Abstract
Allergy to the short ragweed (Ambrosia artemisiifolia) pollen is a major health problem. The ragweed allergen repertoire has been recently expanded with the identification of Amb a 11, a new major allergen belonging to the cysteine protease family. To better characterize Amb a 11, a recombinant proform of the molecule with a preserved active site was produced in Escherichia coli, refolded, and processed in vitro into a mature enzyme. The enzymatic activity is revealed by maturation following an autocatalytic processing resulting in the cleavage of both N- and C-terminal propeptides. The 2.05-Å resolution crystal structure of pro-Amb a 11 shows an overall typical C1A cysteine protease fold with a network of molecular interactions between the N-terminal propeptide and the catalytic triad of the enzyme. The allergenicity of Amb a 11 was confirmed in a murine sensitization model, resulting in airway inflammation, production of serum IgEs, and induction of Th2 immune responses. Of note, inflammatory responses were higher with the mature form, demonstrating that the cysteine protease activity critically contributes to the allergenicity of the molecule. Collectively, our results clearly demonstrate that Amb a 11 is a bona fide cysteine protease exhibiting a strong allergenicity. As such, it should be considered as an important molecule for diagnosis and immunotherapy of ragweed pollen allergy.
Keywords: allergen, cysteine protease, immunotherapy, protein processing, structure-function, ragweed
Introduction
Exposure to pollen from the short ragweed (Ambrosia artemisiifolia) is a major cause of severe type I allergy. The prevalence of IgE sensitization is on the rise (1–3) due to the continuous spreading of the plant, originally native to North America, within several European countries (4, 5). As of today, 10 short ragweed pollen allergens have been registered by the International Union of Immunological Societies (6). Among them, two are considered as major allergens, including the pectate lyase Amb a 1 and the cysteine protease Amb a 11, which are recognized by serum IgE of 90% (7, 8) and 66% (9), respectively, of ragweed pollen-allergic patients. We recently identified natural Amb a 11 as a 28-kDa glycosylated protein displaying amino acid sequence identities with other allergens belonging to the C1A cysteine protease family, such as Der p 1 and Der f 1 (from house dust mite Dermatophagoides sp.; 24.4 and 26.0% identity, respectively), the actinidin Act d 1 (from Actinidia deliciosa; 37% identity), and papain (from Carica papaya; 34.6% identity) molecules (9). The Amb a 11 gene (GenBankTM accession number KF528831) codes for a precursor molecule of 364 residues encompassing both N- and C-terminal propeptides and preceded by a signal sequence of 22 amino acids (UniProt accession V5LU01_AMBAR) (9).
To better understand the biochemical and structural basis for Amb a 11 allergenicity, we produced in the present study a recombinant proform of Amb a 11 (pro-rAmb a 11)4 in Escherichia coli and determined its crystal structure. Herein, we demonstrate that Amb a 11 is a bona fide cysteine protease and that it triggers a strong allergic inflammation in a murine sensitization model. As such, we confirm the importance of this molecule in ragweed pollen allergy.
Experimental Procedures
Recombinant Protein Expression in E. coli
The cDNA encoding pro-rAmb a 11 preceded by a N-terminal 21-residue-long His tag (MGSSHHHHHHSSGLVPRGSHM) was cloned in the pET-15b vector (Merck Millipore, Molsheim, France), and the resulting plasmid was used to transform BL21(DE3) E. coli (Agilent, Les Ulis, France). Expression and pellet lysis were done as described previously (10). Inclusion bodies were washed in lysis buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm EDTA) and then solubilized in 50 mm Tris-HCl, pH 8.0, 8 m urea at 4 °C overnight. The soluble fraction was obtained after a 1:2 dilution (v/v) in a buffer containing 50 mm Tris-HCl, pH 8.0, 1 m NaCl, 8 m urea, 40 mm imidazole and centrifugation at 1,500 × g for 30 min. For the purification and refolding steps according to Choudhury et al. (11), pro-rAmb a 11 was purified by immobilized metal affinity chromatography on a HisTrap HP column (GE Healthcare) under denaturing conditions (50 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 20 mm imidazole, 7 m urea) and then eluted with an 0.02–0.5 m imidazole gradient. Eluted fractions containing pro-rAmb a 11 were pooled and incubated with 10 mm DTT at 37 °C for 1 h. The protein solution was diluted to a 50 μg/ml maximum concentration in a refolding buffer (50 mm Tris-HCl, pH 8.5, 0.5 m NaCl, 5 mm EDTA, 1 mm GSH, 0.1 mm GSSG, 0.5 m arginine, 10% glycerol) and stirred at 4 °C overnight. Samples were then concentrated and dialyzed against PBS (Life Technologies). For the sake of clarity, sequence numbering is done using natural Amb a 11 as the reference. The numbering of the recombinant protein used in the study thus starts at +2. The cDNA encoding the pro-Amb a 11 ΔCT variant, lacking the C-terminal Lys-371 to Leu-386 propeptide, was cloned by PCR using PfuUltraII polymerase (Agilent) and the forward 5′-AGACATATGTTCCATTACCATGAGAGAGAGCTCGAA-3′ and reverse 5′-AGACTCGAGTCAAAATTTGGGGTCCTTGGGTGC-3′ primers. A C155S active site variant (pro-rAmb a 11 C155S) was obtained by site-directed mutagenesis performed with the QuikChange II site-directed mutagenesis kit (Agilent) according to the manufacturer's instructions using the forward 5′-GGATGTGGAAGTAGTTGGGCGTTTGCCGC-3′ and the reverse 5′-GCGGCAAACGCCCAACTACTTCCACATCC-3′ primers. Nucleotidic sequences of all constructs were confirmed by sequencing (Beckman Coulter Genomics, Takeley, UK). The two variants were expressed and purified as described above for pro-rAmb a 11 molecules.
The cDNAs encoding the Amb a 11 and Der p 1 N-terminal propeptides were cloned in the pET-15b and pET-24a(+) vectors, respectively. Expression in E. coli and pellet lysis were done as described above. Both soluble propeptides were purified by immobilized metal affinity chromatography on a HisTrap FF crude column (GE Healthcare) and eluted with an imidazole gradient. Fractions containing Amb a 11 or Der p 1 propeptides were pooled, dialyzed against PBS, and concentrated.
In Vitro Activation of Recombinant Pro-rAmb a 11 and Characterization by Mass Spectrometry
For in vitro activation (i.e. the maturation process revealing the cysteine protease activity), wild-type and pro-rAmb a 11 variant molecules were dialyzed overnight against 20 mm sodium acetate, pH 5.0, and then incubated at 40 °C for 2 h. To remove residual pro-rAmb a 11 and cleaved propeptides, samples were first subjected to immobilized metal affinity chromatography using a HisTrap FF crude column previously equilibrated in PBS and eluted with imidazole. Fractions containing mature rAmb a 11 were then pooled and applied onto a Superdex 75 10/300 GL column (GE Healthcare) equilibrated with 20 mm sodium acetate, pH 5.0. Purified mature recombinant Amb a 11 (termed rAmb a 11) was then dialyzed against PBS. Maturation cleavage sites were characterized by MALDI-TOF MS. To this end, 1 μg of rAmb a 11 was purified using ZipTip C4 pipette tips (Merck Millipore), eluted with 2 μl of a 70% acetonitrile, 0.1% trifluoroacetic acid (TFA) solution, and mixed with 2 μl of 2,5-dihydroxyacetophenone matrix solution (Bruker, Bremen, Germany) prepared according to the manufacturer's instructions. Subsequently, 2 μl of this mixture were spotted on an AnchorChip MALDI target and dried in ambient air. Spectra were acquired on an Autoflex Speed mass spectrometer (Bruker) in a linear positive mode with a method optimized for high resolution in the 5–20-kDa m/z range using the LP 5–20-kDa parameter file. External quadratic calibration was performed prior to acquisition using Bruker's Protein Calibration Standard 1.
Protease Activity Measurement
The rAmb a 11 molecule was first preincubated with 20 mm l-cysteine (Sigma). The protease activity was measured with an Enzcheck protease assay kit (Life Technologies) using green fluorescent casein-Bodipy FL as a substrate according to the supplier's instructions. For optimum pH determination, casein or a Boc-Val-Leu-Lys-AMC (7-amino-4-methylcoumarin) synthetic peptide (Bachem, Bubendorf, Switzerland) was used as a substrate to measure protease activity in a 50 mm polybuffer (mixture of 50 mm citrate, phosphate, potassium chloride, sodium acetate, Tris buffers) with pH values adjusted from 2.0 to 12.0. E64 (10 μm), leupeptin (100 μm), AEBSF (1 mm), PMSF (1 mm), bestatin (10 μm), and pepstatin A (1 μm) (all purchased from Sigma) were used as protease inhibitors in those experiments. For inhibition assays using propeptides, rAmb a 11 or nDer p 1 was incubated with either a recombinant N-terminal propeptide of Amb a 11, a synthetic C-terminal propeptide of Amb a 11 (Proteogenix, Schiltigheim, France), or a recombinant N-terminal propeptide of Der p 1 at a molar ratio of 1:1. Fluorescence intensities were monitored in 96-well plates using a FluoroMax-4 apparatus (Horiba, Les Ulis, France) with excitation and emission wavelengths set at 485/530 nm and 380/450 nm for casein-Bodipy FL and Boc-Val-Leu-Lys-AMC, respectively.
