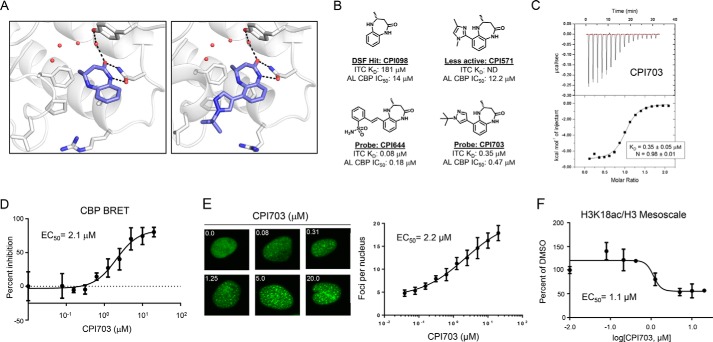
The binding mode of the series was established with co-crystal structures of the CPI098 fragment (1.65 Å resolution) and a more advanced derivative CPI703 (1.86 Å resolution) bound to the CBP bromodomain. The lactam carbonyl of the scaffold formed key H-bonding interactions with Asn1168 and, through water, with Tyr1125 (Fig. 2A, left panel). The lactam NH provided a further interaction with Asn1168, helping position the 4-methyl substituent toward the water pocket, in the space normally occupied by the acetyl methyl group of an acetyl lysine ligand (Protein Data Bank code 3P1C). Interestingly, only the R enantiomer of CPI098 was observed in the electron density, despite use of a racemic mixture for crystallization. Assaying individual enantiomers confirmed that this enantiomer was substantially more potent (∼50-fold).
FIGURE 2.
Discovery of potent, selective, and cell-active CBP/EP300 bromodomain inhibitors. A, co-crystal structures of CPI098 (left panel) and CPI703 (right panel) with the CBP bromodomain. The compounds bind in the acetyllysine recognition site, making key hydrogen bonding interactions with Asn1168 and, through water, with Tyr1125. B, structures of CPI098, CPI703, CPI644, and CPI571. The ITC KD and AlphaLISA IC50 (AL) values are provided. C, ITC analysis of CPI703. CPI703 binds stoichiometrically with n = 0.98 and a KD of 0.35 μm. D, cellular potency of CPI703 using NanoBRET. A dose-response curve of CPI703 using the isolated CBP bromodomain and histone H3.3 with a calculated EC50 of 2.1 μm is shown. E, cellular potency of CPI703 determined by the CBP bromo dot assay. Representative images show ZsG-CBP protein aggregates at the indicated concentrations (left). The dose-dependent increase in the number and intensity of these nuclear foci was used to determine the EC50 of CPI703 (2.2 μm, right). F, cellular potency of CPI703 in an H3K18ac Mesoscale assay. The H3K18ac signal was normalized to total H3 signal, and the percentage change relative to the DMSO control was plotted. ND, not detected; DSF, differential scanning fluorimetry.
Analysis of interactions observed in the complex structure supported structure-activity relationships among compounds with modifications to the CPI098 core. Exploring substitutions around the benzene ring, we determined that a variety of substituents at C6 increased potency (a detailed structure-activity relationship account for this series will be described elsewhere). Of note were two molecules, CPI703 and CPI644, which had IC50 values of 0.47 and 0.18 μm, respectively, in the AlphaLISA CBP assay (Figs. 1, B and C, and 2B). A co-crystal structure of CPI703 bound to CBP revealed favorable interactions between the C6 tert-butyl-pyrazole of the compound and the LPF shelf of the bromodomain (Fig. 2A, right panel), accounting for the significant boost in potency. Importantly, these compounds retained selectivity over the BET family and other bromodomain-containing proteins (Fig. 1, B and C). We chose CPI571 (Fig. 2B) as a negative control compound, because it lacked activity on CBP but otherwise demonstrated physicochemical properties very similar to CPI703 and CPI644. The S-enantiomer, CPI644-(−), was found to be significantly less potent than CPI644, with an IC50 value of 6.0 μm in the peptide AlphaLISA assay. The potencies of these compounds were corroborated by ITC, using the isolated bromodomain of CBP (Figs. 1D and 2, B and C, and data not shown). Extensive profiling by differential scanning fluorimetry using 19 different isolated bromodomains confirmed the selectivity of CPI703 and CPI644 for the CBP bromodomain (Table 1).
TABLE 1.
Differential scanning fluorimetry binding data for CPI703, CPI571, CPI644, and CPI644-(−) on isolated bromodomains
The values ± S.D. are shown.
| Protein | CPI703 | CPI571 | CPI644 | CPI644-(−) |
|---|---|---|---|---|
| ASH1L | 0.00 ± 0.73 | −0.35 ± 0.40 | −0.64 ± 0.25 | −0.23 ± 0.88 |
| BAZ1A | −0.12 ± 0.20 | 0.00 ± 0.00 | −0.12 ± 0.20 | −0.23 ± 0.20 |
| BAZ2B | −0.12 ± 0.40 | 0.23 ± 0.53 | 0.12 ± 0.00 | −0.23 ± 0.35 |
| BPTF | 0.46 ± 0.40 | 0.46 ± 0.53 | 0.58 ± 0.20 | 0.46 ± 0.72 |
| BRD1 | 0.23 ± 0.00 | 0.46 ± 0.20 | 0.46 ± 0.20 | 0.23 ± 0.00 |
| BRD4 | 0.70 ± 0.35 | 0.00 ± 0.00 | 1.39 ± 0.70 | 0.12 ± 0.20 |
| BRD8 | 0.52 ± 0.25 | 0.46 ± 0.20 | 0.87 ± 0.25 | 0.35 ± 0.35 |
| BRD9 | 0.93 ± 0.53 | 0.46 ± 0.53 | 0.70 ± 0.35 | 0.23 ± 0.20 |
| BRG1 | 0.00 ± 0.00 | 0.35 ± 0.35 | 0.12 ± 0.20 | 0.12 ± 0.40 |
| BRPF1 | 0.23 ± 0.00 | −0.12 ± 0.00 | −0.12 ± 0.35 | −0.46 ± 0.35 |
| BRPF3 | 0.35 ± 0.35 | 0.00 ± 0.49 | 0.46 ± 0.20 | 0.35 ± 0.60 |
| CBP | 5.80 ± 1.39 | 0.58 ± 0.00 | 8.70 ± 1.22 | 1.28 ± 0.35 |
| CECR2 | 0.06 ± 0.25 | 0.06 ± 0.25 | 0.12 ± 0.20 | 0.93 ± 0.00 |
| GCN5 | 0.00 ± 0.20 | 0.00 ± 0.20 | −0.12 ± 0.20 | −0.12 ± 0.20 |
| MLL | 0.00 ± 0.20 | 0.58 ± 0.35 | 0.70 ± 0.53 | 0.46 ± 0.20 |
| PCAF | 0.81 ± 0.20 | 0.35 ± 0.35 | −0.23 ± 0.53 | 0.00 ± 1.04 |
| TAF1 BD1-BD2 | 0.35 ± 0.00 | 0.46 ± 0.20 | 0.23 ± 0.20 | 0.35 ± 0.00 |
| TRIM24 | 0.23 ± 0.20 | 0.23 ± 0.20 | 0.23 ± 0.20 | 0.00 ± 0.00 |
| WDR9 | 0.23 ± 0.20 | 0.12 ± 0.00 | 0.12 ± 0.00 | 0.12 ± 0.00 |
Recent work has shown that a subset of small molecule kinase inhibitors show additional binding and functional inhibition of BET bromodomains (26). To test whether benzodiazepinone CBP/EP300 bromodomain inhibitors show similar polypharmacology, we profiled the in vitro activity of CPI644 against a broad panel of serine/threonine and tyrosine protein kinases. At a concentration of 1 μm (well above the IC50 for bromodomain binding), CPI644 showed negligible activity against all of the kinases profiled (supplemental Table S1), suggesting that the phenotypic effects of CBP/EP300 bromodomain inhibitors (see below) are not the result of direct kinase inhibition.
