Abstract
The hepatic hormone hepcidin is the master regulator of systemic iron homeostasis. Its expression level is adjusted to alterations in iron levels, inflammatory cues, and iron requirements for erythropoiesis. Bone morphogenetic protein 6 (BMP6) contributes to the iron-dependent control of hepcidin. In addition, TGF-β1 may stimulate hepcidin mRNA expression in murine hepatocytes and human leukocytes. However, receptors and downstream signaling proteins involved in TGF-β1-induced hepcidin expression are still unclear. Here we show that TGF-β1 treatment of mouse and human hepatocytes, as well as ectopic expression of TGF-β1 in mice, increases hepcidin mRNA levels. The hepcidin response to TGF-β1 depends on functional TGF-β1 type I receptor (ALK5) and TGF-β1 type II receptor (TβRII) and is mediated by a noncanonical mechanism that involves Smad1/5/8 phosphorylation. Interestingly, increasing availability of canonical Smad2/3 decreases TGF-β1-induced hepcidin regulation, whereas the BMP6-hepcidin signal was enhanced, indicating a signaling component stoichiometry-dependent cross-talk between the two pathways. Although ALK2/3-dependent hepcidin activation by BMP6 can be modulated by each of the three hemochromatosis-associated proteins: HJV (hemojuvelin), HFE (hemochromatosis protein), and TfR2 (transferrin receptor 2), these proteins do not control the ALK5-mediated hepcidin response to TGF-β1. TGF-β1 mRNA levels are increased in mouse models of iron overload, indicating that TGF-β1 may contribute to hepcidin synthesis under these conditions. In conclusion, these data demonstrate that a complex regulatory network involving TGF-β1 and BMP6 may control the sensing of systemic and/or hepatic iron levels.
Keywords: bone morphogenetic protein (BMP), iron, liver, SMAD transcription factor, transforming growth factor beta (TGF-β), ALK5, HJV, hepcidin
Introduction
The liver-produced small peptide hormone hepcidin has emerged as a central regulator of systemic iron homeostasis (1). Hepcidin maintains serum iron levels by controlling dietary iron uptake from duodenal enterocytes and iron release from iron recycling macrophages. It binds to the cellular iron exporter Ferroportin (Fpn)5 to trigger its internalization and degradation (1). Hepcidin expression is regulated by several signals, including plasma iron levels, hepatic iron stores, inflammation, erythropoietic activity, and hypoxia (2). Plasma iron levels and hepatic iron stores activate BMP6-ALK2/3-Smad1/5/8 signaling through a yet poorly defined mechanism to transcriptionally modulate hepcidin synthesis (3–5). Notably, hepcidin can be mildly increased upon chronic iron loading of BMP6 knock-out mice, suggesting that the BMP6 pathway does not completely account for the iron response of hepcidin (4). In addition, inflammatory stimuli such as IL6 induce hepcidin expression in human (6) and mouse (7) hepatocytes via the JAK/STAT signaling pathway (8, 9).
The monitoring of iron levels in hepatocytes involves the membrane-bound HJV that is mutated in patients with a juvenile subtype of hereditary hemochromatosis (HH) (10). It acts as a coreceptor for BMP most probably upon heterodimerization with the BMP type I receptors ALK2 and/or ALK3 to regulate hepcidin expression (11). The MHC class I-like protein HFE and the TfR2 are mutated in patients with a less aggressive subtype of HH (12, 13) and are involved in the sensing of transferrin-bound iron (14–16). Mice with hepatocyte-specific deficiency of HFE, TfR2, or HJV show decreased hepcidin levels and an iron overload phenotype (17–19). Interestingly, Smad1/5/8 phosphorylation is attenuated not only in mice with HJV deficiency but also in mice and patients that lack HFE or TfR2. Therefore, it was proposed that HFE and TfR2 may form a functional complex with HJV to stimulate hepcidin expression (5, 20, 21). Indeed, expression of recombinant HFE, TfR2, and HJV in cultured cells reveals formation of a protein complex (22). Whether the HH-associated proteins participate in hepcidin activation by TGF-β1 has not been reported.
TGF-β is produced by nonparenchymal liver cells including hepatic stellate cells, liver sinusoidal endothelial cells, Kupffer cells, and hepatic immune cells (23). TGF-β is a critical factor to maintain tissue homeostasis under healthy conditions but further plays an important role in liver diseases, starting from mild liver injury and inflammation up to end stage cirrhosis and hepatocellular carcinoma (24). TGF-β expression is increased in response to liver damage and enhances hepatocyte destruction, as well as hepatic stellate cell activation, resulting in myofibroblast generation and extracellular matrix deposition (25). In HH patients, immunostaining of TGF-β1 is markedly increased in liver biopsies and normalized following phlebotomy, suggesting that it may contribute to progressive hepatic injury in HH patients (26). Whether or not TGF-β1 is induced by iron accumulation or liver damage in HH patients remains unclear.
TGF-β superfamily ligands activate heteromeric complexes of type I and II serine/threonine kinase receptors. Canonical TGF-β1 signaling is mediated via TGF-β1 type I receptor, also known as ALK5, which phosphorylates Smad2 and Smad3 at C-terminal SXS motifs to facilitate their activation and transcriptional activity (23). Although it is generally accepted that TGF-β1 induces Smad2/3 phosphorylation, several studies have shown that TGF-β1 can also induce Smad1/5/8 phosphorylation via different type I receptors, depending on the cellular context (27–29). In murine hepatocytes (30) and hepatic stellate cells (24), TGF-β1 induces both Smad2/3 and Smad1/5/8 phosphorylation in a time-dependent manner. Previous findings suggested that TGF-β1 may stimulate hepcidin mRNA expression in mouse hepatocytes (31) and human leukocytes (32), but not in human hepatic carcinoma cell lines (33). However, receptors and downstream signaling proteins involved in TGF-β1-induced hepcidin expression have not been investigated.
Here, we show that TGF-β1 regulates hepcidin transcription in hepatocytic cell lines, primary mouse and human hepatocytes, and mice. We show that the TGF-β1 response of hepcidin is mediated by ALK5- and Smad1/5/8-mediated downstream signaling.
Experimental Procedures
Reagents and Cell Line
Human recombinant TGF-β1 was obtained from Peprotech (Hamburg, Germany), BMP6 from R&D (Minneapolis, MN), SB431542 from Sigma-Aldrich, and LDN1931542 from Selleckchem (Houston, TX). Compounds were dissolved at 10 mm in DMSO as stock solutions and stored at −20 °C. The compounds were diluted in normal saline before each experiment and applied in the concentrations indicated. HuH7 cells were obtained from Japanese Collection of Research Bioresources and cultured according to the online instructions.
Human Primary Hepatocytes
Non-neoplastic tissue samples from liver resections were obtained from patients undergoing partial hepatectomy for metastatic liver tumors of colorectal cancer (from University Hospital Regensburg), cholangiocarcinoma with primary sclerosing cholangitis, and cholangiocarcinoma without primary sclerosing cholangitis (from University Hospital of Munich, Campus Grosshadern). Primary human hepatocytes were isolated and cultivated according to the guidelines of the charitable state-controlled foundation Human Tissue and Cell Research (Regensburg, Germany) with informed patient consent approved by the local ethical committee of the University of Regensburg. All experiments involving human tissues and cells have been carried out in accordance to the Code of Ethics of the World Medical Association (Declaration of Helsinki).
Cell Culture and Transient Transfections
Primary murine hepatocytes were isolated from livers of C56BL/6JRj mice by a two-step portal vein collagenase perfusion method as described (34). The cells were plated on collagen-coated 12-well plates at a density of 180,000 cells/well in Williams' medium E, supplemented with 10% fetal bovine serum, 2 mm l-glutamine, and 100 nm dexamethasone. After attachment of cells, medium was changed to serum-free Williams' medium E supplemented with 2 mm l-glutamine and 100 nm dexamethasone. After overnight incubation in 5% CO2 at 37 °C, medium was changed to serum-free Williams' medium E supplemented with 2 mm l-glutamine. Recombinant TGF-β1 and/or BMP6 were added to the serum-free culture medium for the indicated times and concentrations. Transfection of hepatocytes was achieved with RNAimax (Invitrogen) according to the manufacturer's instructions. siRNAs targeting mouse ALK2 (162106), ALK5 (186714), ALK3 (100331), HJV (163123), HFE (158943), TfR2 (72123), Smad2 (156215), and Smad5 (155237) were purchased from Invitrogen, and Smad1 (SI00177072) was from Qiagen. siRNAs targeting human HJV (MU-018751-02), HFE (MU-011051-02), and TfR2 (MU-009686-02) were purchased from GE Dharmacon (Lafayette, CO).
Ectopic AdTGF-β1 Expression in Mice
109 pfu of AdTGF-β1223/225 (porcine) or control virus DL70–3 (35), kindly provided by Prof. Dr. Jack Gauldie, in 100 μl of formulation buffer (2.5% glycerol (w/v), 25 mm NaCl, and 20 mm Tris-HCl, pH 8.0) were injected into the tail vein of male C56BL/6JRj mice. The animals were euthanized 4 days after injection. Heparinized blood was collected by cardiac puncture. The animal experiments were approved by the district government of Niedersachsen, Germany. All efforts were made to minimize the number of animals and their suffering.
Induction of Systemic Iron Overload in Mice
Systemic iron overload was induced in C57BL/6JRj female mice by i.p. administration of 5 mg of iron dextran solution (100 μl of a 50 mg/ml solution, containing 50,000 ppm iron) twice a week beginning from week 5 until week 7 of age. As a control, mice received an injection of 0.5 mg of Dextran 5 solution (100 μl of a 5 mg/ml solution). Mice were sacrificed for analyses at 8 weeks of age. All mouse breeding and animal experiments were approved by and conducted in compliance with the guidelines of the Institutional Animal Care and User Committee of the European Molecular Biology Laboratory (31).