Secondary Structure Analysis of rAmb a 11
To obtain the synchrotron radiation circular dichroism (SRCD) spectra, pro-rAmb a 11, pro-rAmb a 11 ΔCT, and rAmb a 11 were dialyzed against 10 mm Tris-H2SO4, pH 8.5, 150 mm Na2SO4. Measurements were carried out on the DISCO beamline at the SOLEIL synchrotron (Gif-sur-Yvette, France). Four spectral scans were recorded in the 170–265-nm range with 1-nm steps after subtracting signals obtained with the dialysis buffer. The resulting spectra were calibrated with d-10-camphorsulfonic acid using the CDtool software (12) and normalized to protein concentrations. Ratios of secondary structures were determined using the Selcon program (13) in Dichroweb (14). For all samples, spectra were analyzed down to 180 nm, corresponding to a photomultiplier high tension below half of its total variation.
Crystallization and Structure Determination of Pro-rAmb a 11
Crystals were grown at 20 °C using the vapor diffusion method. A first series of crystals was obtained after dialysis of purified pro-rAmb a 11 in 20 mm Tris-HCl, pH 8.5, 150 mm NaCl, 5% (v/v) glycerol. Crystallization conditions were screened using the NeXtal PEGs II Suite (Qiagen, Courtaboeuf, France). A sitting drop containing 0.5 μl of a 10 mg/ml pro-rAmb a 11 solution mixed with an equal volume of a precipitant solution containing 100 mm trisodium citrate, 20% (w/v) polyethylene glycol (PEG) 4000, 20% isopropanol yielded a medium resolution-diffracting crystal of 0.16-mm length after 1 week. A second series of crystals was generated using pro-rAmb a 11 further purified by size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare). The hanging drop contained 3 μl of an 8.6 mg/ml pro-rAmb a 11 solution mixed with 3 μl of a precipitant solution containing 100 mm trisodium citrate, 24% PEG 4000, 30% isopropanol. A large crystal of 1.5-mm length obtained after 3 weeks under those conditions was used to establish a high resolution data set. Prior to data collection, crystals were soaked in a 20% (v/v) glycerol cryoprotectant solution and frozen in liquid nitrogen. Diffraction data were collected at 100 K on the PROXIMA 2A beamline at the SOLEIL synchrotron and then processed and scaled using the XDS program (15). Crystal structures were determined by performing molecular replacements with Phaser (16) and using either the monomer of the vignain from Ricinus communis (Protein Data Bank code 1S4V) (17) or the propapain mutant from Carica papaya (Protein Data Bank code 3TNX) (18) as search models for the medium and high resolution data sets, respectively. Both pro-rAmb a 11 models were refined with non-crystallographic symmetry restraints and TLS (translation/libration/screw motion) using Buster 2.10 (19). Electron density maps were evaluated using the Coot model building software (20). The resulting medium resolution model contains residues Met-22 to Lys-352 and Pro-355 to Leu-386 in monomer A and residues His-21 to Ser-114 and Asn-120 to Pro-350 in monomer B. The high resolution model contains residues Ser-5 to Ser-12, His-21 to Ser-114, and Lys-121 to Asn-346 for monomer A and residues His-21 to Ile-115 and Asp-122 to Pro-348 for monomer B. Molecular graphic images were generated using the PyMOL software, version 1.7.6. The Protein Data Bank codes for the medium and high resolution models are 5EGW and 5EF4, respectively.
Mice, Culture Media, Reagents, and Antibodies
BALB/c female mice (6–8 weeks old) were obtained from Charles River (L'Arbresle, France). The experimental protocol was approved by Stallergenes' ethics committee, and animal handling was performed according to international regulations. Dulbecco's PBS and Roswell Park Memorial Institute (RPMI) 1640 medium were purchased from Life Technologies. For mouse in vitro experiments, the culture medium consisted of RPMI 1640 medium supplemented with 10% fetal calf serum, 1% l-glutamine, 200 units/ml penicillin, 200 μg/ml streptomycin (all from Life Technologies). Endotoxin removal from pro-rAmb a 11 was performed with Pierce high capacity endotoxin removal columns (Thermo Fischer Scientific, Waltham, MA) according to the manufacturer's instructions. Residual endotoxin concentrations determined with the LAL dosage kit (Thermo Fischer Scientific) were below 0.1 endotoxin unit/μg of protein. To assess inflammatory cells in bronchoalveolar lavages (BALs), the following monoclonal antibodies (mAbs) labeled with either fluorescein isothiocyanate (FITC), phycoerythrin, allophycocyanin, or phycoerythrin-allophycocyanin were used: anti-Gr1 (clone RB6-8C5) mAb from BD Biosciences; anti-B220 (clone RA3-6B2), anti-CD3 (clone 145-2C11), anti-CD4 (clone GK1.5), anti-CD8 (clone 53-6.7), anti-CD11b (clone M1/70), anti-CD11c (clone N418), anti-CD19 (clone 1D3), anti-FcϵRI (clone MAR-1), and anti-ICOS (clone C398.4A) mAbs from eBiosciences (San Diego, CA); anti-T1/ST2 (clone DJ8) mAb from MD Bioscience (St. Paul, MN); and anti-CCR3 (clone 83101) mAb from R&D Systems (Lille, France). Corresponding isotype-matched mAbs were used as controls.
Measurements of Airway Inflammation and Immune Responses in Amb a 11-sensitized Mice
For sensitization, mice were immunized intraperitoneally (i.p.) on days 0 and 14 with 10 μg of either pro-rAmb a 11 or rAmb a 11 activated with 20 mm l-cysteine (Sigma), subsequently inhibited or not with 10 μm E64, adsorbed on 2 mg of aluminum hydroxide (Life Technologies), and administered in 100 μl of Dulbecco's PBS. From days 21 to 24, a daily 20-min aerosol challenge was performed with a short ragweed pollen extract (with a dose equivalent to 10 mg of Amb a 1) and 100 μg of LPS (Invivogen, Toulouse, France). Measurements of airway hyperresponsiveness (AHR) were performed by whole body plethysmography (Buxco, Winchester, UK) after exposure to increasing doses (i.e. 6.125, 12.5, 25, and 50 mg/ml) of methacholine (Sigma). Results were expressed as enhanced pause indexes as described previously (21).
To assess inflammatory cells in BALs, mice were anesthetized with an i.p. administration of 50 mg/kg pentobarbital plus 10 mg/kg xylazine (Centravet, Maisons-Alfort, France). BALs were performed with 3 × 400 μl of Dulbecco's PBS, and samples were centrifuged at 500 × g at 4 °C for 5 min. Cells were resuspended in Dulbecco's PBS and stained at 4 °C for 15 min with specific mAbs as mentioned above. Acquisitions and analyses were performed with a FACS Verse apparatus (BD Biosciences) and FlowJo software (Ashland, OR), respectively. Eosinophils were measured in BALs as percentages of CCR3+ CD3− B220− cells among other immune cells, including neutrophils (defined as Gr1high CD3− CD19−cells), lymphocytes (CD3+ or CD19+ cells), and alveolar macrophages (large autofluorescent cells). Type 2 innate lymphoid cells (ILC2s) were identified as side scatter (SSC) low, lineage-negative (Lin−: CD3− CD4− CD8− CD11b− CD11c− CD19− FcϵRI−) cells expressing ICOS and T1/ST2 (SSClow Lin− ICOS+ T1/ST2+).
Allergen-specific IgE and IgG1 antibody responses were assessed in sera obtained from blood samples (collected by cardiac puncture under anesthesia) after centrifugation at 10,000 × g for 10 min. Specific antibody responses were analyzed by ELISA after overnight incubation of serum samples at 4 °C in 96-well plates previously coated overnight with pro-rAmb a 11 at 10 μg/ml. Plates were washed and revealed with anti-mouse IgE (Gentaur, Kampenhout, Belgium) or IgG1 (BD Biosciences) HRP-labeled antibodies. After further washes, specific antibody binding was detected at 405 nm on a Multiskan Ascent spectrophotometer (Thermo Labsystems, Waltham, MA) using 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (Roche Applied Science) as a substrate.