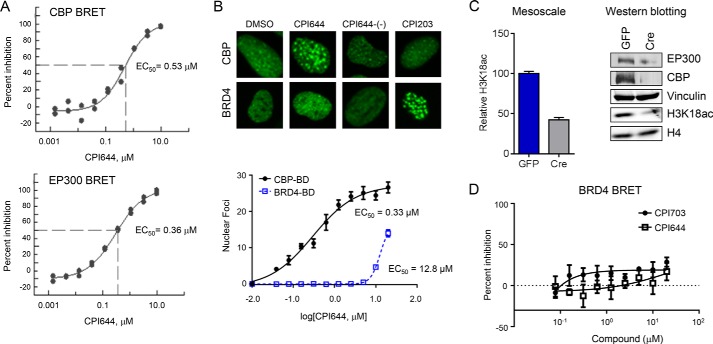
To characterize CBP-bromodomain inhibition in cells, we utilized several orthogonal assays. First, we utilized NanoBRET technology (Promega) to directly monitor the interaction between the isolated CBP bromodomain and histone H3.3. CPI703 and CPI644 inhibited CBP bromodomain binding in a dose-dependent manner with cellular EC50 values of 2.1 and 0.53 μm, respectively (Fig. 2D and Fig. 3A). Inhibition of EP300 bromodomain in cells occurred at similar concentrations (Fig. 3A). Next, we established a complementary assay to monitor the interaction between chromatin and a fluorescent fusion protein containing the CBP bromodomain. This chimeric protein was expressed as a fusion of the CBP bromodomain with a ZsG fluorescent tag (in the context of BRD9 protein i.e. the bromodomain of BRD9 was replaced with that of CBP) (27). Chromatin release was measured by conversion of the largely homogeneous nuclear localization of the ZsG-CBP protein to a more pronounced dot-like pattern (Figs. 2E and 3B). Dot formation occurred in a compound-specific and dose-dependent manner, resulting from displacement of the fluorescently tagged protein from chromatin and its subsequent aggregation (Figs. 2E and 3B). The number and intensity of these foci were measured by high content imaging, and analysis afforded a quantitative determination of target engagement by inhibitor (see “Experimental Procedures”). The number of dots increased upon titration of the inhibitor, yielding a sigmoidal curve when plotted as a function of inhibitor concentration. Cellular EC50 values calculated using this assay (Fig. 2E; CPI703 = 2.2 μm; CPI644 = 0.33 μm) correlated extremely well with the potencies measured by NanoBRET.
FIGURE 3.
Cellular characterization of CBP/EP300 probe molecules. A, cellular potency of CPI644 using NanoBRET. A dose-response of CPI644 using the isolated bromodomain of CBP (top panel) or EP300 (bottom panel) and histone H3.3 with calculated EC50 values of 0.53 and 0.36 μm are shown. B, cellular potency of CPI644 determined by the CBP-bromo or BRD4 (full-length protein) dot assays. Representative images show ZsG-CBP and ZsG-BRD4 protein aggregates from 5.0 μm (CPI644 and CPI644-(−)) and 0.3 μm (CPI203) treatment (top panel). The dose-dependent increase in the number and intensity of these nuclear foci was used to determine the EC50 of CPI644 in the CBP (0.33 μm) or BRD4 (12.8 μm) dot assays (bottom panel). C, GFP (control) or Cre (knock-out) cells (25) were utilized to probe H3K18ac by Mesoscale (left panel). The relative change in H3K18ac signal was plotted setting the GFP control cells to 100%. The right panel shows a Western blot using the indicated antibodies in the GFP (control) or Cre (knock-out) cells (25). D, cellular potency of CPI703 or CPI644 using BRD4 and H3.3 NanoBRET. No appreciable inhibition was observed.
CBP and EP300 modulate gene expression in part through the acetylation of H3K18 and H3K27 (28), and the bromodomain has been linked to its transcriptional activity (29). We therefore sought to determine whether pharmacologic inhibition of CBP/EP300 bromodomains would impact histone acetylation. Given that histone-acetyl antibodies are not as specific as histone-methyl antibodies (30), we established a quantitative antibody-based cellular assay using the Mesoscale technology (Meso Scale Discovery). To determine the specificity of the assay, we utilized CBP/EP300 double knock-out mouse embryonic fibroblasts (28). Histone mass spectrometry confirmed a specific diminution in H3K18ac following double deletion of CBP and EP300, whereas H3K27ac was not detectable (data not shown). Thus, we focused on H3K18ac for future experiments. Double knock-out of CBP/EP300 resulted in a >50% diminution in the H3K18ac signal, providing a benchmark for the Mesoscale assay (Fig. 3C). Titration of CPI703 resulted in a dose-dependent reduction in H3K18ac with a calculated EC50 of 1.1 μm, providing evidence that small molecule bromodomain inhibition blocks CBP/EP300-dependent histone acetylation (Fig. 2F). Taken together, the EC50 values of CPI703 and CPI644 yielded highly congruent cellular potencies from a number of orthogonal, independent cellular assays (Figs. 2 and 3). Moreover, similar to the biochemical analysis, the selectivity of CPI703 and CPI644 for the bromodomain of CBP over BRD4 was corroborated in cells (Fig. 3, B and D).