Adenoviral Transduction of Primary Murine Hepatocytes
AdLacZ, AdSmad1, AdSmad2, AdSmad3, AdSmad5, AdTβRIIDN, AdALK5CA, and AdALK5DN were kindly provided by C. Heldin (Ludwig Institution for Cancer Research, Uppsala, Sweden) (36). Infectivity was determined using the rapid titer kit from BD Biosciences. Briefly, HEK293 cells were infected with serial dilutions of adenovirus. After appearance of the hexon proteins, the cells were fixed and treated with a hexon protein-specific antibody and HRP conjugate antibody and developed with DAB Substrate. The positive cells turn brown so that they can be easily counted under a 20× objective. 50–100 infectious units/cell of a single virus clone were used, and more than 90% of hepatocytes were infected.
Hematology and Plasma Biochemistry for Iron Quantification
Hematological parameters were determined using the ABC ScilVet analyzer (ABX Diagnostics). Plasma iron concentrations and unsaturated iron binding capacity were assessed using SFBC (ThermoScientific) and UIBC (Biolabo) kits. Transferrin saturation was calculated using the formula SFBC/(SFBC + UIBC) × 100.
RNA Isolation and Real Time Quantitative PCR
Total RNA was isolated using the InviTrap spin universal RNA mini kit (Stratec, Berlin, Germany), according to the manufacturer's instruction. First strand cDNA was synthesized from 500 ng of total RNA by MLV reverse transcriptase (Thermo Fisher) with oligo(dT) primer, following the manufacturer's protocol. Real time quantification of transcripts was performed on the ABI prism 7700 sequence detection system using fast start DNA master SYBR Green 1 (Applied Biosystems). Quantitative real time RT-PCR data are presented as fold change relative to control. Relative quantification was performed using the comparative Ct or ddCt method (18). The target gene was first normalized to a reference housekeeping gene Ppia (peptidylprolyl isomerase A, also known as cyclophilin A) or Gapdh and then presented as the difference from the control treatment within each experiment.
Immunoblotting
Protein lysates were obtained by homogenizing snap-frozen tissue in radioimmune precipitation assay buffer supplemented with protease and phosphatase inhibitors (Roche). Equal amounts of cell lysate or tissue lysate were subjected to standard immunoblot analysis, using antibodies: anti-phospho-Smad1/3 (ab52903; Abcam), anti-phospho-Smad1/5/8 (ab3848; Millipore), anti-Smad1 (9743; Cell Signaling), anti-Smad3 (9513, Cell Signaling), and anti-β-actin (a5441; Sigma). Secondary antibodies were goat anti-rabbit IgG-HRP or goat anti-mouse IgG-HRP (Santa Cruz). The band intensity were quantified using the Gel-Pro software (Med Cybernetics, Inc., Silver Spring, MD)
Luciferase Assay
Generation of the luciferase reporter constructs containing a 2,762-bp fragment of the human hepcidin promoter and derivative constructs with mutations in the nearby BMP-responsive element (BMP_RE1_2.7kb), distal BMP-responsive element (BMP_RE2_2.7kb), or the proximal STAT binding site (STAT_BS_2.7kb) was described previously (37). HuH7 cells were seeded onto 24-well plates. 250 ng of reporter constructs was transfected using Lipofectamine 2000 (Invitrogen). The cells were stimulated with TGF-β1 (10 ng/ml), BMP6 (20 ng/ml), or IL6 (40 ng/ml) for 3 h after incubation under serum-depleted conditions for 24 h. Luciferase activity was normalized to protein concentration.
Statistics
The results are expressed as means ± S.D. Student's t test or one-way ANOVA followed by Tukey's post hoc test were used for determining statistical significance. Real time PCR and immunoblot analyses were performed at least three independent murine hepatocyte preparations. In real time PCR assay, three replicate samples (technical replicates) are analyzed.
Results
TGF-β1 Is Activated by Iron Overload and Induces Hepcidin mRNA Expression in Primary Hepatocytes and Mouse Liver
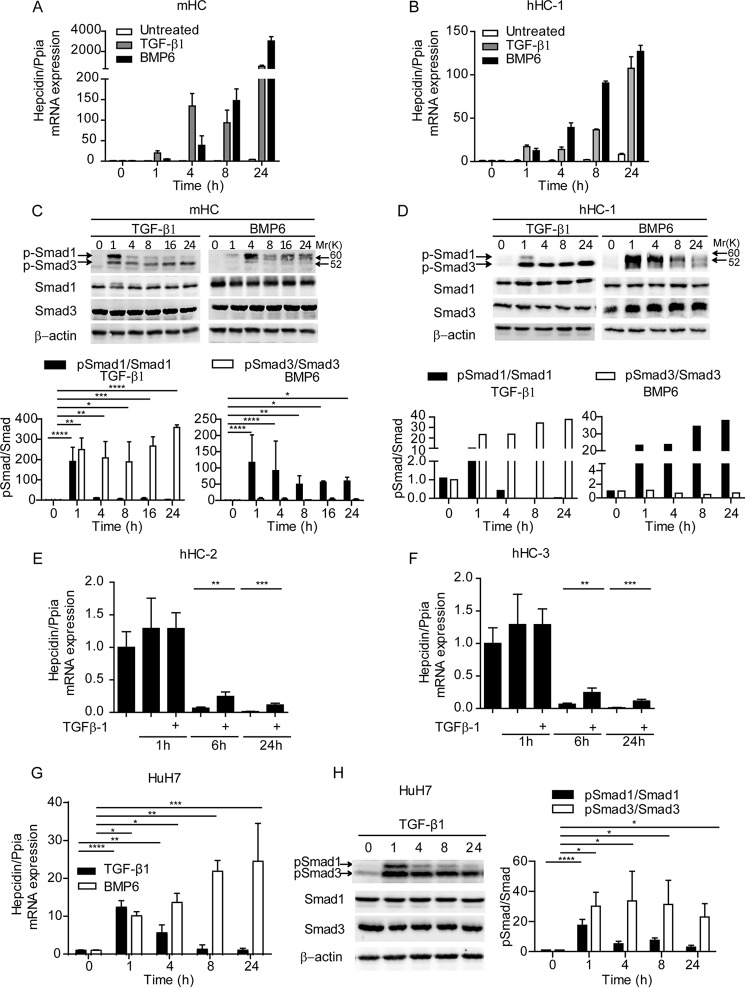
To assess whether TGF-β1 activates hepcidin mRNA expression, we treated mouse hepatocytes (mHC) with TGF-β1 (5 ng/ml) for 1, 4, 8, and 24 h. TGF-β1 rapidly induced hepcidin mRNA levels ∼20-fold at the 1-h time point followed by a time-dependent increase up to over 500-fold at the 24-h time point (Fig. 1A). TGF-β1 caused phosphorylation of Smad3 throughout the time course, whereas phosphorylation of Smad1 was only transiently enhanced at the 1-h time point (Fig. 1C). We speculate that either sustained low level Smad1 phosphorylation or other noncanonical pathways triggered by TGF-β1 may be responsible for hepcidin activation at late time points. In contrast, BMP6 induced Smad1 phosphorylation with a maximum increase at the 4-h time point. A similar response to TGF-β1 was observed in human hepatocytes, isolated from three different patients after liver resection (Fig. 1, B, D, E, and F). Distinct from primary hepatocytes, TGF-β1 only transiently induced hepcidin mRNA expression in the human HCC cell line HuH7, even though the kinetics of R-Smad phosphorylation were comparable with those in hepatocytes (Fig. 1, G and H). For most of the remaining study, we used primary hepatocytes to mimic more physiological conditions.
FIGURE 1.
TGF-β1 activates hepcidin mRNA expression in primary hepatocytes. Different preparations of primary hepatocytes from mice or human vary in their amplitude of cellular responsiveness to TGF-β1 (or any other cytokine), although the same tendency of the response is highly preserved. Valuable comparisons of the magnitude of the responses are restricted to the same preparation of primary mouse hepatocytes. As a result, hepcidin mRNA was analyzed by quantitative real time PCR from three independent murine hepatocyte preparations, whereby one representative data set is shown. Cultures of mHC (A and C) and human hepatocytes (hHC) (B, D, E, and F) were treated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for indicated times. A, B, E, and F, hepcidin mRNA was analyzed by quantitative real time PCR. C and D, total protein and phosphorylated protein levels of Smad1 and Smad3 were analyzed by immunoblot. Protein analyses were performed at least three times. Representative immune blots (upper panels) and quantification of phosphorylation of Smad1 and Smad3 relative to total Smad1 and Smad3 in three different experiments (lower panels) are shown. G and H, HuH7 cells were treated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for the indicated periods of time. Hepcidin mRNA was analyzed by quantitative real time PCR and phosphorylation, and protein levels of Smad1 and Smad3 were measured by Western blotting. H, representative immunoblot (left panels) and quantification of p-Smad1/Smad1 and p-Smad3/Smad3 (right panels) are shown. The results are expressed as means ± S.D. E and F, Student's t test was used for determining statistical significance (n = 3): *, p < 0.05; **, p < 0.01. C, G, and H, one-way ANOVA, followed by Tukey's post hoc test, was used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
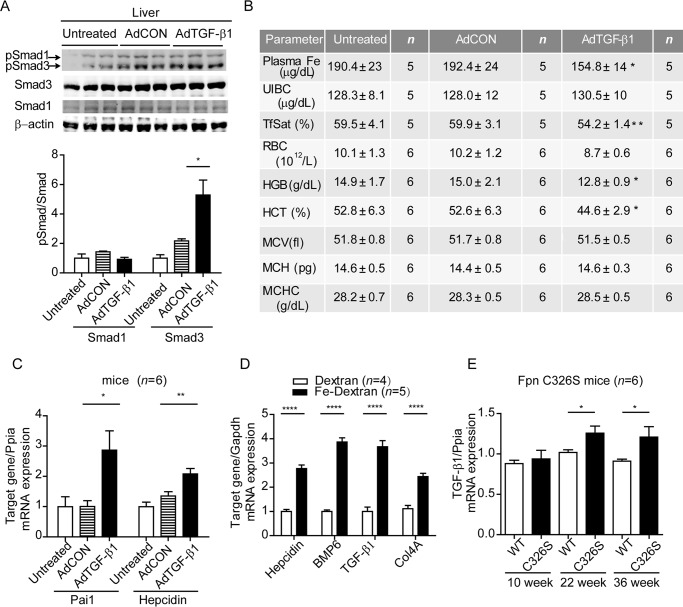
To test whether TGF-β1 induces hepcidin in wild-type mice, adenovirus encoding constitutively active TGF-β1223/225 or DL70–3 control virus was injected into C56BL/6JRj mice through the tail vein. Constitutively active TGF-β1223/225 is preferentially expressed in hepatocytes after systemic intravenous adenovirus vector injections (38, 39). 4 days after injection, hepatic porcine TGF-β1223/225 mRNA was highly induced in the AdTGF-β1223/225 injected group but was undetectable in the untreated and control virus-injected group (AdCON) (data not shown). Consistently, hepatic TGF-β1 signaling was activated, as evidenced by increased phosphorylation of Smad3 (Fig. 2A) and increased mRNA levels of the hepatic target gene Pai1 (Fig. 2C). We further observed a small increase of hepcidin mRNA levels in the liver, indicating that the hepcidin response to TGF-β1 is preserved in vivo (Fig. 2C). Consistent with increased hepcidin mRNA expression, we observe a decrease in plasma iron levels (192.4 ± 24 to 154.8 ± 14 μg/dl; p = 0.027) and transferrin saturation (from 59.92 to 54.20%; p = 0.0099) in the TGF-β1223/225 virus-injected group as compared with the control virus injected group. As a result, hemoglobin values and hematocrit were also down-regulated 4 days after virus injection (Fig. 2B).