Lungs and spleens were harvested, and single cell preparations were obtained to assess T cell responses. To this aim, one lobe obtained from a lung was incubated for 1 h in RPMI 1640 medium supplemented with 75 units/ml collagenase. After blocking residual enzymatic activity with 5 mm EDTA in PBS, lung tissues were dissociated in PBS. Both spleen and lung cells were filtered through a 70-μm sieve, washed twice in PBS before resuspension in culture medium, and plated at 106 cells/well in 96-well plates. Cells were stimulated with pro-rAmb a 11 (10 μg/ml) or culture medium alone. After 72 h at 37 °C in 5% CO2, IL-5 and IL-13 levels were measured in culture supernatants using a MagPix Multiplex assay according to the manufacturer's instructions (Merck Millipore).
Statistical Analyses
Statistical analyses were performed using Prism 6 software (GraphPad, La Jolla, CA). Results are expressed as means ± S.E. Statistical differences between groups were assessed by using the non-parametric Kruskal-Wallis test with subsequent Dunnett's multiple analyses when comparing treated mice with naive mice. Statistical analyses of data on the inhibition of protease activity by E64 were done using the non-parametric Mann-Whitney t test. p values less than 0.05 were considered as significant: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Results
Maturation of Amb a 11 Is Autocatalytic and Independent of the C-terminal Propeptide
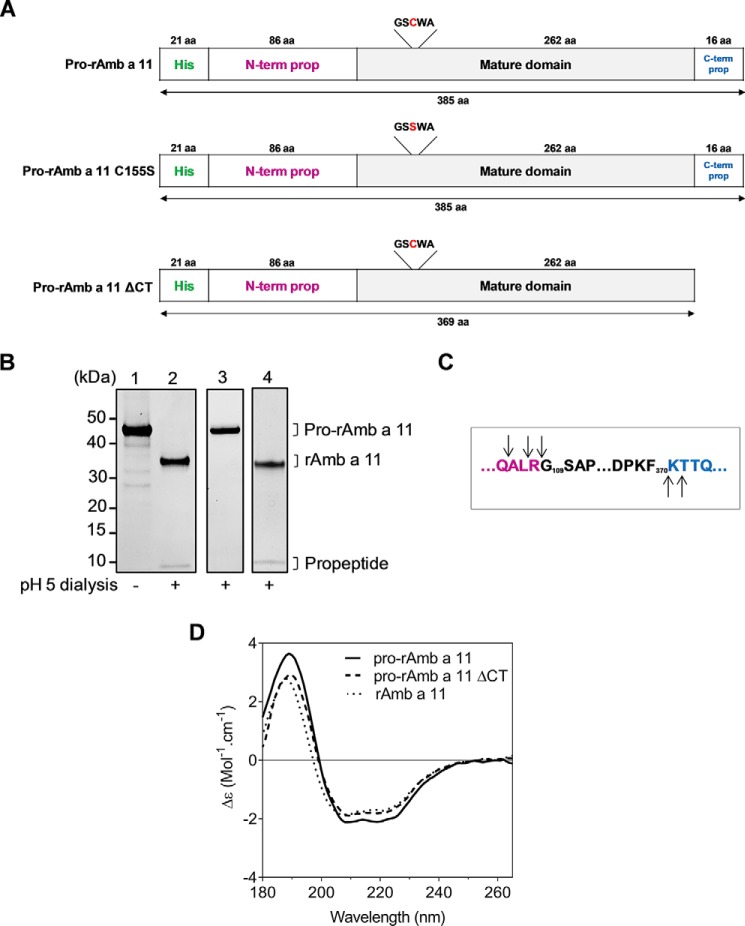
A His-tagged pro-rAmb a 11 molecule, encompassing both the N- and C-terminal propeptides (Fig. 1A, upper panel), was expressed in E. coli and refolded from inclusions bodies as described under “Experimental Procedures”. Pro-rAmb a 11 was converted into rAmb a 11 at pH 5.0 with a shift in the apparent molecular mass from 45 to 35 kDa detected by SDS-PAGE (Fig. 1B, lanes 1 and 2). Cleavage of both propeptides was confirmed by mass spectrometry analysis with up to three additional amino acid residues observed in N or C termini when compared with the natural molecule nAmb a 11 (Fig. 1C). We subsequently compared the in vitro activation of either wild-type pro-rAmb a 11, pro-rAmb a 11 C155S (with a mutated Cys-155 catalytic residue to inactivate the enzyme), or pro-rAmb a 11 ΔCT (with a deletion of the C-terminal propeptide) molecules (Fig. 1A). Mutation of cysteine Cys-155 to a serine completely abolished the in vitro maturation (Fig. 1B, lane 3), indicating that the latter relies upon an autocatalytic mechanism. In contrast, the maturation of pro-rAmb a 11 ΔCT at acidic pH was unaffected (Fig. 1B, lane 4), establishing that the C-terminal propeptide has no major role in the processing of the proform.
FIGURE 1.
In vitro maturation of pro-Amb a 11. A, schematic representation of pro-rAmb a 11, pro-rAmb a 11 C155S, and pro-rAmb a 11 ΔCT molecules with His tag shown in green, N-terminal propeptide (N-term prop) in magenta, mature domain in black, and C-terminal propeptide (C-term prop) in blue. aa, amino acids. B, SDS-PAGE analysis of pro-rAmb a 11 before (lane 1) and after activation at pH 5.0 (lane 2), pro-rAmb a 11 C155S (lane 3), and pro-rAmb a 11 ΔCT (lane 4) after dialysis at pH 5.0. C, MALDI-TOF MS analysis of cleavage sites after maturation of pro-rAmb a 11. Cleavage sites are indicated with arrows. D, SRCD spectra of pro-rAmb a 11, pro-rAmb a 11 ΔCT, and rAmb a 11.
Secondary structures of pro-rAmb a 11, pro-rAmb a 11 ΔCT, and rAmb a 11 were compared by SRCD. As shown in Fig. 1D, SRCD spectra of these various forms of Amb a 11 are similar irrespective of the presence of the N- and C-terminal propeptides, thus indicating that the overall fold of Amb a 11 does not change significantly following maturation. For instance, the α-helical contents estimated for all forms were always between 27 and 29%.
The Cysteine Protease Activity of Amb a 11 Is Specifically Inhibited by Its N-terminal Propeptide
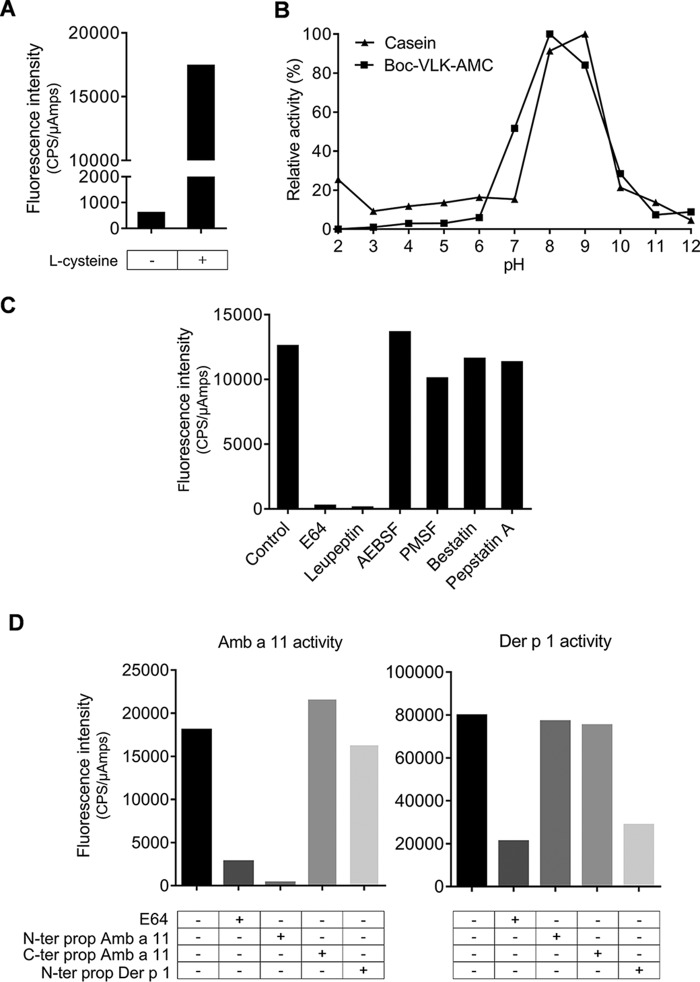
We confirmed that rAmb a 11 exhibits a strong protease activity enhanced by l-cysteine (Fig. 2A), mainly detectable between pH 8.0 and 9.0 (Fig. 2B). This enzymatic activity is inhibited by E64 as well as leupeptin (acting as cysteine and cysteine/serine protease inhibitors, respectively) but not by other class-specific inhibitors (Fig. 2C). We also investigated whether the N- and C-terminal propeptides of Amb a 11 as well as the N-terminal propeptide of Der p 1 were able to inhibit the enzymatic activity of rAmb a 11. A clear inhibition of the protease activity mediated by rAmb a 11 was only observed in the presence of its N-terminal propeptide (Fig. 2D, left panel). Similarly, the nDer p 1 protease activity was specifically inhibited by its own N-terminal propeptide but not by that from Amb a 11 (Fig. 2D, right panel). Taken together, these observations demonstrate a strong specificity in the inhibition of the enzymatic activity of allergenic cysteine proteases such as Amb a 11 and Der p 1 by their respective N-terminal propeptides.