CBP/EP300 Bromodomains Are Required for Human Treg Differentiation
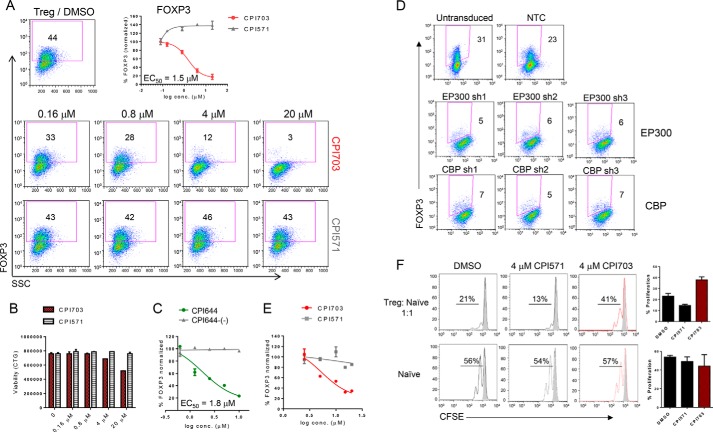
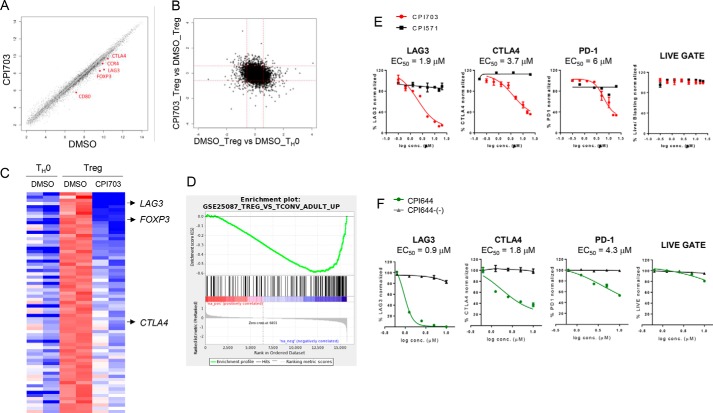
Tregs are marked by expression of the forkhead transcription factor FOXP3, essential for the Treg differentiation program. Loss of function mutations in FOXP3 lead to severe and systemic autoimmunity in both mice and humans (1, 2, 31). Treg-specific deletion of EP300 or CBP in mice results in decreased Treg function and reduced tumor growth in cancer models (21, 22), although the specific role for the bromodomain has not been determined. We asked whether CBP/EP300 bromodomains are involved in modulating similar phenotypes in human cells and specifically whether bromodomain inhibition could result in impaired Treg differentiation. Incubation with CPI703 reduced Treg differentiation in a dose-dependent manner, as enumerated by FOXP3+ cells in the cultures (Fig. 4A). Curve fitting established an IC50 of ∼1.5 μm for FOXP3 inhibition by CPI703 (Fig. 4A). Importantly, this calculated IC50 is highly consistent with the previously established potency of CPI703 in orthogonal cellular assays (Fig. 2, D–F). This effect was also specific, because the inactive control CPI571 had no impact on Treg numbers (Fig. 4A). Notably, up-regulation of CD25 was unaffected by treatment with CPI703 (data not shown), suggesting that naïve T cells undergo normal activation but are unable to specifically differentiate into the Treg lineage. Further, CBP/EP300 bromodomain inhibition did not significantly affect Treg viability (Fig. 4B). Similar results were obtained using our other CBP/EP300 bromodomain inhibitor CPI644, which strongly reduced the percentage of FOXP3+ cells in differentiating Treg cultures, whereas its inactive control CPI644-(−) had no effect (Fig. 4C). To further rule out potential off-target effects of CPI703 or CPI644, we used multiple lentiviral shRNA constructs to selectively knock down expression of CBP or EP300 in human Tregs differentiated from CD4+CD45RA+ naïve T cells in vitro (supplemental Table S2). Puromycin selection for 7 days under Treg differentiation conditions (anti-CD3/CD28 stimulation, TGFβ, and IL-2) resulted in homogeneously transduced populations of Tregs, as demonstrated by GFP expression (supplemental Fig. S1A). All three independent hairpins (for either gene) brought about at least a 50% reduction in the level of the corresponding endogenous protein (supplemental Fig. S1B), and consequently, completely suppressed Treg differentiation (Fig. 4D). To determine whether CBP/EP300 bromodomains could regulate FOXP3 expression in established Tregs, we added CPI703 after naïve human T cells were differentiated into Tregs for 4 days in vitro. This regimen also resulted in a dose-dependent inhibition in the percentage of FOXP3+ cells (Fig. 4E), suggesting an important contribution of CBP/EP300 bromodomains in maintenance of the Treg lineage. Indeed, in vitro differentiated Tregs exhibited reduced suppressive capability when co-cultured with carboxyfluorescein succinimidyl ester-labeled naïve CD4+ T cells in the presence of CPI703, as evidenced by a ∼3-fold increase in naïve T cell proliferation in co-cultures relative to the inactive compound (Fig. 4F, upper panels); this points toward down-modulation of Treg function as a result of CBP/EP300 bromodomain inhibition. Proliferation of naïve T cells alone was minimally affected in the presence of the inhibitor (Fig. 4F, lower panels).
FIGURE 4.
Pharmacological inhibition of CBP/EP300 bromodomains impairs human Treg differentiation. A, human naïve T cells were differentiated into Tregs in the presence of CPI703, the inactive analogue CPI571, or DMSO for 4 days and monitored for FOXP3 expression by flow cytometry. B, cell viability was assessed by CellTitreGlo. C, human naïve T cells were differentiated into Tregs in the presence of CPI644 or the inactive isomer CPI644-(−) and monitored for FOXP3 expression by flow cytometry on day 4. D, phenotypic recapitulation with CREBBP or EP300 shRNA-mediated knockdown. Flow cytometric analysis of transduced cultures with three independent hairpins per target (EP300 or CBP) indicated almost complete absence of FOXP3+ cells. Untransduced and non-targeting control hairpin are shown as comparison. E, human naïve T cells were differentiated into Tregs for 3 days and then treated with either CPI703 or CPI571 as indicated. FOXP3 was measured by FACS after 4 additional days. F, inhibition of Treg suppressive function by CPI703. In vitro differentiated Tregs were added to carboxyfluorescein succinimidyl ester-labeled naïve T cells at a 1:1 ratio (top); naïve T cells activated in the absence of added Tregs are shown in the bottom panel. Compounds were added at 4 μm, and proliferation was measured by FACS on day 3. The gray-shaded histograms correspond to unstimulated cells; at right, the percentage of proliferation indicates the percentage of cells undergoing one or more divisions. CFSE, carboxyfluorescein succinimidyl ester; conc., concentration; NTC, non-targeting control.