FIGURE 2.
TGF-β1 activates hepcidin mRNA expression in wild-type mice and is induced in murine iron-overload models. A, AdTGF-β1223/225 or AdCON viral vectors were injected into the tail vein of male C56BL/6JRj mice, and 4 days later, hepatic protein and phosphorylation levels of Smad1 and Smad3 were analyzed by Western blotting (upper panel). Quantification of Smad1 and Smad3 phosphorylation relative to total Smad1 and Smad3, respectively, are expressed as means ± S.D. (lower panel). One-way ANOVA, followed by Tukey's post hoc test, was used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. B, at the same time point we analyzed plasma iron levels and hematological parameters. UIBC, unsaturated iron binding capacity; TfSat, transferrin saturation; RBC, red blood cell counts; HGB, hemoglobin; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration. The data are shown as means ± S.D. Student's t test was used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C, total RNA from the liver of mice injected with adenovirus expressing TGF-β1223/225 (AdTGF-β1223/225) or a control virus (AdCON) as well as untreated mice were analyzed by quantitative real time PCR analysis for hepcidin, Pai1, or porcine TGF-β1. The results are expressed as means ± S.D. One-way ANOVA, followed by Tukey's post hoc test, was used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. D, hepatic TGF-β1 mRNA expression was analyzed by quantitative real time PCR in C57BL/6J mice administered i.p. with 5 mg of iron dextran solution or 5 mg of Dextran 5 solution (100 μl of 5 mg/ml solution). E, hepatic TGF-β1 mRNA expression in FPN C326S mutant mice of 10, 20, and 36 weeks of age compared with age and sex matched wild-type controls. F and G, Student's t test was used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
We next investigated whether TGF-β1 mRNA expression is altered in mouse models of iron overload and thus may contribute to maintaining iron homeostasis. Hepatic TGF-β1 mRNA expression is increased in mice injected with iron dextran compared with dextran-injected control mice (31). In addition, increased mRNA levels of the principal TGF-β1 target gene Col4a1, as well as of hepcidin and BMP6, were detected (Fig. 2D). Furthermore, mice with an engineered point mutation in the Fpn locus (FpnC326S) that show severe iron overload caused by a disrupted hepcidin/ferroportin regulatory circuitry (40) show significantly increased hepatic TGF-β1 mRNA expression at 22 and 32 weeks of age (Fig. 2E). Lipid peroxidation is a consequence of iron overload in Fpn C326S mice (40). Thus, we cannot exclude the possibility that liver disease-associated factors are responsible for increased TGF-β1 mRNA expression. However, no major liver pathologies are observed in these mouse models (e.g. alanine-aminotranferase and aspartate-aminotransferase activity are not increased) because mice are resistant to progressive liver disease induced by iron accumulation (40). Although the nature of the exact stimulus is not clear yet, TGF-β mRNA levels are robustly increased in response to hepatic iron overload, suggesting that elevated hepcidin levels stimulated by TGF-β1 may contribute to reducing iron overload.
TGF-β1 and BMP6 Independently Activate Hepcidin mRNA Expression
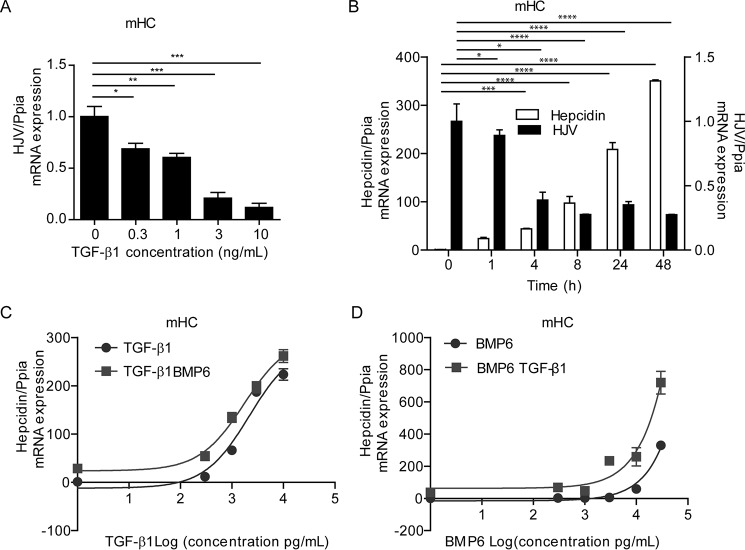
In mouse hepatocytes, the TGF-β1 response of hepcidin is paralleled by a strong dose- and time-dependent decrease in HJV mRNA levels (Fig. 3, A and B), whereas HFE and TfR2 mRNA levels are not affected (data not shown). Because HJV acts as a coreceptor for BMP6, reduced HJV mRNA levels in response to TGF-β1 treatment might restrain BMP6-induced hepcidin mRNA expression. Therefore, we next investigated how TGF-β1 and BMP6 influence each other to activate hepcidin mRNA expression. We treated mHC with 0.3, 1, 3, and 10 ng/ml TGF-β1, with or without 20 ng/ml BMP6 (Fig. 3C) or 0.3, 1, 3, 10, and 30 ng/ml BMP6 with or without 5 ng/ml TGF-β1 (Fig. 3D). We choose these concentrations based on two considerations: (i) 5 ng/ml TGF-β1 (ED50 is 1.5 ng/ml) or 20 ng/ml BMP6 (ED50 is 26.5 ng/ml) efficiently activated hepcidin mRNA expression in mouse hepatocytes; to investigate the interaction of TGF-β1 and BMP6, we selected low cytokine concentrations, which allowed for the activation of Smad1/5/8 signaling to hepcidin without showing a saturated response; and (ii) higher concentrations were avoided because they may not be physiologically relevant. We show that application of increasing amounts of TGF-β1 increased hepcidin mRNA expression, which was further enhanced by addition of BMP6 (Fig. 3C). Vice versa, treatment of mHC with increasing concentrations of BMP6 with or without a constant amount of TGF-β1 (5 ng/ml) showed that addition of TGF-β1 enhanced BMP6-dependent hepcidin induction to a similar extent as a single TGF-β1 application (Fig. 3D). We only observe mild synergism of the two cytokines at the highest BMP6 concentration. These results indicate that despite the fact that TGF-β1 reduces HJV mRNA levels, TGF-β1 treatment did not exhibit antagonistic effects on BMP6-dependent hepcidin induction in mHC within the dose range applied.
FIGURE 3.
TGF-β1 and BMP6 control hepcidin mRNA expression in an additive manner. Mouse hepatocytes were treated with the indicated doses of TGF-β1 (5 ng/ml) for 24 h, and hepcidin (A–D) and HJV (A and B) mRNA levels were measured by quantitative real time PCR. A, results are expressed as means ± S.D. of three independent experiments. B, one representative data set from three different experiments is shown. One-way ANOVA, followed by Tukey's post hoc test, was used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C and D, hepcidin mRNA levels in mHC treated with the indicated doses of TGF-β1 with or without additional BMP6 treatment (20 ng/ml) (C) or treated with the indicated doses of BMP6 with or without additional TGF-β1 treatment (5 ng/ml) (D). Hepcidin mRNA was analyzed by quantitative real time PCR from three independent hepatocyte preparations, whereby one representative data set is shown.