FIGURE 2.
Protease activity of Amb a 11. A, protease activity of rAmb a 11 was monitored with (+) and without (−) l-cysteine preactivation. B, relative activity profiles under various pH conditions using casein (▴) or Boc-VLK-AMC (■) substrates. C, protease activities were measured in the presence of different protease inhibitors specific for either cysteine (E64), cysteine and serine (leupeptin), serine (AEBSF and PMSF), metallo- (bestatin), and aspartic (pepstatin A) proteases. D, protease activities of rAmb a 11 (left panel) and nDer p 1 (right panel) in the presence of either N- (N-ter prop) or C-terminal propeptides (C-ter prop) of Amb a 11 or the N-terminal propeptide of Der p 1. Results are expressed as counts per second/microamperes (CPS/μAmps).
The Crystal Structure of Pro-Amb a 11 Is Typical of C1A Cysteine Proteases
Pro-rAmb a 11 was crystallized in orthorhombic P212121 and trigonal P3121 space groups, leading to 2.70-Å medium and 2.05-Å high resolution structures, respectively. Data collection and refinement statistics are presented in Table 1. Two pro-rAmb a 11 molecules forming non-crystallographic dimers were observed in the asymmetric unit of both crystal structures (Fig. 3, A and B). Each monomer has an ellipsoidal shape with approximate dimensions of 60 × 45 × 30 Å. The N-terminal propeptide (up to Arg-108) contains five α-helices and is in close contact with the mature domain (starting at Gly-109), which comprises six α-helices and a β-sheet formed from six antiparallel β-strands. The N-terminal part of the longest helix (residues Cys-155 to Thr-172) contains the catalytic cysteine (Cys-155) in the vicinity of two additional residues forming the canonical catalytic triad of C1A cysteine proteases including both His-289 with 5.7 Å between α-carbons and Asn-310 with 8.6 Å between α-carbons (Fig. 3B).
TABLE 1.
Data collection and refinement statistics
| Medium resolution data set | High resolution data set | |
|---|---|---|
| Protein Data Bank code | 5EGW | 5EF4 |
| Space group | P212121 | P3121 |
| Asymmetric unit | 1 dimer | 1 dimer |
| Unit cell (Å) | ||
| a | 83.5 | 117.7 |
| b | 89.6 | 117.7 |
| c | 104.1 | 101.7 |
| α = β (°) | 90.0 | 90.0 |
| γ (°) | 90.0 | 120.0 |
| Resolution (Å) | 44.8–2.70 | 41.0–2.05 |
| Observed reflections | 162,322 (16,378)a | 502,584 (48,875) |
| Unique reflections | 21,824 (3,033) | 51,287 (7,310) |
| Completeness (%) | 99.0 (93.7) | 99.8 (99.1) |
| I/σ(I) | 12.90 (1.25) | 10.7 (2.08) |
| CC1/2b | 99.8 (64.8) | 99.6 (60.8) |
| Rsym (%) | 11.3 (99.0) | 18.7 (117.6) |
| Rcryst (%) | 17.9 | 17.0 |
| Rfree (%) | 21.8 | 19.9 |
| r.m.s.d. bond lengths (Å) | 0.010 | 0.010 |
| r.m.s.d. bond angles (°) | 1.23 | 1.00 |
| B average value (protein; Å2) | 76.7 | 30.6 |
a Numbers in parentheses represent values in the highest of the 10 resolution shells: 2.65–2.77 Å (medium resolution data set) or 2.05–2.16 Å (high resolution data set).
b CC1/2 stands for a percentage of correlation between intensities from random half-data set. This parameter is better suited than Rsym for determining the high resolution limit (55).
FIGURE 3.
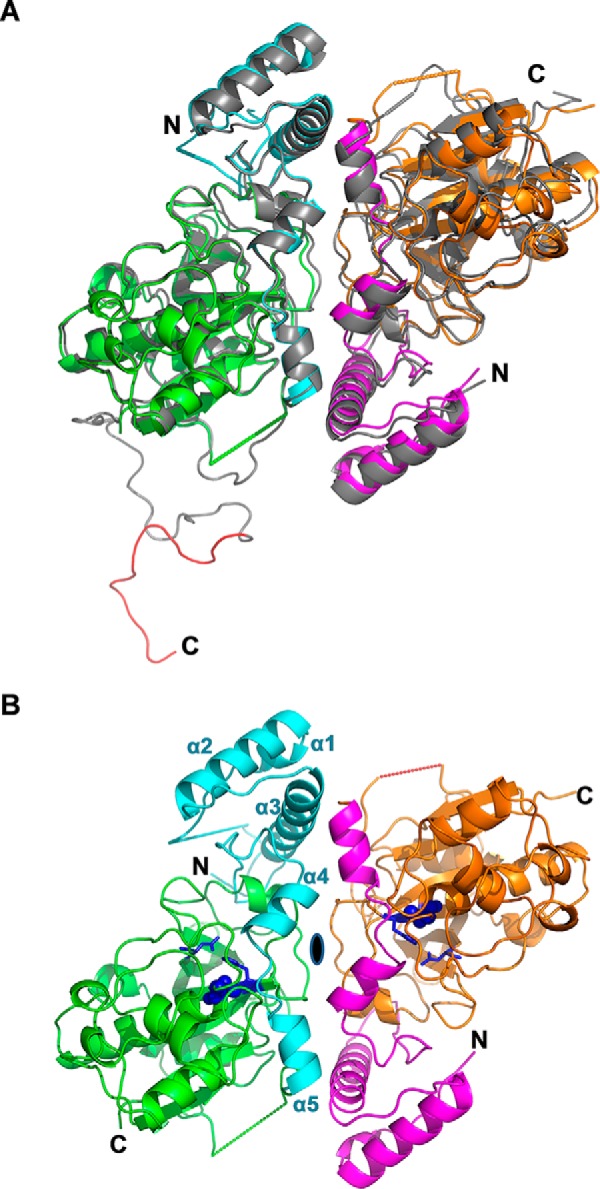
Overall fold of pro-Amb a 11. A, superposition of the pro-rAmb a 11 high resolution and medium resolution structures (ribbon representations) viewed along the 2-fold non-crystallographic symmetry axis. In the high resolution structure, monomer A (left) and monomer B (right) are shown in green and orange with the N-terminal propeptide in cyan and magenta, respectively. Dotted lines connect residues that delineate a segment for which no clear density was observed. The medium resolution structure is shown in gray with N and C termini labeled. The C-terminal propeptide (monomer A, medium resolution) is red. B, high resolution structure of the pro-rAmb a 11 dimer. The 2-fold non-crystallographic symmetry axis is indicated as a black ellipsis. The catalytic triad is labeled in purple-blue with the Cys-155 shown in spheres and His-289 and Asn-310 in sticks. The N and C termini of the protein and helices α1–α5 of the N-terminal propeptide are labeled.