CBP/EP300 Bromodomain Inhibition Alters the Human Treg Transcriptional Signature
To delineate molecular mechanisms underlying the phenotypic and functional impact of CBP/EP300 bromodomain inhibition, we performed full-genome transcriptional profiling, comparing samples from Treg cultures treated either with 4 μm CPI703 or DMSO (4 μm was chosen to ensure robust target coverage, ∼IC75). As an additional control we cultured naïve T cells in the absence of differentiating cytokines (hereafter referred to as TH0). Double-pairwise comparisons ([CPI703-treated Treg versus DMSO-treated Treg] versus [DMSO-treated Treg versus DMSO-treated TH0] (Figs. 5, A and B) revealed a bias toward preferential inhibition by CPI703 of genes that are up-regulated in Tregs compared with those up-regulated in TH0 cells. Scatter plot analysis of Treg cells treated with CPI703 compared with DMSO controls (Fig. 5A) showed more genes down-regulated than up-regulated in compound-treated Tregs, suggesting that bromodomain function is necessary for full CBP/EP300-mediated transcriptional activation. FOXP3 was among the genes significantly down-regulated, as expected (Fig. 5, A and C), serving as an internal control to validate the analysis. Of the genes up-regulated in Tregs (at least 1.5-fold over TH0), 69 genes were inhibited 1.5-fold or more at the transcript level upon CPI703 treatment (Fig. 5C and supplemental Table S3). In addition to FOXP3, other genes that are thought to play important roles in Treg function, such as LAG3, CTLA4, CCR4, and BTLA were up-regulated in Tregs compared with TH0 cells and down-regulated in Tregs upon CBP/EP300 bromodomain inhibition (Fig. 5C and supplemental Table S3). Another important co-inhibitory receptor, TIM-3 (encoded by HAVCR2), was significantly reduced in Tregs upon compound treatment (∼70% down; p < 0.005), although not strongly up-regulated in Tregs compared with TH0 cells (data not shown). GSEA identified the Treg versus Tconv curated gene set (GSE15659_TREG_VS_TCONV_DN) as significantly coincident with the signature elicited by CPI703 treatment (Fig. 5D), confirming the impact of CPI703 on Treg transcription. These transcriptional data for key mediators of Treg function were also recapitulated by flow cytometric analysis. Indeed, levels of LAG-3, CTLA-4, and, to a lesser extent, PD-1, were reduced in a dose-dependent manner by CPI703 and CPI644 (Fig. 5, E and F), with no significant impact on cell viability. Dose-dependent reduction in transcription of FOXP3, LAG3, CTLA4, and PDCD1 (PD-1) was also corroborated by quantitative RT-PCR (data not shown). Thus, we conclude that CBP/EP300 bromodomain inhibition results in the suppression of a network of genes that transcriptionally define Treg cells and their biological functions.
FIGURE 5.
CBP/EP300 bromodomain inhibition alters the human Treg transcriptional signature. A and B, human naïve T cells were cultured with anti-CD3/CD28 and TGF-β/IL-2 (Treg) or anti-CD3/CD28 alone (TH0) and incubated with DMSO or 4 μm CPI703 for 4 days, and whole genome gene expression was evaluated on the Affymetrix exon array platform. Each probe set is shown as a point in the scatter plot; log fold change calculated as difference between mean values of RMA expression levels for each condition. Probe sets are shown in black if a Student's t test on the two pairs of values shows a difference with p < 0.05; otherwise the points are in gray. C, probe sets in the heat map were sorted by fold-change of transcripts in Tregs when treated with CPI703. The top row corresponds to the probe set with the greatest loss of expression with CPI703. The deepest red and blue colors represent values of +0.5 or greater, and −1.5 or less, respectively. D, the complete list of annotated probe sets along with the difference of the average RMA expression values in Tregs with CPI703 treatment versus DMSO treatment was used as input to GSEA. Gene set GSE25087_TREG_VS_TCONV_ADULT_UP showed significant down-regulation in our data set. The enrichment score was −0.5891071; the normalized enrichment score and p values were −1.8608235, and 0.0, respectively (the p value was reported by the GSEA preranked analysis program as being 0.0, based on random permutations). E and F, flow cytometric analysis for the expression of the indicated markers on Tregs treated with CPI703 (and CPI571) (E) and CPI644 (and CPI644-(−)) (F). conc., concentration.
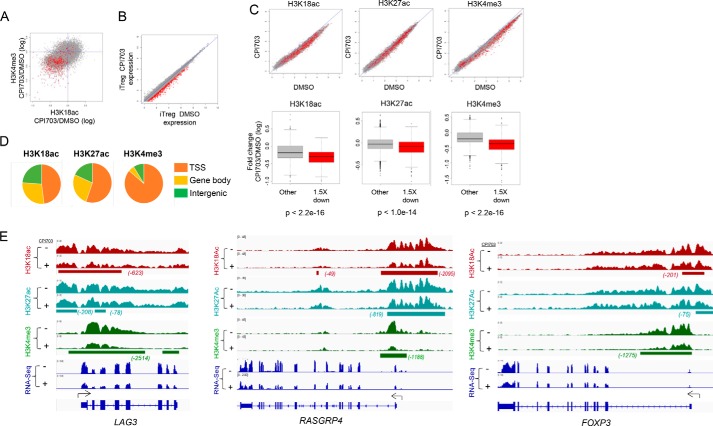
CPI703 Modulates Acetylation in Human Tregs
Mechanistically, CBP/EP300 bromodomain inhibition may facilitate reprogramming of Treg cells by altering the histone acetyl landscape. To address this, we combined global RNA-seq with chromatin immunoprecipitation followed by ChIP-seq to investigate molecular changes in chromatin induced by CBP/EP300 bromodomain inhibition and their association with transcriptional changes. In keeping with the role of CBP/EP300 as transcriptional activators, we found a strong positive correlation between the activation mark H3K4me3 and CBP/EP300-mediated H3K18ac (Fig. 6A) and H3K27ac (not shown) across the genome of human Treg cells. RNA-seq analysis showed that CPI703-mediated CBP/EP300 bromodomain inhibition resulted in the down-regulation of 482 genes by 1.5-fold or more (Fig. 6B). Importantly, CBP/EP300 bromodomain inhibition resulted in diminished H3K18 and H3K27 acetylation (Fig. 6C), similar to previous findings in a different cellular context (Fig. 2F). Consistent with these effects of CPI703 on transcription and CBP/EP300-mediated histone acetylation, we observed a significant concomitant reduction in global H3K4me3 (Fig. 6C). Correlation analysis indicated a strong statistical association between reduced H3K18ac (and H3K27ac) with genes that were down-regulated in their expression by 1.5-fold or more (Fig. 6C). From these data we conclude that optimal deposition of acetyl marks on chromatin by CBP/EP300 necessitates full activity of their bromodomains. H3K18ac, H3K27ac, and H3K4me3 marks were localized primarily at the transcription start sites (TSS; Fig. 6D), in agreement with their role in transcriptional activation. Individual analysis of target loci confirmed this observation, with H3K4me3 being almost exclusively restricted to the TSS, whereas H3K18ac and H3K27ac were highly enriched at the TSS with modest extension toward the gene bodies (Fig. 6, D and E, and supplemental Fig. S2). Furthermore, integrated ChIP-seq signal analysis using the SICER interval analysis algorithm demonstrated that CPI703 treatment reduces H3K18ac, and to a lesser extent H3K27ac, and that this reduction takes place almost exclusively at the TSS regions (Fig. 6E and supplemental Fig. S2), with a minimal impact on histones located 3′ of the TSS. Reduction of H3K18ac is evident around the TSS of exemplar loci such as LAG3, FOXP3, and RASGRP4 (Fig. 6E) (see also HAVCR2 and IRF7; supplemental Fig. S2), matching a corresponding decrease in H3K4me3 and reduction in exon RNA accumulation. We did not, however, observe a global expulsion of EP300 from chromatin as a consequence of bromodomain inhibition, either in Tregs or in other cellular contexts (data not shown). These results are consistent with recent publications exploring the function of CBP/EP300 bromodomain inhibition in cancer cell line contexts (32, 33). The most prevalent functional effect of CBP/EP300 bromodomain inhibition was reduced acetylation of H3K18 and H3K27 (and other proteins; see below). Together, our ChIP-seq and RNA-seq studies indicate that CPI703 inhibits optimal acetylation of histones at the transcriptional start sites of target loci, suggesting a novel mechanistic basis for the role of CBP/EP300 bromodomains in human Treg cells.