ALK5 but Not ALK1/2/3 Kinase Activity Is Required for TGF-β1-dependent Hepcidin Expression in mHC
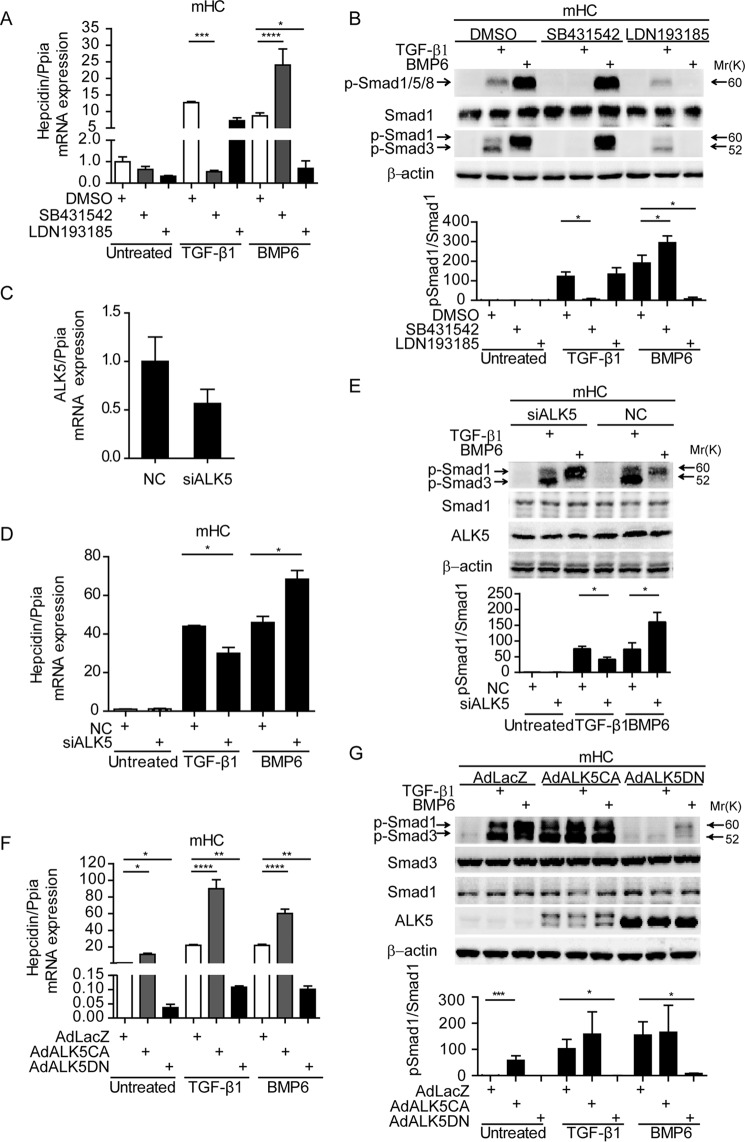
To assess which TGF-β1 family signaling receptors are required for TGF-β1-dependent hepcidin transcription, we cultured mHC in presence of TGF-β1 or BMP6 for 1h together with the ALK inhibitors SB431542 (targeting ALK4/5/7) (41) or LDN193189 (targeting ALK2/3) (42). SB431542 treatment completely blocked hepcidin induction by TGFβ-1 and even mildly enhanced BMP6-induced hepcidin expression (Fig. 4A). Importantly, SB431542 treatment prevented TGF-β1-induced Smad1/Smad3 phosphorylation, whereas Smad1 phosphorylation induced by BMP6 was enhanced, which correlated with elevated hepcidin mRNA expression (Fig. 4B). Inhibition of ALK2/3 by LDN193189 significantly altered neither basal hepcidin levels nor the TGF-β1 response of hepcidin (Fig. 4A). This suggests that hepcidin activation by BMP6 and TGF-β1 are separable. LDN193189 completely abolished BMP6 triggered Smad1 phosphorylation, whereas TGF-β1-induced Smad1/3 phosphorylation remained unaffected (Fig. 4B). Likewise, diminished ALK5 expression by siRNA-mediated knockdown to ∼50% (Fig. 4C) decreased hepcidin mRNA expression, as well as p-Smad1/3 induction (Fig. 4, D and E). These results, in combination with the fact that ALK4/7 only phosphorylates Smad2/3, suggest that TGF-β1-induced Smad1 phosphorylation and hepcidin activation are ALK5-dependent.
FIGURE 4.
ALK5 kinase activity is required for TGF-β1-dependent Smad1 phosphorylation and hepcidin expression in mHC. A and B, mHC were treated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h in the presence or absence of 5 μm SB431542, 1 μm LDN193185, or DMSO. C–E, RNAi directed against ALK5 was performed for 48 h in mHC. Transfected cells were either left untreated or were stimulated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h. F and G, mHC were infected with adenovirus expressing constitutively active (ALK5CA), dominant negative (ALK5DN) ALK5, or a LacZ control. After infection, cells were treated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h or were left untreated. Hepcidin mRNA was analyzed by quantitative real time PCR from three independent hepatocyte preparations, whereby one representative data set is shown (A, D, and F), and whole cell lysate was analyzed by immunoblots, using antibodies targeting either total or phosphor-Smad1/3 or β-actin. (B, E, and G). Representative immunoblots (upper panels) and quantification of p-Smad1/Smad1 (lower panels) are shown. One-way ANOVA, followed by Tukey's post hoc test, was used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. NC, control siRNA.
To further test whether ALK5 by itself (homodimer) might be able to activate Smad1 phosphorylation and hepcidin mRNA expression in hepatocytes, we transduced mHC with adenoviruses expressing constitutively active ALK5 (ALK5CA) or dominant negative ALK5 (ALK5DN). Overexpression of ALK5CA induced basal hepcidin mRNA levels by ∼11-fold (Fig. 4F) and activated phosphorylation of Smad1 by ∼58-fold (Fig. 4G). Vice versa, overexpression of ALK5DN inhibited basal hepcidin levels (25-fold to 4% of LacZ control), as well as hepcidin induction by TGF-β1 and BMP6. Phosphorylation of Smad1 and Smad3 induced by TGF-β1 treatment were abrogated by ALK5DN. Distinct from reducing ALK5 kinase activity by SB431542 or RNAi, ALK5DN overexpression significantly inhibited basal levels and BMP6-induced hepcidin gene expression. This may be explained by “stoichiometric disturbance” of TGF-β/BMP family receptor complexes. Briefly, in addition to the expected homodimerization of ALK5DN and heterodimerization with ALK5 (ALK5DN-ALK5), ALK5DN may also form heterodimers with ALK2/3 (ALK5DN-ALK2/3), whereby only the latter would impact on BMP ligand-induced hepcidin expression. Such an effect is not expected upon application of the ALK5 kinase inhibitor SB431542 or RNAi against ALK5.
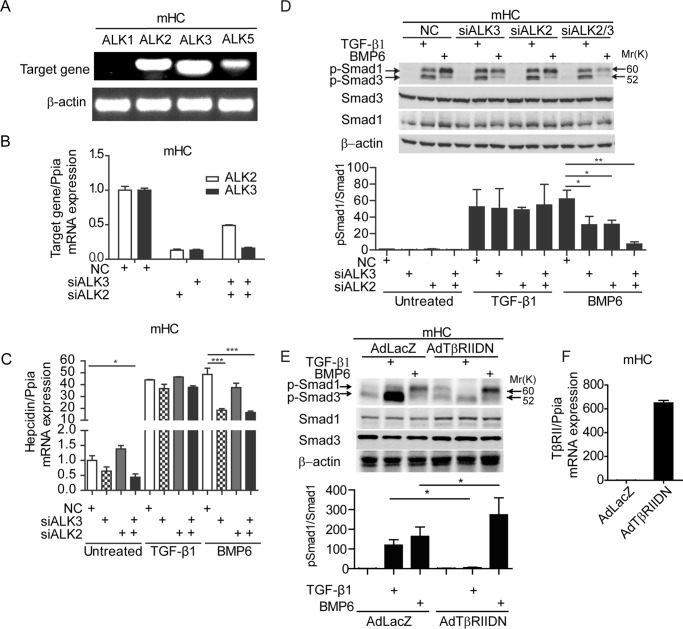
Previous reports demonstrate that ALK5 by itself is unable to induce phosphorylation of Smad1 and that this activity requires formation of heterodimers with ALK1/2/3 (24, 29, 43). To analyze participation of ALK1, ALK2, and ALK3 in TGF-β1-dependent hepcidin induction, we first assessed mRNA expression levels in mHC (Fig. 5A). Although ALK2 and ALK3 mRNA levels were well detectable in hepatocytes, ALK1 mRNA expression was almost not present in our setting. We therefore focused our attention on ALK2 and ALK3, which were reported to play a redundant role in formation of heterodimers with ALK5 to induce Smad1 phosphorylation (29). Depletion of ALK2 and/or ALK3 by siRNA did not affect basal hepcidin expression. BMP6-dependent hepcidin induction was reduced to 38% by siALK3, 77% by siALK2, and 34% by siALK2/3 compared with control siRNA, consistent with previous observations in mice (44). By contrast, the TGF-β1 response of hepcidin was not affected by depletion of ALK2 and/or ALK3 (Fig. 5, B and C). The knockdown of ALK2, ALK3, or both reduced phosphorylated Smad1 levels in response to BMP6 treatment, whereas p-Smad1 remained unaffected in TGF-β1-treated cells (Fig. 5D). These data suggest that ALK2 and ALK3 are not directly involved in TGF-β1-mediated Smad1 phosphorylation and hepcidin activation in hepatocytes.
FIGURE 5.
TGF-β1-p-Smad1-hepcidin regulation is ALK1/2/3-independent and TβRII-dependent. A, relative mRNA levels of ALK1, ALK2, ALK3, and ALK5 in murine primary hepatocytes. The upper band represents the target gene PCR product, and the lower band represents β-actin. B–D, RNAi directed against ALK2, ALK3, or both was performed for 48 h in mHC. Transfected cells were either left untreated or were stimulated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h. E, mHC were infected with adenovirus expressing dominant negative TβRII (TβRIIDN) or a LacZ control. After infection, cells were treated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h or were left untreated. C and F, hepcidin (C) and TβRII (F) mRNA was analyzed by quantitative real time PCR from three independent hepatocyte preparations, whereby one representative data set is shown, and whole cell lysate was analyzed by immunoblots. D and E, Student's t test was used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. NC, control siRNA.
To further examine whether TβRII participates in TGF-β1-induced Smad1/5 phosphorylation in hepatocytes, we transduced mHC with an adenovirus expressing dominant negative TβRII (TGβRIIDN) that lacks kinase activity. Homodimerization or heterodimerization with wild-type TβRII is expected to interfere with Smad activation upon ligand binding. In contrast to ALK5DN, DNTBRII is specific for the TGF-β1 signaling pathway and does not impact on BMP6-induced hepcidin expression (supplemental Fig. S1). Phosphorylation of Smad1 and Smad3 induced by TGF-β1 treatment were abrogated upon TβRIIDN expression (Fig. 5E). Similar to the observations with inhibiting ALK5 with SB431542, TβRIIDN enhanced BMP6-mediated phosphorylation of Smad1. This suggests that activation of ALK5-Smad1/5 is TβRII-dependent in mouse hepatocytes.