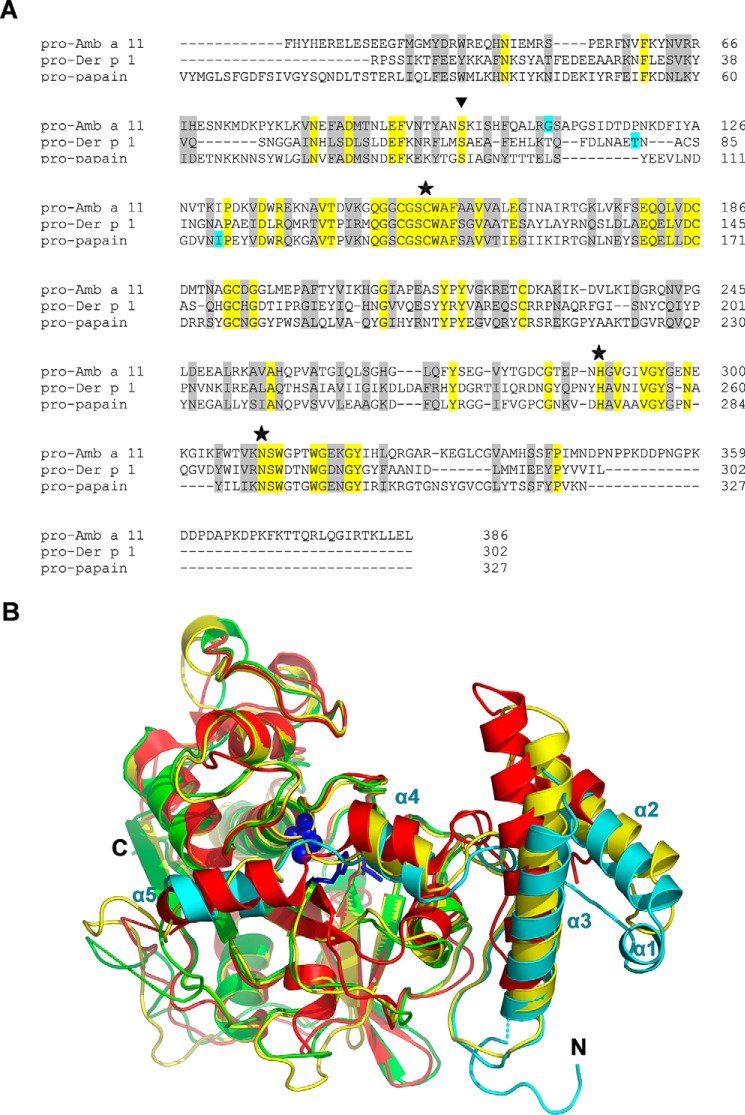
Alignment of primary sequences of Amb a 11, Der p 1, and papain cysteine protease zymogens reveals conserved amino acid residues in the mature domain as well as, to a lesser extent, in the N-terminal propeptides (Fig. 4A) (22). When crystal structures of pro-rAmb a 11, pro-Der p 1 (Protein Data Bank code 1XKG) (23), and propapain (Protein Data Bank code 3TNX) (18) were superposed (Fig. 4B), only minor differences were observed in the N-terminal propeptide regions. The calculated root mean square deviation (r.m.s.d.) between pro-rAmb a 11 and pro-Der p 1 propeptides was 0.46 (73 residues, 99 atoms) and 0.74 Å (96 residues, 408 atoms) between pro-rAmb a 11 and propapain propeptides. Furthermore, the structure of the mature domain of pro-rAmb a 11 is very similar to those of Der p 1 (Protein Data Bank code 2AS8; r.m.s.d., 1.20 Å; 222 residues; 857 atoms) (24), papain (Protein Data Bank code 9PAP; r.m.s.d., 0.68 Å; 232 residues; 1,100 atoms) (25), and vignain (Protein Data Bank code 1S4V; r.m.s.d., 0.52 Å; 224 residues; 1,195 atoms) (17) (data not shown) in accordance with our previous modeling data (9). Altogether, these results indicate that Amb a 11 has a typical C1A cysteine protease fold.
FIGURE 4.
Sequence and structure comparison of pro-Amb a 11 and other allergenic cysteine proteases. A, sequence alignment of pro-Amb a 11 (V5LU01), pro-Der p 1 (P08176), and propapain (P00784) with Clustal Omega. Conserved residues are labeled in yellow, and semi-conserved residues are in gray. The first amino acid of the different mature forms is highlighted in blue. Each of the residues of the catalytic triad (i.e. Cys-155, His-289, and Asn-310) is identified with a star. A triangle indicates the Ser-99 residue of the N-terminal Amb a 11 propeptide. B, superposition of the crystal structures of pro-rAmb a 11 (cyan and green as in Fig. 3), pro-Der p 1 (red; Protein Data Bank code 1XKG), and propapain (yellow; Protein Data Bank code 3TNX). The N- and C termini of pro-rAmb a 11 and helices α1–α5 of its N-terminal propeptide are labeled.
Interestingly, the crystal structure of Amb a 11 revealed that the two C-terminal α-helices (α4, residues from Asn-89 to Tyr-96, and α5, residues from Ile-101 to Arg-108) from the N-terminal propeptide together with the loop connecting them form a lid that covers the active site. As shown in Fig. 5, a network of hydrogen bonds connects Cys-155 and His-289 in the catalytic site with Ser-99 from the propeptide. Specifically, Cys-155 is H-bonded via its γ-sulfur with the terminal oxygen of Ser-99 and the main-chain nitrogen of Gly-290. In addition, H-bonds are also observed between a water molecule and both the γ-oxygen from Ser-99, His-289, the carbonyl atom of Tyr-96, and the terminal nitrogen of Gln-149. The latter also interacts with the side-chain oxygen of Ser-311 (Fig. 5). Importantly, Ser-99, Gln-149, His-289, and Ser-311 residues are conserved in some but not all allergenic C1A cysteine proteases (Fig. 4A).
FIGURE 5.
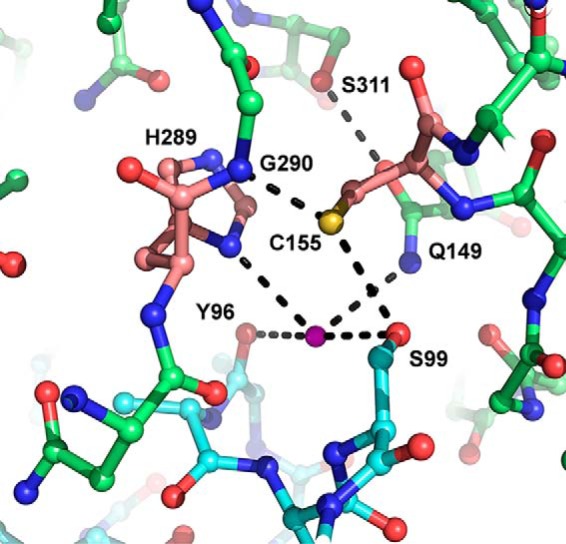
Network of hydrogen bonds between catalytic site and N-terminal propeptide. Residues are shown in ball-and-sticks with residues of the N-terminal propeptide in cyan. The magenta sphere represents a water molecule. Catalytic residues are pink with hydrogen bonds (maximal accepted length, 3.2 Å) shown as black dotted lines.
Amb a 11 Induces Airway Hyperresponsiveness, Lung Inflammation, and Th2 Responses in Sensitized Mice
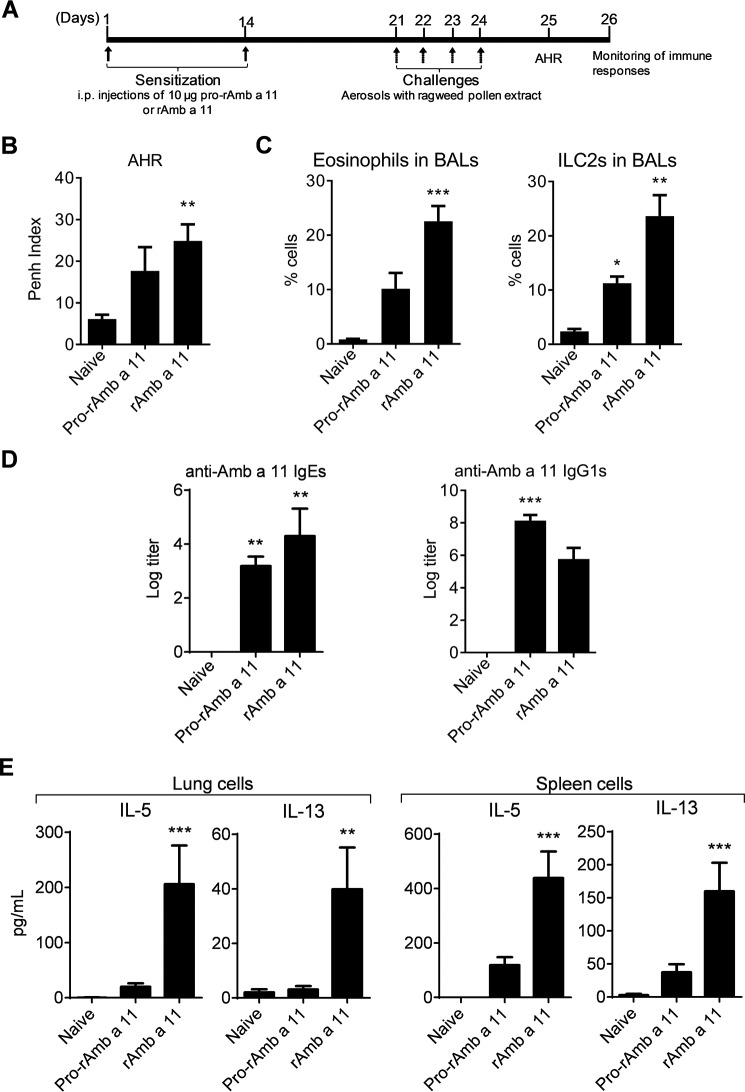
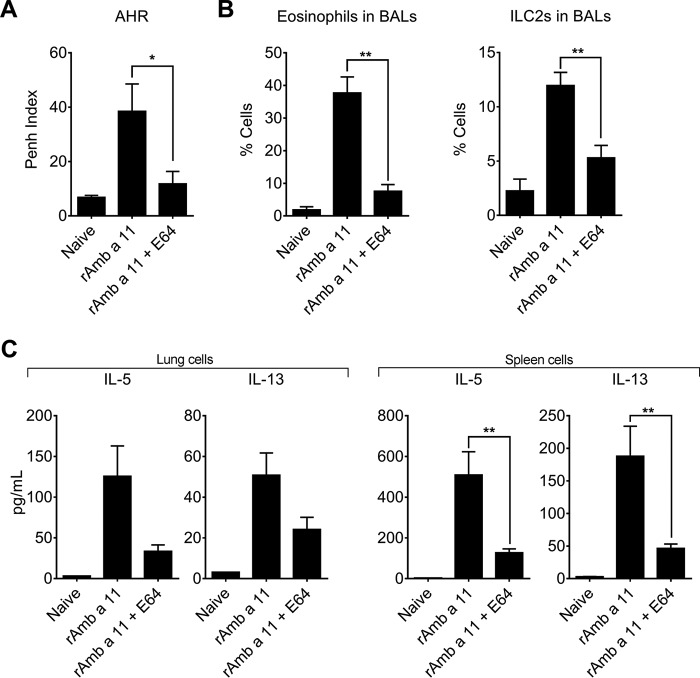
Whereas the identification of Amb a 11 as an allergen stems from (i) the presence of specific IgEs directed to this molecule in sera from 66% of ragweed pollen-allergic patients and (ii) its cysteine protease activity (9), we proceeded to investigate further the allergenicity of Amb a 11 in a murine sensitization model. To this aim, BALB/c mice were sensitized with either pro-rAmb a 11 or rAmb a 11 prior to aerosol challenges with a ragweed pollen extract (Fig. 6A). When compared with naive animals, mice sensitized to either pro-rAmb a 11 or rAmb a 11 exhibited both a clear AHR, eosinophil and ILC2 infiltrates in BALs, and strong Amb a 11-specific serum IgE, IgG1, and Th2 responses (Fig. 6, B–E). Of note, both AHR and inflammatory cell infiltrates were markedly higher in mice sensitized to rAmb a 11 when compared with pro-rAmb a 11 (Fig. 6, B and C). In agreement with the latter results, we also observed a higher secretion of IL-5 and IL-13 by lung and spleen T cells from rAmb a 11-sensitized mice when compared with animals receiving pro-rAmb a 11 (Fig. 6E). To confirm the role of the protease activity on Amb a 11 allergenicity, we performed additional experiments by irreversibly inhibiting the enzymatic activity of Amb a 11 with E64. As shown in Fig. 7, mice sensitized with E64-inhibited rAmb a 11 exhibited significantly lower AHR, eosinophil and ILC2 infiltrates in BALs, and Amb a 11-specific Th2 responses when compared with rAmb a 11-treated animals.
FIGURE 6.
Murine model of sensitization to Amb a 11. A, study design. B, AHR was assessed by whole body plethysmography at day 25 and expressed as enhanced pause (Penh) index values when using a 50 mg/ml dose of methacholine. C, percentages of eosinophils and ILC2s evaluated in BALs by flow cytometry. D, serum IgE and IgG1 antibody responses assessed by ELISA. E, levels of IL-5 and IL-13 were assessed in culture supernatants of lung and spleen cells after in vitro stimulation with pro-rAmb a 11 (10 μg/ml) by using a multiplex cytokine quantification assay. Background levels of cytokines produced by non-activated cells were subtracted. Results are expressed as mean values ±S.E. (error bars) with n = 6 mice per group. Statistical differences between groups were assessed using the non-parametric Kruskal-Wallis test with subsequent Dunnett's multiple analyses when comparing treated mice with naive mice. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 7.