FIGURE 6.
CBP/EP300 bromodomain inhibition alters the chromatin state and reduces H3K18Ac and H3K27Ac at key target loci in human Tregs. A, each point in the scatter plot represents the regularized log2 fold change with treatment (CPI703 versus DMSO) of average ChIP-seq signal at a transcript start site for the given signal (H3K18ac on the x axis and H3K4me3 on the y axis). Points in red represent transcripts showing a loss of expression, as defined by a regularized log2 fold change of at least 1.5 down. B, each point in the scatter plot shows the regularized log expression level of one transcript for CPI703 versus DMSO treatment. Points in red represent transcripts showing a loss of expression, as defined by a regularized log2 fold change of at least 1.5 down. C, scatter plots in the top row show the effect of CPI703 versus DMSO on histone mark signals near TSSs. Each point is one TSS, and red points represent transcripts showing an expression loss of at least 1.5 in regularized log2 space. The box and whiskers plots in the bottom row summarize the change in regularized log2 histone mark ChIP-seq signal for down-regulated (red) and other (gray) transcripts; p values from Student's t test comparing mark changes for down-regulated versus other transcripts. D, fraction of ChIP-seq signal in different genomic regions. SICER version 1.1 was run to identify regions of occupancy. All SICER intervals within 1000 bp of a TSS were annotated as TSS. SICER intervals overlapping the gene body but not TSS were annotated as gene body binding. All other SICER intervals were annotated as intergenic. Total SICER-reported signal in the intervals was summed for each category and summarized in this figure. E, visualization in IGV of ChIP-seq signal at the LAG3, RASGRP4, and FOXP3 loci. Intervals of differential occupancy are shown beneath each pair of treated and control tracks.
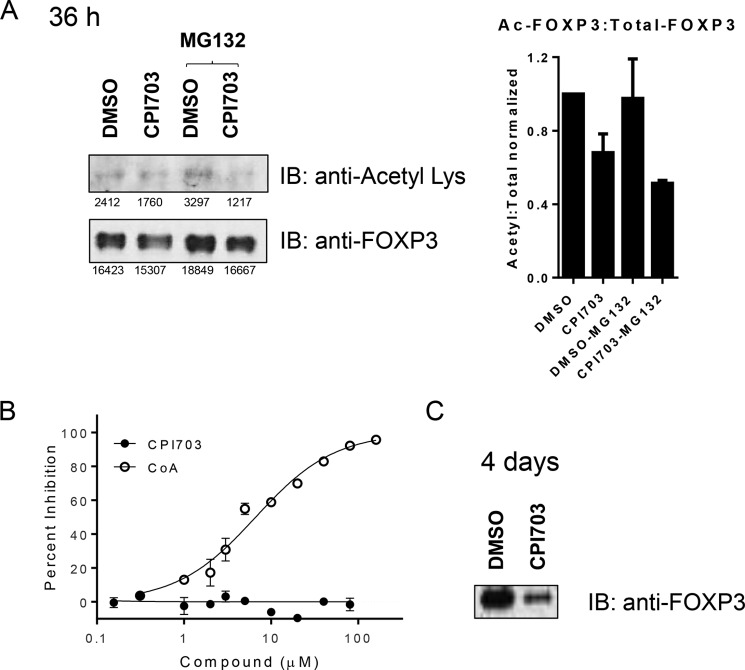
Additionally, stabilization of FOXP3 protein by EP300-mediated acetylation has been previously reported (34). Therefore we asked whether CBP/EP300 bromodomains might also play a significant role in this process, similar to their effects on histone acetylation. We immunoprecipitated FOXP3 early during Treg differentiation (36 h), followed by immunoblotting with a pan-acetyl antibody. Because FOXP3 is known to be degraded via the proteasome pathway (34), we used the proteasomal inhibitor, MG132, to minimize changes in acetylated FOXP3 levels caused by mere changes in total FOXP3 protein. CPI703 treatment led to a clear reduction in the ratio of acetylated FOXP3 to total FOXP3 (Fig. 7A), indicating a role for CBP/EP300 bromodomains in modulating FOXP3 acetylation. Importantly, CPI703 does not directly inhibit EP300 acetyltransferase activity (Fig. 7B). CPI703 treatment over a longer time period (4 days) led to a marked reduction in FOXP3 protein levels, as observed previously by FACS (Figs. 4 and 7C), potentially reflecting an increased susceptibility to proteasomal degradation caused by reduction in protein acetylation. Thus, in addition to a transcriptional effect, these data suggest another level of FOXP3 regulation by CBP/EP300 bromodomains via direct FOXP3 acetylation. Taken together, our results highlight at least two separate, but related, mechanisms underlying the novel role of CBP/EP300 bromodomains in human Treg differentiation and function.
FIGURE 7.
CBP/EP300 bromodomain inhibition modulates FOXP3 acetylation. A, human Tregs were differentiated in the presence of 4 μm CPI703 for 36 h. FOXP3 was immunoprecipitated using anti-Foxp3 magnetic beads and immunoblotted with a pan-acetyl Lys antibody or anti-Foxp3. MG132 was added to reduce proteasome-induced FOXP3 degradation. Quantification for each band is indicated below, and the ratio of acetylated to total FOXP3 is shown in the right panel (average of three independent experiments). B, acetyltransferase assay using full-length EP300 HAT. In the same experiment the reaction product CoA inhibited with IC50 6.2 ± 1.2 μm (+S.E.). The data are the averages of two replicates ± S.E. C, human Tregs where differentiated as in A, for 4 days, and FOXP3 immunoprecipitated as in A and immunoblotted with anti-Foxp3. IB, immunoblotting.