HFE, TfR2, and HJV Stimulate BMP6 but Not TGF-β1-mediated Signaling to Hepcidin
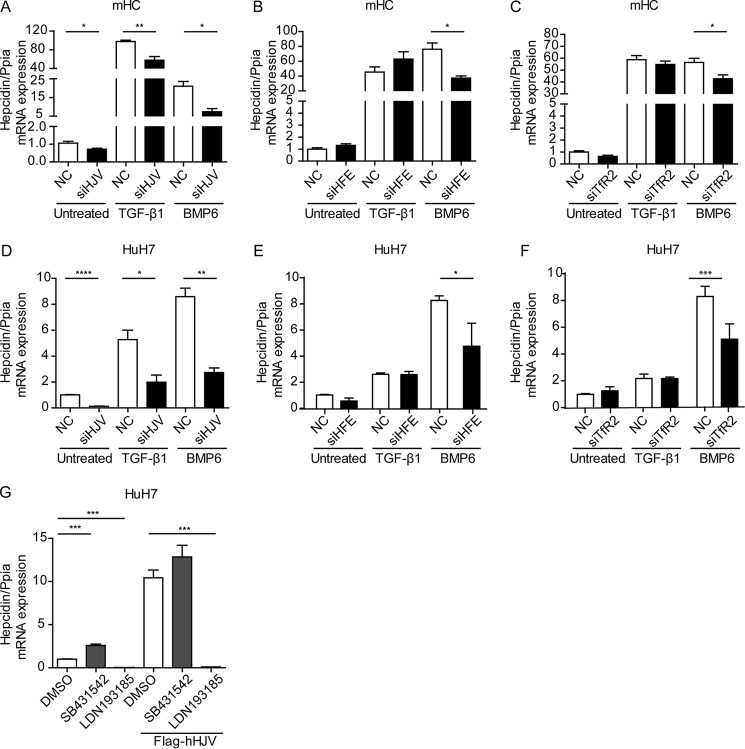
HJV is a coreceptor for BMP signaling that is mutated in a juvenile subtype of HH. Patients and mice with HJV deficiency show low hepcidin mRNA expression levels. We investigated whether HJV modulates the hepcidin response to TGF-β1. Mouse hepatocytes were transfected with siRNA against HJV, and hepcidin mRNA expression was assessed following TGF-β1 and BMP6 treatment. As expected, the knockdown of HJV inhibited basal hepcidin expression (∼3–4-fold), as well as its response to BMP6 (∼2-fold). Interestingly, the TGF-β1 response of hepcidin is also diminished by HJV depletion (∼2-fold) (Fig. 6A). However, this is likely caused by a reduction in basal hepcidin levels. In addition to HJV, HFE, and TfR2 are critical upstream regulators of hepcidin expression that are mutated in patients with different subtypes of HH. The selective gene knockdown of HFE or TfR2 inhibited BMP6-induced hepcidin expression, whereas basal hepcidin levels and the TGF-β1 response of hepcidin remained unaffected (Fig. 6, B and C). Similar results were obtained in HuH7 cells (Fig. 6, D–F). In addition, we overexpressed hHJV in HuH7 cells and show that hepcidin mRNA expression is elevated. However, hHJV-induced hepcidin expression could only be blunted by the BMP inhibitor LDN193189 and not by the TGF-β1 inhibitor SB431542 (Fig. 6G). This finding is consistent with the notion that at least in HuH7 cells, hepcidin induced by overexpression of HJV is ALK2/3-dependent. Taken together, our data indicate that HJV, HFE, and TfR2 predominantly affect BMP-induced hepcidin levels, and this response is mediated by ALK2/3 rather than ALK5.
FIGURE 6.
HJV, HFE, and TfR2 control the hepcidin response to BMP but not to TGF-β1. A–C, siRNAs against HJV (A), HFE (B), and TfR2 (C) or a scrambled control were transfected into murine hepatocytes for 48 h. Transfected cells were either left untreated or were stimulated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h. Hepcidin mRNA was analyzed by quantitative real time PCR from at least three independent hepatocyte preparations, whereby one representative data set is shown. Student's t test was used to determine statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. D–F, HuH7 cells were transfected with an siRNA pool against HJV, HFE, and TfR2 for 72 h. Transfected cells were either left untreated or stimulated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h. Hepcidin mRNA expression was analyzed from three independent experiments. Student's t test was used to determine statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. G, HuH7 cells were transfected with a plasmid expressing FLAG-tagged hHJV for 48 h. Transfected cells were either left untreated or treated with 5 μm SB431542 or 1 μm DN193185 for 24 h. Hepcidin mRNA expression was analyzed. The results are expressed as means ± S.D. of three different experiments. One-way ANOVA followed by Tukey's post hoc test were used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. NC, control siRNA.
The Hepcidin Response to TGF-β1 Is Smad1/5-dependent
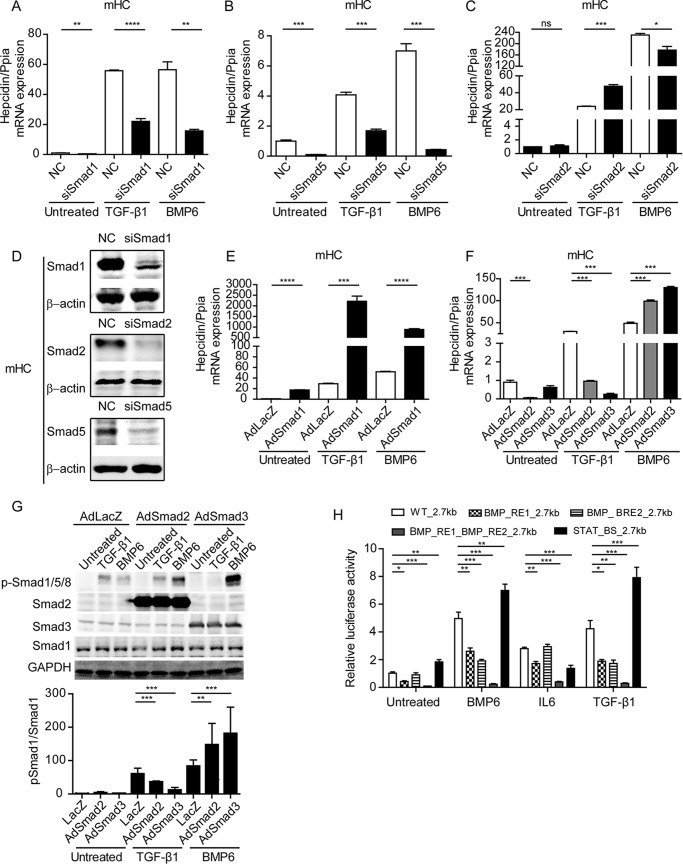
As shown in Fig. 1C, TGF-β1 induced a transient phosphorylation of Smad1 and a persistent phosphorylation of Smad3. We next asked about the functional contribution of Smad proteins to the TGF-β1 response of hepcidin. Smad6 selectively inhibits Smad1/5/8-dependent signaling, whereas Smad7 is a negative regulator of Smad1/5/8 and Smad2/3 signaling. Overexpression of Smad6 or Smad7 strongly reduced basal hepcidin levels and abolished the induction of hepcidin by BMP6 and TGF-β1, which provides evidence that Smad1/5/8 are important in TGF-β1-induced hepcidin expression (31). To better understand this finding, we individually decreased expression of Smad1, Smad2, or Smad5 by siRNA-mediated knockdown before the addition of either TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) (Fig. 7, A–D). RNAi against Smad1 or Smad5 reduced basal hepcidin mRNA levels, as well as its response to TGF-β1 or BMP6 treatment. Basal hepcidin mRNA levels remained unaffected by the knockdown of Smad2, whereas the TGF-β1 response was mildly enhanced, and the BMP6 response was mildly reduced (Fig. 7C). mHC were next infected by viral vectors encoding Smad1, Smad2, or Smad3 expression cassettes and were stimulated with either TGF-β1 or BMP6 for 1 h. Overexpression of Smad1 enhanced expression of hepcidin with or without BMP6 or TGF-β1 treatment, further corroborating the critical role of Smad1 in stimulating the hepcidin response (Fig. 7E). By contrast, overexpression of Smad2 but not of Smad3 reduced basal hepcidin levels, whereas overexpression of Smad2 or Smad3 decreased the hepcidin response to TGF-β1 and stimulated the hepcidin response to BMP6 (Fig. 7F). Phospho-Smad1 levels correlate with the hepcidin response to TGF-β1 and BMP6 treatment in that they are reduced or activated, respectively (Fig. 7G). These data illustrate that Smad2/3 overexpression may interfere specifically with the TGF-β1-induced ALK5 or the BMP6-induced ALK2/3 signaling pathways in that they control the phosphorylation of Smad1.
FIGURE 7.
TGF-β1-controlled activation of hepcidin is Smad1/5-dependent. A–D, mHC were transfected with siRNA against Smad1 (A), Smad5 (B), or Smad2 (C) for 48 h and stimulated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h. D, whole cell lysate was analyzed by immunoblot using antibodies targeting Smad1, Smad2, Smad5, or β-actin. E and F, mHC were infected with adenoviruses (multiplicity of infection, 50) expressing a LacZ control (AdLacZ), Smad1 (AdSmad1), Smad2 (AdSmad2), or Smad3 (AdSmad3), and cells were either left untreated or were stimulated with TGF-β1 (5 ng/ml) or BMP6 (20 ng/ml) for 1 h. G, whole cell lysates were analyzed by immunoblot using antibodies targeting phosphorylated Smad1/5/8, total Smad1, Smad2, Smad3, or GAPDH. A–C, F, and E, hepcidin mRNA was analyzed by quantitative real time PCR from three independent hepatocyte preparations, whereby one representative data set is shown. Student's t test (A–C and E) and one-way ANOVA followed by Tukey's post hoc test (F) were used for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. H, HuH7 cells were transfected with reporter constructs. Luciferase expression is driven by the wild-type hepcidin promoter (WT_2.7kb) or promoter derivatives with mutated or deleted transcription factor binding sites (BMP_RE1_2.7kb, BMP_RE2_2.7kb, BMP_RE1_ BMP_RE2_2.7kb, and STAT_BS_2.7kb). Transfected cells were treated with BMP6 (20 ng/ml), TGF-β1 (10 ng/ml), or IL6 (40 ng/ml) for 3 h. After treatment, the cells were lysed, and luciferase activity was measured and normalized to total protein concentration. The results are expressed as fold change of luciferase activity after normalization. The data are reported as means ± S.D. from at least three independent experiments. Statistical significance was determined with the one-way ANOVA tests and followed by Tukey's post hoc test for determining statistical significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. NC, control siRNA.