Impact of the inhibition of protease activity on Amb a 11 allergenicity. A, AHR was assessed by whole body plethysmography at day 25 in the presence of a 50 mg/ml dose of methacholine and expressed as enhanced pause (Penh) index values. B, percentages of eosinophils and ILC2s evaluated in BALs by flow cytometry. C, levels of IL-5 and IL-13 were assessed by using a multiplex cytokine quantification assay in culture supernatants of lung and spleen cells after in vitro stimulation with pro-rAmb a 11 (10 μg/ml). Background levels of cytokines produced by non-activated cells were subtracted. Results are expressed as mean values ± S.E. (error bars) with n = 6 mice per group. Statistical differences between rAmb a 11 and E64-inhibited rAmb a 11 were assessed using the non-parametric Mann-Whitney t test. *, p < 0.05; **, p < 0.01.
Discussion
Allergens in short ragweed pollen grains are known to cause severe type I respiratory allergies with a growing prevalence of IgE sensitization in both North America and Europe (1–3). Although as of today 10 short ragweed pollen allergens have been officially registered by International Union of Immunological Societies (6), we recently reported the identification of seven novel candidate allergens following an extensive characterization of the short ragweed pollen transcriptome and proteome (26). Among those molecules, only two are considered as major allergens, namely, the pectate lyase Amb a 1 and the cysteine protease Amb a 11 (9), thought to contribute jointly to most of the ragweed pollen allergenicity. Whereas Amb a 1 has been extensively characterized (27), the structure, activity, and allergenicity of Amb a 11 are not yet fully documented.
To further study the latter allergen, we produced a recombinant proform of Amb a 11 in E. coli as inclusion bodies and successfully refolded the zymogen molecule in vitro as confirmed by structural analyses. The in vitro maturation of pro-rAmb a 11 occurs at acidic pH and leads to a mature and enzymatically active Amb a 11 molecule devoid of both N- and C-terminal propeptides. Cleavage sites are consistent with boundaries determined within the natural allergen with some additional residues identified at both termini in agreement with similar observations reported previously for plant-derived recombinant Der p 1 (28). In vitro activation of pro-rAmb a 11 is clearly mediated by an autocatalytic mechanism dependent on the cysteine 155 located in the active site that is the hallmark of C1A cysteine proteases (29, 30). However, as several other proteases have been reported to be present in the ragweed pollen (31, 32), we cannot exclude that the latter can as well contribute to the processing of the zymogen into active Amb a 11 as documented for other cysteine proteases (33).
We describe herein the first crystal structures ever reported for a ragweed pollen allergen. With a few exceptions, most published crystal structures of cysteine protease zymogens, including pro-Der p 1 (23), were obtained from proteins with a mutated catalytic cysteine (18, 34–37). In this study, we determined the high resolution crystal structure of pro-rAmb a 11 with a preserved active site, confirming an overall fold typical of C1A cysteine proteases such as Der p 1 and papain. As a consequence, we could document the contribution of multiple hydrogen bonds in the interaction between critical residues in the active site (Cys-155 and His-289) and other residues within the N-terminal propeptide (Tyr-96 and Ser-99). Of note, no hydrogen bonds similar to the ones reported herein between Cys-155 and Ser-99 have been observed in the published crystal structures of procaricain (Protein Data Bank code 1PCI) and procathepsin K (Protein Data Bank code 7PCK) that retain their catalytic cysteine (36, 37). The potential role of the hydrogen bonds observed in pro-rAmb a 11 in maintaining the N-terminal propeptide in the vicinity of the active site cleft remains to be clarified. In our hands, mutation of Ser-99 in the propeptide did not impair the maturation capacity of pro-rAmb a 11. In addition, our failure to obtain a molecule with protease activity following expression and refolding of mature rAmb a 11 devoid of its N-terminal propeptide (data not shown) is consistent with a potential chaperone function of the latter in the overall folding of the molecule as demonstrated previously for other cysteine proteases (38–42).
As of today, the role of the C-terminal domain of Amb a 11 still remains elusive. During our analysis of Amb a 11 crystals, the C-terminal propeptide (Lys-371 to Leu-386) as well as the 20 upstream residues (Pro-351 to Phe-370) were only observed in monomer A of the medium resolution structure as a region without any α-helix or β-strand, protruding away from the catalytic site, with consequently no significant interaction with the core domain. Based on results obtained with the pro-rAmb a 11 ΔCT variant, we conclude that this C-terminal propeptide does not inhibit the protease activity of rAmb a 11, nor is it required for proper folding and maturation of the molecule. Such observations differ from evidence that the C-terminal propeptide of other plant cysteine proteases, like Act d 1 and ervatamin C, can regulate either the folding or the enzymatic activity of corresponding proforms (43, 44). Of note, the presence of a KLLEL motif at the end of this C-terminal propeptide, resembling the (K/H)DEL motif observed in plant cysteine proteases such as vignain (17, 45, 46), strongly suggests a role for this propeptide in the retention within the endoplasmic reticulum and intracellular transport of the molecule.