Modulation of T Helper Subsets by CBP/EP300 Bromodomain Inhibition
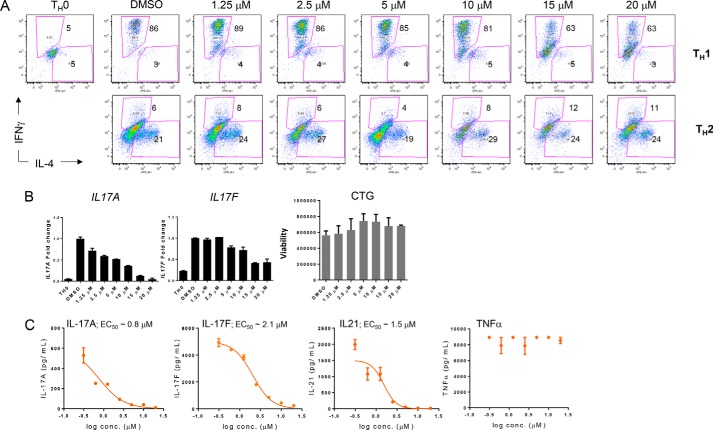
Given the importance of CBP/EP300 bromodomain function within the Treg lineage, we sought to ascertain the specificity of our observed effects vis-à-vis the effect of CPI703 on differentiation of naïve T cells into other TH cell lineages, namely TH1, TH2, and TH17. TH1 and TH2 cell differentiation were largely unperturbed by CPI703, because the percentage of cells producing their canonical cytokines, IFNγ and IL-4, respectively, were unchanged (except for a modest reduction of IFNγ+ TH1 cells at 15 and 20 μm concentrations; Fig. 8A, top panels). However, CBP/EP300 bromodomain inhibition significantly affected TH17 differentiation (Fig. 8, B and C). Both transcript and protein for the characteristic TH17 cytokines, IL-17A, IL-17F, and IL-21, were suppressed by CPI703 with no effect on TNFα production or cell viability (Fig. 8, B and C).
FIGURE 8.
CBP/EP300 bromodomain inhibition does not affect TH1 and TH2 differentiation while strongly suppressing cytokine production by TH17 cells. A, human naïve T cells were differentiated under TH1 or TH2 conditions for 7 days, rested for 2 days, restimulated with PMI/ionomycin for 5 h, and analyzed by flow cytometry. B, human naïve T cells were differentiated under TH17 conditions for 4 days, RNA was isolated and analyzed by qPCR for the indicated transcripts, and cell viability was assessed by CellTitreGlo. C, human naïve T cells were differentiated under TH17 conditions for 7 days, and the culture media were analyzed by Luminex for the indicated analytes. conc., concentration.
Discussion
Chemical probes allow for temporal, titratable, and domain-specific manipulation of molecular targets in human cells. In the current report, we introduce a series of potent and selective CBP/EP300 bromodomain inhibitors with low micromolar cellular activity using numerous cell systems and contexts. Using these inhibitors, we discover novel effects of CBP/EP300 bromodomain modulation on the biology of human Treg cells.
Recent reports have shown that mice with EP300 or CBP-deficient Tregs exhibit enhanced effector responses, reduced peripheral Treg generation, and Treg apoptosis, resulting in rapid allograft rejection and decreased tumor growth. CBP/EP300 HAT inhibition phenocopies the consequences of genetic ablation of either protein in mice (21, 22). Here we show that lentivirus-mediated knockdown of CBP or EP300 in human Tregs severely and non-redundantly inhibited FOXP3 up-regulation with a decrease in cell viability, likely because of a loss of global CBP/EP300-dependent acetylation. In contrast, specific inhibition of CBP/EP300 bromodomains by treatment of differentiating Tregs with CPI703 or CPI644, led to a marked reduction in FOXP3 expression both at protein and transcript levels, with no appreciable effect on cell viability. CBP/EP300 are known to interact with over 300 protein partners and are among the best connected protein “hubs” in the intracellular transcriptional network (19, 35). Given their ubiquitous distribution and the importance of their HAT domains in maintaining basal acetylation levels, we propose that bromodomain inhibition represents a more nuanced and less cytotoxic means for querying the importance of context-specific CBP/EP300 scaffolding function in primary cells and a viable option for therapeutic intervention.
Our results point to the requirement for CBP/EP300 bromodomains in the differentiation of human Tregs from naïve T cells. Transcriptional profiling of inhibitor-treated Tregs reveals significant down-regulation of genes encoding crucial Treg factors (FOXP3), and receptors such as LAG-3, CTLA-4, and TIM-3, which participate in cognate Treg responder cell interactions (36–38). CPI703 treatment also reduced transcript levels of other co-inhibitory receptors such as B- and T-lymphocyte attenuator (BTLA) and CD160 (39, 40), as well as the chemokine receptor CCR4, known to be expressed on the highly suppressive effector Treg subset which comprises the bulk of tumor-infiltrating lymphocytes in cancer patients (4). In accordance with this effect on the Treg transcriptome, CPI703 dampened Treg-mediated suppression of naïve T cell proliferation in Treg + responder T cell co-culture assays, underscoring important functional effects of the altered transcriptional signature elicited by CBP/EP300 bromodomain inhibition. As chromatin regulators, the important role of bromodomain function in the optimal acetylation of histones at target loci was perhaps not unexpected. The robust association between reduced acetylation, namely H3K18ac, and gene transcription upon treatment with CPI703 suggests a mechanistic framework to understand how these chromatin reader domains contribute to gene expression. Furthermore, molecular discrimination between H3K18 and H3K27 acetylation might provide additional layers of nuanced transcriptional control (at least in human Tregs), because we observed a generally stronger effect of CPI703 on H3K18ac than on H3K27ac. It is worth noting that we did not observe complete elimination of H3K18ac, which could be the consequence of partial inhibition of CBP/EP300 HAT activity, or a limitation of the reagents used, as double knock-out of CBP/EP300 in mouse embryonic fibroblasts appeared to reduce H3K18ac by ∼60%.
Additionally, CBP/EP300 bromodomains could impact optimal acetylation of chromatin associated transcription factors, including FOXP3. EP300-mediated acetylation of FOXP3 has been shown to contribute to its stability (34), and it is conceivable that bromodomain-dependent binding to histones maximizes the probability of CBP/EP300-mediated FOXP3 acetylation in cells. Indeed our data on the regulation of FOXP3 acetylation by CPI703 lends support to this hypothesis. In sum, we propose that the precise positioning of CBP/EP300 at TSS-bound transcriptional complexes is critically aided by their bromodomains, allowing optimal H3K18 acetylation (and potentially of transcription factors, such as FOXP3), to promote gene activation at these sites.
In agreement with a recent report (41), we found a significant disruption of TH17 cytokine production by CBP/EP300 bromodomain inhibition. In addition to the well understood role for TH17 cells and associated cytokines in autoimmune and inflammatory processes, their role in tumor immunology has garnered increased interest, especially their pro-tumorigenic effects (42, 43). In that context, there could be a potential synergistic effect of CBP/EP300 bromodomain inhibition within the tumor microenvironment, by a “double hit” to Tregs and TH17 cells, thereby reversing both suppressive and pro-tumorigenic pathways. The lack of effect on IFNγ+ TH1 cells suggests that at least this anti-tumor effector subset might be refractory to CBP/EP300 inhibition, thus leaving a major cytotoxic pathway intact for tumor attack.