To identify the elements within the hepcidin promotor that control TGF-β1-induced hepcidin transcription, we transiently transfected HuH7 cells with reporter constructs driven by the hepcidin promoter. The promoter constructs either contain the wild-type 2.7-kb genomic region upstream of the transcription start site or a promoter with mutations in BMP_RE1_2.7kb or/and BMP_RE2_2.7kb or deletions of the STAT3 binding site (STAT_BS_2.7kb) (45). As expected, TGF-β1, BMP6, and IL6 treatment increased the luciferase activity of the WT promoter construct. The luciferase activity of the BMP_RE1_2.7kb reporter construct was significantly lower compared with the wild-type promoter under basal conditions. Additionally, the inducibility by TGF-β1, BMP6, and IL6 treatment was diminished. By contrast, luciferase activity of the BMP_RE2_2.7kb reporter construct was reduced compared with the wild-type reporter construct following TGF-β1 and BMP6 stimulation, whereas the IL6 response remained unaffected. The hepcidin promotor BMP_RE1_BMP_RE2_2.7kb showed strongly reduced luciferase activity in all conditions analyzed. Mutations in the STAT binding site impaired the response to IL6 and enhanced the sensitivity to BMP6 and TGF-β1 (Fig. 7H). Taken together, our data demonstrate that both BMP6 and TGF-β1 activate hepcidin expression in a Smad1/5-dependent manner and that the same elements of the hepcidin promoter are required for TGF-β1- and BMP6-mediated hepcidin promoter activation.
Discussion
In addition to the well established BMP-mediated signaling that increases hepatic hepcidin levels, this study establishes TGF-β1 as an effective activator of hepcidin mRNA expression both in vitro and in vivo. Expression of BMP6 and TGF-β1 is induced in iron-loaded hepatocytes (26), and thus, TGF-β1 superfamily members may contribute to the hepcidin-controlled reduction of dietary iron uptake. In contrast to BMP6-controlled hepcidin expression, which requires the hepatocytic cell surface receptors ALK2 and ALK3, we show that TGF-β1-dependent hepcidin induction is mediated by ALK5. Despite different requirements for receptors, BMP6 and TGF-β1 signaling to hepcidin converges at the level of Smad1/5 phosphorylation and the necessity for BMP-REs located in the hepcidin promoter (45). The effects of TGF-β1 and BMP6 on hepcidin induction are separable and act in an additive manner.
Smad1/5 Phosphorylation Is Critical for the Hepcidin Response to TGF-β1
It is now well established that TGF-β1 treatment can cause phosphorylation of Smad1/5/8 in various cell types. Although mechanisms may differ, the kinetic profiles are similar: although TGF-β1-induced Smad2/3 phosphorylation is stable over time, Smad1/5/8 phosphorylation is more transient (27, 29, 43, 46, 47), which is confirmed by our data. Despite the transient nature of Smad1/5/8 phosphorylation by TGF-β1, it was shown to stimulate migration (46) and anchorage-independent growth (29) in a breast cancer model system or to promote endothelial cells proliferation, migration, and blood vessel formation (43). The reported functions of TGF-β1-Smad1/5/8 signaling seem different from the canonical TGF-β1-Smad2/3 signaling, and opposing effects have been reported (48). The underlying mechanisms are unclear.
Here we show that TGF-β1-Smad1/5/8 signaling is responsible for hepcidin activation in hepatocytes. Treatment with either TGF-β1 or BMP6 increases hepcidin expression in hepatocytes for at least 24 h. Despite an early and only transient induction of phosphorylation of Smad1/5 by TGF-β1, Smad1/5 levels are most critical for the TGF-β1-dependent hepcidin response. Knockdown of Smad1 or Smad5 reduces basal hepcidin levels and blunts TGF-β-induced hepcidin expression. In addition to phosphorylation of Smad1/5/8, TGF-β1 treatment induces canonical Smad2/3 phosphorylation. However, Smad2/3 seem to counteract hepcidin stimulation, because Smad2 depletion by RNAi stimulated TGF-β1-dependent hepcidin mRNA expression, whereas Smad2 or Smad3 overexpression showed an inhibitory effect. Smad2 and Smad3 exert their effect on hepcidin by controlling phosphorylation levels of Smad1/5/8, strengthening the critical role of Smad1/5/8 in the regulation of hepcidin transcription. Our data argue against formation of a complex of Smad1 and Smad2 or Smad3 to activate hepcidin levels. The inhibitory role of Smad2/3 on Smad1/5/8 may further explain why phosphorylation of Smad1/5/8 is only transiently increased as a consequence of TGF-β1 treatment. It requires further investigation to assess whether the cross-talk of these two pathways only exists in hepatocytes, how activated Smad2/3 causes a dephosphorylation of Smad1/5/8, and how these opposing signaling pathways both triggered by TGF-β1 are coordinated with each other.
Previous studies suggested that phosphorylated Smad3 induced by TGF-β1 inhibits BMP signaling (49). Our data contrast these findings as the overexpression of Smad2 or Smad3 enhanced phosphorylation of Smad1/5/8 and hepcidin mRNA levels following BMP6 treatment. These contradictory data may result from different cellular contexts, which may dictate the nature of the TGF-β1 response or the stoichiometrically artificial overexpression of Smads upon adenoviral infection, e.g. overrepresented Smad2/3 may have occupied most of the TGF-β1 or activin receptors so that more Smad1 was available for BMP receptor signaling.
TGF-β1 Stimulated Smad1/5/8 Phosphorylation and Hepcidin Induction Is Dependent on ALK5 and Independent of BMP Receptors
TGF-β1-triggered Smad1/5/8 phosphorylation is mediated by ALK1/ALK5 complexes in chondrocytes and endothelial cell (43, 50, 51), and heterodimers of ALK5 and ALK2/ALK3 were identified in epithelial cells, epithelium-derived tumor cells, and fibroblasts (29). However, some results contradicting the involvement of BMP receptors were reported (27, 46). Our study shows a direct role of ALK5 in TGF-β-Smad1/5/8 phosphorylation and hepcidin induction. ALK5 kinase activity is required for TGF-β1 induced phosphorylation of Smad1/5/8 (27), and ALK5CA is able to phosphorylate Smad1/5/8. We further show that ALK1/2/3 is unlikely to directly mediate TGF-β1-stimulated Smad1/5/8 phosphorylation and hepcidin induction. ALK1 is expressed at very low levels in hepatocytes, and ALK2 or ALK3 depletion by RNAi does not block TGF-β1-stimulated Smad1/5/8 phosphorylation and hepcidin induction. Because ALK2 and ALK3 were reported to form heterodimers with ALK5 in a redundant manner, we performed siRNA knockdown of ALK2 and ALK3 together. The double knockdown of ALK2 and ALK3 substantially reduced BMP6 induction of Smad1/5/8 phosphorylation but did not affect induction of Smad1/5/8 phosphorylation and hepcidin expression by TGF-β1. In contrast to data reported by Steinbicker et al. (44), we did not observe a statistically significant reduction of basal hepcidin mRNA levels after knockdown of ALK2 and/or ALK3 in mouse hepatocytes. Differences may be explained by the RNAi approach applied in our study, where residual expression of ALK2/3 may remain, whereby Steinbicker et al. studied hepatocytes from ALK2fl/fl Alb-Cre and ALK3fl/fl Alb-Cre mice. Further differences may arise from remaining BMP/Smad1 signaling activity in 6-h starved (0.1% fetal calf serum) hepatocytes (44) compared with hepatocytes that were serum-starved for 24 h (this study). In future experiments, it would be interesting to test whether ALK2/3 knock-out mice sustain the ability to induce hepcidin in response to TGF-β1.
The HH-associated Proteins HFE, TfR2, and HJV Interfere with BMP/Smad Signaling but Not with the TGF-β1 Response of Hepcidin
Proteins mutated in HH attenuate signaling via BMP receptors and decrease Smad1/5/8 phosphorylation (5, 20, 21). HJV was identified as a coreceptor for BMP signaling, and HFE and TfR2 act together with HJV to maintain high hepcidin levels. In liver cells, BMP2, BMP4, and BMP6 are endogenous ligands for HJV, which utilizes ALK2 or ALK3 to regulate BMP signaling and hepcidin expression (11, 52). HFE inhibits ALK3 ubiquitination and proteasomal degradation, thus increasing ALK3 protein availability to maintain hepcidin expression (53). It was suggested that TfR2 could interact with HFE to regulate hepcidin expression through the BMP-HJV pathway, but the precise molecular mechanisms remain unclear. Here, we tested whether HFE, TFR2, or HJV interfere with the TGF-β1 response of hepcidin.
Knockdown of HJV but not HFE and TfR2 reduces basal hepcidin expression, whereas knockdown of HJV, HFE, or TfR2 restrained BMP6-dependent hepcidin induction, consistent with previous findings that HFE or TfR2 knock-out mice exhibit reduced Smad1 phosphorylation and hepcidin expression (20, 21). Only knockdown of HJV diminished TGF-β1-mediated hepcidin induction, which, however, may be explained by the effect of HJV RNAi on basal hepcidin expression. Consistent with this notion, only the BMP signaling inhibitor LDN193185 instead of the TGF-β signaling inhibitor SB431542 prevented the increase of hepcidin in response to HJV overexpression in HuH7 cells. These experiments suggest that the iron-sensing complex, consisting of HFE, TfR2, and HJV in hepatocytes enhances signaling via the BMP receptor but leaves signaling via TGF-β1 receptors unaffected.