In parallel, we also further investigated the allergenicity of Amb a 11. Our previously published data established a 66% prevalence of IgE reactivity against the natural Amb a 11 allergen in allergic patients (9). Although not shown, nAmb a 11 from ragweed pollen extract, pro-rAmb a 11, and rAmb a 11 reacted similarly with serum IgEs from ragweed pollen-allergic patients as well as sheep polyclonal and murine monoclonal antibodies, clearly indicating that IgE and IgG epitopes are preserved in the recombinant molecules. Interestingly, administration of proteolytically active rAmb a 11 in mice elicits a strong allergic inflammation (including AHR, eosinophil infiltrates in BALs, and Amb a 11-specific Th2 responses), whereas sensitization with pro-rAmb a 11 or E64-inhibited rAmb a 11 results in lower allergic responses. This observation is fully consistent with documented evidence that cysteine protease activities of Der p 1 and Der f 1 promote airway epithelium disruption (47, 48), immune cell activation (49, 50), and Th2 inflammation (51–54).
Collectively, our results confirm that the major ragweed pollen allergen Amb a 11 has typical structural features of C1A cysteine proteases and as such has a strong allergenic activity. We thus conclude that Amb a 11 should be considered as a key component of the ragweed pollen for diagnosis and immunotherapy purposes and that the properly folded and well characterized rAmb a 11 molecule described herein represents a valuable tool in this regard.
Author Contributions
R. G., V. B.-L. F., L. M., P. B., and P. M. conceived and designed the experiments. R. G., S. A., J. J., N. B., L. B., M. L. M., F. J., and P. Z. performed the experiments. D. K., M. S., and P. B. analyzed the structural data. R. G., V. B.-L. F., L. M., P. B., and P. M. wrote the paper. V. B.-B., V. B.-L. F., L. M., and P. B. critically reviewed the manuscript.
Acknowledgment
We thank Dr. Franck Wien from SOLEIL synchrotron for valuable help in the SRCD experiment.
This work was supported in part by Stallergenes. R. G., S. A., J. J., N. B., L. B., M. L. M., V. B.-B., V. B.-L. F., L. M., and P. M. are employees at Stallergenes SAS.
The atomic coordinates and structure factors (codes 5EGW and 5EF4) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- pro-rAmb a 11
- recombinant proform of Amb a 11
- AHR
- airway hyperresponsiveness
- BAL
- bronchoalveolar lavage
- CCR
- CC chemokine receptor
- CD
- cluster of differentiation
- ICOS
- inducible T-cell co-stimulator
- ILC2
- type 2 innate lymphoid cell
- nAmb a 11
- natural Amb a 11
- nDer p 1
- natural Der p 1
- pro-rAmb a 11 C155S
- variant with an active site mutation
- pro-rAmb a 11 ΔCT
- variant lacking the C-terminal propeptide
- rAmb a 11
- mature recombinant Amb a 11
- Boc
- t-butoxycarbonyl
- AMC
- 7-amino-4-methylcoumarin
- AEBSF
- 4(2aminoethyl)benzenesulfonyl fluoride
- SRCD
- synchrotron radiation circular dichroism
- r.m.s.d.
- root mean square deviation.
References
- 1. Arbes S. J. Jr., Gergen P. J., Elliott L., and Zeldin D. C. (2005) Prevalences of positive skin test responses to 10 common allergens in the US population: results from the third National Health and Nutrition Examination Survey. J. Allergy Clin. Immunol. 116, 377–383 [DOI] [PubMed] [Google Scholar]
- 2. D'Amato G., Cecchi L., Bonini S., Nunes C., Annesi-Maesano I., Behrendt H., Liccardi G., Popov T., and van Cauwenberge P. (2007) Allergenic pollen and pollen allergy in Europe. Allergy 62, 976–990 [DOI] [PubMed] [Google Scholar]
- 3. Burbach G. J., Heinzerling L. M., Röhnelt C., Bergmann K. C., Behrendt H., Zuberbier T., and GA(2)LEN study (2009) Ragweed sensitization in Europe—GA(2)LEN study suggests increasing prevalence. Allergy 64, 664–665 [DOI] [PubMed] [Google Scholar]
- 4. Oswalt M. L., and Marshall G. D. (2008) Ragweed as an example of worldwide allergen expansion. Allergy Asthma Clin. Immunol. 4, 130–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith M., Cecchi L., Skjøth C. A., Karrer G., and Šikoparija B. (2013) Common ragweed: a threat to environmental health in Europe. Environ. Int. 61, 115–126 [DOI] [PubMed] [Google Scholar]
- 6. Bordas-Le Floch V., Groeme R., Chabre H., Baron-Bodo V., Nony E., Mascarell L., and Moingeon P. (2015) New insights into ragweed pollen allergens. Curr. Allergy Asthma Rep. 15, 63. [DOI] [PubMed] [Google Scholar]
- 7. Asero R., Wopfner N., Gruber P., Gadermaier G., and Ferreira F. (2006) Artemisia and Ambrosia hypersensitivity: co-sensitization or co-recognition? Clin. Exp. Allergy 36, 658–665 [DOI] [PubMed] [Google Scholar]
- 8. Gadermaier G., Wopfner N., Wallner M., Egger M., Didierlaurent A., Regl G., Aberger F., Lang R., Ferreira F., and Hawranek T. (2008) Array-based profiling of ragweed and mugwort pollen allergens. Allergy 63, 1543–1549 [DOI] [PubMed] [Google Scholar]
- 9. Bouley J., Groeme R., Le Mignon M., Jain K., Chabre H., Bordas-Le Floch V., Couret M. N., Bussières L., Lautrette A., Naveau M., Baron-Bodo V., Lombardi V., Mascarell L., Batard T., Nony E., et al. (2015) Identification of the cysteine protease Amb a 11 as a novel major allergen from short ragweed. J. Allergy Clin. Immunol. 136, 1055–1064 [DOI] [PubMed] [Google Scholar]
- 10. Bussières L., Bordas-Le Floch V., Bulder I., Chabre H., Nony E., Lautrette A., Berrouet C., Nguefeu Y., Horiot S., Baron-Bodo V., Van Overtvelt L., De Conti A. M., Schlegel A., Maguet N., Mouz N., et al. (2010) Recombinant fusion proteins assembling Der p 1 and Der p 2 allergens from Dermatophagoides pteronyssinus. Int. Arch. Allergy Immunol. 153, 141–151 [DOI] [PubMed] [Google Scholar]
- 11. Choudhury D., Roy S., Chakrabarti C., Biswas S., and Dattagupta J. K. (2009) Production and recovery of recombinant propapain with high yield. Phytochemistry 70, 465–472 [DOI] [PubMed] [Google Scholar]
- 12. Lees J. G., Smith B. R., Wien F., Miles A. J., and Wallace B. A. (2004) CDtool—an integrated software package for circular dichroism spectroscopic data processing, analysis, and archiving. Anal. Biochem. 332, 285–289 [DOI] [PubMed] [Google Scholar]
- 13. van Stokkum I. H., Spoelder H. J., Bloemendal M., van Grondelle R., and Groen F. C. (1990) Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem. 191, 110–118 [DOI] [PubMed] [Google Scholar]
- 14. Whitmore L., and Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Storoni L. C., McCoy A. J., and Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 17. Than M. E., Helm M., Simpson D. J., Lottspeich F., Huber R., and Gietl C. (2004) The 2.0 Å crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm. J. Mol. Biol. 336, 1103–1116 [DOI] [PubMed] [Google Scholar]
- 18. Roy S., Choudhury D., Aich P., Dattagupta J. K., and Biswas S. (2012) The structure of a thermostable mutant of pro-papain reveals its activation mechanism. Acta Crystallogr. D Biol. Crystallogr. 68, 1591–1603 [DOI] [PubMed] [Google Scholar]
- 19. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., and Womack T. O. (2011) BUSTER, version 2.1.0, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 20. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21. Razafindratsita A., Saint-Lu N., Mascarell L., Berjont N., Bardon T., Betbeder D., Van Overtvelt L., and Moingeon P. (2007) Improvement of sublingual immunotherapy efficacy with a mucoadhesive allergen formulation. J. Allergy Clin. Immunol. 120, 278–285 [DOI] [PubMed] [Google Scholar]
- 22. Novinec M., and Lenarčič B. (2013) Papain-like peptidases: structure, function, and evolution. BioMol Concepts 4, 287–308 [DOI] [PubMed] [Google Scholar]
- 23. Meno K., Thorsted P. B., Ipsen H., Kristensen O., Larsen J. N., Spangfort M. D., Gajhede M., and Lund K. (2005) The crystal structure of recombinant proDer p 1, a major house dust mite proteolytic allergen. J. Immunol. 175, 3835–3845 [DOI] [PubMed] [Google Scholar]