Collectively, our results suggest that CBP/EP300 bromodomains are capable of fine-tuning transcriptional programs across different biological pathways that converge on specific T cell states. Potent, selective, and cell-active chemical probes that disrupt CBP/EP300 bromodomain interactions with chromatin therefore constitute crucial starting points for the development of pharmacologic agents. By impairing Treg differentiation and function, such agents may reverse dampened effector responses in the context of cancer and repotentiate anti-tumor immunity, thus complementing more established cancer immunotherapies, particularly those aimed at enhancing tumor eradication by reversal of cytotoxic CD8+ T cell exhaustion through antibody-mediated checkpoint inhibition (44–46).
Author Contributions
S. G., M. C., A. S., P. J. D., D. M., G. H., D. D.-N., E. C., and T. R. designed, executed, and analyzed biological experiments. A. T., A. C., F. A. R., V. T., and T. C. designed and supervised small molecule inhibitors. A. T., F. A. R., J.-C. H., J. E. A., A. G. C., S. R. M., S. F. B., B. K. A, and R. J. S. designed and supervised drug discovery studies. H.-R. H., P. S., J. A. M., and A. R. C. designed and executed target engagement cell-based assays. H. J., F. P., and J. W. S. designed, executed, and analyzed crystallography experiments. E. P., H.-R. H., L. Z., V. G. W., R. J. S., and R. T. C. designed, executed, and analyzed biochemistry experiments. R. J. S. and R. T. C. supervised biochemistry studies. C. H. and B. M. B. conducted and analyzed bioinformatics analysis of gene transcription and ChIP-seq studies. S. G., J. L. G., and J. M. L. supervised the immunology section of the work. R. J. S. conceived the drug discovery section and wrote the chemical probe section of the manuscript. J. M. L. conceived the biology section, supervised the work, and wrote the immunology section of the manuscript. All authors discussed the results and commented on the manuscript at all stages.
Supplementary Material
A majority of the authors are employees of Constellation or Genentech, which have filed a joint patent in the area.

This article contains supplemental text, supplemental Tables S1 and S2, and supplemental Figs. S1–S5.
The atomic coordinates and structure factors (codes 4YK0 and 5DBM) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Treg
- regulatory T cell
- HAT
- histone acetyltransferase
- BRD
- bromodomain
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- ITC
- isothermal titration calorimetry
- ZsG
- ZsGreen
- GSEA
- gene set enrichment analysis
- RNA-seq
- RNA sequencing
- ChIP-seq
- deep ChIP sequencing
- TSS
- transcript start site.
References
- 1. Sakaguchi S., Ono M., Setoguchi R., Yagi H., Hori S., Fehervari Z., Shimizu J., Takahashi T., and Nomura T. (2006) Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol. Rev. 212, 8–27 [DOI] [PubMed] [Google Scholar]
- 2. Zheng Y., and Rudensky A. Y. (2007) Foxp3 in control of the regulatory T cell lineage. Nat. Immunol. 8, 457–462 [DOI] [PubMed] [Google Scholar]
- 3. Savage P. A., Malchow S., and Leventhal D. S. (2013) Basic principles of tumor-associated regulatory T cell biology. Trends Immunol. 34, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sugiyama D., Nishikawa H., Maeda Y., Nishioka M., Tanemura A., Katayama I., Ezoe S., Kanakura Y., Sato E., Fukumori Y., Karbach J., Jäger E., and Sakaguchi S. (2013) Anti-CCR4 mAb selectively depletes effector-type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc. Natl. Acad. Sci. U.S.A. 110, 17945–17950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Curiel T. J., Coukos G., Zou L., Alvarez X., Cheng P., Mottram P., Evdemon-Hogan M., Conejo-Garcia J. R., Zhang L., Burow M., Zhu Y., Wei S., Kryczek I., Daniel B., Gordon A., Myers L., Lackner A., Disis M. L., Knutson K. L., Chen L., and Zou W. (2004) Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10, 942–949 [DOI] [PubMed] [Google Scholar]
- 6. Kono K., Kawaida H., Takahashi A., Sugai H., Mimura K., Miyagawa N., Omata H., and Fujii H. (2006) CD4+CD25high regulatory T cells increase with tumor stage in patients with gastric and esophageal cancers. Cancer Immunol. Immunother. 55, 1064–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. deLeeuw R. J., Kost S. E., Kakal J. A., and Nelson B. H. (2012) The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin. Cancer Res. 18, 3022–3029 [DOI] [PubMed] [Google Scholar]
- 8. West N. R., Kost S. E., Martin S. D., Milne K., Deleeuw R. J., Nelson B. H., and Watson P. H. (2013) Tumour-infiltrating FOXP3+ lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br. J. Cancer 108, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dougan M., and Dranoff G. (2009) Immune therapy for cancer. Annu. Rev. Immunol. 27, 83–117 [DOI] [PubMed] [Google Scholar]
- 10. Mellman I., Coukos G., and Dranoff G. (2011) Cancer immunotherapy comes of age. Nature 480, 480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Curiel T. J. (2007) Tregs and rethinking cancer immunotherapy. J. Clin. Invest. 117, 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishikawa H., and Sakaguchi S. (2014) Regulatory T cells in cancer immunotherapy. Curr. Opin. Immunol. 27, 1–7 [DOI] [PubMed] [Google Scholar]
- 13. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 14. Portela A., and Esteller M. (2010) Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068 [DOI] [PubMed] [Google Scholar]
- 15. Shahbazian M. D., and Grunstein M. (2007) Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 16. Josling G. A., Selvarajah S. A., Petter M., and Duffy M. F. (2012) The role of bromodomain proteins in regulating gene expression. Genes 3, 320–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen J., Ghazawi F. M., and Li Q. (2010) Interplay of bromodomain and histone acetylation in the regulation of p300-dependent genes. Epigenetics 5, 509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J. P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Müller S., Pawson T., Gingras A. C., Arrowsmith C. H., and Knapp S. (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J., and Li Q. (2011) Life and death of transcriptional co-activator p300. Epigenetics 6, 957–961 [DOI] [PubMed] [Google Scholar]