Possible Mechanisms Mediating TGF-β1-dependent HJV mRNA Down-regulation
Our data show that TGF-β1 treatment reduced HJV mRNA levels. The following mechanisms could be explored in future studies: (i) TGF-β1 treatment is known to induce microRNA expression (54, 55), which may affect mRNA stability of HJV, a known target of mir-122 (56); (ii) TGF-β1 may decrease HJV transcription through epigenetic silencing. Although no CpG islands were identified in human and mouse HJV promoters, previous reports suggest that TGF-β1 may activate nuclear protein deacetylases, which may result in histone deacetylation and affect the transcription of proteins (57); (iii) TGF-β1 may inhibit HJV promoter activity directly or indirectly (e.g. TGF-β1-activated Smads may bind to the HJV promoter). Bioinformatic analysis identified four putative Smad3/4 binding elements between −2533 bp and −1825 bp of the putative mHJV promoter region (PROMO online software; factors predicted within a dissimilarity margin less or equal than 5%). Alternatively, TGF-β1 may induce other transcriptional regulators that inhibit HJV transcription. Future experiments will have to address mechanisms how TGF-β1 regulates HJV expression, an important mechanism for fine-tuning hepcidin signaling.
The Role of TGF-β1-mediated Hepcidin Regulation
Hepcidin levels increase in response to plasma and tissue iron overload to reduce dietary iron uptake (12). Molecular mechanisms underlying iron-controlled hepcidin activation were identified in studies of human genetic diseases, such as HH, or in mouse models associated with iron overload. BMP6 and the canonical Smad1/5/8 signaling pathway are at the core of hepcidin regulation in response to iron. In this study, we show that TGF-β1 mRNA levels are increased in mouse models of iron overload and that TGF-β1 contributes to hepatocyte hepcidin activation via an ALK5 and Smad1/5-dependent signaling pathway. We speculate that TGF-β1 may contribute to the iron-mediated hepcidin response and may be the factor that mediates increased hepcidin levels in iron-loaded BMP6 KO mice (4).
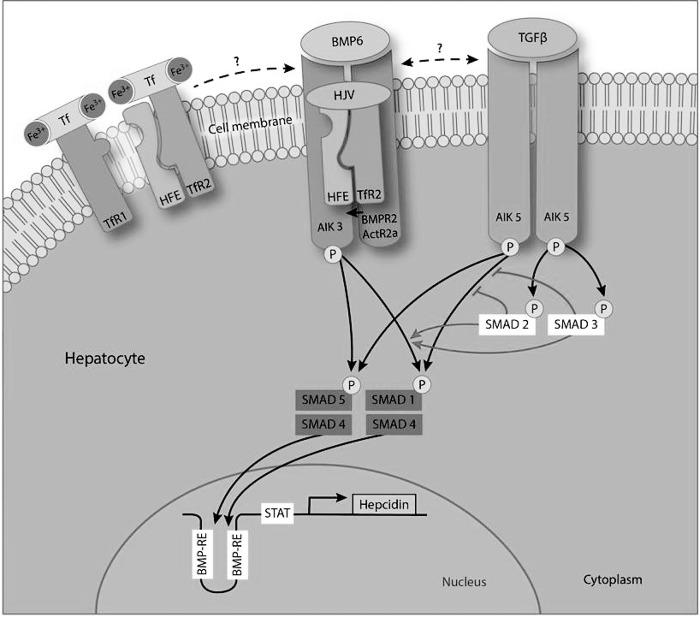
In conclusion, we show that hepcidin activation by TGF-β1 requires ALK5, contrasting the ALK2/3-mediated BMP response of hepcidin. Although both pathways are separable at the ligand and receptor level, they converge at the level of Smad1/5 to control hepcidin transcription via the previously identified BMP-responsive elements in the hepcidin promoter. TGF-β1 and BMP6 act in an additive way to regulate hepcidin expression (Fig. 8).
FIGURE 8.
Model of BMP and TGF-β1 controlled hepcidin activation. TGF-β1 binds to serine and threonine kinase receptors ALK5 (TβRI) and TβRII on the cell surface. The activated ALK5/TβRII complex continuously recruits and phosphorylates Smad2/3 and transiently recruits and phosphorylates Smad1/5 in hepatocytes. BMP6 activates ALK3 (BMPRIA) and BMPRII and continuously phosphorylates Smad1/5. Despite only transient induction, p-Smad1/5 levels are critical for TGF-β1-dependent hepcidin up-regulation. The same elements in the hepcidin promoter sequence are required for TGF-β1- and BMP6-mediated hepcidin promoter activation. Canonical Smad2/3 activation interferes with TGF-β1-induced hepcidin regulation by reducing phosphorylation of Smad1/5, whereas the BMP6-hepcidin signal is enhanced in this setting because of an increase in availability of Smad1/5 signaling components.
Author Contributions
S. C., S. D., and M. U. M. designed the experiments. S. C. carried out most of the experiments. T. F. carried out the virus injection and tissue sample collection. V. S. M. carried out the induction of systemic iron overload in mice. S. A. provided the data of FpnC326S mice. T. S. W. provided the primary human hepatocytes. K. B.-H. and J. A. started the project and joined the discussion. S. C., V. S. M., S. D., and M. U. M. prepared the manuscript.
Supplementary Material
This work was supported by the Federal Ministry of Education and Research (Germany) in Virtual Liver Network Projects 0315755 and 0315761. This work was also supported by a grant from the European Union FP7 Marie Curie International Training Network IT-Liver (to T. F.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1.
- Fpn
- ferroportin
- BMP
- bone morphogenetic protein
- HH
- hereditary hemochromatosis
- mHC
- primary mouse hepatocyte
- TβRII
- TGF-β1 type II receptor
- SFBC
- plasma iron concentration
- UIBC
- unsaturated iron binding capacity
- ANOVA
- analysis of variance.
References
- 1. Ganz T. (2013) Systemic iron homeostasis. Physiol. Rev. 93, 1721–1741 [DOI] [PubMed] [Google Scholar]
- 2. Hentze M. W., Muckenthaler M. U., Galy B., and Camaschella C. (2010) Two to tango: regulation of mammalian iron metabolism. Cell 142, 24–38 [DOI] [PubMed] [Google Scholar]
- 3. Thompson M. D., Wickline E. D., Bowen W. B., Lu A., Singh S., Misse A., and Monga S. P. (2011) Spontaneous repopulation of β-catenin null livers with β-catenin-positive hepatocytes after chronic murine liver injury. Hepatology 54, 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramos E., Kautz L., Rodriguez R., Hansen M., Gabayan V., Ginzburg Y., Roth M. P., Nemeth E., and Ganz T. (2011) Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology 53, 1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ryan J. D., Ryan E., Fabre A., Lawless M. W., and Crowe J. (2010) Defective bone morphogenic protein signaling underlies hepcidin deficiency in HFE hereditary hemochromatosis. Hepatology 52, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 6. Nemeth E., Rivera S., Gabayan V., Keller C., Taudorf S., Pedersen B. K., and Ganz T. (2004) IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J. Clin. Invest. 113, 1271–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee P., Peng H., Gelbart T., Wang L., and Beutler E. (2005) Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proc. Natl. Acad. Sci. U. S. A. 102, 1906–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wrighting D. M., and Andrews N. C. (2006) Interleukin-6 induces hepcidin expression through STAT3. Blood 108, 3204–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verga Falzacappa M. V., Vujic Spasic M., Kessler R., Stolte J., Hentze M. W., and Muckenthaler M. U. (2007) STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood 109, 353–358 [DOI] [PubMed] [Google Scholar]
- 10. Lee P. L., Beutler E., Rao S. V., and Barton J. C. (2004) Genetic abnormalities and juvenile hemochromatosis: mutations of the HJV gene encoding hemojuvelin. Blood 103, 4669–4671 [DOI] [PubMed] [Google Scholar]
- 11. Xia Y., Babitt J. L., Sidis Y., Chung R. T., and Lin H. Y. (2008) Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood 111, 5195–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nemeth E., Roetto A., Garozzo G., Ganz T., and Camaschella C. (2005) Hepcidin is decreased in TFR2 hemochromatosis. Blood 105, 1803–1806 [DOI] [PubMed] [Google Scholar]
- 13. Bridle K. R., Frazer D. M., Wilkins S. J., Dixon J. L., Purdie D. M., Crawford D. H., Subramaniam V. N., Powell L. W., Anderson G. J., and Ramm G. A. (2003) Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet 361, 669–673 [DOI] [PubMed] [Google Scholar]
- 14. Goswami T., and Andrews N. C. (2006) Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J. Biol. Chem. 281, 28494–28498 [DOI] [PubMed] [Google Scholar]
- 15. Bennett M. J., Lebrón J. A., and Bjorkman P. J. (2000) Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature 403, 46–53 [DOI] [PubMed] [Google Scholar]
- 16. Chen J., Chloupková M., Gao J., Chapman-Arvedson T. L., and Enns C. A. (2007) HFE modulates transferrin receptor 2 levels in hepatoma cells via interactions that differ from transferrin receptor 1-HFE interactions. J. Biol. Chem. 282, 36862–36870 [DOI] [PubMed] [Google Scholar]
- 17. Ahmad K. A., Ahmann J. R., Migas M. C., Waheed A., Britton R. S., Bacon B. R., Sly W. S., and Fleming R. E. (2002) Decreased liver hepcidin expression in the Hfe knockout mouse. Blood Cells Mol. Dis. 29, 361–366 [DOI] [PubMed] [Google Scholar]
- 18. Muckenthaler M., Roy C. N., Custodio A. O., Miñana B., deGraaf J., Montross L. K., Andrews N. C., and Hentze M. W. (2003) Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat. Genet. 34, 102–107 [DOI] [PubMed] [Google Scholar]
- 19. Wallace D. F., Summerville L., and Subramaniam V. N. (2007) Targeted disruption of the hepatic transferrin receptor 2 gene in mice leads to iron overload. Gastroenterology 132, 301–310 [DOI] [PubMed] [Google Scholar]
- 20. Corradini E., Garuti C., Montosi G., Ventura P., Andriopoulos B. Jr., Lin H. Y., Pietrangelo A., and Babitt J. L. (2009) Bone morphogenetic protein signaling is impaired in an HFE knockout mouse model of hemochromatosis. Gastroenterology 137, 1489–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corradini E., Rozier M., Meynard D., Odhiambo A., Lin H. Y., Feng Q., Migas M. C., Britton R. S., Babitt J. L., and Fleming R. E. (2011) Iron regulation of hepcidin despite attenuated Smad1,5,8 signaling in mice without transferrin receptor 2 or Hfe. Gastroenterology 141, 1907–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. D'Alessio F., Hentze M. W., and Muckenthaler M. U. (2012) The hemochromatosis proteins HFE, TfR2, and HJV form a membrane-associated protein complex for hepcidin regulation. J. Hepatol. 57, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 23. Massagué J. (2012) TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiercinska E., Wickert L., Denecke B., Said H. M., Hamzavi J., Gressner A. M., Thorikay M., ten Dijke P., Mertens P. R., Breitkopf K., and Dooley S. (2006) Id1 is a critical mediator in TGF-β-induced transdifferentiation of rat hepatic stellate cells. Hepatology 43, 1032–1041 [DOI] [PubMed] [Google Scholar]