- 24. de Halleux S., Stura E., VanderElst L., Carlier V., Jacquemin M., and Saint-Remy J. M. (2006) Three-dimensional structure and IgE-binding properties of mature fully active Der p 1, a clinically relevant major allergen. J. Allergy Clin. Immunol. 117, 571–576 [DOI] [PubMed] [Google Scholar]
- 25. Kamphuis I. G., Kalk K. H., Swarte M. B., and Drenth J. (1984) Structure of papain refined at 1.65 Å resolution. J. Mol. Biol. 179, 233–256 [DOI] [PubMed] [Google Scholar]
- 26. Bordas-Le Floch V., Le Mignon M., Bouley J., Groeme R., Jain K., Baron-Bodo V., Nony E., Mascarell L., and Moingeon P. (2015) Identification of novel short ragweed pollen allergens using combined transcriptomic and immunoproteomic approaches. PLoS One 10, e0136258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gadermaier G., Hauser M., and Ferreira F. (2014) Allergens of weed pollen: an overview on recombinant and natural molecules. Methods 66, 55–66 [DOI] [PubMed] [Google Scholar]
- 28. Burtin D., Chabre H., Olagnier B., Didierlaurent A., Couret M. N., Comeau D., Wambre E., Laparra H., Van Overtvelt L., Montandon F., Batard T., Jonval V., Lorphelin A., Merle C., Berrouet C., Parry L., Gomord V., Van Ree R., and Moingeon P. (2009) Production of native and modified recombinant Der p 1 molecules in tobacco plants. Clin. Exp. Allergy 39, 760–770 [DOI] [PubMed] [Google Scholar]
- 29. Brömme D., Nallaseth F. S., and Turk B. (2004) Production and activation of recombinant papain-like cysteine proteases. Methods 32, 199–206 [DOI] [PubMed] [Google Scholar]
- 30. Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., and Turk D. (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta 1824, 68–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gunawan H., Takai T., Ikeda S., Okumura K., and Ogawa H. (2008) Protease activity of allergenic pollen of cedar, cypress, juniper, birch and ragweed. Allergol. Int. 57, 83–91 [DOI] [PubMed] [Google Scholar]
- 32. Gunawan H., Takai T., Kamijo S., Wang X. L., Ikeda S., Okumura K., and Ogawa H. (2008) Characterization of proteases, proteins, and eicosanoid-like substances in soluble extracts from allergenic pollen grains. Int. Arch. Allergy Immunol. 147, 276–288 [DOI] [PubMed] [Google Scholar]
- 33. Wiederanders B. (2003) Structure-function relationships in class CA1 cysteine peptidase propeptides. Acta Biochim. Pol. 50, 691–713 [PubMed] [Google Scholar]
- 34. Kaulmann G., Palm G. J., Schilling K., Hilgenfeld R., and Wiederanders B. (2006) The crystal structure of a Cys25 → Ala mutant of human procathepsin S elucidates enzyme-prosequence interactions. Protein Sci. 15, 2619–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stack C. M., Caffrey C. R., Donnelly S. M., Seshaadri A., Lowther J., Tort J. F., Collins P. R., Robinson M. W., Xu W., McKerrow J. H., Craik C. S., Geiger S. R., Marion R., Brinen L. S., and Dalton J. P. (2008) Structural and functional relationships in the virulence-associated cathepsin L proteases of the parasitic liver fluke, Fasciola hepatica. J. Biol. Chem. 283, 9896–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sivaraman J., Lalumière M., Ménard R., and Cygler M. (1999) Crystal structure of wild-type human procathepsin K. Protein Sci. 8, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Groves M. R., Taylor M. A., Scott M., Cummings N. J., Pickersgill R. W., and Jenkins J. A. (1996) The prosequence of procaricain forms an α-helical domain that prevents access to the substrate-binding cleft. Structure 4, 1193–1203 [DOI] [PubMed] [Google Scholar]
- 38. Chevigné A., Barumandzadeh R., Groslambert S., Cloes B., Dehareng D., Filée P., Marx J. C., Frère J. M., Matagne A., Jacquet A., and Galleni M. (2007) Relationship between propeptide pH unfolding and inhibitory ability during ProDer p 1 activation mechanism. J. Mol. Biol. 374, 170–185 [DOI] [PubMed] [Google Scholar]
- 39. Demidyuk I. V., Shubin A. V., Gasanov E. V., and Kostrov S. V. (2010) Propeptides as modulators of functional activity of proteases. Biomol. Concepts 1, 305–322 [DOI] [PubMed] [Google Scholar]
- 40. Eder J., and Fersht A. R. (1995) Pro-sequence-assisted protein folding. Mol. Microbiol. 16, 609–614 [DOI] [PubMed] [Google Scholar]
- 41. Wiederanders B., Kaulmann G., and Schilling K. (2003) Functions of propeptide parts in cysteine proteases. Curr. Protein Pept. Sci. 4, 309–326 [DOI] [PubMed] [Google Scholar]
- 42. Nandana V., Singh S., Singh A. N., and Dubey V. K. (2014) Procerain B, a cysteine protease from Calotropis procera, requires N-terminus pro-region for activity: cDNA cloning and expression with pro-sequence. Protein Expr. Purif. 103, 16–22 [DOI] [PubMed] [Google Scholar]
- 43. Paul W., Amiss J., Try R., Praekelt U., Scott R., and Smith H. (1995) Correct processing of the kiwifruit protease actinidin in transgenic tobacco requires the presence of the C-terminal propeptide. Plant Physiol. 108, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dutta S., Choudhury D., Dattagupta J. K., and Biswas S. (2011) C-terminal extension of a plant cysteine protease modulates proteolytic activity through a partial inhibitory mechanism. FEBS J. 278, 3012–3024 [DOI] [PubMed] [Google Scholar]
- 45. Schmid M., Simpson D., Kalousek F., and Gietl C. (1998) A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta 206, 466–475 [DOI] [PubMed] [Google Scholar]
- 46. Okamoto T., Minamikawa T., Edward G., Vakharia V., Herman E., and Okomoto T. (1999) Posttranslational removal of the carboxyl-terminal KDEL of the cysteine protease SH-EP occurs prior to maturation of the enzyme. J. Biol. Chem. 274, 11390–11398 [DOI] [PubMed] [Google Scholar]
- 47. Deb R., Shakib F., Reid K., and Clark H. (2007) Major house dust mite allergens Dermatophagoides pteronyssinus 1 and Dermatophagoides farinae 1 degrade and inactivate lung surfactant proteins A and D. J. Biol. Chem. 282, 36808–36819 [DOI] [PubMed] [Google Scholar]
- 48. Henriquez O. A., Den Beste K., Hoddeson E. K., Parkos C. A., Nusrat A., and Wise S. K. (2013) House dust mite allergen Der p 1 effects on sinonasal epithelial tight junctions. Int. Forum Allergy Rhinol. 3, 630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reubsaet L., Meerding J., Giezeman R., de Kleer I., Arets B., Prakken B., Beekman J., and van Wijk F. (2013) Der p 1-induced CD4+ FOXP3+ GATA3+ T cells have suppressive properties and contribute to the polarization of the TH2-associated response. J. Allergy Clin. Immunol. 132, 1440–1444 [DOI] [PubMed] [Google Scholar]
- 50. Yi M. H., Kim H. P., Jeong K. Y., Kim C. R., Kim T. Y., and Yong T. S. (2015) House dust mite allergen Der f 1 induces IL-8 in human basophilic cells via ROS-ERK and p38 signal pathways. Cytokine 75, 356–364 [DOI] [PubMed] [Google Scholar]
- 51. Comoy E. E., Pestel J., Duez C., Stewart G. A., Vendeville C., Fournier C., Finkelman F., Capron A., and Thyphronitis G. (1998) The house dust mite allergen, Dermatophagoides pteronyssinus, promotes type 2 responses by modulating the balance between IL-4 and IFN-γ. J. Immunol. 160, 2456–2462 [PubMed] [Google Scholar]
- 52. Gough L., Sewell H. F., and Shakib F. (2001) The proteolytic activity of the major dust mite allergen Der p 1 enhances the IgE antibody response to a bystander antigen. Clin. Exp. Allergy 31, 1594–1598 [DOI] [PubMed] [Google Scholar]
- 53. Chapman M. D., Wünschmann S., and Pomés A. (2007) Proteases as Th2 adjuvants. Curr. Allergy Asthma Rep. 7, 363–367 [DOI] [PubMed] [Google Scholar]
- 54. Cunningham P. T., Elliot C. E., Lenzo J. C., Jarnicki A. G., Larcombe A. N., Zosky G. R., Holt P. G., and Thomas W. R. (2012) Sensitizing and Th2 adjuvant activity of cysteine protease allergens. Int. Arch. Allergy Immunol. 158, 347–358 [DOI] [PubMed] [Google Scholar]
- 55. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]