- 20. Bedford D. C., Kasper L. H., Fukuyama T., and Brindle P. K. (2010) Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics 5, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Y., Wang L., Predina J., Han R., Beier U. H., Wang L. C., Kapoor V., Bhatti T. R., Akimova T., Singhal S., Brindle P. K., Cole P. A., Albelda S. M., and Hancock W. W. (2013) Inhibition of p300 impairs Foxp3+ T regulatory cell function and promotes antitumor immunity. Nat. Med. 19, 1173–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y., Wang L., Han R., Beier U. H., Akimova T., Bhatti T., Xiao H., Cole P. A., Brindle P. K., and Hancock W. W. (2014) Two histone/protein acetyltransferases, CBP and p300, are indispensable for Foxp3+ T-regulatory cell development and function. Mol. Cell. Biol. 34, 3993–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muller S., Filippakopoulos P., and Knapp S. (2011) Bromodomains as therapeutic targets. Exp. Rev. Mol. Med. 13, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taylor A. M., Côté A., Hewitt M. C., Pastor R., Leblanc Y., Nasveschuk C. G., Romero F. A., Crawford T. D., Cantone N., Jayaram H., Setser J., Murray J., Beresini M. H., de Leon Boenig G., Chen Z., Conery A. R., Cummings R. T., Dakin L. A., Flynn E. M., Huang O. W., Kaufman S., Keller P. J., Kiefer J. R., Lai T., Li Y., Liao J., Liu W., Pardo E., Lu H., Tsui V., Wang J., Wang Y., Xu Z., Yan F., Yu D., Zawadzke L., Zhu X., Zhu X., Sims R. J. 3rd, Cochran A. G., Bellon S., Audia J. E., Magnuson S., and Albrecht B. K. (2016) Fragment-based discovery of a selective and cell-active benzodiazepinone CBP/EP300 bromodomain inhibitor (CPI-637). ACS Med. Chem. Lett., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mertz J. A., Conery A. R., Bryant B. M., Sandy P., Balasubramanian S., Mele D. A., Bergeron L., and Sims R. J. 3rd (2011) Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U.S.A. 108, 16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ciceri P., Müller S., O'Mahony A., Fedorov O., Filippakopoulos P., Hunt J. P., Lasater E. A., Pallares G., Picaud S., Wells C., Martin S., Wodicka L. M., Shah N. P., Treiber D. K., and Knapp S. (2014) Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat. Chem. Biol. 10, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang H. R., Sims R. J., and Bellon S. F. (2014) Fusion proteins and methods for identifying bromodomain inhibiting compounds. WIPO Patent WO2014144303
- 28. Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y., and Ge K. (2011) Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manning E. T., Ikehara T., Ito T., Kadonaga J. T., and Kraus W. L. (2001) p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21, 3876–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rothbart S. B., Lin S., Britton L. M., Krajewski K., Keogh M. C., Garcia B. A., and Strahl B. D. (2012) Poly-acetylated chromatin signatures are preferred epitopes for site-specific histone H4 acetyl antibodies. Sci. Rep. 2, 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rudensky A. Y. (2011) Regulatory T cells and Foxp3. Immunol. Rev. 241, 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Conery A. R., Centore R. C., Neiss A., Keller P. J., Joshi S., Spillane K. L., Sandy P., Hatton C., Pardo E., Zawadzke L., Bommi-Reddy A., Gascoigne K. E., Bryant B. M., Mertz J. A., and Sims R. J. (2016) Bromodomain inhibition of the transcriptional coactivators CBP/EP300 as a therapeutic strategy to target the IRF4 network in multiple myeloma. eLife 5, e10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Picaud S., Fedorov O., Thanasopoulou A., Leonards K., Jones K., Meier J., Olzscha H., Monteiro O., Martin S., Philpott M., Tumber A., Filippakopoulos P., Yapp C., Wells C., Che K. H., Bannister A., Robson S., Kumar U., Parr N., Lee K., Lugo D., Jeffrey P., Taylor S., Vecellio M. L., Bountra C., Brennan P. E., O'Mahony A., Velichko S., Müller S., Hay D., Daniels D. L., Urh M., La Thangue N. B., Kouzarides T., Prinjha R., Schwaller J., and Knapp S. (2015) Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for leukemia therapy. Cancer Res. 75, 5106–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Loosdregt J., Vercoulen Y., Guichelaar T., Gent Y. Y., Beekman J. M., van Beekum O., Brenkman A. B., Hijnen D. J., Mutis T., Kalkhoven E., Prakken B. J., and Coffer P. J. (2010) Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood 115, 965–974 [DOI] [PubMed] [Google Scholar]
- 35. Goodman R. H., and Smolik S. (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev. 14, 1553–1577 [PubMed] [Google Scholar]
- 36. Jain N., Nguyen H., Chambers C., and Kang J. (2010) Dual function of CTLA-4 in regulatory T cells and conventional T cells to prevent multiorgan autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 107, 1524–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang C. T., Workman C. J., Flies D., Pan X., Marson A. L., Zhou G., Hipkiss E. L., Ravi S., Kowalski J., Levitsky H. I., Powell J. D., Pardoll D. M., Drake C. G., and Vignali D. A. (2004) Role of LAG-3 in regulatory T cells. Immunity 21, 503–513 [DOI] [PubMed] [Google Scholar]
- 38. Anderson A. C. (2014) Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol. Res. 2, 393–398 [DOI] [PubMed] [Google Scholar]
- 39. Cai G., Anumanthan A., Brown J. A., Greenfield E. A., Zhu B., and Freeman G. J. (2008) CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat. Immunol. 9, 176–185 [DOI] [PubMed] [Google Scholar]
- 40. Cai G., and Freeman G. J. (2009) The CD160, BTLA, LIGHT/HVEM pathway: a bidirectional switch regulating T-cell activation. Immunol. Rev. 229, 244–258 [DOI] [PubMed] [Google Scholar]
- 41. Hammitzsch A., Tallant C., Fedorov O., O'Mahony A., Brennan P. E., Hay D. A., Martinez F. O., Al-Mossawi M. H., de Wit J., Vecellio M., Wells C., Wordsworth P., Müller S., Knapp S., and Bowness P. (2015) CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc. Natl. Acad. Sci. U.S.A. 112, 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zou W., and Restifo N. P. (2010) T(H)17 cells in tumour immunity and immunotherapy. Nat. Rev. Immunol. 10, 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coffelt S. B., Kersten K., Doornebal C. W., Weiden J., Vrijland K., Hau C. S., Verstegen N. J., Ciampricotti M., Hawinkels L. J., Jonkers J., and de Visser K. E. (2015) IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brahmer J. R., Tykodi S. S., Chow L. Q., Hwu W. J., Topalian S. L., Hwu P., Drake C. G., Camacho L. H., Kauh J., Odunsi K., Pitot H. C., Hamid O., Bhatia S., Martins R., Eaton K., Chen S., Salay T. M., Alaparthy S., Grosso J. F., Korman A. J., Parker S. M., Agrawal S., Goldberg S. M., Pardoll D. M., Gupta A., and Wigginton J. M. (2012) Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Topalian S. L., Hodi F. S., Brahmer J. R., Gettinger S. N., Smith D. C., McDermott D. F., Powderly J. D., Carvajal R. D., Sosman J. A., Atkins M. B., Leming P. D., Spigel D. R., Antonia S. J., Horn L., Drake C. G., Pardoll D. M., Chen L., Sharfman W. H., Anders R. A., Taube J. M., McMiller T. L., Xu H., Korman A. J., Jure-Kunkel M., Agrawal S., McDonald D., Kollia G. D., Gupta A., Wigginton J. M., and Sznol M. (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hodi F. S., O'Day S. J., McDermott D. F., Weber R. W., Sosman J. A., Haanen J. B., Gonzalez R., Robert C., Schadendorf D., Hassel J. C., Akerley W., van den Eertwegh A. J., Lutzky J., Lorigan P., Vaubel J. M., Linette G. P., Hogg D., Ottensmeier C. H., Lebbé C., Peschel C., Quirt I., Clark J. I., Wolchok J. D., Weber J. S., Tian J., Yellin M. J., Nichol G. M., Hoos A., and Urba W. J. (2010) Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.