- 25. Dooley S., and ten Dijke P. (2012) TGF-β in progression of liver disease. Cell Tissue Res. 347, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Houglum K., Ramm G. A., Crawford D. H., Witztum J. L., Powell L. W., and Chojkier M. (1997) Excess iron induces hepatic oxidative stress and transforming growth factor β1 in genetic hemochromatosis. Hepatology 26, 605–610 [DOI] [PubMed] [Google Scholar]
- 27. Wrighton K. H., Lin X., Yu P. B., and Feng X. H. (2009) Transforming growth factor β can stimulate Smad1 phosphorylation independently of bone morphogenic protein receptors. J. Biol. Chem. 284, 9755–9763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bharathy S., Xie W., Yingling J. M., and Reiss M. (2008) Cancer-associated transforming growth factor β type II receptor gene mutant causes activation of bone morphogenic protein-Smads and invasive phenotype. Cancer Res. 68, 1656–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Daly A. C., Randall R. A., and Hill C. S. (2008) Transforming growth factor β-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol. Cell. Biol. 28, 6889–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weng H. L., Ciuclan L., Liu Y., Hamzavi J., Godoy P., Gaitantzi H., Kanzler S., Heuchel R., Ueberham U., Gebhardt R., Breitkopf K., and Dooley S. (2007) Profibrogenic transforming growth factor-β/activin receptor-like kinase 5 signaling via connective tissue growth factor expression in hepatocytes. Hepatology 46, 1257–1270 [DOI] [PubMed] [Google Scholar]
- 31. Vujić Spasić M., Sparla R., Mleczko-Sanecka K., Migas M. C., Breitkopf-Heinlein K., Dooley S., Vaulont S., Fleming R. E., and Muckenthaler M. U. (2013) Smad6 and Smad7 are co-regulated with hepcidin in mouse models of iron overload. Biochim. Biophys. Acta 1832, 76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Armitage A. E., Eddowes L. A., Gileadi U., Cole S., Spottiswoode N., Selvakumar T. A., Ho L. P., Townsend A. R., and Drakesmith H. (2011) Hepcidin regulation by innate immune and infectious stimuli. Blood 118, 4129–4139 [DOI] [PubMed] [Google Scholar]
- 33. Babitt J. L., Huang F. W., Xia Y., Sidis Y., Andrews N. C., and Lin H. Y. (2007) Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J. Clin. Invest. 117, 1933–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong J., Fujii S., Li H., Nakabayashi H., Sakai M., Nishi S., Goto D., Furumoto T., Imagawa S., Zaman T. A., and Kitabatake A. (2005) Interleukin-6 and mevastatin regulate plasminogen activator inhibitor-1 through CCAAT/enhancer-binding protein-delta. Arterioscler. Thromb. Vasc. Biol. 25, 1078–1084 [DOI] [PubMed] [Google Scholar]
- 35. Sime P. J., Xing Z., Graham F. L., Csaky K. G., and Gauldie J. (1997) Adenovector-mediated gene transfer of active transforming growth factor-β1 induces prolonged severe fibrosis in rat lung. J. Clin. Invest. 100, 768–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fujii M., Takeda K., Imamura T., Aoki H., Sampath T. K., Enomoto S., Kawabata M., Kato M., Ichijo H., and Miyazono K. (1999) Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol. Biol. Cell 10, 3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verga Falzacappa M. V., Casanovas G., Hentze M. W., and Muckenthaler M. U. (2008) A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. J. Mol. Med. 86, 531–540 [DOI] [PubMed] [Google Scholar]
- 38. Fechner H., Haack A., Wang H., Wang X., Eizema K., Pauschinger M., Schoemaker R., Veghel R., Houtsmuller A., Schultheiss H. P., Lamers J., and Poller W. (1999) Expression of coxsackie adenovirus receptor and alphav-integrin does not correlate with adenovector targeting in vivo indicating anatomical vector barriers. Gene Ther. 6, 1520–1535 [DOI] [PubMed] [Google Scholar]
- 39. Shayakhmetov D. M., Li Z. Y., Ni S., and Lieber A. (2004) Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J. Virol. 78, 5368–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Altamura S., Kessler R., Gröne H. J., Gretz N., Hentze M. W., Galy B., and Muckenthaler M. U. (2014) Resistance of ferroportin to hepcidin binding causes exocrine pancreatic failure and fatal iron overload. Cell Metab. 20, 359–367 [DOI] [PubMed] [Google Scholar]
- 41. Inman G. J., Nicolás F. J., Callahan J. F., Harling J. D., Gaster L. M., Reith A. D., Laping N. J., and Hill C. S. (2002) SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 62, 65–74 [DOI] [PubMed] [Google Scholar]
- 42. Sanvitale C. E., Kerr G., Chaikuad A., Ramel M. C., Mohedas A. H., Reichert S., Wang Y., Triffitt J. T., Cuny G. D., Yu P. B., Hill C. S., and Bullock A. N. (2013) A new class of small molecule inhibitor of BMP signaling. PLoS One 8, e62721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goumans M. J., Valdimarsdottir G., Itoh S., Rosendahl A., Sideras P., and ten Dijke P. (2002) Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO J. 21, 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steinbicker A. U., Bartnikas T. B., Lohmeyer L. K., Leyton P., Mayeur C., Kao S. M., Pappas A. E., Peterson R. T., Bloch D. B., Yu P. B., Fleming M. D., and Bloch K. D. (2011) Perturbation of hepcidin expression by BMP type I receptor deletion induces iron overload in mice. Blood 118, 4224–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Casanovas G., Mleczko-Sanecka K., Altamura S., Hentze M. W., and Muckenthaler M. U. (2009) Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J. Mol. Med. 87, 471–480 [DOI] [PubMed] [Google Scholar]
- 46. Liu I. M., Schilling S. H., Knouse K. A., Choy L., Derynck R., and Wang X. F. (2009) TGFβ-stimulated Smad1/5 phosphorylation requires the ALK5 L45 loop and mediates the pro-migratory TGFβ switch. EMBO J. 28, 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nurgazieva D., Mickley A., Moganti K., Ming W., Ovsyi I., Popova A., Sachindra, Awad K., Wang N., Bieback K., Goerdt S., Kzhyshkowska J., and Gratchev A. (2015) TGF-β1, but not bone morphogenetic proteins, activates Smad1/5 pathway in primary human macrophages and induces expression of proatherogenic genes. J. Immunol. 194, 709–718 [DOI] [PubMed] [Google Scholar]
- 48. Goumans M. J., Valdimarsdottir G., Itoh S., Lebrin F., Larsson J., Mummery C., Karlsson S., and ten Dijke P. (2003) Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFβ/ALK5 signaling. Mol. Cell 12, 817–828 [DOI] [PubMed] [Google Scholar]
- 49. Grönroos E., Kingston I. J., Ramachandran A., Randall R. A., Vizán P., and Hill C. S. (2012) Transforming growth factor β inhibits bone morphogenetic protein-induced transcription through novel phosphorylated Smad1/5-Smad3 complexes. Mol. Cell. Biol. 32, 2904–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Finnson K. W., Parker W. L., Chi Y., Hoemann C. D., Goldring M. B., Antoniou J., and Philip A. (2010) Endoglin differentially regulates TGF-β-induced Smad2/3 and Smad1/5 signalling and its expression correlates with extracellular matrix production and cellular differentiation state in human chondrocytes. Osteoarthritis and cartilage 18, 1518–1527 [DOI] [PubMed] [Google Scholar]
- 51. Finnson K. W., Parker W. L., ten Dijke P., Thorikay M., and Philip A. (2008) ALK1 opposes ALK5/Smad3 signaling and expression of extracellular matrix components in human chondrocytes. J. Bone Miner. Res. 23, 896–906 [DOI] [PubMed] [Google Scholar]
- 52. Xia Y., Yu P. B., Sidis Y., Beppu H., Bloch K. D., Schneyer A. L., and Lin H. Y. (2007) Repulsive guidance molecule RGMa alters utilization of bone morphogenetic protein (BMP) type II receptors by BMP2 and BMP4. J. Biol. Chem. 282, 18129–18140 [DOI] [PubMed] [Google Scholar]
- 53. Wu X. G., Wang Y., Wu Q., Cheng W. H., Liu W., Zhao Y., Mayeur C., Schmidt P. J., Yu P. B., Wang F., and Xia Y. (2014) HFE interacts with the BMP type I receptor ALK3 to regulate hepcidin expression. Blood 124, 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kato M., Putta S., Wang M., Yuan H., Lanting L., Nair I., Gunn A., Nakagawa Y., Shimano H., Todorov I., Rossi J. J., and Natarajan R. (2009) TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat. Cell Biol. 11, 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Butz H., Rácz K., Hunyady L., and Patócs A. (2012) Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol. Sci. 33, 382–393 [DOI] [PubMed] [Google Scholar]
- 56. Castoldi M., Vujic Spasic M., Altamura S., Elmén J., Lindow M., Kiss J., Stolte J., Sparla R., D'Alessandro L. A., Klingmüller U., Fleming R. E., Longerich T., Gröne H. J., Benes V., Kauppinen S., et al. (2011) The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Invest. 121, 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Warburton D., Shi W., and Xu B. (2013) TGF-β-Smad3 signaling in emphysema and pulmonary fibrosis: an epigenetic aberration of normal development? Am. J. Physiol. Lung Cell. Mol. Physiol. 304, L83–L85 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.